Abstract
Background
Antibiotic resistance is an increasing phenomenon in many bacterial pathogens including uropathogenic Escherichia coli. Hypothetical anti-virulent agents could be a solution, but first clear virulence associated gene-pool of antibiotic resistant isolates have to be determined. The aim of this study is to investigate the significant associations between genes encoding VFs with antibiotic resistance and phylogenetic groups in UPEC isolates.
Results
The majority of 248 UPEC isolates belonged to phylogenetic group B2 (67.3%). The maximum and minimum resistance was attributed to amoxicillin (90.3%) and both fosfomycin and imipenem (1.6%) respectively. 11.3% of isolates were resistant to all antibiotic agents except that of imipenem, nitrofurantoin and fosfomycin. These highly resistant isolates were placed only in group B2 and D. The most prevalent virulence gene was ompA (93.5%). The hlyA was the only virulence gene that was significantly more prevalent in the highly resistant isolates. The ompA, malX and hlyA genes were obviously more abundant in the antibiotic resistant isolates in comparison to susceptible isolates. The papC gene was associated with amoxicillin resistance (p-value = 0.006, odds ratio: 26.00).
Conclusions
Increased resistance to first line drugs prescribed for UTIs were detected in CA-UPEC isolates in our study.. Minimal resistance was observed against nitrofurantoin, fosfomycin and imipenem. Therefore, they are introduced for application in empirical therapy of UTIs. Fosfomycin may be the most effective antibiotic agent against highly resistant UPEC isolates. The presence of the ompA, malX and hlyA genes were significantly associated with resistance to different antibiotic agents. We assume that the ability of UPEC isolates to upgrade their antibiotic resistance capacity may occurs in compliance with the preliminary existence of specific virulence associated genes. But, more investigation with higher number of bacterial isolates, further virulence associated genes and comparison of gene pools from CA-UPEC isolates with HA-UPEC are proposed to confirm these finding and discovering new aspects of this association.
Keywords: UPEC, Virulence associated genes, Antibiotic resistance, Phylogenetic groups
Background
Urinary tract infection (UTI) is a widespread medical condition that involves both men and women and can occur frequently in different ages [1]. About 150 million new case of UTI worldwide are reported every year that most of them are women [2]. Many bacterial pathogens are known as the causative agents of UTI, but uropathogenic Escherichia coli (UPEC) is accounted as the most common agent (80–90%) of UTIs [2–4]. UPEC isolates are divided into the four main phylogenetic groups (A, B1, B2, and D) based on the some genomic sequences [5]. The most virulent UPEC strains are categorized in B2 and to a lesser extent in group D2. The A and B1 groups are less virulent and usually originate from fecal samples instead [6].
Establishment of UTI by UPEC is a multistep process that starts from overcoming of epithelial immune system and successful colonization of UPEC in the urinary epithelium. This process could be continue to the host tissue injury and even outspreading of bacteria to the blood circulation. Many virulence-factor associated genes are responsible for the production of virulence factors that are involved in each step. Structural factors such as outer membrane proteins, fimbriae and flagella are involved in the colonization process. The OmpA is a bacterial outer membrane protein that is involved in hole infection process including; adhesion, invasion, intracellular survival and immune system evasion of UPEC as well as many other pathogenic bacteria [7]. OmpA promotes some critical steps such as initial binding to bladder epithelium, chronic persistence and post invasion pathogenesis during UPEC infection [8]. Several types of surface exposed apparatus were detected in UPEC that some of the most known of them are type I fimbriae, P fimbriae and S fimbriae. Adhesions such as type 1 fimbriae of UPEC are encoded by the fim operon. Fim proteins have critical roles in assembly, folding and stabilizing of type I fimbriae. FimC chaperon is characterized to support correct folding of structural subunits to protrude from cell membrane, stabilizing them in periplasm and preventing them from unpredicted interactions [9]. The P fimbriae has heteropolymeric structure that is encoded by pap operon. This operon is part of a pathogenicity island that contains variable genes including papA-K [2]. The PapC protein on the E. coli outer membrane is thought to be associated in outgoing translocation of pilin subunits through outer membrane and surface assembly [10]. Attachment of UPEC to renal epithelium via P fimbriae induces local inflammation, which subsequently leads to pain in urinary system [11]. The S fimbriae encoded by sfa genes which are highly homologous with foc operon that contains coding sequences of the Fic fimbriae proteins and these coding sequences could be exchanged between these two gene clusters. Therefore, the same obligatory genes like focC and focD exist in both gene clusters [12].
Agents responsible in iron-acquisition system and secreted toxins of UPEC are mainly enrolled in intracellular survival, immune system evasion and host tissue damage process [2]. In UPEC strains, iron is obtained from surrounding environment by production of various sidrophores, like aerobactin, salmochelin and yersiniabactin which are encoded by iuc, iro and irp gene clusters respectively [13, 14]. HlyA (α-haemolysin) is a secreted lipoprotein toxin that is able to disrupt host nucleated cells by pore forming activity. In addition, it can induce apoptosis in urinary tract epithelium and immune system associated cells [11, 15]. There are many Pathogenicity-associated islands (PAIs) accumulated in extra-intestinal E. coli strains and they carry different virulence associated genes, such as P fimbriae, Type I fimbriae, hemolysin, iron acquisition proteins, some bacteriocins and malX gene. The malX gene is part of a PAI that is associated with UPEC and it is known as phosphotransferase system enzyme II that uses glucose and maltose as the main substrates [16, 17].
