Abstract
In the present work, the succinic acid (SA), L-pyroglutamic acid (L-PGA), N-phenyl-thioacetamide (N-NPTA), 2-amino-5-chloropyridine hydrogen succinate (ACPS), epigallocatechine Gallate (EGCG) or KDH and, selenomethionine (SeM) compounds have been proposed as potential antiviral candidates to treatment of COVID-19 based on B3LYP/6-311++G∗∗ calculations and molecular docking. Solvation energies, stabilization energies, topological properties have been evaluated as function of acceptors and donors groups present in their structures. ACPS presents the higher reactivity in solution possibly because has the higher nucleophilicity and elecrophilicity indexes while KDH evidence the higher solvation energy probably due to the higher quantity of donors and acceptors groups. NBO studies show that KDH is the most stable in solution. Mapped MEP surfaces have evidenced stronger nucleophilic and electrophilic sites in ACPS, in agreement with the three C=O and two N–H and O–H groups present in this species while KDH has only a C=O group but a total of 19 acceptors and donors groups. From the above studies for six species we can propose that the better potential antiviral candidate to treatment of COVID-19 is ACPS and then, KDH. For a better prediction of the antiviral and anti-inflammatory properties of the proposed compounds, molecular docking calculations were performed by using four structures of COVID-19. Docking results were discussed basing on binding affinities and the interaction types among ligands and different amino acid residues, indicating the powerful ability of KDH and then ACPS ligands on front of the novel coronavirus disease especially for the first and the fourth species (6LU7, 7BTF).
Keywords: Pharmaceutical chemistry, Theoretical chemistry, Quantum chemical calculation, COVID-19 treatment, Molecular docking simulation, ACPS, KDH
Pharmaceutical chemistry, Theoretical chemistry, Quantum chemical calculation; COVID-19 treatment; Molecular docking simulation; ACPS; KDH
1. Introduction
December 2019 winsted the awfully outbreak of pneumonia of mysterious cause in Wuhan. This febrile respiratory illness makes the city of Wuhan in China trend in all social media and attracts the focus of global attention of entire world. This new epidemy and its emergency has awakened the echoes of “severe acute respiratory syndrome coronavirus 2” (SARS-CoV-2) SARS-CoV from almost two decades ago. In 2002–2003, SARS-CoV affected 8 096 people, causing severe pulmonary infections and 774 deaths (case fatality ratio: 10%). Even if coronavirus2019-nCoVappears less virulent at this point with 2.16% mortality percentage which is weak compared to SARS-CoV but is one of the most terrified epidemy that the world had seen. Recently, Zhu et al. [1] succeeded to identify and characterize the novel 2019-nCoV. The viral genome has been sequenced and compared to other similar. These findings reveal that the new 2019-nCoV shows 75–80% match to the SARS-CoV and even more closely related to several bat coronaviruses [2]. Two weeks later the first 2019-nCoV protein structure has been published. The 5th of February Zihe Rao and Haitao Yang's research team at Shanghai Tech University identify “the crytal structure of 2019-nCoV main protease in complex with an inhibitor N3” [3]. The inhibitor N3 as the authors suggested, was published for the first time in 2005 by Yang et al. in a study on wide-spectrum anti-Corona virus drug design [4].
Despite the availability of the initial data on the epidemiology, clinical consequences of the 2019-nCoV infections and the virus' genetic sequence, all this information remain insufficient to face this new virus. The main questions remain without answers, as scientist still ignoring the real origin of the virus, the exact duration of its transmission from one patient to another as well as its ability to infect other animal hosts. In this context, many researchers are currently working on the search and design of new antiviral agents for the treatment of COVID-19 [5, 6, 7, 8, 9, 10, 11, 12] or on the possible use of those known compounds of broad-spectrum [13, 14]. In this sense, structures of compounds clinically used as antiviral drugs for the treatment of numerous infections including their activity spectral, mechanisms of action, principal indications and administration have been carefully studied by Clerk [19, 20]. Therefore, biologically those compounds are known while only for some of them the structural, electronic, topological and vibrational properties were studied combining experimental results with theoretical calculations derived from the density functional theory (DFT) [15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26].
This work has the purpose of proposing six candidate compounds for the treatment of COVID-19 based on some important properties predicted by computational DFT calculations and specific molecular docking studies. We think that this can be an important contribution in order to find the best antiviral candidate to treat COVID-19. Hence, succinic acid (SA), L-pyroglutamic acid (L-PGA), N-phenyl-thioacetamide (N-NPTA), 2-amino-5-chloropyridine hydrogen succinate (ACPS), epigallocatechine Gallate (EGCG) or KDH and, selenomethionine (SeM) compounds were proposed as potential candidate antiviral drugs for the treatment of the novel 2019-nCoV based on DFT calculations and of molecular docking. The structures of those compounds were optimized in gas phase and aqueous solution by using the B3LYP/6-311++G∗∗ level of theory and their properties were evaluated taking into account the number of acceptors (N and O atoms) and donors (N–H or OH groups) groups present in their structures. These parameters together with the solvation energy values in aqueous solution and their reactivities are of great importance taking into account that these groups are necessary to analyze their oral bioavailability and absorptions, as suggested by Veber et al. [27] and Lipinski et al. [28]. Hence, the obtained resulted were compared with other published for antiviral compounds and, in particular with niclosamide, whose potential use to treatment of COVID-19 was suggested recently by Xu et al. [7].
2. Methodology calculations
The GaussView program was used to model the initial structures of succinic acid (SA), L-pyroglutamic acid (L-PGA), N-phenyl-thioacetamide (N-NPTA), 2-amino-5-chloropyridine hydrogen succinate (ACPS), epigallocatechine Gallate (EGCG) or KDH and, selenomethionine (SeM) while their optimizations in gas phase and aqueous solution were performed with the Revision A.02 of Gaussian program [29, 30] by using the functional hybrid B3LYP and the 6-311++G∗∗ basis set [31, 32]. Here, only the most stable structures of each compound are presented. In solution all calculations were performed with the IEFPCM and universal solvation methods because both schemes consider the solvent effects [33, 34, 35]. The volumes and its variations in aqueous solution were computed with the Moldraw program [36]. The mapped molecular electrostatic potential (MEP) and topological properties were calculated at the same level of theory with the versions 09 and 2000 of Gaussian and AIM programs, respectively [30, 37, 38]. The version 3.1 of NBO program was used to compute the stabilization energies of six species in both media [46]. The frontier orbital and some descriptors were also calculated in both media and with useful equations reported in the literature for different species containing similar groups [16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 40, 41, 42, 43, 44, 45]. However, docking simulations of the several complexes were carried out via iGEMDOCK [46] and Autodock Vina [47] softwares. The visual schematic representation of ligands inside coronaviruses active sites were plotted throughtout discovery studio visualizer [48].
3. Results and discussion
3.1. Optimizations in gas phase and aqueous solutions
Structures of six antiviral candidates proposed compounds can be seen in Figure 1 while the calculated total and corrected energies by ZPVE energies (E), dipole moments (μ) and volumes (V) of those six species in gas phase and aqueous solution by using B3LYP/6-311++G∗∗ level of theory are presented in Table 1. The E values are presented in that table in crescent order while in Figure 2 are given the variations observed in the dipole moments and volumes for the six species.
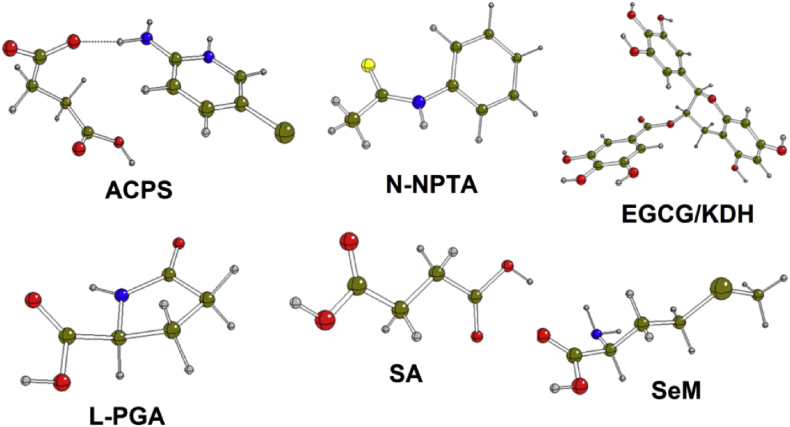
Figure 1.
Optimized structures of six studied compounds: Succinic acid (SA); L-pyroglutamic acid (L-PGA); N-phenyl-thioacetamide (N-NPTA); 2-amino-5-chloropyridine hydrogen succinate (ACPS); Epigallocatechine Gallate (EGCG) or KDH; Selenomethionine (SeM).
Table 1.
Calculated total and corrected by ZPVE energies (E), dipole moments (μ) and volumes (V) of six antiviral candidates proposed in gas phase and aqueous solution by using B3LYP/6-311++G∗∗ level of theory.
| B3LYP/6-311++G∗∗ Method | ||||||||
|---|---|---|---|---|---|---|---|---|
| Species | GAS |
PCM |
||||||
| E (Hartrees) | E/ZPVE (Hartrees) | μ (D) | V (Å3) | E (Hartrees) | E/ZPVE (Hartrees) | μ (D) | V (Å3) | |
| SA | -457.1214 | -457.0160 | 0.00 | 117.7 | -457.1428 | -457.0152 | 0.53 | 114.4 |
| L-PGA | -475.3499 | -475.2248 | 4.96 | 125.7 | -475.3766 | -475.2227 | 7.64 | 125.2 |
| N-NPTA | -763.3409 | -763.1883 | 4.16 | 160.2 | -763.3536 | -763.1873 | 6.89 | 160.7 |
| ACPS | -1220.4569 | -1220.2546 | 9.48 | 243.7 | -1220.5171 | -1220.2227 | 24.09 | 259.5 |
| KDH | -1677.1395 | -1676.7622 | 3.06 | 425.5 | -1677.1979 | -1676.7601 | 4.48 | 426.3 |
| SeM | -2804.0450 | -2803.8814 | 2.23 | 167.0 | -2804.0664 | -2803.8798 | 3.94 | 166.6 |
Succinic acid (SA); L-pyroglutamic acid (L-PGA); N-phenyl-thioacetamide (N-NPTA); 2-amino-5-chloropyridine hydrogen succinate (ACPS); Epigallocatechine Gallate (EGCG) or KDH; Selenomethionine (SeM).
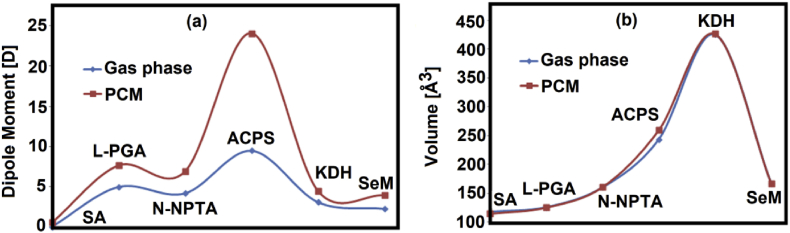
Figure 2.
Variations observed in the dipole moment (a) and volume (b) values of six proposed antiviral compounds in gas phase and aqueous solution by using the B3LYP/6-311++G∗∗ method. Succinic acid (SA); L-pyroglutamic acid (L-PGA); N-phenyl-thioacetamide (N-NPTA); 2-amino-5-chloropyridine hydrogen succinate (ACPS); Epigallocatechine Gallate (EGCG) or KDH; Selenomethionine (SeM).
The E values for the six proposed species show that when they are corrected by zero point vibrational energy (ZPVE) in both media decreasing in the values is observed. However, the dipole moment and volume values don't follow a defined tendency and, only we observed that the dipole moment values in solution increase in relation to the values in gas phase, as observed in Figure 2a. Here, a very important resulted is the high dipole moment value observed in the μ value of ACPS in solution because it change of 9.48 D in gas phase to 24.09 D in solution. Probably, the presences of thirteen donor and acceptor groups justify the high hydration of this species due to the formation of H bonds of those bonds with water molecules. In relation to the volumes practically remaining constants in both media, with exception of V of ACPS which increase slightly in solution possibly due to its high dipole moment in this medium. Consequently, the studies in solution are important and, in special, the determination of solvation energies of all species in aqueous solution. Thus, in Table 2 are summarized the corrected solvation energies (ΔGC/ZPVE) and uncorrected by ZPVE energies (ΔGC) and, volumes variations (ΔV) of six species proposed in aqueous solution by using B3LYP/6-311++G∗∗ level of theory.
Table 2.
Corrected solvation energies (ΔGC/ZPVE) and uncorrected by ZPVE energies (ΔGC) and, volumes variations (ΔV) of nine species proposed in aqueous solution by using B3LYP/6-311++G∗∗ level of theory.
| Species | ΔGun (kJ/mol) | ΔGne (kJ/mol) | ΔGC (kJ/mol) | ΔV (Å3) | ΔGC/ZPVE (kJ/mol) |
|---|---|---|---|---|---|
| SA | -56.13 | 24.41 | -80.54 | -3.3 | -26.51 |
| L-PGA | -70.03 | 20.02 | -90.05 | -0.5 | -25.53 |
| N-NPTA | -33.31 | 14.04 | -47.35 | 0.5 | -16.66 |
| ACPS | -157.90 | 31.98 | -189.88 | 15.8 | -115.65 |
| KDH | -153.18 | 49.16 | -202.34 | 0.8 | -54.67 |
| SeM | -56.13 | 17.97 | -74.10 | -0.4 | -22.17 |
Succinic acid (SA); L-pyroglutamic acid (L-PGA); N-phenyl-thioacetamide (N-NPTA); 2-amino-5-chloropyridine hydrogen succinate (ACPS); Epigallocatechine Gallate (EGCG) or KDH; Selenomethionine (SeM).
Analyzing the values from Table 2 it is observed the high uncorrected ΔGC value for KDH in aqueous solution (-202.34 kJ/mol) while the corrected by ZPVE ΔGC/ZPVE value is very low as compared with the uncorrected (-54.67 kJ/mol). Here, the high ΔGC value of KDH in solution must be attributed to the nineteen acceptors and donors groups present in its structure which evidently generate its higher hydration in aqueous solution. Probably, these groups in solution are hydrated with water molecules. Other possible good candidate of five proposed correspond to ACPS with a ΔGC value of -189.88 kJ/mol while the remaining candidates present enough lower values than that observed for KDH. When the volume values are evaluated from Table 2 we observed that KDH presents in solution a value much smaller than the observed for ACPS in solution. Thus the higher volume variation in solution is predicted for ACPS (15.8 Å3). Previous studies on some antiviral agents we found that a high or low ΔGC or ΔGC/ZPVE values not necessarily is related to a low reactivity [24]. Hence, the presence of total numbers of acceptors and donors groups is necessary to investigate in the six proposed species. Thus, in Table 3 are presented the uncorrected solvation energies by ZPVE energies (ΔGC) and total numbers of N–H and O–H groups and N and O atoms present in six antiviral porposed species in aqueous solution by using the hybrid B3LYP/6-311++G∗∗ method. Here, in N-phenyl-thioacetamide (N-NPTA) the C=S group was considered as a C=O group although we know that the electronegativity of O atom is higher than the corresponding to S atom.
Table 3.
Uncorrected solvation energies by ZPVE energies (ΔGC) and numbers of N–H and O–H groups and N and O atoms present in six antiviral proposed species in aqueous solution by using the hybrid B3LYP/6-311++G∗∗ method.
| Nº | Species | ΔGC | N–H | O–H | O | C=O | N | Total | Groups | Rings |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SA | -80.54 | 2 | 4 | 2 | 8 | ||||
| 2 | L-PGA | -90.05 | 1 | 1 | 3 | 2 | 1 | 8 | R5 | |
| 3 | N-NPTA | -47.35 | 1 | 1 | 1 | 3 | C=S | R6 | ||
| 4 | ACPS | -189.88 | 3 | 1 | 4 | 3 | 2 | 13 | Cl | R6 |
| 5 | KDH | -202.34 | 8 | 10 | 1 | 19 | 4 R6 | |||
| 6 | SeM | -74.10 | 2 | 1 | 2 | 1 | 1 | 7 | Se |
Succinic acid (SA); L-pyroglutamic acid (L-PGA); N-phenyl-thioacetamide (N-NPTA); 2-amino-5-chloropyridine hydrogen succinate (ACPS); Epigallocatechine Gallate (EGCG) or KDH; Selenomethionine (SeM).
