Abstract
Malarial rhythmic fevers are the consequence of the synchronous bursting of red blood cells (RBCs) upon completion of the malaria parasite asexual cell cycle. Here we hypothesized that an intrinsic clock in the parasite underlies the 24-hour-based rhythms of RBC bursting. We show that parasite rhythms are flexible and lengthen to match the rhythms of hosts with long circadian periods. We also show that malaria rhythms persist even when host food intake is evenly spread across 24 hours suggesting that host feeding cues are not required for synchrony. Moreover, we find that the parasite population remains synchronous and rhythmic even in an arrhythmic clock mutant host. Thus, we propose that parasite rhythms are generated by the parasite, possibly to anticipate its circadian environment.
One Sentence Summary
Host rhythms are not necessary for malaria rhythmicity.
Main Text
Multiple daily rhythms have been described in malaria infections, including fevers, host-seeking behavior of the mosquito vector, oocyst burden, gametocyte numbers and blood-stage schizogony timing (1–3). In the blood, erythrocytes (red blood cells or RBCs) burst approximately 24, 48 or 72 hours after invasion, depending on the Plasmodium species (4). These periodic blood-stage asexual cell cycle rhythms are accompanied by robust rhythms in gene expression (5–8). It has been proposed that host rhythms drive the daily rhythms of malaria parasite, since shifting the normal circadian rhythms of the host leads to a change in timing of cell-cycle rhythms in the parasite (9–12). This idea is also supported by the observation that malaria parasites lose their populational synchrony in culture, where host rhythms are absent (13). However, virtually all living organisms have intrinsic circadian clocks that can anticipate daily changes of their environment (14), and we previously showed that the parasite that causes sleeping sickness has an intrinsic circadian clock (15). Because the malaria cell cycle and gene expression are intimately linked with daily rhythms, we hypothesized that, instead of merely responding to host cues, the malaria parasite also has an internal timekeeping mechanism.
Malaria parasite rhythms persist in constant darkness
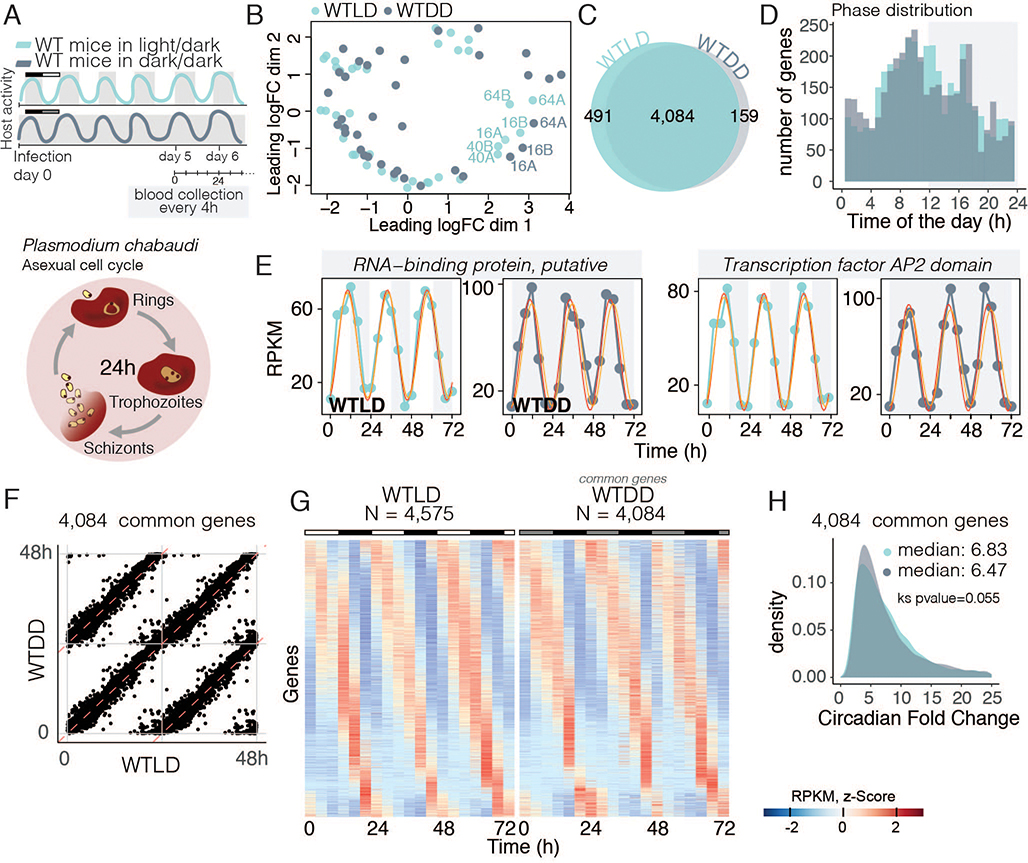
The 24-hour light/dark cycle is the primary cue used by the host to entrain its circadian rhythms to the environment. Thus, to dissect which cues are essential for malaria parasite synchrony, we first assessed the effects of light/dark cycles on parasite rhythms. We probed the transcriptomes of parasites every 4 hours for three days from wild-type (WT) mice housed in regular light/dark (LD; 12 hours:12 hours) conditions or in complete darkness (DD; Fig. 1A). Consistent with previous reports, when exposed to regular light/dark cycles Plasmodium chabaudi populations were synchronized with robust daily rhythms in both the asexual cycle and gene expression (6, 12). Malaria gene expression was strikingly rhythmic; as unbiased multidimensional scaling analysis identified similar patterns of expression across the >5,000 genes for samples taken 24 hours apart (e.g.,16 hours, 40 hours, 64 hours clustering together) independent of the host LD or DD lighting conditions (WTLD and WTDD, Fig. 1B). Using statistical tests for ~24-hour periodicity (detailed in the Methods, Fig. S1), we found that, out of the 5,244 genes expressed by blood-stage parasites, over 4,000 were cycling in both lighting conditions (>80% of the transcriptome, Fig. 1C–D, Data Table 1). This is a slightly higher number of genes than described previously for LD conditions (6), possibly because of a combination of higher sensitivity of sequencing technique, multiple cycles assessed per condition, and robust statistical methods. Importantly, the phase at which these common cycling genes peaked and their median circadian fold-change (6.5-fold) was maintained in both LD and DD conditions, indicating that malaria rhythms persist in constant darkness, and suggesting that lighting cues are not an important signal for this intracellular parasite within its rhythmic host (Fig. 1C–H), consistent with our prior research on the sleeping sickness parasite (15).
Fig. 1.