Antibiotic resistance is an expanding phenomenon in many bacterial pathogens including UPEC [18]. Now today antibiotic resistance is a world spread problem and uncontrolled use of antibiotic agents can increase the antibiotic resistance in many countries including Iran. Therefore successful prescription of antibiotic agents will be limited day by day. Use of other alternatives such as presumptive anti-virulence agents could be a solution, but it will be operational if precise gene-pool that encodes virulence factors in resistant isolate and any association between these virulence associated genes and antibiotic resistance be investigated. In the present study we tried to investigate the antibiotic resistance patterns of community acquired UPEC isolates as well as the association between phylogenetic origin and virulence associated genes with antibiotic resistance in these isolates.
Results
Antibiotic resistance of different phylogenetic groups
All E. coli isolates were categorized into the four phylogenetic groups as follows: 167(67.3%) Group B2, 53(21.4%) Group D, 16(6.5%) Group A and 12(4.8%) Group B1.
Data obtained from disk diffusion test showed various frequency of antibiotic resistance including, 90.3% amoxicillin, 67.7% trimethoprim-sulfamethoxazole, 61.3% cephalexin, 59.7% ceftriaxone, 58.1% cefotaxime, 43.5% ciprofloxacin, 41.9% azithromycin, 40.3% ceftazidime, 27.4% gentamycin and minimum antibiotic resistance was detected against fosfomycin (1.6%), nitrofurantoin (3.2%) and imipenem (1.6%).
Totally 28(11.3%) out of 248 isolates were highly resistant. Resistance against amoxicillin, trimethoprim-sulfamethoxazole, cephalexin, ceftriaxone, cefotaxime, ciprofloxacin, azithromycin, ceftazidime and gentamycin were detected in these isolates. Twelve (42.9%) and 4 (14.3%) of these highly resistant isolates were resistant to nitrofurantoin and imipenem respectively, but resistance against fosfomycin was not observed in them.
Various resistance patterns were detected in four phylogenetic groups. Resistance to fosfomycin and imipenem were not identified in groups A and B1. All fosfomycin resistant isolates were placed in group B2. Resistance to gentamycin was not observed in group A. All group A and group B1 isolates were resistant to amoxycillin.
Logistic regression analysis showed no significant association between resistance or susceptibility to antibiotic agents with phylogenetic origins of UPEC isolates, except that of azithromycin. The Group B2 (p-value = 0.004, odds ratio: 0.195) and D (p-value = 0.003, odds ratio: 0.160) origins were associated with azithromycin susceptibility.
The bulk of the highly resistant isolates (82.1%) were placed in group B2 and they were absent from B1 and A groups (p-value =0.05, the Chi-square analysis), (detailed data not shown).
Association of UPEC virulence genes with antibiotic resistance profile
The most prevalent virulence gene was ompA gene that was detected in 232(93.5%) of UPEC isolates. Prevalence of other six virulence genes were as follows: fimC 124(50%), irp2 156 (62.9%), malX 184(74.2%), papC 204(82.3%), sfa/focCD 52(21%) and hlyA 76(30.6%).
All seven virulence genes were found in four phylogenetic groups. The carriage of papC and sfa/focCD were significantly associated with group B2 (Tables 1 and 2).
Table 1.
Prevalence of seven virulence genes in various phylogenetic groups
| Gene | Phylogenetic group n (%) | |||
|---|---|---|---|---|
| A (n = 16) | B1 (n = 12) | B2 (n = 167) | D (n = 53) | |
| ompA | 16 (100) | 8 (66.7) | 156 (93.4) | 52 (98.1) |
| fimC | 10 (62.5) | 6 (50) | 86 (41.5) | 22 (41.5) |
| irp2 | 12 (75) | 8 (66.7) | 108 (64.7) | 28 (52.8) |
| malX | 9 (56.3) | 7 (58.3) | 127 (76) | 41 (77.4) |
| papC | 9 (56.3) | 7 (58.3) | 144 (86.2) | 44 (83) |
| sfa/foc | 3 (18.8) | 1 (8.3) | 42 (25.1) | 6 (11.3) |
| hly | 6 (37.5) | 2 (16.7) | 50 (29.9) | 18 (34) |
Bold n (%): Binary logistic regression analysis showed significant association (p ≤ 0.05)
Table 2.
Binary logistic regression analysis of virulence-associated genes as predictor of phylogenetic origins
| Phylogenetic origin | ||||||||
|---|---|---|---|---|---|---|---|---|
| B2 | D | B1 | A | |||||
| OR | P | OR | P | OR | P | OR | P | |
| ompA | 0.858 | 0.796 | 5.149 | 0.117 | 0.111 | 0.002 | 2.092 | 0.998 |
| fimC | 1.452 | 0.191 | 0.591 | 0.104 | 0.610 | 0.454 | 1.271 | 0.682 |
| irp2 | 0.933 | 0.823 | 0.697 | 0.269 | 0.667 | 0.630 | 4.585 | 0.022 |
| malX | 1.079 | 0.830 | 1.255 | 0.547 | 0.679 | 0.613 | 0.566 | 0.385 |
| papC | 2.172 | 0.024 | 1.078 | 0.870 | 0.298 | 0.052 | 0.065 | 0.000 |
| Sfa | 2.366 | 0.025 | 0.380 | 0.038 | 0.413 | 0.424 | 0.966 | 0.962 |
| hlyA | 0.696 | 0.256 | 0.950 | 0.892 | 1.208 | 0.834 | 3.860 | 0.064 |
OR odds ratio, p p-value; Bold p: Significant association (p ≤ 0.05) was shown by binary logistic regression analysis; When p ≤ 0.05, then OR > 1 means that virulence associated gene is introduced as predictor of that phylogenetic origin, but OR < 1 means that virulence associated gene is a predictor of other phylogenetic origins
Phylogenetic group A isolates showed reduced carriage of papC in comparison to the other groups. Instead, carriage of irp2 was significantly associated with this group (Tables 1 and 2).
