Abstract
Hepatoblastoma (HB) is the most common malignant liver tumor in children. Abnormal activation of the Wnt/β-catenin signaling pathway plays an important role in the formation and development of HB. Genes in HB show a global hypomethylation change, accompanied by hypermethylation of specific tumor suppressor genes (TSGs). This article reviews the hypermethylation changes in several TSGs, such as RASSF1A, SOCS1, APC, HHIP, and P16, and analyzes the pathways and mechanisms of TSGs regulating gene expression. The role of the methylation-regulating enzymes DNA methyltransferases (DNMTs) and ten-eleven translocation (TET) family members enzymes in the methylation changes of HB was analyzed, and it was speculated that the occurrence of HB is partly due to the obstruction of liver differentiation in the early stage of differentiation. The origin cells may be incompletely differentiated hepatocytes remaining in the liver of children after birth. Therefore, further studying the role of methylation regulating enzymes in methylation changes in HB is a promising future research direction.
Keywords: Methylation, Hepatoblastoma, DNMTs, TETs, UHRF
Background
Hepatoblastoma (HB) is the most common malignant liver tumor in children, accounting for more than 80% of all pediatric malignant liver tumors and 1% of all pediatric tumors. The incidence rate is 1.5 ppm, and the annual growth rate is 2.7% [1, 2]. The histological types of HB are diverse but mainly include epithelial cell types: pure fetal type, embryonic type, mixed type, and small-cell undifferentiated type. The varieties of different histological types suggest that the tumor cells are in different stages of differentiation and that the clinical characteristics are different. HB may originate from primary hepatoblasts or from less differentiated liver stem cells or human fetal liver multipotent progenitor cells (hFLMPCs) [3]. A meta-analysis revealed similarities between HB, early mouse embryonic liver, and hepatocyte-derived stem/progenitor cells, suggesting that HB may originate from hepatic precursor cells [4]. HB cells can express CD44, CD90, CD133, and other cancer stem cell markers, suggesting that cancer stem cells also exist in HB [5].
The specific mechanism for the transformation of normal hepatocytes into HB is unknown. Cells may begin to transform in an early stage of liver differentiation, or the transformation may be caused by gene mutation after liver development completed. The Wnt/β-catenin signaling pathway plays a key role in the process of tumorigenesis. The catenin beta 1 gene (CTNNB1) encodes the β-catenin protein, and mutations in CTNNB1 occur widely in HB. Mutations in CTNNB1 are closely related to HB and are most likely the original aberrations leading to tumorigenesis. In normal hepatocytes, the β-catenin N-terminal serine and threonine residues encoded by CTNNB1 exon 3 are phosphorylated by the APC regulator of WNT signaling pathway/Axin/ glycogen synthase kinase 3 beta (APC/Axin/GSK3β) complex and then recognized by the ubiquitin-conjugating complex and ubiquitinated. The protein is subsequently degraded by the proteasome, which reduces the β-catenin protein concentration in the cytoplasm and inactivates the Wnt/β-catenin signaling pathway [6]. Mutation of the CTNNB1 gene causes the β-catenin protein to be unable to be degraded, abnormally aggregate in the cytoplasm, and ectopically transferred to the nucleus [7]. In the nucleus, β-catenin associates with the T cytokine/lymphocyte enhancement factor binding factor (TCF/LEF) family. Transcription factors bind and recruit coactivators (BCL9, CBP/300, Pygo, etc.) to activate the transcription of downstream target genes [8]. Approximately 60–80% of HBs have CTNNB1 mutations, including point mutations and deletions in exon 3 [9]. Other genes with mutations in HBs include APC (20.51%), AXIN1 (1.67%), AXIN2 (3.75%), and leucine-rich repeat-containing G protein-coupled receptor 6 (LGR6) (12.5%) [10].
In the 1980s, Feinberg and Vogelstein first linked the epigenetic inheritance of genes to cancer [11]. Epigenetics alters the spatial structure of genes by modifying genes involved in processes such as DNA methylation, histone deacetylation, and RNA methylation, regulating gene transcription and translation and affecting gene expression. The expression of proteins is different between different organs, and the protein expression within the same organ is different at different developmental stages. Epigenetics plays an important role in this process. In the past few decades, an increasing number of studies have shown that epigenetic changes are widely involved in the occurrence, invasion, and metastasis of cancer [12] and promote tumorigenesis and tumor development by inactivating tumor suppressor genes (TSGs) and activating oncogenes. HB gene mutations are simple, and whole-exon sequencing shows that there are only 2.9 mutations per HB tumor on average [13], which is much lower than 35–66 mutations per tumor in liver cancer [14–16], indicating that HBs are not only caused by the accumulation of gene mutations; in addition, most HBs occur in children, suggesting that epigenetic mechanisms that control development play an important role in the occurrence and development of HB. This article reviews the role of DNA methylation in HB occurrence, diagnosis, and treatment.
Changes in DNA methylation in HB
HB is a rare tumor, and there is sparse literature on DNA methylation. In the PubMed database, only 97 articles were returned from a search using the keywords “hepatoblastoma” and “methylation”, which is far lower than the 2513 articles returned using the keywords “hepatocellular carcinoma” and “methylation”. This shows that methylation has been far less studied in HB than in other tumors. There are a large number of differentially methylated regions (DMRs) and differentially methylated sites (DMSs) in HB and normal liver cells. Using the HM450K methylation chip to analyze HB tissue and adjacent normal liver tissue, 1359 DMSs were detected, involving 765 genes. Of these DMSs, 58% were hypomethylated, and most of the hypermethylated sites were located in the CpG islands in the promoter region of the genes. These 1359 CpG sites overlapped highly with CpG sites that were altered during liver development, suggesting that some of the HBs were caused by methylation changes in the early stages of liver development [17]. According to GO and KEGG analyses of the different genes, the three most affected physiological processes were cell adhesion, blood coagulation, and nervous system development, and the three most affected cellular functions were protein binding, nucleotide binding and ATP binding [18]. The lower overall methylation levels in HB cells than in normal hepatocytes have also been confirmed by other experiments [18]. These data show that methylation in HB is characterized by low levels of global DNA methylation and high levels of methylation of specific genes, which is consistent with the methylation characteristics of other tumors [19].
DNA methylation changes, such as hypomethylation of oncogenes and hypermethylation of TSGs, regulate gene expression, acting as an inactivator of TSGs and an activator of oncogenes [20]. The regulation of imprinted genes involved in methylation also plays a role in tumorigenesis in HB [21]. DNA methylation regulates gene expression and affects the expression of downstream genes through various pathways, playing a role in inhibiting cancer cell apoptosis, promoting cancer cell proliferation and cell cycle dysfunction, and promoting tumor metastasis (Table 1).
Table 1.