Circadian host cues and not environmental changes synchronize the parasite population. A. Schematic representation of the experimental design in which mice were kept either under light/dark cycles (WTLD – light blue) or in complete darkness (WTDD - grey) where their internal rhythms is the only time cue for the parasites. Activity represents the daily rhythm profile of host activity. Diagram of the asexual cell-cycle stages known to be synchronous among the parasite population throughout the day. B. Multidimensional scaling assessment of parasite gene expression within each host shows samples that were collected 24 hours apart clustering together as if they were replicates, even when considering all >5,000 genes (cycling or not, using this unbiased method). This creates a circular shape, characteristic of very rhythmic datasets. Samples were collected every 4 hours for three consecutive days starting on day 5 post-infection. WTLD, n = 108 mice, WTDD, n=36 mice. The annotation 16A refers to gene expression data from the parasites collected at 16 hours after lights on (ZT16, light blue) or from the equivalent time point in DD (CT16, grey), replicate A. Similar annotations are used for the remaining data points. C. The number of parasite cycling genes with a 24-hour period, showing that gene expression in the parasite is rhythmic in both conditions. D. Phase (time of peak of expression during the day) is maintained across conditions. E. Example of two of the top 20 cycling genes in both conditions include the RNA-binding protein (PCHAS_0508500) and the transcription factor with AP2 domain (PCHAS_0110100). F. Phase of the common cycling genes is perfectly correlated. Phase correlation is double plotted. G. Heatmaps sorted by phase of genes cycling within the WTLD host, showing that in WTDD their profile is virtually unchanged. Each row is one gene whose gene expression is z-scored. H. Circadian foldchange for the common cycling genes across conditions does not change upon isolation of environmental cues. Ks means Kolmogorov-Smirnov test.
Malaria parasites actively synchronize their rhythms with the host
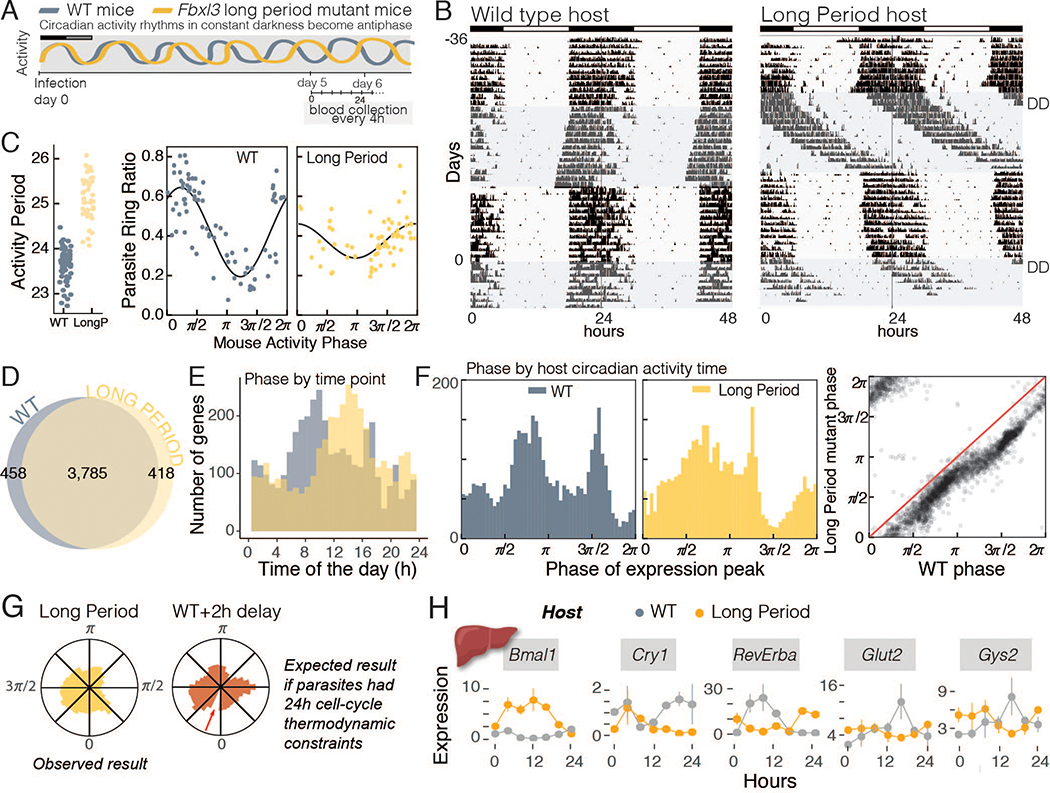
Although these results showed that malaria parasites remain synchronized in a host with normal circadian rhythms, we wondered whether malaria parasites, similar to many other organisms, can entrain to periods that are not exactly 24 hours (16). If so, this would suggest that their cell-cycle is not locked to a 24-hour thermodynamic constraint, and, instead, that they may have their own intrinsic rhythms which can be modulated by host cues. Previous studies have shown that the timing (‘phase’) of malaria parasite rhythms can adjust to host rhythms when hosts experience jetlag (9–12). However, instead of just the timing, we aimed to directly test the plasticity of their rhythms (‘how long’ a cycle would last, i.e., ‘period’) in a host with an abnormal circadian period (17, 18). Therefore, we challenged the plasticity of parasite rhythms by infecting both wild-type (WT) and long-period Fbxl3 (Long Period) mutant mice maintained under constant darkness (DD) and assessed the effects on asexual cell-cycle and transcriptional rhythms. Under DD conditions, the period of WT mice activity rhythms was 23.7 hours (± 0.3h, n = 34 mice), whereas that of Long Period mice was 25.7 hours (± 0.7h, n = 37 mice; Fig. 2A–B), which caused activity onset to shift approximately 2 hours later each day. Thus, after 6–7 days in DD, Long Period mice were in opposite phase to WT animals (Fig. 2A). Remarkably, instead of staying locked to a 24-hour asexual cycle, malaria parasites in Long Period hosts slowed their cell cycle to match the longer period rhythms of the host. When these data were analyzed relative to the length and timing of the host activity period, it was clear that the timing of parasite cell division matched the host (WT or Long Period) activity rhythms (Fig. 2C). Gene expression rhythms were also longer, with significant overlap between the cycling genes from parasites in either host type (Fig. 2D, Fig. S2A–B, Data Table 1) and with the phase of gene expression matching host activity rhythms (Fig. 2E–F). This was a gradual slowing down (Fig. 2F), which suggests that there is not a thermodynamic constraint imposed on the 24-hour cell-cycle period and that parasites were not completing the 24-hour cell division and then waiting for a host cue to resume the next cycle of replication (Fig. 2G and S2C–D). Instead, distribution of phases of cycling genes was spread out rather uniformly across each cycle. Further characterization of the phase alignment hinted at an active, intrinsic phenomenon, since, although the timing of gene expression was aligned with the host’s activity, the two were not perfectly correlated (Fig. 2F, right panel). The parasite peak gene expression in the Long Period mice lagged behind (0.64 radians, i.e. 10% of the cycle) and the amplitude of the asexual cycle was not as high as within a WT host (Fig. 2C, reduction of 62% in amplitude), as might be expected from a parasite that is actively adjusting (i.e., gradually delaying) their internal rhythm to the longer period. These findings suggest that the parasite asexual cell cycle and gene expression may not be simply driven by the host rhythms but are generated by the parasite. Also, importantly, the parasite’s rhythmicity is not merely a consequence of the length of its cell cycle, as it can be actively synchronized to the period length of the host.