Prevalence of the ompA gene in group B1 was significantly less than the other groups (Tables 1 and 2).
The binary logistic regression analysis showed that hlyA was the only virulence gene that was significantly more prevalent in the highly resistant isolates showing resistance to all antibiotic agents except that of imipenem, nitrofurantoin and fosfomycin (p-value = 0.000, odds ratio: 8.000), (detailed data not shown).
Prevalence of different virulence genes in antibiotic resistant isolates were studied separately for each antibiotic agents. Among 9 investigated virulence genes, sfa/focCD, irp2, and fimC were absent in isolates that were resistant to imipenem. We did not observe sfa/focCD gene in nitrofurantoin resistant isolates and hlyA, papC, malX, irp2 and fimC were absent from fosfomycin resistant isolates (Table 3).
Table 3.
Frequency of different virulence genes in antibiotic resistant isolates in comparison to susceptible isolates
| Antibiotic agents | R or S (n) | Virulence genes n (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| hlyA | sfa/foc | papC | malX | Irp2 | fimC | ompA | ||
| Azithromycin | R(140) | 52 (37.1) | 24 (17.1) | 116 (82.9) | 104 (74.3) | 92 (65.7) | 68 (48.6) | 136 (97.1) |
| S(108) | 24 (22.2) | 28 (25.9) | 88 (81.5) | 80 (74.1) | 64 (59.3) | 56 (51.9) | 96 (88.9) | |
| Amoxicillin | R(228) | 72 (31.6) | 44 (19.3) | 192 (84.2) | 172 (75.4) | 136 (69.6) | 112 (49.1) | 216 (94.7) |
| S(20) | 4 (20) | 8 (40) | 12 (60) | 12 (60) | 20 (100) | 12 (60) | 16 (80) | |
| Imipenem | R(8) | 8 (100) | 0 (0) | 8 (100) | 8 (100) | 0 (0) | 0 (0) | 8 (100) |
| S(240) | 68 (28.3) | 52 (21.7) | 196 (81.7) | 176 (73.3) | 156 (65) | 124 (51.7) | 228 (93.3) | |
| Gentamycin | R(68) | 28 (41.2) | 4 (5.9) | 60 (88.2) | 60 (88.2) | 40 (58.8) | 40 (58.8) | 60 (88.2) |
| S(180) | 48 (26.7) | 48 (26.7) | 144 (80) | 124 (68.9) | 116 (64.4) | 84 (46.7) | 172 (95.6) | |
| Ciprofloxacin | R(116) | 48 (41.4) | 24 (20.7) | 92 (79.3) | 100 (86.2) | 68 (58.6) | 48 (41.4) | 112 (96.6) |
| S(132) | 28 (21.2) | 28 (21.2) | 112 (84.8) | 84 (63.6) | 88 (66.7) | 76 (57.6) | 120 (90.9) | |
| Cefotaxime | R(164) | 56 (34.1) | 32 (19.5) | 136 (82.9) | 128 (78) | 100 (61) | 72 (43.9) | 156 (95.1) |
| S(84) | 20 (23.8) | 20 (23.8) | 68 (81) | 56 (66.7) | 56 (66.7) | 52 (61.9) | 76 (90.5) | |
| Fosfomycin | R(4) | 0 (0) | 4 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (100) |
| S(244) | 76 (31.1) | 48 (19.7) | 204 (83.6) | 184 (75.4) | 156 (63.9) | 124 (50.8) | 228 (93.4) | |
| Cephalexin | R(184) | 68 (37) | 40 (21.7) | 156 (84.8) | 140 (76.1) | 108 (58.7) | 80 (43.5) | 180 (97.8) |
| S(64) | 8 (12.5) | 12 (18.8) | 48 (75) | 44 (68.8) | 48 (75) | 44 (68.8) | 52 (81.3) | |
| Ceftazidime | R(112) | 40 (35.7) | 20 (17.9) | 92 (82.1) | 92 (82.1) | 64 (57.1) | 48 (42.9) | 108 (96.4) |
| S(136) | 36 (26.5) | 32 (23.5) | 112 (82.4) | 92 (67.6) | 92 (67.6) | 76 (55.9) | 124 (91.2) | |
| Ceftriaxone | R(160) | 56 (35) | 32 (20) | 136 (85) | 128 (80) | 100 (62.5) | 68 (42.5) | 152 (95) |
| S(88) | 20 (22.7) | 20 (22.7) | 68 (77.3) | 56 (63.6) | 56 (63.6) | 56 (63.6) | 80 (90.9) | |
| Nitrofurantoin | R(24) | 16 (66.7) | 0 (0) | 16 (66.7) | 24 (100) | 8 (33.3) | 8 (33.3) | 24 (100) |
| S(224) | 60 (26.8) | 188 (83.3) | 160 (71.4) | 148 (66.1) | 148 (51.8) | 116 (51.8) | 208 (92.9) | |
| Cotrimoxazole | R(176) | 52 (29.5) | 32 (18.2) | 144 (81.8) | 128 (72.7) | 108 (61.4) | 84 (47.7) | 164 (93.2) |
| S(72) | 24 (33.3) | 20 (27.8) | 60 (83.3) | 56 (77.8) | 48 (66.7) | 40 (55.6) | 68 (94.4) | |
R Resistant, S Sensitive
The fimC, irp2 and sfa/focCD genes were significantly more prevalent in isolates sensitive to different class of antibiotic agents in comparison to resistant isolates. On the contrary, ompA, malX and hlyA genes were obviously more abundant in the isolates resistant to different antibiotic agents (Table 4). The papC gene was associated with amoxicillin resistance (p-value = 0.006, odds ratio: 26.00). Prevalence of different virulence genes among resistant isolates is presented in Table 3.
