Abstract
After serving the Dictyostelium community for many years, the first version of dictyBase (Chisholm et al. 2006; Fey et al. 2006) was in need of a decisive update. The original dictyBase software was not adaptable to more current demands such as handling the import of large-scale data from recently sequenced genomes, keeping up with changes in the Gene Ontology (GO), or handling the automatic annotation of over 20,000 new strains. Therefore, we have embarked on a complete overhaul of dictyBase. The new infrastructure will allow the introduction of new data, such as more expressive GO annotations and Dictyostelium disease orthologs. A modern user interface aims to streamline usage of the database including orders from the Dicty Stock Center (DSC). New displays will allow novel views including the combination of data in two new tools. With the underlying software infrastructure now in place, dictyBase software engineers and curators are currently adding the user interfaces, new tools and content pages for the evolving Version 2 of dictyBase. This review highlights the emerging status of the new dictyBase, updated pages and annotations that will soon be available in the new environment, an overview of our annotation procedures, and plans to involve the community in curation efforts.
Keywords: dictyBase, Dicty Stock Center, Dictyostelium, overhaul, update
Introduction
dictyBase, the model organism database for Dictyostelium discoideum and related species, has provided the central Dictyostelium web portal for researchers, teachers and students since 2002. The DSC, a resource of Dictyostelium strains and plasmids, relocated to Northwestern University in 2009 and is now tightly integrated with dictyBase. (Fey et al. 2013). Because the first version of dictyBase software was not expandable and adaptable to house additional new large-scale data available from researchers, we have now created a completely new database housed in a cloud environment. The update includes building a modern web interface using the JavaScript library React, first created by Facebook Inc. Our new software environment is easily scalable to host numerous genomes of new species and to incorporate large novel datasets provided by the research community. We have already created an efficient user interface to allow dictyBase curators to directly add and edit web page content by logging into the HTML5 websites. This will also allow interested users to participate in certain annotations in the future. Furthermore, the new dictyBase websites adapt to any screen size and thus may be used on all mobile devices (see Figure 6).
Fig. 6. New DSC frontpage in mobile view.
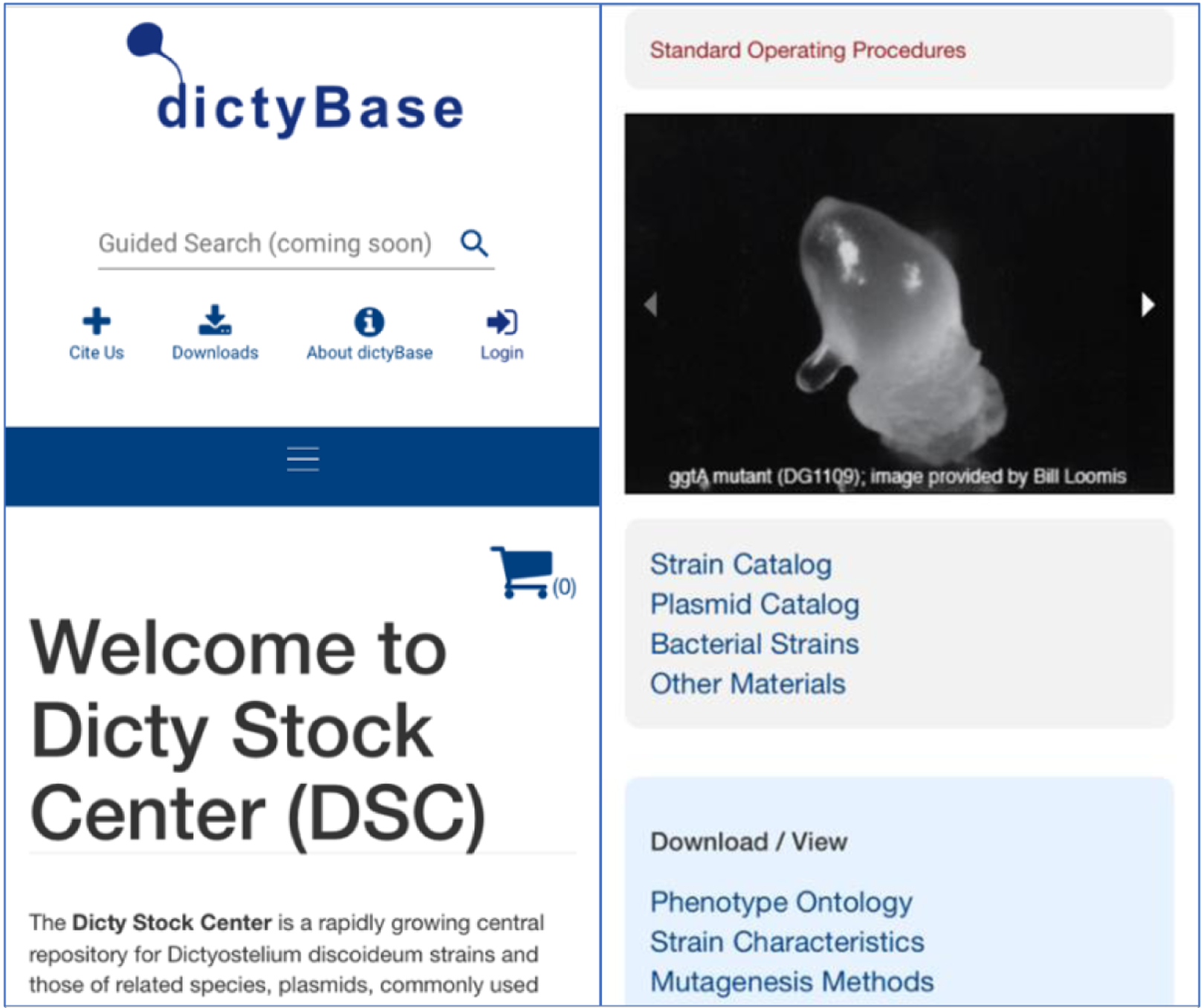
The new DSC hub, shown by two screen shots of partial page views when scrolling down on a mobile phone. Mobile users will be soon able to view and search for materials, and place orders via their device in an intuitive way. The left panel shows the top ofthe page while the right panel is a view near the bottom. Note the link to the SOPs on the upper right (red).
dictyBase V2 will house and display new data, for example the annotation extensions added as context to GO terms and disease ortholog annotations. At the time of writing, we are internally testing and creating user interfaces for the new database, expecting a first public beta release later in 2019. Finally, dictyBase and the DSC also subscribe to the FAIR sharing principle, standing for findability, accessibility, interoperability, and reusability. dictyBase is public and free to use, annotations are available for download, and the new code may be used to create another database while our software endpoint also allows the extraction of data. All new dictyBase code is open source and available on GitHub.
The sections below will provide a general description of the current organization of dictyBase and highlight several emerging interfaces and data displays. The focus here is to inform dictyBase and DSC users about new and upcoming data displays, annotations and tools. Technical details will be later described in a separate publication. dictyBase and DSC V2 is a work in progress (Basu et al. 2015) and continuous updates will import additional data as well as provide new displays and downloads in timely future releases.
1. dictyBase V2, evolving
The new dictyBase front page has an updated web technology and look. It retains some features of the original such as a top toolbar with pulldown menus which provide quick access to various features of interest to dictyBase users. Also as before, recent Dictyostelium publications are displayed. Several new features on the front page include buttons that serve as quick links to newer items, currently including access to updated tools (see section 2) and an example of a gene page that is under development. Soon recently curated genes and the latest available DSC items will be updated automatically. The update also improves maintenance: for example, dictyNews are now tweeted only and automatically appear on our front page in the upper right corner. Thus, the dynamic new page will offer regular updates and simplify maintenance. The new dictyBase search is not yet functional; however, many pages from the top bar now have updated web content. Furthermore, the top bar links to the new DSC front page, which is the gateway for all Stock Center content (see section 3).
dictyBase primarily houses the manually annotated Dictyostelium discoideum genome, which consists of over 13,000 genes including over 600 annotated pseudogenes (Fey et al 2009), and nearly 500 identified ncRNAs (Sucgang et al. 2003; Aspegren et al. 2004). Each gene has an individual gene summary page that contains all currently available gene and gene product information, as well as diverse annotations and links to other internal and external resources. The new gene page will be added section by section as they are completed by our software engineers. At the time of writing, the publicly available example gene page displays the updated GO section, illustrated in Figure 1 below.
Fig. 1. Biological Process GO annotation with extensions.