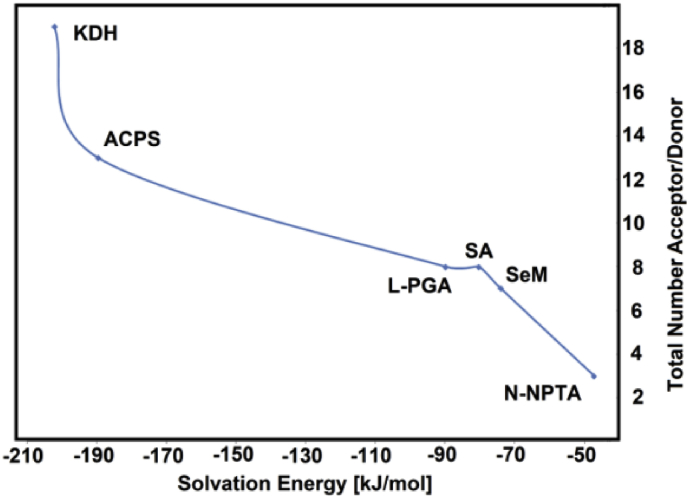
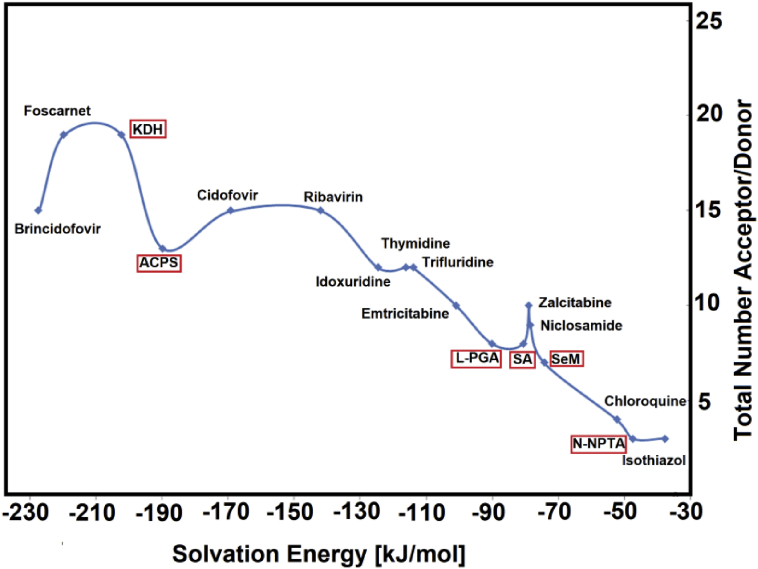
Hence, when the ΔGC values are reordered we observed that the total number of acceptor/donor groups increase according increase the ΔGC values but the observed relationship is nor lineal, as was also observed in the study of potential antiviral niclosamide [26]. The graphic obtained is given in Figure 3. Then, the total number of acceptor/donor groups together with the corresponding ΔGC values for the six compounds proposed here are compared with those observed for the antiviral isothiazol, chloroquine, niclosamide, zalcitabine, emtricitabine, trifluridine, thymidine, idoxuridine, ribavirirn, cidofovir, foscarnet and brincidofovir agents [16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26] in Table 4. The results are presented in Figure 4.
Figure 3.
Total number of acceptor and donors groups of six antiviral candidates as function of corrected solvation energy values calculated in aqueous solution by using B3LYP/6-311++G∗∗ level of theory. Succinic acid (SA); L-pyroglutamic acid (L-PGA); N-phenyl-thioacetamide (N-NPTA); 2-amino-5-chloropyridine hydrogen succinate (ACPS); Epigallocatechine Gallate (EGCG) or KDH; Selenomethionine (SeM).
Table 4.
Uncorrected solvation energies by ZPVE energies (ΔGC) and numbers of N–H and O–H groups and N and O atoms present in ten antiviral species in aqueous solution by using the hybrid B3LYP method.
| Nº | Species | ΔGC | Total Acceptor/donor |
|---|---|---|---|
| 1 | Isothiazolb, | -37,51 | 3 |
| 2 | N-NPTAa | -47,35 | 3 |
| 3 | Chloroquinea | -52,06 | 4 |
| 4 | SeMa | -74,10 | 7 |
| 5 | Niclosamidea | -78,43 | 9 |
| 6 | Zalcitabinec | -78,92 | 10 |
| 7 | SAa | -80,54 | 8 |
| 8 | L-PGAa | -90,05 | 8 |
| 9 | Emtricitabined | -100,88 | 10 |
| 10 | Trifluridinee | -113,85 | 12 |
| 11 | Thymidinef | -116,16 | 12 |
| 12 | Idoxuridineg,# | -124,50 | 12 |
| 13 | Ribavirinh | -141,85 | 15 |
| 14 | Cidofoviri | -169,21 | 15 |
| 15 | ACPSa | -189,88 | 13 |
| 16 | KDHa | -202,34 | 19 |
| 17 | Foscarnetj | -219,64 | 19 |
| 18 | Brincidofoviri | -227,34 | 15 |
Figure 4.
Total number of acceptor and donors groups of six antiviral candidates as function of corrected solvation energy values calculated in aqueous solution by using B3LYP/6-311++G∗∗ level of theory. Succinic acid (SA); L-pyroglutamic acid (L-PGA); N-phenyl-thioacetamide (N-NPTA); 2-amino-5-chloropyridine hydrogen succinate (ACPS); Epigallocatechine Gallate (EGCG) or KDH; Selenomethionine (SeM).
Figure 4 shows the increase in the total number of acceptors and donors groups with the increase in the ΔGC values from the antiviral isothiazol to brincidofovir and, where it is observed that KDH and ACPS present the most negative values of ΔGC, than the other four proposed compounds and, also these values are closer than foscarnet, cidofovir and brincidofovir. The observed differences can be because the ΔGC values in the compared antiviral agents were predicted with the B3LYP/6-31G∗ method. Obviously, the presence of 13 acceptors and donors groups in ACPS and of 19 ones in KDH justify those ΔGC values.
3.2. Frontier orbitals and chemical quantum descriptors
The purpose of this work is to propose candidates in order to find that of higher reactivity against COVID-19 and, for these reasons, the frontier orbitals and gap values from long time are known parameters to predict the reactivities and behaviours of all type of species. Here, these orbitals were calculated for the six compounds in gas phase and aqueous solution by using the B3LYP/6-311++G∗∗ method [16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 37, 38, 39, 40, 41, 42, 43]. Thus, the gap values were calculated from the differences between the frontier orbitals and, then, with the gap values and by using equations reported in the literature are calculated the chemical potential (μ), electronegativity (χ), global hardness (η), global softness (S), global electrophilicity index (ω) and global nucleophilicity index (Ε) descriptors by using the functional hybrid B3LYP with the 6-311++G∗∗ basis set. Table 5 shows the HOMO, LUMO, gap and descriptors for the six proposed compounds in the two media. Gap values are calculated as the difference between the EHOMO and ELUMO. Hence, a higher gap indicates lower reactivity while a low value a higher reactivity. The deep evaluation of gap values revel two important resulted, one of which, is the great change in the gap value that present ACPS when change the medium and, the same gap values that present SeM in both media.
Table 5.
Calculated HOMO-LUMO, gap values and chemical potential (μ), electronegativity (χ), global hardness (η), global softness (S), global electrophilicity index (ω) and global nucleophilicity index (Ε) of six antiviral proposed compounds by using the hybrid B3LYP level of theory.
| B3LYP/6-311++G∗∗ | ||||||
|---|---|---|---|---|---|---|
| Gas phase | ||||||
| Orbitals | ACPS | KDH | L-PGA | SeM | N-NPTA | SA |
| HOMO | -6.4573 | -6.0273 | -7.1893 | -6.0137 | -5.8804 | -8.0954 |
| LUMO | -2.1715 | -1.3633 | -0.9197 | -0.5633 | -1.6191 | -0.5687 |
| ∣GAP∣ | 4.2858 | 4.6640 | 6.2696 | 5.4504 | 4.2613 | 7.5267 |
| Descriptors | ||||||
| χ | -2.1429 | -2.3320 | -3.1348 | -2.7252 | -2.1307 | -3.7634 |
| μ | -4.3144 | -3.6953 | -4.0545 | -3.2885 | -3.7498 | -4.3321 |
| η | 2.1429 | 2.3320 | 3.1348 | 2.7252 | 2.1307 | 3.7634 |
| S | 0.2333 | 0.2144 | 0.1595 | 0.1835 | 0.2347 | 0.1329 |
| ω | 4.3432 | 2.9278 | 2.6220 | 1.9841 | 3.2996 | 2.4933 |
| Ε | -9.2453 | -8.6174 | -12.7100 | -8.9618 | -7.9894 | -16.3030 |
| Aqueous solution | ||||||
| Orbitals | ACPS | KDH | L-PGA | SeM | N-NPTA | SA |
| HOMO | -4,5525 | -6,0709 | -7,0695 | -6,0246 | -5,7144 | -8,0355 |
| LUMO | -3,6246 | -1,3633 | -0,9197 | -0,5742 | -1,5320 | -0,6095 |
| ∣GAP∣ | 0,9279 | 4,7076 | 6,1498 | 5,4504 | 4,1824 | 7,4260 |
| Descriptors | ||||||
| (eV) | ACPS | KDH | L-PGA | SeM | N-NPTA | SA |
| χ | -0.4640 | -2.3538 | -3.0749 | -2.7252 | -2.0912 | -3.7130 |
| μ | -4.0886 | -3.7171 | -3.9946 | -3.2994 | -3.6232 | -4.3225 |
| η | 0.4640 | 2.3538 | 3.0749 | 2.7252 | 2.0912 | 3.7130 |
| S | 1.0777 | 0.2124 | 0.1626 | 0.1835 | 0.2391 | 0.1347 |
| ω | 18.0151 | 2.9350 | 2.5947 | 1.9973 | 3.1388 | 2.5160 |
| Ε | -1.8969 | -8.7493 | -12.2830 | -8.9915 | -7.5768 | -16.0494 |
χ = - [E(LUMO)- E(HOMO)]/2; μ = [E(LUMO) + E(HOMO)]/2; η = [E(LUMO) - E(HOMO)]/2;S = ½η; ω = μ2/2η; Ε = μ∗η; Succinic acid, SA; L-pyroglutamic acid, L-PGA; N-phenyl-thioacetamide N-NPTA; 2-amino-5-chloropyridine hydrogen succinate, ACPS; Epigallocatechine Gallate (EGCG) KDH; Selenomethionine, SeM.
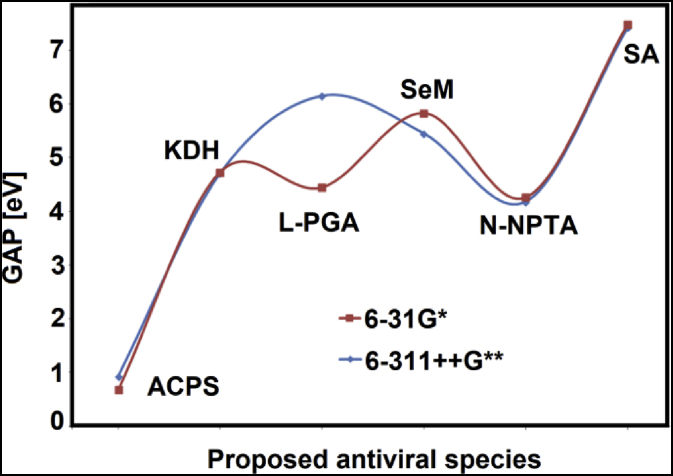
Here, for ACPS in gas phase the change observed in the gap value from 4.2858 eV to 0.9279 eV in solution implies an expected high reactivity for this species in aqueous solution which could be attributed to high electrophilicity index predicted in this medium (18.0151 eV). This resulted in ACPS is in agreement with the strong increase in the dipole moment value of 9.48 D in gas phase to 24.09 D in aqueous solution while in KDH it is observed a change in the dipole moment from 3.06 D in gas phase to 4.48 D in water. On the other hand, the same gap values (5.4504 eV) are observed for SeM in both media, in agreement with the dipole moment values where a null value is observed in gas phase and 0.53 D in solution. When the gap values predicted for these six species in aqueous solution by using the 6-311++G∗∗ basis set are compared with those reported for other antiviral agents by using 6-31G∗ basis set some differences are observed. Here, the gap values for cidofovir and brincidofovir in aqueous solution were calculated by using 6-31G∗ basis set together with the corresponding to the proposed six species in solution in order to see the effect of basis set on the gap values.
The comparisons among all species can be seen in Table 6 while the variations among species calculated with both basis sets are clearly observed in Figure 5. The comparisons between gap values of species computed with different basis sets show that the gap values only for ACPS and L-PGA increase when increase the size basis set while for the other four species decrease the gap values increasing, this way, the reactivities of species. Then, the comparisons for six species by using both B3LYP/6-31G∗ and B3LYP/6-311++G∗∗ levels of theory are shown in Figure 6. Quickly, the higher variation in the gap value can be seen for L-PGA while for SA the values remain practically constant with both methods.
Table 6.
Calculated gap values of six antiviral proposed compounds in aqueous solution compared with values reported for other antiviral agents in the same medium by using the hybrid B3LYP level of theory.
| Species | GAP (eV) |
|
|---|---|---|
| B3LYP Method | ||
| 6-31G∗ | 6-311++G∗∗ | |
| Niclosamide | 3,7225 | |
| Chloroquinea | 4,4571 | |
| Isothiazolb | 4,507 | |
| Zalcitabinec | 5,3595 | |
| Emtricitabined | 4,9336 | |
| Trifluridinee | 5,5876 | |
| Thymidinef | 5,4748 | |
| Idoxuridineg,# | 5,0162 | |
| Ribavirinh | 5,8428 | |
| Cidofoviri | 5,2015 | |
| Foscarnetj | 4,7387 | |
| Brincidofoviri | 5,2987 | |
| ACPS | 0,6777 | 0,9279 |
| KDH | 4,7343 | 4,7076 |
| L-PGA | 4,4472 | 6,1498 |
| SeM | 5,8300 | 5,4504 |
| N-NPTA | 4,2638 | 4,1824 |
| SA | 7,4856 | 7,4260 |
Succinic acid, SA; L-pyroglutamic acid, L-PGA; N-phenyl-thioacetamide N-NPTA; 2-amino-5-chloropyridine hydrogen succinate, ACPS; Epigallocatechine Gallate (EGCG) KDH; Selenomethionine, SeM.
This work.
From Ref [16].
From Ref [17].
From Ref [20].
From Ref [18].
From Ref [19].
From Ref [23].
From Ref [24].
From Ref [22].
Idoxuridine calculated by using B3LYP/3-21G∗ calculations.
Figure 5.
Variations in the gap values for the proposed six species in solution by using the 6-311++G∗∗ basis set with the corresponding observed for twelve antiviral predicted in the same medium at B3LYP/6-31G∗level of theory. Succinic acid, SA; L-pyroglutamic acid, L-PGA; N-phenyl-thioacetamide N-NPTA; 2-amino-5-chloropyridine hydrogen succinate, ACPS; Epigallocatechine Gallate (EGCG) KDH; Selenomethionine, SeM.
Figure 6.
Variations in the gap values predicted for the proposed six species in aqueous solution by using both B3LYP/6-31G∗ and B3LYP/6-311++G∗∗ levels of theory. Succinic acid, SA; L-pyroglutamic acid, L-PGA; N-phenyl-thioacetamide N-NPTA; 2-amino-5-chloropyridine hydrogen succinate, ACPS; Epigallocatechine Gallate (EGCG) KDH; Selenomethionine, SeM.
These studies show that with both methods ACPS has the most low gap value; hence, it is the most reactive species, as compared with the other ones. Consequently, the reactivity order predicted with the 6-311++G∗∗ basis set follow the tendency: ACPS > N-NPTA > KDH > SeM > L-PGA > SA while the trend change when the other method is employed to ACPS > N-NPTA > L-PGA > KDH > SeM > SA. In relation to the descriptors, the higher electrophilicity and nucleophilicity indexes observed for ACPS in solution could justify its higher reactivity in this medium. Besides, the above studies have evidenced a higher dipole moment value for ACPS in aqueous solution (24.09 D) and a higher expansion of volume in the same medium (15.8 Å3). Evidently, the presences of other atoms or rings in the structures of proposed species have influence on the stabilities of same in solution.
3.3. NBO and AIM studies
The inter- and intra-molecular H-bonding interactions can be characterized by using natural bonding orbital (NBO) analysis [36, 39]. Hence, the nature and characteristics of bonds and involved energies, the electronic exchanges, the hydrogen bonds between the donor-acceptor compounds and the transfer reactions can be studied with that theory. In the NBO analysis, the interaction between donor-acceptor is characterized by a significant energy E(2). The higher E(2) value, the greater interaction between the acceptors and the electron donors. This energy is given by the following expression:
The terms qi, F (i. j) and εi. εj represent, respectively, the occupation of the orbital i, the Fock matrix element outside diagonal and finally the diagonal elements.
Here, the stabilities of six proposed species were investigated in gas phase and aqueous solution analyzing the energies of acceptor-donors interactions by using the Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis and with the topological properties calculated at the 6-311++G∗∗ level of theory with the versions 3.1 and 2000 of NBO and AIM program [37, 38, 39].
Thus, the main stabilization energies for the six species in the two media calculated with the B3LYP/6-311++G∗∗ method are shown in Table 7.
Table 7.
