Abstract
Background
Fibrinogen-like protein 2 (FGL2) is a member of the fibrinogen-like protein family and possesses important regulatory functions in both innate and adaptive immune responses. FGL2 is overexpressed in glioma, and its expression level is negatively associated with the prognosis of glioma patients. However, the diagnostic value of FGL2 is unknown in breast carcinoma.
Material/Methods
We comprehensively analyzed the expression pattern of FGL2 in breast cancer. Several online databases – TCGA, Oncomine, GEPIA, Kaplan-Meier plotter, and PrognoScan – were used in this study.
Results
Based on the TCGA dataset and Oncomine database, we found that the expression level of FGL2 was remarkably lower in breast cancer compared with adjacent normal tissues. Clinical data showed that the expression level of FGL2 was significantly associated with radiation therapy, PR status, and tumor stage. Bioinformatics analysis of the GEPIA, Kaplan-Meier plotter, and PrognoScan databases showed that lower FGL2 expression levels were associated with a worse prognosis in breast cancer patients. Furthermore, the expression level of FGL2 was positively correlated with the immune cell infiltrations in breast cancer, especially those cells with high antitumor activities. GO, KEGG, and GSEA analyses also validated that FGL2 was closely related to genes involved in the immune response, signal transduction, and T cell receptor signaling pathway in breast cancer.
Conclusions
The results demonstrated that high expression of FGL2 is a useful marker for breast cancer treatment and appears to be correlated with enhanced antitumor activities in breast cancer patients.
MeSH Keywords: Fibrinogen; Lymphocytes, Tumor-Infiltrating; Prognosis
Background
Breast cancer is a common invasive cancer that is the second leading cause of cancer-related deaths in women [1]. It is estimated that more than 268 600 women were diagnosed with breast carcinoma in the USA in 2019 [2]. In women, breast cancer is responsible for 30% of all new cancer diagnoses in the USA. In East Asia, the morbidity rate of breast cancer rapidly increased in the last decade (3). In contrast to Western countries, more than 40% of cases occur in women under 50 years old in East Asia. Thus, breast cancer has become a heavy economic and social burden worldwide. Risk factors of breast cancer include genetic factors, environmental exposure, dietary factors, and increasing age [4]. BRCA mutation is correlated with higher morbidity in breast cancer [5]. Other significant gene mutations include CDH1, CHEK2, NF1, PALB2, and PTEN. A multidisciplinary approach is useful in the treatment of breast cancer [6], and molecular subtypes should be considered in a multidisciplinary setting. The therapeutic targets and clinically relevant biomarkers are HER2, ER and PR in breast cancer. Trastuzumab (Herceptin), a monoclonal antibody to HER2, has been demonstrated to prolong disease-free survival (DFS) in HER2-positive breast cancer patients [8]. Next-generation sequencing (NGS) and mutation analysis may provide new therapeutic targets to benefit breast cancer patients [9].
Fibrinogen-like protein 2 (FGL2) belongs to the fibrinogen-like protein family [10], which is mainly expressed in macrophages, T cells, endothelial cells, and tumor cells. FGL2 is a vital immune regulator of both innate and adaptive responses, with pleiotropic effects [11]. It is usually confirmed to possess multifunctional activities and immune regulatory functions in inflammation, allograft rejection, and abortion. Furthermore, low levels of FGL2 can lead ti impaired Treg cell activities [12]. Alterations in FGL2 structure or expression level are correlated with deadly viral infections such as HIV and coronavirus [13,14]. Previous studies have demonstrated that upregulated FGL2 levels were associated with shorter overall survival in glioblastoma (GBM) patients [15]. However, the relationship between breast cancer and FGL2 has not been thoroughly explored.
In our study, we utilized bioinformatics methods to analyze the FGL2 expression pattern in breast cancer. TCGA datasets and the Oncomine database were employed to contrast FGL2 expression in breast cancer and normal adjacent tissues. The TCGA dataset was analyzed to assess the relationship between FGL2 and clinical characteristics of breast cancer patients. We used the online datasets PrognoScan, GEPIA, and Kaplan-Meier plotter to evaluate the prognostic potential of FGL2 in breast cancer, and the TIMER dataset was used to analyze the relationship between FGL2 and immune marker sets and tumor-infiltrating lymphocytes. The STRING website was utilized for FGL2 protein–protein interaction network screening. GO and KEGG calculates on the DAVID website were used to identify the main biological signatures of FGL2-correlated genes in breast cancer, and the GSEA dataset was used to analyze the biological functions of FGL2. The framework and concise content of this study are shown in Figure 1.
Figure 1.
Schematic diagram of the study.
Material and Methods
Patients and samples
The transcriptome profiles were downloaded from TCGA dataset. Multiple samples were used to avoid clinical and race bias in different studies. The expression level of the desired mRNA was normalized by R language for further analysis.
Oncomine database analysis
In this study, the mRNA expression differences of FGL2 between cancers and adjacent normal tissues were investigated with the Oncomine databases. The threshold P-value was determined as 0.001. The threshold fold change was determined to be 2.0. The threshold gene rank was the top 10%.
GEPIA database analysis
The prognostic potential of FGL2 in breast cancer was evaluated in the GEPIA database (http://gepia.cancerpku.cn/). GEPIA is a new interactive website based on genotype-tissue expression (GTEx) and TCGA data.
PrognoScan database analysis
The prognostic potential of FGL2 was validated in the PrognoScan dataset. Cox P-value <0.05 was considered as the threshold value.
Kaplan-Meier plotter dataset
In this study, the association between FGL2 expression and prognosis of breast cancer was tested by Kaplan-Meier plotter. The log-rank P-value and hazard ratio (HR) with 95% confidence intervals were also calculated.
TIMER database analysis
The TIMER database was utilized to visualize the correlation of FGL2 expression with immune cell infiltration in breast cancer. The infiltrated immune cells included CD8+ T cells, macrophages, B cells, CD4+ T cells, neutrophils, and dendritic cells. Furthermore, the associations between FGL2 and gene markers of tumor-infiltration immune cells were investigated with correlation modules.
Protein–protein interaction network construction
The STRING database was used to identify the protein–protein interactions of FGL2. The number of edges, number of nodes, average node degree, and PPI enrichment value can be evaluated by the STRING database. Nodes in the network were classified by their corresponding role in biological processes. Edges were classified based on the molecular function of FGL2. The connectivity degree of each protein node was calculated, and the top hub nodes were determined in the PPI network.
Functional enrichment analyses
Biological process (BP) of GO and KEGG analysis were performed in the DAVID website (https://david.ncifcrf.gov/). KEGG pathway analysis can be used to understand high-level functions and utilities of the biological system, linking genomic information to higher-order functional information. GO enrichment analysis stores graphical representations of cellular processes, including membrane transport, signal transduction, metabolism, and cell cycle.
Gene set enrichment analysis (GSEA)
GSEA is a statistical approach to assess whether the genes from particular pathways or other predefined gene sets show concordant differences or statistical significance between biological states. We used the hallmark gene set download from the GSEA website and 28 immune gene sets as the annotate data to analyze the different expression levels of FGL2 enrichment score in different pathways or immune cells. Gene sets with an FDR <10% and a nominal P-value <0.05 were determined as significant.