Changes in DNA methylation and the effects on HB
| Gene | Methylation status of promoter | Possible mechanisms | Effects on HB |
|---|---|---|---|
| RASSF1A | hypermethylation | Hippo pathway |
metastasis; poor responsiveness to chemotherapy; With poor prognosis; |
| SOCS1 [22–24] | hypermethylation | JAK/STAT pathway |
Tumor formation and growth; Liver regeneration |
| APC [25] | hypermethylation | Wnt/β-Catenin pathway | Tumorigenesis |
| HHIP [26, 27] | hypermethylation | Hh pathway |
Tumor growth; Angiogenesis; |
| P16 | hypermethylation | Cell cycle | Tumorigenesis or no effect |
| MT1G | hypermethylation | PI3K/Akt pathway | With poor prognosis |
| NDRG2 [28] | hypermethylation | ERK1/2 pathway; |
With poor prognosis; Metastasis |
| GPR180 | hypermethylation | Unknown | Metastasis |
| MST1R | hypermethylation | Unknown | Metastasis |
| OCIAD2 [29] | hypermethylation | AKT pathway |
metastasis; Tumor growth and invasion |
| PARP6 | hypermethylation | Unknown | metastasis |
| AFP | hypomethylation | Hippo pathway |
Diagnostic biomarker; Prognostic biomarker; Follow-up indicator; Chemotherapy responsiveness indicator |
| KLK4 | hypomethylation | Unknown | Tumorigenesis |
| MYCN | hypomethylation | Unknown | Tumor growth |
| PLAG1 | Loss of imprinting | IGF2 pathway |
With poor prognosis; Tumorigenesis |
| H19/IGF2 | Loss of imprinting | IGF2 pathway | Tumorigenesis |
| IGFBP3 | hypermethylation | IGF2 pathway | Tumorigenesis |
DNA methylation as an inactivator of TSGs
At present, DNA methylation in HB has been found mainly to regulate the expression of TSGs, and many TSGs have been studied.
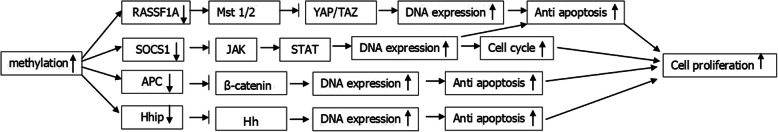
Ras association domain family member 1A (RASSF1A) is a widely studied TSG that is inhibited and inactivated in lung, breast, kidney, bladder and ovarian cancers, and DNA methylation plays an important role in gene inactivation [30]. In HB, the methylation rate of the RASSF1A gene is 33.8 to 44.3%, and RASSF1A gene methylation is directly related to distant metastasis and poor prognosis [31, 32]. The incidence of RASSF1A gene methylation is high, and the overall survival rate and event-free survival rate associated with high RASSF1A gene methylation are lower than those associated with low RASSF1A gene methylation [33]. Compared with patients with low methylation levels of the RASSF1A gene, patients with high methylation levels of the RASSF1A gene have a high incidence of distant metastasis and a low overall survival rate and event-free survival rate [31–33]. RASSF1A promoter methylation also predicts low responsiveness to chemotherapy [33]. Methylation of the promoter region of the RASSF1A gene silences the gene and inhibits the macrophage stimulating 1/2 (Mst1/2) protein, thereby activating the core protein Yes-associated protein/Tafazzin (YAP/TAZ) of the Hippo pathway; this inhibits the apoptosis of cancer cells and promotes the proliferation of cancer cells [34, 35], and the Hippo pathway plays an important role in the formation of HB [36] (Fig. 1).
Fig. 1.
Increased DNA methylation results in decreased expression of multiple genes, increased cell proliferation through different pathways, and caused tumor formation or metastasis
The suppressor of cytokine signaling 1 (SOCS1) gene has a dual opposite, it can function as either a tumor suppressor or a tumor promoter. SOCS1 gene was overexpressed in human breast cancer, epidermis and neuronal tumors and showed the characteristics of an oncogene [37–39]. Studies have shown that in HB, hypermethylation in the CpG region of the SOCS1 gene promoter leads to decreased SOCS1 gene expression in nearly half of the cases and decreased levels of the suppressor of cytokine signaling 1/ Jun activating binding protein 1/ STAT-induced STAT inhibitor-1 (SOCS1/JAB1/SSI-1) protein complex, activating the the Janus kinase-signal transducers and activators of transcription (JAK/STAT) signaling pathway and inhibiting apoptosis of cancer cells [40]. Other results also showed that the methylation level in the promoter region of the SOCS1 gene in HB increased [33, 41], indicating that abnormal DNA methylation increases SOCS1 silencing and plays an important role in the development of HB (Fig. 1).
In the canonical Wnt/β-catenin pathway, the degradation of β-catenin protein depends on the function of APC, Axin1/2, casein kinase 1 (CK1), and GSK3 protein complexes [42]. A decrease in APC promotes HB development by reducing the degradation of β-catenin through two mechanisms. One mechanism is mutation or deletion [43], as when the APC gene is mutated, APC cannot play its functional role, and the degradation of β-catenin protein is reduced. The second mechanism involves DNA hypermethylation, which inhibits the expression of APC and causes the APC gene to lose its function [41]. Familial adenomatous polyposis (FAP) is mainly caused by mutations in the APC gene, and the incidence of HB in patients with FAP is greatly increased [44] (Fig. 1).
The Hedgehog (Hh) pathway plays an important role in regulating the development of endoderm-derived tissues during embryogenesis. When embryonic development is complete, Hedgehog-interacting protein (HHIP) inhibits the Hh pathway, leading to the disappearance of or a reduction in Hh signaling pathway activity [45]. The hypermethylation of the HHIP gene promoter region results in nonexpression of HHIP protein; as such, the inhibition of the Hh pathway disappears, and the abnormal activation of the Hh pathway leads to the occurrence of various tumors [46, 47]. In HB, the HHIP gene promoter region is hypermethylated in 26% of patients [48]. In in vitro experiments, treatment of HB cell lines with the DNA demethylating drug 5-azacytidine restored HHIP protein expression, increased tumor cell apoptosis, and reduced tumor activity, indicating that abnormal activation of the Hh pathway caused by hypermethylation of the HHIP gene plays an important role in the occurrence and development of HB [48] (Fig. 1).
The P16 gene (CDKN2/MTS-1/INK4A) is a TSG that regulates the cell cycle, and its transcription product, P16 protein, is an inhibitor of cyclin-dependent kinases 4 (CDK4) and 6 (CDK6) [49]. It has been proven that in many tumors, methylation of the promoter region of the P16 gene leads to nonexpression of the P16 gene, which is involved in tumor formation [50]. Whether the P16 gene methylation is involved in HB formation is still controversial. Studies have reported abnormal methylation in the P16 gene promoter region in 12 of 24 HB patients, with significantly reduced P16 protein expression, suggesting that the P16 protein is involved in the tumorigenesis of HB [51]. Some scholars also performed methylation-specific PCR (MSP) and found that there was no hypermethylation of the P16 gene promoter in 27 HB tissues, and changes in the CDKN2A gene family were not related to the occurrence of HB [52, 53]. The role of the P16 gene in HB needs further experimental confirmation.