Fig 2.
Parasite rhythms are not locked to a 24-hour period, but are instead plastic and adjust to a different period length. A. Schematic representation of the experimental design in which WT mice or a long circadian period mutant were infected in complete darkness (DD) where rhythms shift to the opposite phase after 6 days. Blood was collected every 4 hours for three consecutive days starting on day 5 post-infection. B. Running-wheel activity actograms to monitor circadian behavior upon release in complete darkness (shaded areas of the actogram). Mice were recorded for 36 days to characterize their phenotypes prior to infection (day 0, second period in DD, shaded area). C. Internal circadian period of each mouse (left panel). Asexual cycle timing was converted from collection time (from blood collected at standard local time) to match the circadian time of activity period of the host (n= 62 WT mice, n= 43 long period mice, from 3 independent experiments). D. Number of cycling genes of the parasite in each host. E. Phase of expression of cycling genes based on the clock (local) time of collection (ZT0 in the previous light/dark schedule, with 0 hours representing time at which lights were turned on prior to constant darkness), representative of day 6 post-infection (blood from 36 mice per group, from 2 independent biological replicates for each timepoint) and F. after matching to the circadian time of the host (reliable circadian onset identifiable for n=35 WT and n=29 long period mice), showing their alignment to host rhythms. For WT 2π is roughly 23.7 hours for each mouse; and for Long Period 2π is roughly 25.7 hours. Right panel shows the correlation of the phases among genes. G. Phase plots for parasite gene expression when infecting the Long Period mutants. On the left is represented the observed phase (peak expression) of the cycling genes, and on the right is the expected results for the phase of parasite gene expression when infecting Long Period mutants if the parasite rhythms were driven by a 24-hour thermodynamic constraint cell-cycle, i.e., would be a similar phase plot to when infecting WT mice with a 2 hour delay to resume cycle. H. Host liver gene expression of clock genes and two metabolic genes. Each time point is composed of 3–4 mice per condition, with line connecting them being only for visualization purposes. Error bars represent standard deviation.
Malaria rhythms do not require host feeding rhythms
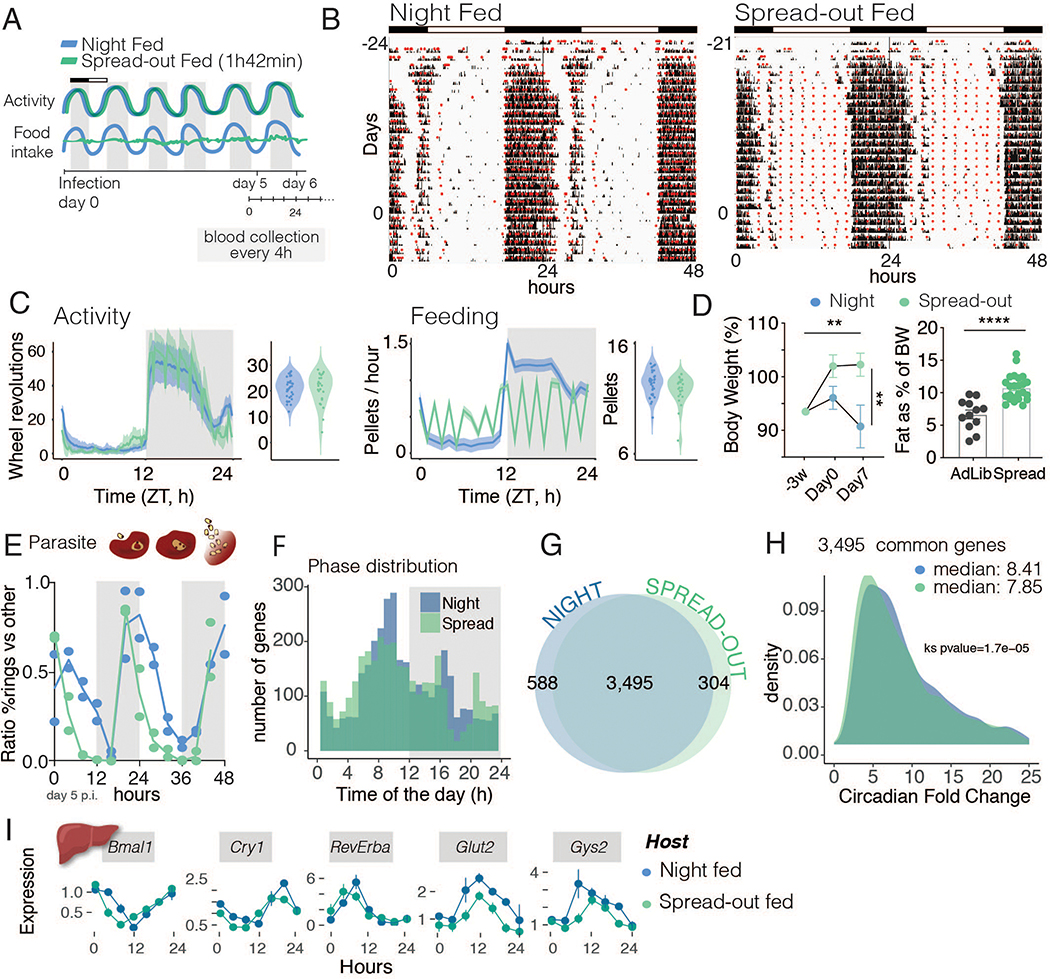
Together, our first two experiments demonstrated that lighting cues are not necessary for parasite rhythms and that there is plasticity in parasite rhythmicity, but these findings do not exclude the possibility of other non-photic host cues (19–21). Feeding rhythms have been proposed to drive rhythms of the asexual cell cycle of the parasite (19, 20), since the timing of parasite cell-cycle division is reversed in animals fed in the daytime. Because both WT and Long Period mutant mice in DD maintain their circadian feeding rhythms and liver gene expression (Fig. 2H and S2B), we next wanted to test whether feeding rhythms are necessary for parasite cell-cycle and transcriptional rhythms. Infected mice were housed in automated feeders (22) in LD conditions that either restricted the timing of food intake to the dark when mice usually eat (‘night fed mice’) or, in order to abolish feeding rhythms, had food access evenly distributed throughout the 24-hour day (‘spread-out fed mice’, Fig. 3A). The total amount of food provided was the same in both conditions (14 × 300 mg food pellets in 24 hours, Fig. 3B–C). Regardless of when food was available, mice still chose to run during the dark phase (Fig. 3B–C). As predicted, the metabolic challenge of feeding with the unnatural spread-out pattern led to a significant body weight and body fat percentage increase in the spread-out fed group relative to nighttime fed conditions (23, 24), even though food consumption and running wheel activity were similar (Fig. 3C–D). This reinforces that the experimental design was successful in abolishing feeding rhythms. If feeding rhythms drive the synchrony of malaria, abolishing feeding rhythms should make the probability of finding parasites at any stage of the cycle at any time of the day the same (i.e., the rhythmicity of the parasite population would be lost, even if each parasite still takes ~24 hours to divide). Instead, we found that even during spread-out feeding conditions, the asexual cell-cycle rhythms of malaria parasites were maintained (Fig. 3E, Fig. S2E–F), with >90% of cycling genes in the spread-out condition being shared with regular night-feeding (Fig. 3F–H, Data Table 1). These data clarify that the host cue that parasites align to is not the actual food intake rhythm, and, notably, feeding rhythms do not drive the oscillations in the parasite population, contrary to what has been proposed (19). Other downstream cue(s) that may change upon feeding manipulations (such as body temperature, metabolites or hormones) and that do not change in the absence of feeding rhythms are likely used by parasites as a time cue (Fig. 3I). The amplitude of the oscillation of the common cycling genes was slightly reduced (6% reduction) with spread-out feeding, implying that a minor percentage of parasite transcripts rely on rhythmic feeding cues (Fig. 3H, Fig. S2F). Together, these data suggest that, although rhythms in host food intake may be a cue for malaria cell-cycle timing, they are not necessary for malaria rhythmicity.
