Abstract
Significance: Generalized selenoprotein deficiency has been associated with mutations in SECISBP2, SEPSECS, and TRU-TCA1-1, 3 factors that are crucial for incorporation of the amino acid selenocysteine (Sec) into at least 25 human selenoproteins. SECISBP2 and TRU-TCA1-1 defects are characterized by a multisystem phenotype due to deficiencies of antioxidant and tissue-specific selenoproteins, together with abnormal thyroid hormone levels reflecting impaired hormone metabolism by deiodinase selenoenzymes. SEPSECS mutations are associated with a predominantly neurological phenotype with progressive cerebello-cerebral atrophy.
Recent Advances: The recent identification of individuals with defects in genes encoding components of the selenocysteine insertion pathway has delineated complex and multisystem disorders, reflecting a lack of selenoproteins in specific tissues, oxidative damage due to lack of oxidoreductase-active selenoproteins and other pathways whose nature is unclear.
Critical Issues: Abnormal thyroid hormone metabolism in patients can be corrected by triiodothyronine (T3) treatment. No specific therapies for other phenotypes (muscular dystrophy, male infertility, hearing loss, neurodegeneration) exist as yet, but their severity often requires supportive medical intervention.
Future Directions: These disorders provide unique insights into the role of selenoproteins in humans. The long-term consequences of reduced cellular antioxidant capacity remain unknown, and future surveillance of patients may reveal time-dependent phenotypes (e.g., neoplasia, aging) or consequences of deficiency of selenoproteins whose function remains to be elucidated. The role of antioxidant therapies requires evaluation. Antioxid. Redox Signal. 33, 481–497.
Keywords: SECISBP2, SEPSECS, TRU-TCA1-1, selenium, selenoprotein deficiency, thyroid hormone metabolism
Introduction
Selenium (Se) is an essential micronutrient that is incorporated as the amino acid selenocysteine (Sec) into human selenoproteins, encoded by 25 separate genes. Most selenoproteins function as oxidoreductases, with the Sec residue involved in catalytic activity. Selenoproteins have diverse functions ranging from maintenance of redox potential; regulation of redox-sensitive biochemical pathways; protection of genetic material, proteins, and membranes from oxidative damage; metabolism of thyroid hormones; regulation of gene expression; and control of protein folding (Table 1). However, the function of several selenoproteins is unknown (57).
Table 1.
Selenoproteins
| Selenoprotein | Main known function | Subcellular localization | Expression pattern |
|---|---|---|---|
| GPX1 | Oxidoreductase protection against oxidative stress | Cytoplasmic | Most tissues |
| GPX2 | Oxidoreductase protection against oxidative stress | Nuclear and cytoplasmic | Gastrointestinal tract, bone marrow, immune system, liver, gallbladder, kidney, and urinary bladder |
| GPX3 | Oxidoreductase protection against oxidative stress | Secreted | Most tissues, high in kidney, thyroid, adipose |
| GPX4 | Oxidoreductase protection against oxidative stress | Nucleus and mitochondria | Most tissues, high in testis, adipose tissue |
| GPX6 | Oxidoreductase protection against oxidative stress | Predicted secreted | Testis, epididymis, olfactory system |
| TXNRD1 | Oxidoreductase protection against oxidative stress | Nuclear and cytoplasmic | Ubiquitous |
| TXNRD2 | Oxidoreductase protection against oxidative stress | Cytoplasmic and mitochondria | Ubiquitous |
| TXNRD3 | Oxidoreductase protection against oxidative stress | Intracellular | Most tissues, high in testis |
| DIO1 | Oxidoreductase thyroid hormone metabolism | Intracellular membrane-associated | Kidney, liver, and thyroid gland |
| DIO2 | Oxidoreductase thyroid hormone metabolism | Intracellular membrane-associated | Low in several tissues, high in thyroid, esophagus, cervix, ectocervix, pituitary, endometrium, and brain |
| DIO3 | Oxidoreductase thyroid hormone metabolism | Intracellular membrane-associated | Several tissues, high in cervix, uterine, placenta, and urinary bladder |
| MSRB1 | Oxidoreductase, Methionine Sulfoxide Reduction | Nuclear and cytoplasmic | Ubiquitous |
| SELENOF | Oxidoreductase ER-associated protein folding control | Endoplasmic reticulum | Ubiquitous |
| SELENOH | Unknown Oxidoreductase dna/rna binding motif | Nuclear | Ubiquitous |
| SELENOI | Oxidoreductase phospholipid biosynthesis | Transmembrane | Ubiquitous |
| SELENOK | ER-associated protein folding control | ER, plasma membrane | Ubiquitous |
| SELENOM | Unknown | Nuclear and perinuclear | Ubiquitous |
| SELENON | Oxidoreductase redox-related calcium homeostasis | Endoplasmic reticulum | Ubiquitous |
| SELENOO | Protein AMPylation activity | Mitochondria | Ubiquitous |
| SELENOP | Transport/oxidoreductase | Secreted, cytoplasmic | Most tissues, high in liver and small intestine |
| SELENOS | Oxidoreductase ER-associated protein folding control | Endoplasmic reticulum | Ubiquitous |
| SELENOT | Unknown oxidoreductase | Endoplasmic reticulum | Ubiquitous |
| SELENOV | Unknown | Intracellular | Thyroid, parathyroid, testis, and brain |
| SELENOW | Oxidoreductase protection against oxidative stress | Intracellular | Ubiquitous |
| SEPHS2 | Selenophosphate synthesis | Intracellular | Ubiquitous, high in liver kidney |
SELENOP, selenoprotein P; SEPHS2, selenophosphate synthetase 2.
Biosynthesis of selenoprotein requires a UGA codon within its messenger RNA (mRNA) to be recoded as the amino acid Sec, preventing its recognition as a premature stop, possibly targeting the transcript for nonsense-mediated decay (57, 79). This process is achieved via a unique Sec-insertion machinery, comprising cis-acting SEleniumCysteine Insertion Sequence (SECIS) elements located in the 3′-UTR (untranslated region) of selenoprotein mRNAs and the UGA codon, interacting with trans-acting factors (SECIS binding protein 2 [SECISBP2], Sec tRNA [transfer RNA], specific eukaryotic elongation factor [EEFSEC], and Sec-tRNA[Ser]Sec) (Fig. 1) (4, 24, 33, 58, 85).
FIG. 1.
Pathway of selenocysteine synthesis and its incorporation into selenoproteins. Sec is synthesized on its own tRNA (tRNA[Ser]Sec), which undergoes maturation through sequential modifications, with initial attachment of serine by SARS resulting in Ser-tRNA[Ser]Sec. Subsequent phosphorylation of this serine residue by PSTK generates O-Phosphoseryl-tRNA[Ser]Sec. Finally, SEPSECS catalyzes the acceptance of a selenophosphate, generated from selenide and ATP by SEPHS2, resulting in Sec-tRNA[Ser]Sec. An intermediate complex that includes Sec-tRNA[Ser]Sec, TRNAU1AP, and EEFSEC is subsequently formed. This complex is guided by an interaction with SECISBP2 to the SECIS-element of selenoprotein mRNA, ready for incorporation into the nascent polypeptide. Other factors (ribosomal protein L30, eukaryotic initiation factor eIF4a3, nucleolin, …) also have regulatory roles and influence the Sec insertion process. EEFSEC, Sec tRNA-specific eukaryotic elongation factor; mRNA, messenger RNA; PSTK, phosphoseryl-tRNA kinase; SARS, seryl-tRNA synthetase; Sec, selenocysteine; SECIS, SEleniumCysteine Insertion Sequence; SECISBP2, SECIS binding protein 2; SEPHS2, selenophosphate synthetase 2; SEPSECS, O-phosphoserine tRNA:Sec tRNA synthase; tRNA, transfer RNA; TRNAU1AP, tRNA selenocysteine 1 associated protein 1. Color images are available online.
In contrast to other amino acids, selenocysteine does not have an aminoacyl-tRNA synthetase, but is synthesized on its own tRNA, encoded by TRU-TCA1-1 (Fig. 1) (5). This tRNA was originally named the human opal suppressor gene and a truncated pseudogene that is not expressed also exists (68). Delivery to the ribosome and subsequent cotranslational insertion of Sec is mediated by a multiprotein complex (81), which includes EEFSEC and SECISBP2 as well as other factors. The specialized elongation factor EEFSEC (rather than general elongation factors eEf1a and EF-Tu, which delivers all other aa-tRNAs) delivers Sec-tRNA[Ser]Sec to the ribosome acceptor site (85). SECISBP2 interacts with the SECIS element, a stem–loop structure present in the 3′-UTR of every selenoprotein mRNA (12, 13, 23). The SECIS elements within each selenoprotein mRNA are distinct, but share common structural features, consisting of two helices separated by an internal loop, a GA quartet of non-Watson–Crick base pairs, and an apical loop, resulting in the adoption of a “kink-turn” structure (21, 33, 41, 89, 90). All selenoprotein mRNAs contain a single SECIS element, except for selenoprotein P (SELENOP), which has two, tandemly repeated SECIS elements. This configuration coincides with human SELENOP being the only selenoprotein that contains more than one selenocysteine residue (57). The SECIS element is required for Sec incorporation, whereas other motifs in some selenoprotein mRNAs (e.g., Sec redefinition element, proximal stem loop element) have been described as contributing to the translation process (15, 22, 47, 48, 62).
The EEFSEC–Sec-tRNA[Ser]Sec–SECISBP2 complex, bound to SECIS in the selenoprotein mRNA, is believed to be in close proximity to the ribosomal complex, thereby preventing recognition of UGA as a translational stop and poised to mediate Sec incorporation into the polypeptide when the UGA codon is presented. Ribosomal protein L30 (RPL30), tRNA selenocysteine 1 associated protein 1 (TRNAU1AP), eukaryotic translation initiation factor 4A3 (EIF4A3), and nucleolin are other factors that regulate the Sec insertion process (Fig. 1) (49, 57, 79). However, three factors, Sec-tRNA[Ser]Sec, EEFSEC, and SECISBP2, have been shown to be essential and sufficient for Sec incorporation in vitro (42, 59, 63, 82).