There are other TSGs involved in HB formation due to DNA methylation. Methylation levels in the metallothionein-1G (MT1G) gene promoter region were specifically increased, and hypermethylation in the MT1G promoter region was directly associated with poor prognosis, suggesting that MT1G could be used as a diagnostic marker and a potential drug therapeutic target [41]. The N-Myc downstream regulated gene 2 (NDRG2) gene plays the role of the TSG in many tumors [54], and its promoter region in HB presents hypermethylation, which is directly related to the prognosis of tumors [41]. Hypermethylation of the genes G protein-coupled receptor 180 (GPR180), macrophage stimulating 1 receptor (MST1R), OCIA domain containing 2 (OCIAD2), and poly (ADP-ribose) polymerase family member 6 (PARP6) was also involved in the tumorigenesis of HB, and methylation status was significantly correlated with metastasis [55].
DNA methylation is an activator of oncogenes
DNA methylation in HB is characterized by decreased global methylation levels, weakened inhibition of oncogenes, increased expression of oncogenes, and promoted tumor proliferation. However, there are few studies of oncogene methylation in HB. There are many types of proteins with increased expression in HB, such as glypican 3 (GPC3) [56], transforming growth factor-β2 (TGF-β2) [57], CBP/P-300 interacting transactivator 1 (CITED1) [58], nuclear receptor subfamily 5 group A member 2 (LRH-1, NR5A2) [59], PCNA clamp associated factor (NS5ATP9, PCLAF) [60], and serpin family B member 3 (SERPINB3) [61], but most of the literature has focused on the effect of increased protein expression on tumors. The reason for the increase in protein expression is less studied. Nevertheless, it is certain that the increase in some proteins is related to the reduced methylation of related genes (Table 1).
Alpha-fetoprotein (AFP) is the most commonly used marker for the diagnosis and prognosis of liver cancer and HB. Its level is directly related to clinical prognosis and is an important basis for clinical staging. AFP also plays an important role in liver cell growth, differentiation, angiogenesis, apoptosis, and immune regulation [62]. Some experimental results have pointed out that the methylation level of the AFP gene promoter region in HB cells is significantly lower than that in normal liver cells, and the mRNA expression is increased [18], which indicates that gene methylation plays an important role in the upregulation of AFP expression (Table 1).
The kallikrein-related peptidase 4 (KLK4) gene is considered an oncogene and plays an important role in the formation of many tumors, such as gastric cancer and prostate cancer [63]. Studies have shown that in HB, the methylation level of the gene promoter region is reduced, and the expression of KLK4 mRNA and protein is increased [64]. Gene methylation plays an active role in oncogene expression. The MYCN gene is also an important oncogene and plays an important role in the formation of pediatric neuroblastoma and medulloblastoma [65]. In pediatric HB, MYCN is overexpressed, and its promoter region is hypomethylated. The use of the MYCN inhibitors alisertib (MLN8237) and JQ1 can inhibit tumor growth, indicating that the MYCN gene is also involved in the tumorigenic process in HB [66] (Table 1).
Regulation of DNA methylation of imprinted genes
Imprinted genes play an important role in tissue development and differentiation. DNA methylation is an important way to maintain gene imprinting. Loss of imprinting (LOI) caused by changes in methylation is involved in the formation of various tumors.
The human chromosomes include more than 30 imprinted regions. In HB, one study used 12 cases of HB and adjacent normal liver tissues and found that all tumors were abnormally methylated in at least one DMR [21]. Of the 33 DMRs studied, 18 showed abnormal methylation, while 15 did not show abnormal methylation. There is hypomethylation in liver tissues adjacent to the tumor; both hypermethylation and hypomethylation are present in the tumor tissue, and hypermethylation is more frequent. It is speculated that prior to tumorigenesis, some DMRs in adjacent normal liver tissues are hypomethylated. This hypomethylation may be a common phenomenon in embryonic tumors. Hypermethylation only occurs in tumor tissues and may be obtained during tumorigenesis [21]. The most common sites for gene and epigenetic changes are 11p15.5 and 20q13.3. H19 and IGF2, which are closely related to tumors, are located in the 11p15.5 region [21, 67].
Pleomorphic adenoma gene 1 (PLAG1) is a gene located on 8q12.1, and its expression product is a transcription enhancer of the P3 enhancer of the insulin-like growth factor 2 (IGF2) gene. Studies showed that the PLAG1 gene was highly expressed in 19/20 cases of HB, which is 3–12 times higher than the normal liver expression level of children of the same age and similar to that in the embryonic stage, and his high expression is directly related to the poor prognosis of HB [68]. This result shows that the PLAG1 protein promotes tumorigenesis by enhancing the transcription of the H19/IGF2 genes.
H19/IGF2 are currently the most widely studied imprinted genes and play a key role in the development of many tumors. In normal cells, the H19 gene is maternally expressed and paternally imprinted, and the IGF2 gene is paternally expressed and maternally imprinted. The expression of H19 and IGF2 genes is affected by the differentially methylated region (DMR) upstream of H19 and the downstream enhancer of H19 [69]. If the methylation of the DMR region is abnormal, then H19 or IGF2 suffers the LOI, which will lead to the occurrence of many tumors, including HB [70]. The methylation status of the H19 differentially methylated region (DMR), the loss of heterozygosity (LOH), and the expression of the IGF2 allele in 54 HB tumors was analyzed, and the results showed that 12 cases (22%) had LOH, 9 cases (17%) had loss of imprinting (LOI) [71], and IGF2 was overexpressed in HB. These findings indicate that disruption of the H19/IGF2 gene imprinting plays an important role in the formation of some HBs.
Insulin-like growth factor-binding protein 3 (IGFBP3) is a multifunctional protein synthesized by the liver. IGFBP3 functions through the H19/IGF axis. It mainly mediates growth inhibition and induces apoptosis by binding to IGF [72]. Studies have shown that the IGFBP3 gene is highly expressed in normal liver tissue and is low in HB, mainly due to hypermethylation of the promoter region [73]. Treatment of HB cell lines with 5-aza-2-deoxycytidine demethylated the promoter region, resulting in reduced cell colony formation and a reduced ability to invade and metastasize. It was also found that hypermethylation of the IGFBP3 promoter region was related to macrovascular invasion and distant metastasis [73].
Abnormalities of DNA methylation regulatory enzymes in HB
DNA methylation regulatory enzymes mainly include DNA methyltransferases (DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L) and demethylation enzymes (TET1, TET2, and TET3). DNA methyltransferases (DNMTs) increase DNA methylation and ten-eleven translocation (TET) family members enzymes play a demethylating role, reducing DNA methylation levels.
