Abstract
Most species of the genus Bifidobacterium contain the gene cluster PFNA, which is presumably involved in the species-specific communication between bacteria and their hosts. The gene cluster PFNA consists of five genes including fn3, which codes for a protein containing two fibronectin type III domains. Each fibronectin domain contains sites similar to cytokine-binding sites of human receptors. Based on this finding we assumed that this protein would bind specifically to human cytokines in vitro. We cloned a fragment of the fn3 gene (1503 bp; 501 aa) containing two fibronectin domains, from the strain B. longum subsp. longum GT15. After cloning the fragment into the expression vector pET16b and expressing it in E. coli, the protein product was purified to a homogenous state for further analysis. Using the immunoferment method, we tested the purified fragment’s ability to bind the following human cytokines: IL-1β, IL-6, IL-10, TNFα. We developed a sandwich ELISA system to detect any specific interactions between the purified protein and any of the studied cytokines. We found that the purified protein fragment only binds to TNFα.
Keywords: Bifidobacterium, Immunoferment, Analysis, Cytokines, Receptors, PFNA cluster, fn3 gene
Highlights
-
•
Gut microorganisms experience pressure from factors of the host’s immune system.
-
•
Gut bacteria are able to respond to the presence of cytokines in the growth medium.
-
•
Bifidobacteria play a significant role in shaping and sustaining the immune system.
-
•
Bifidobacterial protein binds specifically to human cytokines in vitro.
1. Introduction
Bifidobacteria are the prevalent group of bacteria inhabiting the gastrointestinal tract (GIT) of healthy children and amounting to a few percent of the total gut microbiota in adults [1]. Bifidobacteria are known to play a significant role in shaping and sustaining the immune system [2,3]. Today there is mounting evidence emphasizing the immunomodulatory properties of bifidobacteria, which are due to both their cell components and metabolites [4,5]. Like pathogenic bacteria, bifidobacteria and other commensal microorganisms experience pressure from factors of the host’s immune system. The mechanisms involved in ensuring the survival of commensal microbiota in the midst of an inflammatory process remain poorly understood [6]. The survival of organisms depends foremost on their ability to respond and adapt swiftly to the changing environmental conditions. Their adaptive potential, in turn, is determined by the efficiency of their internal and external signaling pathways. Research in the field of microbial endocrinology established that over the long course of evolution, microorganisms developed sensory systems for the detection of certain molecules produced by the host [[7], [8], [9]]. Thus, microorganisms are capable of recognizing immune system signals and changing their growth rate and other features accordingly. The mechanisms underlying this elegant communication have not been unraveled yet. For instance, it is almost impossible to find any information in the literature regarding the receptors of commensal bacteria for the detection of immune signals. Only a few studies have tackled this issue in pathogenic bacteria [[10], [11], [12], [13], [14], [15], [16], [17], [18]]. Two of those studies have found interleukin binding receptor-like proteins [17,18].
Previously, we identified and partially characterized the gene cluster named PFNA, consisting mainly of five genes: pkb2, fn3, aa-atp, duf58 and tgm [19,20]. These genes code respectively for: 1) a serine-threonine protein kinase (STPK) Pkb2 [[20], [21], [22]]; 2) a lengthy protein molecule FN3 containing motifs inside their FN type III domains similar to those of cytokine receptors [19,23]; 3) a putative MoxR ATPase AAA-ATP; 4) a protein with unknown function named Duf58 containing the annotated domain DUF58; 5) a putative transglutaminase. STPK Pkb2 is a transmembrane protein possibly involved in signal transduction via its external part, which is apparently ligand-binding. Signal transduction systems are the means of communication between bifidobacteria and their environment, often representing a host. Thus, these systems are potentially involved in signal recognition and transduction between bifidobacteria and the immune system. The putative transglutaminase Tgm is a polytopic transmembrane protein similar in structure to many receptors, ion channels and transporters, which makes it also potentially involved in the communication between bifidobacteria and their hosts. The protein encoded by the gene fn3 is involved in adhesion, which was established experimentally in the strain B.bifidum S17 [24,25]. Type III fibronectin domains are widespread among cell adhesion molecules of the superfamily IgSF [26] in eukaryotes. They are also a part of the structure of bacterial proteins involved in adhesion to host epithelial cells due to the binding of fibronectin [27]. The annotated motifs in the FN type III domains are similar to those of the cytokine-binding site of the receptor gp-130 - a transmembrane receptor which is required for signal transduction by a set of cytokines. This implies a likely interaction between the protein FN3 and the cytokines produced by the host’s immune system. The family of ligands of the receptor gp-130 include IL-6 and other cytokines of this group [28]. The goal of this study was to test the hypothesis suggested previously [19,23], which stipulates that the FN3 protein is capable of binding in vitro, components of the immune system, namely cytokines.
2. Materials and methods
2.1. Bacterial strains, plasmid vectors
The following strains were used: E. coli DH5a (F–, Ф 80 ΔlacZΔM15, Δ (lacZYA-argF), U169) (Promega, USA) [29], E. coli BL21 (DE3) (F–, dcm, ompT, hsdS (rB –MB–), gal λ (DE3)) (Novagen, USA) [30] and Bifidobacterium longum subsp. longum GT15 [22] whose genome is completely sequenced and available in GenBank (accession number CP006741).
We used the pET16b (Novagen, USA) [30] expression vector carrying an N-terminal His-Tag sequence to enable protein purification. E. coli strains were cultivated in Luria-Bertani (LB) broth [31]. Ampicillin (150 μg/ml) was used as a selective agent for plasmid carrying cells.
2.2. DNA manipulations
Isolation of plasmid DNA, production of competent E. coli cells, transformation and analysis of recombinant plasmids were carried out using standard methods [31]. Fragment of the fn3 gene (length: 1503 b.p.; BLGT_RS02815) encoding 2 FN3 domains (ranging from position 1494 to 1994 aa) were amplified from the strain B. longum subsp. longum GT15 genomic DNA using the PCK-100 PC R kit (Dialat at PT-50, Russia) in the thermocycler PTC-0150 (MJ Research, Inc., USA) [22]. PCR was carried out with the following oligonucleotides: fn3-N (5′tcgtcatatgccgagccgccactgctc3′) and fn3-C (5′gatcctcgagctactgcttgtgaatggtggt3′). The resulting DNA fragment was cloned into the pET16b expression vector between the NdeI and XhoI restriction sites.