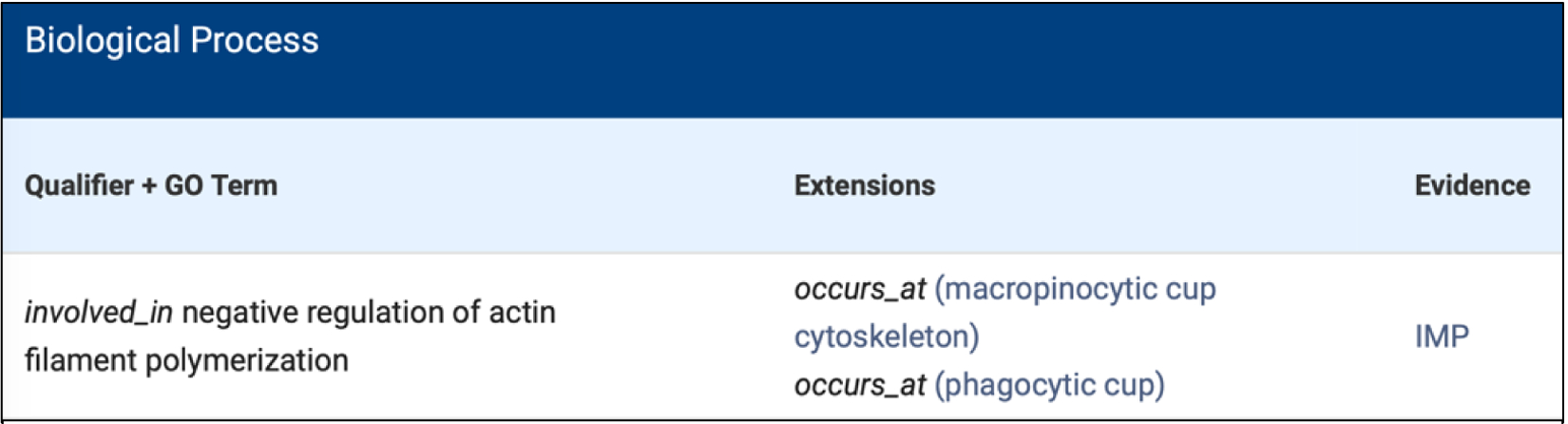
Displayed is one annotation for the GflB rap guanine nucleotide exchange factor. The relations are indicated in italic and with underscores between words, whereas the GO terms are currently linked to the individual term including the definition and ancestor chart in QuickGO (Huntley et al. 2009). Here, GflB is involved_in the negative regulation of actin filament polymerization which occurs_at the macropinocytic cup cytoskeleton and the phagocytic cup. The annotation is inferred from mutant phenotype (IMP).
1.a. General Gene Page overview
The gene page is designed to summarize available annotations for each gene. Each gene page displays information separated into sections that contain genomic and functional annotations. Table 1 lists the major sections of dictyBase gene pages. Availability of permanent sections and those only present when populated with available data (conditional availability) are indicated in column 3. When a section contains manual annotations by dictyBase curators it is indicated in column 4. Sections with increasing annotations such as GO, or Strains and Phenotypes, link out to their specific detailed pages in dictyBase where all data is accessible (see also sections 1.b. and 1.c.). Also, the references link to their own interface and here papers annotated by curators are linked to all genes investigated in that reference, which may be sorted by categories. In addition, we describe below in section 1.d two new additions to the gene page: spatial expression data and human disease gene orthologs.
Table 1.
Major sections of the dictyBase gene page.
| Section Name | Section description | Conditional Availability |
Manual Annotations |
|---|---|---|---|
| General Information | Gene / product names and description | no | yes |
| Genomic Information | Chromosomal info and genomic snapshot | no | partial |
| Gene Product Info | Protein info, curation status, sequences | no | yes |
| Associated Sequences | EST sequences and GenBank entries | yes | no |
| Gene Ontology | GO annotations with highest support | yes | yes |
| Strains & Phenotypes | Strain and phenotype annotations | yes | yes |
| Links | External and internal links | no | partial |
| Summary | Summary of functional data when known | yes | yes |
| Latest References | Newest 5 references describing the gene | yes | no |
1.b. Gene Ontology
The GO is a resource designed to describe the function, biological process and cellular location of gene products through a controlled vocabulary of terms. GO consortium members are trained annotators who meet and communicate regularly to reach a consensus on annotation consistency and develop the ontology further across model organisms and other databases using GO (The Gene Ontology Consortium 2018). This concerted effort produces expert functional GO annotations, manually derived from published literature. These annotations are recognized by their experimental evidence code (e.g. IDA, IMP; see Table 2) and by their link to the reference from which the experimental annotation has been derived. We display the original and easily recognizable 3-letter code for evidence; however internally we store the detailed evidence and conclusion ontology (ECO; Giglio et al. 2019) as there is a direct translation between the two. The experimental annotations provide the ‘gold standard’ for further initiatives such as the phylogenetic annotation project (evidence IBA) in which curators perform annotation inferences across evolutionary related proteins based on the known function of certain proteins within PANTHER families (Gaudet et al. 2011; Mi et al. 2019). In addition, electronic annotations (IEA) are provided by groups such as InterPro and UniProtKB (Camon et al. 2003; Burge et al. 2012). Thus, conserved D. discoideum genes lacking published experimental results, as well as sequenced, but not manually annotated organisms such as other Dictyostelids still receive useful functional information from these GO initiatives. These semi-manual and electronic initiatives continuously improve through the growing gold standard annotations by experts in the field. At the time of this writing, D. discoideum had 75,108 total GO annotations to 9,395 distinct gene products. Among these, 8,261 are experimental gold-standard annotations from the published literature to 1,525 distinct gene products.
Table 2. Selected GO evidence codes:
These codes indicate from which source the annotation was derived. From the top to IEP, these are the experimental (EXP) evidence codes annotated manually from published experiments. IC and codes such as ISO are also annotated by curators. IBA is a manual code added by expert curators using a specialized tool, see text. A complete list of evidence codes is available.
| IDA - Inferred from Direct Assay |
| IMP - Inferred from Mutant Phenotype |
| IGI - Inferred from Genetic Interaction |
| IPI - Inferred from Physical Interaction |
| IEP - Inferred from Expression Pattern |
| IC - Inferred by Curator |
| ISO - Inferred by Sequence Orthology |
| IBA - Inferred from Biological aspect of Ancestor |
| IEA - Inferred from Electronic Annotation |
| ND - Inferred No biological Data available |
The redesigned dictyBase GO interface now contains additional data displaying new relationships between GO terms, and between gene products and GO terms. A few years ago, the GO consortium implemented GO term annotation extensions, which provide more context to a single annotation (Huntley and Lovering 2017). For example, a molecular function GO term such as protein kinase activity can be directly associated with the target protein, or a biological process GO term may be connected to a specific developmental stage during which the process occurs. The relation terms used to connect annotations are also a controlled vocabulary in order to maintain consistency within and between annotating groups. An example of an annotation with extensions on the new GO tab of our gene page is visualized in Figure 1. This is a more recent effort, currently there are 1,222 individual annotation extensions in dictyBase, and 406 distinct gene products are annotated with at least one extension.
The GO field is a constantly developing research area. For several years, curators across the GO consortium have been adding annotation extensions to GO terms (see above). The additional context to GO terms imparted by annotation extensions has now contributed to new ways to consider and display GO terms and their relationships. Thus, the GO consortium has recently implemented the Gene Ontology Causal Activity Model (GO-CAM; The Gene Ontology Consortium, 2018) to allow the representation of comprehensive models of biological pathways. The new GO-CAM curation encourages the annotation of biological pathways, with relations between GO terms added for each step in the pathway (see Figure 2). dictyBase and other curators use a new tool created by the GO consortium (Noctua), to accomplish these GO-CAM annotations. Currently dictyBase curators have made preliminary efforts to annotate some models for training purposes. GO-CAM models center on molecular functions that are part of a biological process, which in turn are part of regulatory or signaling networks. The nature of Dictyostelium research through the ready manipulation of its genome results in abundant functional analyses by mutant phenotypes. Thus, Dictyostelium gene annotations are often to a biological process identified through a mutant phenotype, and therefore data for the molecular function of a gene is not always available. In these cases, annotations are made to the root GO term ‘GO:0003674 molecular function’ as a starting point. When a function is published this annotation can be updated in Noctua, as any current annotations may be updated when new data becomes available. For the new dictyBase, we share with other groups in the GO consortium an interest in importing not only the annotations created in Noctua but also the pathway graphics. This is a project that we and others are developing together with the GO consortium to find a common solution. To this end, the GO consortium recently released a new user interface to view models that are produced for wider consumption and which go directly from the participating groups into the GO pipeline. There are currently no Dictyostelium annotations in production yet, but we will add these in the near future.
