Abstract
Locally advanced rectal cancer (LARC) response to neoadjuvant chemoradiotherapy (nCRT) is very heterogeneous and up to 30% of patients are considered non-responders, presenting no tumor regression after nCRT. This study aimed to determine the ability of pre-treatment T2-weighted based radiomics features to predict LARC non-responders. A total of 67 LARC patients who underwent a pre-treatment MRI followed by nCRT and total mesorectal excision were assigned into training (n = 44) and validation (n = 23) groups. In both datasets, the patients were categorized according to the Ryan tumor regression grade (TRG) system into non-responders (TRG = 3) and responders (TRG 1 and 2). We extracted 960 radiomic features/patient from pre-treatment T2-weighted images. After a three-step feature selection process, including LASSO regression analysis, we built a radiomics score with seven radiomics features. This score was significantly higher among non-responders in both training and validation sets (p < 0.001 and p = 0.03) and it showed good predictive performance for LARC non-response, achieving an area under the curve (AUC) = 0.94 (95% CI: 0.82–0.99) in the training set and AUC = 0.80 (95% CI: 0.58–0.94) in the validation group. The multivariate analysis identified the radiomics score as an independent predictor for the tumor non-response (OR = 6.52, 95% CI: 1.87–22.72). Our results indicate that MRI radiomics features could be considered as potential imaging biomarkers for early prediction of LARC non-response to neoadjuvant treatment.
Keywords: locally advanced rectal cancer, radiomics, neo-adjuvant chemoradiotherapy, non-responders, magnetic resonance imaging
1. Introduction
Colorectal cancer is the third most common cancer worldwide and the second leading cause of oncologic-related mortality around the globe [1]. Rectal cancers account for approximately 30% of the total cases of colorectal malignancies [2]. Currently, the standard-of-care treatment for patients with locally advanced rectal cancer (LARC) is neoadjuvant chemotherapy (nCRT) followed by total mesorectal excision (TME) [3,4,5]. However, the individual response to nCRT is very heterogeneous, ranging from a pathological complete response to no tumor regression or even disease progression [6,7,8,9,10]. The percentage of patients who do not achieve tumor regression after nCRT, defined as non-responders (NR), is reported to be between 7 and 30% [11,12,13,14,15]. Additionally, the potential side effects of nCRT can be very serious such as hematologic, gastrointestinal or dermatologic effects, incontinence or sexual dysfunction [15,16,17,18,19] and 14–27% of patients with LARC who received this regimen developed acute or long-term grade 3–4 toxic effects as reported by Sauer et al. [20]. It is therefore argued that for non-responders the side effects of nCRT may outweigh its benefits, while nCRT does not improve the clinical outcome of these patients [21,22,23]. Early prediction of NR before the beginning of neoadjuvant therapy could be of great value in order to avoid ineffective treatment and to develop a more tailored strategy of care such as a primary surgical intervention or an intensified treatment regimen.
The rectal cancer response to nCRT can be assessed by means of endorectal ultrasound or MRI examination performed at the end of the neoadjuvant treatment [24,25]. However, the final confirmation of the tumor response can only be made by histopathologic examination of the surgical specimens and identifying non-responders before surgery still remains a great challenge. Extensive efforts have been made in various fields, focusing on gene expression, mutations and molecular metabolites as potential noninvasive biomarkers for predicting the response to nCRT in LARC patients [26,27,28,29,30].
In the radiology field, a new method, radiomics, has been developed based on the idea that medical images contain more information than what the human eye can perceive [31]. Radiomics represents a non-invasive, high-throughput post-processing technique, which extracts large amounts of quantitative features from routinely acquired medical images [32,33]. By measuring the distribution and relationships of gray levels within a lesion, radiomics texture features can reveal nonvisual information associated with tumor heterogeneity and the microenvironment; thus providing a detailed and comprehensive characterization of the tumor phenotype [34,35,36]. The heterogeneity of a tumor can be potentially related with its response to treatment and prognosis since heterogeneous tumors are prone to have a more aggressive behavior and increased resistance to treatment [37,38,39]. Since they are high dimensional inputs, radiomics features have been employed in multiple machine learning and deep learning algorithms for developing radiomics signatures useful in the oncology field [40,41,42,43]. Recent studies have demonstrated the efficacy of radiomics features as biomarkers for lesion characterization, therapy guidance and tumor prognosis among various types of cancers, including rectal cancer [44,45,46,47,48].
Based on these previous promising results, we hypothesize that magnetic resonance (MR)-based radiomics features may have a potential role in predicting locally advanced rectal cancer resistance to nCRT. The aim of the present study was to investigate the value of T2 weighted-based radiomics features extracted from baseline MRI for the prediction of non-responding rectal tumors and to develop a radiomics score based on these parameters.
2. Results
2.1. Patients Characteristics
A total of 67 patients (mean age: 60.36 ± 10.89) were included in this study. The 67 patients were split into two groups: training group (44 patients) and validation group (23 patients). In the training group, 17 patients were classified according to the tumor regression grade (TRG) as non-responders (NR) having TRG = 3, while the remaining 27 were classified as responders (R), 5 of them having TRG = 1 and 22 with TRG = 2. In the validation group, 8 patients were identified as non-responders, while 15 subjects were added to the responders group (5 with TRG 1 and 10 with TRG 2).
Patients and tumor characteristics are summarized in Table 1. No significant differences were observed in terms of age, gender, T stage and N stage between responders and non-responders in both training and validation groups. In the training group, tumor length, tumor differentiation grade and mesorectal fascia involvement (MRF) were significantly different among non-responders versus responders, however, this was not confirmed in the validation dataset.
Table 1.
Baseline characteristics of the study population.
| Variable | Training | p Value | Validation | p Value | ||
|---|---|---|---|---|---|---|
| Responders (n = 27) |
Non-Responders (n = 17) |
Responders (n = 15) |
Non-Responders (n = 8) |
|||
| Age (years) | 58.33 ± 1.38 | 56.47 ± 2.94 | 0.572 | 66.40 ± 9.60 | 64.12 ± 16.10 | 0.722 |
| Gender | 0.185 | 0.057 | ||||
| Male | 22 (66.7%) | 11 (33.3%) | 5 (45.5%) | 6 (54.5%) | ||
| Female | 5 (45.5%) | 6 (54.5%) | 10 (83.3%) | 2 (16.7%) | ||
| Tumor length (cm) | 58.81 ± 17.99 | 68.88 ± 11.60 | 0.04 * | 62.06 ± 15.01 | 61.50 ± 13.89 | 0.929 |
| Tumor differentiation grade | 0.01 * | 0.149 | ||||
| Well differentiated | 15 (83.3%) | 3 (16.7%) | 5 (83.3%) | 1 (16.7%) | ||
| Moderately differentiated | 11 (55.0%) | 9 (45.0%) | 9 (69.2%) | 4 (30.8%) | ||
| Poor differentiated | 1 (16.7%) | 5 (83.3%) | 1 (25.0%) | 3 (75.0%) | ||
| Clinical tumor stage (cT) | 0.907 | 0.779 | ||||
| T2 | 4 (66.7%) | 2 (33.3%) | 2 (50%) | 2 (50%) | ||
| T3 | 19 (59.4%) | 13 (40.6%) | 11 (68.8%) | 5 (31.2%) | ||
| T4 | 4 (66.7%) | 2 (33.3%) | 2 (66.7%) | 1 (33.3%) | ||
| Clinical nodal stage (cN) | 0.3 | 0.757 | ||||
| N1 | 8 (72.7%) | 3 (27.3%) | 8 (66.7%) | 4 (33.3%) | ||
| N2 | 19 (57.6%) | 14 (42.4%) | 7 (63.6%) | 4 (36.4%) | ||
| MRF | 0.024 * | 0.679 | ||||
| Positive | 4 (33.3%) | 8 (66.7%) | 10 (62.5%) | 6 (37.5%) | ||
| Negative | 23 (71.9%) | 9 (28.1%) | 5 (71.4%) | 2 (28.6%) | ||
* Statistically significant p < 0.05; results are presented as the mean ± standard deviation or number (%).
2.2. Feature Selection and Radiomics Score Construction—Training Set
A total of 960 radiomics features were extracted from T2W images for each patient. After the inter-reader agreement evaluation, only features with an intraclass coefficient ≥ 0.75 were included in the further steps (874 features). To develop the radiomics signature, we first performed a univariate analysis of radiomics features between the responders and non-responders groups. 74 features with an adjusted p value < 0.05 were included in the next step (Appendix Table A1).