2.3. Expression of the fn3 gene in E.coli
E. coli BL21 (DE3) cells containing the recombinant plasmid pET16b:fn3 were routinely grown in LB broth at 37 °C until they reached OD (600) of 0.6–0.8. Expression of the fn3 gene was induced by the addition of 1.0 mM isopropyl-β-D-thiogalactoside (IPTG) for 5 h at 28 °C. Cells were precipitated by centrifugation and frozen at −20 °C. To study the expression of fn3, the cells were suspended in a sample buffer containing 62.5 mM Tris-HCl, pH 6.8, 5% glycerol, 2% 2-mercaptoethanol, 0.1% SDS, 0.001% bromphenol blue, then heated at 95 °C for 10 min and analyzed by SDS-PAGE. Protein fractions of E. coli BL21 (DE3) containing an empty pET16b plasmid were used as negative controls.
2.4. Purification of recombinant protein FN3
The biomass of BL21 (DE3) strain containing the pET16b:fn3 plasmid was resuspended in lysis buffer (50 mM NaH2PO4, 5 mM Tris-HCl, 300 mM NaCl, 10 mM imidazole, 1 mM PMSF, 5 mM DTT, pH 8.0) containing lysozyme (1 mg/ml), 0.5% Triton X-100 and 20 mM 2-mercaptoethanol and was sonicated. Insoluble particles were removed by centrifugation (7500 g, 30 min, 4 °C). The resulting lysate was filtered using a 0.22 μm pore size Millex-GP.
Protein isolation and purification were performed using a BioLogic LP chromatography system equipped with a fraction collector (Bio-Rad, USA). The clarified lysate was loaded onto a Bio-Scale™ Mini Profinity™ IMAC Cartridge (5 ml, Bio-Rad, USA), then equilibrated and washed with the lysis buffer, followed by another washing with the buffer (50 mM NaH2PO4, 5 mM Tris-HCl, 300 mM NaCl, 50 mM imidazole, 1 mM PMSF, 5 mM DTT, pH 8.0). The bound protein was eluted from the columns with buffer containing 300 mM imidazole. For further purification of the protein, dialysis was performed in PBS buffer containing 10% glycerol and 1 mM PMSF. The concentration of the isolated protein was measured by Qubit fluorimeter (Invitrogen, USA). The average yield of purified protein was approximately 20 mg per liter of E. coli culture. The purified protein was stored at −80 °C.
2.5. Raising FN3-specific antibodies
The FN3-specific polyclonal antibodies were raised by immunization of rabbits, which was carried out via subcutaneous immunization (5 injections at 2-week intervals) with the recombinant FN3 protein obtained from B. longum subsp. longum GT15 previously. Each immunization consisted of administering 50 μg of the protein mixed with Freund’s adjuvant per rabbit [32]. Complete Freund’s Adjuvant was used in the first injection, and incomplete Freund’s Adjuvant in the second and subsequent injections. The protein was dissolved in saline and emulsified at a 1:1 ratio with Freund’s adjuvant. Two weeks after the last immunization, 30–50 ml of blood was collected from each immunized rabbit. Blood serum was collected and stored at 4 °C after the addition of 0.1% sodium azide (NaN3).
The FN3-specific antibodies were isolated by affinity chromatography. The serum from immunized rabbits was diluted 4-fold in PBS, centrifuged and passed through 4 B CNBr-activated sepharose conjugated to the FN3 protein. Bound to the sorbent the FN3-specific antibodies were eluted and immediately neutralized by adding ammonium hydroxide. The sorbent was fully regenerated by washing the column with four volumes of the PBS and stored at 4°С with 0.1% sodium azide (NaN3) [33].
To concentrate the resulting antibodies, they were salted out with 34,2% ammonium sulfate and dialyzed against PBS. The protein concentration was determined by the Lowry method and the titer of antibodies was quantified using ELISA. The recombinant FN3 protein (5 μg/ml) was coated onto Nunc MaxiSorp flat-bottom 96-well plates in carbonate-bicarbonate coating buffer (CBB) (18 mM Na2CO3, 450 mM NaHCO3, pH 9.5) by incubation at 37 °C for 1 h. The plates were then washed four times with phosphate-buffered solution with 0.05% Tween 20 (PBST). Then 100 μl of polyclonal rabbit IgG were added to the wells at serial 2-fold dilutions starting a concentration of 1 mg/ml, incubated at 37 °C for 1 h and washed four times with PBST. HRP-conjugated goat anti-rabbit IgG (ABCAM) secondary antibodies were added to the wells diluted 1:20000. The plates were incubated for 60 min at 37 °C and washed four times with PBST and developed with 3,3′5,5′-tetramethylbenzidine (TMB). The reactions were stopped with 1 M sulphuric acid. Bacterial aminoglycoside phosphotransferase APHVIII from Streptomyces rimosus ATCC 10970, purified previously at our laboratory, was used as a negative control (coated onto Nunc MaxiSorp flat-bottom 96-well plates under the same conditions as FN3). The absorbance was measured at 450 nm using a Universal Microplate Reader (Biotek, Winooski, VT, USA). Statistical analysis was performed using an unpaired t-test. The resulting solution of polyclonal antibodies was further used in the ongoing reactions.
2.6. Construction of ELISA system to test the ability of FN3 protein to bind various human cytokines
During the study, we opted for the conditions that allowed us to reveal the cytokine binding capacity of the FN3 protein. We employed 3 solid-phase ELISA schemes.