Table 4.
Binary logistic regression analysis of virulence-associated genes as predictor of resistance to various antibiotic agents
| Resistance to antibiotic agents | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Azithromycin | Amoxicillin | Imipenem | Gentamycin | Ciprofloxacin | Cefotaxime | Fosfomycin | Cephalexin | Ceftazidime | Ceftriaxone | Nitrofurantoin | Cotrimoxazole | |||||||||||||
| OR | p | OR | p | OR | p | OR | p | OR | p | OR | p | OR | p | OR | p | OR | p | OR | p | OR | p | OR | p | |
| ompA | 4.919 | 0.010 | 3.386 | 0.151 | 0.000 | 0.999 | 0.493 | 0.201 | 2.012 | 0.253 | 2.010 | 0.197 | 0.872 | 1.000 | 6.493 | 0.002 | 2.791 | 0.092 | 1.612 | 0.392 | 2.406 | 1.000 | 0.816 | 0.745 |
| fimC | 0.313 | 0.753 | 1.504 | 0.498 | 0.000 | 0.995 | 1.726 | 0.080 | 0.563 | 0.039 | 0.482 | 0.008 | 0.233 | 1.000 | 0.377 | 0.003 | 0.616 | 0.074 | 0.457 | 0.005 | 0.342 | 0.045 | 0.648 | 0.136 |
| irp2 | 1.618 | 0.850 | 0.000 | 0.995 | 0.000 | 0.995 | 0.672 | 0.240 | 1.348 | 0.363 | 1.052 | 0.862 | 0.456 | 1.000 | 0.724 | 0.360 | 0.839 | 0.559 | 1.254 | 0.455 | 1.111 | 0.857 | 0.876 | 0.668 |
| malX | 0.939 | 0.855 | 1.842 | 0.457 | 0.000 | 0.999 | 4.700 | 0.000 | 5.170 | 0.000 | 1.609 | 0.118 | 0.000 | 0.994 | 0.878 | 0.736 | 2.235 | 0.011 | 2.058 | 0.017 | 3.121 | 0.996 | 0.764 | 0.425 |
| papC | 0.301 | 0.673 | 26.00 | 0.000 | 2.344 | 1.000 | 1.427 | 0.483 | 0.189 | 0.000 | 0.756 | 0.486 | 0.027 | 1.000 | 1.496 | 0.344 | 0.610 | 0.204 | 0.970 | 0.942 | 0.000 | 0.995 | 1.164 | 0.732 |
| Sfa | 0.514 | 0.041 | 0.115 | 0.001 | 0.79 | 1.000 | 0.151 | 0.001 | 0.697 | 0.287 | 0.584 | 0.115 | 3.208 | 0.993 | 0.752 | 0.478 | 0.523 | 0.053 | 0.600 | 0.137 | 0.000 | 0.997 | 0.578 | 0.094 |
| hlyA | 1.887 | 0.033 | 1.711 | 0.482 | 1.167 | 0.994 | 1.643 | 0.140 | 2.651 | 0.002 | 1.348 | 0.370 | 2.166 | 1.000 | 3.433 | 0.003 | 1.246 | 0.478 | 1.493 | 0.221 | 0.000 | 0.996 | 0.875 | 0.676 |
OR odds ratio, p p-value; Bold p: Significant association (p ≤ 0.05) was shown by binary logistic regression analysis; When p ≤ 0.05, then OR > 1 means that virulence associated gene is introduced as predictor of resistance to antibiotic agent, but OR < 1 means that virulence associated gene is a predictor of susceptibility to antibiotic agent
Discussion
Antibiotic resistance is a globally increasing problem especially in developing countries. This is due to greater access to antibiotic drugs, over-prescription or arbitrary use of antibiotic agents.
The highest resistance were detected against amoxicillin and trimethoprim-sulfamethoxazole in our assessments. High rate of resistance to first line antibiotics that are prescribed to treat uncomplicated UTIs such as amoxicillin and trimethoprim-sulfamethoxazole were also reported in other studies from Iran and other countries [14, 18–21]. This condition is conducive to uncontrolled application of empirical therapy with fluoroquinolones, third generation cephalosporins and even new macrolides like azithromycin. Therefore, increased resistance to these antibiotic agents could be expected as well it is observable in our results.
Aminoglycosides like gentamycin and carbapenem antibiotics are not usually prescribed for outpatients UTIs in Iran in this regard, lower rate resistance against them is observed.