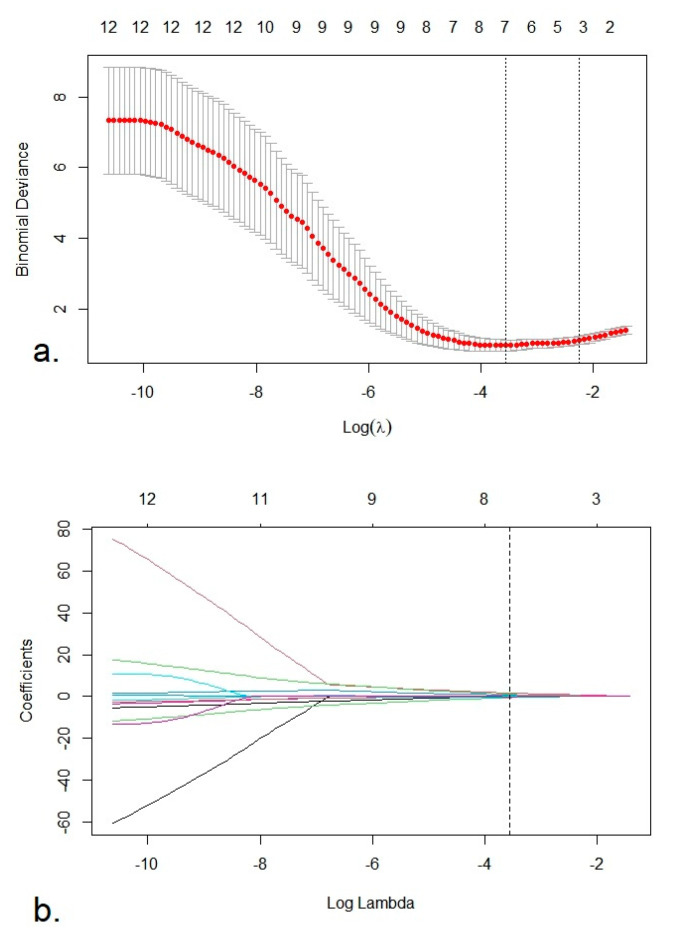
These features were secondly reduced to 12 potential predictors by applying a Spearman correlation analysis, excluding redundant features with correlation coefficients >0.9/<−0.9. The correlation matrix is shown in Appendix Figure A1. Finally, through a least absolute shrinkage and selection operator (LASSO) binary logistic regression method using 10-fold cross-validation, seven radiomics features with non-zero coefficients were selected to construct the radiomics score (Rad-Score). The feature selection process using the LASSO algorithm is shown in Figure 1a,b.
Figure 1.
(a) Selection of the tuning parameter lambda (λ) using 10-fold cross validation. Binomial deviances from the least absolute shrinkage and selection operator (LASSO) regression cross-validation model were plotted as a function of log (λ). The dotted vertical lines were drawn at the optimal λ value based on the minimum criteria and 1 standard error of the minimum criteria. The optimal λ value of 0.028 and log (λ) = −3.57 was selected. (b). LASSO coefficient profiles of the 12 radiomics features. The vertical dotted line was plotted at the optimal λ value, resulting in seven radiomics feature with non-zero coefficients.
The radiomics score was a linear combination of these 7 features, weighted according to their respective LASSO coefficients (presented in Table 2). The equation for calculating the Rad-Score is the following:
where Ci is the coefficient of the ith feature, Xi the ith feature and b the intercept.
Table 2.
List of selected radiomics features and their coefficients for calculation of the radiomics score.
| Variable | Coefficient | 95% CI | |
|---|---|---|---|
| Upper | Lower | ||
| Intercept | −0.875 | ||
| log-sigma-5-0-mm-3D_glszm_SmallAreaEmphasis | 1.621 | −0.460 | 4.428 |
| wavelet-LHL_glcm_Correlation | −0.581 | −3.954 | 3.322 |
| wavelet-LHL_firstorder_10Percentile | 0.660 | −1.536 | 16.268 |
| wavelet-HHL_glcm_MCC | −0.074 | −4.724 | 20.345 |
| wavelet-HHL_glcm_Imc1 | 0.984 | −0.232 | 8.356 |
| wavelet-HHL_firstorder_Kurtosis | −0.144 | −7.629 | 7.938 |
| wavelet-HHL_glszm_SmallAreaHighGrayLevelEmphasis | −0.070 | −4.027 | 24.241 |
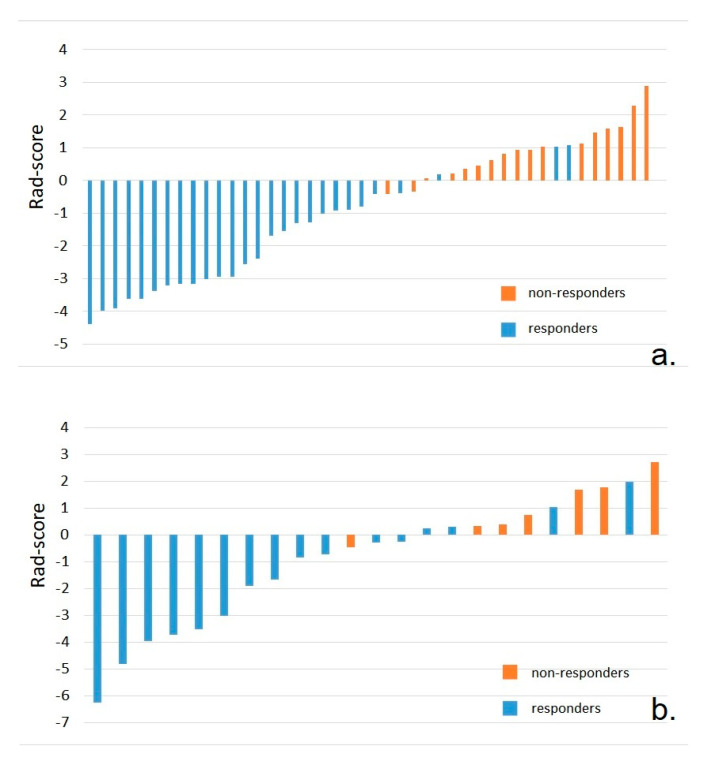
The radiomic score was calculated for each patient (Figure 2a). There was a significant difference of the Rad-Score between non-responders and responders, patients from the first group having higher values (0.92 ± 0.87 vs. −2.00 ± 1.55, p < 0.001).
Figure 2.
(a). Bar charts of the radiomics signature for each patient in the training set. (b). Bar charts of the radiomics signature for each patient in the validation set. The orange bars indicate the patients in the non-responders group, while the blue bars represent patients in the responders group.
2.3. Performance of the Radiomics Score—Training Set
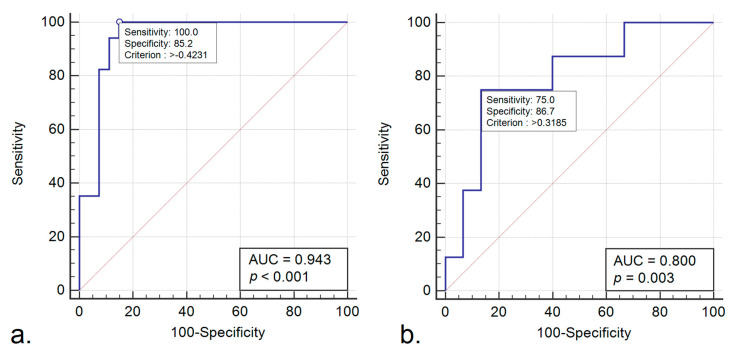
Figure 3a shows the receiver operating characteristic (ROC) curve for the radiomics score in the training set. The radiomics score predicted a rectal cancer non-response to nCRT with an area under the curve (AUC) of 0.94 (95% CI, 0.82–0.99) and accuracy of 91%, resulting in a sensitivity of 100% (95% CI, 80.5–100%) and a specificity of 85.2% (95% CI, 66.3–95.8%) for the cut-off value of −0.42. Additionally, the radiomics signature achieved a positive predictive value (PPV) of 81% (95% CI, 58.1–94.6%) and a negative predictive value (NPV) of 100% (95% CI, 85.2–100%).
Figure 3.
(a). Receiver operating characteristic (ROC) curve of the radiomics score for predicting locally advanced rectal cancer (LARC) non-responders—training set. (b). ROC curve of the radiomics score for predicting LARC non-responders—validation set.
ROC analysis was also performed for each of the seven radiomics features to evaluate their individual diagnostic performance. Table 3 shows the AUC, sensitivity, sensibility, PPV and NPV for the determined cut-off values. However, the performance of the Rad-Score was higher than the performance of each separate feature for distinguishing non-responders.
Table 3.