2.6.1. ELISA Scheme 1
At the first stage of the research, we employed the following ELISA schemesch1: recombinant IL-6, IL-10 and TNFα (Invitrogen, USA) were resuspended according to the manufacturer’s specifications and coated onto Nunc MaxiSorp flat-bottom 96- well plates with Carbonate buffer (pH 9.7) by incubation overnight at 4 °C. After the incubation time, the plates were washed four times with PBST and the recombinant FN3 protein was added 100 μl per well (2 μg/ml). The plates were incubated for 60 min at 37 °C and washed four times with PBST. After washing the plates, 100 μl of polyclonal rabbit IgG were added to the wells at a concentration of 20 ng/ml, and then they were incubated at 37 °C for 1 h and washed four times with PBST. HRP-conjugated goat anti-rabbit IgG (ABCAM, United Kingdom) secondary antibodies were added to the wells diluted 1:20000. Incubation, washing and color development with TMB reagent were performed as described above.
2.6.2. ELISA, Scheme 2
In the second stage, we used 96-well polystyrene flat bottom plates coated with cytokine-specific antibodies of from the human cytokine determination kits (VECTOR-BEST, Russia). We added in each well one at a time, first 100 μl of the cytokines IL6, IL10 or TNFα at a concentration of 250 pg/ml, then 100 μl of the FN3 protein at a concentration of 2 μg/ml and lastly 100 μl of the FN3-specific polyclonal antibodies, obtained as described above, at a concentration of 20 ng/ml. In each case, the plates were incubated for 1 h at 37 °C, followed by washing (4 times) with PBST. Finally, HRP-conjugated goat anti-rabbit IgG (ABCAM, United Kingdom) secondary antibodies were added to the wells diluted 1:20000. Incubation, washing and color development with TMB reagent were performed as described above.
2.6.3. ELISA, Scheme 3
In the final ELISA schemesch3, Nunc MaxiSorp 96-well polystyrene plates were coated with rabbit polyclonal antibodies specific to the FN3 protein as described previously by incubation in carbonate/bicarbonate buffer for 1 h at 37 °C followed by washing with PBST four times. To avoid nonspecific adsorption, the wells were incubated in a 1% casein solution for 1 h at 37 °C. After washing, 100 μl of the FN3 protein solution at a concentration of 2 μg/ml was added into the wells, incubated for 1 h at 37 °C and washed four times with PBST. At the next stage, solutions of the cytokines at a concentration of 250 pg/ml were added, incubated for 1 h at 37 °C and washed as described earlier. Finally, HRP-conjugated goat anti-rabbit IgG (VECTOR-BEST, Russia) secondary antibodies were added to the wells. Incubation, washing and color development with TMB reagent were performed as described above.
In all 3 schemes, the absorbance was measured at 450 nm using the automated uQuant Universal Microplate Reader (BIO-TEK INSTRUMENTS, INC, Winooski, VT, USA). Statistical analysis was performed using an unpaired t-test. Nonspecific interactions between components were assessed by excluding each one once at a time. To reduce nonspecific interaction with various substrates of the protein FN3, which is classified as an adhesive protein, it was introduced into wells in PBST containing 1% casein (Invitrogen, USA).
2.7. Bioinformatic analysis
To search for nucleotide and amino acid sequences we used the databases available on NCBI (http://www.ncbi.nlm.nih.gov/) and UniProt (http://www.uniprot.org/). To calculate the molecular weight and isoelectric point of proteins, we used the ProtParam program (http://web.expasy.org/protparam/).
3. Results
3.1. Bioinformatics analysis of the fn3 gene in B. longum subsp. longum GT15
Previously, based on the analysis of the genomes of 34 species of bifidobacteria, we identified and characterized a species-specific cluster of PFNA genes, which consists of 5–8 genes depending on the species [19,20]. One of the genes found in the PFNA cluster is fn3, which encodes two type III fibronectin domains (FN3 domains) located in the C-terminal region.
Motifs involved in interaction with cytokines have been well characterized in eukaryotic proteins with fibronectin (FN3) domains [34,35]. Cytokines represent essential mediators of cell–cell communication with particularly important roles within the immune system. To this day, there is no account in the literature of receptors capable of interacting with cytokines, including commensal bacteria of the human intestine, among bacterial fibronectin binding proteins. At the same time, FN3-like domains are present in most bacteria, including bifidobacteria. In most cases, cytokine-binding motifs in proteins containing FN3 domains were identified based on their similarity with eukaryotic proteins.
We performed a bioinformatics analysis of the fn3 gene and its corresponding protein in bifidobacteria. All human-derived species of bifidobacteria contained at least one protein with an FN3 domain, while the majority contained two FN3 domains, each containing motifs of cytokine receptors. In the case of B. longum, B. bifidum, B. breve, B. adolescentis, B. dentium, B. catenulatum, B. kashiwanohense and B. pseudocatenulatum, the motifs of cytokine receptors were quite conservative: WS-PS, WS-ES, WS-DS, WS-AS or WS-YS. Contrastingly, in the species B. angulatum, the amino acid sequence of the motif of the cytokine receptor SG-QA in the second FN3 domain, is different from the that of motifs of the other species. The species B. gallicum contained only one FN3 domain marked by the motif of cytokine receptor VS-PS [23].
The FN3 protein that we identified in the strain B. longum subsp. longum GT15 (208,205 kDa) consists of 1994 amino acid residues. The region between amino acids 1 and 1493 does not contain annotated domains and does not show high sequence similarity with proteins of other bacteria. However, the protein contains in the C-terminal region two “type III fibronectin domains” which were located within the amino acids regions 1494–1581 and 1586–1671. Each of these regions contained sites annotated as “Cytokine receptor motif”. The sequences of cytokine receptors in the first “Fibronectin type III domain” of B. longum subsp. longum GT15 was WS-PS, and in the second domain - WS-ES.
3.2. Cloning and expression of the fn3 gene in E. coli
We amplified a fragment of the fn3 gene located between nucleotides 4480 and 5982 (fragment size 1503 bp) encoding two “type III fibronectin domains” ranging from amino acid 1494 to 1994 and cloned it into a pET16b expression vector. The fragment was amplified using PCR and the sequenced genomic DNA of B. longum subsp. longum GT15 as a template. The pET16b expression vector contained a His-Tag linker region for the isolation and purification of proteins.