Evidence indicates that, higher antibiotic resistance rate is expected in hospital acquired bacteria in comparison to community acquired bacteria [22]. But antibiotic resistance increases continually in community acquired bacteria. Our results showed a high prevalence of antibiotic resistance against different antibiotic agents in CA-UPEC isolates. Thus, continuous screening of resistance patterns in CA-bacteria like CA-UPEC is mandatory.
Our finding revealed that, 11.3% of isolates were resistant to all antibiotic agents except that of imipenem, nitrofurantoin and fosfomycin. Therefore, nitrofurantoin, fosfomycin and carbapenem agents like imipenem are suitable candidates for empirical therapy of UTIs in outpatients. None of the highly resistant isolates were resistant to fosfomycin. Hence, fosfomycin is introduced as the most effective antibiotic agent against highly resistant CA-UPEC isolates.
UPEC outer membrane proteins (e.g., OmpA), are highly conserved and common among different strains [2, 8]. This is concomitant with our results indicating the highest prevalence of ompA gene among different virulence genes (Table 1). Statistical analysis showed that the prevalence of ompA was not significantly different between A, B2 and D phylogenetic groups. But, the prevalence of ompA was significantly lower in Group B1 (p-value = 0.003, odds ratio: 0.029).
The papC gene was the second common virulence factor and the most common adhesion among the UPEC isolates in the present study. In this regard, P fimbriae was reported as the second common virulence factor of UPEC Bien et al. [11]. More association of papC with UPEC pathotypes in comparison to other pathotypes has been also reported by Titilawo et al. [23]. High prevalence of P fimbriae associated genes in our study may be attributed to special colonization properties of CA-UPEC isolates that were obtained from this geographical area and show the importance of P fimbriae as the major adhesion in these isolates.
Phylogenetic origins analysis showed that, the most numbers of UPEC isolates belonged to Group B2 (67.3%) and subsequently Group D (21.4%) respectively. Similar results were obtained from other studies [24]. The most of the VFs were not significantly correlated with any of phylogenetic groups, except for papC and sfa/focCD genes which were predominantly more prevalent in Group B2 (Tables 1 and 2). The significant association between the papC and sfa/focCD genes with Group B2 is reasonable, because most of the UPEC isolates are categorized in B2 phylogenetic group. Therefore, papC and sfa/focCD genes are introduced as a predictor of Group B2 origin.
The present study findings showed a strong association between the irp2 gene and Group A origin (Table 2). The irp2 gene of Yersinia is present in different genera of the family Enterobacteriaceae including E. coli, Klebsiella, Citrobacter and others [25]. On the other hand, irp2 is considered as one of the virulence associated genes in diarrheagenic E. coli that is mainly allocated in phylogenetic Group A [26]. Therefore, the irp2 gene is introduced as a predictor for Group A origin. Therefore, uropathogenic ability of some diarrheagenic E. coli strains may be considered as a hypothesis again in our study, as was reported previously [26].
Logistic regression analysis also showed that papC gene is a predictor for amoxicillin resistance (p-value = 0.006, odds ratio: 26.00). A parallel finding has been reported by Karami et al., in which they introduced papC gene as a predictor for ampicillin resistance [6].
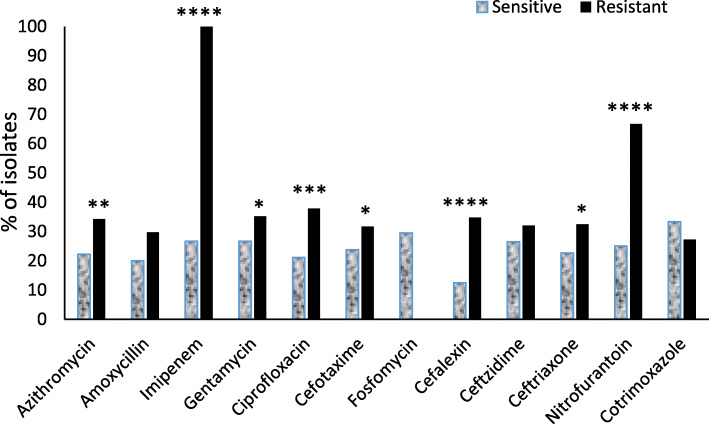
Data obtained from logistic regression analysis showed reduced prevalence of some virulence genes such as irp2, fimC and sfa/focCD in antibiotic resistant isolates in comparison to antibiotic susceptible isolates. On the contrary, the ompA, malX and hlyA genes were obviously more abundant in the isolates resistant to different antibiotic agents (Table 4). Moreover, the isolates that contained all three genes (ompA + malX + hlyA) were significantly (confidence level ≥ 95%) more resistant to azithromycin, imipenem, ciprofloxacin, cephalexin, ceftriaxone, nitrofurantoin, and even with lower confidence level (80–90%) to cefotaxime and gentamycin in comparison to other isolates (Fig. 1). Therefore, the ompA, malX and hlyA genes are introduced as possible predictors of antibiotic resistance in CA-UPEC isolates.
Fig. 1.