Main delocalization energies (in kJ/mol) for the six proposed species in gas and aqueous solution phases by using B3LYP/6-311++G∗∗ calculations.
| B3LYP/6-311++G∗∗a | ||||||||
|---|---|---|---|---|---|---|---|---|
| SA |
SeM |
N-NPTA |
L-PGA |
|||||
| Delocalization | Gas | PCM | Gas | PCM | Gas | PCM | Gas | PCM |
| ΔEπ→π∗ | 494.12 | 507.91 | ||||||
| ΔEn→π∗ | 368.60 | 376.20 | 185.21 | 188.14 | 379.33 | 78.29 | 434.93 | 460.80 |
| ΔEn→σ∗ | 430.92 | 411.85 | 213.97 | 205.53 | 56.89 | 241.14 | 416.62 | 385.69 |
| ΔEσ∗→π∗ | 118.88 | |||||||
| ΔEπ∗→π∗ | 1001.40 | |||||||
| ΔETotal | 799.52 | 788.05 | 399.18 | 393.67 | 930.34 | 1947.62 | 851.55 | 846.49 |
| KDH | ACPS | |||||||
| ΔEπ→π∗ | 217.36 | 277.97 | ||||||
| ΔEσ→σ∗ | 8448.11 | 43977.70 | 79.59 | |||||
| ΔEn→π∗ | 739.61 | 980.38 | ||||||
| ΔEn→σ∗ | 2860.29 | 604.43 | 425.56 | |||||
| ΔEπ∗→π∗ | 5003.46 | 545.61 | 422.18 | |||||
| ΔETotal | 16311.86 | 43977.70 | 2186.6 | 2106.09 | ||||
This work.
The results of Table 7 show that the three SA, SeM and L-PGA species in both media present only two ΔEn→π∗ and ΔEn→σ∗ interactions from lone pairs of O and N atoms toward different antibonding π and σ orbitals. For N-NPTA, other additional ΔEπ→π∗ interaction it is observed in gas phase while in solution other extra two ΔEσ∗→π∗ and ΔEπ∗→π∗ interactions are observed. KDH presents only three ΔEσ→σ∗, ΔEn→σ∗ and ΔEπ∗→π∗ interactions and where surprisingly the ΔEσ→σ∗ interaction has high values, especially in solution. Here, it necessary to clarify that this species present other interactions of more low energies which were not considered here due to the high values of ΔEσ→σ∗ interactions. ACPS shows five interactions in gas phase but only four in aqueous solution where the total energy values have lower values, as compared with the observed for KDH. Hence, KDH is the most stable species in both media and especially in aqueous solution. The higher number of acceptors and donors groups that present this species (19, from Table 4) together with the higher solvation energy value probably justify its higher stability in this medium and, hence, its lower reactivity. According to the total energy value the stability order in both media increase is: KDH > ACPS > N-NPTA > L-PGA > SA > SeM.
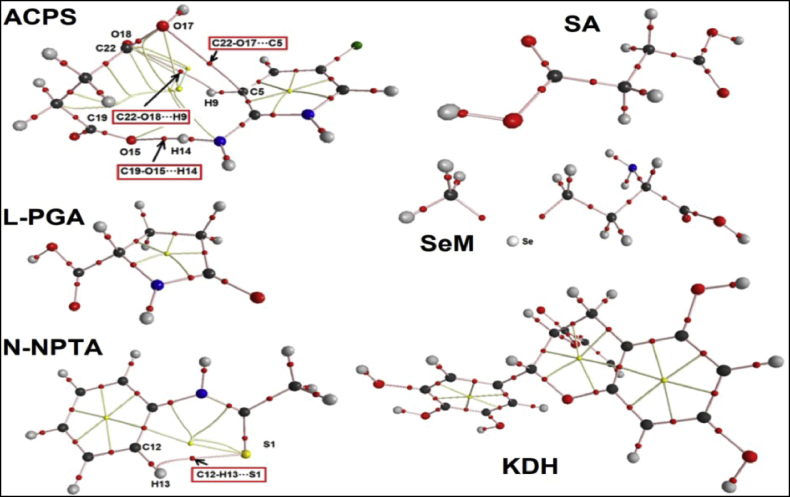
The Bader's theory of atoms in molecules (AIM) is also useful to investigate intra-molecular interactions based on the topological properties calculated in the bond critical point (BCPs) and ring critical points (RCPs) [37, 38]. These properties are the electronic density ρ(r), the Laplacian ∇2ρ(r), the eigenvalues (λ1, λ2, λ3) and the λ1/λ3 ratio. Hence, the properties of RCPs and BCPs can be obtained calculating those parameters. For these reasons, these properties were calculated for the six species here proposed by using B3LYP/6-311++G∗∗ calculations. In this case, we have presented the molecular graphics obtained for all species in aqueous solution in Figure 7. Here, the values of topological properties for those six species were no presented because as observed from Figure 7 in the SA, SeM, L-PGA and KDH species are not observed H bonds interactions and only the RCPs are observed in L-PGA and KDH. Hence, in the structure of N-NPTA is observed the new C12–H13⋅⋅⋅S1 interaction while in ACPS are observed three new C19–O15⋅⋅⋅H14, C22–O17⋅⋅⋅C5 and C22–O18⋅⋅⋅H9 interactions which generate three RCPs. Obviously, in these new H bonds interactions the ratio of λ1/λ3 < 1 and ∇2ρ(r) > 0. Due to those three new H bonds interactions the ACPS species is most stable than the other ones. Thus, these analyses have shown that ACPS in aqueous solution is most stable than the other ones but it resulted don't justify the higher reactivity observed for this species in solution.
Figure 7.
Molecular graphics of Succinic acid, SA; L-pyroglutamic acid, L-PGA; N-phenyl-thioacetamide N-NPTA; 2-amino-5-chloropyridine hydrogen succinate, ACPS; Epigallocatechine Gallate (EGCG) KDH; Selenomethionine, SeM in aqueous solution showing the geometry of all their bond critical points (BCPs) and ring critical points (RCPs) at the B3LYP/6-311++G∗∗ level of theory.
3.4. Molecular electrostatic potentials (MEP)
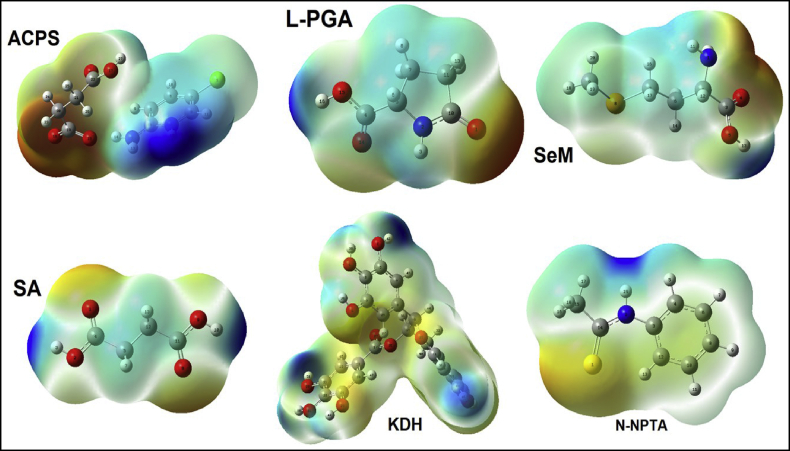
Molecular electrostatic potential (MEP) surfaces, also called as electrostatic potential maps, are 3D surfaces useful to observe the charge distributions of different species [49]. These surfaces can be easily obtained by using the file check during the optimization of a species with the Gaussian 09 program. Hence, the mapped surface allows us a lot of information related to the distribution of the electrostatic potential, the dipole moments of the molecules, the electrophilic and nucleophilic attack sites of the molecules against reactive biological potentials and the possible formation of hydrogen bonding interactions In this study, these mapped MEP surfaces were obtained for the six species in aqueous solution but by using the B3LYP/6-311++G∗∗ method for ACPS, L-PGA, SeM and SA while with the B3LYP/6-31G∗ method for KDH and N-NPTA. Hence, the graphic of those surfaces can be seen in Figure 8.
Figure 8.
Calculated electrostatic potential surfaces on the molecular surfaces of Succinic acid, SA; L-pyroglutamic acid, L-PGA; N-phenyl-thioacetamide N-NPTA; 2-amino-5-chloropyridine hydrogen succinate, ACPS; Epigallocatechine Gallate (EGCG) KDH; Selenomethionine, SeM in aqueous solution. Color ranges ±0.070 a.u. B3LYP functional with 6–311++G∗∗and 6-31G∗ basis sets. Isodensity value of 0.004.
We observed that the strong red colorations, which indicate nucleophilic sites, are observed on O atoms of C=O bonds and these are most evident in ACPS than L-PGA; SeM and SA while these colorations are weak in KDH and N-NPTA. These results are in agreement with the numbers of acceptors and donors groups present in this species because from Tables 3 and 4 the ACPS species has three C=O bonds and evidence a total of 13 acceptors and donors groups while KDH has only a C=O group but a total of 19 acceptors and donors groups. On the other hand, the intense blue colours, indicative of electrophilic sites, are also observed on the two N–H and O–H bonds of ACPS, where it is observed a most large region, as compared with the observed in the other species due to the two closer N–H bonds. In all species, the blue colorations are distributed in different regions due to the different positions of N–H and O–H groups, as observed in Figure 8. With this study we can easily justify why ACPS is most reactive than the other ones.
3.5. Molecular docking studies
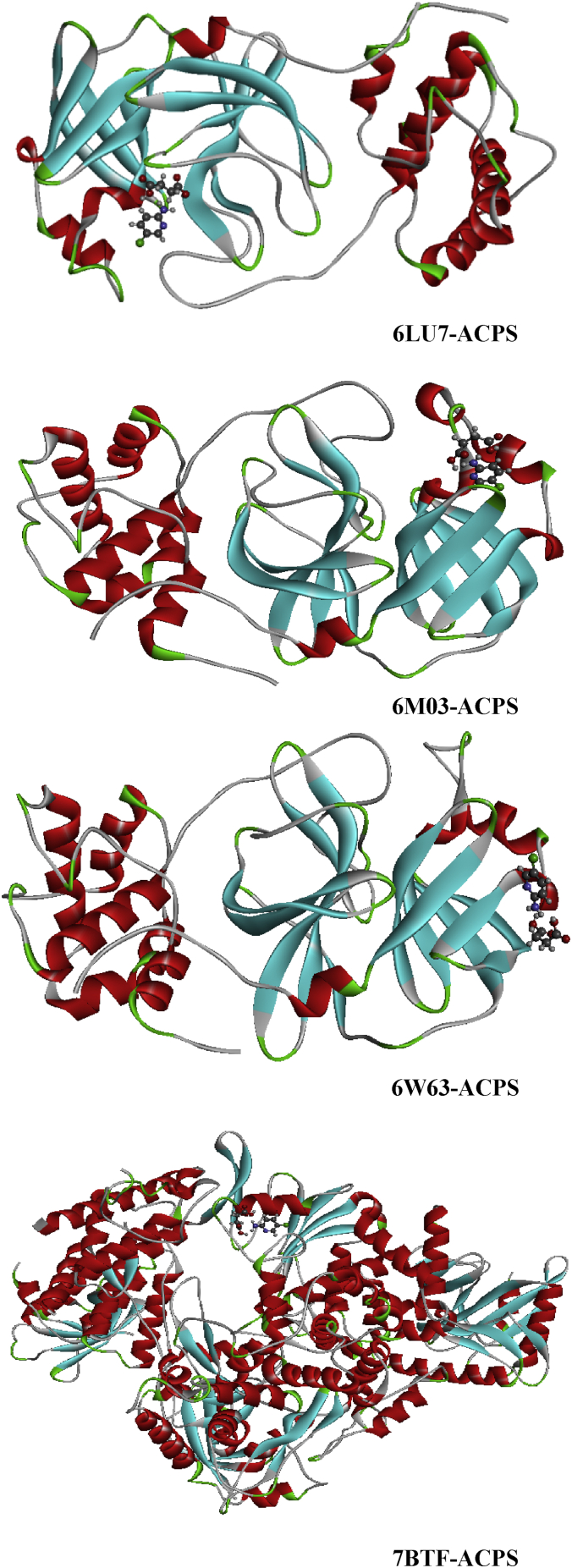
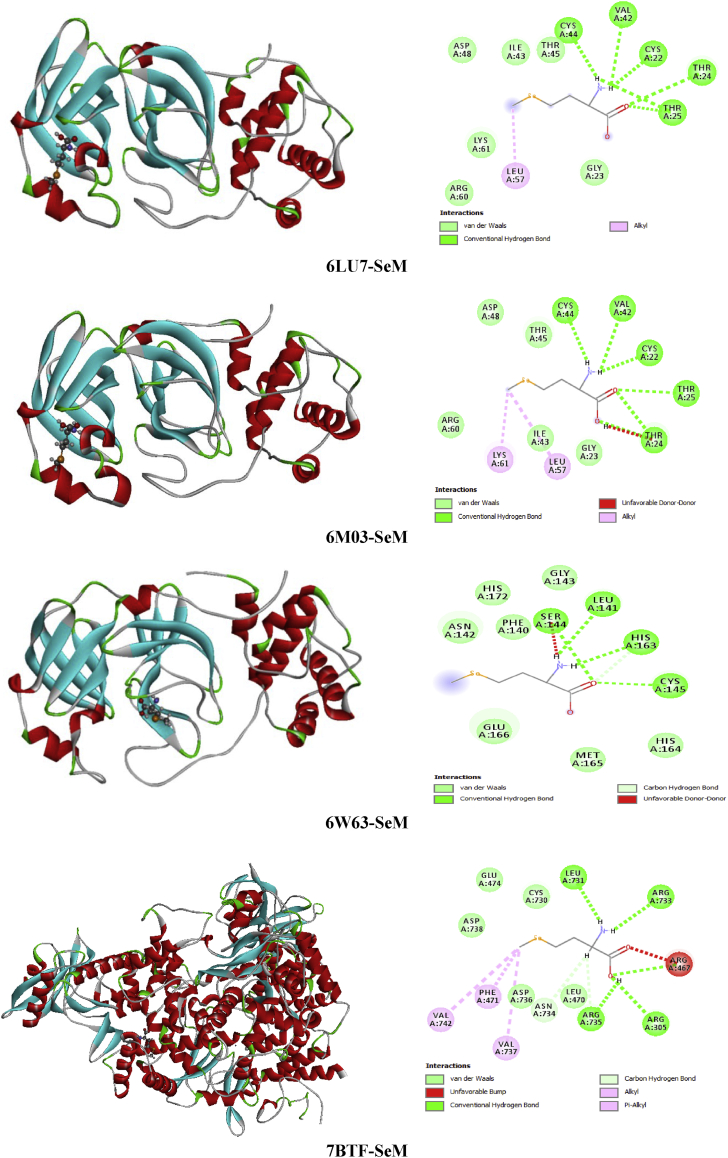
Protein-ligand interactions are the most important part in drug design industry. To analyze and to investigate these interactions, molecular docking analysis was made using the following accuracy setting; population size (800), generations (80) and number of solutions (10). The goal of this section is to evaluate the inhibitory effect of the six species the succinic acid (SA), L-pyroglutamic acid (L-PGA), N-phenyl-thioacetamide (N-NPTA), 2-amino-5-chloropyridine hydrogen succinate (ACPS), epigallocatechine Gallate (EGCG) or KDH and, selenomethionine (SeM) against the four structures 6LU7, 6M03, 6W63 and 7BTF of COVID-19 or 2019-nCoV which obtained from RCSB PDB [50]. It is worthy to mention that the novel 2019-nCoV has the same symptoms as a flue. It can cause fatigue, fever, and cough, which may be accompanied by nasal congestion, runny nose, headache, expectoration and diarrhea. In the light of these informations we made our choice. The selection of ligands was principally built on their anti-inflammatory and anti-viral activities. The candidate compounds were classed in two categories; heterocyclic compounds (SA, L-PGA, KDH) and organic amines (NPTA, ACPS, SeM). Calculation results and the binding interactions of the docked compounds in the various viruses were illustrated in Tables 8 and 9. These lattes demonstrate the molecular docking calculation of the binding affinities and scores of each complex with the help of iGEMDOCK and Auto Dock protocols. The protein-ligand complexes were calculated by measuring various non-polar and polar interactions, for instance van der Waal's interactions, H-bonding interaction, electrostatic interactions, and hydrophobic interactions for each ligand. As clearly seen in Table 8, the greater part of the binding score is van der Waals energies, the H-bond energies for the six studied compounds are significant but weaker than VDW ones. Whereas, Table 9 illustrate the binding score of each complex calculated by using Auto Dock Vina program. This table show that Epigallocatechine Gallate (KDH) possesses the most higher score with the different chosen COVID-19 virus especially the latest structure of this disease 6W63 (-10.4 kcal/mol) and 7BTF (-9.5 kcal/mol).
-
a.
Succinic acid (SA):
Table 8.