Individual diagnostic performance of the selected radiomics features.
| Variable | Cut-Off Value |
AUC | Accuracy (%) |
Se (%) |
Sp (%) |
PPV (%) |
NPV (%) |
|---|---|---|---|---|---|---|---|
| logsigma5_0mm_3D_glszm_SmallAreaEmphasis | −0.24 | 0.80 | 72.7 | 94.1 | 59.3 | 59.3 | 94.1 |
| wavelet-LHL_glcm_Correlation | −0.58 | 0.74 | 65.6 | 100 | 44.4 | 53.1 | 100.0 |
| wavelet_LHL_firstorder_10Percentile | −0.28 | 0.71 | 75.0 | 94.1 | 62.9 | 61.5 | 94.4 |
| wavelet-HHL_glcm_MCC | 0.34 | 0.69 | 63.6 | 88.2 | 48.1 | 51.7 | 86.7 |
| wavelet_HHL_glcm_Imc1 | 0.01 | 0.75 | 75.0 | 88.2 | 66.7 | 62.5 | 90.0 |
| wavelet_HHL_firstorder_Kurtosis | 0.33 | 0.69 | 68.2 | 70.6 | 66.7 | 57.1 | 78.3 |
| wavelet_HHL_glszm_SmallAreaHighGrayLevel Emphasis | 0.13 | 0.71 | 68.2 | 82.3 | 59.3 | 56.0 | 84.2 |
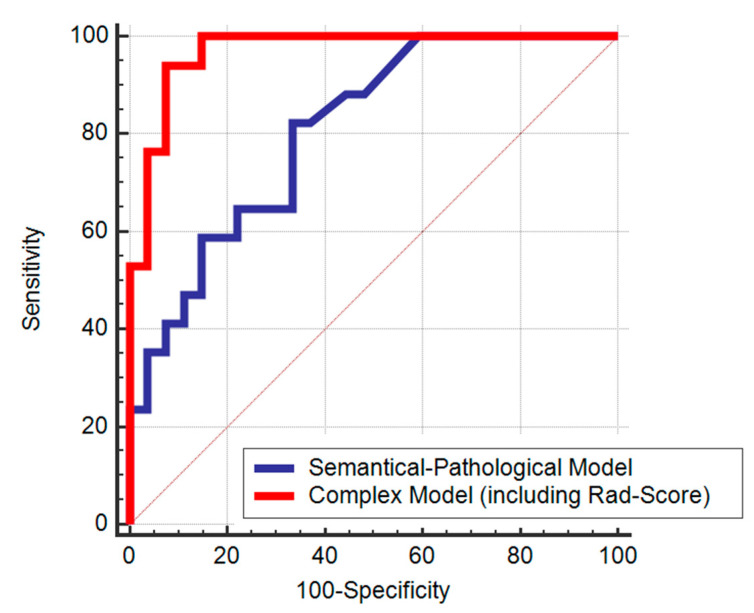
Using the variables with significant difference among responders and non-responders in the training group: tumor length, tumor differentiation grade and MRF status, we conducted a multivariate logistic regression analysis to develop a semantical-pathological model for the prediction of LARC non-response (Table 4). Afterwards, we constructed a complex model, adding the radiomics score to the semantical-pathological model (Table 5). In the complex model, the Rad-Score was identified as an independent predictor of the LARC lack of response to neoadjuvant treatment (OR = 6.52, CI: 1.87–22.72). The ROC curves of the semantic-pathological model and the complex model are shown in Figure 4. Adding the radiomics score to the first model improved its performance for the differentiation of non-responders (AUC = 0.97 (95% CI, 0.66–0.91) vs. AUC = 0.80 (95% CI, 0.87–0.99), p = 0.007).
Table 4.
Multivariate logistic regression analysis for the prediction of the non-responders―semantical-pathological model.
| Variable | Coefficient | Std. Error | p Value | Odds Ratio (OR) | 95% CI | |
|---|---|---|---|---|---|---|
| Upper | Lower | |||||
| Tumor length | 0.03 | 0.02 | 0.23 | 1.03 | 0.98 | 1.08 |
| Tumor differentiation grade—poorly differentiated | −2.67 | 1.23 | 0.10 | 0.07 | 0.05 | 1.30 |
| MRF status—positive | −1.38 | 0.84 | 0.30 | 0.25 | 0.06 | 0.77 |
| Constant | 0.906 | 2.14 | 0.67 | 2.47 | ||
Table 5.
Multivariate logistic regression analysis for the prediction of non-responders―complex model.
| Variable | Coefficient | Std. Error | p Value | Odds Ratio (OR) | 95% CI | |
|---|---|---|---|---|---|---|
| Upper | Lower | |||||
| Tumor length | 0.008 | 0.06 | 0.889 | 1.0008 | 0.90 | 1.13 |
| Tumor differentiation grade—poorly differentiated | −4.561 | 3.30 | 0.167 | 0.10 | 0.00 | 7.92 |
| MRF status—positive | −0.904 | 1.52 | 0.551 | 0.40 | 0.02 | 7.92 |
| Rad-Score | 1.876 | 0.64 | 0.003 * | 6.52 | 1.87 | 22.72 |
| Constant | 4.11 | 5.14 | 0.42 | 61.17 | ||
* Statistically significant p < 0.05.
Figure 4.
ROC curves of the semantical-pathological and complex models score for predicting LARC non-responders.
2.4. Validation of the Radiomics Score
In the validation set, the Rad-Scores of non-responders patients were significantly higher than the scores of responders (0.52 ± 1.74 vs. −1.62 ± 2.36, p = 0.03; Figure 2b). The performance of the Rad-Score for discrimination of non-responders was confirmed in the validation set, yielding an AUC = 0.80 (95% CI, 0.58–0.94) (Figure 3b). For the previous cut-off value of −0.42 (established in the training group), in the validation group there were recorded sensitivity = 75% (95% CI, 34.9–96.8%), specificity = 60% (95% CI, 32.3–83.7%), PPV = 50% (95% CI, 21.1–78.9%), NPV = 81.8% (95% CI, 48.2–97.7%) and accuracy of 65%.
3. Discussion
Our study evaluated the ability of radiomic features extracted from T2-weighted images to help differentiate non-responders (NR) from responders. In recent years, there was an increased interest in the field of radiomics for predicting the rectal cancer response to nCRT, with numerous research being conducted in this respect. The majority of studies focused on predicting the pathological complete response, using a single MRI sequence (T2-WI or apparent diffusion coefficient (ADC) maps) or a multiparametric approach [49,50,51,52,53,54,55]. Additionally, there are several studies that investigated the performance of radiomic features to discriminate good responders [56,57,58,59,60]. In contrast to our research, these investigations either used other pathologic classifications for the quantification of tumor regression grade such as Dworak [57] or Mandard [59] or the authors divided their study population different from our approach, including in the non-responders’ group both patients with a TRG score of 2 and 3 [56,58,60]. In our research, we used the Ryan TRG classification and we considered patients with a TRG score of 3 as non-responders, while patients with TRG 2 were included in the responders’ group. We chose this split method since TRG 3 is equivalent to significant fibrosis outgrown by cancer/no fibrosis with extensive residual cancer, meaning that the neoadjuvant treatment had little to no effect on the rectal tumor, while TRG 2 represents residual cancer outgrown by fibrosis, indicating that the rectal tumor tissue achieved a moderate degree of regression after nCRT [61]. To the best of our knowledge, there are only a few published papers that studied the performance of radiomics features to predict the resistance of rectal cancer to nCRT [62,63,64] and all of them applied the same criteria of defining the rectal cancer non-response as in this present paper. However, two of them used only parameters extracted from the ADC maps for the development of the prediction model [62,63]. In the study by Zhou et al., the authors built a multiparametric prediction model, including features from T2 weighted images, diffusion weighted images and contrast-enhanced T1-weighted images [64].
As well as these previous studies, we analyzed only the pre-treatment, baseline MRI scans. For the prediction of non-responders, it is important to identify these patients in an early phase, before or shortly after the beginning of nCRT in order to be able to adjust their regimen or to offer them alternative treatment options. However, contrary to the above-mentioned investigations, we selected only T2-WI images for our radiomic analysis and model construction. High-resolution T2-weighted sequence is the most important one for the assessment and local staging of rectal cancer since it offers a good visualization of rectal wall layers and it provides good contrast between tumor, surrounding fat and mesorectal fascia [65]. Therefore, it is an essential sequence included in the standard rectal MR protocol and it is always performed when evaluating rectal tumors [66]. Moreover, when compared to images obtained by another sequence, T2-WI images have better stability in appearance. With respect to diffusion weighted images, these can be affected by distortion or by susceptibility artifacts, which may alter tumor segmentation and feature extraction [67,68]. The contrast-enhanced T1-WI sequence is not routinely included in the MRI protocol for rectal cancer staging. The potential of radiomics features extracted from MRI T2-weighted images for predicting a pathological complete response of rectal cancer was demonstrated in several recent studies, which reported promising results of their radiomics models with AUCs ranging from 0.69 to 0.93 [51,52,57,69,70,71]. In contrast to MRI, a recent study had demonstrated that radiomics features extracted from CT images showed no predictive power for complete pathological response in LARC [72], while another research showed that MRI T2-WI radiomics model performed better than CT radiomics model for predicting the LARC response to nCRT [73].
In addition to the previous papers that investigated the performance of radiomics for the prediction of non-response in LARC, our radiomics analysis was performed using the whole tumor volume rather than using only a single section. Although segmenting a volume of interest (VOI) is a time-consuming process, we believe that selection of a single slice might not be very representative, and the information obtained from a 3D lesion might be more reliable for the characterization of the entire tumor.
Regarding the image preprocessing and feature extraction, we extracted radiomics features acquired from both unfiltered and filtered images, applying Laplacian of Gaussian (LoG) filters and Wavelet filters. The majority of the features included in our final radiomic score were obtained from filtered images using the wavelet filters. Wavelet filters are useful for signal denoising and there are several radiomics studies that applied them for different purposes [74,75,76], including in the field of rectal cancer [72,77,78]. In a recent research of He et al., which aimed to develop an MRI-based radiomics signature for tumor grading of rectal carcinoma, the most relevant features included in their classifier were derived from wavelet-filtered images [77]. Additionally, in the predictive model constructed by Liang et al. for the prediction of metachronous liver metastasis in patients with rectal cancer, their optimal selected feature model based on T2-WI consisted only of parameters extracted from filtered images using wavelet filters [78]. Additionally, wavelet filters have proved to be useful even for preprocessing CT images, a significant proportion of the selected features from the recent study of Hamerla et al. for the differentiation of complete LARC responders being obtained from wavelet-filtered images [72].