Fig. 2. GO CAM model.
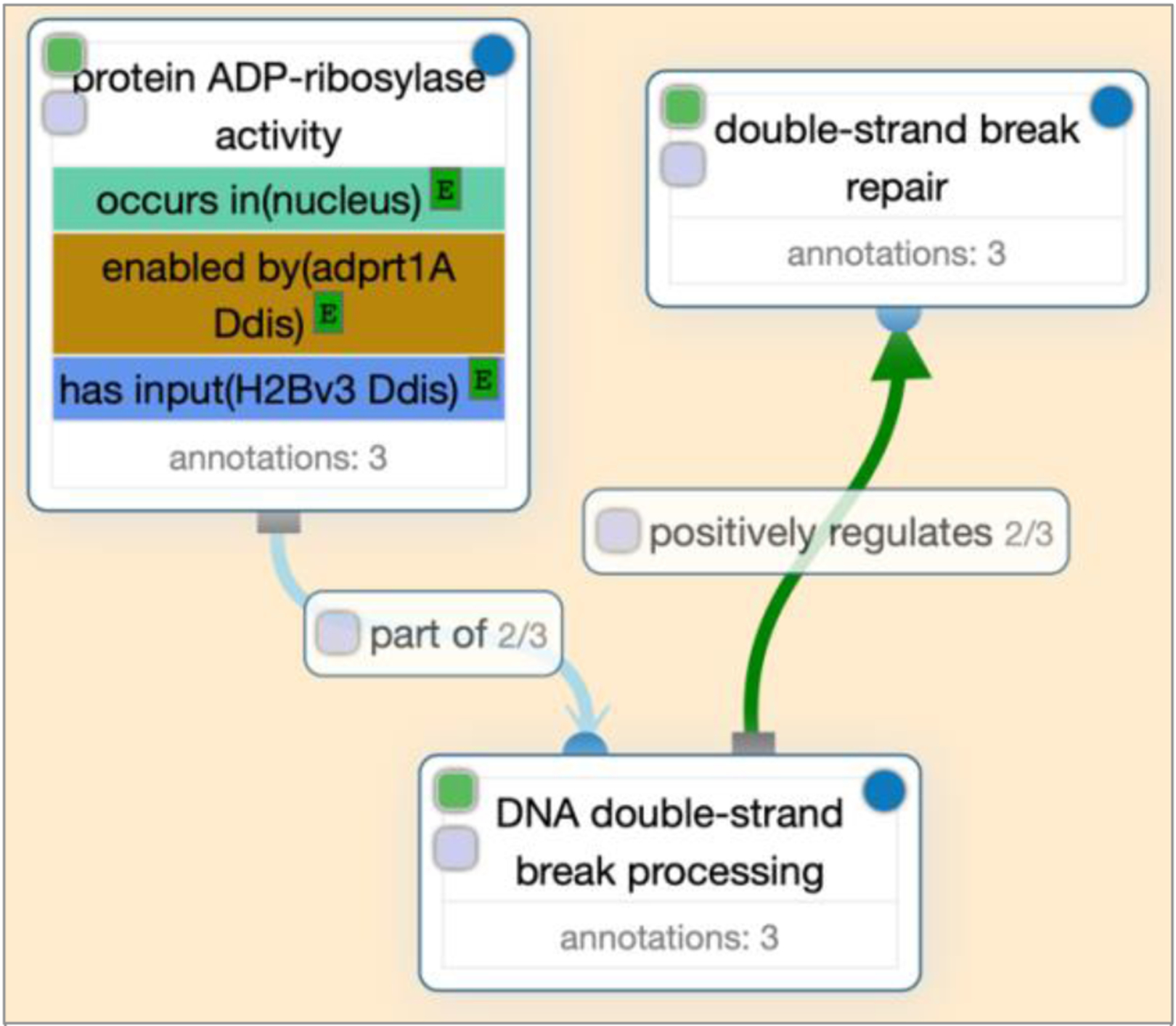
A GO-CAM in Noctua that contains selected core annotations from two references to the D. discoideum gene adprt1A, involved in the process of DNA double-strand break repair. Adprt1A enables ADP-ribosylase activity on the histone H2Bv3 that occurs in the nucleus and this function is part of the double-strand break processing, which positively regulates the repair.
1.c. Strains, Plasmids, and Phenotypes
Because Dictyostelium typically lives and develops as a haploid organism, knockout mutations are easy and very efficient for non-essential genes. Furthermore, conditional knockouts and RNAi have been effectively utilized, and tagging or expressing fluorescent and other labels is also regularly applied, leading to a large and diverse collection of mutant strains. Numerous gene functions have been detected by mutant analyses, and therefore, strain and phenotype annotations are a major focus of dictyBase literature curation.
Strain curation at dictyBase is accomplished largely from the published literature during paper annotation. We have developed standardized methods and controlled vocabulary terms for strain curation, including a ‘strain descriptor’ to handle the lack of uniformity in strain nomenclature; a list of useful strain characteristics (e.g. overexpressor, blasticidin resistant), and a list of the genetic modifications that a strain might have (endogenous deletion, insertion, etc.). Curators also have added strain IDs into the parental strain field for many years, and now the parental strains will be linked in the strain record. Also, researchers may submit strains to the DSC before the paper describing the mutants is publicly available. Strain deposits are accompanied by submission forms that help DSC staff to annotate the submitted strains, making them available to order from the DSC. Thus, if strains are already in the DSC, phenotypes and any necessary modifications to the strain annotation are added by curators when the paper is published. Alternatively, strains may be annotated from the literature before the strain has actually been submitted to the DSC which expedites work for the DSC as the mutant strains are already in the database. This demonstrates the close relationship between the DSC and dictyBase curators.
Plasmids are typically exclusively annotated when submitted to the DSC. We encourage submission of plasmids, as this is a cost-effective way to share materials. Curators add the plasmid used for transformation during strain curation with the exact name stored in the database, if available. In the new dictyBase, the matching plasmid name on the strain page will be linked to the plasmid record (Figure 3).
Fig. 3. dictyBase Strain and phenotype annotation.
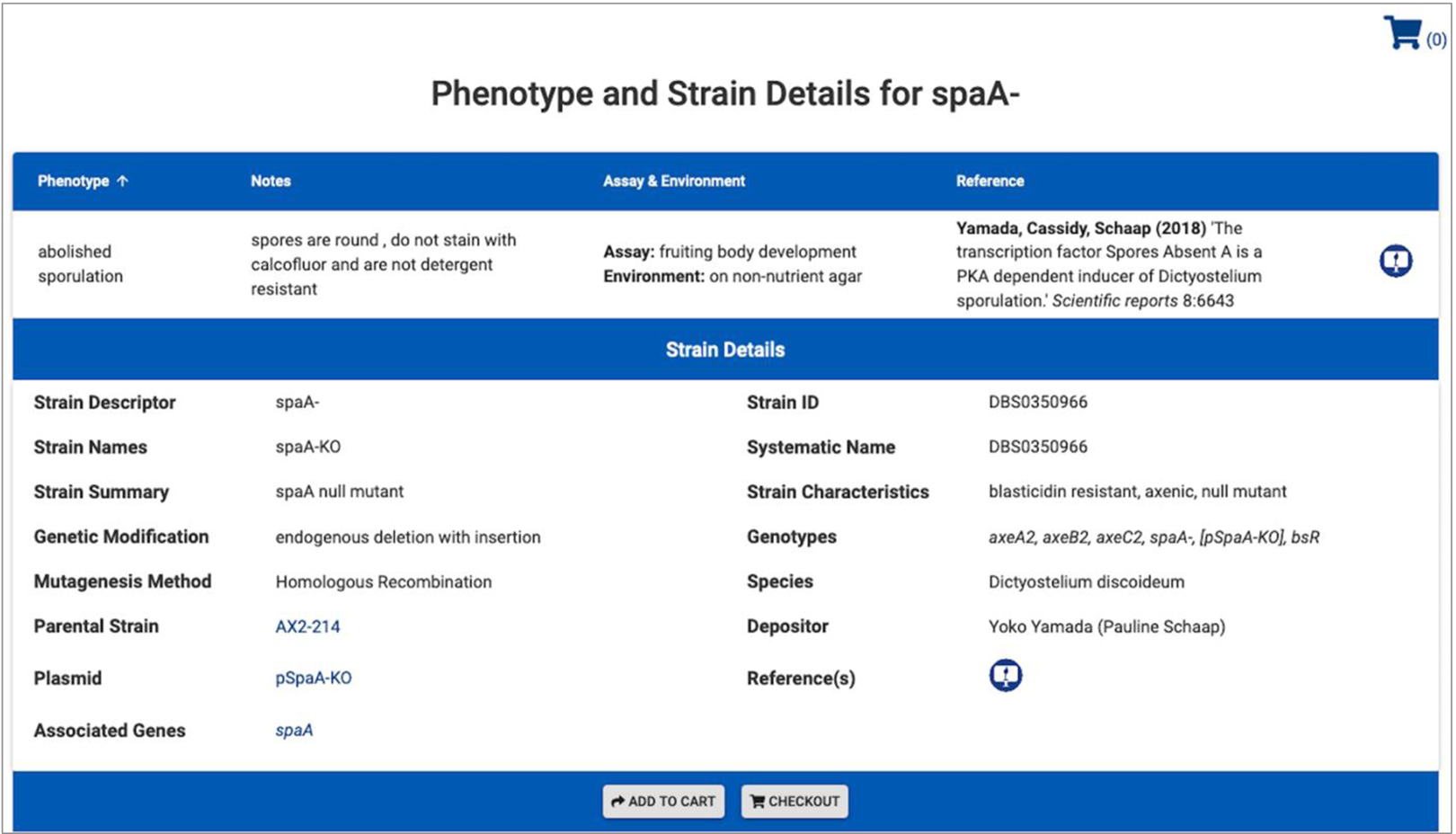
Strain and phenotype annotations have been imported into the new database. New links have been established on the strain page from both the plasmid and the parental strain to their respective pages in the database (both bottom left column). Phenotypes are annotated with a term from the Phenotype Ontology (here: abolished sporulation), a Note to further describe the phenotype, and the Assay used and Environment the experiment has been performed in. A full literature citation is included. Literature symbols link to the reference in dictyBase. Note, strain annotations as displayed in this view are not yet public but will be released soon.