The biological importance of selenoproteins is highlighted by the fact that both Trsp (mouse tRNA[Ser]Sec) and Secisbp2 null mice are embryonically lethal (19, 78). Mutations in individual human selenoproteins and their consequences have recently been reviewed in detail elsewhere (35): selenoprotein N (myopathy), glutathione peroxidase 4 (respiratory failure and skeletal defects), thioredoxin reductase 2 (associated with familial glucocorticoid deficiency and dilated cardiomyopathy), and thioredoxin reductase 1 (generalized epilepsy); two reports describe patients with a complex, hereditary spastic paraplegia and mutations in SELENOI, which catalyzes the transfer of phosphoethanolamine from CDP-ethanolamine to diacylglycerol to produce phosphatidylethanolamine (2, 46). Here, we describe mutations in genes (SEPSECS, SECISBP2, and TRU-TCA1-1) encoding three components of the selenocysteine insertion pathway, affecting general incorporation of selenocysteine into selenoproteins and their clinical consequences.
SECISBP2
SECISBP2 is an obligated limiting factor for selenoprotein synthesis, as first shown by the absence of selenoprotein synthesis in SECISBP2-depleted cell lysates, with restoration of production by repletion with SECISBP2 (23, 24). Human SECISBP2 is a large (854 amino acids, 120-kDa) protein, with the first 400, amino (N-) terminal, residues being dispensable for its function in vitro (25, 26) (Fig. 2). In contrast, the carboxy-terminal (C-terminal) region (amino acids 399–784) is both necessary and sufficient for SECIS-binding and Sec incorporation in vitro and contains several functional domains. The Sec incorporation domain (SID), located centrally in SECISBP2, is not essential for SECIS binding but required for Sec incorporation. The RNA-binding domain (RBD) contains an L7Ae-type RNA interaction motif identified in a large family of ribosomal proteins (e.g., RPL30, SUP1, eRF-1, and 15.5-kD/Snu13p) (4, 5, 16, 25, 26), which interacts with the “kink-turn” structure adopted by the SECIS-element. The RBD mediates interaction with the SECIS element (34, 89) and 28S ribosomal RNA (25, 49, 56, 59). A domain amino-terminal (N-terminal) to the L7Ae module, referred to as either the bipartite, SID, or K-rich region, is involved in specific recognition of SECIS elements and other regulatory RNA motifs, thereby also controlling selenoprotein expression levels (17, 30, 83). SECISBP2 also contains several other functional motifs, as shown in Figure 2 (69).
FIG. 2.
Genomic organization of SECISBP2 and functional domains of SECISBP2 with the position of human mutations. (A) The organization of the SECISBP2 gene (top), with naturally occurring aminoterminal splice variants, each containing distal exons 8–17 shown next; the functional domains of SECISBP2 protein with the location of human mutations superimposed is shown (bottom). Arrowheads denote the location of ATG codons, which could function as alternative sites for the initiation of translation. Functional domains in SECISBP2 protein: N-terminal domain (1–399); minimal functional protein (shaded gray, 399–784); SID (399–517); minimal RBD (517–784); Lysine-rich domain involved in RNA specificity and ribosome binding (517–544); L7Ae homology module (620–745); NLS (380–390); redox-sensitive CRD (584–854); and two NES (NES1: 634–657; NES2: 756–770) (69). (B, C) Model of the L7ae RBD of SECISBP2. The position of wild-type residues (E679, C691) (B) and corresponding mutations (E679D, C691R) at these locations (C) is shown. The model was generated by using the phyre2 web portal, which predicts and analyzes protein structures based on homology/analogy to solved protein crystal structures (51). CRD, cysteine-rich domain; NES, nuclear export signals; NLS, nuclear localization signal; N-terminal, amino-terminal; phyre2, protein homology/analogy recognition engine 2; RBD, RNA-binding domain; SID, Sec incorporation domain. The figures were generated with MacPyMOL Molecular Graphics System, Schrödinger, LLC.
Alternative splicing events in the 5′-region of human SECISBP2, with use of alternative initiation of translation from downstream ATG start codons in exons 2, 3a, 3b, 5, and 7, generates five different protein isoforms, each containing a varying N-terminal protein sequence (Fig. 2) (24, 70). These alternate splicing events alter content of the dispensable N-terminal region, but not the essential C-terminal domain, within protein isoforms. Nevertheless, it is possible that the alternately spliced isoforms do play a role in regulation of SECISBP2-dependent Sec incorporation and selenoprotein expression in vivo. During protein synthesis, dynamic interaction of SECISBP2 with the ribosome and SECIS element is essential for recruitment of the EEFsec/Sec-tRNA[Ser]sec complex to the UGA codon, enabling the incorporation of Sec into the polypeptide.
Homozygous or compound heterozygous mutations in SECISBP2 have been described in 13 individuals from 11 families (Table 2). Disruption of SECISBP2 function prevents appropriate Sec incorporation into selenoproteins during their biosynthesis, resulting in a multisystem disorder due to deficiency of diverse selenoproteins (Table 3). The biochemical signature that identifies SECISBP2-deficient patients consists of low circulating selenium (reflecting low plasma SELENOP and GPX3) and abnormal thyroid hormone levels due to diminished activity of deiodinases (Table 3) (31, 75). Most cases present in childhood due to growth retardation with raised circulating free thyroxine (FT4), normal to low free triiodothyronine (FT3), and raised reverse triiodothyronine (T3) levels, reflecting deficiency of all three deiodinase enzymes.
Table 2.
Human SECISBP2 Mutations: Genetics and Effect
| Family | Gene mutation | Predicted protein change | Alleles affected | Suggested mechanism | Ethnicity | References |
|---|---|---|---|---|---|---|
| A | c.1619 G > A | R540Q | Homozygous | Predicted to affect SECIS and ribosome binding | Saudi Arabian | (31) |
| B | c.1312A>T | K438X | Compound heterozygous | Premature stop, no/decreased full-length protein | Irish/Kenyan | (31) |
| c.IVS8ds +29 G > A | fs431X | |||||
| C | c.382 C > T | R128X | Homozygous | Premature stop, no full-length protein | Ghanaian | (28) |
| D | c.358 C > T | R120X | Compound heterozygous | Premature stop, no full-length protein | Brazilian | (9) |
| c.2308 C > T | R770X | Premature stop, no full-length protein | ||||
| E | c.668delT | F223fs255X | Compound heterozygous | Premature stop, no full-length protein | British | (75) |
| c.IVS7 -155, T > A | fs295X+fs302X | Premature stop, no or decreased full-length protein | ||||
| F | c. 2017T>C | C691R | Compound | Predicted to affect SECIS and ribosome binding, increased degradation | British | (75) |
| 1–5 Intronic SNPs | fs65X+fs76X | Heterozygous | Premature stop, no full-length protein/splice variants affected | |||
| G | c.1529_1541dup CCAGCGCCCCACT | M515fs563X | Compound heterozygous | Premature stop, no full-length protein | Japanese | (43) |
| c.235 C > T | Q79X | |||||
| H | c.2344 C > T | Q782X | Compound heterozygous | Premature stop, no full-length protein | Turkish | (32) |
| c.2045–2048 delAACA | K682fs683X | |||||
| I | c.660 C > T | R197X | Compound heterozygous | Premature stop, no full-length protein/splice variants affected | Argentinian | (37) |
| c.2108 G > T or C | E679D | Predicted to affect SECIS and ribosome binding | ||||
| J | c.800_801insA | K267Kfs*2 | Homozygous | Premature stop, no full-length protein | Turkish | (20) |
| K | c.283delT | T95Ifs31* | Compound heterozygous | Premature stop, no full-length protein | N/A | (55) |
| c.589C>T | R197X |
N/A, not available; SECIS, SEleniumCysteine Insertion Sequence; SNP, single nucleotide polymorphism.
Table 3.