Our radiomics model mainly consisted of first-order features derived from histogram and second order features derived from the grey-level co-occurrence matrix (GLCM). First-order features describe the distribution of voxel intensities within a region/volume of interest, without considering spatial interactions [34]. Second order or textural features that characterize the spatial relationship between voxels are calculated from different matrices. GLCM quantifies the frequency of specific gray values along a distance or direction [79].
As for the predictive performance of our constructed radiomics model, it achieved AUCs of 0.94 and 0.80 in the training and validation datasets, both indicators showing a good performance. Our results are in concordance with Zhou et al., their radiomics score yielding an AUC of 0.822 [64]. However, their model was a multiparametric one and it included only one feature from T2-WI [64]. In their research, Zhou et al. also developed a single-modality, T2-WI based radiomics signature, which achieved an AUC of 0.602 and 0.630 in the training and validation cohorts, both values being lower than the AUC obtained in the present paper. One possible explanation for these discrepant results can be the different pre-processing and feature extraction algorithm, our study applying in addition wavelet filters, while Zhou et al. extracted the radiomics features only from original and LoG-filtered images. Moreover, our observations are similar with the findings of Liu et al. and Yang et al., which reported AUCs of 0.908 and 0.83 respectively for predicting resistant rectal adenocarcinoma using ADC maps as a basis for feature extraction [62,63]. Additionally, our study was relatively in line with the prior investigations of Yi et al. and Shi et al., whose radiomics models achieved AUCs of 0.90 and 0.91 respectively for distinguishing good responders from non-good responders, although their classification of non-responders was slightly different than ours [57,58]. In our research, the multivariate analysis indicated that the radiomics score was the only independent predictor for the differentiation of LARC non-responders, having an odds ratio of 6.52.
Although our results were significant, this study had some limitations. First, this was a single-institution, retrospective study with a small sample size of patients. Our statistical approach included only one classification method: the binary logistic regression method and more advanced classifiers may provide better prediction performance. Therefore, further larger studies, preferably prospective and multicentric, are needed to overcome these limitations and to validate the reported data in order to provide a better generalization and to assess the potential for clinical translation of our proposed radiomics signature.
4. Materials and Methods
4.1. Study Population
The Institutional Review Board of Regional Institute of Gastroenterology and Hepatology “Prof. Dr Octavian Fodor” Cluj-Napoca approved this retrospective, HIPAA (Health Insurance Portability and Accountability Act)-compliant study and waived the requirement for written informed consent (IRB 5337/22.04.2020). We performed a retrospective analysis in our electronic medical database for patients diagnosed with rectal cancer who underwent an MR examination for initial tumor staging between January 2017 and May 2019. The inclusion criteria were patients diagnosed with locally advanced rectal cancer on pre-treatment (baseline) MR examination who underwent long-course neoadjuvant CRT followed by total mesorectal excision. The exclusion criteria were as follows: patients who did not complete the standard nCRT (12 patients), patients who did not undergo surgical resection (5 patients), patients without available tumor regression grading information on the pathological record (6 patients) and MR examinations with insufficient quality for proper analysis (9 patients). Our final study population consisted of 67 eligible patients, who were divided into a training group (44 patients) and into a validation group (23 patients).
4.2. Image Acquisition
All patients underwent rectum MRI scans 1 or 2 weeks before the start of chemoradiation. The examinations were performed in a single institution, using a 1.5 Tesla MRI scanner (Symphony TIM upgrade, Siemens AG, Erlangen, Germany) with an 8-channel phased array body coil. The protocol included three T2 weighted turbo spin-echo (TSE) sequences in the sagittal, oblique-axial high-resolution and oblique-coronal high-resolution planes. DWI images were obtained in axial planes using EPI sequences at three b-values (b50, b400 and b 800 s/mm2) and restriction of diffusion was quantified by the ADC value. The parameters of the MRI sequences are provided in Table 6. No bowel preparation was received prior to the MRI examination.
Table 6.
MRI parameters.
| MRI Parameter | TSE T2-Weighted Image | DWI | ||
|---|---|---|---|---|
| Sagittal | HR Coronal Oblique | HR Axial Oblique | ||
| TR (ms) | 3500 | 3500 | 4000 | 5800 |
| TE (ms) | 91 | 91 | 80 | 96 |
| Slice no | 28 | 25 | 25 | 30 |
| Bandwidth (Hz/pixel) | 391 | 391 | 391 | 1132 |
| FOV (mm) | 220 | 220 | 200 | 250 |
| Slice thickness (mm) | 3 | 4 | 3 | 4 |
| Matrix | 256 × 256 | 256 × 256 | 256 × 256 | 136 × 160 |
| Acquisition time (min) | 4 | 5.5 | 6 | 4.5 |
4.3. Reference Standard
Surgically resected specimens were histopathologically examined and tumor regression grading (TRG) was established according to the criteria proposed by Ryan et al. [80]: TRG 1 = no viable cancer cells, or single cells, or small groups of cancer cells; TRG 2 = residual cancer outgrown by fibrosis and TRG 3 = significant fibrosis outgrown by cancer, or no fibrosis with extensive residual cancer.
4.4. Preprocessing, Segmentation and Feature Extraction
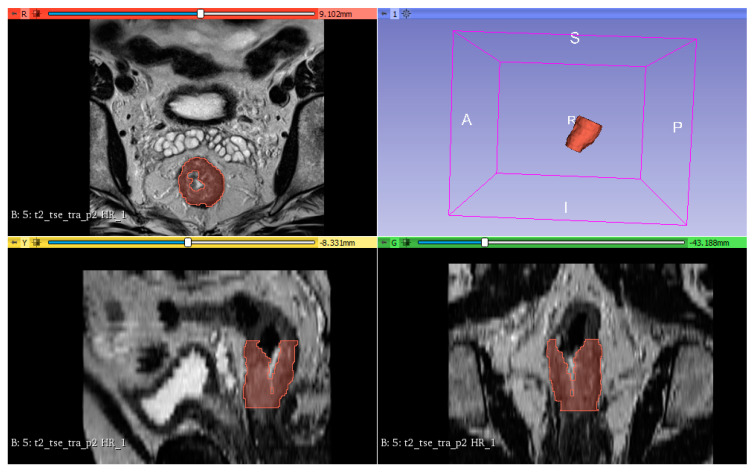
All oblique-axial high-resolution T2-WI TSE image were retrieved from a picture archiving and communication system (PACS, Carestream, Canada) for image segmentation. Three radiologists: one radiology resident (Bianca Petresc) and two senior radiologists with 10 and 8 years of experience (Cosmin Caraiani and Andrei Lebovici) in gastrointestinal MRI reviewed all images and reached a consensus about the tumor location. Afterwards, the radiology resident and one senior radiologist (C.C.) have independently segmented the whole tumor volume, by manually delineating the lesion on each consecutive slide, excluding the uninvaded rectal wall and the intestinal lumen. The segmentations were then independently reviewed by the other senior radiologist (Andrei Lebovici) and adjustments were made by agreement when necessary. All the three radiologists were blinded to the pathological results. The 3D segmentation of the tumors was performed using a designated, open source software 3D Slicer, version 4.10.2 (available at: https://www.slicer.org/). Figure 5 shows an example of tumor segmentation.
Figure 5.
An example of rectal tumor VOI segmentation.
Prior to radiomic feature extraction, all MR images were pre-processed by the method proposed by van Griethuysen et al. [81] for noise reduction, intensity normalization and discretization. Six categories of radiomics features were extracted: 14 shape features, 18 first-order features, 22 gray level co-occurrence matrix (GLCM) features, 16 gray-level size zone matrix (GLSZM) features, 16 gray-level run length matrix (GLRLM) features and 14 gray-level dependence matrix (GLDM) features. Features were automatically extracted from images with and without preprocessing filters. The filters included Laplacian of the Gaussian (LoG) filter with sigma values of 3.0 and 5.0 mm and wavelet filter, using either a low band-pass filter or a high-band pass filter in x, y, z directions. Finally, a total of 934 radiomic features were obtained. PyRadiomics version 2.1.2. [82] was used for pre-processing and feature extraction and detailed information about the PyRadiomics configuration is provided in Supplementary File S1.