In HB, studies have found that DNMT1, DNMT3A, UHRF1, TET1, and TET2 are all upregulated and 5hmC is increased [74]. The DNMT1/UHRF1 complex is key for methylation maintenance. Increased expression of DNMT1, DNMT3A and UHRF1 proteins can cause hypermethylation of the gene promoter region, which may play a key role in the hypermethylation of the promoter region of HB TSGs [74]. Other experiments have also confirmed that UHRF1 expression in HB is increased [75], TSG promoter region methylation is increased, and gene expression is decreased. After knockout of UHRF1, TSG was re-expressed, indicating that UHRF1 is a key protein promoting hypermethylation and inhibiting the expression of TSGs, which can lead to hypermethylation of TSGs and inhibit expression. There are also data showing that the overexpression of UHRF1 damages the correlation between UHRF1 and DNMT1 [76], making DNMT1 unstable and ectopic [77], and inhibiting the activity of DNMT3A [78], which may also be an important reason for the global hypomethylation of tumors.
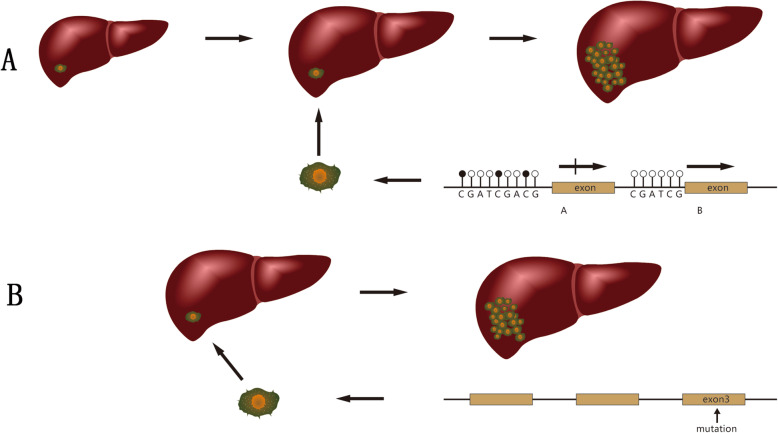
In hematological tumors and most adult solid tumors, including liver cancer, TET gene mutations leading to loss of gene function and reduced 5hmC content are important causes of tumorigenesis [79–81] and indicate a poor prognosis [82]. TET acts as a TSG. However, in HB, both TET and 5hmC increase, which indicates that the tumorigenic mechanisms of TET and 5hmC in HB and adult tumors are different. Data indicate that during embryonic development, TET enzymes are highly expressed and inhibit differentiation [83, 84]. Therefore, we can speculate that the occurrence of HB is due to the obstruction of liver differentiation in the early stage of differentiation (Fig. 2). The origin of HB is the incompletely differentiated hepatocytes remaining in the liver of children. This is consistent with the conclusions obtained from a whole-genome methylation scan of HB [17], and it can also explain why HB includes many different histological types of cells.
Fig. 2.
Two mechanisms of hepatoblastoma formation. a: Hepatic differentiation is blocked during the embryonic stage, and DNA methylation stays in the early stages of liver differentiation. Incompletely differentiated cells remain in the fetal liver after birth, and abnormal proliferation of the cells causes hepatoblastoma. b: The mutation of exon 3 of CTNNB1 gene in fetal liver cells causes β-Catenin accumulation, and abnormal cell proliferation leads to HB
Diagnosis and treatment of DNA methylation aberrations
DNA methylation changes, as a general change in tumor cells, can cause tumor cells to show specific changes that make them different from ordinary cells. In the initial diagnosis of HB, imaging screening and biopsy-based pathological diagnoses are mainly used. These methods are more reliable and specific than other methods. DNA methylation can be used as an index for screening for tumor recurrence and metastasis during postoperative review and as an index for judging prognosis. According to the literature reports, the RASSF1A, UHRF1, MT1G, and NDRG2 promoter regions are hypermethylated. The degree of methylation is related to postoperative recurrence and distant metastasis and can be used as a marker for diagnosis and prognosis [31, 41, 54, 73, 85].
For the treatment of DNA methylation aberrations, because DNA methylation is achieved by the function of DNMTs, DNMTs are a therapeutic target for DNA demethylation and restoration of TSG functions. The most studied demethylating drugs at present are mainly DNMT inhibitors [86]. The mechanism of antitumor action of DNMT inhibitors is mainly to inhibit the activity of DNMTs so that the duplicated DNA is not methylated, and all alleles would be demethylated so that the expression of TSGs would be restored and they could play their role in inhibiting tumor growth, achieving antitumor therapy [87]. Currently, the most widely used DNMT inhibitors are decitabine (5-aza-2′-deoxycytidine) and azacytidine [88], but there is no clinical information on the treatment of HB with either drug.
Conclusion and prospective directions
HB is a tumor that proliferates because hepatocyte differentiation is blocked in early development. Mutations of the CTNNB1 gene can be detected in most tumors, and DNA methylation plays an important role in the occurrence and development of tumors. Hypermethylation reduces the expression of specific TSGs, while global hypomethylation increases the expression of other genes, promoting tumor development through various pathways. The dysfunction of a variety of DNA methylation regulatory enzymes plays a key role in the hypermethylation of TSGs and the hypomethylation of oncogenes, especially the UHRF1 protein.
According to the review in this article, DNA methylation can affect the formation and metastasis of hepatoblastoma through different cell signaling pathways, and the metastasis of hepatoblastoma is an important factor affecting the prognosis. Therefore, further research on the mechanism of DNA methylation in hepatoblastoma formation and metastasis has important basic and clinical significance.
At present, various tumor cells show global hypomethylation and hypermethylation of specific TSGs. It is speculated that this wide range of methylation changes is accomplished by DNA methylation regulatory enzymes. Therefore, further research on the regulatory mechanism of DNA methylation regulatory enzymes and their role in tumor development should be a future direction. Second, the mechanism by which DNA methylation regulatory enzymes enhance the methylation of TSGs and reduce the methylation of other genes and whether other proteins play a role in this process needs to be further studied. Third, the main factors influencing the long-term prognosis of HB are vascular invasion and distant metastasis [89]. The methylation level of multiple TSGs is closely related to vascular invasion and distant metastasis, which can be studied with large sample sizes.
Acknowledgements
Not applicable.