Human SECISBP2 Mutations: A Multisystem Disorder with a Thyroid Signature
| Family | [Se] | TT4 | FT4 | TT3 | rT3 | TSH | Growth and skeletal | Musculoskeletal | Neurocognitive | Hearing | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | L | H | H | L | H | H | Short stature, DBA | N/A | Normal mental development | Normal | N/A |
| B | L | H | H | N/L | H | N | Short stature, DBA | N/A | N/A | Normal | N/A |
| C | L | H | H | N/L | H | N | Short stature, DBA | N/A | N/A | N/A | |
| D | L | H | H | N/L | H | H | Short stature, DBA, kyphoscoliosis | Hypotonia, hip girdle weakness, spirometry: reduced expiratory and inspiratory flow, fatty infiltration of muscle | Impaired mental development and motor coordination | Bilateral sensorineural loss | Failure to thrive, bilateral clinodactyly, asymmetric leg length, peripheral sensory neuropathy, and increased fat mass |
| E | L | N/A | H | N | N/A | N | Short stature, genu valgus | Lumbar spinal rigidity, reduced axial and neck strength, spirometry: reduced vital capacity, nocturnal hypoventilation, fatty infiltration of muscle | Developmental delay | Bilateral sensorineural loss, vertigo | Azoopsermia, Raynauds disease, photosensitivity, mild lymphopenia and reduced red cell mass, low insulin and high adiponectin levels, favorable blood lipid profile, and low intrahepatic lipid |
| F | L | N/A | H | L | N/A | N | Short stature | Proximal and axial myopathy, lumbar rigidity, fatty infiltration of adductor muscles | Developmental delay | Bilateral sensorineural loss | Failure to thrive, eosinophilic colitis, increased fat mass, high adiponectin levels, and hypoglycemia with low fasting insulin |
| G | L | N/A | H | N/L | N/A | N/H | Short stature, DBA | Fatty infiltration of muscles | Delayed motor and intellectual development, IQ 70 | Bilateral mild conductive loss rotatory vertigo | Failure to thrive, hypoplastic thyroid gland, no photosensitivity, and increased fat mass |
| H | L | H | H | L | H | N/H | Short stature | N/A | Mental and motor retardation, IQ 50 | N/A | N/A |
| I | N/A | H | H | L | H | N | DBA | N/A | N/A | N/A | Failure to thrive |
| J | L | H | H | L | H | N/H | Decreased growth velocity from 13 years | Muscle weakness, fatty infiltration of the muscles | Normal early development | Normal | Right-eye ptosis, attention-deficit disorder with poor school performance, impaired growth hormone response, obese (body mass index 29.5), and impaired OGTT |
| K | N/A | H | H | L | H | N | Short stature, DBA | N/A | Developmental delay | N/A | Failure to thrive |
| L | L | H | H | L | H | N | Delayed growth | Hypotonia, myopathy with fatty infiltration of muscles, and ataxic gait | Developmental delay | Bilateral sensorineural loss | Dysarthic speech, tapered fingers, seizures, and increased fat mass |
DBA, delayed bone age; FT4, free thyroxine; H, high; IQ, intelligence quotient; L, low; N, normal; N/H, high-normal; N/L, low normal; OGTT, oral glucose tolerance test; rT3, reverse triiodothyronine; Se, selenium; T3, triiodothyronine; T4, thyroxine; TSH, thyroid stimulating hormone; TT3, total triiodothyronine; TT4, total thyroxine; TSH, thyroid stimulating; hormone.
Muscle weakness, due to progressive rigid spine muscular dystrophy, affecting axial and proximal limb muscles with raised creatine kinase (CK) levels and fatty infiltration on imaging, is similar to that seen in patients with mutations in selenoprotein N. In one patient (proband E), male azoospermic infertility was described, reflecting loss of testis selenoproteins (mitochondrial GPX4, thioredoxin reductase, and selenoprotein V) required for spermatogenesis (75).
Significantly decreased expression levels of antioxidant selenoenzymes are associated with increased levels of cellular reactive oxygen species (ROS). Clinical consequences of raised cellular ROS include skin photosensitivity, progressive sensorineural hearing loss, and possibly increased total body adipose tissue mass paradoxically associated with enhanced systemic insulin sensitivity (75).
Reduced red blood cell and total lymphocyte counts, with impaired mononuclear cell cytokine secretion and T cell proliferation (similar to findings in T cell-specific Trsp null mice) (80), were recorded in one case, proband E (75). Although other hematological and immune cell phenotypes have not been formally evaluated, immunodeficiency is neither a reported feature in other SECISBP2 mutation cases nor seen in mouse models of selenoprotein deficiency. Additional age-dependent phenotypes such as neurodegeneration, premature aging, or neoplasia may emerge but have not been described hitherto.
Oral selenium supplementation in some SECISBP2 patients raised total serum Se levels, but without clinical effect (9, 20, 28) or altering synthesis (circulating GPX's, SELENOP) or action (thyroid hormone metabolism) of selenoproteins (77). Treatment of probands C and F with T3 alone (thyroxine [T4] was not effective in one case) or in combination with growth hormone (proband G) resulted in an improvement in growth, development, and bone maturation. Treatment of proband G with a combination of alpha tocopherol (vitamin E) and T3 resulted in the most promising response, with decreased serum levels of lipid peroxidation products, altered FT4 and FT3 concentrations, and increased circulating white blood cells and neutrophils, all of which were reversed after treatment withdrawal (74). These observations suggest that treatment with antioxidants, to counteract the effects of elevated cellular ROS, is the best available therapeutic option for this disorder.
Most SECISBP2 mutations identified to date cause premature stop codons, resulting in the absence of full-length SECISBP2 protein. However, elegant minigene experiments have shown that, for premature stops located in the N-terminal part of the protein, initiation of translation from alternative, downstream ATG codons in exons 5 (Met233) and 7 (Met300) permits low-level synthesis of shorter SECISBP2 isoforms (28, 75). Some premature stop mutations are situated downstream of Met300 and may completely eliminate synthesis of functional protein. However, stop mutations (e.g., R770X, Q782X), distal to the RBD, might generate C-terminally truncated proteins whose RNA binding and nuclear localization functions remain, partially, intact (Fig. 2). In a patient with defective mRNA splicing due to an intronic mutation IVS8ds +29G>A (31), it has been shown that levels of normally spliced transcript are only reduced by 50% and if a similar mechanism operates with other splice site mutations, this suggests some preservation of normally spliced SECISBP2 mRNA in such cases.
Three missense SECISBP2 mutations (R540Q, C691R, and E679D) have been described: The R540Q mutation localizes to the K-rich region within the RBD (Fig. 2), with the R540Q mutant exhibiting reduced binding to SECIS elements in GPX1 and DIO2 mRNAs, correlating with diminished GPX1 and DIO2 enzyme activity in patient-derived primary cells and the mouse model. Detailed analyses suggest that R540Q mutant SECISBP2 fails to bind only a subset of SECIS-elements, consistent with the K-rich region mediating the recognition of specific (type I and type II) SECIS elements (14); a mouse model revealed a possible tissue-specific pattern of SECISBP2 protein stability, correlating with varying loss or preservation of expression of different selenoproteins (14, 31, 96).
The C691R SECISBP2 mutation, also located in its RBD, is expected to affect RNA binding. Homology modeling, based on the crystal structure of the spliceosomal 15.5 kDa protein (87), suggests that the mutation of cysteine to a bulky and charged arginine residue (75) may destabilize its hydrophobic core, disrupting local protein structure (Fig. 2B, C). In vitro assays, showing enhanced proteasomal degradation of the C691R mutant SECISBP2 protein, confirmed this (75). A mouse model suggests that the C691R mutant SECISBP2 is unable to bind RNA and is non-functional (96). The E679D SECISBP2 mutation is predicted to be deleterious (PolyPhen-2 algorithm score of 0.998) and also located in the RBD and may, therefore, affect its RNA binding function, but this has not been investigated in detail (37) (Fig. 2).
Knowledge that knockout (KO) of Secisbp2 in mice is embryonic lethal (78), with no evidence for an alternative Sec-incorporation mechanism in humans, suggests that there is some residual SECISBP2 activity in all patients. All patients described to date are expected to harbor at least one allele that directs the synthesis of SECISBP2 at either reduced levels or that is only partially functional, in combination with either a mutant or a shorter form of the protein synthesized from Met300 (Table 2). A limited number of patients, mostly compound heterozygous for different SECISBP2 mutation combinations, with limited knowledge of phenotypes in heterozygous relatives, have been described, making it difficult to assess the effect of a specific SECISBP2 mutation or its correlation with the severity of phenotype. However, since SECISBP2 is rate limiting for Sec incorporation, a significant reduction in functional SECISBP2 protein levels will result in diminished but not complete loss of selenoprotein synthesis. A further variable is that differences in the architecture of SECIS elements within different selenoprotein mRNAs may dictate the extent to which reduced SECISBP2 protein limits their biosynthesis and expression levels in vivo, as suggested by elegant experiments testing SECISBP2 with luciferase reporter genes containing different SECIS elements (60, 84). Future studies, undertaking ribosomal RNA (96) or selenoprotein expression profiling in secisbp2 mutant mouse models and in vitro reconstitution experiments with different mutant SECISBP2 proteins and SECIS element containing luciferase reporter genes (84), may help us better understand the effect of specific SECISBP2 mutations. In turn, greater understanding of how different mutations affect selenoprotein expression may enable better prediction of clinical outcome or targeting of therapy in patients.
TRU-TCA1-1
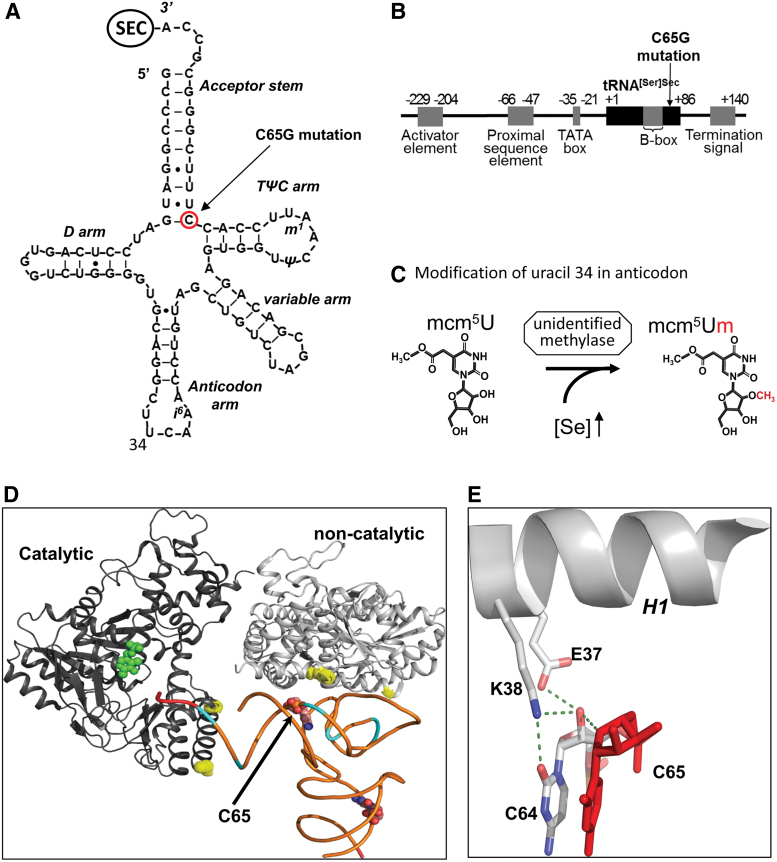
Selenocysteine is synthesized on its own tRNA (Fig. 1), tRNA[Ser]Sec (encoded by TRU-TCA1-1) that has several unique features, being longer (90 nucleotides vs. usual 78), with an atypical long acceptor- and D-stem with a few modified bases (57) that distinguishes it from other tRNAs (Fig. 3A). Its promoter region, containing both tRNA and U small nuclear RNA (snRNA) gene regulatory elements (18, 39, 44) forming a new class of RNA polymerase III transcribed genes, also differs from other tRNA genes (Fig. 3B) and consists of four regulatory elements: an activator element, containing an SPH motif, but is octamer independent; the proximal sequence element; an extended TATA-motif, essential for efficient transcription; and an internal B box (64, 65).