Statistical analysis
The discrepancies in FGL2 expression were evaluated by t test. Spearman correlation was analyzed by R language. The SPSS 20.0 software was used to measure the data. Log-rank and Cox regression analysis were applied to investigate the prognostic value of FGL2. P<0.05 represented statistical significance.
Results
The mRNA expression levels of FGL2 in breast cancer
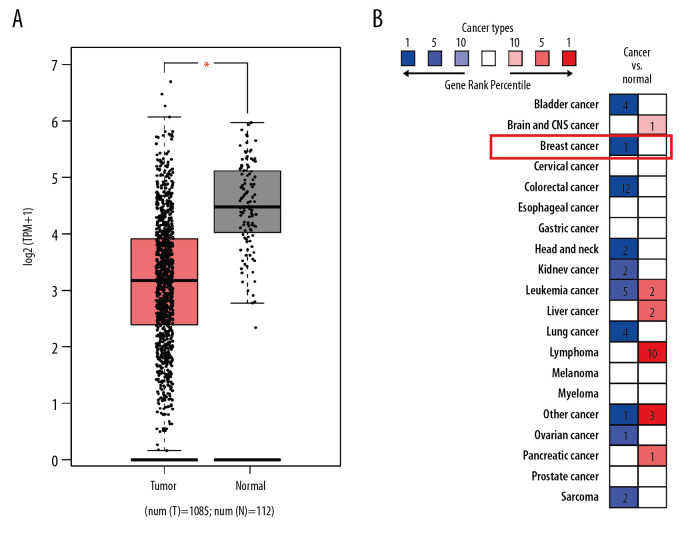
Bioinformatics databases were utilized to evaluate the differential expression of FGL2 between breast cancer and adjacent normal tissues. TCGA database indicated that FGL2 was prominently lower in breast invasive carcinoma (BRCA) compared with that in matched normal tissue (Figure 2). The Oncomine database (Study Accession: EGAS00000000083) also showed that FGL2 expression was lower in breast cancer compared with adjacent normal tissue (P-value <1.01E-12, fold change=−2.578, Gene rank in top 1%). These results indicate that decreased FGL2 expression may be conducive to the tumorigenesis of breast cancer.
Figure 2.
FGL2 expression levels in breast cancer. (A) FGL2 expression levels are significantly lower in breast cancer tissue than levels in adjacent normal tissues in the TCGA database. The left bar represents FGL2 levels in breast cancer tissue. The right bar represents FGL2 levels in matched normal data. (* P<0.05). (B) FGL2 expression in different cancers compared with adjacent normal tissues in the Oncomine database. Threshold (P-value): 1E-4. Threshold (fold change): 2. Threshold (gene rank): top 10%.
The correlation between FGL2 and the clinical characteristics of breast cancer patients
The TCGA database was used to assess the correlation between FGL2 expression and the clinical characteristics in breast cancer. Data of 1104 patients were collected from the TCGA dataset. As shown in Table 1, FGL2 expression was found to be significantly correlated with sex (P=0.004), radiation therapy (P=0.004), and PR status (P=0.013). Furthermore, FGL2 expression was also significantly different among tumor stages (P=0.038).
Table 1.
Association of FGL2 with the clinical pathological characteristics of breast cancer patients derived from the TCGA database.
| Variables | Clinical pathological | Number | Mean±SD | P |
|---|---|---|---|---|
| Sex | Female | 1091 | 2.89±1.06 | 0.004 |
| Male | 12 | 2.00±0.89 | ||
| Radiation therapy | No | 439 | 2.77±1.05 | 0.004 |
| Yes | 553 | 2.96±1.05 | ||
| Age | <60 | 589 | 2.89±1.07 | 0.713 |
| ≥60 | 513 | 2.86±1.04 | ||
| HER2 status | Negative | 290 | 2.92±1.01 | 0.496 |
| Positive | 69 | 2.83±0.95 | ||
| ER status | Negative | 212 | 2.87±1.18 | 0.458 |
| Positive | 727 | 2.93±1.01 | ||
| PR status | Negative | 308 | 2.79±1.13 | 0.013 |
| Positive | 628 | 2.98±1.00 | ||
| T stage | T1 | 281 | 3.02±1.04 | 0.038 |
| T2 | 641 | 2.83±1.07 | ||
| T3 | 138 | 2.93±1.06 | ||
| T4 | 40 | 2.58±0.94 | ||
| N stage | N0 | 519 | 2.88±1.11 | 0.28 |
| N1 | 366 | 2.85±1.01 | ||
| N2 | 120 | 2.95±1.03 | ||
| N3 | 78 | 3.07±0.94 | ||
| M stage | M0 | 917 | 2.87±1.06 | 0.134 |
| M1 | 22 | 2.53±0.93 | ||
| TNM stage | I | 182 | 3.00±1.08 | 0.055 |
| II | 628 | 2.83±1.84 | ||
| III | 250 | 2.95±1.00 | ||
| IV | 20 | 2.45±0.95 |
Prognostic potential of FGL2 in breast cancer
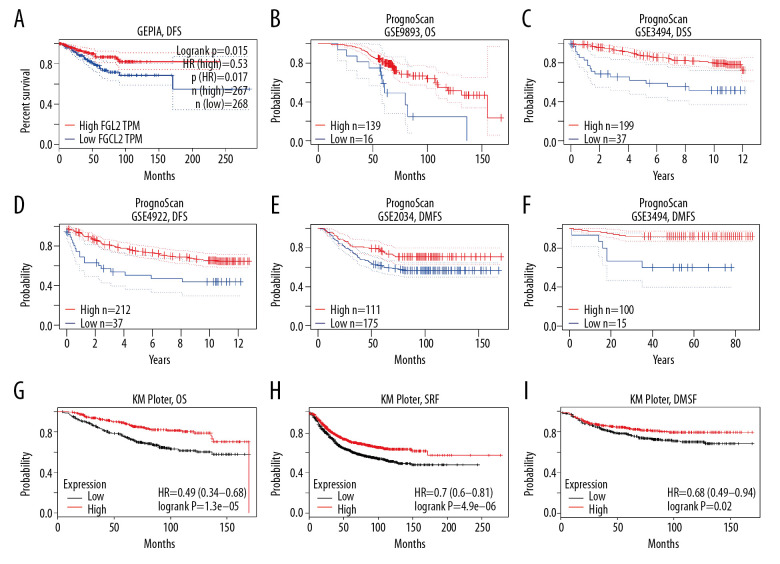
Three datasets (the GEPIA dataset, PrognoScan dataset, and KM plotter dataset) were utilized to explore the association between FGL2 level and breast carcinoma survival rate (Figure 3). In the GEPIA dataset, a lower FGL2 expression was correlated with a worse prognosis of disease-free survival (DFS) in breast cancer (DFS HR=0.53, log-rank P=0.015, cutoff-high=75%). In the PrognoScan database, 5 cohorts were analyzed, and the results showed that lower FGL2 was associated with poor prognosis of breast cancer patients (OS HR=0.59, 95% CI=0.35 to 0.99, Cox P=0.045139; DSS HR=0.60, 95% CI=0.37 to 0.99, Cox P=0.043958; DFS HR=0.73, 95% CI=0.58 to 0.94, Cox P=0.012526; DMFS HR=0.67, 95% CI=0.49 to 0.92, Cox P=0.013025; DMFS HR=0.29, 95% CI=0.14 to 0.60, Cox P=0.000852). Furthermore, the prognostic potential of FGL2 in breast cancer was also confirmed in the KM plotter dataset. Lower FGL2 was related to a unfavorable prognosis of breast cancer (OS HR=0.49, 95% CI=0.35 to 0.68, Cox P=1.3e-05; RFS HR=0.7, 95% CI=0.60 to 0.81, Cox P=4.9e-06; DMFS HR=0.68, 95% CI=0.49 to 0.94, Cox P=0.02).