Prevalence of isolates which contained three virulence associated genes (ompA + malX + hlyA), in sensitive and resistant isolates. Prevalence (%) of isolates which contained three virulence associated genes (ompA + malX + hlyA) in sensitive isolates (white columns) in comparison to resistant isolates (black columns) have shown separately for any of the tested antibiotic agents. *p < 0.2, **p < 0.05, ***p < 0.01 and ****p < 0.001 (Fisher’s exact test analysis)
The lower prevalence of genes encoding type 1 fimbriae, P fimbriae and also hlyA gene in quinolone-resistant isolates in comparison to susceptible isolates were reported in some investigations [27–29]. Our finding confirmed them about type1 fimbriae and genes encoding P fimbriae, but it’s in contrary with the results attributed to hly genes. The logistic regression analysis showed that, the prevalence of this gene was significantly more prevalent in ciprofloxacin resistant isolates in comparison to susceptible isolates (Tables 3 and 4). This contradiction is reasonable because, the gradual evolutionary mechanisms responsible for antibiotic resistance in more virulent isolate could be arises any time and independently from any changes in genes encoding VFs, just in consequence of the continuous encounter with antibiotic agents. This finding is in line with other studies reported previously [30–32].
Conclusion
The UPEC isolates have very versatile gene pools contains some virulence associated genes that might be more intrinsic in comparison to antibiotic resistance genes. The results obtained from the present study showed that some surface exposed adhesions and iron-acquisition system are obviously associated with antibiotic susceptibility. On the contrary, ompA and some of the other pathogenicity-island related genes predominantly are associated with antibiotic resistance. We believe that antibiotic resistance acquisition may occur in compliance with the preliminary existence of specific genetic background including genes encoding VFs such as ompA, malX and hlyA in CA-UPEC isolates and descriptive reasons maybe revealed later. Although, our results showed an association between antibiotic resistance pattern and some virulence associated genes, but more investigation with higher number of bacterial isolates are needed to confirm these findings and the comparison of gene pools from CA-UPEC isolates with HA-UPEC could be more beneficial in this respect. Studies on further virulence associated genes and some resistance genes like ESBLs could be resulted in the discovery of new aspects of this association.
Methods
Bacterial isolates and culture conditions
All bacterial isolates were obtained from urine samples of symptomatic outpatients who referred to medical laboratories of Zabol, southeast of Iran, during 2018–2019. Patients with a history of antibiotic use in the recent month were excluded. Agar plates containing the harvested bacteria were transferred to the microbiology laboratory of medical faculty regularly. Bacterial isolates were inoculated primarily on EMB agar plates and incubated at 37 °C for 24 h. The grown colonies with metallic shine appearance were confirmed by biochemical tests. All isolates were inoculated into the 1.5 ml microtubes containing Muller-Hinton broth with 20% glycerol and preserved at − 70 °C for subsequent use.
Antibiotic susceptibility assessment
Antibiotic susceptibility profiles were determined by standard disc diffusion test, Kirby-Bauer method, according to CLSI guidelines [33]. Susceptibility and resistance criteria were estimated based on CLSI M100-S27 protocol [34]. ATCC E.coli 25,922 was used as control strain in disk diffusion susceptibility test. Twelve antibiotic disk in different classes were used including β-lactames (amoxicillin 30 μg, cephalexin 30 μg, ceftriaxone 30 μg, cefotaxime 30 μg, ceftazidime 30 μg), aminoglycosides (gentamicin 10 μg), phosphonic antibiotics (fosfomycin 200 μg), nitrofuran antibiotics (nitrofurantoin 300 μg), carbapenems (imipenem 10 μg), sulfonamides (trimethoprim-sulfamethoxazole 25 μg), quinolones (ciprofloxacin 5 μg) and macrolides (azithromycin 15 μg), that all were from Rosco Diagnostica (Denmark), were used in susceptibility assessments.
DNA extraction and PCR conditions
All UPEC isolates were cultured in Luria-Bertani broth (Merck, Germany) and incubated for 24 h at 37 °C. Genomic DNA was prepared from harvested bacteria by boiling lysis method. Briefly, bacterial suspensions in distilled water were boiled at 95 °C in water bath for 10 min. After centrifugation, the supernatant was stored at − 20 °C. Aliquots of 2.5 μl template DNA were used for PCR [24, 35]. The sequences that used as primer were acquired from two previous studies [5, 36]. All primers were purchased from (Bioneer, South Korea). Polymerase chain reactions were conducted by Biometra termocycler (T-Gradient thermoblock, Germany).
Phylogenetic classification
Genomic DNA was extracted from all E. coli isolates and PCR amplification was performed with specific primers incorporated to chuA and yjaA genes and TspE4.C2 sequences. Each isolate was allocated into one of the four phylogenetic groups (A, B, B2 & D) based on the existence of the PCR products as described earlier by Clermount, et al. [5]. In summary, the phylogenetic groups were assigned according to the following genotypes: group B2 (chuA+/yjaA+), group D (chuA+/yjaA−), group B1 (chuA−/TspE4.C2+) and group A (chuA−/TspE4.C2−).
Statistical analysis
Data analysis was performed by SPSS software version 18. The Chi-square and Fisher’s exact test were used to compare the association of genes encoding VFs with different variables such as antibiotic resistance and phylogenetic groups. Binary logistic regression analysis was used to assess the role of virulence genes and phylogenetic origin as predictors of resistance to different antibiotic agents. The p-value ≤0.05 was considered as significant.
Supplementary information
Additional file 1. Table of primers & PCR conditions
Acknowledgments
The authors wish to thank the staff and technicians at microbiology laboratory, Zabol University of medical sciences for their friendly cooperation. We also thanks Dr. Amin Beigzadeh for his kindly cooperation in revision step.