Abbreviations
- APC
APC regulator of WNT signaling pathway
- CITED1
CBP/P-300 interacting transactivator 1
- CK1
Casein kinase 1
- CTNNB1
Catenin beta 1 gene
- DMR
Differentially methylated region
- DMS
Differentially methylated sites
- DNMT
DNA methyltransferase
- GPC3
Glypican 3
- GPR180
G protein-coupled receptor 180
- GSK3β
Glycogen synthase kinase 3 beta
- HB
Hepatoblastoma
- Hh
Hedgehog
- HHIP
Hedgehog-interacting protein
- IGF
Insulin-like growth factor
- IGFBP3
Insulin-like growth factor-binding protein 3
- JAB1
Jun activating binding protein 1
- JAK/STAT
The Janus kinase-signal transducers and activators of transcription
- LGR6
Leucine-rich repeat-containing G protein-coupled receptor 6
- LOI
Loss of imprinting
- Mst
Macrophage stimulating
- MST1R
Macrophage stimulating 1 receptor
- MT1G
Metallothionein-1G
- NDRG2
N-Myc downstream regulated gene 2
- NR5A2
Nuclear receptor subfamily 5 group A member 2
- OCIAD2
OCIA domain containing 2
- PARP6
Poly (ADP-ribose) polymerase family member 6
- PLAG1
Pleomorphic adenoma gene 1
- RASSF1A
Ras association domain family member 1A
- SOCS1
Suppressor of cytokine signaling 1
- SSI-1
STAT-induced STAT inhibitor-1
- TET
Ten-eleven translocation
- TGF-β2
Transforming growth factor-β2
- TSG
Tumor suppressor gene
- YAP/TAZ
Yes-associated protein/Tafazzin
Authors’ contributions
Conception and literature search: Hongyu Shen, Qingtao Yan, Jing Zhang; Manuscript writing and final approval: Gang Shen and Huixian Liu. The author(s) read and approved the final manuscript.
Funding
Funding source: Weifang Science and Technology Development Project, Weifang Science and Technology Bureau, 2016YX001 and 2019YX007.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Weifang People’s Hospital before it started.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lim I, Bondoc AJ, Geller JI, Tiao GM. Hepatoblastoma-The Evolution of Biology, Surgery, and Transplantation. Children (Basel) 2018;6(1):1. doi: 10.3390/children6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kehm RD, Osypuk TL, Poynter JN, Vock DM, Spector LG. Do pregnancy characteristics contribute to rising childhood cancer incidence rates in the United States? Pediatr Blood Cancer. 2018;65(3):10.1002/pbc.26888. . [DOI] [PMC free article] [PubMed]
- 3.Bell D, Ranganathan S, Tao J, Monga SP. Novel advances in understanding of molecular pathogenesis of Hepatoblastoma: a Wnt/β-catenin Perspect ive. Gene Expr. 2017;17(2):141–54. [DOI] [PMC free article] [PubMed]
- 4.Bhusari S, Pandiri AR, Nagai H, Wang Y, Foley J, Hong HH, Ton TV, DeVito M, Shockley KR, Peddada SD, et al. Genomic profiling reveals unique molecular alterations in Hepatoblastomas and adjacent hepatocellular carcinomas in B6C3F1 mice. Toxicol Pathol. 2015;43(8):1114–1126. doi: 10.1177/0192623315599853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahnassy AA, Fawzy M, El-Wakil M, Zekri AR, Abdel-Sayed A, Sheta M. Aberrant expression of cancer stem cell markers (CD44, CD90, and CD133) contributes to disease progre ssion and reduced survival in hepatoblastoma patients: 4-year survival data. Transl Res. 2015;165(3):396–406. doi: 10.1016/j.trsl.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Han ZG. Mutational landscape of hepatoblastoma goes beyond the Wnt-β-catenin pathway. Hepatology. 2014;60(5):1476–8. [DOI] [PubMed]
- 7.Sharma M, Jamieson C, Johnson M, Molloy MP, Henderson BR. Specific armadillo repeat sequences facilitate beta-catenin nuclear transport in live cells via direct binding to nucleoporins Nup62, Nup153, and RanBP2/Nup358. J Biol Chem. 2012;287(2):819–831. doi: 10.1074/jbc.M111.299099. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Wang W, Smits R, Hao H, He C. Wnt/β-Catenin signaling in liver cancers. Cancers (Basel). 2019;11(7):926. [DOI] [PMC free article] [PubMed]
- 9.Dubbink HJ, Hollink I, Avenca VC, Wang W, Liu P, Doukas M, van Noesel MM, Dinjens W, Wagner A, Smits R. A novel tissue-based ss-catenin gene and immunohistochemical analysis to exclude familial adenomatous polyposis among children with hepatoblastoma tumors. Pediatr Blood Cancer. 2018;65(6):e26991. doi: 10.1002/pbc.26991. [DOI] [PubMed] [Google Scholar]
- 10.Perugorria MJ, Olaizola P, Labiano I, Esparza-Baquer A, Marzioni M, Marin J, Bujanda L, Banales JM. Wnt-beta-catenin signalling in liver development, health and disease. Nat Rev Gastroenterol Hepatol. 2019;16(2):121–136. doi: 10.1038/s41575-018-0075-9. [DOI] [PubMed] [Google Scholar]
- 11.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 12.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in Cancer and aging. Cancer Res. 2016;76(12):3446–3450. doi: 10.1158/0008-5472.CAN-15-3278. [DOI] [PubMed] [Google Scholar]
- 13.Eichenmüller M, Trippel F, Kreuder M, Beck A, Schwarzmayr T, Häberle B, Cairo S, Leuschner I, von Schweinitz D, Strom TM, et al. The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J Hepatol. 2014;61(6):1312–1320. doi: 10.1016/j.jhep.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, Deng Q, Wang Q, Li KY, Dai JH, Li N, Zhu ZD, Zhou B, Liu XY, Liu RF, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44(10):1117–1121. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 15.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathw ays in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleary SP, Jeck WR, Zhao X, Chen K, Selitsky SR, Savich GL, Tan TX, Wu MC, Getz G, Lawrence MS, et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology. 2013;58(5):1693–1702. doi: 10.1002/hep.26540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maschietto M, Rodrigues TC, Kashiwabara AY, de Araujo E, Marques AT, da Costa CML, da Cunha IW, RVL D, Cypriano M, Brentani H, et al. DNA methylation landscape of hepatoblastomas reveals arrest at early stages of liver differentiation and cancer-related alterations. Oncotarget. 2017;8(58):97871–97889. doi: 10.18632/oncotarget.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui X, Liu B, Zheng S, Dong K, Dong R. Genome-wide analysis of DNA methylation in hepatoblastoma tissues. Oncol Lett. 2016;12(2):1529–1534. doi: 10.3892/ol.2016.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soozangar N, Sadeghi MR, Jeddi F, Somi MH, Shirmohamadi M, Samadi N. Comparison of genome-wide analysis techniques to DNA methylation analysis in human cancer. J Cell Physiol. 2018;233(5):3968–3981. doi: 10.1002/jcp.26176. [DOI] [PubMed] [Google Scholar]