Figure 3.
Survival rates for FGL2 in breast cancer. The GEPIA database, PrognoScan database, and Kaplan-Meier plotter database were used to analyze the prognostic potential of FGL2 in breast cancer. (A) In the GEPIA database, a lower expression level of FGL2 was associated with a poor prognosis of disease-free survival (DFS) in breast cancer. (B–F) In the PrognoScan database, GSE9893, GSE3494, GSE4922, GSE2034, and GSE19615 were used to evaluate overall survival (OS), disease-specific survival (DSS), disease-free survival (DFS), and distant metastasis-free survival (DMFS), respectively. (G–I) The Kaplan-Meier plotter database was used to measure OS, relapse-free survival (RFS), and DMFS related to FGL2 in breast cancer.
The relationship between FGL2 and immune status in the breast cancer tumor microenvironment
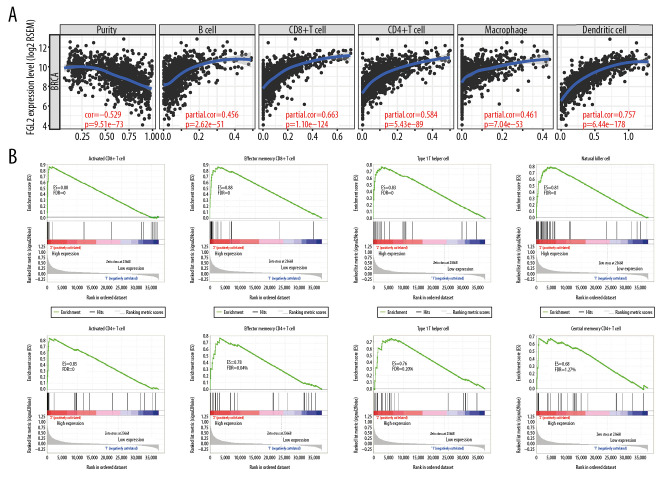
A higher density of tumor-infiltrating lymphocytes (TILs) is an important parameter leading to better prognoses and therapeutic effects in the treatment of cancer. In this study, the TIMER dataset was utilized to investigate the relationship between FGL2 and TILs. The results showed that FGL2 was positively correlated with infiltrating levels of B cells (r=0.456, P=2.62e-51), CD8+ T cells (r=0.663, P=1.10e-124), CD4+ T cells (r=0.584, P=5.43e–89), macrophages (r=0.461, P=7.04e-53), and DCs (r=0.757, P=6.44e-178) in BRCA (Figure 4A). These results suggest that FGL2 has critical roles in immune cell infiltration in breast cancer.
Figure 4.
The correlation between FGL2 expression and immune status in the tumor microenvironment. (A) The FGL2 expression level was positively correlated with infiltrating levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and DCs in BRCA. (B) The FGL2 expression level was positively correlated with infiltrating levels of antitumor immune cells, such as activated CD8+ T cells, effector memory CD8+ T cells, Th1 cells, NK cells, activated CD4+ T cells, effector memory CD4+ T cells, NKT cells, and central memory CD4+ T cells. (ES – enrichment score; FDR – false discovery rate).
Further analyses using the GSEA tool demonstrated that FGL2 was positively correlated with infiltrating levels of antitumor immune cells, such as activated CD8+ T cells (P=0), effector memory CD8+ T cells (P=0), type 1 T helper (Th1) cells (P=0), natural killer (NK) cells (P=0), activated CD4+ T cells (P=0), effector memory CD4+ T cells (P=0), natural killer T (NKT) cells (P=0.003), and central memory CD4+ T cells (P=0.014) (Figure 4B). These results show that higher FGL2 expression level is correlated with strong antitumor activities in breast cancer.
Apart from TILs, we also investigated the relationship between FGL2 expression and immune marker sets of diverse immune infiltrating cells. Detailed correlations between FGL2 and immune marker genes in breast cancer are listed in the Supplementary Tables 1 and 2. Generally speaking, FGL2 was positively correlated with immune marker sets in the TIMER database (Supplementary Table 1) and GEPIA database (Supplementary Table 2). HLA class II histocompatibility antigens are important factors in the antitumor immune response by presenting tumor-specific antigens to CD4+ T cells after phagocytosis of tumors by antigen-presenting cells (APCs). We found that 4 HLA class II genes (HLA-DPA1, HLA-DRA, HLA-DPB1, and HLA-DQB1) were positively correlated with FGL2 expression levels. Class II transcription activator (CIITA), which is responsible for HLA class II gene transcription, is also positively correlated with FGL2. Thus, these results showed that FGL2 can participate in the activities of antigen processing against tumors.
Protein–protein interactions related to FGL2 in the STRING database
The STRING website was utilized to screen the protein–protein interactions related to FGL2. As shown in Figure 5, 21 nodes and 108 edges were filtered in the PPI network complex. Network nodes indicated proteins, and edges indicated the associations between protein and protein. The 10 most significant nodes were: MS4A6A, FGB, FGG, FGA, FGL1, F2, TYROBP, THBS1, FN1, and SERPINF2. In the network nodes, MS4A6A was correlated with FGL2 (score: 0.801). As a multimeric receptor complex, MS4A6A is involved in signal transduction. The results indicated that FGL2 contributes to signal transduction in the immune response correlated with MS4A6A.
Figure 5.