To study the expression of the cloned fragment, we cultivated the E. coli BL21 (DE3) strain containing the recombinant plasmid pET16b:fn3 in LB liquid medium supplemented with IPTG inducer. Then the cells were pelleted by centrifugation, resuspended in sample buffer and analyzed by SDS-PAGE. The molecular weight of the expressed recombinant protein matched the calculated molecular weight of the protein FN3 fragment from position 1494 to 1994 amino acid (54 kDa). The maximum expression of the fn3 gene was achieved in E. coli BL21 (DE3) cells.
3.3. Isolation of the recombinant protein FN3 for generation of antibodies
To verify if we could isolate the protein under native conditions, we optimized the protein extraction protocol. The biomass of E. coli BL21 (DE3) containing the recombinant plasmid pET16b:fn3 was thawed at 4 °C, resuspended in native lysis buffer, and incubated at 4 °C for 1 h. Then, the suspension was subjected to ultrasonic disintegration, preventing the solution from heating, and centrifuged to separate into a soluble fraction and a precipitate. Analysis of the isolated fractions showed that the target protein is in soluble form, which makes it possible to isolate the recombinant FN3 protein under native conditions.
The FN3 recombinant protein was purified from E. coli BL21 (DE3) biomass via chromatography under native conditions onto Bio-Scale Mini IMAC cartridge. The protein yield was 20 mg (concentration 2 mg/ml), the purity of the recombinant protein was confirmed by SDS-PAGE (Supplementary Fig. S1). Mass spectrometric analysis confirmed that the isolated protein is a fragment of the FN3 protein belonging to B. longum subsp. longum GT15.
3.4. Study of the specific binding of the recombinant FN3 protein to cytokines and other substrates
3.4.1. Production and characterization of FN3-specific polyclonal antibodies
To develop a sandwich ELISA assay that would allow us to check the ability of the recombinant FN3 protein to bind human cytokines, we raised polyclonal antibodies specific to the recombinant FN3 protein. We obtained the FN3-specific polyclonal antibodies from the serum of immunized rabbits. The process of antibody production and isolation is described in the materials and methods section. The resulting affinity purified and concentrated preparation contained 1 mg of FN3-specific rabbit polyclonal antibodies per 1 mL and was highly active – specific interaction with the FN3 protein was detected even after 1:820000 dilution. We used the bacterial aminoglycoside phosphotransferase APHVIII, purified previously at our laboratory, as a negative control. The produced rabbit polyclonal antibodies interacted with APHVIII only in the smallest dilutions (1: 400), which is usually the case for nonspecific binding. Adding rabbit nonspecific γ-globulins instead of the polyclonal antibodies did not cross-react either with the FN3 protein adsorbed to the wells of a 96-well polystyrene plate (see. Supplementary appendix Tables S1–S3).
3.4.2. Nonspecific binding of recombinant FN3 protein to various substrates
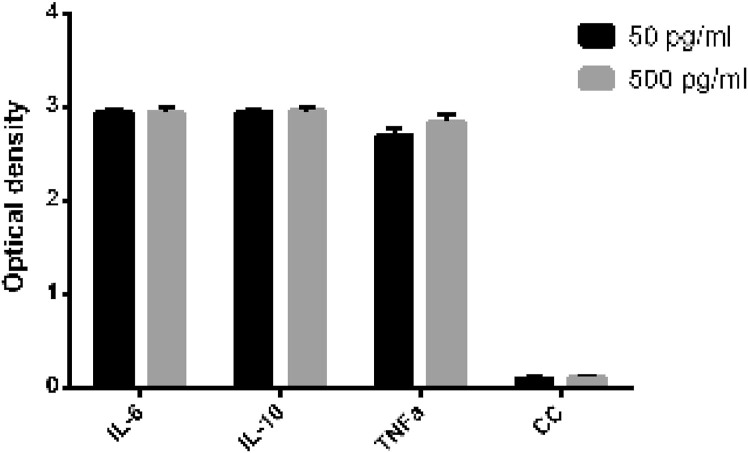
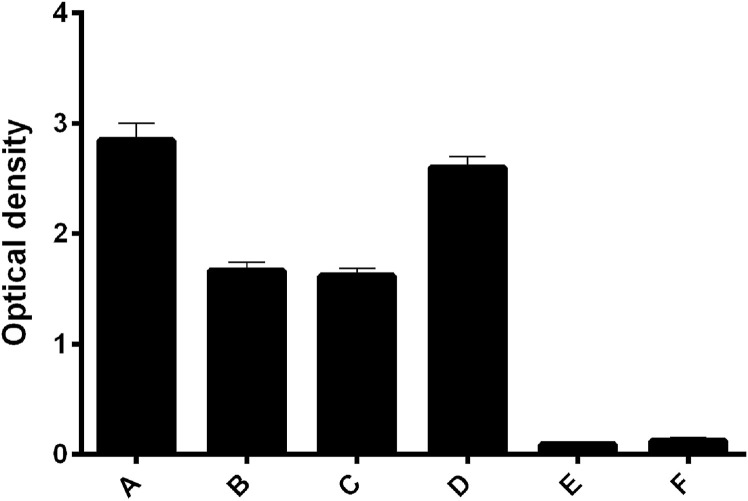
To conduct an initial evaluation of the cytokine-binding capacity of the FN3 protein, we first proceeded with ELISA Scheme 1, described in the materials and methods. When conducting this study, we had to carry out a number of preliminary experiments, the results of which are not shown. The studied fragment of the FN3 protein reacted with cytokines in the same way. Assuming that the capacity of the constructed system with different cytokines may vary, we had to evaluate the effect of dose dependence. It turned out that Scheme 1 was not suitable for our task since the FN3 protein generated a strong positive reaction with all the cytokines, and the reaction intensity did not correlate with the concentration of cytokines bound to the solid phase (Fig. 1 ). Further testing showed (Scheme 2 ELISA) that the FN3 protein binds nonspecifically to virtually any substrate attached to a solid surface, including the antibody-cytokine complex, antibodies to cytokines, and even to non-treated polystyrene (Fig. 2 ). All while nonspecific interaction of other reaction components with each other and with the solid surface was excluded experimentally (see materials and methods).