- 20.Paska AV, Hudler P. Aberrant methylation patterns in cancer: a clinical view. Biochem Med (Zagreb) 2015;25(2):161–176. doi: 10.11613/BM.2015.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rumbajan JM, Maeda T, Souzaki R, Mitsui K, Higashimoto K, Nakabayashi K, Yatsuki H, Nishioka K, Harada R, Aoki S, et al. Comprehensive analyses of imprinted differentially methylated regions reveal epigenetic and genetic c haracteristics in hepatoblastoma. BMC Cancer. 2013;13:608. doi: 10.1186/1471-2407-13-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagnyukova TV, Tryndyak VP, Muskhelishvili L, Ross SA, Beland FA, Pogribny IP. Epigenetic downregulation of the suppressor of cytokine signaling 1 (Socs1) gene is associated with the STAT3 activation and development of hepatocellular carcinoma induced by methyl-deficiency in rats. Cell Cycle. 2008;7(20):3202–3210. doi: 10.4161/cc.7.20.6816. [DOI] [PubMed] [Google Scholar]
- 23.Seki E, Kondo Y, Iimuro Y, Naka T, Son G, Kishimoto T, Fujimoto J, Tsutsui H, Nakanishi K. Demonstration of cooperative contribution of MET- and EGFR-mediated STAT3 phosphorylation to liver regeneration by exogenous suppressor of cytokine signalings. J Hepatol. 2008;48(2):237–245. doi: 10.1016/j.jhep.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 24.Zhao RC, Zhou J, He JY, Wei YG, Qin Y, Li B. Aberrant promoter methylation of SOCS-1 gene may contribute to the pathogenesis of hepatocellular carcinoma: a meta-analysis. J BUON. 2016;21(1):142–151. [PubMed] [Google Scholar]
- 25.Zhang L, Jin Y, Zheng K, Wang H, Yang S, Lv C, Han W, Yu Y, Yang Y, Geng D, et al. Whole-genome sequencing identifies a novel variation of WAS gene coordinating with heterozygous germline mutation of APC to enhance hepatoblastoma oncogenesis. Front Genet. 2018;9:668. doi: 10.3389/fgene.2018.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eichenmuller M, Gruner I, Hagl B, Haberle B, Muller-Hocker J, von Schweinitz D, Kappler R. Blocking the hedgehog pathway inhibits hepatoblastoma growth. Hepatology. 2009;49(2):482–490. doi: 10.1002/hep.22649. [DOI] [PubMed] [Google Scholar]
- 27.Olsen CL, Hsu PP, Glienke J, Rubanyi GM, Brooks AR, et al. BMC Cancer. 2004;4:43. doi: 10.1186/1471-2407-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Y, Ma J, Wu L, Wang Q, Li X, Li X, Zhang Y, Zhang J, Yao L, Zhang J, et al. Hyperthermia-induced NDRG2 upregulation inhibits the invasion of human hepatocellular carcinoma via suppressing ERK1/2 signaling pathway. PLoS One. 2013;8(4):e61079. doi: 10.1371/journal.pone.0061079. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Wu D, Yang X, Peng H, Guo D, Zhao W, Zhao C, Zhou X. OCIAD2 suppressed tumor growth and invasion via AKT pathway in Hepatocelluar carcinoma. Carcinogenesis. 2017;38(9):910–919. doi: 10.1093/carcin/bgx073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malpeli G, Innamorati G, Decimo I, Bencivenga M, Nwabo Kamdje AH, Perris R, Bassi C. Methylation dynamics of RASSF1A and its impact on cancer. Cancers. 2019;11(7):959. doi: 10.3390/cancers11070959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugawara W, Haruta M, Sasaki F, Watanabe N, Tsunematsu Y, Kikuta A, Kaneko Y. Promoter hypermethylation of the RASSF1A gene predicts the poor outcome of patients with hepatoblasto ma. Pediatr Blood Cancer. 2007;49(3):240–249. doi: 10.1002/pbc.21031. [DOI] [PubMed] [Google Scholar]
- 32.Honda S, Miyagi H, Suzuki H, Minato M, Haruta M, Kaneko Y, Hatanaka KC, Hiyama E, Kamijo T, Okada T, et al. RASSF1A methylation indicates a poor prognosis in hepatoblastoma patients. Pediatr Surg Int. 2013;29(11):1147–1152. doi: 10.1007/s00383-013-3371-z. [DOI] [PubMed] [Google Scholar]
- 33.Honda S, Haruta M, Sugawara W, Sasaki F, Ohira M, Matsunaga T, Yamaoka H, Horie H, Ohnuma N, Nakagawara A, et al. The methylation status of RASSF1A promoter predicts responsiveness to chemotherapy and eventual cure in hepatoblastoma patients. Int J Cancer. 2008;123(5):1117–1125. doi: 10.1002/ijc.23613. [DOI] [PubMed] [Google Scholar]
- 34.Schroeder MC, Halder G. Regulation of the hippo pathway by cell architecture and mechanical signals. Semin Cell Dev Biol. 2012;23(7):803–811. doi: 10.1016/j.semcdb.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev. 2014;94(4):1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 36.Tao J, Calvisi DF, Ranganathan S, Cigliano A, Zhou L, Singh S, Jiang L, Fan B, Terracciano L, Armeanu-Ebinger S, et al. Activation of β-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;147(3):690–701. [DOI] [PMC free article] [PubMed]
- 37.Zhang J, Li H, Yu JP, Wang SE, Ren XB. Role of SOCS1 in tumor progression and therapeutic application. Int J Cancer. 2012;130(9):1971–1980. doi: 10.1002/ijc.27318. [DOI] [PubMed] [Google Scholar]
- 38.Raccurt M, Tam SP, Lau P, Mertani HC, Lambert A, Garcia-Caballero T, Li H, Brown RJ, McGuckin MA, Morel G, et al. Suppressor of cytokine signalling gene expression is elevated in breast carcinoma. Br J Cancer. 2003;89(3):524–532. doi: 10.1038/sj.bjc.6601115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laner-Plamberger S, Wolff F, Kaser-Eichberger A, Swierczynski S, Hauser-Kronberger C, Frischauf AM, Eichberger T. Hedgehog/GLI signaling activates suppressor of cytokine signaling 1 (SOCS1) in epidermal and neural tumor cells. PLoS One. 2013;8(9):e75317. doi: 10.1371/journal.pone.0075317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagai H, Naka T, Terada Y, Komazaki T, Yabe A, Jin E, Kawanami O, Kishimoto T, Konishi N, Nakamura M, et al. Hypermethylation associated with inactivation of the SOCS-1 gene, a JAK/STAT inhibitor, in human hepa toblastomas. J Hum Genet. 2003;48(2):65–69. doi: 10.1007/s100380300008. [DOI] [PubMed] [Google Scholar]