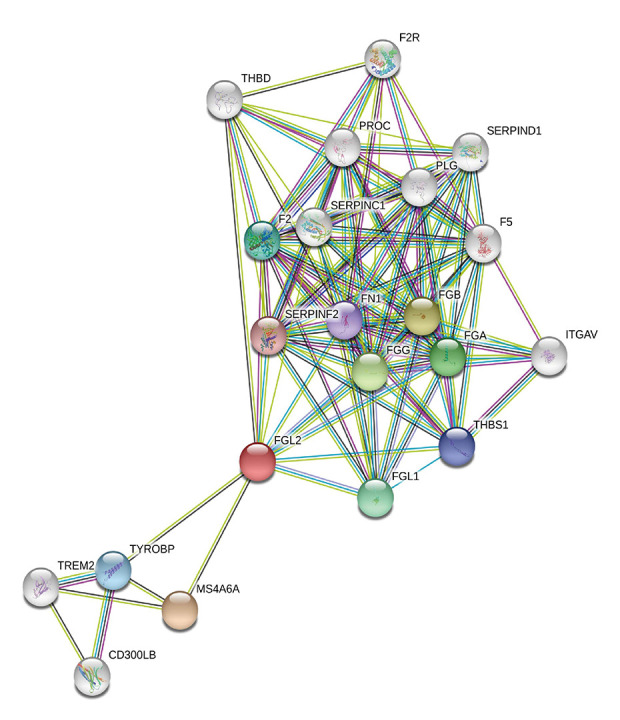
Modular analysis of the protein–protein interaction (PPI) network complex. In the STRING database, 21 proteins were filtered in the network complex. The PPI module consisted of 21 nodes and 108 edges. The average local clustering coefficient was 0.78. The PPI enrichment P-value was less than 1.0e-16. The top 5 predicted functional partners were MS4A6A (score: 0.801), FGB (score: 0.781), FGG (score: 0.776), FGA (score: 0.768), and FGL1 (score: 0.748). The blue and purple edges are considered known interactions. Green, red, and deep blue edges are predicted interactions. Orange, black, and light blue represent text mining, coexpression, and protein homology, respectively.
Functional and pathway enrichment analysis
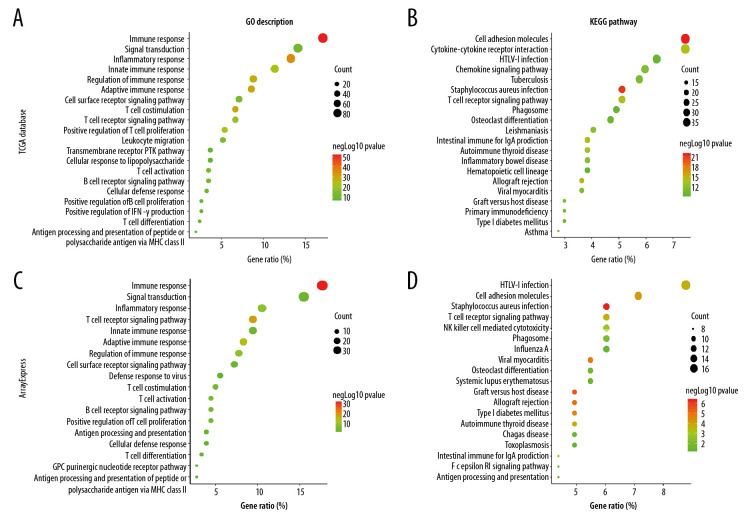
Functional and pathway enrichment analyses were performed to investigate the main biological signatures of FGL2 in breast cancer. We analyzed the related genes by Spearman correlation analysis (|r| >0.6 and P<0.01). In total, 479 genes were identified in the TCGA dataset. Then, we performed GO and KEGG analysis in the DAVID website. GO analysis showed that FGL2-corralated genes were mainly enriched in immune response, signal transduction, leukocyte migration, T cell costimulation/differentiation/proliferation/activation, B cell receptor signaling pathway, and B cell proliferation (Figure 6A). KEGG pathway analysis showed that FGL2-corralated genes were mostly associated with cell adhesion molecules in breast cancer. Additionally, KEGG pathway analysis suggested that FGL2-correlated genes were also associated with other canonical immune pathways, including cytokine-cytokine receptor interaction, chemokine signaling pathway, and T cell receptor signaling pathway (Figure 6B).
Figure 6.
GO analysis and KEGG pathway analysis of FGL2-related genes in breast cancer. (A) GO analysis of FGL2 in breast cancer with the TCGA database. FGL2 was correlated with the immune response, signal transduction, and inflammatory response. (B) KEGG pathway analysis of FGL2 in breast cancer with the TCGA database. FGL2 was correlated with cell adhesion molecules and cytokine-cytokine receptor interactions. (C) GO analysis of FGL2 in breast cancer with the ArrayExpress dataset. FGL2 was correlated with the immune response, signal transduction, and inflammatory response. (D) KEGG pathway analysis of FGL2 in breast cancer with the ArrayExpress dataset. FGL2 was correlated with HTLV-1 infection and cell adhesion molecules.
To validate the results calculated in the DAVID website, we used a microarray dataset to investigate the biological signatures of FGL2 in breast cancer on the website of the European Bioinformatics Institute (EMBL-EBI). In total, 185 genes were found in the microarray meta-dataset of breast cancer (E-MTAB-6703). GO analysis and KEGG pathway analysis showed similar results to TCGA dataset analyses (Figure 6C, 6D). These results demonstrated that FGL2 was positively correlated with immune signaling pathways.
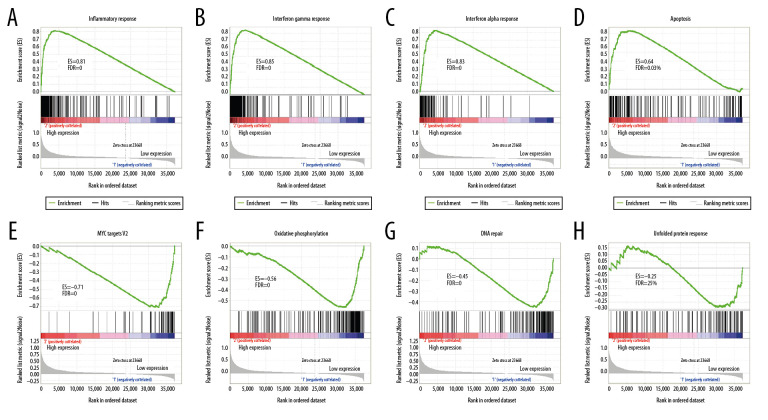
Gene set enrichment analysis (GSEA)
For validation, we used GSEA analysis to predict the biological function of FGL2 in breast cancer. As shown in Figure 7, GSEA suggested that increased FGL2 expression was positively associated with the inflammatory response in breast cancer. The results also indicated that FGL2 was positively correlated with inflammatory signaling pathways, including the IFN-γ response, IFN-α response, and apoptosis. FGL2 was negatively correlated with MYC targets V2, oxidative phosphorylation, DNA repair, and unfolded protein response.
Figure 7.
GSEA was used to predict the biological functions of FGL2 in breast cancer. (A) Inflammatory response, (B) Interferon-gamma response, (C) Interferon-alpha response, (D) Apoptosis, (E) MYC targets V2, (F) Oxidative phosphorylation (G) DNA repair, (H) Unfolded protein response. (ES – enrichment score; FDR – false discovery rate).