The binding scores of the docked ligands in COVID-19 active sites by using iGEMDOCK program.
| Ligands | COVID-19 viruses | Interaction energies (kcal/mol) | EH-bond | EVDW | EElectrost |
|---|---|---|---|---|---|
| SA | 6LU7 | -66.94 | -24.62 | -40.00 | -2.31 |
| 6M03 | -62.65 | -29.92 | -30.96 | -1.76 | |
| 6W63 | -63.75 | -17.44 | -40.32 | 5.98 | |
| 7BTF | -67.52 | -24.58 | -37.23 | -5.71 | |
| L-PGA | 6LU7 | -70.98 | -26.75 | -42.75 | -1.48 |
| 6M03 | -72.39 | -27.49 | -44.50 | -0.40 | |
| 6W63 | -81.44 | -33.84 | -46.82 | -0.77 | |
| 7BTF | -73.58 | -17.06 | -54.24 | -2.27 | |
| N-PTA | 6LU7 | -55.68 | -3.5 | -52.18 | 0 |
| 6M03 | -55.39 | -3.5 | -51.89 | 0 | |
| 6W63 | -59.68 | -8.20 | -51.48 | 0 | |
| 7BTF | -74.50 | -9.59 | -64.90 | 0 | |
| ACPS | 6LU7 | -100.70 | -31.64 | -65.02 | -4.03 |
| 6M03 | -76.77 | -30.64 | -46.12 | 0 | |
| 6W63 | -93.68 | -26.70 | -60.11 | -6.86 | |
| 7BTF | -99.01 | -22.93 | -74.02 | -2.04 | |
| KDH | 6LU7 | -135.65 | -37.18 | -98.47 | 0 |
| 6M03 | -121.67 | -41.57 | -80.09 | 0 | |
| 6W63 | -122.12 | -27.79 | -94.33 | 23.36 | |
| 7BTF | -129.13 | -58.71 | -70.42 | 0 | |
| SeM | 6LU7 | -67.76 | -30.09 | -37.67 | 0 |
| 6M03 | -62.86 | -25.34 | -37.51 | 0 | |
| 6W63 | -71.49 | -24.18 | -44.99 | -2.31 | |
| 7BTF | -81.87 | -27.86 | -49.99 | -4.00 |
Table 9.
Autodock binding affinities of the candidate compounds.
| Protein | Binding score (kcal/mol) |
|||||
|---|---|---|---|---|---|---|
| SA | L-PGA | NPTA | ACPS | KDH | SeM | |
| 6LU7 | -3.9 | -4.4 | -4.2 | -4.2 | -8.3 | -4.4 |
| 6M03 | -4.2 | -4.5 | -4.8 | -3.8 | -7.4 | -4.7 |
| 6W63 | -4.7 | -4.3 | -4.7 | -4.4 | -10.4 | -4.9 |
| 7BTF | -4.2 | -5.0 | -4.6 | -3.9 | -9.5 | -4.9 |
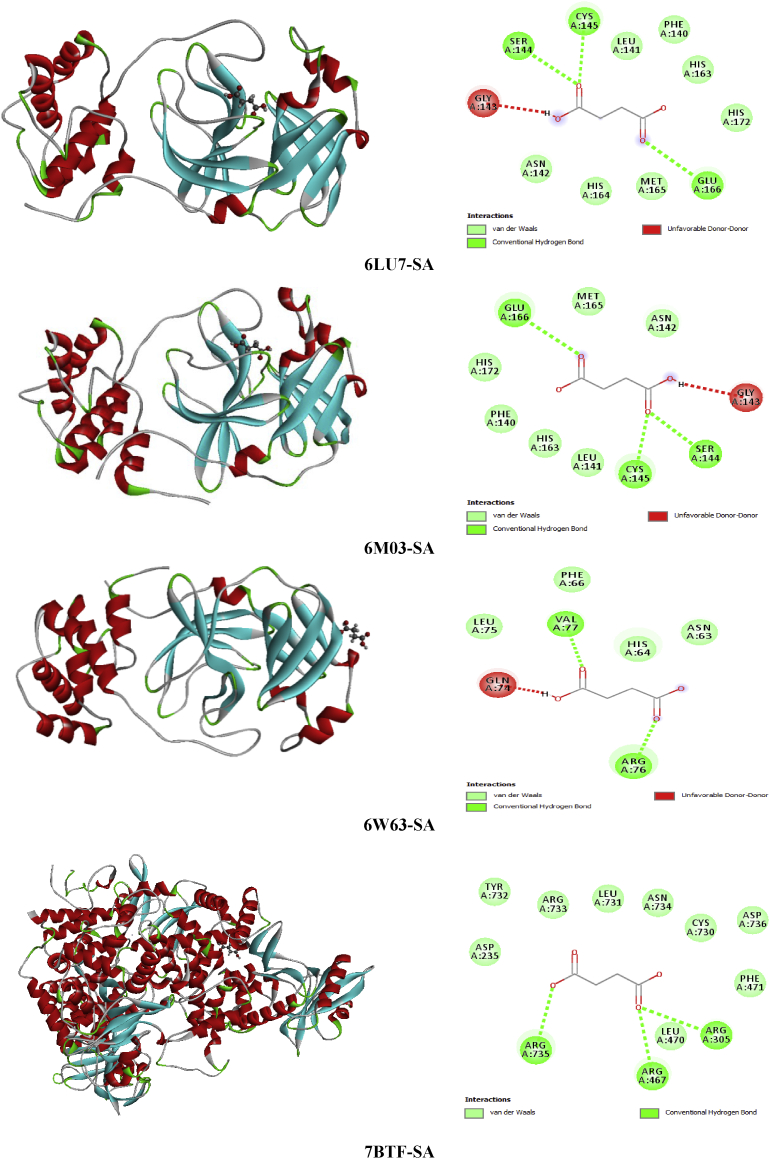
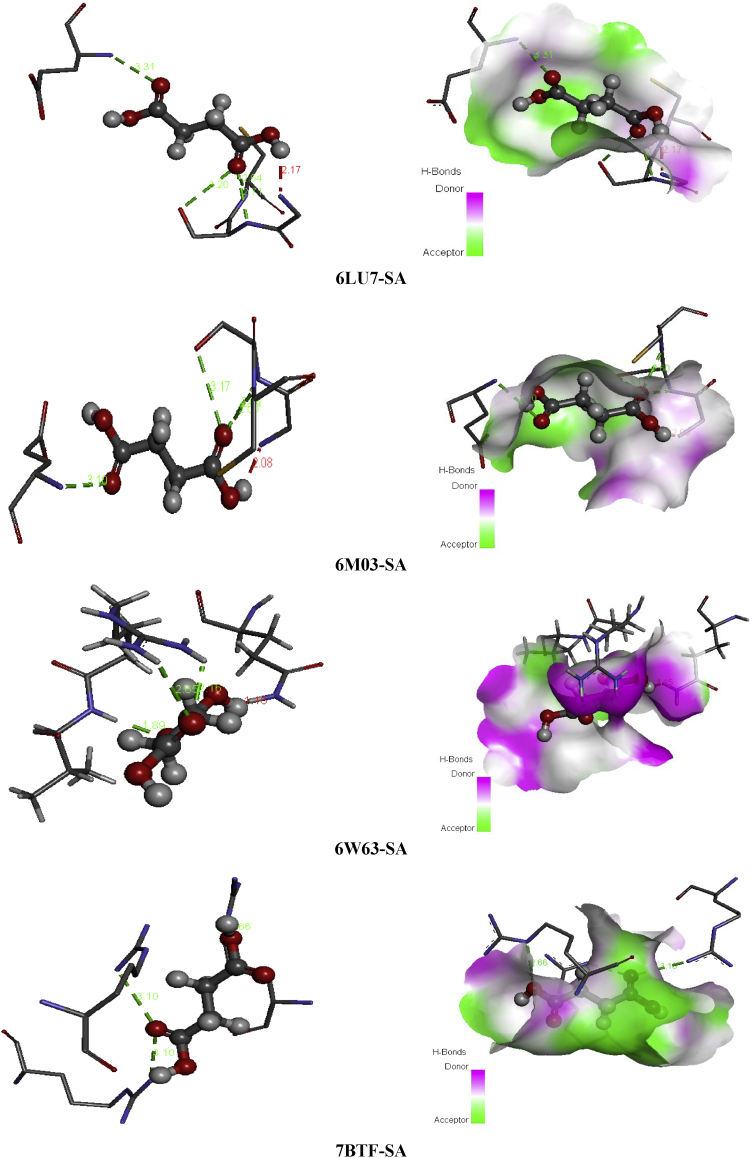
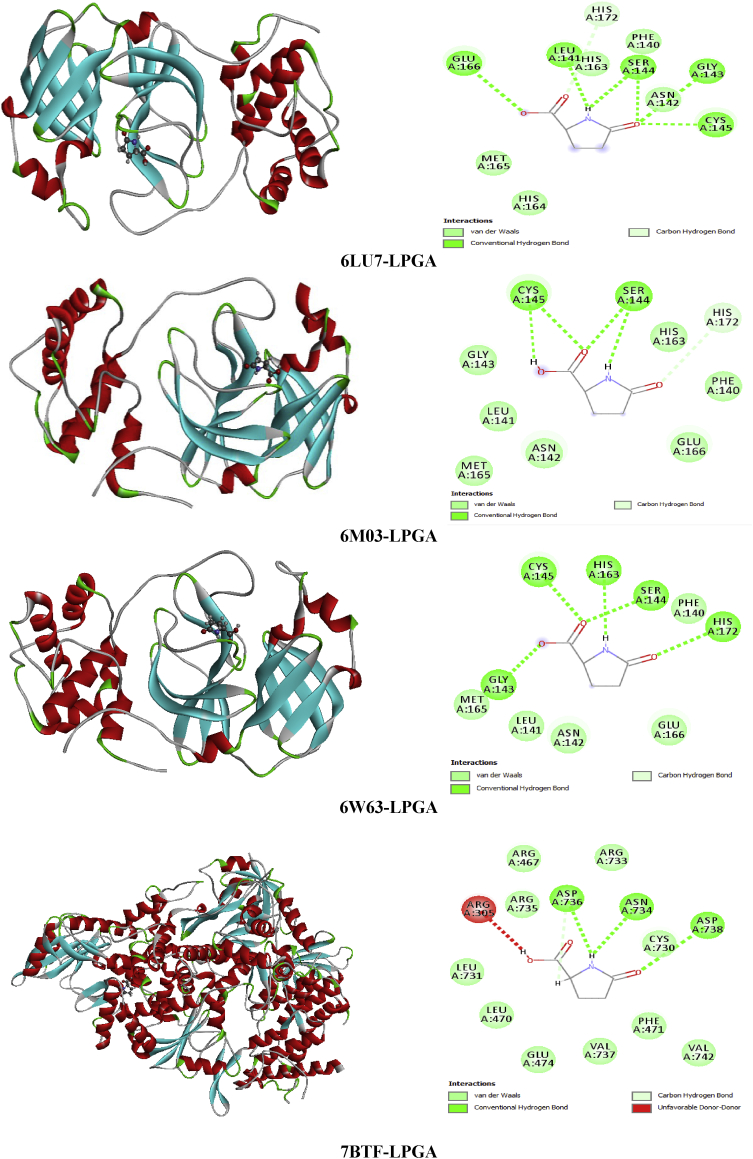
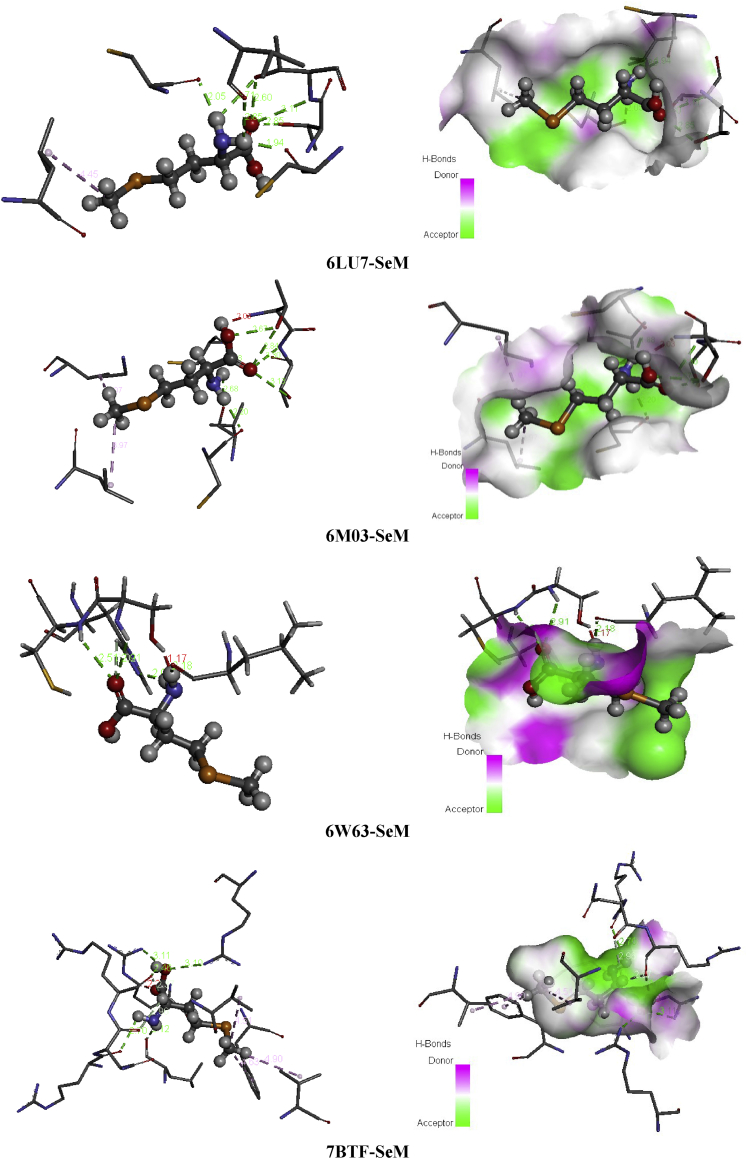
Succinic acid is known with its wide range of applications in pharmaceutical, medical and chemical. It can be used in the chemical industry for the synthesis of lubricants, cleansers and surfactants [51]; as a pH regulator in the food industry [52], flavoring agent and antimicrobial agent. It is also employed as an additive in vitamin formulation and antibiotic in the pharmaceutical industry [53, 54]. Referring to these benefits mentioned above SA seems to be a good inhibitor candidate against the coronavirus epidemic. For this reason, molecular docking simulation of Succinic acid (SA) has been performed with several coronaviruses. Docking study demonstrate the binding affinities of SA to the target coronavirus. The Table 8 collects the total energy of the docked compound and COVID-2019 viruses. According to these finding results, the SA interact better with 7BTF than that the other viruses, indicating score = -67.52 kcal/mol, EH-bond = -24.58 kcal/mol, EVDW = -37.23 kcal/mol and EElectro = -5.71 kcal/mol. The interaction energies of 6M03 and 6W63 are slightly weaker compared to 7BTF but it presents the greatest hydrogen bond -29.92 kcal/mol (6M03) and VDW interaction -40.32 kcal/mol (6W63). The best positions of the title ligand within the active site of the several viruses are mapped in Figure 9 with 2D plots and the various interactions between amino residues and SA atoms are tabulated in Table 10. For 6LU7, the oxygen atoms of carbonyl groupment (O1,O8) engaged to A:GLY:166, A:SER:144 and A:CYS:145 via three strong conventional hydrogen bonds with distances 3.31, 2.71, 2.63 Å. In 6M03 and 6W63 viruses, the O1 and O8 atoms are involved in strong conventional H-bonds (3 in 6M03 and 2 in 6W63) with A:SER:144, A:CYS:145, A:GLY:166 for 6M03 and A:ARG:76, A:VAL:77 for 6W63 with bond lengths ranging 2.64–3.09 Å (6M03), 1.88–2.15 Å (6W63). In the case of 7BTF, the O1 and O9 of the carboxylic functional group are bridged to A:ARG:305, A:ARG:467 and A:ARG:735 by hydrogen bonded interactions, having distances 3.09, 3.10 and 2.65 Å. In addition, 2 dimensional map of the candidate compound prove the presence of VDW interactions with the help of numerous amino acid residues. However, the Figure 10 illustrates the electron donor-acceptor contacts within the diverse complexes in term of H-bond surface. The oxygen atoms O8, O1 and O9 of SA play the role of an electron donor in the various systems except in 7BTF, the O1 atom of carbonyl group is considered as electron acceptor.
-
b.
L-pyroglutamic acid:
Figure 9.
The best poses of SA in 6LU7, 6M03, 6W63 and 7BTF viruses along with 2D visual representation.
Table 10.
Residues-ligand interactions for Succinic acid and the different COVID-19 constructions.
| Virus | Residues | Atoms | Distance (Å) | Category | Type |
|---|---|---|---|---|---|
| 6LU7 | A:SER:144 | O8 | 2.71 | H-bond | Conventional hydrogen bond |
| A:CYS:145 | 2.63 | ||||
| A:GLY:166 | O1 | 3.31 | |||
| 6M03 | A:SER:144 | O1 | 2.64 | H-bond | Conventional hydrogen bond |
| A:CYS:145 | 2.60 | ||||
| A:GLU:166 | O8 | 3.09 | |||
| 6W63 | A:ARG:76 | O1 | 2.15 | H-bond | Conventional hydrogen bond |
| A:VAL:77 | O8 | 1.88 | |||
| 7BTF | A:ARG:305 | O1 | 3.09 | H-bond | Conventional hydrogen bond |
| A:ARG:467 | 3.10 | ||||
| A:ARG:735 | O9 | 2.65 |
Figure 10.
Non covalent interactions and 3D H-bond surface of SA in 6LU7, 6M03, 6W63 and 7BTF.