FIG. 3.
Genomic and structural organization of Sec-tRNA[Ser]Sec showing the position of human mutation. (A) The primary structure of human Sec-tRNA[Ser]Sec is shown in a cloverleaf model, with the location of C65G TRU-TCA1–1 mutation identified in the patient indicated (circled red). The acceptor stem constitutes paired 5′ and 3′ terminal bases, with the D arm, the anticodon arm, the variable arm, and the TψC arm depicted. Mammalian Sec-tRNA[Ser]Sec undergoes post-transcriptional modification at positions 34 (mcm5U or mcm5Um), 37 (i6A), 55 (ψ), and 58 (m1A). (B) Schematic of the TRU-TCA1-1 gene showing the coding region (black boxes) and regulatory elements (gray boxes) (43), with the location of C65G mutation identified in the patient. (C) The two Sec-tRNA[Ser]Sec isoforms, containing either mcm5U or mcm5Um modifications of the uracil at position 34 in the anticodon arm, differ from each other by a single methyl group on the 2′-O-ribosyl moiety. This reaction is catalyzed by an unknown methylase, and abundance of the mcm5Um isoform increases with selenium concentration (19). (D) Crystal structure showing catalytic and non-catalytic dimers of the SEPSECS tetramer bound to tRNA[Ser]Sec (71), with the position of the C65 nucleotide indicated. Other nucleotides in tRNA[Ser]Sec (cyan) and amino acids in SEPSECS (yellow) involved in RNA–protein interaction are also highlighted, as are the nucleotides (red) toward the position of Sec and the pyridoxal-5-phosphate substrate (green) within the catalytic domain. (E) A close-up of the structure around C65, showing H-bonds (dashed green lines) formed between C64 and the C64–C65 backbone with residues (E37 and K38) situated in helix 1 (H1) of the non-catalytic dimer of SEPSECS. mcm5U, 5-methoxycarbonyl-methyluridine; mcm5Um, 5-methoxycarbonylmethyl-2′-O-methyluridine. Color images are available online.
Two major isoforms of Sec-tRNA[Ser]Sec have been identified, containing either 5-methoxycarbonyl-methyluridine (mcm5U) or its methylated form 5-methoxycarbonylmethyl-2′-O-methyluridine (mcm5Um) at position 34 (Fig. 3C). Uridine 34 is located in the anticodon loop and its methylation may contribute to stabilization of the codon–anticodon interaction (19, 27, 45, 79). Methylation at uridine 34 is executed by a methylase whose identity is not known and the relative abundance of the two Sec-tRNA[Ser]Sec isoforms is known to be influenced by systemic Se-status, increasing selenium levels resulting in more mcm5Um isoform (19, 27, 45). Each Sec-tRNA[Ser]Sec subtype has a different role, with synthesis of cellular housekeeping selenoproteins (e.g., TXNRD1, TXNRD3, GPX4) being dependent on the mcm5U isoform; whereas the expression of cellular, stress-related selenoproteins (e.g., GPX1, GPX3, SELENOW) requires the mcm5Um isoform (19, 79). Methylation of mcm5U, the final step in post-transcriptional maturation of Sec-tRNA[Ser]Sec, is dependent on correct aminoacylation (53, 54), intact secondary/tertiary structure (54), and other, prior, base modifications (Fig. 3A) of the tRNA (27).
A single patient, homozygous for a single nucleotide change (C65G) in TRU-TCA1-1 has been identified (Fig. 3A) (76). The proband exhibits a similar phenotype to that seen in SECISBP2 mutation patients. However, a comparison of cellular selenoprotein expression profiles in the two disorders has revealed differences, with the expression of relative housekeeping selenoproteins (e.g., TXNRDs, GPX4) being more preserved than in SECISBP2 cases. In contrast, the expression of stress-related selenoproteins (e.g., GPX1, GPX3) was similarly reduced in both disorders. In primary cells from the TRU-TCA1-1 mutation patient, lower total tRNA[Ser]Sec expression with a disproportionately greater diminution in Sec-tRNA[Ser]Sec mcm5Um levels was observed, with decreased i6A modification at position 37 suggesting that its post-transcriptional maturation is impaired. The mutation had no effect on tRNA[Ser]Sec aminoacylation with serine or Sec synthesis or its interaction with O-phosphoserine tRNA:Sec tRNA synthase (SEPSECS). Low levels of tRNA[Ser]Sec in the proband were insufficient to direct normal synthesis of stress-related selenoproteins, but they were not rate limiting for adequate synthesis of some housekeeping selenoproteins and similar, differential preservation of selenoprotein synthesis has been observed in murine tRNAsec mutant models (19, 57).
The human SEPSECS–tRNA[Ser]Sec structure shows that C65 is situated in the acceptor arm, next to C64 in the TΨC–arm (Fig. 3D). C64 interacts with Lys38 and Glu37 in helix 1 of the non-catalytic part of the SEPSECS moiety (Fig. 3E), raising the possibility that the C65G mutation could affect the stability of the Sec-tRNA[Ser]Sec–SEPSECS complex and the selenoprotein synthesis.
In summary, these observations indicate that reduction in Sec-tRNA[Ser]Sec levels, with particular deficiency of the Sec-tRNA[Ser]Sec mcm5Um subtype, contributes to the selective pattern of selenoprotein deficiency seen in the proband. The precise mechanism mediating the reduction in mutant Sec-tRNA[Ser]Sec presence remains unclear, with defective post-transcriptional modification of mutant Sec-tRNA[Ser]Sec or instability of the mutant Sec-tRNA[Ser]Sec–SEPSECS complex being possibilities.
SEPSECS
Human SEPSECS was initially identified as an autoantigen (soluble liver antigen/liver pancreas) in autoimmune hepatitis (52). Subsequent studies in which mammalian cell extracts were treated with autoimmune hepatitis patients' serum showed that SEPSECS co-precipitated with Sec-tRNA[Ser]Sec, as part of a ribonucleoprotein complex (40). This led to the identification of SEPSECS as the enzyme that catalyzes the conversion of O-phosphoserine-tRNA[Ser]Sec to Sec-tRNA[Ser]Sec, using selenophosphate as a donor substrate (81, 93) (Fig. 1).
Crystal structures of the archaeal and murine Sepsecs apo-enzymes as well as human wild type and mutant SEPSECS complexed with Sec-tRNA[Ser]Sec have been solved, suggesting that SEPSECS is a distinct member of the fold type I family of the pyridoxal phosphate-dependent enzyme family (7, 38, 68a, 73). The human structure shows a complex containing an SEPSECS tetramer binding two Sec-tRNA[Ser]Sec molecules through their long acceptor-TΨC arms, with the non-catalytic SEPSECS dimer mediating RNA–protein interactions that stabilize the complex and the CCA end of Ser-tRNA[Ser]Sec residing in the active site of the catalytic SEPSECS dimer (Figs. 3D and 4B). The conversion of Ser-tRNA[Ser]Sec to Sec-tRNA[Ser]Sec by SEPSECS is pyridoxal-5-phosphate (PLP) cofactor dependent, with the proposed mechanism involving a conformational change in SEPSECS on Ser-tRNA[Ser]Sec binding, enabling the phosphoserine of Ser-tRNA[Ser]Sec to be oriented correctly for conversion to occur (68a).
FIG. 4.
Genomic and structural organization of SEPSECS with the positions of the human mutations. (A) The organization of human SEPSECS gene (top) and schematic of SEPSECS protein (bottom) with the location of human mutations superimposed. Arrowheads denote the location of the ATG start codon. (B) Crystal structure showing a single catalytic–non-catalytic dimer from the complex bound to tRNA[Ser]Sec, with the position of all the human SEPSECS point mutations superimposed (71). Mutations associated with early onset (red) or late onset (yellow) disease and the pyridoxal-5-phosphate (cyan) substrate are highlighted. Color images are available online.
Homozygous and compound heterozygous mutations in SEPSECS (Table 4, Figs. 4, and 5) are associated with profound intellectual disability, global developmental delay, spasticity, epilepsy, and hypotonia with progressive microcephaly due to cortical and cerebellar atrophy on magnetic resonance imaging (1). Additional phenotypes described in other patients include axonal neuropathy, optic atrophy, and early onset epileptic encephalopathy with burst suppression (6, 67, 72). The timing of presentation in patients with SEPSECS mutations is variable, ranging from severe prenatal onset to delayed postnatal presentation and a mild, late onset phenotype in three patients (50, 86). This disorder is classified as autosomal recessive pontocerebellar hypoplasia type 2D (PCH2D, OMIM No. 613811), also known as progressive cerebellocerebral atrophy (PCCA) (1, 11). Mice homozygous for a SEPSECS mutation (Y334C) present with Sedaghatian-type spondylometaphyseal dysplasia and die shortly after birth, in contrast to humans with the same mutation. This divergence in phenotype may be due to species differences, dietary environment, or the genetic background of the mouse line used (36).
Table 4.