- 41.Sakamoto LH, DE Camargo B, Cajaiba M, Soares FA, Vettore AL. MT1G hypermethylation: a potential prognostic marker for hepatoblastoma. Pediatr Res. 2010;67(4):387–393. doi: 10.1203/PDR.0b013e3181d01863. [DOI] [PubMed] [Google Scholar]
- 42.Russell JO, Monga SP. Wnt/β-catenin signaling in liver development, homeostasis, and pathobiology. Annu Rev Pathol. 2018;13:351–78. [DOI] [PMC free article] [PubMed]
- 43.Mazzoni SM, Fearon ER. AXIN1 and AXIN2 variants in gastrointestinal cancers. Cancer Lett. 2014;355(1):1–8. doi: 10.1016/j.canlet.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trobaugh-Lotrario AD, Lopez-Terrada D, Li P, Feusner JH. Hepatoblastoma in patients with molecularly proven familial adenomatous polyposis: clinical characteristics and rationale for surveillance screening. Pediatr Blood Cancer. 2018;65(8):e27103. doi: 10.1002/pbc.27103. [DOI] [PubMed] [Google Scholar]
- 45.Beachy PA, Hymowitz SG, Lazarus RA, Leahy DJ, Siebold C. Interactions between hedgehog proteins and their binding partners come into view. Genes Dev. 2010;24(18):2001–2012. doi: 10.1101/gad.1951710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tada M, Kanai F, Tanaka Y, Tateishi K, Ohta M, Asaoka Y, Seto M, Muroyama R, Fukai K, Imazeki F, et al. Down-regulation of hedgehog-interacting protein through genetic and epigenetic alterations in human h epatocellular carcinoma. Clin Cancer Res. 2008;14(12):3768–3776. doi: 10.1158/1078-0432.CCR-07-1181. [DOI] [PubMed] [Google Scholar]
- 47.Taniguchi H, Yamamoto H, Akutsu N, Nosho K, Adachi Y, Imai K, Shinomura Y. Transcriptional silencing of hedgehog-interacting protein by CpG hypermethylation and chromatic struc ture in human gastrointestinal cancer. J Pathol. 2007;213(2):131–139. doi: 10.1002/path.2216. [DOI] [PubMed] [Google Scholar]
- 48.Eichenmüller M, Gruner I, Hagl B, Häberle B, Müller-Höcker J, von Schweinitz D, Kappler R. Blocking the hedgehog pathway inhibits hepatoblastoma growth. Hepatology. 2009;49(2):482–490. doi: 10.1002/hep.22649. [DOI] [PubMed] [Google Scholar]
- 49.Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer. 2012;130(8):1715–1725. doi: 10.1002/ijc.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clinica Chimica Acta. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Shim Y-H, Park H-J, Choi MS, Kim JS, Kim H, Kim JJ, Jang J-J, Yu E. Hypermethylation of the p16 gene and lack of p16 expression in hepatoblastoma. Mod Pathol. 2003;16(5):430–436. doi: 10.1097/01.MP.0000066799.99032.A7. [DOI] [PubMed] [Google Scholar]
- 52.Iolascon A, Giordani L, Moretti A, Basso G, Borriello A, Della Ragione F. Analysis of CDKN2A, CDKN2B, CDKN2C, and cyclin ds gene status in hepatoblastoma. Hepatology. 1998;27(4):989–995. doi: 10.1002/hep.510270414. [DOI] [PubMed] [Google Scholar]
- 53.Harada K, Toyooka S, Maitra A, Maruyama R, Toyooka KO, Timmons CF, Tomlinson GE, Mastrangelo D, Hay RJ, Minna JD, et al. Aberrant promoter methylation and silencing of the RASSF1A gene in pediatric tumors and cell lines. Oncogene. 2002;21(27):4345–4349. doi: 10.1038/sj.onc.1205446. [DOI] [PubMed] [Google Scholar]
- 54.Hu W, Fan C, Jiang P, Ma Z, Yan X, Di S, Jiang S, Li T, Cheng Y, Yang Y. Emerging role of N-myc downstream-regulated gene 2 (NDRG2) in cancer. Oncotarget. 2016;7(1):209–223. doi: 10.18632/oncotarget.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Honda S, Minato M, Suzuki H, Fujiyoshi M, Miyagi H, Haruta M, Kaneko Y, Hatanaka KC, Hiyama E, Kamijo T, et al. Clinical prognostic value of DNA methylation in hepatoblastoma: four novel tumor suppressor candidates. Cancer Sci. 2016;107(6):812–819. doi: 10.1111/cas.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ortiz MV, Roberts SS, Glade Bender J, Shukla N, Wexler LH. Immunotherapeutic targeting of GPC3 in pediatric solid embryonal tumors. Front Oncol. 2019;9:108. doi: 10.3389/fonc.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dropmann A, Dediulia T, Breitkopf-Heinlein K, Korhonen H, Janicot M, Weber SN, Thomas M, Piiper A, Bertran E, Fabregat I, et al. TGF-beta1 and TGF-beta2 abundance in liver diseases of mice and men. Oncotarget. 2016;7(15):19499–19518. doi: 10.18632/oncotarget.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy AJ, de Caestecker C, Pierce J, Boyle SC, Ayers GD, Zhao Z, Libes JM, Correa H, Walter T, Huppert SS, et al. CITED1 expression in liver development and hepatoblastoma. Neoplasia. 2012;14(12):1153–1163. doi: 10.1593/neo.12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin J, Jin J, Woodfield SE, Patel RH, Jin NG, Shi Y, Liu B, Sun W, Chen X, Yu Y, et al. Targeting LRH-1 in hepatoblastoma cell lines causes decreased proliferation. Oncol Rep. 2019;41(1):143–53. [DOI] [PMC free article] [PubMed]
- 60.Quan M, Liu S, Wang Q, Li G, Zhang Y, Feng S, Liang J, Cheng J. NS5ATP9 Promotes Beclin 1-dependent starvation-induced autophagy of hepatoblastoma cells. J Cell Biochem. 2015;116(8):1574–1582. doi: 10.1002/jcb.25111. [DOI] [PubMed] [Google Scholar]
- 61.Turato C, Buendia MA, Fabre M, Redon MJ, Branchereau S, Quarta S, Ruvoletto M, Perilongo G, Grotzer MA, Gatta A, et al. Over-expression of SERPINB3 in hepatoblastoma: a possible insight into the genesis of this tumour? Eur J Cancer. 2012;48(8):1219–1226. doi: 10.1016/j.ejca.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 62.Meng W, Bai B, Bai Z, Li Y, Yue P, Li X, Qiao L. The immunosuppression role of alpha-fetoprotein in human hepatocellular carcinoma. Discov Med. 2016;21(118):489–494. [PubMed] [Google Scholar]
- 63.Kontos CK, Chantzis D, Papadopoulos IN, Scorilas A. Kallikrein-related peptidase 4 (KLK4) mRNA predicts short-term relapse in colorectal adenocarcinoma patients. Cancer Lett. 2013;330(1):106–112. doi: 10.1016/j.canlet.2012.11.036. [DOI] [PubMed] [Google Scholar]