Fig 3.
Feeding rhythms are not required for parasite rhythmicity. A. Schematic representation of the experimental design in which WT mice were either fed at nighttime or the same amount of food was spread throughout the day, thereby abolishing daily rhythms in food intake. Blood was collected every 4 hours for two consecutive days, starting on day 5 post-infection. B. Running-wheel activity actograms to monitor activity (in black) and food intake (in red circles). C. Average daily profile of activity for the n=26 nighttime fed mice and n=24 spread-out fed mice. Spread-out feeding had no effect on the timing or total amount of activity. However, with spread out feeding, food intake lost daily rhythms despite the same total consumption. D. Left panel shows body weight comparison, Two-Way ANOVA, time factor and feeding condition both p < 0.01. Right panel shows body fat composition, Mann-Whitney, p < 0.0001. E. Parasite asexual cell-cycle stage over time. F. Phase (peak time) of parasite daily gene expression. G. Number of cycling genes in the parasite from host in each feeding condition. H. Circadian fold change of the common cycling genes across feeding conditions. Ks means Kolmogorov-Smirnov test. I. Host liver gene expression of clock genes and two metabolic genes. Each time point is composed of 3–4 mice per condition, with line connecting them being only for visualization purposes. Error bars represent standard deviation. Two-way ANOVA shows an effect of time for all genes p < 0.0001, and an effect of feeding condition in Cry1 (p < 0.05), Glut2 (p < 0.0001) and Gys2 (p < 0.001).
Malaria rhythms persist in an arrhythmic host
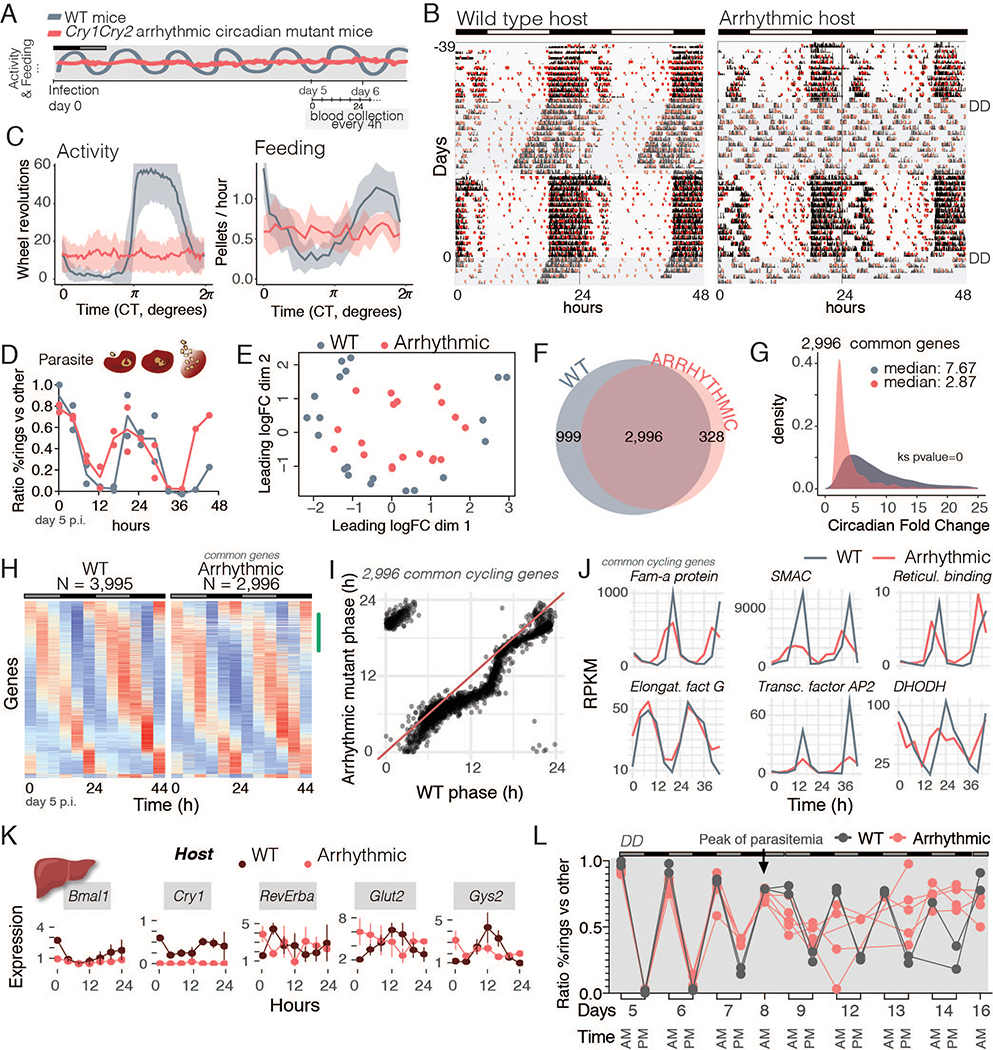
Finally, we asked whether malaria rhythms could persist within a completely arrhythmic host. If parasites do not have clocks, we would predict that they would very rapidly lose synchrony in the absence of host rhythms; however, if parasites have clocks we would predict that parasites maintain synchrony or slowly lose it over time. To test this, we entrained WT and Cry1/Cry2 arrhythmic clock mutant mice in a LD 12 hours:12 hours cycle, then infected and released them into DD. Arrhythmic mutant mice lose both their wheel-activity and feeding rhythms immediately under DD (25) (Fig. 4A–C and Fig. S3A). The arrhythmic mutants showed higher body fat percentage than WT mice in DD (as in WT mice under spread-out feeding conditions) despite having lower body weight (Fig. 3D and Fig. S3B). Remarkably, both malaria asexual cell-cycle rhythms and transcriptional rhythms persisted with ~24 hours rhythmicity for 5 to 7 days after infection, even in the arrhythmic host (Fig. 4D–E and Fig. S3C). Unbiased multidimensional scaling detected a circular pattern representing a similarity in gene expression across timepoints that varied throughout the day and returned to an initial value as the day ended, similar to what is observed in WT (Fig. 4E and Fig. 1B). This result indicates that parasite gene expression remains rhythmic across 24 hours, even without host cues.