Discussion
FGL2 is a member of the fibrinogen superfamily of proteins, which are mainly expressed by inflammatory cells, including macrophages, CD4+ T, CD8+ T, and endothelial cells [10]. FGL2 can oligomerize into a tetramer by forming interchain disulfide bonds. Previous studies have demonstrated that FGL2 exhibits pleiotropic effects in the immune system. Liu et al. found that FGL2 contributes to complement activation and coagulation in virus-induced fulminant hepatitis [16]. Sun et al. showed that FGL2 regulates the functions of T cells in cirrhotic patients with hepatocellular carcinoma [17]. Khattar et al. reported that mice deficient in FGL2 have increased early viral replication of acute viral hepatitis [18]. Owing to the important role of FGL2 in immune regulation, this protein has attracted increasing attention in recent years.
Cancer is a type of disease involving abnormal cell growth correlated with abnormal immune system function. Researchers have focused on the relationship between FGL2 and several types of cancers. Birkhauser et al. found that a mouse renal carcinoma model had elevated expression levels of FGL2, CCL1, CXCL9, and HMGB1 [19]. These differentially-expressed genes contribute to altered tumor microenvironment in renal carcinoma. Yan et al. reported that FGL2 promotes the development of glioblastoma multiforme (GBM) by inhibiting CD103+ dendritic cell differentiation [20]. However, the expression and significance of FGL2 in breast cancer have not been investigated.
In our study, we examined FGL2 expression in breast cancer using the TCGA and the Oncomine datasets. By utilizing the TCGA database, we demonstrated that FGL2 expression was remarkably lower in breast invasive carcinoma (BRCA) compared with normal tissues. The same result was verified in the Oncomine database. We also estimated the correlation between FGL2 and clinical features of breast cancer. FGL2 levels were significantly correlated with radiation therapy and PR status. In addition, FGL2 was associated with tumor (T) stages in breast cancer. These results suggested that FGL2 has potential as a diagnostic biomarker of breast cancer. In addition, we estimated the correlation between the FGL2 expression level and survival prognosis of breast cancer with the GEPIA, PrognoScan, and KM plotter datasets. The results showed that lower FGL2 expression level was correlated with poor breast cancer prognosis, which indicated that low FGL2 expression can act an independent risk factor for poor survival prognosis of breast cancer patients.
Another aspect in our study is the relationship between FGL2 and immune status in the tumor microenvironment of breast cancer. Our results demonstrated that FGL2 level was positively correlated with infiltrating levels of B cells, macrophages, CD8+ T cells, CD4+ T cells, and DCs in breast cancer. Moreover, FGL2 expression was positively correlated with infiltrating levels of antitumor cells, such as effector memory CD8+ T cells, activated CD8+ T cells, Th1 cells, natural killer cells, activated CD4+ T cells, effector memory CD4+ T cells, natural killer T cells, and central memory CD4+ T cells. In brief, these results indicate that FGL2 is positively correlated with antitumor activities in breast cancer.
Furthermore, we evaluated the protein–protein interaction related to FGL2 in the STRING database. The PPI network complex showed that FGL2 was correlated with MS4A6A, FGB, FGG, FGA, FGL1, F2, TYROBP, THBS1, FN1, and SERPINF2. MS4A6A is a member of the membrane-spanning 4A gene family and participates in signal transduction as a factor in a multimeric receptor complex [21]. FGB together with FGG and FGA polymerizes to form an insoluble fibrin matrix, which participates in cell migration [22]. FGL1 is a member of the fibrinogen family of proteins, which also includes FGL2 [23]. F2 functions include inflammation and wound healing. TYROBP is a transmembrane signaling polypeptide involved in signal transduction [24]. THBS1 mediates cell-to-matrix and cell-to-cell interactions [25]. FN1 is involved in cell adhesion and migration processes [26]. SERPINF2 is a member of the serpin family of serine protease inhibitors. FGL2 is correlated with signal transduction, cell adhesion, cell-to-cell interactions, and inflammation by interacting with other proteins. We also used GO and KEGG analysis to analyze biological functions and related pathways of FGL2-correlated genes in breast cancer. GO analysis indicated that FGL2-correlated genes were involved in the immune response, inflammatory response, innate immune response, regulation of immune response, and adaptive immune response in breast cancer. This result showed that FGL2 is closely correlated with immune activities in breast cancer. In addition, GO analysis in this study revealed that FGL2-correlated genes participate in signal transduction, cell surface receptor signaling pathway, T cell costimulation, T cell receptor signaling pathway, and cell adhesion. MHC class II molecules present antigens to T cells and activate T cells [27]. The biological processes of MHC class II molecules include signal transduction, the cell surface receptor signaling pathway, cell adhesion, T cell costimulation, and the T cell receptor signaling pathway. Therefore, these results are in agreement with the conclusion that FGL2 is positively correlated with MHC class II for antigens expressed in breast cancer.
Finally, we performed GSEA analysis to investigate the biological functions of FGL2 associated with breast cancer. FGL2 was positively correlated with the inflammatory response. Increased FGL2 enhanced the inflammatory response in breast cancer, including the IFN-γ response, IFN-α response, complement, and apoptosis. IFN-γ is the signature cytokine of Th1 cells and is also produced by NK cells. IFN-γ is necessary for CD4+ T cell-mediated antitumor immunity, partially by the upregulation of HLA class II expression. IFN-γ is also responsible for inhibiting angiogenesis in tumors. IFN-α can activate CD8+ central memory T cells, CD4+ effector memory T cells, and NK cells. IFN-α treatment has moderate success and a clinical response for renal carcinoma, melanoma, and breast cancer. CD8+ T cells can induce tumor apoptosis via Fas/FasL interactions. FGL2 is negatively correlated with the MYC targets V2, oxidative phosphorylation, DNA repair, and unfolded protein response, which are all involved in tumorigenesis.
Conclusions
In summary, we showed that FGL2 expression was significantly lower in breast cancer compared with adjacent normal tissues and that FGL2 levels were significantly associated with radiation therapy, PR status, and tumor (T) stage. Lower FGL2 expression was correlated with poor prognosis of breast carcinoma. FGL2 was positively correlated with antitumor immune cells infiltration in breast cancer. PPI analysis showed that FGL2 was related with signal transduction, cell adhesion, cell-to-cell interactions, and inflammation by interacting with other proteins. GO, KEGG, and GSEA analyses also demonstrated that FGL2 expression was positively associated with activation of CD8+ T cells, Th1 cells, NK cells, CD4+ T cells, and NKT cells, which may be regulated via the IFN-γ response, IFN-α response, complement, and apoptosis pathways. Our findings indicate that FGL2 is a promising biomarker for clinical prediction in human breast cancer, and high expression levels of FGL2 are positively associated with enhanced antitumor activities.
Supplementary Data
Supplementary Table 1.