Fig. 1.
Interaction of FN3 protein with cytokines adsorbed to polystyrene plates. The legend indicates the concentration of cytokines used for solid phase adsorption. CC – «conjugate » control (no polyclonal rabbit antibodies against the FN3 protein fragment; secondary antibodies added after the cytokines).
Fig. 2.
Nonspecific interaction of the FN3 protein with various substrates adsorbed onto the wells of the plate. A. Analyzed cytokines (IL-6, IL-10, TNFα — the values yielded by the three cytokines were relatively similar and grouped together); B. Cytokine-specific antibodies + cytokine complex; C. Cytokine-specific antibodies; D. Blank wells without any adsorbed molecules; E. Negative control 1 (without the FN3 protein); F. Negative control 2 (without FN3-specific antibodies).
To reduce nonspecific binding to substrates, 1% casein was added to the solution of the FN3 protein. The FN3 protein was not able to bind specifically to immobilized cytokines (either to directly adsorbed cytokines or to cytokines attached to a trap of antibodies). This could be explained by the fact that adsorption onto a solid surface can lead to the blockage of the binding sites of FN3-molecule. Moreover, a strong nonspecific interaction between the FN3 protein and the solid surface (see Fig. 2) could hinder any specific interaction.
3.4.3. Specific cytokine binding activity of the FN3 protein
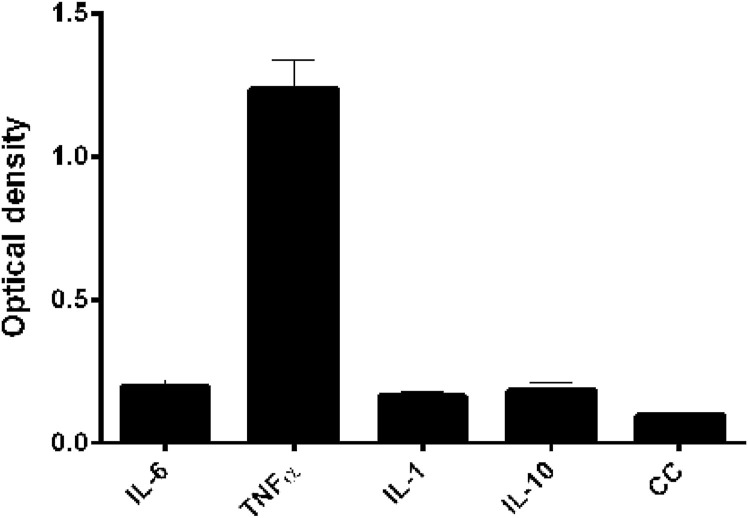
To exclude the possibility of blockage of the cytokine-binding motif of the FN3 protein and its nonspecific adsorption onto the solid surface, we employed ELISA Scheme 3 described in the materials and methods section: The FN3-specific rabbit polyclonal antibodies were first adsorbed onto the solid surface after which the FN3 protein was added as a “second layer”. Then, we added into the wells the cytokine solutions and the peroxidase conjugates from the commercial kits VECTOR-BEST for detecting cytokines. This design revealed a specific interaction between the FN3 protein and the cytokine TNFα, but not with IL-6, IL-1, or IL-10 (Fig. 3 ). This observation suggests the possibility that the FN3 protein of B. longum subsp. longum GT15 binds human cytokines, namely, the pro-inflammatory cytokine TNFα. As a control for TNFα, we used a cytokine that does not bind specifically the FN3 protein in our ELISA scheme, IL-6.
Fig. 3.
Specific interaction of FN3 protein with cytokines. CC – «conjugate » control (no cytokines; secondary antibodies added after the FN3 protein fragment). The values yielded by secondary antibodies against the cytokines IL-6, IL-10, IL-1 and TNFα were relatively similar and grouped together.
4. Discussion
Bifidobacteria as strictly anaerobic bacteria are considered to be some of the oldest representatives of Actinobacteria [36]. As they coevolved alongside their hosts (insects, birds, and mammals), they appear to have developed a species-specific gene cluster (PFNA operon), which allowed them to adapt better to their niches and interact with the host’s immune system [19,20]. The PFNA cluster is unique to the genus Bifidobacterium since it is absent in other genera [20]. Previously, we demonstrated that the genes of the PFNA cluster exhibit unusually high level of interspecific divergence [20], suggesting that the functions of this operon are species-specific.
Today, the functional role of most of the genes that make up the PFNA operon and their products remains unknown. Nevertheless, at least two genes of this operon (pkb2 and tgm) are potential components of a signal system that accounts for a bidirectional communication between the host cells and different species of bifidobacteria [19]. Moreover, another gene of the operon, fn3, codes for a protein with high adhesive properties since it has been shown to be involved in the bacterial adhesion to intestinal mucus [24,25]. In this study, we established that a fragment of FN3 protein bound to various substrates including not coated polystyrene plates thereby exhibiting high non-specific adhesion in vitro. Another important feature of the FN3 protein is the presence of two type III fibronectin domains in its structure [23]. Each of these domains contains motifs similar to cytokine-binding sites of human receptors. The logic behind evaluating the cytokine binding ability of an isolated fragment of a FN3 protein containing two FN3-like domains rests on two facts.
First, as noted above, these domains contain motifs similar in structure to cytokine binding regions of cytokine receptors [23]. The ability of FN3-containing proteins to bind cytokines has not been studied to date, which is why we focused on the adhesive properties of these proteins. The presence of cytokine binding domains in FN3-containing proteins has been mentioned before [37].