Fig 4.
Malaria rhythmicity is not driven by the host. A. Schematic representation of the experimental design in which WT mice or arrhythmic mutants were infected and released into constant darkness (DD). Blood was collected every 4 hours for two consecutive days, starting on day 5 post-infection. B. Running-wheel activity actograms to monitor activity (in black) and food intake (in red circles). Mice were recorded for 36 days to characterize their phenotypes prior to infection (day 0, shaded area represents second period in DD). C. Average circadian profile of activity for WT (n= 19) and arrhythmic mice (n=18) in DD. Arrhythmic mice lost both activity and feeding rhythms upon darkness. D. Parasite asexual cell-cycle stage across time. E. Multidimensional scaling assessment of parasite gene expression within each host shows rhythmicity (circular pattern) of gene expression even in an arrhythmic mutant. F. Number of cycling genes in the parasite from within each host. G. Circadian fold-change of the common cycling genes across conditions. Amplitude of the ~3,000 genes that remained cycling was compared with a Kolmogorov-Smirnov test (Ks). H. Heatmaps sorted by phase of genes cycling within the WT host, where the profile of parasite gene expression in the arrhythmic host is virtually unchanged. Each row is one gene whose expression is z-scored. The green mark on the side of the heatmap represents the phase (0–8 hours) which lost many of the 1,000 genes that stopped cycling within the arrhythmic host. I. Phase (time of peak of expression during the day) was maintained across conditions. J. Top 25 common cycling genes showing examples that drop amplitude and some whose profile is unaffected. Fam-a protein (PCHAS_0100100), schizont membrane associated cytoadherence protein (SMAC) (PCHAS_0101300), reticulocyte binding protein (PCHAS_0101100), elongation factor G (PCHAS_0101900), transcription factor AP2 (PCHAS_0103600) and DHODH, dihydroorotate dehydrogenase (PCHAS_0102800). K. Host liver gene expression of clock genes and two metabolic genes. Each time point is composed of 3–4 mice per condition, with line connecting them being only for visualization purposes. Error bars represent standard deviation. L. WT (n = 2) and arrhythmic mutant mice (n= 5) were kept in constant darkness (DD) for over a week prior to infection and during the remainder of the experiment. Mutant arrhythmicity was confirmed with running wheel activity and food intake patterns. Mice were infected and synchrony was assessed at two time points: at previous lights ON (6 AM, light grey) and previous lights OFF (6 PM, dark grey) schedule for 16 days after the infection. Parasites infecting WT mice showed significant rhythmicity before and after the day 8 (peak of parasitemia, JTK and LS circadian algorithms, days 5–8 and 12–14, p < 0.05), whereas parasites infecting arrhythmic mice were only rhythmic prior to day 8 (JTK and LS cycling analysis for days 5–8, p < 0.05; for days 12–14, N.S.). Statistics?
We identified ~1,000 genes that ceased cycling within arrhythmic hosts, suggesting that these are genes that cycled due to external host signals (Fig. 4F and Fig. S3E). However, the vast majority of genes (~3,000, or 60%) remained cycling in both WT and arrhythmic hosts, suggesting that these genes are driven by the internal timekeeping mechanism of the parasite rather than driven by host circadian rhythms (Fig. 4F, Data Table 1). A small number of genes (328) whose rhythms were revealed upon the absence of host rhythms suggest that some host rhythms counterbalance these gene expression pattern of the parasite. There was a clear reduction in amplitude of the common cycling genes in parasites from arrhythmic hosts (Fig. 4H and Fig. S3D), as would be expected for any population of cells when environmental cues are removed and similar to what has been described for other circadian systems (15, 26, 27). Despite the abundance of research describing absence of rhythms in these arrhythmic mutants (28, 29) it is always complex to prove the negative, that there are no remaining rhythms that could maintain parasite population synchrony. However, if arrhythmic mutant mice were to have any residual rhythms able to set the time of (entrain) the parasite population, then the amplitude of the parasite rhythms should remain similar to when parasites infect a WT host. Instead, we observed the amplitude decreased with time, which is consistent with parasite desynchronizing. This has been well-described in mammalian cells, where in the absence of entraining signals endogenous clocks slowly drift apart (15, 26, 27).
The overall daily rhythms of the transcriptional cascade were similar in WT and arrhythmic hosts. Most of the cycling genes that ceased cycling had maximum expression at the beginning of the cycle (~0–8 hours, Fig. 4H and Fig. S3D), while those that remained cycling showed little change in their relative phase of expression (Fig. 4I). Despite the decreased amplitude of the oscillation of some genes, their overall expression profiles were unchanged (Fig. 4G and 4J). Among the top 25 common cycling genes (likely internally driven by the parasite timekeeping mechanism), were the Fam-a proteins, which is one family of Plasmodium-specific proteins exported into the RBC that are involved in the transporting and uptake of host phosphatidylcholine for parasite membrane synthesis (30). Other examples include the schizont membrane associated protein (SMAC), which is associated with sequestration in the vasculature (31, 32), and reticulocyte binding protein, which is associated with invasion of RBCs (33). As an essential control, we also assessed whether molecular rhythms (in addition to wheel and feeding behavior rhythms) were abolished in the arrhythmic mutant hosts. As expected, arrhythmic hosts showed no rhythms in either clock gene expression or metabolic pathways (Fig. 4K).