FGL2 was positively correlated with most immune marker sets in the TIMER database.
| Description | Gene markers | Cor | P | Description | Gene markers | Cor | P |
|---|---|---|---|---|---|---|---|
| T cell (general) | CD3D | 0.589 | 6.66E-94 | CTL (cytotoxic T lymphocytes) | CD8A | 0.61 | 2.93E-102 |
| CD3E | 0.621 | 3.80E-107 | CD8B | 0.486 | 4.19E-60 | ||
| CD2 | 0.658 | 2.29E-124 | GZMB | 0.45 | 1.12E-50 | ||
| B cell | CD19 | 0.362 | 2.53E-32 | Dendritic cell | HLA-DPB1 | 0.65 | 2.23E-120 |
| CD79A | 0.386 | 1.37E-36 | HLA-DQB1 | 0.472 | 2.83E-56 | ||
| CD79B | 0.341 | 1.59E-28 | HLA-DRA | 0.758 | 2.83E-186 | ||
| CD22 | 0.225 | 6.93E-13 | HLA-DPA1 | 0.774 | 2.36E-199 | ||
| M1 macrophage | INOS | 0.003 | 9.33E-01 | DEC-205 | 0.411 | 9.88E-42 | |
| CIITA | 0.648 | 2.39E-119 | BDCA-1 | 0.46 | 2.96E-53 | ||
| IRF5 | 0.289 | 1.49E-20 | BDCA-4 | 0.292 | 5.93E-21 | ||
| COX2 | 0.196 | 4.34E-10 | BDCA-2 | 0.466 | 8.39E-55 | ||
| M2 macrophage | CD163 | 0.568 | 6.67E-86 | CD11c | 0.547 | 1.03E-78 | |
| IRF4 | 0.591 | 1.22E-94 | Th1 | CD38 | 0.555 | 2.26E-81 | |
| VSIG4 | 0.476 | 2.84E-57 | T-bet | 0.597 | 3.80E-97 | ||
| MS4A4A | 0.655 | 9.05E-123 | STAT4 | 0.612 | 4.38E-103 | ||
| TAM | CCL2 | 0.431 | 2.84E-46 | STAT1 | 0.483 | 3.35E-59 | |
| CCL5 | 0.529 | 1.02E-72 | IFN-γ | 0.519 | 1.19E-69 | ||
| CD68 | 0.562 | 9.57E-84 | TNF-α | 0.224 | 9.45E-13 | ||
| IL10 | 0.551 | 5.84E-80 | Th2 | GATA3 | −0.107 | 7.48E-04 | |
| Neutrophils | CD66b (CEACAM8) | 0.031 | 3.33E-01 | IL13 | 0.184 | 5.48E-09 | |
| CD15 | 0.327 | 2.91E-26 | STAT6 | 0.044 | 1.63E-01 | ||
| CD11b (ITGAM) | 0.517 | 6.19E-69 | Tfh | BCL6 | 0.108 | 6.77E-04 | |
| CCR7 | 0.49 | 4.27E-61 | CD200 | 0.358 | 1.79E-31 | ||
| Natural killer cell | NKp46 | 0.457 | 1.99E-52 | IL21 | 0.387 | 7.16E-37 | |
| NKp44 | 0.165 | 1.61E-07 | ICOS | 0.604 | 0.09E-100 | ||
| NKp30 | 0.473 | 1.32E-56 | Th17 | STAT3 | 0.138 | 1.19E-05 | |
| FCGR3A | 0.592 | 4.66E-95 | IL17A | 0.16 | 3.69E-07 | ||
| FCGR3B | 0.325 | 6.52E-26 | IL1A | 0.295 | 1.86E-21 | ||
| NKG2A (KLRC1) | 0.468 | 3.30E-55 | IL1B | 0.444 | 2.40E-49 | ||
| KIR2DL1 | 0.277 | 5.07E-19 | CCL20 | 0.119 | 1.73E-04 | ||
| KIR2DL3 | 0.312 | 7.11E-24 | Treg | FOXP3 | 0.493 | 4.48E-62 | |
| KIR3DL1 | 0.337 | 8.71E-28 | CCR8 | 0.562 | 6.60E-84 | ||
| TGFβ | 0.209 | 2.95E-11 |
Supplementary Table 2.
FGL2 was positively correlated with most immune marker sets in the GEPIA database.
| Description | Gene markers | Cor | P | Description | Gene markers | Cor | P |
|---|---|---|---|---|---|---|---|
| T cell (general) | CD3D | 0.51 | 0 | CTL (cytotoxic T lymphocytes ) | CD8A | 0.64 | 0 |
| CD3E | 0.56 | 0 | CD8B | 0.44 | 0 | ||
| CD2 | 0.66 | 0 | GZMB | 0.45 | 0 | ||
| B cell | CD19 | 0.23 | 1.40E-14 | Dendritic cell | HLA-DPB1 | 0.65 | 0 |
| CD79A | 0.28 | 0 | HLA-DQB1 | 0.39 | 0 | ||
| CD79B | 0.26 | 0 | HLA-DRA | 0.74 | 0 | ||
| CD22 | 0.19 | 2.00E-10 | HLA-DPA1 | 0.76 | 0 | ||
| M1 Macrophage | INOS | −0.021 | 4.90E-01 | DEC-205 | 0.44 | 0 | |
| CIITA | 0.54 | 0 | BDCA-1 | 0.48 | 0 | ||
| IRF5 | 0.34 | 0 | BDCA-4 | 0.39 | 0 | ||
| COX2 | −0.022 | 4.70E-01 | BDCA-2 | 0.38 | 0 | ||
| M2 Macrophage | CD163 | 0.45 | 0 | CD11c | 0.53 | 0 | |
| IRF4 | 0.44 | 0 | Th1 | CD38 | 0.55 | 0 | |
| VSIG4 | 0.46 | 0 | T-bet | 0.65 | 0 | ||
| MS4A4A | 0.64 | 0 | STAT4 | 0.64 | 0 | ||
| TAM | CCL2 | 0.35 | 0 | STAT1 | 0.5 | 0 | |
| CCL5 | 0.5 | 0 | IFN-γ | 0.47 | 0 | ||
| CD68 | 0.53 | 0 | TNF-α | 0.15 | 5.80E-07 | ||
| IL10 | 0.56 | 0 | Th2 | GATA3 | −0.16 | 2.40E-07 | |
| Neutrophils | CD66b (CEACAM8) | 0.049 | 1.10E-01 | IL13 | 0.26 | 0 | |
| CD15 | 0.41 | 0 | STAT6 | 0.24 | 1.80E-15 | ||
| CD11b (ITGAM) | 0.31 | 0 | Tfh | BCL6 | 0.18 | 3.70E-09 | |
| CCR7 | 0.2 | 2.60E-11 | CD200 | 0.43 | 0 | ||
| Natural killer cell | NKp46 | 0.59 | 0 | IL21 | 0.53 | 0 | |
| NKp44 | 0.02 | 5.10E-01 | ICOS | 0.64 | 0 | ||
| NKp30 | 0.3 | 0 | Th17 | STAT3 | 0.27 | 0 | |
| FCGR3A | 0.59 | 0 | IL17A | 0.097 | 1.40E-03 | ||
| FCGR3B | 0.078 | 1.10E-02 | IL1A | 0.039 | 2.00E-01 | ||
| NKG2A (KLRC1) | 0.35 | 0 | IL1B | 0.37 | 0 | ||
| KIR2DL1 | 0.021 | 4.90E-01 | CCL20 | 0.019 | 5.20E-01 | ||
| KIR2DL3 | 0.32 | 0 | Treg | FOXP3 | 0.54 | 0 | |
| KIR3DL1 | 0.34 | 0 | CCR8 | 0.44 | 0 | ||
| TGFβ | 0.25 | 0 |
Footnotes
Conflicts of interest
None.