Human SEPSECS Mutations: Genetics and Effect
| Family | Gene mutation | Predicted protein change | Alleles affected | Suggested mechanism | Ethnicity | Phenotype | References |
|---|---|---|---|---|---|---|---|
| A, B | c.1001A>G | Y334C | Homozygous | Affects folding and reduced catalytic activity | Jewish/Iraqi | PCH2D | (1) |
| C, D | c.715G>A | A239T | Compound heterozygous | Affects folding and reduced catalytic activity | Iraqi/Moroccan | PCH2D | (1) |
| c.1001A>G | Y334C | ||||||
| E, F, G | c.974C>G | T325S | Compound heterozygous | Affects folding and reduced catalytic activity | Finnish | PCH2D | (6) |
| c.1287C>A | Y429X | Premature stop, no full-length protein | |||||
| H | c.1001A>C | Y334H | Homozygous | Predicted to affect folding and reduce catalytic activity | Arabian | PCH2D | (72) |
| I | c.77delG | R26Pfs*42 | Compound heterozygous | Premature stop, no full-length protein | Japanese | Late onset PCH2D | (50) |
| c.356A>G | N119S | Predicted reduced catalytic activity/RNA binding | |||||
| J | c.356A>G | N119S | Compound heterozygous | Predicted reduced catalytic activity/RNA binding | Japanese | Late onset PCH2D | (50) |
| c.467G>A | R156Q | Predicted reduced catalytic activity | |||||
| K | c.1A>G | M1V | Compound heterozygous | No ATG start, absence of full-length protein, | N/A | PCH2D | (97) |
| c.388 + 3A>G | G130Vfs*35 | premature stop, no full-length protein | |||||
| L | c.1466A>T | D489V | Homozygous | Likely pathogenic | Jordan | Developmental Delay/intellectual Disability | (61) |
| M | c.1027–1120del | E343Lfs*2 | Homozygous | Premature stop, no full-length protein | N/A | Neurodegenerative disease | (3) |
| N | c.176C>T | A59V | Homozygous | Predicted to affect folding, dimerization and reduce activity | N/A | EOEE-BS; PCH2D | (67) |
| O | c. 1321G>A | G441R | Homozygous | Predicted to reduce catalytic activity | N/A | Late onset PCH2D | (86) |
| P | N/A | N/A | N/A | N/A | N/A | PCH2D | (8) |
EOEE-BS, early onset epileptic encephalopathy with burst suppression; PCH2D, pontocerebellar hypoplasia type 2D.
The effect of SEPSECS mutations on selenoprotein expression has only been studied in four patients (Family E, G, F; 6). In brain tissue levels of TXNRD1, TXNRD2, GPX1, and GPX4 proteins are reduced, correlating with increased cellular oxidative stress. However, selenoprotein deficiency is not generalized with normal TXNRD levels in a patient's fibroblast and muscle cells, suggesting that residual SEPSECS activity preserves selenoprotein synthesis in some tissues or existence of alternative pathway(s). Consistent with findings in human tissues Sepsecs mutant mice exhibit decreased Gpx4 expression in neurons but not hepatocytes (36).
Although the selenium content of brain is not high (95), it is kept very stable (10, 66), exemplified by the fact that in systemic selenium deficiency circulating SELENOP delivers this trace element preferentially to this organ at the expense of other tissues (66). This may explain why a reduction in SEPSECS activity could have a greater effect on brain development and function compared with other tissues.
It is remarkable that an overt central nervous system (CNS) phenotype is not reported in patients with SECISBP2 and TRU-TCA1-1 defects, but with most individuals with these mutations being children the possibility of a late onset neurological phenotype cannot be discounted. However, a neurodegenerative phenotype has been described in brain-specific Gpx4, Trsp, and Secisbp2 KO mouse models, suggesting that Se deficiency and/or reduced redox capacity has a larger impact on the mouse brain compared with the human brain (78, 91, 92).
Biochemical hallmarks of selenoprotein deficiency in SECISBP2 and TRU-TCA1-1 mutation cases include low circulating selenium and abnormal thyroid hormone levels, reflecting deficiency of circulating selenoproteins (SEPP, GPx3) or all three deiodinase enzymes, respectively (31, 75). In SEPSECS mutation patients, low serum selenium is not reported to be part of the phenotype and thyroid status has only been partially investigated in four patients, documenting either normal thyroid hormone (Family H; 72) or normal T4 but elevated thyroid stimulating hormone (TSH) levels (Family E, G, F; 6). However, more detailed investigation, with measurement of T3 or reverse T3 levels, which would be abnormal with decreased deiodinase enzyme activity, has not been undertaken in SEPSECS mutation cases (6). Myopathic features with raised CK levels, abnormal mitochondria, cytoplasmic bodies, and increased lipid accumulation in muscle have been documented in one SEPSECS mutation case (Family H; 72), with broad-based gait and postural instability suggesting muscle weakness in another patient (Family O, 86). Similar findings have been noted in adult SECISBP2 mutation patients, reflecting deficiency of selenoprotein N and altered redox capacity in skeletal muscle (75). Overall, these observations suggest that some SEPSECS mutation patients can exhibit phenotypes associated with more global deficiency of selenoproteins. It is also possible that severity of neurological problems in patients has precluded detailed investigation and ascertainment of non-neurological phenotypes.
The availability of the crystal structure of human SEPSECS–tRNA[Ser]Sec (68a, 73) (Figs. 4 and 5), together with an SEPSECS activity assay using an Escherichia coli strain lacking endogenous Sec-synthase (SelA) activity (94), has enabled detailed studies of some SEPSECS (A239T, Y334C, T325S, and Y429X) mutations (1, 6, 73). These pathogenic variants were found to be less soluble than WT protein in vitro, with loss of functional activity. The A239T mutant SEPSECS failed to form stable tetramers, possibly as a result of a steric clash destabilizing two helices (H8–H9) within the enzyme's core. Y334C and T325S SEPSECS mutants are predicted to fold similar to wild-type SEPSECS in the crystal structure and retain binding to tRNA[Ser]Sec (73) but affect its catalytic pocket, reducing enzyme activity (1, 6, 73). Mutation of Tyr334 to Histidine is also recorded (6), and in silico analysis predicts that this variant is likely to have a similar deleterious effect as the Y334C mutation (Fig. 5). The premature stop SEPSECS mutant (Y429X) is insoluble and inactive (6, 73), and three other premature stop mutants (Table 4) can be expected to have a similar effect.
FIG. 5.
Detailed views comparing wild-type SEPSECS crystal structure and mutated amino acids modeled in the SEPSECS crystal structure that cause either early onset (red) or late onset (yellow) disease (71). Hydrogen bonds (dotted green lines), pyridoxal-5-phosphate substrate (cyan), tRNA (orange), and H2O (blue) are shown. The helices (H), beta-sheets (β), amino acids, and nucleotides involved in hydrogen bond networks or that are part of the active catalytic domain are labeled. Crystal structures for T325S and Y334C SEPSECS mutants are available and in the panels with these mutations an overlay of wild type (gray) and mutant (cyan) is shown (73). The effect of these mutations is described in detail in section SEPSECS in main text. The model was generated by using the phyre2 web portal, which predicts and analyzes protein structures based on homology/analogy recognition to solve protein crystal structures (51). The figures were generated with MacPyMOL Molecular Graphics System, Schrödinger, LLC. Color images are available online.
In silico analysis predicts that the A59V SEPSECS mutation results in steric hindrance destabilizing helix 2 and helix 3, possibly affecting its catalytic function and/or dimerization (Fig. 5). The N119S SEPSECS mutation is a conservative amino acid substitution with a probable small effect, weakening an H-bond network in the non-catalytic dimer and possibly affecting RNA interaction. Likewise, the mutation of Arg156 to Glutamine in SEPSECS is a conservative change, predicted to perturb local structure minimally and possibly reducing activity of the catalytic site (Fig. 5). The G441R SEPSECS mutation, situated close to the catalytic site, changes the side chain of this amino acid from small and neutral to large and polar, but as Gly441 is situated within a loop, its possible effect on catalytic activity might be limited. Absence of two SEPSECS mutations (M1V, D489V) from crystal structures precludes in silico analyses: M1V affects the first methionine in SEPSECS and unless an alternative start codon (e.g., position 61) is used, no protein will be generated; the D489V mutation changes the size and charge of this amino acid and can, therefore, be expected to have a major impact on protein function/stability.
Three SEPSECS mutation patients (patient I [R26Pfs*42–N119S], J [N119S–R156Q], and O [G441R]) presented with late onset PCH2D, with a progressive but milder degree of CNS atrophy (50, 86). In silico analyses suggest that these mutations have a less deleterious effect on SEPSECS function (Fig. 5), but it is also conceivable that environmental factors or patients' genetic background may have modulated their phenotype. Future studies need to investigate the relationship between mutations, their effect on SEPSECS protein function, and general expression of selenoproteins in different tissues and patient phenotypes in more detail.
Conclusions
SECISBP2, Sec-tRNA[Ser]Sec, and SEPSECS are essential components of the selenoprotein biosynthesis pathway. Unsurprisingly, in patients, harboring mutations in any of these genes, the expression of most members of the selenoproteome is affected, sometimes in a tissue-specific manner, resulting in a complex, multisystem phenotype. The combination of the nature of the gene defect, genetic/ethnic background of individuals, and environmental factors (e.g., selenium and/or iodine status) might also contribute to inter-individual differences in phenotypes. Further complexity is due to the fact that most patients harbor compound heterozygous mutations with monoallelic mutations in individuals having no reported phenotype. This makes it difficult to assess the effect of a particular mutation on the Sec-incorporation pathway. Although a substantial body of knowledge regarding the individual functions of SECISBP2, Sec-tRNA[Ser]Sec, and SEPSECS exists, we do not have a comprehensive understanding of the Sec-insertion pathway. Nevertheless, most observations in patients with gene defects accord with our current knowledge of this pathway. However, it is interesting that mutations in SECISBP2 and TRU-TCA1-1 present with similar phenotypes (growth retardation and myopathy together with abnormal thyroid function), whereas the dominant phenotype in SEPSECS mutation cases is PCCA.