4.5. Feature Selection and Statistical Analysis
The inter-reader agreement was evaluated using the intraclass coefficient (ICC) between the features extracted from the radiology resident’s segmentation and the senior radiologist’s segmentation. Only features with an ICC ≥ 0.75 were selected for further feature selection process, resulting in a total of 874 features. All radiomics extracted features were normalized by transforming the data into standardized ranges across all subjects with a mean of 0 and an SD of 1 (z-score transformation). To control overfitting in our radiomics model, we used three feature selection steps. First, we performed a univariate analysis using the Mann–Whitney U-test to identify the features with significant difference between the responders and non-responders groups. The Benjamini–Hochberg (BH) method was used to adjust for multiple testing. BH-adjusted p values less than 0.05 were considered significant. Secondly, the Spearman correlation analysis was used to reduce redundancy. This was conducted between any 2 features and when the Spearman coefficient was >0.9/< − 0.9 the feature with the higher p-value in the univariate analysis was eliminated. Finally, regularized multivariate logistic regression analysis with the least absolute shrinkage and selection operator (LASSO) conducted by a 10 cross-fold cross-validation was applied to the previously selected features. The final selected features were combined into a radiomics score, which was calculated by a linear combination of the selected features weighted by their respective LASSO coefficients. Receiver operating characteristic (ROC) curve analysis was conducted and area under the curve (AUC), sensitivity, specificity and accuracy were calculated to evaluate the performance of the radiomics score for the prediction of non-responders in both training and validation sets. Multivariate analysis using binary logistic regression (enter method) was performed to identify independent predictors of non-responders, including as independent variables the patients’ and tumors’ characteristics and the radiomics score. p values < 0.05 were considered statistically significant. All statistical analysis was performed using commercially available software SPSS Statistics for Windows, version 18.0 (SPSS Inc., Chicago, IL, USA) and free available R software version 3.6.3 using the “glmnet”, “selectiveinference” and “corrplot” packages.
5. Conclusions
Our study has shown that radiomics features extracted from pre-treatment T2-weighted images may play a role as a potential imaging biomarker to predict rectal cancer resistance to neoadjuvant CRT. However, to confirm its ability for the prediction of LARC non-response, our proposed radiomics model needs to be externally validated in larger, multicentric studies.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/7/1894/s1, Supplementary File S1: PyRadiomics Configuration.
Appendix A
Table A1.
List of selected radiomics features by univariate analysis for classification of non-responders vs. responders.
| Feature | p Value * |
|---|---|
| log-sigma-5-0-mm-3DglszmSizeZoneNonUniformityNormalized | 0.001 |
| log-sigma-5-0-mm-3Dglszm_SizeZoneNonUniformity | 0.013 |
| log-sigma-5-0-mm-3DglszmGrayLevelNonUniformity | 0.044 |
| log-sigma-5-0-mm-3Dglszm_SmallAreaEmphasis | 0.001 |
| wavelet-LHL_glcm_Contrast | 0.035 |
| wavelet-LHLglcmDifferenceEntropy | 0.033 |
| wavelet-LHLglcmInverseVariance | 0.044 |
| wavelet-LHLglcmIdm | 0.044 |
| wavelet-LHLglcmCorrelation | 0.009 |
| wavelet-LHLglcmSumEntropy | 0.049 |
| wavelet-LHLglcmImc2 | 0.033 |
| wavelet-LHLglcmImc1 | 0.020 |
| wavelet-LHLglcmDifferenceAverage | 0.031 |
| wavelet-LHLglcmId | 0.042 |
| wavelet-LHLgldmDependenceEntropy | 0.033 |
| wavelet-LHLgldmSmallDependenceEmphasis | 0.042 |
| wavelet-LHLgldmDependenceNonUniformityNormalized | 0.044 |
| wavelet-LHLfirstorderInterquartileRange | 0.037 |
| wavelet-LHLfirstorderUniformity | 0.037 |
| wavelet-LHLfirstorderRobustMeanAbsoluteDeviation | 0.042 |
| wavelet-LHLfirstorderEntropy | 0.047 |
| wavelet-LHLfirstorder10Percentile | 0.020 |
| wavelet-LHLglrlmGrayLevelNonUniformityNormalized | 0.042 |
| wavelet-LHLglrlmRunVariance | 0.044 |
| wavelet-LHLglrlmRunEntropy | 0.047 |
| wavelet-LHLglszmGrayLevelVariance | 0.047 |
| wavelet-LHLglszmGrayLevelNonUniformityNormalized | 0.031 |
| wavelet-LHLglszmZonePercentage | 0.047 |
| wavelet-LLHgldmDependenceEntropy | 0.014 |
| wavelet-LLHglszmGrayLevelNonUniformityNormalized | 0.031 |
| wavelet-LLHglszmSizeZoneNonUniformity | 0.049 |
| wavelet-HHLglcmJointAverage | 0.039 |
| wavelet-HHLglcmSumAverage | 0.039 |
| wavelet-HHLglcmContrast | 0.042 |
| wavelet-HHLglcmDifferenceEntropy | 0.047 |
| wavelet-HHLglcmDifferenceVariance | 0.037 |
| wavelet-HHLglcmAutocorrelation | 0.037 |
| wavelet-HHLglcmSumEntropy | 0.049 |
| wavelet-HHLglcmMCC | 0.037 |
| wavelet-HHLglcmSumSquares | 0.039 |
| wavelet-HHLglcmClusterProminence | 0.021 |
| wavelet-HHLglcmImc2 | 0.010 |
| wavelet-HHLglcmImc1 | 0.005 |
| wavelet-HHLglcmDifferenceAverage | 0.047 |
| wavelet-HHLglcmClusterTendency | 0.033 |
| wavelet-HHLgldmGrayLevelVariance | 0.031 |
| wavelet-HHLgldmHighGrayLevelEmphasis | 0.023 |
| wavelet-HHLgldmSmallDependenceHighGrayLevelEmphasis | 0.029 |
| wavelet-HHLgldmDependenceNonUniformityNormalized | 0.047 |
| wavelet-HHLgldmLargeDependenceEmphasis | 0.042 |
| wavelet-HHLgldmLargeDependenceLowGrayLevelEmphasis | 0.039 |
| wavelet-HHLgldmDependenceVariance | 0.039 |
| wavelet-HHLfirstorderMeanAbsoluteDeviation | 0.049 |
| wavelet-HHLfirstorderMaximum | 0.049 |
| wavelet-HHLfirstorderRootMeanSquared | 0.035 |
| wavelet-HHLfirstorderMinimum | 0.029 |
| wavelet-HHLfirstorderRange | 0.033 |
| wavelet-HHLfirstorderVariance | 0.031 |
| wavelet-HHLfirstorderKurtosis | 0.039 |
| wavelet-HHLglrlmGrayLevelVariance | 0.031 |
| wavelet-HHLglrlmGrayLevelNonUniformityNormalized | 0.044 |
| wavelet-HHLglrlmRunVariance | 0.047 |
| wavelet-HHLglrlmLongRunEmphasis | 0.047 |
| wavelet-HHLglrlmShortRunHighGrayLevelEmphasis | 0.027 |
| wavelet-HHLglrlmShortRunEmphasis | 0.047 |
| wavelet-HHLglrlmLongRunHighGrayLevelEmphasis | 0.031 |
| wavelet-HHLglrlmRunPercentage | 0.047 |
| wavelet-HHLglrlmRunEntropy | 0.042 |
| wavelet-HHLglrlmHighGrayLevelRunEmphasis | 0.021 |
| wavelet-HHLglrlmRunLengthNonUniformityNormalized | 0.047 |
| wavelet-HHLglszmGrayLevelVariance | 0.026 |
| wavelet-HHLglszmSmallAreaHighGrayLevelEmphasis | 0.019 |
| wavelet-HHLglszmZonePercentage | 0.049 |
| wavelet-HHLglszmHighGrayLevelZoneEmphasis | 0.023 |
* Adjusted p-values for multiple tests (Benjamini–Hochberg method).
Figure A1.
Correlation matrix of the radiomics features using Spearman coefficients of correlation.