From several decades ago, heterocyclic compound was catching the interest of many scientists all over the globe due to their diverse area of application, especially their critical role in drug discovery. Despite this long history, chemistry of heterocyclic compound was showing an undeniable interest to study. It was and remains a fascinating field of research which need to be deeply investigated. Referring to previous studies, many uses of heterocyclic compounds as antiallergic [55], analgesic [56], anti-inflammatory, antidiabetic [57] and Ocular hypotensive activities [58]. One of the most motivating cyclical amino L-pyroglutamic acid (L-PGA) which has several pharmaceutical properties. For this reason it considered as a potent molecule in the pharmaceutical drug industry. Also L-PGA possess numerous neurologist interest since it competence to treat Alzheimer disease. Regarding to their antibacterial activity [59], antioxidant activity [60], anti-inflammatory activity on RAW 264.7 macrophages [61], the L-PGA and its derivatives were in the front line of interest of many chemist and biologist. In the present work, we have been using this compound to evaluate its capacity in the inhibition of COVID-19 by using four types of coronaviruses (6LU7, 6M03, 6W63 and 7BTF). The molecular docking results reported in Table 8 showed that the binding energies of L-PGA in the following coronaviruses; 6LU7, 6M03, 6W63 and 7BTF are -70.98, -72.93, -81.44 and -73.58 kcal/mol, respectively. As clearly seen from this table, the L-PGA-6W63 complex possess the biggest interaction energy (-81.44 kcal/mol) compared with the other complexes, demonstrating the highest hydrogen bonding (-33.84 kcal) and Van der Waals (-46.82) interactions. In 6LU7 virus, the L-PGA has the lowest binding ligand -70.98 kcal/mol with the weaker VDW (-70.98 kcal/mol) interaction. Whereas, the 7BTF virus has the smallest H-bond with the stronger VDW and electrostatic interactions which found to be -17.06, 54.24 and -2.27 kcal/mol. In the title ligand, the oxygen atom of the carboxylic and keto groupment as well as the nitrogen atom of the pyrroliuimwere the most important groups for hydrogen bonds formation. The best poses of L-PGA in 6LU7, 6M03, 6W63 and 7BTF viruses along with 2D visual representation were provided in Figure 11, interactions as well as the associating bond lengths are summarized in Table 11. Concerning the 6LU7, the A:SER:144, A:GLU:143 and A:CYS:145 form hydrogen bonding interactions with O1 oxygen atom of imidazole ring with the subsequent distances; 2.66, 3.17, 2.60 Å. The hydrogen atom related to nitrogen one enter in H-bond with A:LEU:141 (2.05 Å) and A:SER:144 (2.37 Å). Then O15 and O14 atoms of carboxylic group are involved with A:GLU:166 and A:HIS:172 residues in hydrogen bonds, indicating bond lengths 3.10 and 3.60 Å. For 6M03-L-PGA, the A:SER:144 and A:CYS:145 are included in H-bonds with atoms (O14, H16) of carboxylic group as well imidazole ring (H3) with distance range 2.36–2.99 Å. The other residue, A:HIS:172, participates with O1 (3.47 Å) oxygen atom in weak carbon hydrogen bond. In addition, the docked compound on 6W63 possess five hydrogen bond category; two bonds are performed between A:GLY:143, A:HIS:163 and H16, two among A:SER:144, A:CYS:145 and O14. The last one between A:HIS:172 and the oxygen atom of imidazole ring. For the next virus (7BTF), the hydrogen H16 of hydroxyl group forms four hydrogen bond interactions with A:ARG:305, A:ASP:734 and A:ASP:736. The other H-bond is formed between O1 (3.11 Å) and A:ASP:738. On the other hand, the 2D representation shows the existing of van der Waals interactions via almost the same residues for the three first viruses; A:PHE:140, A:ASN:142…, and A:GLU:474, A:VAL:737.... for 7BTF. In the H-bond surface (Figure 12), the oxygen atom O14 of carbonyl group play the role of electron donor in 6M03 and 6W63, and electron acceptor in 6LU7. The hydrogen atom H16 of the hydroxyl group participates as an electron acceptor in 6M03 and 6W63. While in 7BTF and 6LU7 as an electron donor. The oxygen atom O15 of the same group is considered as electron donor in 6LU7. Then, the O1 attached to imidazole ring contribute like an electron acceptor in 6M03 and like electron donor in the other viruses. The H3 of this ring is an electron acceptor in 6M03 and 6LU7.
-
c.
N-phenyl-thioacetamide NPTA:
Figure 11.
The best poses of L-PGA in 6LU7, 6M03, 6W63 and 7BTF viruses along with 2D visual representation.
Table 11.
Residues-ligand interactions for L-PGA acid and the different COVID-19 constructions.
| Virus | Residues | Atoms | Distance (Å) | Category | Type |
|---|---|---|---|---|---|
| 6LU7 | A:GLU:166 | O15 | 3.10 | H-bond | Conventional H-bond |
| A:LEU:141 | H3 | 2.05 | |||
| A:SER:144 | O1 | 2.66 | |||
| H3 | 2.37 | ||||
| A:GLU:143 | O1 | 3.17 | |||
| A:CYS:145 | O1 | 2.60 | |||
| A:HIS:172 | O14 | 3.60 | Carbon H-bond | ||
| 6M03 | A:SER:144 | O14 | 2.36 | H-bond | Conventional H-bond |
| H3 | 2.94 | ||||
| A:CYS:145 | O14 | 2.60 | |||
| H16 | 2.99 | ||||
| A:HIS:172 | O1 | 3.47 | Carbon H-bond | ||
| 6W63 | A:GLU:143 | H16 | 2.21 | H-bond | Conventional H-bond |
| A:SER:144 | O14 | 2.26 | |||
| A:CYS:145 | O14 | 1.95 | |||
| A:HIS:172 | O1 | 2.53 | Carbon H-bond | ||
| A:HIS:163 | H16 | 1.91 | Conventional H-bond | ||
| 7BTF | A:ARG:305 | H16 | 2.14 | Other | Unfavourable donor-donor |
| A:ASP:738 | O1 | 3.11 | H-bond | Conventional H-bond | |
| A:ASP:734 | H16 | 2.26 | |||
| A:ASP:736 | H16 | 2.22 | Carbon H-bond | ||
| H16 | 2.73 |
Figure 12.
Non covalent interactions and 3D H-bond surface of NPTA in 6LU7, 6M03, 6W63 and 7BTF.
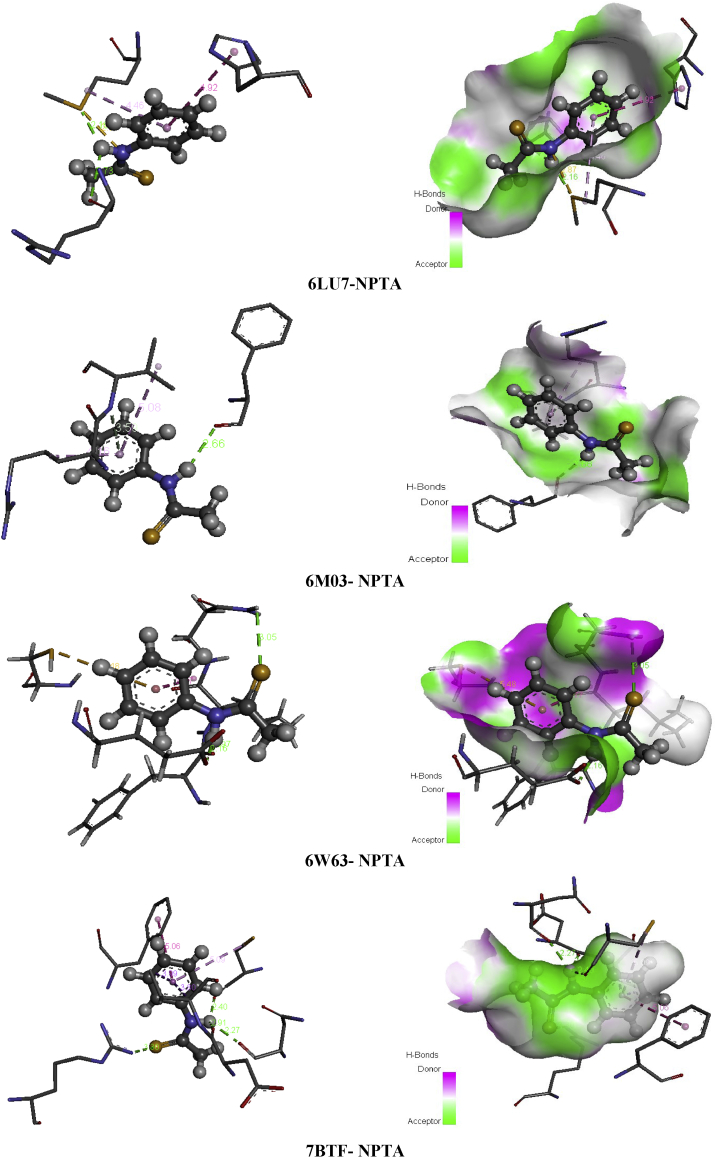
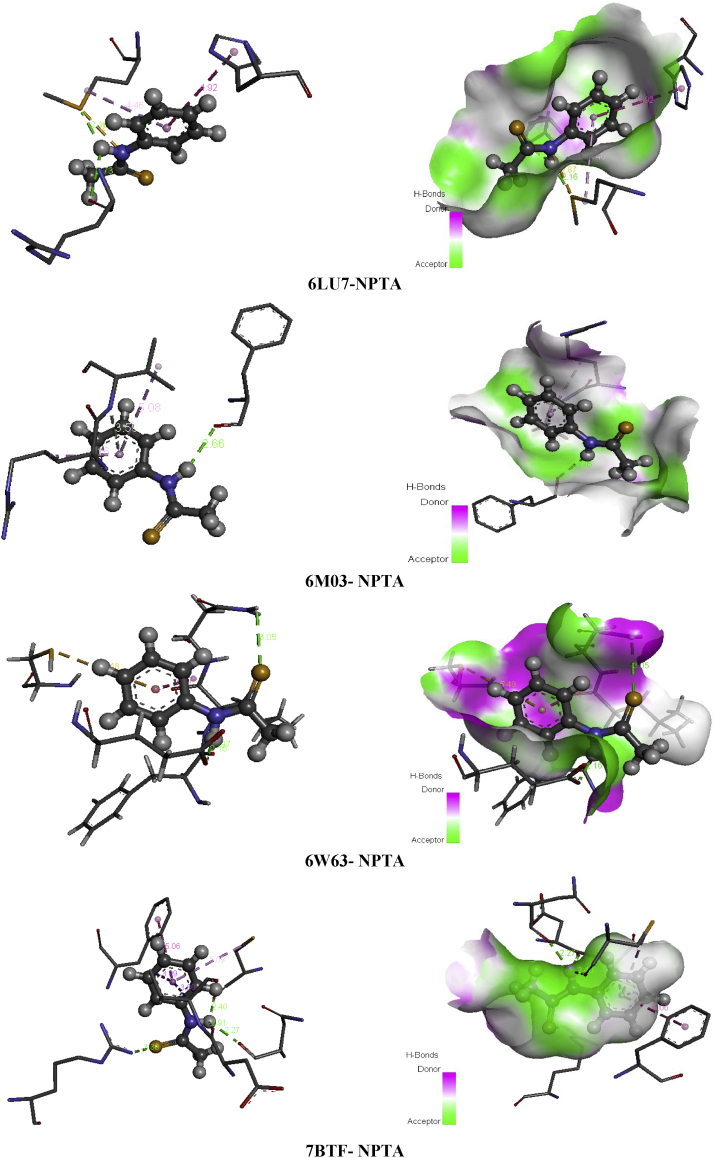
Heterocyclic compounds chemistry paid much attention since are considered as antidiuretic, anti-inflammatory, anticonvulsant and anti-cancer [62]. Thioacetamide derivatives possess significant medicinal properties ant it useful in tumor treatment [63, 64, 65, 66], it also considered as α-glucosidase inhibitor [67]. Building on their importance, we serve to test the inhibitory effect of the ligand under investigation namely N-phenyl-thioacetamide (NPTA) against COVID-19 disease. Wherefore, molecular docking study of the title compound was made with 6LU7, 6M03, 6W63 and 7BTF coronaviruses. The total binding energies of the N-phenyl-thioacetamide (NPTA) compound with four 2019-ncov structure are collected in Table 8. This latter make out that our ligand presents the best score with 7BTF which about -74.50 kcal/mol with H-bond energy (-9.59 kcal/mol) and strong VDW bond (-64.90 kcal/mol). The other structures of COVID-19 present weaker binding affinities -59.68 (6W63), -55.68 (6LU7) and -55.39 (6M03). The best poses NPTA/2019-nCoV interactions in 3D and 2D diagrams are plotted in Figure 13.
Figure 13.
The best poses of SA in 6LU7, 6M03, 6W63 and NPTA viruses along with 2D visual representation.
Table 12 present the interactions existing in each complex with distances. Concerning 6LU7, two conventional H-bonds were formed between A:MET:165, A:ARG:188 and H19 with distances 2.15 and 2.73 Å. The phenyl ring involves with A:MET:165 and A:HIS:41 in two hydrophobic interactions (pi-alkyl and pi-pi T-shaped). Likewise, the nitrogen N2 atom interact with A:MET:165 amino residue via sulfur-X interaction. For 6M03, hydrogen bond, two pi-alkyl interactions were observed among the three A:VAL:77, A:ARG:76, A:VAL:77 amino acids and phenyl ring. However, the hydrogen atoms H19 linked to nitrogen N2 present a conventional H-bond with A:PHE:66, having 2.65 Å bond length. In 6W63-NPTA complex, the S1 and H19 atoms were exhibited three hydrogen bonding with A:ASN:142, A:PHE:140, A:GLU:166, indicating bond lengths 3.04, 1.96, 2.16 Å. The phenyl ring was bridged to A:CYS:145 and A:LEU:141 throughout pi-sulfur and amide-pi stacked, respectively. For 7BTF virus, the amino acid residues A:CYS:730, A:ASN:734, A:Asp:736 were found to be implicated in the formation of conventional H-bonds with the following distances; 2.39, 2.27, 2.90 Å. Then the phenyl ring of the title compound hydrophobic bonded to A:CYS:730, A:VAL:737 and A:PHE:471 (pi-alkyl, pi-sigma, pi-pi T-shaped) with distance in the region 3.80–5.08 Å. The sulfur atom form conventional hydrogen bond with A:ARG:305 residue demonstrate 3.29 Å. It should be mentioned that van der Waals interactions in all complexes are widely founded compared to the other bonds, this is in good agreement with energetically results. In Figure 14, the H19, N2 atoms participates as an electron acceptor in all systems, while the sulphur atom is considered like electron donor and the phenyl ring as electron donor-acceptor simultaneously.
-
d.
2-amino-5-chloropyridine hydrogen succinate:
Table 12.
Residues-ligand interactions for NPTA and the different COVID-19 constructions.
| Virus | Residues | Atoms | Distance (Å) | Category | Type |
|---|---|---|---|---|---|
| 6LU7 | A:MET:165 | H19 | 2.15 | H-bond | Conventional H-bond |
| N2 | 2.87 | Other | Sulfur-X | ||
| Phenyl ring | 4.46 | Hydrophobic | pi-alkyl | ||
| A:ARG:188 | H19 | 2.73 | H-bond | Conventional H-bond | |
| A:HIS:41 | Phenyl ring | 4.29 | Hydrophobic | pi-pi T-shaped | |
| 6M03 | A:PHE:66 | H19 | 2.65 | H-bond | Conventional H-bond |
| A:VAL:77 | Phenyl ring | 3.58 | H-bond | Pi-donor hydrogen bond | |
| 5.08 | Hydrophobic | pi-alkyl | |||
| A:ARG:76 | 4.16 | pi-alkyl | |||
| 6W63 | A:ASN:142 | S1 | 3.04 | H-bond | Conventional H-bond |
| A:PHE:140 | H19 | 1.96 | |||
| A:GLU:166 | H19 | 2.16 | |||
| A:CYS:145 | Phenyl ring | 5.47 | Other | pi-sulfur | |
| A:LEU:141 | 3.76 | Hydrophobic | Amide-pi stacked | ||
| 7BTF | A:ARG:305 | S1 | 3.29 | H-bond | Conventional H-bond |
| A:CYS:730 | H19 | 2.39 | H-bond | Conventional H-bond | |
| Phenyl ring | 5.08 | Hydrophobic | pi-alkyl | ||
| A:ASN:734 | H19 | 2.27 | H-bond | Conventional H-bond | |
| A:ASP:736 | H19 | 2.90 | H-bond | Conventional H-bond | |
| A:VAL:737 | Phenyl ring | 3.80 | Hydrophobic | pi-sigma | |
| A:PHE:471 | 5.05 | pi-pi T-shaped |
Figure 14.
Non covalent interactions and 3D H-bond surface of NPTA in 6LU7, 6M03, 6W63 and 7BTF.