- 64.Liu B, Cui X, Zheng S, Dong K, Dong R. Aberrant KLK4 gene promoter hypomethylation in pediatric hepatoblastomas. Oncol Lett. 2017;13(3):1360–1364. doi: 10.3892/ol.2017.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roussel MF, Stripay JL. Epigenetic drivers in pediatric medulloblastoma. Cerebellum. 2018;17(1):28–36. doi: 10.1007/s12311-017-0899-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eberherr C, Beck A, Vokuhl C, Becker K, Häberle B, Von Schweinitz D, Kappler R. Targeting excessive MYCN expression using MLN8237 and JQ1 impairs the growth of hepatoblastoma cells. Int J Oncol. 2019;54(5):1853–1863. doi: 10.3892/ijo.2019.4741. [DOI] [PubMed] [Google Scholar]
- 67.Chitragar S, Iyer VK, Agarwala S, Gupta SD, Sharma A, Wari MN. Loss of heterozygosity on chromosome 11p15.5 and relapse in hepatoblastomas. Eur J Pediatr Surg. 2011;21(1):50–53. doi: 10.1055/s-0030-1267208. [DOI] [PubMed] [Google Scholar]
- 68.Van Dyck F, Declercq J, Braem CV, Van de Ven WJM. PLAG1, the prototype of the PLAG gene family: versatility in tumour development (review) Int J Oncol. 2007;30(4):765–774. [PubMed] [Google Scholar]
- 69.Sasaki H, Ishihara K, Kato R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J Biochem. 2000;127(5):711–715. doi: 10.1093/oxfordjournals.jbchem.a022661. [DOI] [PubMed] [Google Scholar]
- 70.Brouwer-Visser J, Huang GS. IGF2 signaling and regulation in cancer. Cytokine Growth Factor Rev. 2015;26(3):371–377. doi: 10.1016/j.cytogfr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 71.Honda S, Arai Y, Haruta M, Sasaki F, Ohira M, Yamaoka H, Horie H, Nakagawara A, Hiyama E, Todo S, et al. Loss of imprinting of IGF2 correlates with hypermethylation of the H19 differentially methylated regi on in hepatoblastoma. Br J Cancer. 2008;99(11):1891–1899. doi: 10.1038/sj.bjc.6604754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ranke MB. Insulin-like growth factor binding-protein-3 (IGFBP-3). Best Pract Res Clin Endocrinol Metab. 2015;29(5):701–11. [DOI] [PubMed]
- 73.Regel I, Eichenmüller M, Joppien S, Liebl J, Häberle B, Müller-Höcker J, Vollmar A, von Schweinitz D, Kappler R. IGFBP3 impedes aggressive growth of pediatric liver cancer and is epigenetically silenced in vascular invasive and metastatic tumors. Mol Cancer. 2012;11:9. doi: 10.1186/1476-4598-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rivas MP, Aguiar T, Fernandes GR, Caires-Júnior LC, Goulart E, Telles-Silva KA, Cypriano M, de Toledo S, Rosenberg C, Carraro DM, et al. TET Upregulation Leads to 5-Hydroxymethylation Enrichment in Hepatoblastoma. Front Genet. 2019;10:553. doi: 10.3389/fgene.2019.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beck A, Trippel F, Wagner A, Joppien S, Felle M, Vokuhl C, Schwarzmayr T, Strom TM, von Schweinitz D, Längst G, et al. Overexpression of UHRF1 promotes silencing of tumor suppressor genes and predicts outcome in hepatobl astoma. Clin Epigenet. 2018;10:27. doi: 10.1186/s13148-018-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pacaud R, Brocard E, Lalier L, Hervouet E, Vallette FM, Cartron PF. The DNMT1/PCNA/UHRF1 disruption induces tumorigenesis characterized by similar genetic and epigenetic signatures. Sci Rep. 2014;4:4230. doi: 10.1038/srep04230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson RT, et al. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25(2):196–209. doi: 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jia Y, Li P, Fang L, Zhu H, Xu L, Cheng H, Zhang J, Li F, Feng Y, Li Y, et al. Negative regulation of DNMT3A de novo DNA methylation by frequently overexpressed UHRF family proteins as a mechanism for widespread DNA hypomethylation in cancer. Cell Discov. 2016;2:16007. doi: 10.1038/celldisc.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu Y-C, Ling Z-Q. The role of TET family proteins and 5-hydroxymethylcytosine in human tumors. Histol Histopathol. 2014;29(8):991–997. doi: 10.14670/HH-29.991. [DOI] [PubMed] [Google Scholar]
- 80.Necula LG, Mambet C, Albulescu R, Diaconu CC. Epigenetics in gastric carcinogenesis: TET genes as important players. J Immunoassay Immunochem. 2015;36(5):445–455. doi: 10.1080/15321819.2015.1017402. [DOI] [PubMed] [Google Scholar]
- 81.Liu J, Jiang J, Mo J, Liu D, Cao D, Wang H, He Y, Wang H. Global DNA 5-Hydroxymethylcytosine and 5-Formylcytosine contents are decreased in the early stage of hepatocellular carcinoma. Hepatology. 2019;69(1):196–208. doi: 10.1002/hep.30146. [DOI] [PubMed] [Google Scholar]
- 82.Chen Z, Shi X, Guo L, Li Y, Luo M, He J. Decreased 5-hydroxymethylcytosine levels correlate with cancer progression and poor survival: a systematic review and meta-analysis. Oncotarget. 2017;8(1):1944–1952. doi: 10.18632/oncotarget.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30(7):733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rawłuszko-Wieczorek AA, Siera A, Jagodziński PP. TET proteins in cancer: Current ‘state of the art’. Crit Rev Oncol Hematol. 2015;96(3):425–436. doi: 10.1016/j.critrevonc.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 85.Beck A, Trippel F, Wagner A, Joppien S, Felle M, Vokuhl C, Schwarzmayr T, Strom TM, von Schweinitz D, Langst G, et al. Overexpression of UHRF1 promotes silencing of tumor suppressor genes and predicts outcome in hepatoblastoma. Clin Epigenetics. 2018;10:27. doi: 10.1186/s13148-018-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pan Y, Liu G, Zhou F, Su B, Li Y. DNA methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med. 2018;18(1):1–14. doi: 10.1007/s10238-017-0467-0. [DOI] [PubMed] [Google Scholar]
- 87.Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15(12):3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gnyszka A, Jastrzebski Z, Flis S. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res. 2013;33(8):2989–2996. [PubMed] [Google Scholar]
- 89.Qiao G-L, Li L, Cheng W, Ge J, Zhang Z, Wei Y. Predictors of survival after resection of children with hepatoblastoma: A single Asian center experience %J European journal of surgical oncology : the journal of the European Society of Surgical Oncology and the British Association of. Surg Oncol. 2014;40(11):1533–1539. doi: 10.1016/j.ejso.2014.07.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.