Author Contributions
Conceptualization, B.P., A.L., C.C. and M.M.B.; methodology, B.P., A.L. and C.C.; formal analysis, B.P. and D.S.F.; investigation, B.P., A.L. and C.C.; resources, F.G., A.L., C.C. and D.S.F.; data curation, B.P.; writing—original draft preparation, B.P.; writing—review and editing, F.G., A.L. and C.C.; visualization, B.P.; supervision, M.M.B.; project administration, M.M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fedewa S.A., Ahnen D.J., Meester R.G.S., Barzi A., Jemal A. Colorectal Cancer Statistics, 2017. CA Cancer J. Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 3.van Gijn W., Marijnen C.A.M., Nagtegaal I.D., Kranenbarg E.M.K., Putter H., Wiggers T., Rutten H.J.T., Påhlman L., Glimelius B., van de Velde C.J.H. Preoperative Radiotherapy Combined with Total Mesorectal Excision for Resectable Rectal Cancer: 12-Year Follow-up of the Multicentre, Randomised Controlled TME Trial. Lancet Oncol. 2011;12:575–582. doi: 10.1016/S1470-2045(11)70097-3. [DOI] [PubMed] [Google Scholar]
- 4.Benson A.B., Venook A.P., Al-Hawary M.M., Cederquist L., Chen Y.J., Ciombor K.K., Cohen S., Cooper H.S., Deming D., Engstrom P.F., et al. Rectal Cancer, Version 2.2018, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2018;16:874–901. doi: 10.6004/jnccn.2018.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van de Velde C.J.H., Boelens P.G., Borras J.M., Coebergh J.W., Cervantes A., Blomqvist L., Beets-Tan R.G.H., van den Broek C.B.M., Brown G., van Cutsem E., et al. EURECCA Colorectal: Multidisciplinary Management: European Consensus Conference Colon & Rectum. Eur. J. Cancer. 2014;50:1.e1–1.e34. doi: 10.1016/j.ejca.2013.06.048. [DOI] [PubMed] [Google Scholar]
- 6.Kong J.C., Guerra G.R., Warrier S.K., Lynch A.C., Michael M., Ngan S.Y., Phillips W., Ramsay G., Heriot A.G. Prognostic value of tumour regression grade in locally advanced rectal cancer: A systematic review and meta-analysis. Colorectal Dis. 2018;20:574–585. doi: 10.1111/codi.14106. [DOI] [PubMed] [Google Scholar]
- 7.Vironen J., Juhola M., Kairaluoma M., Jantunen I., Kellokumpu I. Tumour Regression Grading in the Evaluation of Tumour Response after Different Preoperative Radiotherapy Treatments for Rectal Carcinoma. Int. J. Colorectal Dis. 2005;20:440–445. doi: 10.1007/s00384-004-0733-y. [DOI] [PubMed] [Google Scholar]
- 8.Bujko K., Kolodziejczyk M., Nasierowska-Guttmejer A., Michalski W., Kepka L., Chmielik E., Wojnar A., Chwalinski M. Tumour Regression Grading in Patients with Residual Rectal Cancer after Preoperative Chemoradiation. Radiother. Oncol. 2010;95:298–302. doi: 10.1016/j.radonc.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Ryan J.E., Warrier S.K., Lynch A.C., Heriot A.G. Assessing Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer: A Systematic Review. Colorectal Dis. 2015;17:849–861. doi: 10.1111/codi.13081. [DOI] [PubMed] [Google Scholar]
- 10.Park I.J., You Y.N., Agarwal A., Skibber J.M., Rodriguez-Bigas M.A., Eng C., Feig B.W., Das P., Krishnan S., Crane C.H., et al. Neoadjuvant Treatment Response as an Early Response Indicator for Patients with Rectal Cancer. J. Clin. Oncol. 2012;30:1770–1776. doi: 10.1200/JCO.2011.39.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connell M.J., Colangelo L.H., Beart R.W., Petrelli N.J., Allegra C.J., Sharif S., Pitot H.C., Shields A.F., Landry J.C., Ryan D.P., et al. Capecitabine and Oxaliplatin in the Preoperative Multimodality Treatment of Rectal Cancer: Surgical End Points from National Surgical Adjuvant Breast and Bowel Project Trial R-04. J. Clin. Oncol. 2014;32:1927–1934. doi: 10.1200/JCO.2013.53.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fokas E., Liersch T., Fietkau R., Hohenberger W., Beissbarth T., Hess C., Becker H., Ghadimi M., Mrak K., Merkel S., et al. Tumor Regression Grading after Preoperative Chemoradiotherapy for Locally Advanced Rectal Carcinoma Revisited: Updated Results of the CAO/ARO/AIO-94 Trial. J. Clin. Oncol. 2014;32:1554–1562. doi: 10.1200/JCO.2013.54.3769. [DOI] [PubMed] [Google Scholar]
- 13.Gérard J.P., Azria D., Gourgou-Bourgade S., Martel-Lafay I., Hennequin C., Etienne P.L., Vendrely V., François E., de La Roche G., Bouché O., et al. Clinical Outcome of the ACCORD 12/0405 PRODIGE 2 Randomized Trial in Rectal Cancer. J. Clin. Oncol. 2012;30:4558–4565. doi: 10.1200/JCO.2012.42.8771. [DOI] [PubMed] [Google Scholar]
- 14.Fokas E., Strobel P., Fietkau R., Ghadimi M., Liersch T., Grabenbauer G.G., Hartmann A., Kaufmann M., Sauer R., Graeven U., et al. Tumor regression grading after preoperative chemoradiotherapy as a prognostic factor and individual-level surrogate for disease-free survival in rectal cancer. J. Natl. Cancer Inst. 2017;109:109. doi: 10.1093/jnci/djx095. [DOI] [PubMed] [Google Scholar]
- 15.Carlomagno C., Farella A., Bucci L., D’Armiento F.P., Pesce G., Pepe S., Cannella L., Pacelli R., de Stefano A., Solla R., et al. Neo-adjuvant treatment of rectal cancer with capecitabine and oxaliplatin in combination with radiotherapy: A phase II study. Ann. Oncol. 2009;20:906–912. doi: 10.1093/annonc/mdn719. [DOI] [PubMed] [Google Scholar]
- 16.Fokas E., Allgäuer M., Polat B., Klautke G., Grabenbauer G.G., Fietkau R., Kuhnt T., Staib L., Brunner T., Grosu A.L., et al. Randomized Phase II Trial of Chemoradiotherapy plus Induction or Consolidation Chemotherapy as Total Neoadjuvant Therapy for Locally Advanced Rectal Cancer: CAO/ARO/AIO-12. J. Clin. Oncol. 2019;37:3212–3222. doi: 10.1200/JCO.19.00308. [DOI] [PubMed] [Google Scholar]
- 17.Wolff H.A., Conradi L.C., Beissbarth T., Leha A., Hohenberger W., Merkel S., Fietkau R., Raab H.R., Tschmelitsch J., Hess C.F., et al. Gender Affects Acute Organ Toxicity during Radiochemotherapy for Rectal Cancer: Long-Term Results of the German CAO/ARO/AIO-94 Phase III Trial. Radiother. Oncol. 2013;108:48–54. doi: 10.1016/j.radonc.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Herman J.M., Narang A.K., Griffith K.A., Zalupski M.M., Reese J.B., Gearhart S.L., Azad N.S., Chan J., Olsen L., Efron J.E., et al. The quality-of-life effects of neoadjuvant chemoradiation in locally advanced rectal cancer. Int. J. Radiat. Oncol. Biol. Phys. 2013;85:e15–e19. doi: 10.1016/j.ijrobp.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loos M., Quentmeier P., Schuster T., Nitsche U., Gertler R., Keerl A., Kocher T., Friess H., Rosenberg R. Effect of Preoperative Radio(Chemo)Therapy on Long-Term Functional Outcome in Rectal Cancer Patients: A Systematic Review and Meta-Analysis. Ann. Surg. Oncol. 2013;20:1816–1828. doi: 10.1245/s10434-012-2827-z. [DOI] [PubMed] [Google Scholar]
- 20.Sauer R., Becker H., Hohenberger W., Rödel C., Wittekind C., Fietkau R., Martus P., Tschmelitsch J., Hager E., Hess C.F., et al. Preoperative versus Postoperative Chemoradiotherapy for Rectal Cancer. N. Engl. J. Med. 2004;351:1731–1740. doi: 10.1056/NEJMoa040694. [DOI] [PubMed] [Google Scholar]
- 21.Rödel C., Martus P., Papadoupolos T., Füzesi L., Klimpfinger M., Fietkau R., Liersch T., Hohenberger W., Raab R., Sauer R., et al. Prognostic Significance of Tumor Regression after Preoperative Chemoradiotherapy for Rectal Cancer. J. Clin. Oncol. 2005;23:8688–8696. doi: 10.1200/JCO.2005.02.1329. [DOI] [PubMed] [Google Scholar]
- 22.Huh J.W., Kim H.C., Kim S.H., Park Y.A., Cho Y.B., Yun S.H., Lee W.Y., Park H.C., Choi D.H., Park J.O., et al. Tumor Regression Grade as a Clinically Useful Outcome Predictor in Patients with Rectal Cancer after Preoperative Chemoradiotherapy. Surgery. 2019;165:579–585. doi: 10.1016/j.surg.2018.08.026. [DOI] [PubMed] [Google Scholar]
- 23.Lee Y.C., Hsieh C.C., Chuang J.P. Prognostic Significance of Partial Tumor Regression after Preoperative Chemoradiotherapy for Rectal Cancer: A Meta-Analysis. Dis. Colon Rectum. 2013;56:1093–1101. doi: 10.1097/DCR.0b013e318298e36b. [DOI] [PubMed] [Google Scholar]
- 24.Gersak M.M., Badea R., Graur F., al Hajjar N., Furcea L., Dudea S.M. Endoscopic Ultrasound for the Characterization and Staging of Rectal Cancer. Current State of the Method. Technological Advances and Perspectives. Med. Ultrason. 2015:227–234. doi: 10.11152/mu.2013.2066.172.gsk. [DOI] [PubMed] [Google Scholar]