In all patients, some phenotypes (e.g., photosensitivity, age-dependent hearing loss, and neurodegeneration) are clearly progressive, perhaps reflecting the absence of antioxidant selenoenzymes and resulting in cumulative oxidative damage to DNA, proteins, and membrane lipids and dysfunction of redox-dependent signaling pathways. Some phenotypes can clearly be linked to deficiency of specific selenoproteins (e.g., abnormal thyroid function and DIO1,2,3; low plasma Se and SELENOP, GPX3; azoospermia and SELENOV, GPX4, and TXRND3; myopathy; and SELENON).
However, the precise role of many selenoproteins in human biological processes is unknown, making the identification of causal links between altered expression of specific selenoproteins and human disease a particular challenge. Further, the role of individual selenoproteins needs to be analyzed in the context of a complex cellular biochemical environment, where antagonistic, additive, and synergistic effects can occur. Recent advances, including analyzing selenoprotein expression in mouse models, RNA ribosome profiling (96) in vitro technologies such as CRISPR-Cas9-VLP (88), dissection of SECIS and other functional RNA elements (22, 62) using luciferase-based reporter assays (84), and modeling using crystal structures (e.g., SEPSECS–Sec-tRNA[Ser]Sec, EEFSEC) (29), can help us better understand the complex Sec-insertion pathway in general and more specifically the effect of mutations in genes within this pathway and their consequences on selenoprotein expression. This knowledge will provide the essential basis for understanding the pathogenesis of human disease due to generalized or specific selenoprotein deficiencies and may enable the identification of therapies targeted at specific processes (e.g., oxidative stress) in which selenoproteins play a key role.
Abbreviations Used
- CK
creatine kinase
- CNS
central nervous system
- C-terminal
carboxy-terminal
- EEFSEC
Sec tRNA specific eukaryotic elongation factor
- FT3
free triiodothyronine
- FT4
free thyroxine
- KO
knockout
- mcm5U
5-methoxycarbonyl-methyluridine
- mcm5Um
5-methoxycarbonylmethyl-2′-O-methyluridine
- mRNA
messenger RNA
- NMD
nonsense-mediated decay
- N-terminal
amino-terminal
- PCCA
progressive cerebellocerebral atrophy
- PCH2D
pontocerebellar hypoplasia type 2D
- RBD
RNA-binding domain
- ROS
reactive oxygen species
- RPL30
ribosomal protein L30
- Se
selenium
- Sec
selenocysteine
- SECIS
SEleniumCysteine Insertion Sequence
- SECISBP2
SECIS binding protein 2
- SELENOP
selenoprotein P
- SEPSECS
O-phosphoserine tRNA:Sec tRNA synthase
- SID
Sec incorporation domain
- T3
triiodothyronine
- T4
thyroxine
- tRNA
transfer RNA
- UTR
untranslated region
Funding Information
E.S. and K.C. are supported by the Wellcome Trust (210755/Z/18/Z), and K.C. is supported by the National Institute of Health Research Cambridge Biomedical Centre.
References
- 1. Agamy O, Ben Zeev B, Lev D, Marcus B, Fine D, Su D, Narkis G, Ofir R, Hoffmann C, Leshinsky-Silver E, Flusser H, Sivan S, Söll D, Lerman-Sagie T, and Birk OS. Mutations disrupting selenocysteine formation cause progressive cerebello-cerebral atrophy. Am J Hum Genet 87: 538–544, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahmed MY, Al-Khayat A, Al-Murshedi F, Al-Futaisi A, Chioza BA, Pedro Fernandez-Murray J, Self JE, Salter CG, Harlalka GV, Rawlins LE, Al-Zuhaibi S, Al-Azri F, Al-Rashdi F, Cazenave-Gassiot A, Wenk MR, Al-Salmi F, Patton MA, Silver DL, Baple EL, McMaster CR, and Crosby AH. A mutation of EPT1 (SELENOI) underlies a new disorder of Kennedy pathway phospholipid biosynthesis. Brain 140: 547–554, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA, Faqeih E, Alhashem A, Bashiri FA, Al-Owain M, Kentab AY, Sogaty S, Al Tala S, Temsah MH, Tulbah M, Aljelaify RF, Alshahwan SA, Seidahmed MZ, Alhadid AA, Aldhalaan H, AlQallaf F, Kurdi W, Alfadhel M, Babay Z, Alsogheer M, Kaya N, Al-Hassnan ZN, Abdel-Salam GM, Al-Sannaa N, Al Mutairi F, El Khashab HY, Bohlega S, Jia X, Nguyen HC, Hammami R, Adly N, Mohamed JY, Abdulwahab F, Ibrahim N, Naim EA, Al-Younes B, Meyer BF, Hashem M, Shaheen R, Xiong Y, Abouelhoda M, Aldeeri AA, Monies DM, and Alkuraya FS. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep 10: 148–161, 2015 [DOI] [PubMed] [Google Scholar]
- 4. Allmang C, Carbon P, and Krol A. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA 8: 1308–1318, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Allmang C and Krol A. Selenoprotein synthesis: UGA does not end the story. Biochimie 88: 1561–1571, 2006 [DOI] [PubMed] [Google Scholar]
- 6. Anttonen AK, Hilander T, Linnankivi T, Isohanni P, French RL, Liu Y, Simonović M, Söll D, Somer M, Muth-Pawlak D, Corthals GL, Laari A, Ylikallio E, Lähde M, Valanne L, Lönnqvist T, Pihko H, Paetau A, Lehesjoki AE, Suomalainen A, and Tyynismaa H. Selenoprotein biosynthesis defect causes progressive encephalopathy with elevated lactate. Neurology 85: 306–315, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Araiso Y, Palioura S, Ishitani R, Sherrer RL, O'Donoghue P, Yuan J, Oshikane H, Domae N, Defranco J, Söll D, and Nureki O. Structural insights into RNA-dependent eukaryal and archaeal selenocysteine formation. Nucleic Acids Res 36: 1187–1199, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arrudi-Moreno M, Fernández-Gómez A, and Peña-Segura JL. A new mutation in the SEPSECS gene related to pontocerebellar hypoplasia type 2D. Med Clin (Barc) 7753: 30592–30595, 2019 [DOI] [PubMed] [Google Scholar]
- 9. Azevedo MF, Barra GB, Naves LA, Ribeiro Velasco LF, Godoy Garcia Castro P, de Castro LC, Amato AA, Miniard A, Driscoll D, Schomburg L, and de Assis Rocha Neves F. Selenoprotein-related disease in a young girl caused by nonsense mutations in the SBP2 gene. J Clin Endocrinol Metab 95: 4066–4071, 2010 [DOI] [PubMed] [Google Scholar]
- 10. Behne D, Hilmert H, Scheid S, Gessner H, and Elger W. Evidence for specific selenium target tissues and new biologically important selenoproteins. Biochim Biophys Acta 966: 12–21, 1988 [DOI] [PubMed] [Google Scholar]
- 11. Ben-Zeev B, Hoffman C, Lev D, Watemberg N, Malinger G, Brand N, and Lerman-Sagie T. Progressive cerebellocerebral atrophy: a new syndrome with microcephaly, mental retardation, and spastic quadriplegia. J Med Genet 40: e96, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berry M, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, and Larsen PR. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′-untranslated region. Nature 353: 273–276, 1991 [DOI] [PubMed] [Google Scholar]
- 13. Berry M, Banu L, Harney J, and Larsen P. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J 12: 3315–3322, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bubenik JL and Driscoll DM. Altered RNA binding activity underlies abnormal thyroid hormone metabolism linked to a mutation in selenocysteine insertion sequence-binding protein 2. J Biol Chem 282: 34653–34662, 2007 [DOI] [PubMed] [Google Scholar]
- 15. Bubenik J, Miniard A, and Driscoll DM. Alternative transcripts and 3′UTR elements govern the incorporation of Selenocysteine into Selenoprotein S. PLoS One 8: e62102, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Caban K and Copeland PR. Size matters: a view of selenocysteine incorporation from the ribosome. Cell Mol Life Sci 63: 73–81, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Caban K, Kinzy SA, and Copeland PR. The L7Ae RNA binding motif is a multifunctional domain required for the ribosome-dependent Sec incorporation activity of Sec insertion sequence binding protein 2. Mol Cell Biol 27: 6350–6360, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carbon P and Krol A. Transcription of the Xenopus laevis selenocysteine tRNA(Ser)Sec gene: a system that combines an internal B box and upstream elements also found in U6 snRNA genes. EMBO J 10: 599–606, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carlson BA, Yoo M, Tsuji PA, Gladyshev VM, and Hatfield DL. Mouse models targeting selenocysteine tRNA expression for elucidating the role of selenoproteins in health and development. Molecules 14: 3509–3527, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Çatli G, Fujisawa H, Kirbiyik Ö, Mimoto MS, Gençpinar P, Özdemir TR, Dündar BN, and Dumitrescu AM. A novel homozygous selenocysteine insertion sequence binding protein 2 (SECISBP2, SBP2) gene mutation in a Turkish boy. Thyroid 28: 1221–1223, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chapple CE, Guigó R, and Krol A. SECISaln, a web-based tool for the creation of structure-based alignments of eukaryotic SECIS elements. Bioinformatics 25: 674–675, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cockman EM, Narayan V, Willard B, Shetty SP, Copeland PR, and Driscoll DM. Identification of the selenoprotein S positive UGA recoding (SPUR) element and its position dependent activity. RNA Biol 21: 1–15, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Copeland P and Driscoll D. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J Biol Chem 274: 25447–25454, 1999 [DOI] [PubMed] [Google Scholar]