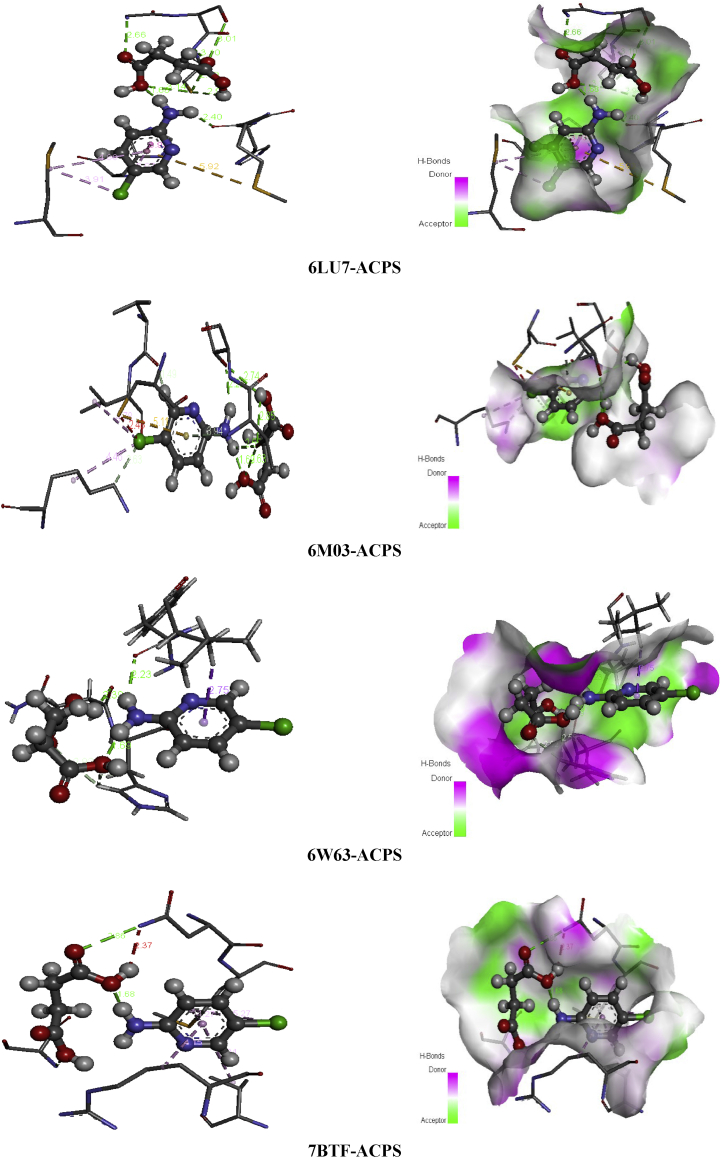
Pyridine and its derivatives are the most important compounds in the field of heterocyclic chemistry [68]. The succinic acid is a precursor for many organic materials with diverse industrial uses [69]. Its derivatives are largely exploited in medicinal, chemicals and pharmaceutical purposes [70]. One of succinic acid derivate is the 2-amino-5-chloropyridinium hydrogen succinate (ACPS) is our candidate and our aim in this section will be evaluating its inhibitory activity against 2019-nCoV by using four arrangements of coronavirus epidemic. Table 8 summarizes the interaction energies, the increasing order of the energy score of ASPC ligand on front of the four viruses is 6LU7>7BTF>6W63>6M03 with interaction energies -100.70, -99.01, -93.68, -76.77 kcal/mol. As shown, the 6LU7 present the greatest score with the strongest H-bond energy (-31.64 kcal/mol). The 7BTF demonstrate significant binding energy about -99.01 kcal/mol accompanied with the highest VDW interaction -70.42 kcal/mol which could mainly assign to an electronic delocalization within the studied compound. The docking calculation results of 2-amino-5-chloropyridine hydrogen succinate (ACPS) compound with the various 2019-nCoV structures are depicted in Figure 15. This figure presents the best docked poses of our ligand in 6LU7, 6M03, 6W63 and 7BTF. The interaction between ACPS and several amino acid residues in 2019-nCoV is reported in Table 13. For 6LU7, the carboxylic groups of succunic acid (O16, O18, O15) participate with A:GLY:143, A:SER:144, A:CYS:145 in four conventional H-bonds with the following bond lengths: 2.66, 3.00, 2.59, 3.13 Å. The hydrogen atom H27 of hydroxyl group as well as the H13 atom linked nitrogen N3 are implicated in hydrogen bonding interactions, indicating bond lengths 2.39, 2.94 Å. The residues A:MET:165, A:HIS:41, A:MET:49 form with 2-amino-5-chloropyridinium groupment pi-sulfur, pi-pi stacked and pi-alkyl bonds, respectively, with distance range between 3.92-5.91 Å. Then the chlorine atom Cl1 (3.91 Å) involve in alkyl interaction with A:MET:49. Related to 6M03, We notice the presence of carbon H-bond as well hydrophobic bonded between the residues A:LYS:61, A:ILE:43 and CL1 with distances 2.62 and 4.77 Å. The hydrogen atom of 2-amino-5-chloropyridinium group (H13,H11) are bridged to A:THR:25, A:VAL:42 via two H-bonds. In addition, the pi-donor H-bond and pi-sulfur interaction were founded among A:THR:24, A:CYS:22 and 2-amino-5-chloropyridinium ring with approximately donor-acceptor distance 3.93 and 5.11 Å. The interaction between H27 and A:THR:25, A:THR:24 were achieved by two hydrogen bonds, having 2.84 and 2.73 Å as distance. In 6W63-ACPS, the bond lengths of the subsequent hydrogen bonded interactions A:PHE:66/H13, A:ASN:63/H27, A:HIS:64/(O15,O18) are respectively 2.23, 2.39, 2.55, 2.92 Å. The pi-sigma is trained among A:LEU:67 amino acid residue and 2-amino-5-chloropyridinium ring with 2.75 Å. In 7BTF-ACPS complex, the latest ring exhibited pi-alkyl interactions with A:MET:601, A:ARG:583, A:VAL:588 with a distance equal to 4.26, 4.15, 5.25 Å. While the chlorine atom of the same group showed alkyl linkage with the amino acids A:MET:601 (5.27 Å). As we can notice, the carbonyl and the hydroxyl group of succinic acid is engaged in two conventional hydrogen bond with A:SER:592 (3.11 Å) and A:ASN:600 (2.86 Å). From Figure 16, the oxygen atom of carboxylic groups contribute as an electron donor except O15 which an electron acceptor, the hydrogen atoms (H13,H27) are an electron acceptor. The 2-amino-5-chloropyridinium ring plays the role of an electron donor (6LU7, 7BTF) and electron acceptor (6M03, 6W63). While the chlorine atom Cl1 participate as an acceptor for 6LU7, as donor for 6W63 and 6W03.
-
e.
Epigallocatechine Gallate (EGCG or KDH)
Figure 15.
The best docked poses of our ligand in 6LU7, 6M03, 6W63 and 7BTF.
Table 13.
Residues-ligand interactions for ACPS and the different COVID-19 constructions.
| Virus | Residues | Atoms | Distance (Å) | Category | Type |
|---|---|---|---|---|---|
| 6LU7 | A:GLY:143 | O16 | 2.66 | H-bond | Conventional H-bond |
| A:SER:144 | O18 | 3.00 | |||
| A:CYS:145 | O18 | 2.59 | |||
| O15 | 3.13 | ||||
| A:HIS:164 | H13 | 2.39 | |||
| A:CYS:145 | H27 | 2.94 | |||
| A:MET:165 | 2-amino-5-chloropyridinium ring | 5.91 | Other | pi-sulfur | |
| A:HIS:41 | 3.92 | Hydrophobic | pi-pi stacked | ||
| Cl1 | 5.06 | Alkyl | |||
| A:MET:49 | Cl1 | 3.91 | pi-alkyl | ||
| 2-amino-5-chloropyridinium ring | 4.88 | ||||
| 6M03 | A:THR:25 | H13 | 2.22 | H-bond | Conventional H-bond |
| H27 | 2.73 | ||||
| A:THR:24 | H27 | 2.84 | |||
| A:LYS:61 | Cl1 | 2.62 | Carbon H-bond | ||
| A:VAL:42 | H11 | 1.49 | |||
| A:THR:24 | 2-amino-5-chloropyridinium ring | 3.93 | pi-donor H-bond | ||
| A:CYS:22 | 5.11 | Other | pi-sulfur | ||
| A:ILE:43 | Cl1 | 4.77 | Hydrophobic | Alkyl | |
| A:LYS:61 | Cl1 | 4.46 | |||
| 6W63 | A:PHE:66 | H13 | 2.23 | H-bond | Conventional H-bond |
| A:ASN:63 | H27 | 2.39 | |||
| A:HIS:64 | O15 | 2.55 | Carbon H-bond | ||
| O18 | 2.92 | ||||
| A:LEU:67 | 2-amino-5-chloropyridinium ring | 2.75 | Hydrophobic | pi-sigma | |
| 7BTF | A:SER:592 | O17 | 3.11 | H-bond | Conventional H-bond |
| A:ASN:600 | O16 | 2.86 | |||
| A:MET:601 | Cl1 | 5.27 | Hydrophobic | Alkyl | |
| 2-amino-5-chloropyridinium ring | 4.26 | pi-alkyl | |||
| A:ARG:583 | 4.15 | ||||
| A:VAL:588 | 5.24 |
Figure 16.
Non covalent interactions and 3D H-bond surface of ACPS in 6LU7, 6M03, 6W63 and 7BTF.
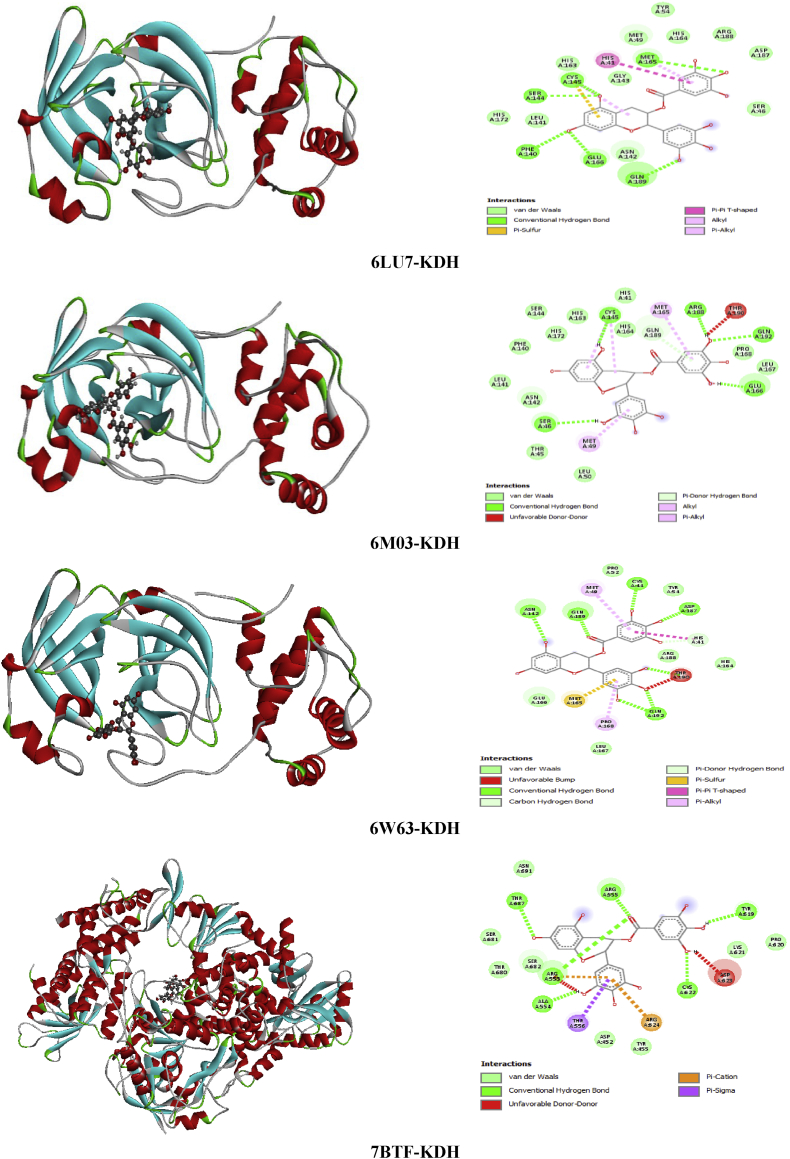
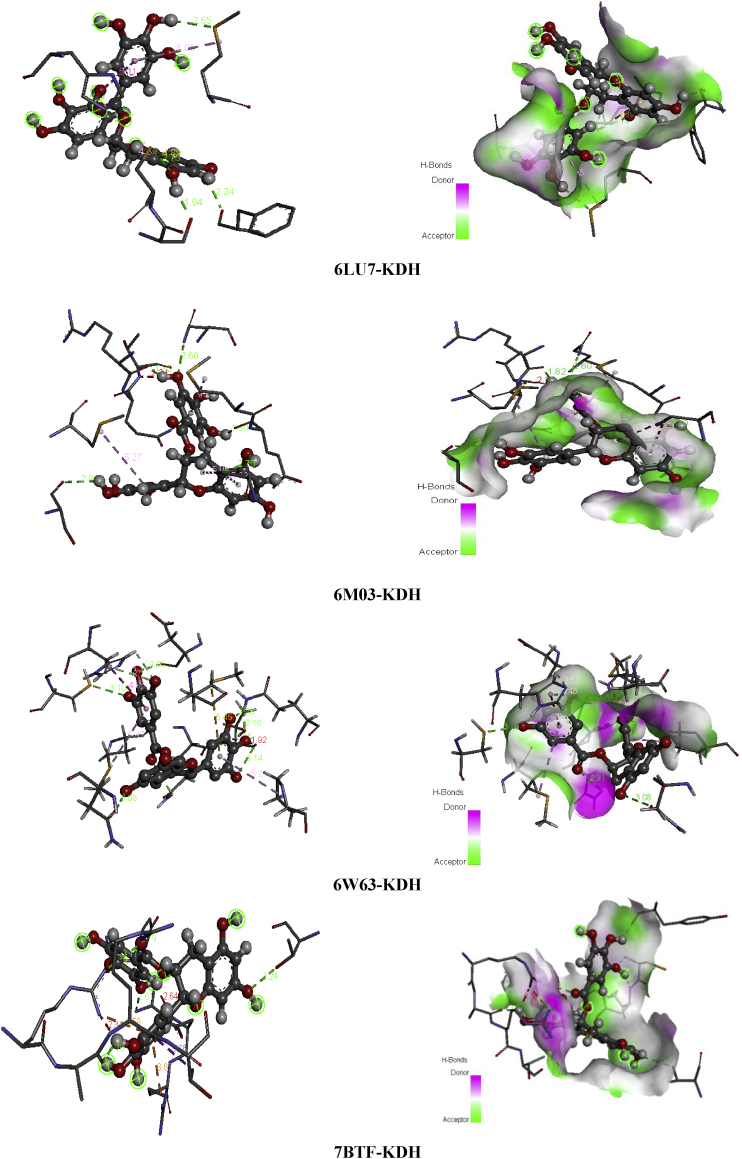
The epigallocatechin gallate (EGCG) is the most bioactive ingredient of green tea. EGCG is known with its diverse physiological activities, for instance its anti -oxidation and anti -inflammatory activities [71, 72, 73, 74]. For its natural source and its major activities EGCG seems to be one a good candidate as an inhibitor for the novel 2019-nCoV. The docking simulation results (Table 8) showed that KDH ligand has the highest binding energies with the different viruses compared to the other five ligands. The total energies of KDH with 6LU7, 6M03, 6W63 and 7BTF are found to be -135.65, -121.67, -122.12 and -129.13 kcal/mol, respectively. This ligand has the best energy with the first structure of COVID-19 (6LU7) and the second stronger score with the last structure published of novel coronavirus (7BTF). The H-bond energy of KDH is -58.71 kcal/mol (7BTF)/-37.18 kcal (6LU7) and the VDW energy is -70.42 kcal/mol (7BTF)/-98.47 kcal/mol (6LU7). We have mention that the other viruses have a strong interaction energy value with powerful hydrogen bonding and Van der Waals interactions. The binding site accompanied with 2 dimensional diagram of KDH in 6LU7, 6M03, 6W63 and 7BTF are represented in Figure 17. The intermolecular interactions type among KDH and residues of each virus with bond lengths are presented in Table 14. Relating to 6LU7, The A:MET:165, A:CYS:145, A:HIS:41 amino residues formed hydrophobic bond category with trihydroxybenzoate rings. The O1, H48, H50 and O31 atoms interact with A:GLN:189, A:PHE:140, A:MET:165 and A:GNL:189 training four hydrogen bond type. The distance between these interactions are 2.67, 2.23, 2.64, 2.98 Å. For 6M03-KDH, the trihydroxybenzoate rings are involved in two hydrophobic interactions with A:MET:149 and A:MET:165 with 5.27 and 4.26 Å, only the ring which containing C30, C18 carbons forms H-bond with A:GLN:189 (2.91Å). The A:CYS:145 amino acid acts with H41 (2.70 Å) and trihydroxybenzoate ring (5.06 Å) forming hydrogen and hydrophobic interactions. In addition, there are four other H-bonds connecting, respectively, O33, H42, H49, H51 and A:GLU:192, A:SER:46, A:GLY:166, A:ARG:188, including bond lengths 2.60, 2.94, 1.60, 1.82 Å. Concerning the 6W63-KDH complex, there are eight conventional hydrogen bonds between A:GLN:189/O27, A:THR:190/(O4,O6), A:GLN:192/(O1,O4), A:ASN:142/O24, A:ASP:187/O31, A:CYS:44/O33 with bond lengths in the range 2.16–3.14 Å. The A:MET:165, A:PRO:168, A:MET:49 implicate π-sulfur and two hydrogen bonds. The O29 and trihydroxybenzoate ring interact with A:HIS:41, having distance 2.53 and 4.95 Å. The 2D 7BTF plot show that, the trihydroxybenzoate ring of KDH molecule is implicated in 2 π-cation,π-donor H-bond with A:ARG:553 (3.59 Å) and A:ARG:624 (3.87 Å) amino groupments of coronavirus, and in hydrophobic interaction with A:THR:556 (3.75 Å). The oxygen atom O27 is involved in two H-bond interactions with A:ARG:553 and A:ARG:555 residues, having distance approximately 3.16 and 2.79 Å. Thus, the following interactions; A:CYS:622/O29, A:THR:687/O23, A:ALA:554/O6 and A:TYR:619/H50 form four hydrogen bonds with 2.60, 3.23, 1.70, 2.32 Å bond lengths. However, the four complexes mark the formation of lot VDW interactions in term of frequent amino acid residues. This is good correlation with the VDW energies previously seen. Then, H-bond plots of Figure 18 make out that all hydrogen atoms are considered as electron acceptor and the oxygen atoms join as electron donor be expecting O29 (acceptor). The phenyl rings of the title ligand may be electron donor and electron acceptor in the same time.