Phenotypes associated with each strain are also annotated during literature curation. Mutant analysis is an important tool in Dictyostelium research and mutant phenotypes are abundant. Since phenotypes are attached to strains, and the strains are then linked to genes, strain curation always precedes phenotype curation. This explains why curators add a large amount of strains that are not available in the DSC, or which might only become available later. Phenotypes are annotated with terms from our phenotype ontology (DDPHENO), a controlled vocabulary that is constantly being expanded as new phenotypes are described. The phenotype ontology is based on the standardized and widely-used Entity-Quality system. There are two parts to the phenotype ontology: the entity (E) changed in the mutant, and a quality (Q) describing that modification. For example, a ‘small spore’ phenotype qualifies the spore (E) as having ‘decreased size’ (Q) resulting in the phenotype term ‘decreased spore size’. See Figure 3 for a strain annotation in the new dictyBase that contains one annotated phenotype. Recently a phenotype reconciliation project has been initiated by a group of ontology experts and curators with the goal of unifying and standardizing all model organism phenotype ontologies. Specifically, this effort aims to reconcile logical definitions across species, while keeping phenotypes that are specific to each organism, similar to genera-specific GO terms. We recently began working with this group to integrate our DDPHENO ontology into the project.
In the near future we also plan to add historic DSC phenotype annotations. Until the end of 2008, the DSC was located at Columbia University under the direction of the late Jakob Franke. Initially, his team investigated phenotypes and annotated them in dictyBase as free text. Many years later, curators at dictyBase translated these free text annotations into phenotype ontology terms. This project resulted in nearly 400 new phenotype annotations. In addition, approximately 100 strains in the DSC that are not published and thus have no annotations, will receive for the first time one or more phenotype annotations.
1.d. Novel annotations
In expectation of the new gene page and the new genome browser (see section 2.a. below) dictyBase curators have been annotating new data types offline for the past couple of years. Previously, while curating publications, curators typically captured gene and protein names, strains, phenotypes, and functional information through the GO. However, we have recently started to curate spatial expression data when genes or proteins were reported to be expressed in specific life cycle stages and cell types. Furthermore, we have also manually annotated Dictyostelium orthologs of human disease genes. These new annotations are currently in the pipeline to be added to the gene page after all existing annotation sections have been built.
Spatial Expression: When spatial expression has been experimentally shown in a paper, curators annotate terms using the Dictyostelium Anatomy Ontology (Gaudet et al. 2008). For example, if a protein was shown to be expressed in the prespore region of the migrating slug an annotation will be made to the term ‘prespore region of the migratory slug’ with the ID DDANAT:0000033. The annotation contains the gene name, the anatomy term and ID, the PubMed reference, and indicates if protein or RNA expression has been tested. To date, over 290 annotations have been annotated by curators. Those annotations will be imported into the new database soon and displayed on the gene page. Furthermore, we plan to solicit annotations of spatial expression from the user community. See also section 2.b. below for a graphical representation of these annotations in dictyAccess.
Disease Orthologs: Dictyostelium has a large number of orthologs to human genes, and a significant number of the human genes have been identified as genes causing disease when mutated. dictyBase curators have currently annotated 46 Dictyostelium orthologs with the respective Disease Ontology term (DO; Kibbe et al. 2014), mostly from published literature when authors analyzed the orthologous relationship. Curators also do their own analysis before annotating and a few annotations came from these analyses and will be cited by a dictyBase reference. The disease orthologs will be displayed near the top of the gene page, in a new section as shown in this example for gene deeJ:
Disease: Parkinson’s disease - PMID: 28819044 - H.s. ortholog: Q99497
The disease name will link to the entry in the DO, the PubMed ID to the dictyBase reference link, and the human ortholog to UniProtKB, where typically a summary of the disease and further informative links are available.
After adding the manually created disease annotations we plan to accelerate these annotations by implementing a semi-automatic approach. In a first step, we will use an automated pipeline to associate Dictyostelium and human disease genes through orthologous mapping. This will create two sets of associations, one with human disease genes that are associated with OMIM diseases (Online Mendelian Inheritance in Man; Amberger et al. 2015) and the other with human orthologs of Dictyostelium genes. The intersection of the two sets, human-disease genes and human-Dicty orthologs, will provide an initial list of Dictyostelium disease gene annotations. The result will contain Dictyostelium and human gene identifiers, associated disease names, OMIM IDs, and ortholog type (1:1, or 1:many). OMIM will then be mapped to DO terms. Curators will spot check 1:1 ortholog mappings and as time permits, the one to many hits will be assessed. This will increase disease annotations considerably.
2. Tools to view genomes and annotations
The complete database overhaul based on new and improved software will also allow the implementation of novel or updated tools for viewing and browsing the genomes and other annotations housed in dictyBase. Below are described the first new tools we plan to include in the new dictyBase environment: an updated genome browser and the dictyAccess tool for a novel graphical view.
2.a. New genome Browser
The genome browser is an essential tool for visual access to Dictyostelid genomes at dictyBase. For many years we have used GBrowse as our browsing tool. However, GBrowse has limitations particularly in handling multiple genomes and next generation deep sequencing reads. The new dictyBase genome browser is based on JBrowse (Buels et al. 2016; Hofmeister and Schmitz 2018), a fast genome browser built with JavaScript and HTML5. JBrowse, as its predecessor GBrowse, has been developed by GMOD, the Generic Model Organism Database. JBrowse is able to display genomic annotations, sequences and quantitative data directly from our database and is scalable to house multiple genomes and quantitative data. We have created a first instance of JBrowse containing data for D. discoideum and the three additional species we currently house, D. purpureum, D. fasciculatum, and P. pallidum. Handling the data tracks, such as changing the track order, or zooming in and out, and all other data view manipulations are fast and intuitive in JBrowse. For example, JBrowse allows rapidly zooming in to the deepest level to view the actual genome sequence. Figure 4 displays a window of our current JBrowse instance.
Fig. 4. JBrowse genomic view.