- 24. Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, and Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J 19: 306–314, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Copeland PR, Stepanik VA, and Driscoll DM. Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol Cell Biol 21: 1491–1498, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Jesus LA, Hoffmann PR, Michaud T, Forry EP, Small-Howard A, Stillwell RJ, Morozova N, Harney JW, and Berry MJ. Nuclear assembly of UGA decoding complexes on selenoprotein mRNAs: a mechanism for eluding nonsense-mediated decay? Mol Cell Biol 26: 1795–1805, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Diamond AM, Choi IS, Crain PF, Hashizume T, Pomerantz SC, Cruz R, Steer CJ, Hill KE, Burk RF, McCloskey JA, and Hatfield DL. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec). J Biol Chem 268: 14215–14223, 1993 [PubMed] [Google Scholar]
- 28. Di Cosmo C, McLellan N, Liao XH, Khanna KK, Weiss RE, Papp L, and Refetoff S. Clinical and molecular characterization of a novel selenocysteine insertion sequence-binding protein 2 (SBP2) gene mutation (R128X). J Clin Endocrinol Metab 94: 4003–4009, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dobosz-Bartoszek M, Pinkerton MH, Otwinowski Z, Chakravarthy S, Söll D, Copeland PR, and Simonović M. Crystal structures of the human elongation factor eEFSec suggest a non-canonical mechanism for selenocysteine incorporation. Nat Commun 7: 12941, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Donovan J, Caban K, Ranaweera R, Gonzalez-Flores JN, and Copeland PR. A novel protein domain induces high affinity selenocysteine insertion sequence binding and elongation factor recruitment. J Biol Chem 283: 35129–35139, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dumitrescu AM, Liao XH, Abdullah MS, Lado-Abeal J, Majed FA, Moeller LC, Boran G, Schomburg L, Weiss RE, and Refetoff S. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat Genet 37: 1247–1252, 2005 [DOI] [PubMed] [Google Scholar]
- 32. Dumitrescu AM and Refetoff S. The syndromes of reduced sensitivity to thyroid hormone. Biochim Biophys Acta 1830: 3987–4003, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, and Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J 19: 4796–4805, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fletcher JE, Copeland PR, Driscoll DM, and Krol A. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA 7: 1442–1453, 2001 [PMC free article] [PubMed] [Google Scholar]
- 35. Fradejas-Villar N. Consequences of mutations and inborn errors of selenoprotein biosynthesis and functions. Free Radic Biol Med 1: 206–214, 2018 [DOI] [PubMed] [Google Scholar]
- 36. Fradejas-Villar N, Reuter U, Stein V, and Schweizer U. Mutated selenocysteine synthase creates a Sedaghatian-type spondylometaphyseal dysplasia mouse model. Abstract book O75 page 76, the 11th International Symposium in Biology and Medicine Se2017, Stockholm, August 1 3–17, 2017 [Google Scholar]
- 37. Fu J, Liao X, Menucci MB, Dumitrescu AM, and Weiss RE. Thyroid hormone metabolism defect caused by novel compound heterozygous mutations in the SBP2 gene. 6th International Congress of Endocrinol Abstract 0508, 2014
- 38. Ganichkin OM, Xu XM, Carlson BA, Mix H, Hatfield DL, Gladyshev VN, and Wahl MC. Structure and catalytic mechanism of eukaryotic selenocysteine synthase. J Biol Chem 283: 5849–5865, 2008 [DOI] [PubMed] [Google Scholar]
- 39. Geiduschek EP and Tocchini-Valentini GP. Transcription by RNA polymerase III. Annu Rev Biochem 57: 873–914, 1988 [DOI] [PubMed] [Google Scholar]
- 40. Gelpi C, Sontheimer EJ, and Rodriguez-Sanchez JL. Autoantibodies against a serine tRNA-protein complex implicated in cotranslational selenocysteine insertion. Proc Natl Acad Sci U S A 89: 9739–9743, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grundner-Culemann E, Martin GW 3rd, Harney JW, and Berry MJ. Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA 5: 625–635, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gupta N, Demong LW, Banda S, and Copeland PR. Reconstitution of selenocysteine incorporation reveals intrinsic regulation by SECIS elements. J Mol Biol 425: 2415–2422, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hamajima T, Mushimoto Y, Kobayashi H, Saito Y, and Onigata K. Novel compound heterozygous mutations in the SBP2 gene: characteristic clinical manifestations and the implications of GH and triiodothyronine in longitudinal bone growth and maturation. Eur J Endocrinol 166: 757–764, 2012 [DOI] [PubMed] [Google Scholar]
- 44. Hatfield D, Gladyshev V, Park JM, Park SI, Chittum H, Huh J, Carlson B, Kim M, Moustafa M, and Lee BJ. Biosynthesis of selenocysteine and its incorporation into protein as the 21st amino acid. In: Comprehensive Natural Products Chemistry, edited by Kelly JW. Oxford, UK: Elsevier Science, 1999, Vol. 4, pp. 353–380 [Google Scholar]
- 45. Hatfield D, Lee BJ, Hampton L, and Diamond AM. Selenium induces changes in the selenocysteine tRNA[Ser]Sec population in mammalian cells. Nucleic Acids Res 19: 939–943, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Horibata Y, Elpeleg O, Eran A, Hirabayashi Y, Savitzki D, Tal G, Mandel H, and Sugimoto H. EPT1 (selenoprotein I) is critical for the neural development and maintenance of plasmalogen in humans. J Lipid Res 59: 1015–1026, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Howard MT, Aggarwal G, Anderson CB, Khatri S, Flanigan KM, and Atkins JF. Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. EMBO J 24: 1596–1607, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Howard MT, Moyle MW, Aggarwal G, Carlson BA, and Anderson CB. A recoding element that stimulates decoding of UGA codons by Sec tRNA[Ser]Sec. RNA 13: 912–920, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Howard MT and Copeland PR. New directions for understanding the codon redefinition required for selenocysteine incorporation. Biol Trace Elem Res 192: 18–25, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Iwama K, Sasaki M, Hirabayashi S, Ohba C, Iwabuchi E, Miyatake S, Nakashima M, Miyake N, Ito S, Saitsu H, and Matsumoto N. Milder progressive cerebellar atrophy caused by biallelic SEPSECS mutations. J Hum Genet 61: 527–531, 2016 [DOI] [PubMed] [Google Scholar]
- 51. Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kernebeck T, Lohse AW, and Grötzinger. A bioinformatical approach suggests the function of the autoimmune hepatitis target antigen soluble liver antigen/liver pancreas. J Hepatol 34: 230–233, 2001 [DOI] [PubMed] [Google Scholar]
- 53. Kim JY, Carlson BA, Xu XM, Zeng Y, Chen S, Gladyshev VN, Lee BJ, and Hatfield DL. Inhibition of selenocysteine tRNA[Ser]Sec amino acylation provides evidence that amino acylation is required for regulatory methylation of this tRNA. Biochem Biophys Res Commum 409: 814–819, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim LK, Matsufuji T, Matsufuji S, Carlson BA, Kim SS, Hatfield DL, and Lee BJ. Methylation of the ribosyl moiety at position 34 of selenocysteine tRNA[Ser]Sec is governed by both primary and tertiary structure. RNA 6: 1306, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Korwutthikulrangsri M, Raimondi C, and Dumitrescu AM.. Novel compound heterozygous SBP2 gene mutations in a boy with developmental delay and failure to thrive. 13th IWRTH Doorn, the Netherlands abstract book p22, 2018
- 56. Kossinova O, Malygin A, Krol A, and Karpova G. The SBP2 protein central to selenoprotein synthesis contacts the human ribosome at expansion segment 7L of the 28S rRNA. RNA 20: 1046–1056, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Labunskyy VM, Hatfield D L, and Gladyshev VN. Selenoproteins: molecular pathways and physiological roles. Physiol Rev 94: 739–777, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee BJ, Worland PJ, Davis JN, Stadtman TC, and Hatfield DL. Identification of a selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon, UGA. J Biol Chem 264: 9724–9727, 1989 [PubMed] [Google Scholar]
- 59. Lescure A, Fagegaltier D, Carbon P, and Krol A. Protein factors mediating selenoprotein synthesis. Curr Protein Pept Sci 3: 143–151, 2002 [DOI] [PubMed] [Google Scholar]
- 60. Low SC, Grundner-Culemann E, Harney JW, and Berry MJ. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J 19: 6882–6890, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Makrythanasis P, Nelis M, Santoni FA, Guipponi M, Vannier A, Béna F, Gimelli S, Stathaki E, Temtamy S, Mégarbané A, Masri A, Aglan MS, Zaki MS, Bottani A, Fokstuen S, Gwanmesia L, Aliferis K, Bustamante Eduardo M, Stamoulis G, Psoni S, Kitsiou-Tzeli S, Fryssira H, Kanavakis E, Al-Allawi N, Sefiani A, Al Hait S, Elalaoui SC, Jalkh N, Al-Gazali L, Al-Jasmi F, Bouhamed HC, Abdalla E, Cooper DN, Hamamy H, and Antonarakis SE. Diagnostic exome sequencing to elucidate the genetic basis of likely recessive disorders in consanguineous families. Hum Mutat 36: 1203–1210, 2014 [DOI] [PubMed] [Google Scholar]