Source of support: This work was supported by the National Natural Science Foundation of China [grant numbers 31500704 and 81904142] and the research program of Beijing University of Chinese Medicine [grant numbers 1000041510049, BUCM-2019-JCRC006, and 2019-JYB-TD013]
References
- 1.Elmore JG. Screening for breast cancer. JAMA. 2005;293:1245–56. doi: 10.1001/jama.293.10.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. A Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Fan L, Strasser-Weippl K, Li J-L, et al. Breast cancer in China. Lancet Oncol. 2014;15(7):279–89. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 4.Winters S, Martin C. Breast cancer epidemiology, prevention, and screening. Prog Mol Biol Transl Sci. 2017;151:1–32. doi: 10.1016/bs.pmbts.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Afghahi A, Kurian AW. The changing landscape of genetic testing for inherited breast cancer predisposition. Curr Treat Options Oncol. 2017;18(5):27. doi: 10.1007/s11864-017-0468-y. [DOI] [PubMed] [Google Scholar]
- 6.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–50. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 7.Matsen CB, Neumayer LA. Breast cancer: A review for the general surgeon. JAMA Surg. 2013;148:971–79. doi: 10.1001/jamasurg.2013.3393. [DOI] [PubMed] [Google Scholar]
- 8.Pernas S, Barroso Sousa R, Tolaney SM. Optimal treatment of early stage HER2-positive breast cancer. Cancer. 2018;124:4455–66. doi: 10.1002/cncr.31657. [DOI] [PubMed] [Google Scholar]
- 9.Rossing M, Sørensen CS, Ejlertsen B, Nielsen FC. Whole genome sequencing of breast cancer. APMIS. 2019;127:303–15. doi: 10.1111/apm.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Liu Y, Chen F. Soluble fibrinogen like protein 2 (sFGL2), the novel effector molecule for immunoregulation. Oncotarget. 2017;8:3711–23. doi: 10.18632/oncotarget.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang G. Physiological functions and clinical implications of fibrinogen-like 2: A review. World J Clin Infect Dis. 2013;3:37–46. doi: 10.5495/wjcid.v3.i3.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shalev I. Targeted deletion of fgl2 leads to impaired regulatory T cell activity and development of autoimmune glomerulonephritis. J Immunol. 2008;180:249–60. doi: 10.4049/jimmunol.180.1.249. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh Y, Chen CWS, Schmitz SH, et al. Candidate genes associated with susceptibility for SARS-coronavirus. Bull Math Biol. 2010;72:122–32. doi: 10.1007/s11538-009-9440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Smith AJ, Schacker TW, et al. Microarray analysis of lymphatic tissue reveals stage-specific, gene expression signatures in HIV-1 infection. J Immunol. 2009;183:1975–82. doi: 10.4049/jimmunol.0803222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan J, Kong L, Hu J, et al. FGL2 as a multimodality regulator of tumor-mediated immune suppression and therapeutic target in gliomas. J Natl Cancer Inst. 2015;107(8) doi: 10.1093/jnci/djv137. djv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Tan Y, Zhang J, et al. C5aR, TNF-a, and FGL2 contribute to coagulation and complement activation in virus-induced fulminant hepatitis. J Hepatol. 2015;62:354–62. doi: 10.1016/j.jhep.2014.08.050. [DOI] [PubMed] [Google Scholar]
- 17.Sun Y, Xi D, Ding W, et al. Soluble FGL2, a novel effector molecule of activated hepatic stellate cells, regulates T-cell function in cirrhotic patients with hepatocellular carcinoma. Hepatol Int. 2014;8:567–75. doi: 10.1007/s12072-014-9568-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khattar R, Luft O, Yavorska N, et al. Targeted deletion of FGL2 leads to increased early viral replication and enhanced adaptive immunity in a murine model of acute viral hepatitis caused by LCMV WE. PLoS One. 2013;8:e72309. doi: 10.1371/journal.pone.0072309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birkhäuser FD, Koya RC, Neufeld C, et al. Dendritic cell-based immunotherapy in prevention and treatment of renal cell carcinoma. J Immunother. 2013;36:102–11. doi: 10.1097/CJI.0b013e31827bec97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan J, Zhao Q, Gabrusiewicz K, et al. FGL2 promotes tumor progression in the CNS by suppressing CD103+ dendritic cell differentiation. Nat Commun. 2019;10:488. doi: 10.1038/s41467-018-08271-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Y, Buckley TR, Tu L, et al. Structural organization of the human MS4A gene cluster on chromosome 11q12. Immunogenetics. 2001;53:357–68. doi: 10.1007/s002510100339. [DOI] [PubMed] [Google Scholar]
- 22.Espitia Jaimes C, Fish RJ, Neerman-Arbez M. Local chromatin interactions contribute to expression of the fibrinogen gene cluster. J Thromb Haemost. 2018;16:2070–82. doi: 10.1111/jth.14248. [DOI] [PubMed] [Google Scholar]
- 23.Nagdas SK, Winfrey VP, Olson GE. Two fibrinogen-like proteins, FGL1 and FGL2, are disulfide-linked subunits of oligomers that specifically bind nonviable spermatozoa. Int J Biochem Cell Biol. 2016;80:163–72. doi: 10.1016/j.biocel.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Tomasello E, Vivier E. KARAP/DAP12/TYROBP: Three names and a multiplicity of biological functions. Eur J Immunol. 2005;35:1670–77. doi: 10.1002/eji.200425932. [DOI] [PubMed] [Google Scholar]
- 25.Polymeropoulos MH. XHRD: Dinucleotide repeat polymorphism at the human thrombospondin gene (THBS1) Nucleic Acids Res. 1990;18:7467. doi: 10.1093/nar/18.24.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staunton D. MCAA: Preparation of recombinant fibronectin fragments for functional and structural studies. Methods Mol Biol. 2009;522:73–99. doi: 10.1007/978-1-59745-413-1_5. [DOI] [PubMed] [Google Scholar]
- 27.Unanue ER, Turk V, Neefjes J. Variations in MHC class II antigen processing and presentation in health and disease. Annu Rev Immunol. 2016;34:265–97. doi: 10.1146/annurev-immunol-041015-055420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
FGL2 was positively correlated with most immune marker sets in the TIMER database.