Abbreviations
- UTI
Urinary tract infection
- UPEC
Uropathogenic Escherichia coli
- VF
Virulence Factor
- CLSI
Clinical and Laboratory Standards Institute
- ATCC
American Type Culture Collection
- EMB
Eosin Methylene Blue
- PCR
Polymerase Chain Reaction
- CA-UPEC
Community Acquired Uropathogenic Escherichia coli
- HA-UPEC
Hospital Acquired Uropathogenic Escherichia coli
- PAI
Pathogenicity Associated Islands
Authors’ contributions
The basic idea was from OT, He contributed to the designing of the work, actual steps of methods, obtaining, analyzing and interpretation of the data and writing of the original draft of manuscript. Resources were provided by OT. ZY cooperated in obtaining the bacterial isolates, contributed to designing and performing the assessments, she also reviewed the final manuscript. MS contributed to collecting the bacterial isolates, designing and performing the assessments, she also reviewed the final manuscript. All authors have involved in revision process and all read and approved the submitted version of the manuscript.
Funding
This study was supported by vice chancellor for research and technology, Zabol University of medical sciences. Majority of the experiments were performed in research laboratories of Zabol University of medical sciences and experiment resources were provided by the funder.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Consent to participate is not applicable for this study because this study does not contain any studies with human participants or animals and the isolates included in the study were obtained from existing clinical collections routinely assembled as part of laboratory practices of clinical laboratories.
The raw data used during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12866-020-01933-1.
References
- 1.Beerepoot MA, Geerlings SE, van Haarst EP, van Charante NM, ter Riet G. Nonantibiotic prophylaxis for recurrent urinary tract infections: a systematic review and meta-analysis of randomized controlled trials. J Urol. 2013;190(6):1981–1989. doi: 10.1016/j.juro.2013.04.142. [DOI] [PubMed] [Google Scholar]
- 2.Terlizzi ME, Gribaudo G, Maffei ME. UroPathogenic Escherichia coli (UPEC) infections: virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front Microbiol. 2017;8:1566. doi: 10.3389/fmicb.2017.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marrs CF, Zhang L, Foxman B. Escherichia coli mediated urinary tract infections: are there distinct uropathogenic E. coli (UPEC) pathotypes? FEMS Microbiol Lett. 2005;252(2):183–190. doi: 10.1016/j.femsle.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 4.Barber AE, Norton JP, Spivak AM, Mulvey MA. Urinary tract infections: current and emerging management strategies. Clin Infect Dis. 2013;57(5):719–724. doi: 10.1093/cid/cit284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clermont O, Bonacorsi S, Bingen E. Rapid and simple 473 determination of the Escherichia coli phylogenetic group. Appl Environ. 2000;474:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karami N, Wold A, Adlerberth I. Antibiotic resistance is linked to carriage of papC and iutA virulence genes and phylogenetic group D background in commensal and uropathogenic Escherichia coli from infants and young children. Eur J Clin Microbiol Infect Dis. 2017;36(4):721–729. doi: 10.1007/s10096-016-2854-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Confer AW, Ayalew S. The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet Microbiol. 2013;163(3–4):207–222. doi: 10.1016/j.vetmic.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson TF, Watts KM, Hunstad DA. OmpA of uropathogenic Escherichia coli promotes postinvasion pathogenesis of cystitis. Infect Immun. 2009;77(12):5245–5251. doi: 10.1128/IAI.00670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schilling JD, Mulvey MA, Hultgren SJ. Structure and function of Escherichia coli type 1 pili: new insight into the pathogenesis of urinary tract infections. J infect Dis. 2001;183(Supplement_1):S36–S40. doi: 10.1086/318855. [DOI] [PubMed] [Google Scholar]
- 10.Norgren M, Baga M, Tennent JM, Normark S. Nucleotide sequence, regulation and functional analysis of the papC gene required for cell surface localization of pap pili of uropathogenic Escherichia coli. Mol Microbiol. 1987;1(2):169–178. doi: 10.1111/j.1365-2958.1987.tb00509.x. [DOI] [PubMed] [Google Scholar]
- 11.Bien J, Sokolova O, Bozko P. Role of Uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int J Nephrol. 2012;2012:681473. doi: 10.1155/2012/681473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riegman N, Kusters R, Van Veggel H, Bergmans H. Van Bergen en Henegouwen P, Hacker J, et al. F1C fimbriae of a uropathogenic Escherichia coli strain: genetic and functional organization of the foc gene cluster and identification of minor subunits. J Bacteriol. 1990;172(2):1114–1120. doi: 10.1128/jb.172.2.1114-1120.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khasheii B, Anvari S, Jamalli A. Frequency evaluation of genes encoding siderophores and the effects of different concentrations of Fe ions on growth rate of uropathogenic Escherichia coli. Iran J Microbiol. 2016;8(6):359–365. [PMC free article] [PubMed] [Google Scholar]
- 14.Paniagua-Contreras GL, Monroy-Perez E, Rodriguez-Moctezuma JR, Dominguez-Trejo P, Vaca-Paniagua F, Vaca S. Virulence factors, antibiotic resistance phenotypes and O-serogroups of Escherichia coli strains isolated from community-acquired urinary tract infection patients in Mexico. J Microbiol Immunol Infect. 2017;50(4):478–485. doi: 10.1016/j.jmii.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Lee JH, Subhadra B, Son YJ, Kim DH, Park HS, Kim JM, et al. Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli strains isolated from patients with urinary tract infections in South Korea. Lett Appl Microbiol. 2016;62(1):84–90. doi: 10.1111/lam.12517. [DOI] [PubMed] [Google Scholar]