- 25.Kalisz K.R., Enzerra M.D., Paspulati R.M. MRI Evaluation of the Response of Rectal Cancer to Neoadjuvant Chemoradiation Therapy. Radiographics. 2019;39:538–556. doi: 10.1148/rg.2019180075. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Aguilar J., Chen Z., Smith D.D., Li W., Madoff R.D., Cataldo P., Marcet J., Pastor C. Identification of a Biomarker Profile Associated with Resistance to Neoadjuvant Chemoradiation Therapy in Rectal Cancer. Ann. Surg. 2011;254:486–493. doi: 10.1097/SLA.0b013e31822b8cfa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia H., Shen X., Guan Y., Xu M., Tu J., Mo M., Xie L., Yuan J., Zhang Z., Cai S., et al. Predicting the Pathological Response to Neoadjuvant Chemoradiation Using Untargeted Metabolomics in Locally Advanced Rectal Cancer. Radiother. Oncol. 2018;128:548–556. doi: 10.1016/j.radonc.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 28.Huh J.W., Lee J.H., Kim H.R. Pretreatment Expression of 13 Molecular Markers as a Predictor of Tumor Responses after Neoadjuvant Chemoradiation in Rectal Cancer. Ann. Surg. 2014;259:508–515. doi: 10.1097/SLA.0b013e31829b3916. [DOI] [PubMed] [Google Scholar]
- 29.Park I.J., Yu Y.S., Mustafa B., Park J.Y., Seo Y.B., Kim G.-D., Kim J., Kim C.M., Noh H.D., Hong S.-M., et al. A Nine-Gene Signature for Predicting the Response to Preoperative Chemoradiotherapy in Patients with Locally Advanced Rectal Cancer. Cancers. 2020;12:800. doi: 10.3390/cancers12040800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dayde D., Tanaka I., Jain R., Tai M.C., Taguchi A. Predictive and Prognostic Molecular Biomarkers for Response to Neoadjuvant Chemoradiation in Rectal Cancer. Int. J. Mol. Sci. 2017;18:573. doi: 10.3390/ijms18030573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambin P., Leijenaar R.T.H., Deist T.M., Peerlings J., de Jong E.E.C., van Timmeren J., Sanduleanu S., Larue R.T.H.M., Even A.J.G., Jochems A., et al. Radiomics: The Bridge between Medical Imaging and Personalized Medicine. Nat. Rev. Clin. Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 33.Mayerhoefer M.E., Materka A., Langs G., Häggström I., Szczypiński P., Gibbs P., Cook G. Introduction to Radiomics. J. Nucl. Med. 2020;61:488–495. doi: 10.2967/jnumed.118.222893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lubner M.G., Smith A.D., Sandrasegaran K., Sahani D.V., Pickhardt P.J. CT Texture Analysis: Definitions, Applications, Biologic Correlates, and Challenges. Radiographics. 2017;37:1483–1503. doi: 10.1148/rg.2017170056. [DOI] [PubMed] [Google Scholar]
- 35.Rogers W., Thulasi Seetha S., Refaee T.A.G., Lieverse R.I.Y., Granzier R.W.Y., Ibrahim A., Keek S.A., Sanduleanu S., Primakov S.P., Beuque M.P.L., et al. Radiomics: From Qualitative to Quantitative Imaging. Br. J. Radiol. 2020;93:20190948. doi: 10.1259/bjr.20190948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capobianco E., Dominietto M. From Medical Imaging to Radiomics: Role of Data Science for Advancing Precision Health. J. Pers. Med. 2020;10:15. doi: 10.3390/jpm10010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dagogo-Jack I., Shaw A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 38.McGranahan N., Swanton C. Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Hardiman K.M., Ulintz P.J., Kuick R.D., Hovelson D.H., Gates C.M., Bhasi A., Rodrigues Grant A., Liu J., Cani A.K., Greenson J.K., et al. Intra-Tumor Genetic Heterogeneity in Rectal Cancer. Lab. Investig. 2016;96:4–15. doi: 10.1038/labinvest.2015.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei L., Osman S., Hatt M., el Naqa I. Machine Learning for Radiomics-Based Multimodality and Multiparametric Modeling. Q. J. Nucl. Med. Mol. Imaging. 2019;63:323–338. doi: 10.23736/S1824-4785.19.03213-8. [DOI] [PubMed] [Google Scholar]
- 41.Cuocolo R., Caruso M., Perillo T., Ugga L., Petretta M. Machine Learning in Oncology: A Clinical Appraisal. Cancer Lett. 2020;481:55–62. doi: 10.1016/j.canlet.2020.03.032. [DOI] [PubMed] [Google Scholar]
- 42.Koenigkam Santos M., Raniery Ferreira Júnior J., Tadao Wada D., Priscilla Magalhães Tenório A., Henrique Nogueira Barbosa M., Mazzoncini De Azevedo Marques P. Artificial Intelligence, Machine Learning, Computer-Aided Diagnosis, and Radiomics: Advances in Imaging towards to Precision Medicine. Radiologia Brasileira. 2019;52:387–396. doi: 10.1590/0100-3984.2019.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papanikolaou N., Matos C., Koh D.M. How to Develop a Meaningful Radiomic Signature for Clinical Use in Oncologic Patients. Cancer Imaging. 2020;20 doi: 10.1186/s40644-020-00311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schick U., Lucia F., Dissaux G., Visvikis D., Badic B., Masson I., Pradier O., Bourbonne V., Hatt M. MRI-derived radiomics: Methodology and clinical applications in the field of pelvic oncology. Br. J. Radiol. 2019;92:20190105. doi: 10.1259/bjr.20190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z., Wang S., Dong D., Wei J., Fang C., Zhou X., Sun K., Li L., Li B., Wang M., et al. The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges. Theranostics. 2019;9:1303–1322. doi: 10.7150/thno.30309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nougaret S., Tibermacine H., Tardieu M., Sala E. Radiomics: An Introductory Guide to What It May Foretell. Curr. Oncol. Rep. 2019;21:70. doi: 10.1007/s11912-019-0815-1. [DOI] [PubMed] [Google Scholar]
- 47.Bi W.L., Hosny A., Schabath M.B., Giger M.L., Birkbak N.J., Mehrtash A., Allison T., Arnaout O., Abbosh C., Dunn I.F., et al. Artificial Intelligence in Cancer Imaging: Clinical Challenges and Applications. CA Cancer J. Clin. 2019;69:127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horvat N., Bates D.D.B., Petkovska I. Novel Imaging Techniques of Rectal Cancer: What Do Radiomics and Radiogenomics Have to Offer? A Literature Review. Abdom. Radiol. 2019;44:3764–3774. doi: 10.1007/s00261-019-02042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z., Zhang X.Y., Shi Y.J., Wang L., Zhu H.T., Tang Z., Wang S., Li X.T., Tian J., Sun Y.S. Radiomics Analysis for Evaluation of Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Clin. Cancer Res. 2017;23:7253–7262. doi: 10.1158/1078-0432.CCR-17-1038. [DOI] [PubMed] [Google Scholar]
- 50.Cui Y., Yang X., Shi Z., Yang Z., Du X., Zhao Z., Cheng X. Radiomics Analysis of Multiparametric MRI for Prediction of Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Eur. Radiol. 2019;29:1211–1220. doi: 10.1007/s00330-018-5683-9. [DOI] [PubMed] [Google Scholar]
- 51.Horvat N., Veeraraghavan H., Khan M., Blazic I., Zheng J., Capanu M., Sala E., Garcia-Aguilar J., Gollub M.J., Petkovska I. MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy. Radiology. 2018;287:833–843. doi: 10.1148/radiol.2018172300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y., Liu W., Pei Q., Zhao L., Güngör C., Zhu H., Song X., Li C., Zhou Z., Xu Y., et al. Predicting Pathological Complete Response by Comparing MRI-Based Radiomics Pre- and Postneoadjuvant Radiotherapy for Locally Advanced Rectal Cancer. Cancer Med. 2019;8:7244–7252. doi: 10.1002/cam4.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang L., Qiu M., Xia C., Li Z., Wang Z., Zhou X., Wu B. Value of High-Resolution DWI in Combination with Texture Analysis for the Evaluation of Tumor Response after Preoperative Chemoradiotherapy for Locally Advanced Rectal Cancer. Am. J. Roentgenol. 2019;212:1279–1286. doi: 10.2214/AJR.18.20689. [DOI] [PubMed] [Google Scholar]
- 54.Shu Z., Fang S., Ye Q., Mao D., Cao H., Pang P., Gong X. Prediction of Efficacy of Neoadjuvant Chemoradiotherapy for Rectal Cancer: The Value of Texture Analysis of Magnetic Resonance Images. Abdom. Radiol. 2019;44:3775–3784. doi: 10.1007/s00261-019-01971-y. [DOI] [PubMed] [Google Scholar]
- 55.Aker M., Ganeshan B., Afaq A., Wan S., Groves A.M., Arulampalam T. Magnetic Resonance Texture Analysis in Identifying Complete Pathological Response to Neoadjuvant Treatment in Locally Advanced Rectal Cancer. Dis. Colon Rectum. 2019;62:163–170. doi: 10.1097/DCR.0000000000001224. [DOI] [PubMed] [Google Scholar]