Second, the ability to bind cytokines has been demonstrated for many microorganisms whether commensal, opportunistic or pathogenic. For instance, the study of Moriel D.G. et al. (2016) [17] demonstrated that the protein IrmA isolated from the uropathogenic E. coli binds the cytokines IL-2, IL-4 and IL-10. Ahlstrand T. et al. (2017) showed that the lipoprotein BilRI in Aggregatibacter actinomycetemcomitans binds the cytokines IL-1β, IL-8, IL-10,TNFα, TGF-1β and IFNγ, which results in the uptake of IL-1β and reduction of extracellular DNA content in the biofilm [38]. Cytokine binding ability was reported for other proteins such as: 1) Caf1A of Yersinia pestis (binds IL-1β, probably participates in the uptake of this cytokine); 2) PilQ and PilE of Neisseria meningitidis (binds IL-8, TNFα, which leads to uptake of cytokines, increased virulence and resistance to complement); 3) OprF of Pseudomonas aeruginosa (binds IFNγ, which leads to increased expression of PA-I lectin and pyocyanin and activation of QS signaling) [18].
These data show that the binding phenomena of various cytokines are very widespread among pathogenic and opportunistic bacteria interacting with the human organism. Given the role of commensal microorganisms such as bifidobacteria in ensuring the homeostasis of the gut microbiota and normal functioning of the mucosal immune system, we expected to detect similar properties in Bifidobacterium proteins. The presence of motifs in FN3-containing proteins similar in structure to cytokine binding sites of mammalian receptors [23,35], encouraged us to opt for FN3-containing proteins as suitable candidates for this role. As mentioned before, one of these proteins is encoded by one of the genes of the cluster PFNA of B. longum. Proceeding from these premises, we aimed at detecting specific interaction between the FN3 protein and human cytokines.
We set out to assess the ability of a fragment of the FN3 protein containing two FN type III domains to bind in vitro to various human cytokines. However, the protein’s strong adhesive properties rendered the task of revealing its specific interaction with cytokines immobilized on a solid surface, impossible.
The fact that it was impossible to detect any specific interaction between the FN3 protein and cytokines attached to a solid surface suggests one of two things:
-
1)
The FN3 protein undergoes conformational changes when adsorbed onto plastic, which blocks its active site responsible for adhesion and cytokine-binding ability;
-
2)
The cytokine-binding motif of the protein is directed inward or is located very close to the adhesion site, which leads to spatial blockage during the adhesion of the FN3 protein to a solid surface. To find out which of these two hypotheses is true, it is necessary to evaluate the crystal structure of the FN3 protein.
We managed to overcome the non-specific interaction between the FN3 protein and solid surfaces by constructing an ELISA sandwich system, in which the FN3 protein, attached to rabbit polyclonal IgG, acted as a cytokine trap. This construction allowed us to detect a specific interaction between the FN3 protein and TNFα, but not with IL-1, IL-6 or IL-10.
A bioinformatic analysis of proteins containing FN type III domains showed that they are present in the majority of bacteria (unpublished data), but only in the genus Bifidobacterium are they a part of a PFNA operon. This could indicate that, over the course of evolution, bacteria of the genus Bifidobacterium used proteins containing FN3 domains as an instrument for the identification of cytokines of the host organism.
It is well known that the components of the immune system can affect the growth of bacteria [[10], [11], [12], [13]], i.e. microorganisms not only affect the host organism, but are also able, in turn, to perceive signals from it and accordingly respond to them. Until recently, such data for bifidobacteria were absent from the literature.
It is self-evident that for bifidobacteria to be affected by the host organism, they need to be able to receive a feedback signal from the human body. One possible pathway is the cytokine system of the body. The FN3 protein could be involved in the detection of such a signal. In addition, the activity of FN type III domains could depend on the concentration of TNFα in the immediate vicinity of bifidobacteria cells, which lead to the uptake of TNFα as it was reported for some pathogens [18]. At the same time, the possibility of TNFα-mediated signal transduction in B. longum cannot be completely ruled out. Other components of the PFNA operon, such as pkb2 and tgm could fulfill this role, but this subject needs to be addressed in a separate study.
Previously, we demonstrated that living cells of B. longum subsp. longum GT15, upon coincubation with THP-1 cells, increased TNFα, IL-8, and IL10 mRNA expression [39]. Such selective ability to induce cytokine production together with the ability to recognize specific cytokines suggest that bifidobacteria in general and B. longum subsp. longum GT15 in particular could be involved in the regulation of inflammation in the intestine as an active element.
The effect of the gut microbiota on the development and maintenance of homeostasis of the immune status in humans could not be overestimated. Probiotic bacteria (probiotics) such as bifidobacteria, are a major component of the gut microbiota, which selectively regulate the expression of pro-inflammatory and anti-inflammatory cytokines [40,41].
The study of the cytokine binding properties of proteins of commensal flora microorganisms is becoming more and more relevant in the emerging epidemiological situation. One of the most significant components of the pathogenesis of the currently spreading severe coronavirus disease 2019 (COVID-19) is uncontrolled inflammation (cytokine “storm”) [[42], [43], [44]]. Selective binding of TNFα, one of the key inflammatory mediators, by a fragment of the FN3 protein B. longum opens up prospects in the development of new drugs aimed at directional inhibition and eliminating this cytokine in uncontrolled inflammation. This question requires a deeper study and further study of the properties of FN3 proteins, including the identification of similar properties in other species of bifidobacteria.
Bifidobacteria, which constitute a major part of the commensal microbiota of animals and humans, are unique in two ways. First, they were among the first obligate anaerobic bacteria to occupy the intestines of host organisms and shape their immune systems. Second, under the natural conditions of childbirth and breastfeeding, bifidobacteria are the first bacteria colonizing the intestines of newborns allowing the former to play an important role in the formation of the immune system. The PFNA operon including the gene fn3 is potentially a unique feature of bifidobacteria. Studying the PFNA operon can enhance our conceptions [41,45] regarding the interaction of bifidobacteria with the host organism.