- 62. Mariotti M, Shetty S, Baird L, Wu S, Loughran G, Copeland PR, Atkins JF, and Howard MT. Multiple RNA structures affect translation initiation and UGA redefinition efficiency during synthesis of selenoprotein P. Nucleic Acids Res 45: 13004–13015, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mehta A, Rebsch CM, Kinzy SA, Fletcher JE, and Copeland PR. Efficiency of mammalian selenocysteine incorporation. J Biol Chem 279: 37852–37859, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Myslinski E, Krol A, and Carbon P. Optimal tRNA(Ser)SeC gene activity requires an upstream SPH motif. Nucleic Acids Res 20: 203–209, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Myslinski E, Schuster C, Huet J, Sentenac A, Krol A, and Carbon P. Point mutations 5′ to the tRNA selenocysteine TATA box alter RNA polymerase III transcription by affecting the binding of TBP. Nucleic Acids Res 21: 5852–5858, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nakayama A, Hill KE, Austin LM., Motley AK, and Burk RF.. All regions of mouse brain are dependent on selenoprotein P for maintenance of selenium. J Nutr 137: 690–693, 2007 [DOI] [PubMed] [Google Scholar]
- 67. Olson HE, Kelly M, LaCoursiere CM, Pinsky R, Tambunan D, Shain C, Ramgopal S, Takeoka M, Libenson MH, Julich K, Loddenkemper T, Marsh ED, Segal D, Koh S, Salman MS, Paciorkowski AR, Yang E, Bergin AM, Sheidley BR, and Poduri A. Genetics and genotype-phenotype correlations in early onset epileptic encephalopathy with burst suppression. Ann Neurol 81: 419–429, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. O'Neill VA, Eden FC, Pratt K, and Hatfield DL. A human opal suppressor tRNA gene and pseudogene. J Biol Chem 260: 2501–2508, 1985 [PubMed] [Google Scholar]
- 68a. Palioura S, Sherrer RL, Steitz TA, Söll D, and Simonovic M. The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science 325: 321–325, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Papp LV, Lu J, Holmgren A, and Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid Redox Signal 9: 775–806, 2007 [DOI] [PubMed] [Google Scholar]
- 70. Papp LV, Wang J, Kennedy D, Boucher D, Zhang Y, Gladyshev VN, Singh RN, and Khanna KK. Functional characterization of alternatively spliced human SECISBP2 transcript variants. Nucleic Acids Res 36: 7192–7206, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. This reference has been deleted
- 72. Pavlidou E, Salpietro V, Phadke R, Hargreaves IP, Batten L, McElreavy K, Pitt M, Mankad K, Wilson C, Cutrupi MC, Ruggieri M, McCormick D, Saggar A, and Kinali M. Pontocerebellar hypoplasia type 2D and optic nerve atrophy further expand the spectrum associated with selenoprotein biosynthesis deficiency. Eur J Paediatr Neurol 20: 483–488, 2016 [DOI] [PubMed] [Google Scholar]
- 73. Puppala AK, French RL, Matthies D, Baxa U, Subramaniam S, and Simonović M. Structural basis for early-onset neurological disorders caused by mutations in human selenocysteine synthase. Sci Rep 6: 32563, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Saito Y, Shichiri M, Hamajima T, Ishida N, Mita Y, Nakao S, Hagihara Y, Yoshida Y, Takahashi K, Niki E, and Noguchi N. Enhancement of lipid peroxidation and its amelioration by vitamin E in a subject with mutations in the SBP2 gene. Lipid Res 56: 2172–2182, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schoenmakers E, Agostini M, Mitchell C, Schoenmakers N, Papp L, Rajanayagam O, Padidela R, Ceron-Gutierrez L, Doffinger R, Prevosto C, Luan J, Montano S, Lu J, Castanet M, Clemons N, Groeneveld M, Castets P, Karbaschi M, Aitken S, Dixon A, Williams J, Campi I, Blount M, Burton H, Muntoni F, O'Donovan D, Dean A, Warren A, Brierley C, Baguley D, Guicheney P, Fitzgerald R, Coles A, Gaston H, Todd P, Holmgren A, Khanna KK, Cooke M, Semple R, Halsall D, Wareham N, Schwabe J, Grasso L, Beck-Peccoz P, Ogunko A, Dattani M, Gurnell M, and Chatterjee K. Mutations in the selenocysteine insertion sequence-binding protein 2 gene lead to a multisystem selenoprotein deficiency disorder in humans. J Clin Invest 120: 4220–4235, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schoenmakers E, Carlson B, Agostini M, Moran C, Rajanayagam O, Bochukova E, Tobe R, Peat R, Gevers E, Muntoni F, Guicheney P, Schoenmakers N, Farooqi S, Lyons G, Hatfield D, and Chatterjee K. Mutation in human selenocysteine transfer RNA selectively disrupts selenoprotein synthesis. J Clin Invest 126: 992–996, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schomburg L, Dumitrescu AM, Liao XH, Bin-Abbas B, Hoeflich J, Köhrle J, and Refetoff S. Selenium supplementation fails to correct the selenoprotein synthesis defect in subjects with SBP2 gene mutations. Thyroid 19: 277–281, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Seeher S, Atassi T, Mahdi Y, Carlson BA, Braun D, Wirth EK, Klein MO, Reix N, Miniard AC, Schomburg L, Hatfield DL, Driscoll DM, and Schweizer U. Secisbp2 is essential for embryonic development and enhances selenoprotein expression. Antioxid Redox Signal 21: 835–849, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shetty SP and Copeland PR. Selenocysteine incorporation: a trump card in the game of mRNA decay. Biochemie 114: 97–101, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shrimali RK, Irons RD, Carlson BA, Sano Y, Gladyshev VN, Park JM, and Hatfield DL. Selenoproteins mediate T cell immunity through an antioxidant mechanism. J Biol Chem 283: 20181–20185, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, Harney JW, Carlson BA, Xu XM, Hatfield DL, and Berry MJ. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol Cell Biol 26: 2337–2346, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Suppmann S, Persson BC, and Böck A. Dynamics and efficiency in vivo of UGA directed selenocysteine insertion at the ribosome. EMBO J 18: 2284–2293, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Takeuchi A, Schmitt D, Chapple C, Babaylova E, Karpova G, Guigo R, Krol A, and Allmang C. A short motif in Drosophila SECIS binding protein 2 provides differential binding affinity to SECIS RNA hairpins. Nucleic Acids Res 37: 2126–2141, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Touat-Hamici Z, Bulteau AL, Bianga J, Jean-Jacques H, Szpunar J, Lobinski R, and Chavatte L. Selenium-regulated hierarchy of human selenoproteome in cancerous and immortalized cells lines. Biochim Biophys Acta Gen Subj 1862: 2493–2505, 2018 [DOI] [PubMed] [Google Scholar]
- 85. Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, and Berry MJ. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep 1: 158–163, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. van Dijk T, Vermeij JD, van Koningsbruggen S, Lakeman P, Baas F, and Poll-The BT. A SEPSECS mutation in a 23-year-old woman with microcephaly and progressive cerebellar ataxia. J Inherit Metab Dis 41: 897–898, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, and Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell 6: 1331–1342, 2000 [DOI] [PubMed] [Google Scholar]
- 88. Vindry C, Guillin O, Mangeot PE, Ohlmann T, and Chavatte L. A versatile strategy to reduce UGA-selenocysteine recoding efficiency of the ribosome using CRISPR-Cas9-viral-like-particles targeting selenocysteine-tRNA[Ser]Sec gene. Cells 8: 574, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Walczak R, Carbon P, and Krol A. An essential non-Watson-Crick base pair motif in 3′UTR to mediate selenoprotein translation. RNA 4: 74–84, 1998 [PMC free article] [PubMed] [Google Scholar]
- 90. Walczak R, Westhof E, Carbon P, and Krol A. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA 2: 367–379, 1996 [PMC free article] [PubMed] [Google Scholar]
- 91. Wirth EK, Bharathi BS, Hatfield D, Conrad M, Brielmeier M, and Schweizer U. Cerebellar hypoplasia in mice lacking selenoprotein biosynthesis in neurons. Biol Trace Elem Res 158: 203–210, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wirth EK, Conrad M, Winterer J, Wozny C, Carlson BA, Roth S, Schmitz D, Bornkamm GW, Coppola V, Tessarollo L, Schomburg L, Köhrle J, Hatfield DL, and Schweizer U. Neuronal selenoprotein expression is required for interneuron development and prevents seizures and neurodegeneration. FASEB J 24: 844–852, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Xu XM, Mix H, Carlson BA, Grabowski PJ, Gladyshev VN, Berry MJ, and Hatfield DL. Evidence for direct roles of two additional factors, SECp43 and soluble liver antigen, in the selenoprotein synthesis machinery. J Biol Chem 280: 41568–41575, 2005 [DOI] [PubMed] [Google Scholar]
- 94. Yuan J, Palioura S, Salazar JC, Su D, O'Donoghue P, Hohn MJ, Cardoso AM, Whitman WB, and Söll D. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc Natl Acad Sci U S A 103: 18923–18927, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zachara BA, Pawluk H, Bloch-Boguslawska E, Sliwka KM, Korenkiewicz J, Skok Z, and Ryc K. Tissue level, distribution, and total body selenium content in healthy and diseased humans in Poland. Arch Environ Health 56: 461–466, 2001 [DOI] [PubMed] [Google Scholar]
- 96. Zhao W, Bohleber S, Schmidt H, Seeher S, Howard MT, Braun D, Arndt S, Reuter U, Wende H, Birchmeier C, Fradejas-Villar N, and Schweizer U. Ribosome profiling of selenoproteins in vivo reveals consequences of pathogenic Secisbp2 missense mutations. J Biol Chem 294: 14185–14200, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhu X, Petrovski S, Xie P, Ruzzo EK, Lu YF, McSweeney KM, Ben-Zeev B, Nissenkorn A, Anikster Y, Oz-Levi D, Dhindsa RS, Hitomi Y, Schoch K, Spillmann RC, Heimer G, Marek-Yagel D, Tzadok M, Han Y, Worley G, Goldstein J, Jiang YH, Lancet D, Pras E, Shashi V, McHale D, Need AC, and Goldstein DB. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med 17: 774–781, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]