- 56.Nie K., Shi L., Chen Q., Hu X., Jabbour S.K., Yue N., Niu T., Sun X. Rectal Cancer: Assessment of Neoadjuvant Chemoradiation Outcome Based on Radiomics of Multiparametric MRI. Clin. Cancer Res. 2016;22:5256–5264. doi: 10.1158/1078-0432.CCR-15-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yi X., Pei Q., Zhang Y., Zhu H., Wang Z., Chen C., Li Q., Long X., Tan F., Zhou Z., et al. MRI-Based Radiomics Predicts Tumor Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Front. Oncol. 2019;9:552. doi: 10.3389/fonc.2019.00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi L., Zhang Y., Nie K., Sun X., Niu T., Yue N., Kwong T., Chang P., Chow D., Chen J.H., et al. Machine learning for prediction of chemoradiation therapy response in rectal cancer using pre-treatment and mid-radiation multi-parametric MRI. Magn. Reson. Imaging. 2019;61:33–40. doi: 10.1016/j.mri.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meng Y., Zhang C., Zou S., Zhao X., Xu K., Zhang H., Zhou C. MRI Texture Analysis in Predicting Treatment Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer. Oncotarget. 2018;9:11999–12008. doi: 10.18632/oncotarget.23813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shayesteh S.P., Alikhassi A., Fard Esfahani A., Miraie M., Geramifar P., Bitarafan-rajabi A., Haddad P. Neo-Adjuvant Chemoradiotherapy Response Prediction Using MRI Based Ensemble Learning Method in Rectal Cancer Patients. Phys. Med. 2019;62:111–119. doi: 10.1016/j.ejmp.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 61.Kim S.H., Chang H.J., Kim D.Y., Park J.W., Baek J.Y., Kim S.Y., Park S.C., Oh J.H., Yu A., Nam B.H. What Is the Ideal Tumor Regression Grading System in Rectal Cancer Patients after Preoperative Chemoradiotherapy? Cancer Res. Treat. 2016;48:998–1009. doi: 10.4143/crt.2015.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu M., Lv H., Liu L.H., Yang Z.H., Jin E.H., Wang Z.C. Locally Advanced Rectal Cancer: Predicting Non-Responders to Neoadjuvant Chemoradiotherapy Using Apparent Diffusion Coefficient Textures. Int. J. Colorectal Dis. 2017;32:1009–1012. doi: 10.1007/s00384-017-2835-3. [DOI] [PubMed] [Google Scholar]
- 63.Yang C., Jiang Z.K., Liu L.H., Zeng M.S. Pre-Treatment ADC Image-Based Random Forest Classifier for Identifying Resistant Rectal Adenocarcinoma to Neoadjuvant Chemoradiotherapy. Int. J. Colorectal Dis. 2020;35:101–107. doi: 10.1007/s00384-019-03455-3. [DOI] [PubMed] [Google Scholar]
- 64.Zhou X., Yi Y., Liu Z., Cao W., Lai B., Sun K., Li L., Zhou Z., Feng Y., Tian J. Radiomics-Based Pretherapeutic Prediction of Non-Response to Neoadjuvant Therapy in Locally Advanced Rectal Cancer. Ann. Surg. Oncol. 2019;26:1676–1684. doi: 10.1245/s10434-019-07300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gourtsoyianni S., Papanikolaou N. Role of Magnetic Resonance Imaging in Primary Rectal Cancer-Standard Protocol and Beyond. Semin. Ultrasound CT MRI. 2016;37:323–330. doi: 10.1053/j.sult.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 66.Horvat N., Rocha C.C.T., Oliveira B.C., Petkovska I., Gollub M.J. MRI of Rectal Cancer: Tumor Staging, Imaging Techniques, and Management. Radiographics. 2019;39:367–387. doi: 10.1148/rg.2019180114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Treiber J.M., White N.S., Steed T.C., Bartsch H., Holland D., Farid N., McDonald C.R., Carter B.S., Dale A.M., Chen C.C. Characterization and Correction of Geometric Distortions in 814 Diffusion Weighted Images. PLoS ONE. 2016;11:e0152472. doi: 10.1371/journal.pone.0152472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tao R., Fletcher P.T., Gerber S., Whitaker R.T. A Variational Image-Based Approach to the Correction of Susceptibility Artifacts in the Alignment of Diffusion Weighted and Structural MRI; Proceedings of the 21st International Conference, IPMI 2009; Williamsburg, VA, USA. 5–10 July 2009; pp. 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Antunes J.T., Ofshteyn A., Bera K., Wang E.Y., Brady J.T., Willis J.E., Friedman K.A., Marderstein E.L., Kalady M.F., Stein S.L., et al. Radiomic Features of Primary Rectal Cancers on Baseline T2-Weighted MRI Are Associated With Pathologic Complete Response to Neoadjuvant Chemoradiation: A Multisite Study. J. Magn. Reson. Imaging. 2020 doi: 10.1002/jmri.27140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cusumano D., Dinapoli N., Boldrini L., Chiloiro G., Gatta R., Masciocchi C., Lenkowicz J., Casà C., Damiani A., Azario L., et al. Fractal-Based Radiomic Approach to Predict Complete Pathological Response after Chemo-Radiotherapy in Rectal Cancer. Radiol. Med. 2018;123:286–295. doi: 10.1007/s11547-017-0838-3. [DOI] [PubMed] [Google Scholar]
- 71.Shaish H., Aukerman A., Vanguri R., Spinelli A., Armenta P., Jambawalikar S., Makkar J., Bentley-Hibbert S., del Portillo A., Kiran R., et al. Radiomics of MRI for Pretreatment Prediction of Pathologic Complete Response, Tumor Regression Grade, and Neoadjuvant Rectal Score in Patients with Locally Advanced Rectal Cancer Undergoing Neoadjuvant Chemoradiation: An International Multicenter Study. Eur. Radiol. 2020 doi: 10.1007/s00330-020-06968-6. [DOI] [PubMed] [Google Scholar]
- 72.Hamerla G., Meyer H.-J., Hambsch P., Wolf U., Kuhnt T., Hoffmann K.-T., Surov A. Radiomics Model Based on Non-Contrast CT Shows No Predictive Power for Complete Pathological Response in Locally Advanced Rectal Cancer. Cancers. 2019;11:1680. doi: 10.3390/cancers11111680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Z.Y., Wang X.D., Li M., Liu X.J., Ye Z., Song B., Yuan F., Yuan Y., Xia C.C., Zhang X., et al. Multi-Modal Radiomics Model to Predict Treatment Response to Neoadjuvant Chemotherapy for Locally Advanced Rectal Cancer. World J Gastroenterol. 2020;26:2388–2402. doi: 10.3748/wjg.v26.i19.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moradmand H., Aghamiri S.M.R., Ghaderi R. Impact of Image Preprocessing Methods on Reproducibility of Radiomic Features in Multimodal Magnetic Resonance Imaging in Glioblastoma. J. Appl. Clin. Med Phys. 2020;21:179–190. doi: 10.1002/acm2.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stanzione A., Cuocolo R., Cocozza S., Romeo V., Persico F., Fusco F., Longo N., Brunetti A., Imbriaco M. Detection of Extraprostatic Extension of Cancer on Biparametric MRI Combining Texture Analysis and Machine Learning: Preliminary Results. Acad. Radiol. 2019;26:1338–1344. doi: 10.1016/j.acra.2018.12.025. [DOI] [PubMed] [Google Scholar]
- 76.Tan X., Ma Z., Yan L., Ye W., Liu Z., Liang C. Radiomics Nomogram Outperforms Size Criteria in Discriminating Lymph Node Metastasis in Resectable Esophageal Squamous Cell Carcinoma. Eur. Radiol. 2019;29:392–400. doi: 10.1007/s00330-018-5581-1. [DOI] [PubMed] [Google Scholar]
- 77.He B., Ji T., Zhang H., Zhu Y., Shu R., Zhao W., Wang K. MRI-Based Radiomics Signature for Tumor Grading of Rectal Carcinoma Using Random Forest Model. J. Cell. Physiol. 2019;234:20501–20509. doi: 10.1002/jcp.28650. [DOI] [PubMed] [Google Scholar]
- 78.Liang M., Cai Z., Zhang H., Huang C., Meng Y., Zhao L., Li D., Ma X., Zhao X. Machine Learning-Based Analysis of Rectal Cancer MRI Radiomics for Prediction of Metachronous Liver Metastasis. Acad. Radiol. 2019;26:1495–1504. doi: 10.1016/j.acra.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 79.Haralick R.M., Shanmugam K. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973;3:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 80.Ryan R., Gibbons D., Hyland J.M.P., Treanor D., White A., Mulcahy H.E., O’Donoghue D.P., Moriarty M., Fennelly D., Sheahan K. Pathological Response Following Long-Course Neoadjuvant Chemoradiotherapy for Locally Advanced Rectal Cancer. Histopathology. 2005;47:141–146. doi: 10.1111/j.1365-2559.2005.02176.x. [DOI] [PubMed] [Google Scholar]
- 81.van Griethuysen J.J.M., Lambregts D.M.J., Trebeschi S., Lahaye M.J., Bakers F.C.H., Vliegen R.F.A., Beets G.L., Aerts H.J.W.L., Beets-Tan R.G.H. Radiomics Performs Comparable to Morphologic Assessment by Expert Radiologists for Prediction of Response to Neoadjuvant Chemoradiotherapy on Baseline Staging MRI in Rectal Cancer. Abdom. Radiol. 2020;45:632–643. doi: 10.1007/s00261-019-02321-8. [DOI] [PubMed] [Google Scholar]
- 82.van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., Beets-Tan R.G.H., Fillion-Robin J.C., Pieper S., Aerts H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.