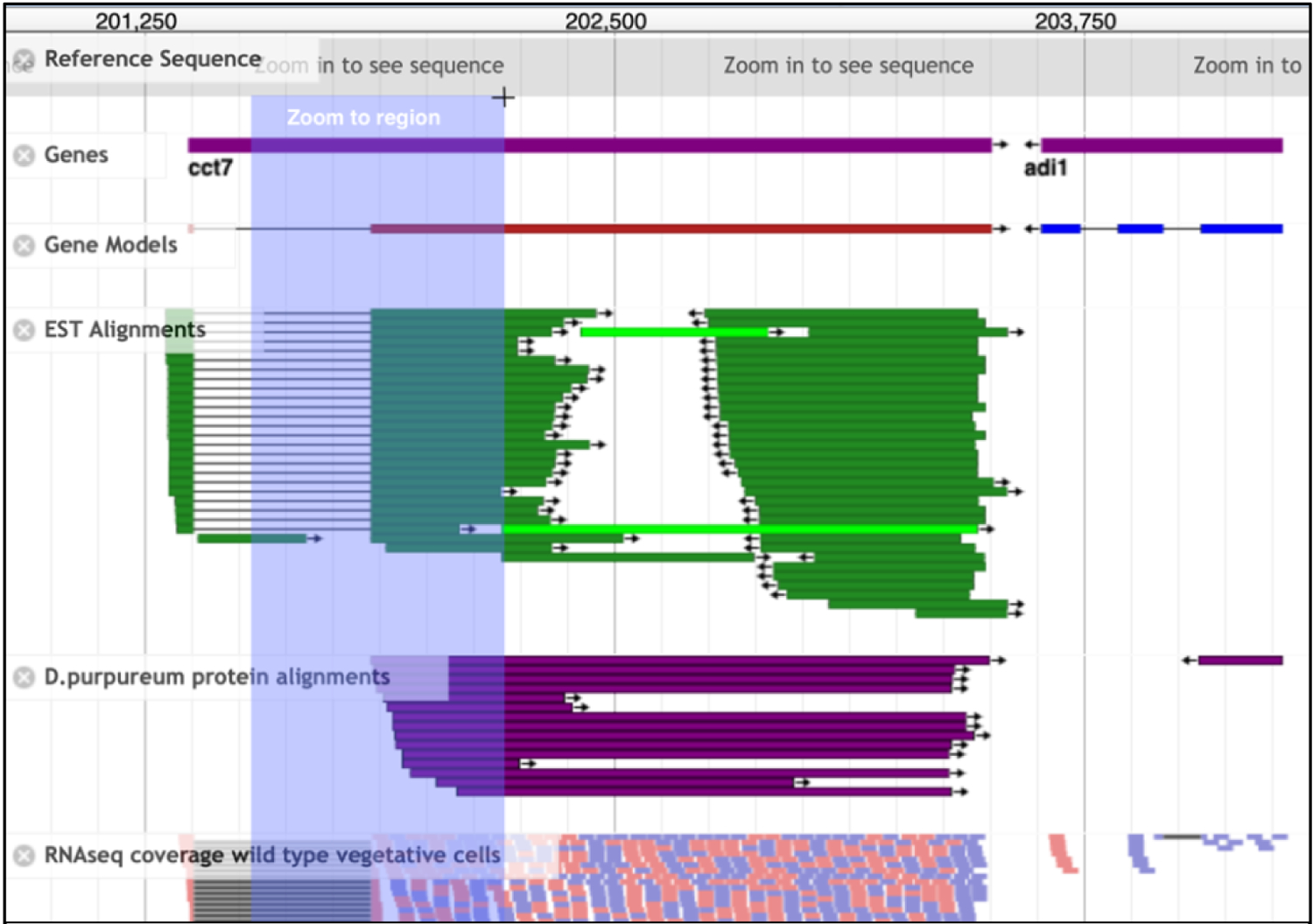
This JBrowse window displays two genes, cct7 and adi1 on chromosome 6 (chromosome number not shown in this view). Tracks selected are listed on the left and include the gene models, a compact view of the ESTs (green) as well as the D. purpureum sequence alignments (purple), and lastly, a partial view of one vegetative wild type RNAseq track. The lilac hue depicts a region highlighted to zoom in by drawing a desired window with your mouse. When releasing the mouse button the zoomed in region appears instantly. Not shown here are the navigation tools on top to zoom in and out, select the chromosome, genome etc., or the side bar to select tracks.
The update to JBrowse as the dictyBase default genome browser will also allow us to deploy WebApollo (Lee et al. 2013), a JBrowse plugin, as our gene model curation tool. Since WebApollo shares the database with JBrowse, new or updated genome annotations will be instantly available for display in JBrowse. This will allow collaborative editing of gene models for logged in users. We plan to open gene model editing to interested users in the community for the other Dictyostelid genomes we house; dictyBase will not have the resources to comprehensively curate the additional new genomes. Furthermore, we plan to add a new track to JBrowse for the GWDI mutant insertion sites and this track will also be incorporated in dictyAccess (see below).
2.b. dictyAccess
A new tool in dictyBase, we have created a circular view in dashboard (Circos display; Krzywinski et al. 2009) that ideally fits a visual representation of annotations allowing display of an overview or unique combinations of data; for example combining data such as gene tracks, RNAseq expression, and certain GO process annotations. The Circos visualization provides a Genomic Bird’s Eye view and the display is a list of images for each of the six chromosomes. Clicking one leads to a larger display that has a zoom control to focus on any particular region and directional mouse control for selecting arbitrary regions. Genomic orientations are separated by colors. There are nearly no limits as to which data may be represented and combined in the Circos display. Figure 5 displays a combination of genomic and expression data which is not yet publicly available. It combines gene tracks, newly annotated spatial expression tracks representing protein and RNA expression to terms that were limited to contain the word ‘stalk’ (e.g. stalk of the mid culminant, prestalk AB core region), and two quantitative RNAseq tracks (a track for the 20-hour developmental time point, and a track where all time points during development, from vegetative cells to mature sorocarp, are combined). Zooming in is seamless and limited only by the computer screen, enabling a relatively easy alignment of expression peaks and genes as seen by the insert of Figure 5. The Circos displays will be generated on the fly with users able to customize their view by selecting tracks and colors. Any data either connected to a location on the chromosome or annotated to a specific gene may be added as a track in the Circos display to create a view of unique data combinations. Currently the publicly available Circos display contains gene tracks and the annotated pseudogenes on separate tracks. Data sets to be selected as tracks will be added regularly. In the future, from any annotated region or gene selected in the track, we will establish a direct link to JBrowse for an alternative linear view. Together, these two tools will provide an integrated graphical visualization of diverse annotations that may be aligned on the chromosome and are linked to chromosomal locations.
Fig. 5. dictyAccess genomic view with selected tracks.
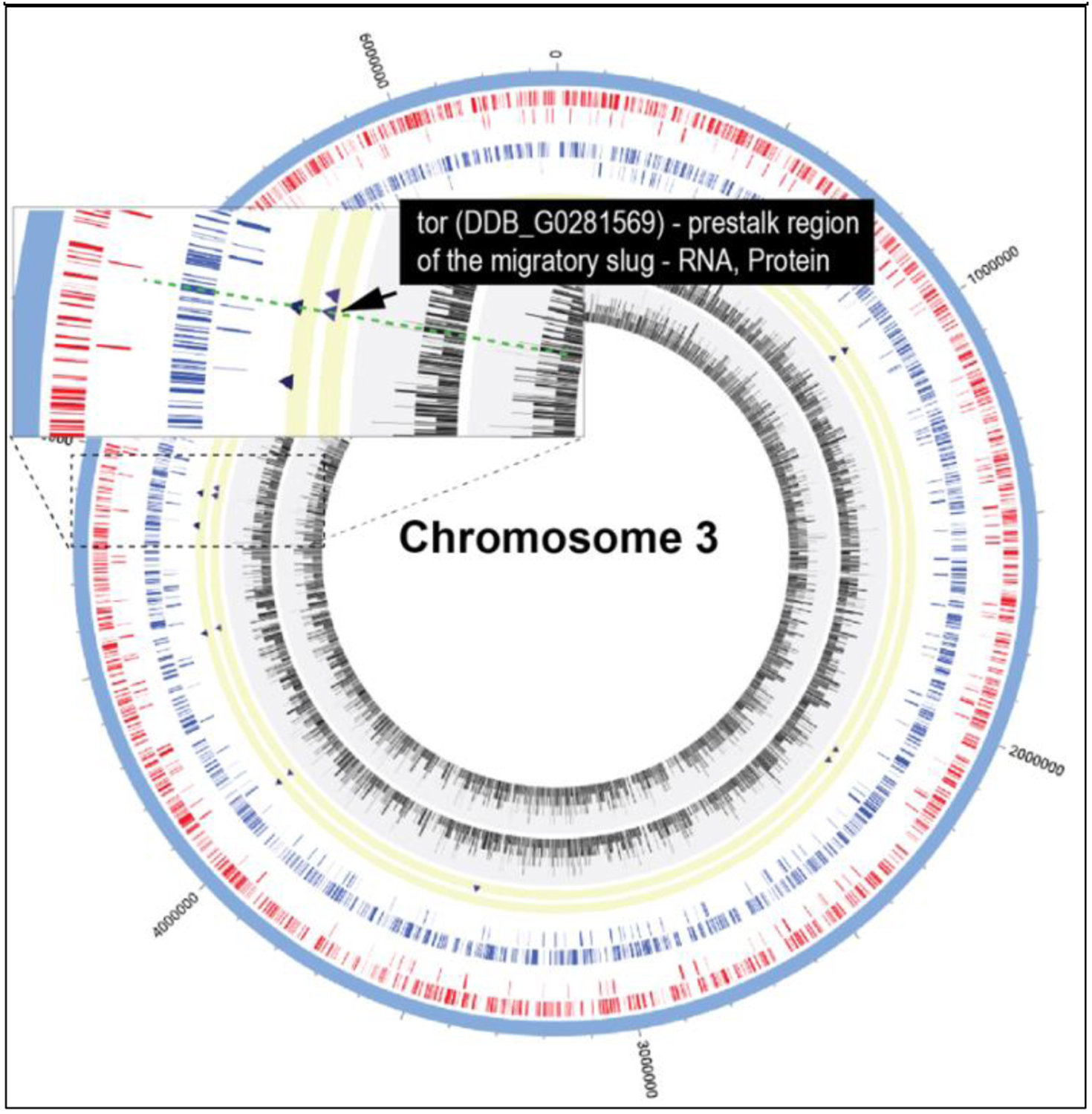
Tracks starting from the outer positive gene track (red); negative gene track (blue). Spatial expression tracks outer: Protein, inner RNA expression (both yellow with purple expression peaks); Next, the RNAseq track of 20 hours time point development (late culminant), followed by the inner most track that combines expression of 24 hours development (both black). The insert represents a zoomed in region of all tracks. Upon mouse-over on the spatial expression track the gene name, the ID, the annotated term and whether it is RNA or Protein expression are shown. As the zoomed in insert shows, the tor gene has an annotation to both RNA and Protein spatial expression. The green line here aligns the tor spatial expression peaks with the blue (negative) track to the actual tor gene and on the quantitative expression tracks (black) aligned are strong peaks on both RNAseq tracks. Note the expression tracks will be available in the near future.