| Description | Gene markers | Cor | P | Description | Gene markers | Cor | P |
|---|---|---|---|---|---|---|---|
| T cell (general) | CD3D | 0.589 | 6.66E-94 | CTL (cytotoxic T lymphocytes) | CD8A | 0.61 | 2.93E-102 |
| CD3E | 0.621 | 3.80E-107 | CD8B | 0.486 | 4.19E-60 | ||
| CD2 | 0.658 | 2.29E-124 | GZMB | 0.45 | 1.12E-50 | ||
| B cell | CD19 | 0.362 | 2.53E-32 | Dendritic cell | HLA-DPB1 | 0.65 | 2.23E-120 |
| CD79A | 0.386 | 1.37E-36 | HLA-DQB1 | 0.472 | 2.83E-56 | ||
| CD79B | 0.341 | 1.59E-28 | HLA-DRA | 0.758 | 2.83E-186 | ||
| CD22 | 0.225 | 6.93E-13 | HLA-DPA1 | 0.774 | 2.36E-199 | ||
| M1 macrophage | INOS | 0.003 | 9.33E-01 | DEC-205 | 0.411 | 9.88E-42 | |
| CIITA | 0.648 | 2.39E-119 | BDCA-1 | 0.46 | 2.96E-53 | ||
| IRF5 | 0.289 | 1.49E-20 | BDCA-4 | 0.292 | 5.93E-21 | ||
| COX2 | 0.196 | 4.34E-10 | BDCA-2 | 0.466 | 8.39E-55 | ||
| M2 macrophage | CD163 | 0.568 | 6.67E-86 | CD11c | 0.547 | 1.03E-78 | |
| IRF4 | 0.591 | 1.22E-94 | Th1 | CD38 | 0.555 | 2.26E-81 | |
| VSIG4 | 0.476 | 2.84E-57 | T-bet | 0.597 | 3.80E-97 | ||
| MS4A4A | 0.655 | 9.05E-123 | STAT4 | 0.612 | 4.38E-103 | ||
| TAM | CCL2 | 0.431 | 2.84E-46 | STAT1 | 0.483 | 3.35E-59 | |
| CCL5 | 0.529 | 1.02E-72 | IFN-γ | 0.519 | 1.19E-69 | ||
| CD68 | 0.562 | 9.57E-84 | TNF-α | 0.224 | 9.45E-13 | ||
| IL10 | 0.551 | 5.84E-80 | Th2 | GATA3 | −0.107 | 7.48E-04 | |
| Neutrophils | CD66b (CEACAM8) | 0.031 | 3.33E-01 | IL13 | 0.184 | 5.48E-09 | |
| CD15 | 0.327 | 2.91E-26 | STAT6 | 0.044 | 1.63E-01 | ||
| CD11b (ITGAM) | 0.517 | 6.19E-69 | Tfh | BCL6 | 0.108 | 6.77E-04 | |
| CCR7 | 0.49 | 4.27E-61 | CD200 | 0.358 | 1.79E-31 | ||
| Natural killer cell | NKp46 | 0.457 | 1.99E-52 | IL21 | 0.387 | 7.16E-37 | |
| NKp44 | 0.165 | 1.61E-07 | ICOS | 0.604 | 0.09E-100 | ||
| NKp30 | 0.473 | 1.32E-56 | Th17 | STAT3 | 0.138 | 1.19E-05 | |
| FCGR3A | 0.592 | 4.66E-95 | IL17A | 0.16 | 3.69E-07 | ||
| FCGR3B | 0.325 | 6.52E-26 | IL1A | 0.295 | 1.86E-21 | ||
| NKG2A (KLRC1) | 0.468 | 3.30E-55 | IL1B | 0.444 | 2.40E-49 | ||
| KIR2DL1 | 0.277 | 5.07E-19 | CCL20 | 0.119 | 1.73E-04 | ||
| KIR2DL3 | 0.312 | 7.11E-24 | Treg | FOXP3 | 0.493 | 4.48E-62 | |
| KIR3DL1 | 0.337 | 8.71E-28 | CCR8 | 0.562 | 6.60E-84 | ||
| TGFβ | 0.209 | 2.95E-11 |
Supplementary Table 2.
FGL2 was positively correlated with most immune marker sets in the GEPIA database.
| Description | Gene markers | Cor | P | Description | Gene markers | Cor | P |
|---|---|---|---|---|---|---|---|
| T cell (general) | CD3D | 0.51 | 0 | CTL (cytotoxic T lymphocytes ) | CD8A | 0.64 | 0 |
| CD3E | 0.56 | 0 | CD8B | 0.44 | 0 | ||
| CD2 | 0.66 | 0 | GZMB | 0.45 | 0 | ||
| B cell | CD19 | 0.23 | 1.40E-14 | Dendritic cell | HLA-DPB1 | 0.65 | 0 |
| CD79A | 0.28 | 0 | HLA-DQB1 | 0.39 | 0 | ||
| CD79B | 0.26 | 0 | HLA-DRA | 0.74 | 0 | ||
| CD22 | 0.19 | 2.00E-10 | HLA-DPA1 | 0.76 | 0 | ||
| M1 Macrophage | INOS | −0.021 | 4.90E-01 | DEC-205 | 0.44 | 0 | |
| CIITA | 0.54 | 0 | BDCA-1 | 0.48 | 0 | ||
| IRF5 | 0.34 | 0 | BDCA-4 | 0.39 | 0 | ||
| COX2 | −0.022 | 4.70E-01 | BDCA-2 | 0.38 | 0 | ||
| M2 Macrophage | CD163 | 0.45 | 0 | CD11c | 0.53 | 0 | |
| IRF4 | 0.44 | 0 | Th1 | CD38 | 0.55 | 0 | |
| VSIG4 | 0.46 | 0 | T-bet | 0.65 | 0 | ||
| MS4A4A | 0.64 | 0 | STAT4 | 0.64 | 0 | ||
| TAM | CCL2 | 0.35 | 0 | STAT1 | 0.5 | 0 | |
| CCL5 | 0.5 | 0 | IFN-γ | 0.47 | 0 | ||
| CD68 | 0.53 | 0 | TNF-α | 0.15 | 5.80E-07 | ||
| IL10 | 0.56 | 0 | Th2 | GATA3 | −0.16 | 2.40E-07 | |
| Neutrophils | CD66b (CEACAM8) | 0.049 | 1.10E-01 | IL13 | 0.26 | 0 | |
| CD15 | 0.41 | 0 | STAT6 | 0.24 | 1.80E-15 | ||
| CD11b (ITGAM) | 0.31 | 0 | Tfh | BCL6 | 0.18 | 3.70E-09 | |
| CCR7 | 0.2 | 2.60E-11 | CD200 | 0.43 | 0 | ||
| Natural killer cell | NKp46 | 0.59 | 0 | IL21 | 0.53 | 0 | |
| NKp44 | 0.02 | 5.10E-01 | ICOS | 0.64 | 0 | ||
| NKp30 | 0.3 | 0 | Th17 | STAT3 | 0.27 | 0 | |
| FCGR3A | 0.59 | 0 | IL17A | 0.097 | 1.40E-03 | ||
| FCGR3B | 0.078 | 1.10E-02 | IL1A | 0.039 | 2.00E-01 | ||
| NKG2A (KLRC1) | 0.35 | 0 | IL1B | 0.37 | 0 | ||
| KIR2DL1 | 0.021 | 4.90E-01 | CCL20 | 0.019 | 5.20E-01 | ||
| KIR2DL3 | 0.32 | 0 | Treg | FOXP3 | 0.54 | 0 | |
| KIR3DL1 | 0.34 | 0 | CCR8 | 0.44 | 0 | ||
| TGFβ | 0.25 | 0 |