-
f.
Selenomethionine:
Figure 17.
The best poses of KDH in 6LU7, 6M03, 6W63 and 7BTF viruses along with 2D visual representation.
Table 14.
Residues-ligand interactions for KDH and the different COVID-19 constructions.
| Virus | Residues | Atoms | Distance (Å) | Category | Type |
|---|---|---|---|---|---|
| 6LU7 | A:GLN:189 | O1 | 2.67 | H-bond | Conventional H-bond |
| A:PHE:140 | H48 | 2.23 | |||
| A:MET:165 | H50 | 2.64 | |||
| Phenyl ring (C32,C20) | 5.04 | Hydrophobic | pi-alkyl | ||
| A:GLN:189 | O31 | 2.98 | H-bond | Carbon H-bond | |
| A:CYS:145 | Phenyl ring (C14,C13) | 4.88 | Other | pi-sulfur | |
| Phenyl ring (C9,C16) | 4.85 | Hydrophobic | Alkyl | ||
| A:HIS:41 | Phenyl ring (C32,C20) | 5.10 | pi-pi T shaped | ||
| 6M03 | A:GLU:192 | O33 | 2.60 | H-bond | Conventional H-bond |
| A:SER:46 | H42 | 2.94 | |||
| A:CYS:145 | H41 | 2.70 | |||
| Phenyl ring (C9,C16) | 5.06 | Hydrophobic | Alkyl | ||
| A:GLY:166 | H49 | 1.60 | H-bond | Conventional H-bond | |
| A:ARG:188 | H51 | 1.82 | |||
| A:GLN:189 | Phenyl ring (C32,C20) | 2.91 | pi-donor H-bond | ||
| A:MET:49 | Phenyl ring (C7,C2) | 5.27 | Hydrophobic | pi-alkyl | |
| A:MET:165 | Phenyl ring (C32,C20) | 4.26 | |||
| 6W63 | A:GLN:189 | O27 | 2.91 | H-bond | Conventional H-bond |
| A:THR:190 | O4 | 2.34 | |||
| O6 | 3.14 | ||||
| A:GLN:192 | O1 | 2.28 | |||
| O4 | 2.16 | ||||
| A:ASN:142 | O24 | 3.07 | |||
| A:ASP:187 | O31 | 2.79 | |||
| A:CYS:44 | O33 | 3.10 | |||
| A:HIS:41 | O29 | 2.53 | Carbon H-bond | ||
| Phenyl ring (C32,C20) | 4.95 | Other | pi-pi Tshaped | ||
| A:MET:165 | Phenyl ring (C7,C2) | 5.45 | Hydrophobic | pi-sulfur | |
| A:PRO:168 | 5.20 | pi-alkyl | |||
| A:MET:49 | Phenyl ring (C32,C20) | 5.41 | |||
| 7BTF | A:ARG:553 | O27 | 3.16 | H-bond | Conventional H-bond |
| Phenyl ring (C7,C2) | 3.59 | H-bond + electrostatic | pi-cation, pi donor H-bond | ||
| A:ARG:555 | O27 | 2.79 | H-bond | Conventional H-bond | |
| A:CYS:622 | O29 | 2.60 | |||
| A:THR:687 | O23 | 3.23 | |||
| A:ALA:554 | O6 | 1.70 | |||
| A:TYR:619 | H50 | 2.32 | |||
| A:ARG:624 | Phenyl ring (C7,C2) | 3.87 | H-bond + electrostatic | pi-cation, pi donor H-bond | |
| A:THR:556 | 3.75 | Hydrophobic | pi-sigma |
Figure 18.
Non covalent interactions and 3D H-bond surface of KDH in 6LU7, 6M03, 6W63 and 7BTF.
Selenium is an indispensable trace element with vital value to human health as well as its functions primarily through selenoproteins [75]. It has been a focus of innumerable research since it first discovery from half a century ago. For 20 years ago, this element has come to occupy the central focus of researchers. Relevant studies proved that its deficiency can be closely related to an increasing risk of many diseases such as cancer [76], Alzheimer's disease [77], and onset of HIV [78]. In 2011, Allen et al. [79] discovered the usefulness role of Selenium in resistance to the pathological effects of a Citrobacter rodentium infection. These discoveries highlight the important role of the cellular Se status in the process of oxidative stress reduction during inflammation. These findings come together made selenomethionine (SeMet) a good candidate to predict its inhibition activity against the new 2019-nCoV. The structural specificity of SeMet makes it more flexible and easier to interact with proteins via its H- bond acceptor and donor. In this context, molecular docking analysis of selenomethionine (SeM) with 6LU7, 6M03, 6W63 and 7BTF are carried out (Table 8). Docking computation discloses that Selenomethionine (SeM) has average binding affinities to the target coronaviruses. The binding affinities of this ligand to 7BTF exhibit score = -81.87 kcal/mol with EH-bond = - 27.86 kcal/mol, EVDW = - 49.99 kcal/mol and EElectro = - 4.00 kcal/mol. Comparing between the four complexes, the 6LU7-SeM possess the weak interaction value -67.76 kcal/mol but it has the highest H-bond energy which found to be -30.09 kcal/mol. In the other hand the two other structures of 2019-nCoV demonstrate total energies weaker than 7BTF one witch equal to -71.49 kcal/mol (6W63)/-62.86 kcal/mol (6M03). The binding mode of SeM ligand in 6LU7, 6M03, 6W63 and 7BTF active sites are reported in Figure 19 with the 2D diagram. The Table 15 attest the different interactions between our ligand and the different viruses via the amino residues with distances (Å). In 6LU7, the oxygen O4 of carbonyl group bridged to A:THR:24 and A:THR:25 with distance 2.85 and 3.11 Å. Then, the title ligand interacts with A:THR:25, A:CYS:44, A:CYS:22, A:VAL:42 via hydrogen atoms H10 and H11 of the amino radical group (NH2), indicating the presence of two conventional hydrogen bonds for each hydrogen atom with distances 2.70 and 2.04 Å (H10), 1.94 and 2.84 Å (H11). The A:LEU:57 was involved in alkyl interaction with the carbon atom C9 with bond length 4.45 Å. In the case of 6M03, two hydrogen bonding interactions were noticed between A:THR:24 and A:THR:25 with donor oxygen atoms (O4,O5) of carboxylic acid with distances equal to 2.84, 2.66, 3.12 Å. The hydrogen atoms of the amino radical group are implicated in three conventional hydrogen bonds with A:CYS:44, A:CYS:22 and A:VAL:42, demonstrating distance in the range 1.88–2.68 Å. The alkyl interaction was established among A:LEU:57 residue and C9 with 3.96 Å donor –acceptor distance. Two H-bonds takes place between the keto groupment (O4) and A:SER:144, A:CYS:145 amino acids with 2.91 and 2.51 Å. Then, two conventional hydrogen bonds are omnipresent between a hydrogen atom of the amino radical (H10 and H11) and A:LEU:141, A:HIS:163. Only A:HIS:163 get in touch with the carbonyl group (O4) in carbon H-bond. For 7BTF virus, the A:ARG:305 and A:ARG:467 residues interact with the hydroxyl group (O5 = 3.09, 3.11 Å) forming two H-bonds. Also the hydrogen atom H13 of hydroxyl group forms three H-bonds (2 conventional H-bond and carbon H-bond) with A:ARG:735, A:ASN:734, A:ARG:735 Å. Then, two conventional H-bonds were shown by H10 of amino radical with A:LEU:731, A:ARG:733 with distance 2.12 and 2.70 Å. Two alkyl and pi-alkyl linkage among C9 and A:VAL:737, A:VAL:742 and A:PHE:471 with 4.51, 4.90, 4.62 Å distances. In addition, from 2D representation, we can notice the attendance of VDW interactions throughout the different residues. These interactions justify the strength of energy value which founded above. The Figure 20 presents the hydrogen surface which identifies the electron acceptor-donor atoms. According to this figure, the oxygen (O4,O5) and the hydrogen (H10, H11, H13) atoms which intervene in the intermolecular interactions between the ligand and viruses are considered as electron donor and electron acceptor, respectively.
Figure 19.
The best poses of SeM in 6LU7, 6M03, 6W63 and 7BTF viruses along with 2D visual representation.
Table 15.
Residues-ligand interactions for SeM and the different COVID-19 constructions.
| Virus | Residues | Atoms | Distance (Å) | Category | Type |
|---|---|---|---|---|---|
| 6LU7 | A:THR:24 | O4 | 2.85 | H-bond | Conventional H-bond |
| A:THR:25 | 2.59 | ||||
| H10 | 2.70 | ||||
| A:CYS:44 | 2.04 | ||||
| A:CYS:22 | H11 | 1.94 | |||
| A:VAL:42 | 2.84 | ||||
| A:LEU:57 | C9 | 4.45 | Hydrophobic | Alkyl | |
| 6M03 | A:THR:24 | O4 | 2.84 | H-bond | Conventional H-bond |
| O5 | 2.66 | ||||
| A:THR:25 | O4 | 3.12 | |||
| A:CYS:44 | H10 | 2.20 | |||
| A:CYS:22 | H11 | 1.88 | |||
| A:VAL:42 | H10 | 2.68 | |||
| A:LEU:57 | C9 | 3.96 | Hydrophobic | Alkyl | |
| 6W63 | A:SER:144 | O4 | 2.91 | H-bond | Conventional H-bond |
| A:CYS:145 | 2.51 | ||||
| A:LEU:141 | H10 | 2.18 | |||
| A:HIS:163 | H11 | 2.09 | |||
| O4 | 3.02 | Carbon H-bond | |||
| 7BTF | A:ARG:305 | O5 | 3.09 | H-bond | Conventional H-bond |
| A:ARG:467 | 3.11 | ||||
| A:LEU:731 | H10 | 2.12 | |||
| A:ARG:733 | 2.70 | ||||
| A:ARG:735 | H13 | 2.46 | |||
| 2.36 | Carbon H-bond | ||||
| A:ASN:734 | 2.96 | ||||
| A:VAL:737 | C9 | 4.51 | Hydrophobic | Alkyl | |
| A:VAL:742 | 4.90 | ||||
| A:PHE:471 | 4.62 | pi-alkyl |
Figure 20.
Non covalent interactions and 3D H-bond surface of SeM in 6LU7, 6M03, 6W63 and 7BTF.
From the above finding, which based on binding affinities, binding score and interactions between ligand and amino acid residues as well as the position of the ligand in coronaviruses active sites, we can conclude that epigallocatechine Gallate (KDH) and then 2-amino-5-chloropyridine hydrogen succinate (ACPS) are promoter inhibitors against COVID-19 epidemic.
4. Conclusions
In the present work, the succinic acid (SA), L-pyroglutamic acid (L-PGA), N-phenyl-thioacetamide (N-NPTA), 2-amino-5-chloropyridine hydrogen succinate (ACPS), epigallocatechine Gallate (EGCG) or KDH and, selenomethionine (SeM) compounds have been proposed as potential antiviral candidates for treatment of COVID-19 combining B3LYP/6-311++G∗∗ calculations in gas phase and aqueous solution with molecular docking. Thus, solvation energies, stabilization energies, mapped molecular electrostatic potential surfaces, topological properties and frontier orbitals of those six species have been evaluated as function of acceptors and donors groups present in their structures. ACPS presents the higher reactivity in solution possibly because has the higher nucleophilicity and elecrophilicity indexes while KDH evidence the higher solvation energy possibly due to the higher quantity of donors and acceptors groups. NBO studies show that KDH is the most stable in solution; hence, it species is not reactive in this medium, as compared with ACPS. Mapped MEP surfaces have evidenced that stronger nucleophilic and electrophilic sites are present in ACPS, in agreement with the three C=O bonds and two N–H and O–H bonds with a total of 13 acceptors and donors groups present in this species while KDH has only a C=O group but a total of 19 acceptors and donors groups. From the above studies for six species we can propose that the better potential antiviral candidates to treatment of COVID-19 are ACPS and then, KDH. Molecular docking computation demonstrates that the candidate compounds KDH and in the second order ACPS are effective species in the treatment of COVID-19 disease. It possesses the highest interaction energies especially for the first and last structures of this virus (6LU7 and 7BTF) with abundant interaction types (H-bond, VDW, hydrophobic....). The four remained studied compounds showed also an important total energy which makes them an important candidate to study. Finally, we hope that our contribution can help to develop a rigorous solution to this worldwide concern.
Declarations
Author contribution statement
Abir Sagaama: Conceived and designed the analysis; Wrote the paper.
Silvia Antonia Brandan, Noureddine Issaoui: Conceived and designed the analysis; Analyzed and interpreted the data; Wrote the paper.
Takoua Ben Issa: Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
This work was supported by the Ministry of Higher Education and Scientific Research of Tunisia.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J.…Niu P. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W.…Chen H.D. 2020. Discovery of a Novel Coronavirus Associated with the Recent Pneumonia Outbreak in Humans and its Potential Bat Origin. [Google Scholar]
- 3.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y.…Duan Y. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020:1–5. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 4.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W.…Hilgenfeld R. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10) doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teengam Prinjaporn, Siangproh Weena, Tuantranont Adisorn, Vilaivan Tirayut, Chailapakul Orawon, Henry Charles S. Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB, and HPV oligonucleotides. Anal. Chem. 2017;89:5428–5435. doi: 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon Ji-seong, Kim Gyudong, Jarhad Dnyandev B., Kim Hong-Rae, Shin Young-Sup, Qu Shuhao, Sahu Pramod K., Kim Hea Ok, Lee Hyuk Woo, Wang Su Bin, Kong Yun Jeong, Chang Tong-Shin, Ogando Natacha S., Kovacikova Kristina, Snijder Eric J., Posthuma Clara C., van Hemert Martijn J., Jeong Lak Shin. Design, synthesis, and anti-RNA virus activity of 6′-fluorinated-aristeromycin analogues. J. Med. Chem. 2019;62:6346–6362. doi: 10.1021/acs.jmedchem.9b00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Jimin, Shi Pei-Yong, Li Hongmin, Zhou Jia. Broad Spectrum Antiviral Agent Niclosamide and its Therapeutic Potential. ACS Infect. Dis. 2020;6(5):909–915. doi: 10.1021/acsinfecdis.0c00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiang Dao Feng, Bigley Andrew N., Desormeaux Emily, Narindoshvili Tamari, Raushel Frank M. Enzyme-catalyzed kinetic resolution of chiral precursors to antiviral prodrugs. Biochemistry. 2019;58:3204–3211. doi: 10.1021/acs.biochem.9b00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Shu-Lin, Wang Zhi-Gang, Xie Hai-Yan, Liu An-An, Lamb Don C., Pang Dai-Wen. Single-virus tracking: from imaging methodologies to virological applications. Chem. Rev. 2020;120:1936–1979. doi: 10.1021/acs.chemrev.9b00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Chao, Zhao Lei, Xia Shuai, Zhang Tianhong, Cao Ruiyuan, Liang Guodong, Li Yue, Meng Guangpeng, Wang Weicong, Shi Weiguo, Zhong Wu, Jiang Shibo, Liu Keliang. De Novo design of α-helical lipopeptides targeting viral fusion proteins: a promising strategy for relatively broad-spectrum antiviral drug discovery. J. Med. Chem. 2018;61:8734–8745. doi: 10.1021/acs.jmedchem.8b00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Łoczechin Aleksandra, Séron Karin, Barras Alexandre, Giovanelli Emerson, Belouzard Sandrine, Chen Yen-Ting, Metzler-Nolte Nils, Boukherroub Rabah, Dubuisson Jean, Szunerits Sabine. Functional carbon quantum dots as medical count, ermeasures to human coronavirus. ACS Appl. Mater. Interfaces. 2019;11:42964–42974. doi: 10.1021/acsami.9b15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sofian Gatfaoui, Abir Sagaama, Noureddine Issaoui, Thierry Roisnel, Houda Marouani, Headlining non-covalent interactions of 1-ethylpiperazine-1,4-diium bis(nitrate) with COVID-19: synthesis, experimental and theoretical study, Solid State Sciences, Accepted.