Author contributions
Conceptualization V·N.D., I·N.D.; methodology I·N.D.; validation M.G.A., D.A.M., I·N·C., N.A.S., M.V.G., K·K·B.; formal analysis I·N.D., M.S.D.; investigation M.G.A., D.A.M., I·N·C., N.A.S., M.V.G., K·K·B., M.S.D.; writing—original draft preparation I·N.D., V·N.D.; writing—review and editing M.G.A., D.A.M, M.S.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by the State assignment “Genetic technologies in biology, medicine, agricultural and environmental activities” No. 0112-2019-0002.
Declaration of competing interest
The authors declare no conflict of interest.
Handling Editor: Dena Lyras
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.anaerobe.2020.102247.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1: Isolation and purification of recombinant FN3 protein. Table S1. Optical density of serum samples of rabbits immunized with the FN3 protein fragment bound to a specific antigen. Table S2 Optical density of affinity-purified rabbit antibodies specific to the FN3 protein fragment bound to specific and non-specific antigens. Table S3 Optical density of affinity-purified rabbit antibodies specific to the FN3 protein fragment and total IgG of non immunized rabbits against the protein FN3 of B. longum GT15.
References
- 1.O’Callaghan A., van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turroni F., Milani C., Duranti S., Ferrario C., Lugli G.A., Mancabelli L., van Sinderen D., Ventura M. Bifidobacteria and the infant gut: an example of co evolution and natural selection. Cell. Mol. Life Sci. 2018;75:103–118. doi: 10.1007/s00018-017-2672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plaza-Diaz J., Ruiz-Ojeda F.J., Gil-Campos M., Gil A. Mechanisms of action of probiotics. Adv. Nutr. 2019;10:S49–S66. doi: 10.1093/advances/nmy063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Wu Y., Wang Y., Xu H., Mei X., Yu D., Wang Y., Li W. Antioxidant properties of probiotic bacteria. Nutrients. 2017;9:E521. doi: 10.3390/nu9050521. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turroni F., Ventura M., Butto L.F., Duranti S., O’Toole P.W., Motherway M.O., van Sinderen D. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell. Mol. Life Sci. 2014;71:183–203. doi: 10.1007/s00018-013-1318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen T.W., Schofield W.B., Barry N.A., Putnam E.E., Rundell E.A., Trent M.S., Degnan P.H., Booth C.J., Yu H., Goodman A.L. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes D.T., Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat. Rev. Microbiol. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesouhaitier O., Veron W., Chapalain A., Madi A., Blier A.S., Dagorn A., Connil N., Chevalier S., Orange N., Feuilloley M. Gram-negative bacterial sensors for eukaryotic signal molecules. Sensors. 2009;9:6967–6990. doi: 10.3390/s90906967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freestone P. Communication between bacteria and their hosts. Scientifica. 2013;2013:361073. doi: 10.1155/2013/361073. (Cairo) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porat R., Clark B.D., Wolff S.M., Dinarello C.A. Enhancement of growth of virulent strains of Escherichia coli by interleukin-1. Science. 1991;254:430–432. doi: 10.1126/science.1833820. [DOI] [PubMed] [Google Scholar]
- 11.Hogan J.S., Todhunter D.A., Smith K.L., Schoenberger P.S., Sordillo L.M. Growth responses of coliform bacteria to recombinant bovine cytokines. J. Dairy Sci. 1993;76:978–982. doi: 10.3168/jds.S0022-0302(93)77425-1. [DOI] [PubMed] [Google Scholar]
- 12.Denis M., Campbell D., Gregg E.O. Interleukin-2 and granulocyte-macrophage colony-stimulating factor stimulate growth of a virulent strain of Escherichia coli. Infect. Immun. 1991;59:1853–1856. doi: 10.1128/iai.59.5.1853-1856.1991. PMID: 2019445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meduri G.U., Kanangat S., Stefan J., Tolley E., Schaberg D. Cytokines IL-1 beta, IL-6, and TNF-alpha enhance in vitro growth of bacteria. Am. J. Respir. Crit. Care Med. 1999;160:961–967. doi: 10.1164/ajrccm.160.3.9807080. [DOI] [PubMed] [Google Scholar]
- 14.Luo G., Niesel D.W., Shaban R.A., Grimm E.A., Klimpel G.R. Tumor necrosis factor alpha binding to bacteria: evidence for a high-affinity receptor and alteration of bacterial virulence properties. Infect. Immun. 1993;61:830–835. doi: 10.1128/iai.61.3.830-835.1993. PMID: 8381771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L., Estrada O., Zaborina O., Bains M., Shen L., Kohler J.E., Patel N., Musch M.W., Chang E.B., Fu Y.X., Jacobs M.A., Nishimura M.I., Hancock R.E., Turner J.R., Alverdy J.C. Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005;309:774–777. doi: 10.1126/science.1112422. [DOI] [PubMed] [Google Scholar]
- 16.Zav’yalov V.P., Chernovskaya T.V., Navolotskaya E.V., Karlyshev A.V., MacIntyre S., Vasiliev A.M., Abramov V.M. Specific high affinity binding of human interleukin 1 beta by Caf1A usher protein of Yersinia pestis. FEBS Lett. 1995;371:65–68. doi: 10.1016/0014-5793(95)00878-d. [DOI] [PubMed] [Google Scholar]