Malaria clocks are entrained by host cues
There are at least two ways parasites could maintain rhythms in the absence of host cues: i) having an internal clock but unable to mutually synchronize, in which with period variation across the population would lead to slow decay in synchrony (as observed in fibroblasts in culture (26, 27)); or ii) having a clock that allows parasites to release rhythmic signals that can mutually synchronize the parasites with each other. To distinguish between these two possibilities, we housed WT and arrhythmic mutant mice for over a week in darkness to ensure arrhythmicity in the arrhythmic genotype. We then infected them and followed parasite synchrony for 16 days. We observed that parasites in WT mice remained synchronized throughout the protocol. In contrast, in arrhythmic mutant mice synchrony was maintained with a slow decay only until the peak of parasitemia (~8–9 days post infection, JTK and LS p < 0.05, Fig. 4L and Fig. S3F) after which parasite synchrony was lost (JTK and LS p > 0.05, Fig. 4L). These findings are similar to those seen in mammalian cells with intrinsic circadian rhythms that require an external cue to synchronize the population, as over time the small variation in their internal rhythms leads to overall population asynchrony (26, 27). Thus, these observations support hypothesis (i) instead of hypothesis (ii), since the parasite population could not resynchronize upon losing synchrony, showing that parasites are not able to release a signal to mutually synchronize themselves, but rather need to integrate some host cue. Taken together, our findings indicate that malaria rhythms are maintained through internal clocks, but they still require host cues to entrain their rhythms in order to maintain synchrony across the parasite population and to remain synchronous in the long-term.
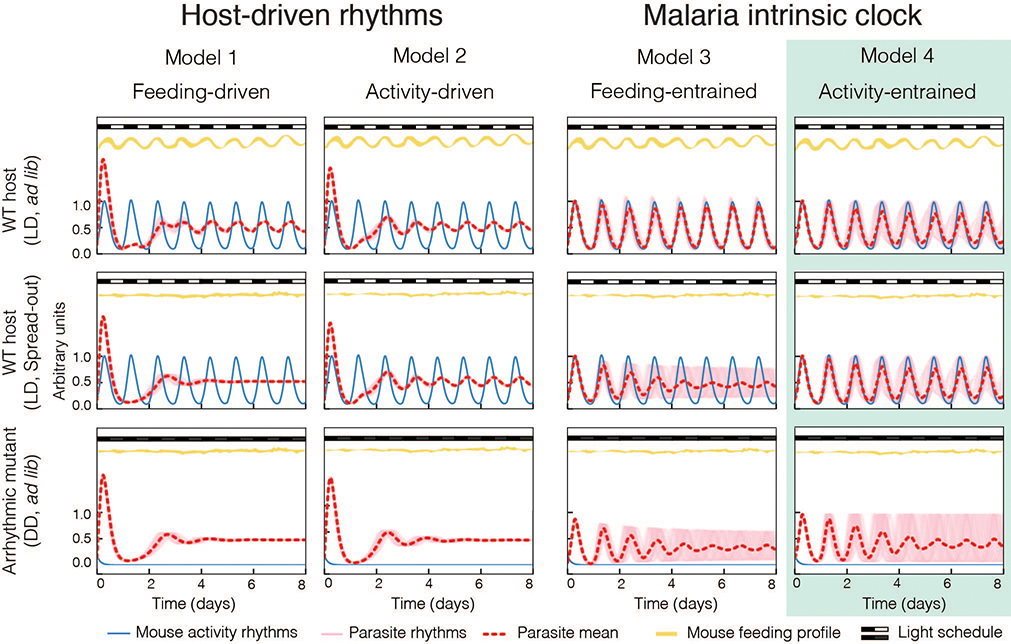
As a complement to our biological experiments, we developed mathematical models of the host and parasite transcriptional systems to test whether the rhythmicity of malaria parasites could be a simple “reactionary” response to host stimuli or whether malaria parasites have an intrinsic timekeeping mechanism (Fig. 5). We produced 4 different models using ordinary differential equations: models 1 and 2 assume that malaria parasites do not have an intrinsic clock and that the parasite rhythms observed are merely responses to host feeding patterns (model 1) or driven by host circadian activity (model 2). Model 1 reflects the current understanding of the field in which the malaria parasite rhythms reflect a simple reactionary response, known as a “just-in-time” response, to feeding/fasting cycles. In models 3 and 4, malaria has its own clock generating the parasite rhythms, with the clock capable of entraining to host feeding (model 3) or activity (model 4) cues to initiate synchrony, thereby reflecting our hypothesis. We applied these models to a population of 100 parasites with a distribution of periods among them of 24.2 hours ±1.3 hour (34). Using these models, we simulated our experimental challenges (WT in LD ad lib. or under spread-out feeding and arrhythmic mutant hosts) and then compared our simulation results to the experimental observations. The direct comparison would be from the experimental data to the ‘parasite mean’ rhythms since experimentally we studied population dynamics.
Fig 5.
Malaria rhythms are intrinsic. Mathematical modeling of the two main hypotheses: “Host-driven rhythms” of malaria parasites, either driven by feeding rhythms (model 1) or activity rhythms (model 2); or “Malaria intrinsic clock” whose rhythms are synchronized with the host clock by integrating feeding (model 3) or/and activity rhythms (model 4). Simulations of model 1–4 recreating the i) ‘normal’ infection WT in light/dark conditions (LD) with food available ad libitum (top row); and the experimental challenges of ii) ‘Spread-out feeding’ experiment in LD in which feeding rhythms were abolished, and mice voluntarily maintained rhythmic activity profile (middle row); and, iii) recreating the ‘Arrhythmic host’ experiment in complete darkness (DD) with food available ad lib., which due to their lack of a functional clock both feeding and activity behaviors became arrhythmic. Model 4 is the model that best recapitulates the observed experimental results.
We began by testing model 1, which predicted that under evenly distributed feeding conditions parasite rhythms would be lost (model 1, WT spread-out and arrhythmic mutant, Fig. 5 middle and bottom row, Fig. S4 and S5). The simulation result did not match what we observed experimentally (Fig. 3E–H and Fig. 4J), thus model 1 is not valid. Model 2 predicts that malaria rhythms would be immediately lost in an arrhythmic host (model 2, Fig. 5 bottom row, Fig. S4 and S5). However, the simulation results do not match what we observed experimentally since rhythmicity of the parasite population is maintained even after 5–7 days (Fig 4 D–J), thus model 2 can also be rejected. Instead, only an intrinsic timekeeping mechanism of the parasite (models 3 or 4) could explain our observed experimental results, including remaining rhythmic in the absence of any rhythmic host cues to the parasite population (Fig. 5). Since upon spread-out feeding, the amplitude of parasite rhythms is predicted to drop significantly if the intrinsic parasite oscillator was entrained by feeding (model 3, middle row), and this does not match our observed results (Fig. 3H), the model that best describes the biophysical oscillation experimentally observed is model 4. Model 4 recapitulates the slow decay in synchrony experimentally observed in arrhythmic hosts (Fig. 4D–J and Fig. 4L) and amplitude that persists upon spread out feeding. Altogether, modeling of the biophysical properties of the host and malaria parasites rhythms further supports the idea that malaria rhythmicity is driven by an intrinsic clock that is entrained by circadian signals from the host.