- 16.Östblom A, Adlerberth I, Wold AE, Nowrouzian FL. Pathogenicity island markers, virulence determinants malX and usp, and the capacity of Escherichia coli to persist in infants' commensal microbiotas. Appl Environ Microbiol. 2011;77(7):2303–2308. doi: 10.1128/AEM.02405-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reidl J, Boos W. The malX malY operon of Escherichia coli encodes a novel enzyme II of the phosphotransferase system recognizing glucose and maltose and an enzyme abolishing the endogenous induction of the maltose system. J Bacteriol. 1991;173(15):4862–4876. doi: 10.1128/jb.173.15.4862-4876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raeispour M, Ranjbar R. Antibiotic resistance, virulence factors and genotyping of Uropathogenic Escherichia coli strains. Antimicrob Resist Infect Control. 2018;7(1):118. doi: 10.1186/s13756-018-0411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neamati F, Firoozeh F, Saffari M, Zibaei M. Virulence genes and antimicrobial resistance pattern in Uropathogenic Escherichia coli isolated from hospitalized patients in Kashan. Iran Jundishapur J Microbiol. 2015;8(2):e17514. doi: 10.5812/jjm.17514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah C, Baral R, Bartaula B, Shrestha LB. Virulence factors of uropathogenic Escherichia coli (UPEC) and correlation with antimicrobial resistance. BMC Microbiol. 2019;19(1):204. doi: 10.1186/s12866-019-1587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perletti G, Magri V, Cai T, Stamatiou K, Trinchieri A, Montanari E. Resistance of uropathogens to antibacterial agents: emerging threats, trends and treatments. Arch Ital Urol Androl. 2018;90(2):85–96. doi: 10.4081/aiua.2018.2.85. [DOI] [PubMed] [Google Scholar]
- 22.Matta R, Hallit S, Hallit R, Bawab W, Rogues A-M, Salameh P. Epidemiology and microbiological profile comparison between community and hospital acquired infections: a multicenter retrospective study in Lebanon. J Infect Public Health. 2018;11(3):405–411. doi: 10.1016/j.jiph.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Titilawo Y, Obi L, Okoh A. Occurrence of virulence gene signatures associated with diarrhoeagenic and non-diarrhoeagenic pathovars of Escherichia coli isolates from some selected rivers in South-Western Nigeria. BMC Microbiol. 2015;15(1):204. doi: 10.1186/s12866-015-0540-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munkhdelger Y, Gunregjav N, Dorjpurev A, Juniichiro N, Sarantuya J. Detection of virulence genes, phylogenetic group and antibiotic resistance of uropathogenic Escherichia coli in Mongolia. J Infect Dev Ctries. 2017;11(1):51–57. doi: 10.3855/jidc.7903. [DOI] [PubMed] [Google Scholar]
- 25.Bach S, de Almeida A, Carniel E. The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol Lett. 2000;183(2):289–294. doi: 10.1111/j.1574-6968.2000.tb08973.x. [DOI] [PubMed] [Google Scholar]
- 26.Abe CM, Salvador FA, Falsetti IN, Vieira MA, Blanco J, Blanco JE, et al. Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol Med Microbiol. 2008;52(3):397–406. doi: 10.1111/j.1574-695X.2008.00388.x. [DOI] [PubMed] [Google Scholar]
- 27.Vila J, Simon K, Ruiz J, Horcajada JP, Velasco M, Barranco M, et al. Are quinolone-resistant uropathogenic Escherichia coli less virulent? J Infect Dis. 2002;186(7):1039–1042. doi: 10.1086/342955. [DOI] [PubMed] [Google Scholar]
- 28.Piatti G, Mannini A, Balistreri M, Schito AM. Virulence factors in urinary Escherichia coli strains: phylogenetic background and quinolone and fluoroquinolone resistance. J Clin Microbiol. 2008;46(2):480–487. doi: 10.1128/JCM.01488-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harwalkar A, Gupta S, Rao A, Srinivasa H. Lower prevalence of hlyD, papC and cnf-1 genes in ciprofloxacin-resistant uropathogenic Escherichia coli than their susceptible counterparts isolated from southern India. J Infect Public Health. 2014;7(5):413–419. doi: 10.1016/j.jiph.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, et al. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62(2):606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hacker J, Bender L, Ott M, Wingender J, Lund B, Marre R, et al. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extra intestinal Escherichia coli isolates. Microb Pathog. 1990;8(3):213–225. doi: 10.1016/0882-4010(90)90048-u. [DOI] [PubMed] [Google Scholar]
- 32.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23(6):1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 33.Wayne P. Performance standards for antimicrobial susceptibility testing. 2011. Clinical and laboratory standards Institute. [PubMed] [Google Scholar]
- 34.CLSI . M100-S27: performance standards for antimicrobial susceptibility testing, 27th informational supplement. Wayne: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 35.Queipo-Ortuño MI, Colmenero JDD, Macias M, Bravo MJ, Morata P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin Vaccine Immunol. 2008;15(2):293–296. doi: 10.1128/CVI.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ewers C, Li G, Wilking H, Kieβling S, Alt K, Antáo E-M, et al. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int J Med Microbiol. 2007;297(3):163–176. doi: 10.1016/j.ijmm.2007.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table of primers & PCR conditions
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.