3. The Dicty Stock Center (DSC)
The DSC has served the Dictyostelium research community since 2002. The DSC has been located at Northwestern University in Chicago since 2009, where dictyBase is also located. The mission of the DSC is the long-term storage and distribution of strains and other research materials used in Dictyostelium research. The Dicty community relies heavily on the DSC to enable their research. This is exemplified by a recently completed project funded by the UK Research Council to generate a Genome Wide Dictyostelium Insertion bank (GWDI strains). The project to make genome-wide null mutants would likely not have been funded without the ability to deposit the strains into the DSC so that they would be broadly available to the research community.
3.a. DSC Overview and Operations
Over the past 10 years the DSC has evolved techniques for the most robust growth of strains and all procedures are governed by Standard Operating Procedures (SOPs). While latest updates about the recovery of the GWDI strains have not yet been incorporated in the current SOPs, the latest available document has been linked from our new DSC home page. As time allows, updated or new methods are incorporated. In this dictyBase update the new DSC home page has been generated to serve as a central gateway for all related links pertaining to the Stock Center (see Figure 6, the DSC home page in mobile view).
Since its inception, the DSC has assisted numerous students, and both new and established researchers. Aside from sending DSC materials, we often answer questions about growth techniques or in which medium customers received the ordered strain(s). Also, the DSC has to recover and grow a large variety of mutants that may have been stored long-term. We strive to achieve optimal growth for all strains. For example, we noticed that low nutrition bacterial plates are superior for recovering diverse mutants compared to plates rich in nutrients. Thus, all strains have the best chance to grow up slowly, and not be overwhelmed by their bacterial food source. The media used by the DSC are marked in our technique pages and described in our SOPs. We also maintain an FAQ page, created from actual user questions, which is especially useful for new DSC users.
3.b. The DSC Collection
The DSC contains over 2,000 regular mutant and wild type strains such as null mutants, tagged strains, natural isolates and a few diploid strains. The DSC also includes strains from numerous Dictyostelid species of the genera Dictyostelium, Acytostelium, and Polysphondylium, which are used for evolutionary research and comparative genomics. Furthermore, the DSC stores several bacterial strains such as E. coli or K. aerogenes as diverse bacterial food sources for Dictyostelium. More recently the DSC received over 21,000 strains through the REMI-seq project. The generated GWDI strains are now being added to the database and may soon be ordered for the first time through the DSC shopping cart. GWDI strain annotations are being added automatically, specified by curators and scripted from the GWDI table available at the REMI-seq project. See how a typical GWDI strain annotation will appear in Figure 7.
Fig. 7. GWDI strain annotation in dictyBase.
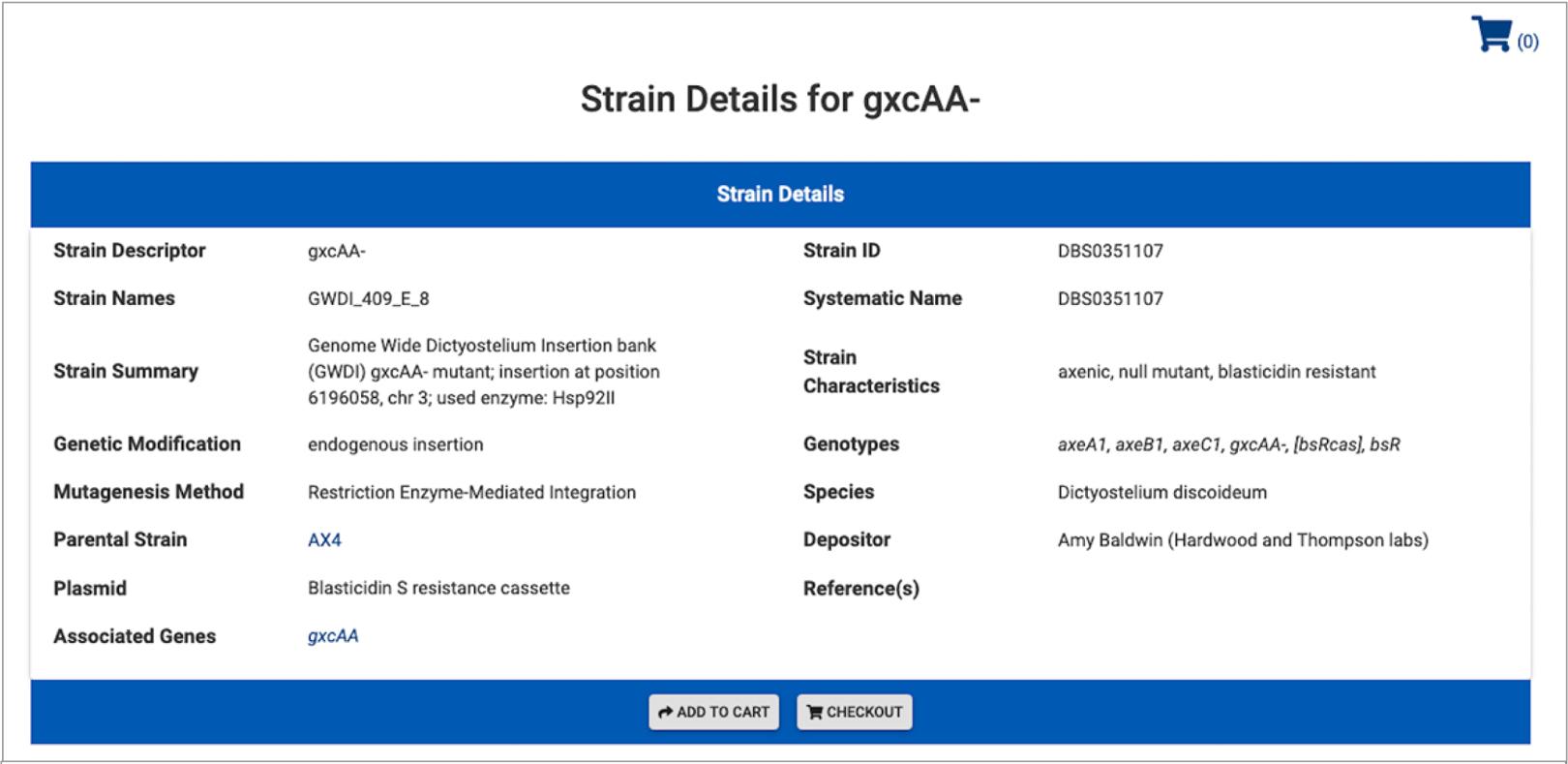
The GWDI strains are added automatically in dictyBase formatted as demonstrated above. Annotations come from the REMI-seq table and from dictyBase controlled vocabularies and formatted genotypes.