Malaria rhythms may be under transcriptional regulation
Finally, to investigate the molecular mechanisms underlying the Plasmodium clock, we searched for putative regulatory regions for the cycling genes. Although we did not find clear homologues between known clock genes and the malaria parasite genome (Data Table 2), we defined several putative regulatory elements that may be responsible for the cycling of sets of genes at specific times of day. Interestingly, the co-expression cluster M identified from our datasets whose 174 genes peak at 18 hours, is enriched (63% of genes, Figs. S6–S7) for an E-box DNA motif, CACGTG, that is a binding site of CLOCK:BMAL1, the transcriptional activator complex in the core circadian clock mechanism in mammals (35). The absence of clock homologues is not surprising, as early eukaryotes are quite divergent, and even across the well-established circadian rhythm models (cyanobacteria, fungi, plants and animals) the genes that control such oscillations are not conserved (14, 36). Nonetheless, the enrichment of specific regulatory motifs may suggest a transcriptional-translation mechanism underlying the oscillations in Plasmodium gene expression.
Conclusions
Our findings show that periodic rhythms are intrinsic to each parasite, although the parasites in a population do not have a means to synchronize themselves to each other without host cues. These findings are consistent with the fact that parasites lose synchrony in culture (13) and with findings from other isolated cell types: individual mammalian cells have slightly different circadian periods losing synchrony with time and thus need to be repeatedly resynchronized when in culture. It is also important to note that, although the most evident malaria parasite rhythm is the asexual cell cycle, clocks can regulate many more aspects of biology (14, 37). This may provide an explanation for malaria parasites having developmental cycles shorter than 24 hours (18-hour cycle for P. yoelii (38) and 21-hour for P. berghei (39)), similarly to cyanobacteria (40) and T. brucei (15). Future studies will need to address what comprises the parasite timekeeping mechanism that allows it to sustain intrinsic rhythms while also receiving and integrating timing cues from the host. In addition, it may be interesting to assess the contribution of non-transcriptional oscillators to parasite rhythms. Peroxiredoxin oxidation rhythms exist in RBCs (which do not perform transcription); however, based our findings it is unlikely that the weak rhythms found in RBCs of arrhythmic mutants (41, 42) could drive oscillations of 60% of the genome of the parasite. Even if they play a minor role they are not able to maintain the synchrony (since there is a decrease in amplitude) nor re-entrain the parasite population once synchrony is lost. These findings are also supported by human malaria parasites showing different period lengths when cultured in the same blood (Smith et al co-submitted). It would also be important to investigate how asexual cell cycle timing is gated by parasite transcriptional rhythms that persist in the absence of host rhythms (43, 44). Since perturbing either the clock mechanism or the integration of host cues may alter the host-parasite interaction, better understanding of the complex host-parasite interaction may improve treatments for malaria.
Supplementary Material
Acknowledgments
We thank Krista Matthews and Meg Phillips for helpful discussions; Nelly Garduño and Guadalupe Martinez for assistance in maintaining mice in the automated feeders and performing smears; Marleen deGroot for breeding the Fbxl3 mutant mice, and Lisa Thomas for breeding both Fbxl3 mutant and Cry1/Cry2 double knockout mice. We also thank Ksenija Slavic, Sofia Marques, Vanessa Zuzarte-Luis, Ângelo F. Chora and Iset Medina Vera for help during WTLD collection. Thanks also to Kimberly Cox for editing the manuscript and Fernando Augusto for the artwork and both for helping us communicate these findings clearly. The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources that have contributed to the research results reported within this paper (URL: http://www.tacc.utexas.edu) and the BioHPC cluster at UTSW.
Funding: F.R.-F. is an Associate, IK is a Lab Manager II and J.S.T is an Investigator in the Howard Hughes Medical Institute. J.H.A. is supported by NIH NIA F32-AG064886 and NIH T32-HLO9701, E.B.K. by NIH R01-HL128538, K24-HL105664, P01-AG009975 and MGH Department of Neurology, M.M.M. by PTDC/BIA-MOL/30112/2017 and PTDC/MED-IMU/28664/2017, and F.R.-F. by NIH NIGMS 1K99GM132557–01;
Footnotes
Competing interests: Authors declare no competing interests.; and
Data and materials availability: All sequencing data is deposited in GEO datasets (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145855) and mathematical modeling available at https://github.com/JohnAbel/Ferreira_malaria_model (Zenodo doi: 10.5281/zenodo.3708087).
References and Notes
- 1.Hawking F, Worms MJ, Gammage K, 24- and 48-hour cycles of malaria parasites in the blood; their purpose, production and control. Trans R Soc Trop Med Hyg 62, 731–765 (1968). [DOI] [PubMed] [Google Scholar]
- 2.Jones MD, Hill M, Hope AM, The circadian flight activity of the mosquito Anopheles Gambiae: phase setting by the light regime. The Journal of experimental biology 47, 503–511 (1967). [DOI] [PubMed] [Google Scholar]
- 3.Pittendrigh CS, Temporal organization: reflections of a Darwinian clock-watcher. Annual review of physiology 55, 16–54 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Hawking F, Worms MJ, Gammage K, Host temperature and control of 24-hour and 48-hour cycles in malaria parasites. Lancet 1, 506–509 (1968). [DOI] [PubMed] [Google Scholar]
- 5.Bozdech Z et al. , The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol 1, E5 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoo R et al. , Integrated analysis of the Plasmodium species transcriptome. EBioMedicine 7, 255–266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Roch KG et al. , Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301, 1503–1508 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Llinas M, Bozdech Z, Wong ED, Adai AT, DeRisi JL, Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res 34, 1166–1173 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taliaferro WH TL, Alteration in the time of sporulation of Plasmodium brasilianum in monkeys by reversal of light and dark. Am J Hyg. 20, 50–59 (1934). [Google Scholar]
- 10.Cambie G LICAG, Niches horaires de trois espèces de Plasmodies coexistant chez un rongeur de Centrafrique. Comptes Rendus de l’Académie des Sciences de Paris 310, 183–188 (1990). [PubMed] [Google Scholar]
- 11.David PH, Hommel M, Benichou JC, Eisen HA, da Silva LH, Isolation of malaria merozoites: release of Plasmodium chabaudi merozoites from schizonts bound to immobilized concanavalin A. Proc Natl Acad Sci U S A 75, 5081–5084 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautret P, Deharo E, Tahar R, Chabaud AG, Landau I, The adjustment of the schizogonic cycle of Plasmodium chabaudi chabaudi in the blood to the circadian rhythm of the host. Parasite 2, 69–74 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Trager W, Jensen JB, Human malaria parasites in continuous culture. Science 193, 673–675 (1976). [DOI] [PubMed] [Google Scholar]