- 17.Moriel D.G., Heras B., Paxman J.J., Lo A.W., Tan L., Sullivan M.J., Dando S.J., Beatson S.A., Ulett G.C., Schembri M.A. Molecular and structural characterization of a novel Escherichia coli interleukin receptor mimic protein. mBio. 2016;7 doi: 10.1128/mBio.02046-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Högbom M., Ihalin R. Functional and structural characteristics of bacterial proteins that bind host cytokines. Virulence. 2017;8:1592–1601. doi: 10.1080/21505594.2017.1363140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyachkova M.S., Chekalin E.V., Danilenko V.N. Positive selection in Bifidobacterium genes drives species-specific host-bacteria communication. Front. Microbiol. 2019;10:2374. doi: 10.3389/fmicb.2019.02374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nezametdinova V.Z., Mavletova D.A., Alekseeva M.G., Chekalina M.S., Zakharevich N.V., Danilenko V.N. Species-specific serine-threonine protein kinase Pkb2 of Bifidobacterium longum subsp. longum: genetic environment and substrate specificity. Anaerobe. 2018;51:26–35. doi: 10.1016/j.anaerobe.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Nezametdinova V.Z., Zakharevich N.V., Alekseeva M.G., Averina O.V., Mavletova D.A., Danilenko V.N. Identification and characterization of the serine/threonine protein kinases in Bifidobacterium. Arch. Microbiol. 2014;196:125–136. doi: 10.1007/s00203-013-0949-8. [DOI] [PubMed] [Google Scholar]
- 22.Zakharevich N.V., Averina O.V., Klimina K.M., Kudryavtseva A.V., Kasianov A.S., Makeev V.J., Danilenko V.N. Complete genome sequence of Bifidobacterium longum GT15: identification and characterization of unique and global regulatory genes. Microb. Ecol. 2015;70:819–834. doi: 10.1007/s00248-015-0603-x. [DOI] [PubMed] [Google Scholar]
- 23.Zakharevich N.V., Nezametdinova V.Z., Averina O.V., Chekalina M.S., Alekseeva M.G., Danilenko V.N. Complete genome sequence of Bifidobacterium angulatum GT102: potential genes and systems of communication with host. Russ. J. Genet. 2019;55:847–864. doi: 10.1134/S1022795419070160. [DOI] [Google Scholar]
- 24.Westermann C. Dissertation to Obtain the Doctoral Degree Dr. Med. Rer. Nat. The Faculty of Natural Sciences of the University of Ulm; 2015. Analysis of potential host-colonization factors in Bifidobacterium bifidum S17. [Google Scholar]
- 25.Westermann C., Gleinser M., Corr S.C., Riedel C.U. A critical evaluation of bifidobacterial adhesion to the host tissue. Front. Microbiol. 2016;7:1220. doi: 10.3389/fmicb.2016.01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preedy V.R. CRC Press; 2016. Adhesion Molecules; p. 534. [Google Scholar]
- 27.Konkel M.E., Larson C.L., Flanagan R.C. Campylobacter jejuni FlpA binds fibronectin and is required for maximal host cell adherence. J. Bacteriol. 2010;192:68–76. doi: 10.1128/JB.00969-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bravo J., Staunton D., Heath J.K., Jones E.Y. Crystal structure of a cytokinebinding region of gp130. EMBO J. 1998;17:1665–1674. doi: 10.1093/emboj/17.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoue H., Nojima H., Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 30.Mierendorf R., Yeager K., Novy R. Innovations. Newslett. Novagen. 1994:11–13. [Google Scholar]
- 31.Sambrook J., Fritsch E.E., Maniatis T. Cold Sprind Harbor Laboratory Press; 1989. Molecular Cloning: a Laboratory Manual; p. 479. [Google Scholar]
- 32.Levkovits I., Pernis B. Academic press; 1981. Immunological Methods; p. 467. [Google Scholar]
- 33.Oliver C., Jamur M.C. Methods in Molecular Biology. Humana Press; 2010. Immunocytochemical methods and protocols; p. 416. [DOI] [PubMed] [Google Scholar]
- 34.Bazan J.F. Structural design and molecular evolution of a cytokine receptor superfamily. Proc. Natl. Acad. Sci. U.S.A. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liongue C., Sertori R., Ward A.C. Evolution of cytokine receptor signaling. J. Immunol. 2016;197:11–18. doi: 10.4049/jimmunol.1600372. [DOI] [PubMed] [Google Scholar]
- 36.Gao B., Paramanathan R., Gupta R.S. Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie Leeuwenhoek. 2006;90:69–91. doi: 10.1007/s10482-006-9061-2. [DOI] [PubMed] [Google Scholar]
- 37.Hymes J.P., Johnson B.R., Barrangou R., Klaenhammer T.R. Functional analysis of an S-Layer-Associated fibronectin-binding protein in lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2016;82:2676–2685. doi: 10.1128/AEM.00024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahlstrand T., Tuominen H., Beklen A., Torittu A., Oscarsson J., Sormunen R., Pöllänen M.T., Permi P., Ihalin R. A novel intrinsically disordered outer membrane lipoprotein of Aggregatibacter actinomycetemcomitans binds various cytokines and plays a role in biofilm response to interleukin-1β and interleukin-8. Virulence. 2017;8:115–134. doi: 10.1080/21505594.2016.1216294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Averina O.V., Ermolenko E.I., Ratushniy A.Yu, Tarasova E.A., Borschev YuYu, Leontieva G.F., Kramskaya T.A., Kotyleva M.P., Danilenko V.N., Suvorov A.N. Influence of probiotics on cytokine production in the in vitro and in vivo systems. Medic. Immunol. (Russia) 2015;17:443–454. [Google Scholar]
- 40.Skelly A.N., Sato Y., Kearney S., Honda K. Mining the microbiota for microbial and metabolite- based immunotherapies. Nat. Rev. Immunol. 2019;19:305–323. doi: 10.1038/s41577-019-0144-5. org/10.1038/s41577-019-0144-5 [DOI] [PubMed] [Google Scholar]
- 41.Delgado S., Sánchez B., Margolles A., Ruas-Madiedo P., Ruiz L. Molecules produced by probiotics and intestinal microorganisms with immunomodulatory activity. Nutrients. 2020;12:391–404. doi: 10.3390/nu12020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W., Zhao Y., Zhang F., Wang Q., Li T., Liu Z., Wang J., Qin Y., Zhang X., Yan X., Zeng X., Zhang S. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): the Perspectives of clinical immunologists from China [published online ahead of print, 2020 Mar 25] Clin. Immunol. 2020;214:108393. doi: 10.1016/j.clim.2020.108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruiz L., Delgado S., Ruas-Madiedo P., Margolles A., Sánchez B. Proteinaceous molecules mediating bifidobacterium-host interactions. Front. Microbiol. 2016;7:1193. doi: 10.3389/fmicb.2016.01193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.