The availability of GWDI mutants increased the number of strains available in the DSC roughly 10 times, and strain orders increased strongly when they became available in April 2018. The GWDI mutants were created by combining restriction enzyme-mediated integration (REMI) mutagenesis (Kuspa and Loomis 1992) with Next-Generation Sequencing (NGS) technology. This approach resulted in a collection of over 21,000 genome-wide Dictyostelium mutants in which insertion sites are known. The collection includes approximately 14,000 distinct genomic loci, of which nearly 70% are intragenic. Approximately 5,500 different genes have at least one insertion. The availability of the GWDI strains will allow researchers to conduct complex phenotyping of protein families and increase the ease with which whole genome phenotypic screens can be conducted. We expect the availability of these strains to greatly enhance and accelerate Dictyostelium research. For example, knockout strains from members of whole gene families that have not been available before may now be ordered. For DSC procedures the availability of GWDI strains means additional, careful work. When ordered, GWDI strains are recovered from a 96 well plate using a sterile needle to recover the frozen plug, which is then placed on a prepared bacterial plate. The well is resealed with foil to protect the neighboring wells, and re-stored at −80 °C. When growth of the recovered cells is robust two new plates are inoculated: one for the user who ordered the strain and another for the DSC as a starter plate for strain storage. Sometimes, a well does not contain a single allele and therefore represents a known mixture of 2 or more mutants. These ‘mini pools’ need special care to give each mutant equal chance to grow. Thus, methods in the DSC are continuously evolving as research demands. Available DSC items are listed in Table 3.
Table 3. DSC materials & fees.
Listed fees are for non-profit institutions. Numbers indicate availability of individual items or aliquots.
| Material | Number | Cost / item |
|---|---|---|
| Reg. strains | 2,041 | $ 30.- |
| GWDI strains | 21,521 | $ 30.- |
| Plasmids | 948 | $ 15.- |
| Antibodies | 15 | $ 40.- |
| Genomic library pools | 29 | $ 40.- |
| GWDI pooled libraries | 80 | $ 100.- |
The DSC houses nearly 950 plasmids, which are an effective alternative when users prefer to create a mutant in their own Dictyostelium parent background. Extrachromosomal vectors expressing fluorescent labels are popular items, as are the large collection of the extrachromosomal pDM plasmids (Veltman et al. 2009) that include simple tagged vectors or inducible expression plasmids, while a large number contain the Gateway system (Thomason et al. 2006; Walhout et al. 2000), which allows genes to be cloned using specific recombinase enzymes. Plasmids are quickly processed in the DSC either as a transformed bacterial plate or as dried DNA on filter paper.
The DSC also contains a diverse collection of additional materials that are limited resources and cannot be regenerated in the DSC as are strains or plasmids (bottom three items in Table 3). Therefore, the prices for these items are somewhat higher. For example, a popular antibody and a stocked cDNA library from the same source (Robinson et al. 2000) have been depleted recently in the DSC. However, we also receive newer special stocks, for example a deep coverage genomic library in E. coli (Rosengarten et al. 2015). Because these items are very diverse and not ordered often, they are not stored in our new database. As before, they are viewable on their own Additional Materials website with all available information and may be ordered by emailing the DSC. Finally, the recently received collection of GWDI pooled libraries is also limited supply and takes significant laboratory resources to work with and utilize. Thus, we advise this material to be ordered only once per laboratory.
3.c. DSC Orders
The DSC serves customers around the world. Approximately 55% of the orders come from the US and 45% from other countries, of these 30% are from Europe and 15% from Asia. 264 plasmids have been ordered in the past year and 486 regular strains. In addition, 261 GWDI strains have been ordered since they became available a year ago. This means that on average every work day in the first year one GWDI strain has been ordered in addition to some regular strains and plasmids. Only a few GWDI mutants have been ordered more than once: currently a total of 246 GWDI individual strains (or multi allelic ‘mini pools’) have been diligently stored in triplicates and tested for growth in the DSC.
In our new database, the ordering process for customers will be streamlined and made less error prone. Soon strains can be searched and added to the shopping cart in a new user interface. The old order form often fails, especially for new and non-U.S. customers, so we created a completely new form. For example, the new form will auto fill country names thus avoiding many previous errors. Furthermore, DSC staff currently needs to remind customers to send payment information, but the new form will request that information before processing the order. Thus, the new order form will streamline the experience of ordering and should reduce email traffic concerning orders. In the near future a login will be available for dictyBase colleagues to autofill their name and address, making ordering even more convenient. When checking out their order and before the final step, a review of the shopping cart containing the list of items including the fee of each item will be displayed, which allows final shopping cart edits before submitting the order (see Figure 8). The new order will automatically drive formatting of two different confirmation emails, one for the user and the other for DSC staff. Users will receive a PDF file as order confirmation containing a list of ordered items, their price, and a link to the online payment information. The items listed will also contain additional information, for example the associated reference. The reference is included to benefit the submitter, as users of the donated materials are encouraged to cite the original paper when publishing DSC materials. The user confirmation also will serve as a receipt. The function as a receipt will reduce email traffic further as many users ask for a quote or receipt before payment, which currently has to be manually created. The order confirmation for the DSC staff will include information such as the drug resistances of the strain(s) and where the item is located (inventory information).
Fig. 8. Reviewing the DSC shopping cart.
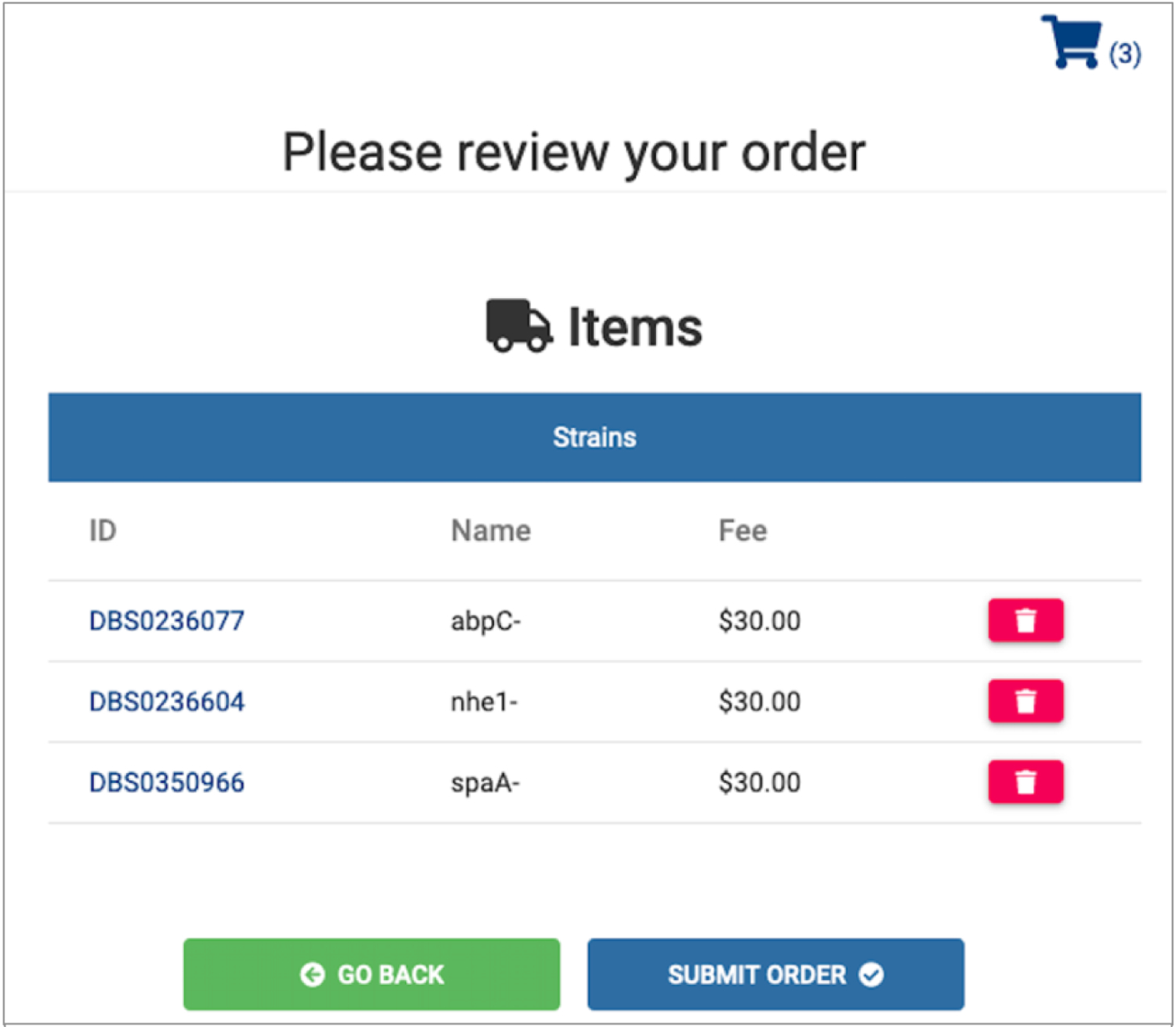
This is the shopping cart view before final checkout. Items may still be deleted here and the Go Back button allows further edits. The price of each item is included which may help final decisions before checking out the order.