- 14.Rijo-Ferreira F, Takahashi JS, Figueiredo LM, Circadian rhythms in parasites. PLoS Pathog 13, e1006590 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rijo-Ferreira F, Pinto-Neves D, Barbosa-Morais NL, Takahashi JS, Figueiredo LM, Trypanosoma brucei metabolism is under circadian control. Nat Microbiol 2, 17032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aton SJ, Block GD, Tei H, Yamazaki S, Herzog ED, Plasticity of circadian behavior and the suprachiasmatic nucleus following exposure to non-24-hour light cycles. J Biol Rhythms 19, 198–207 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Siepka SM et al. , Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129, 1011–1023 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godinho SI et al. , The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316, 897–900 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Prior KF et al. , Timing of host feeding drives rhythms in parasite replication. PLoS Pathog 14, e1006900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirako IC et al. , Daily Rhythms of TNFalpha Expression and Food Intake Regulate Synchrony of Plasmodium Stages with the Host Circadian Cycle. Cell Host Microbe, 13, 796–808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schibler U, Ripperger J, Brown SA, Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms 18, 250–260 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Acosta-Rodriguez VA, de Groot MHM, Rijo-Ferreira F, Green CB, Takahashi JS, Mice under Caloric Restriction Self-Impose a Temporal Restriction of Food Intake as Revealed by an Automated Feeder System. Cell Metab 26, 267–277 e262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohsaka A et al. , High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6, 414–421 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Hatori M et al. , Time-Restricted Feeding without Reducing Caloric Intake Prevents Metabolic Diseases in Mice Fed a High-Fat Diet. Cell Metab 15, 848–860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitaterna MH et al. , Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A 96, 12114–12119 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagoshi E et al. , Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA, Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14, 2289–2295 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaix A, Lin T, Le HD, Chang MW, Panda S, Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock. Cell Metab 29, 303–319 e304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Horst GT et al. , Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Fougere A et al. , Variant Exported Blood-Stage Proteins Encoded by Plasmodium Multigene Families Are Expressed in Liver Stages Where They Are Exported into the Parasitophorous Vacuole. PLoS Pathog 12, e1005917 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fonager J et al. , Reduced CD36-dependent tissue sequestration of Plasmodium-infected erythrocytes is detrimental to malaria parasite growth in vivo. J Exp Med 209, 93–107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cunningham DA et al. , ICAM-1 is a key receptor mediating cytoadherence and pathology in the Plasmodium chabaudi malaria model. Malar J 16, 185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowman AF, Tonkin CJ, Tham WH, Duraisingh MT, The Molecular Basis of Erythrocyte Invasion by Malaria Parasites. Cell Host Microbe 22, 232–245 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Ko CH et al. , Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol 8, e1000513 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koike N et al. , Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science 338, 349–354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell-Pedersen D et al. , Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6, 544–556 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rijo-Ferreira F, Takahashi JS, Genomics of circadian rhythms in health and disease. Genome Med 11, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deharo E, Gautret P, Ginsburg H, Chabaud AG, Landau I, Synchronization of Plasmodium yoelii nigeriensis and P. y. killicki infection in the mouse by means of Percoll-glucose gradient stage fractionation: determination of the duration of the schizogonic cycle. Parasitol Res 80, 159–164 (1994). [DOI] [PubMed] [Google Scholar]
- 39.Deharo E, Coquelin F, Chabaud AG, Landau I, The erythrocytic schizogony of two synchronized strains of plasmodium berghei, NK65 and ANKA, in normocytes and reticulocytes. Parasitol Res 82, 178–182 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Kondo T et al. , Circadian rhythms in rapidly dividing cyanobacteria. Science 275, 224–227 (1997). [DOI] [PubMed] [Google Scholar]
- 41.O’Neill JS et al. , Circadian rhythms persist without transcription in a eukaryote. Nature 469, 554–558 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho CS, Yoon HJ, Kim JY, Woo HA, Rhee SG, Circadian rhythm of hyperoxidized peroxiredoxin II is determined by hemoglobin autoxidation and the 20S proteasome in red blood cells. Proc Natl Acad Sci U S A 111, 12043–12048 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Q, Pando BF, Dong G, Golden SS, van Oudenaarden A, Circadian gating of the cell cycle revealed in single cyanobacterial cells. Science 327, 1522–1526 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong CI et al. , Circadian rhythms synchronize mitosis in Neurospora crassa. Proc Natl Acad Sci U S A 111, 1397–1402 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siepka SM, Takahashi JS, Methods to record circadian rhythm wheel running activity in mice. Methods Enzymol 393, 230–239 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rijo-Ferreira F et al. , Sleeping sickness is a circadian disorder. Nat Commun 9, 62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greischar MA, Reece SE, Savill NJ, Mideo N, The Challenge of Quantifying Synchrony in Malaria Parasites. Trends Parasitol 35, 341–355 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Takahashi JS et al. , ChIP-seq and RNA-seq Methods to Study Circadian Control of Transcription in Mammals. Methods Enzymol 551, 285–321 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin M, Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet 17, 10–12 (2011). [Google Scholar]
- 51.Ritchie ME et al. , limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warnes B. B. Gregory R., Lodewijk Bonebakker, Robert Gentleman, Wolfgang Huber, Andy Liaw TL, Martin Maechler, Arni Magnusson, Steffen Moeller, Venables Marc, S. a. B., in R package version 2.17.0. (2015). [Google Scholar]
- 53.Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB, MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics 32, 3351–3353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hughes ME, Hogenesch JB, Kornacker K, JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25, 372–380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang R, Su Z, Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics 26, i168–174 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thaben PF, Westermark PO, Detecting Rhythms in Time Series with RAIN. J Biol Rhythms 29, 391–400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinz S et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leise TL, Indic P, Paul MJ, Schwartz WJ, Wavelet meets actogram. J Biol Rhythms 28, 62–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gonze D, Bernard S, Waltermann C, Kramer A, Herzel H, Spontaneous synchronization of coupled circadian oscillators. Biophys J 89, 120–129 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herzog ED, Aton SJ, Numano R, Sakaki Y, Tei H, Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J Biol Rhythms 19, 35–46 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Novak B, Tyson JJ, Design principles of biochemical oscillators. Nat Rev Mol Cell Biol 9, 981–991 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Damiola F et al. , Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14, 2950–2961 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andersson JÅ, Johan; Diehl Moritz., CasADi: A symbolic package for automatic differentiation and optimal control. in Recent Advances in Algorithmic Differentiation. Springer, 297–307 (2012). [Google Scholar]
- 64.E Jones TO, P Peterson, SciPy: Open source scientific tools for Python. https://www.scipy.org, (2001).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.