Conclusion
The first major release of the new dictyBase focuses on the DSC: strains including the new GWDI mutants, phenotypes of the strains, cited literature, and the ordering process via a shopping cart with newly generated forms and ordering confirmations. The DSC ordering process is being streamlined and made more efficient; it will work for our users worldwide, and also reduce email traffic due to order problems or quote requests. At the time of writing, this release is approaching the first testing stage. Strains and phenotypes will then populate the new gene page, joining the current display of GO annotations. Together, the new dictyBase will have an efficient and modern user interface, will integrate both current and new datasets, and include new display tools. The gene page that researchers and students are accustomed to viewing is being redesigned and will be combined with new data. Because dictyBase and the DSC interface are tightly integrated and serve a diverse userbase, significant effort is required to design and engineer a new database. For utmost flexibility and fast rendering the new dictyBase is housed in a cloud system. New webpages with the expected content such as Dicty techniques or Dicty labs are continually added as time permits. Finally, the new dictyBase is a work in progress, and database updates will be released regularly.
References
- AMBERGER JS, BOCCHINI CA, SCHIETTECATTE F, SCOTT AF, HAMOSH A (2015). OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 43 (Database issue): D789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASPEGREN A, HINAS A, LARSSON P, LARSSON A, SODERBOM F (2004) Novel non-coding RNAs in Dictyostelium discoideum and their expression during development. Nucleic Acids Res 32 4646–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BASU S, FEY P, JIMENEZ-MORALES D, DODSON RJ, CHISHOLM RL (2015). dictyBase 2015: Expanding data and annotations in a new software environment. Genesis 8: 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUELS R, YAO E, DIESH CM, HAYES RD, MUNOZ-TORRES M, HELT G, GOODSTEIN DM, ELSIK CG, LEWIS SE, STEIN L, HOLMES IH (2016). JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol 17: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURGE S, KELLY E, LONSDALE D, MUTOWO-MUELLENET P, MCANULLA C, MITCHELL A, SANGRADOR-VEGAS A, YONG SY, MULDER N, HUNTER S (2012). Manual GO annotation of predictive protein signatures: the InterPro approach to GO curation. Database (Oxford) 2012:bar068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAMON E, MAGRANE M, BARRELL D, BINNS D, FLEISCHMANN W, KERSEY P, MULDER N, OINN T, MASLEN J, COX A, APWEILER R (2003). The Gene Ontology Annotation (GOA) project: implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res 4: 662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHISHOLM RL, GAUDET P, JUST EM, PILCHER KE, FEY P, MERCHANT SN, KIBBE WA (2006). dictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res 34 (Database issue): D423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FEY P, DODSON RJ, BASU S, CHISHOLM RL (2013). One stop shop for everything Dictyostelium: dictyBase and the Dicty Stock Center in 2012 In Dictyostelium discoideum Development. (Eds Eichinger L and Rivero F). Methods in Molecular Biology, Vol. 983 Humana Press, Totowa, NJ, pp. 59–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FEY P, GAUDET P, CURK T, ZUPAN B, JUST EM, BASU S, MERCHANT SN, BUSHMANOVA YA, SHAULSKY G, KIBBE WA, CHISHOLM RL (2009) dictyBase--a Dictyostelium bioinformatics resource update. Nucleic Acids Res 37 (Database issue): D515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FEY P, GAUDET P, PILCHER KE, FRANKE J, CHISHOLM RL (2006). dictyBase and the Dicty Stock Center In Dictyostelium discoideum Protocols. (Eds Eichinger L and Rivero F). Methods in Molecular Biology™, Vol. 346 Humana Press, pp. 51–74. [DOI] [PubMed] [Google Scholar]
- GAUDET P, WILLIAMS JG, FEY P, CHISHOLM RL (2008). An anatomy ontology to represent biological knowledge in Dictyostelium discoideum. BMC Genomics 9: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAUDET P, LIVSTONE MS, LEWIS SE, THOMAS PD (2011). Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform 5:449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIGLIO M, TAUBER R, NADENDLA S, MUNRO J, OLLEY D, BALL S, MITRAKA E, SCHRIML LM, GAUDET P, HOBBS ET, ERILL I, SIEGELE DA, HU JC, MUNGALL C, CHIBUCOS MC (2019). ECO, the Evidence & Conclusion Ontology: community standard for evidence information. Nucleic Acids Res 47 (D1): D1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOFMEISTER BT, SCHMITZ RJ (2018). Enhanced JBrowse plugins for epigenomics data visualization. BMC Bioinformatics 19 (1) : 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUNTLEY RP, BINNS D, DIMMER E, BARRELL D, O’DONOVAN C, APWEILER R (2009). QuickGO: a user tutorial for the web-based Gene Ontology browser. Database (Oxford). 2009:bap010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUNTLEY RP, LOVERIN RC (2017). Annotation Extensions In The Gene Ontology Handbook (Eds Dessimoz C C and Škunca N). Methods in Molecular Biology, Vol. 1446 . Humana Press, New York, NY, pp 233–243. [Google Scholar]
- KIBBE WA, ARZE C, FELIX V, MITRAKA E, BOLTON E, FU G, MUNGALL CJ, BINDER JX, MALONE J, VASANT D, PARKINSON H, SCHRIML LM (2014). Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res 43 (Database issue): D1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRZYWINSKI M, SCHEIN J, BIROL I, CONNORS J, GASCOYNE R, HORSMAN D, JONES SJ, MARRA MA (2009). Circos: an information aesthetic for comparative genomics. Genome Res 19 (9): 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUSPA A, LOOMIS WF (1992). Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci 89 (18): 8803–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE E, HELT GA, REESE JT, MUNOZ-TORRES MC, CHILDERS CP, BUELS RM, STEIN L, HOLMES IH, ELSIK CG, LEWIS SE (2013). Web Apollo: a web-based genomic annotation editing platform. Genome Biol 14 (8): R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MI H, MURUGANUJAN A, HUANG X, EBERT D, MILLS C, GUO X, THOMAS P (2019). Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat Protoc 3: 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINSON DN, SPUDICH JA (2000). Dynacortin, a genetic link between equatorial contractility and global shape control discovered by library complementation of a Dictyostelium discoideum cytokinesis mutant. J Cell Biol 150 (4): 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSENGARTEN RD, BELTRAN PR, SHAULSKY G (2015). A deep coverage Dictyostelium discoideum genomic DNA library replicates stably in Escherichia coli. Genomics 4: 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUCGANG, CHEN, LIU, LINDSAY, LU, MUZNY, SHAULSKY, LOOMIS, GIBBS & KUSPA (2003) Sequence and structure of the extrachromosomal palindrome encoding the ribosomal RNA genes in Dictyostelium. Nucleic Acids Res 31: 2361–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- THE GENE ONTOLOGY CONSORTIUM (2018). The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47 (D1): D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THOMASON PA, BRAZILL DT, COX EC (2006). A series of Dictyostelium expression vectors for recombination cloning. Plasmid 56 (3): 145–152. [DOI] [PubMed] [Google Scholar]
- VELTMAN DM, AKAR G, BOSGRAAF L, VAN HAASTERT PJ (2009). A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid 61 (2): 110–118. [DOI] [PubMed] [Google Scholar]
- WALHOUT AJ, TEMPLE GF, BRASCH MA, HARTLEY JL, LORSON MA, VAN DEN HEUVEL S, VIDAL M (2000). GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol 328: 575–92. [DOI] [PubMed] [Google Scholar]