- 13.De Clercq E. Antiviral drugs: current state of the art. J. Clin. Virol. 2001;22:73–89. doi: 10.1016/s1386-6532(01)00167-6. [DOI] [PubMed] [Google Scholar]
- 14.De Clercq E. Antiviral drugs in current clinical use. J. Clin. Virol. 2004;30:115–133. doi: 10.1016/j.jcv.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Márquez Maria Belén, Brandán Silvia Antonia. A structural and vibrational investigation on the antiviral deoxyribonucleoside thymidine agent in gas and aqueous solution phases. Int. J. Quant. Chem. 2014;114(3):209–221. [Google Scholar]
- 16.Romani Davide, Márquez María J., Márquez María B., Brandán Silvia A. Structural, topological and vibrational properties of an isothiazole derivatives series with antiviral activities. J. Mol. Struct. 2015;1100:279–289. [Google Scholar]
- 17.Checa María A., Rudyk Roxana A., Chamorro Eduardo E., Brandán Silvia A. Chapter 1, structural and vibrational properties of a reverse inhibitor against the HIV virus, dideoxynucleoside zalcitabine in gas and aqueous solution phases. In: Brandán Silvia A., editor. Descriptors, Structural and Spectroscopic Properties of Heterocyclic Derivatives of Importance for the Health and the Enviromental. Edited Collection, Nova Science Publishers, Inc; 2015. pp. 1–26. [Google Scholar]
- 18.Brandán Silvia Antonia. Structural, topological, electronic and vibrational properties of the antiviral trifluridine agent. their comparison with thymidine. Paripex Indian J. Res. 2017;6(10):346–360. [Google Scholar]
- 19.Romani Davide, Brandán Silvia Antonia. Spectroscopic and structural study of the antiviral idoxuridine agent by using DFT and SCRF calculations, IJSRM. Int. J. Soc. Res. Methodol. 2017;8(1):66–86. 2454-2008. [Google Scholar]
- 20.Romani Davide, Brandán Silvia Antonia. Investigating the structural and vibrational properties of the nucleoside reverse transcriptase inhibitor emtricitabine, IJSRM. Int. J. Soc. Res. Methodol. 2017;8(1):236–277. 2454-2008. [Google Scholar]
- 21.Iramain Maximiliano A., Ledesma Ana E., Romani Davide, Brandán Silvia Antonia. Structural, electronic, topological and vibrational properties of isomers of thymidine. A study combining spectroscopic data with DFT calculations. IJSRM, Int. J. Soc. Res. Methodol. 2017;8(2):197–238. [Google Scholar]
- 22.Iramain Maximiliano A., Brandán Silvia A. Structural and vibrational study on the acid, hexa-hydrated and anhydrous trisodic salts of antiviral drug Foscarnet, Drug Designing & Intellectual Properties. Int. J. 2018;1(3):1–17. [Google Scholar]
- 23.Ladetto María F., Márquez María J., Romani Davide, Brandán Silvia A. Structural and vibrational properties of the antiviral ribavirin drug in gas and aqueous environmental. A complete assignment of their vibrational spectra. J. Adv. Chem. 2019;16:6325–6353. [Google Scholar]
- 24.Romani Davide, Brandán Silvia Antonia. Effect of the side chain on the properties from cidofovir to brincidofovir, an experimental antiviral drug against to Ebola virus disease. Arab. J. Chem. 2019;12:2959–2972. [Google Scholar]
- 25.Romani Davide, Noureddine Olfa, Issaoui Noureddine, Brandán Silvia Antonia. Properties and Reactivities of Niclosamide in Different Media, a Potential Antiviral to Treatment of COVID-19 byUsing DFT Calculations and Molecular Docking. Biointerface Res. Appl. Chem. 2020;10(6):7295–7328. [Google Scholar]
- 26.Issa T.B., Sagaama A., Noureddine I. Computational study of 3-thiophene acetic acid: molecular docking, electronic and intermolecular interactions investigations. Comput. Biol. Chem. 2020;86:107268. doi: 10.1016/j.compbiolchem.2020.107268. [DOI] [PubMed] [Google Scholar]
- 27.Veber D.F., Johnson S.R., Cheng H.-Y., Brian R., Ward K.W., Kopple K.D. Molecualr Properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 28.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development setting. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 29.A.B. Nielsen, A.J. Holder, Gauss View 3.0, User’s Reference, GAUSSIAN Inc., Pittsburgh, PA, 2000–2003.
- 30.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., Nakatsuji H., Caricato M., Li X., Hratchian H.P., Izmaylov A.F., Bloino J., Zheng G., Sonnenberg J.L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J.A., Jr., Peralta J.E., Ogliaro F., Bearpark M., Heyd J.J., Brothers E., Kudin K.N., Staroverov V.N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Rega N., Millam J.M., Klene M., Knox J.E., Cross J.B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R.E., Yazyev O., Austin A.J., Cammi R., Pomelli C., Ochterski J.W., Martin R.L., Morokuma K., Zakrzewski V.G., Voth G.A., Salvador P., Dannenberg J.J., Dapprich S., Daniels A.D., Farkas Ö., Foresman J.B., Ortiz J.V., Cioslowski J., Fox D.J. Gaussian, Inc.; Wallingford CT: 2009. Gaussian 09, Revision A.02. [Google Scholar]
- 31.Becke A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. 1988;A38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 32.Lee C., Yang W., Parr R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. 1988;B37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 33.Miertus S., Scrocco E., Tomasi J. Electrostatic interaction of a solute with a continuum. Chem. Phys. 1981;55:117–129. [Google Scholar]
- 34.Tomasi J., Persico J. Molecular interactions in solution: an overview of methods based on continous distributions of the solvent. Chem. Rev. 1994;94:2027–2094. [Google Scholar]
- 35.Marenich A.V., Cramer C.J., Truhlar D.G. Universal solvation model based on solute electron density and a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J. Phys. Chem. 2009;B113:6378–6396. doi: 10.1021/jp810292n. [DOI] [PubMed] [Google Scholar]
- 36.Ugliengo P. University of Torino, Dipartimento Chimica IFM; Torino, Italy: 1998. Moldraw Program. [Google Scholar]
- 37.Biegler-Köning F., Schönbohm J., Bayles D. AIM2000; A program to analyze and visualize atoms in molecules. J. Comput. Chem. 2001;22:545. doi: 10.1002/jcc.10085. [DOI] [PubMed] [Google Scholar]
- 38.Bader R.F.W. Oxford University Press; Oxford: 1990. Atoms in Molecules, A Quantum Theory. [Google Scholar]
- 39.Glendening E.D., Badenhoop J.K., Reed A.D., Carpenter J.E., Weinhold F. University of Wisconsin; Madison, WI: 1996. NBO 3.1; Theoretical Chemistry Institute. [Google Scholar]
- 40.Kausteklis J., Aleksa V., Iramain M.A., Brandán S.A. Cation-anion interactions in 1-buthyl-3-methyl imidazolium nitrate ionic liquid and their effect on their structural and vibrational properties. J. Mol. Struct. 2018;1164:1–14. [Google Scholar]
- 41.Ben M’leh C., Brandán S.A., Issaoui N., Roisnel T., Marouani H. Synthesis and physic-chemical properties of a novel chromate compound with potential biological applications, bis(2-phenylethylammonium) chromate(VI) J. Mol. Struct. 2019;1185:168–182. [Google Scholar]
- 42.Ruiz Hidalgo J., Neske A., Iramain M.A., Alvarez P.E., Leyton Bongiorno P., Brandán S.A. FT-IR, FT-Raman and UV-visible spectra of motrilin acetogenin isolated from Annona cherimolia. J. Mol. Struct. 2019;1196:508–517. [Google Scholar]
- 43.Kausteklis J., Aleksa V., Iramain M.A., Brandán S.A. DFT study and vibrational assignment of 1-Butyl-3-methylimidazolium trifluoromethanesulfonate ionic liquid by using the FT-Raman spectrum. J. Mol. Struct. 2019;1175:663–676. [Google Scholar]
- 44.Rudyk R.A., Checa M.A., Catalán C.A.N., Brandán S.A. Structural, FT-IR, FT-Raman and ECD studies on the free base, cationic and hydrobromide species of scopolamine alkaloid. J. Mol. Struct. 2019;1180:603–617. [Google Scholar]
- 45.Manzur M.E., Brandán S.A. S(-) and R(+) species derived from antihistaminic promethazine agent: structural and vibrational studies. Heliyon. 2019;5:e02322. doi: 10.1016/j.heliyon.2019.e02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J.M., Chen C.C. GEMDOCK: a generic evolutionary method for molecular docking. Proteins Struct. Funct. Bioinforma. 2004;55:288–304. doi: 10.1002/prot.20035. [DOI] [PubMed] [Google Scholar]
- 47.Trott O., Olson A.J., Comput J. Inside Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Discovery Studio 4.5 Guide, Accelrys Inc., San Diego. 2009. http://www.accelrys.com [Google Scholar]
- 49.Besler B.H., Merz K.M., Jr., Kollman P.A. Atomic charges derived from semi empirical methods. J. Comput. Chem. 1990;11:431–439. [Google Scholar]
- 50.https://www.rcsb.org/
- 51.Carlson A., Coggio B., Lau K., Mercogliano C., Millis J. Industrial production of succinic acid. Chem. Fuels Bio-Based Build. Blocks. 2016;1 [Google Scholar]
- 52.Kumari N., Agrawal U., Mishra A.K., Kumar A., Vasudeva P., Mohanty N.K., Saxena S. Predictive role of serum and urinary cytokines in invasion and recurrence of bladder cancer. Tumor Biol. 2017;39(4) doi: 10.1177/1010428317697552. 1010428317697552. [DOI] [PubMed] [Google Scholar]
- 53.Akhtar J., Idris A., Aziz R.A. Recent advances in production of succinic acid from lignocellulosic biomass. Appl. Microbiol. Biotechnol. 2014;98(3):987–1000. doi: 10.1007/s00253-013-5319-6. [DOI] [PubMed] [Google Scholar]
- 54.Noordover B.A., van Staalduinen V.G., Duchateau R., Koning C.E., van Benthem R.A., Mak M.…van Haveren J. Co-and terpolyesters based on isosorbide and succinic acid for coating applications: synthesis and characterization. Biomacromolecules. 2006;7(12):3406–3416. doi: 10.1021/bm060713v. [DOI] [PubMed] [Google Scholar]
- 55.Martínez-Montero S., Fernández S., Sanghvi Y.S., Theodorakis E.A., Detorio M.A., Mcbrayer T.R.…Ferrero M. Synthesis, evaluation of anti-HIV-1 and anti-HCV activity of novel 2′, 3′-dideoxy-2′, 2′-difluoro-4′-azanucleosides. Bioorg. Med. Chem. 2012;20(23):6885–6893. doi: 10.1016/j.bmc.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Acevedo C.M., Kogut E.F., Lipton M.A. Synthesis and analysis of the sterically constrained L-glutamine analogues (3S, 4R)-3, 4-dimethyl-L-glutamine and (3S, 4R)-3, 4-dimethyl-L-pyroglutamic acid. Tetrahedron. 2001;57(30):6353–6359. [Google Scholar]
- 57.Yoshinari O., Igarashi K. Anti-diabetic effect of pyroglutamic acid in type 2 diabetic Goto-Kakizaki rats and KK-A y mice. Br. J. Nutr. 2011;106(7):995–1004. doi: 10.1017/S0007114511001279. [DOI] [PubMed] [Google Scholar]
- 58.El-Nezhawy A.O., Alrobaian M., Khames A., El-Badawy M.F., Abdelwahab S.F. Design and total synthesis of (-)-codonopsinine,(-)-codonopsine and codonopsinine analogues by O-(2-oxopyrrolidin-5-yl) trichloroacetimidate as amidoalkylating agent with improved antimicrobial activity via solid lipid nanoparticle formulations. Bioorg. Med. Chem. 2019;27(7):1263–1273. doi: 10.1016/j.bmc.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 59.Tan S.W.B., Chai C.L., Moloney M.G. Mimics of pramanicin derived from pyroglutamic acid and their antibacterial activity. Org. Biomol. Chem. 2017;15(8):1889–1912. doi: 10.1039/c6ob02828d. [DOI] [PubMed] [Google Scholar]
- 60.Moutevelis-Minakakis P., Papavassilopoulou E., Michas G., Georgikopoulou K., Ragoussi M.E., Neophytou N.…Hadjipavlou-Litina D. Synthesis, in silico docking experiments of new 2-pyrrolidinone derivatives and study of their anti-inflammatory activity. Bioorg. Med. Chem. 2011;19(9):2888–2902. doi: 10.1016/j.bmc.2011.03.044. [DOI] [PubMed] [Google Scholar]
- 61.Hirai S., Horii S., Matsuzaki Y., Ono S., Shimmura Y., Sato K., Egashira Y. Anti-inflammatory effect of pyroglutamyl-leucine on lipopolysaccharide-stimulated RAW 264.7 macrophages. Life Sci. 2014;117(1):1–6. doi: 10.1016/j.lfs.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 62.Montgomery C.P., Murray B.S., New E.J., Pal R., Parker D. Acc. Chem. Res. 2009;42:925. doi: 10.1021/ar800174z. [DOI] [PubMed] [Google Scholar]
- 63.Wong E., Giandomenico C.M. Chem. Rev. 1999;99:2451. doi: 10.1021/cr980420v. [DOI] [PubMed] [Google Scholar]
- 64.Rodney P.F., Nakayama-Ratchford N., Dai H., Lippard S.J., Am J. Chem. Rev. 2007;129:8438. doi: 10.1021/ja073231f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galanski M., Jakupec M.A., Keppler B.K. Curr. Med. Chem. 2005;12:2075. doi: 10.2174/0929867054637626. [DOI] [PubMed] [Google Scholar]
- 66.Shaw E.L.T. Plat. Met. Rev. 1979;23:90. [Google Scholar]
- 67.Mohammadi-Khanaposhtani M., Rezaei S., Khalifeh R., Imanparast S., Faramarzi M.A., Bahadorikhalili S.…Larijani B. Design, synthesis, docking study, α-glucosidase inhibition, and cytotoxic activities of acridine linked to thioacetamides as novel agents in treatment of type 2 diabetes. Bioorg. Chem. 2018;80:288–295. doi: 10.1016/j.bioorg.2018.06.035. [DOI] [PubMed] [Google Scholar]
- 68.Hemamalini M., Fun H.K. 2-Amino-4-methylpyridinium 4-nitrobenzoate. Acta Crystallogr. E Struct. Rep. Online. 2010;66(2) doi: 10.1107/S1600536810025912. o335–o335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song H., Lee S.Y. Production of succinic acid by bacterial fermentation. Enzym. Microb. Technol. 2006;39(3):352–361. [Google Scholar]
- 70.Sauer C. The Cultural Geography Reader. Routledge; 2008. The morphology of landscape; pp. 108–116. [Google Scholar]
- 71.Li R., Huang Y.G., Fang D., Le W.D. (−)Epigallocatechin gallate inhibits lipopolysaccharide-induced microglial activation and protects against inflammationmediated dopaminergic neuronal injury. J. Neurosci. Res. 2004;78(5):723–731. doi: 10.1002/jnr.20315. [DOI] [PubMed] [Google Scholar]
- 72.Cavet M.E., Harrington K.L., Vollmer T.R., Ward K.W., Zhang J.Z. Anti-inflammatory and anti-oxidative effects of the green tea polyphenol epigallocatechin gallate in human corneal epithelial cells. Mol. Vis. 2011;17:533. [PMC free article] [PubMed] [Google Scholar]
- 73.Zhong Y., Chiou Y.S., Pan M.H., Shahidi F. Anti-inflammatory activity of lipophilic epigallocatechin gallate (EGCG) derivatives in LPS-stimulated murine macrophages. Food Chem. 2012;134(2):742–748. doi: 10.1016/j.foodchem.2012.02.172. [DOI] [PubMed] [Google Scholar]
- 74.Akhtar N., Haqqi T.M. Epigallocatechin-3-gallate suppresses the global interleukin-1beta-induced inflammatory response in human chondrocytes. Arthritis Res. Ther. 2011;13(3):R93. doi: 10.1186/ar3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Allan C.B., Lacourciere G.M., Stadtman T.C. Responsiveness of selenoproteins to dietary selenium. Annu. Rev. Nutr. 1999;19(1):1–16. doi: 10.1146/annurev.nutr.19.1.1. [DOI] [PubMed] [Google Scholar]
- 76.Rayman M.P. The importance of selenium to human health. Lancet. 2000;356(9225):233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 77.Lüth H.J., Holzer M., Gärtner U., Staufenbiel M., Arendt T. Expression of endothelial and inducible NOS-isoforms is increased in Alzheimer’s disease, in APP23 transgenic mice and after experimental brain lesion in rat: evidence for an induction by amyloid pathology. Brain Res. 2001;913(1):57–67. doi: 10.1016/s0006-8993(01)02758-5. [DOI] [PubMed] [Google Scholar]
- 78.Hurwitz B.E., Klaus J.R., Llabre M.M., Gonzalez A., Lawrence P.J., Maher K.J., Schneiderman N. Suppression of human immunodeficiency virus type 1 viral load with selenium supplementation: a randomized controlled trial. Arch. Intern. Med. 2007;167(2):148–154. doi: 10.1001/archinte.167.2.148. [DOI] [PubMed] [Google Scholar]
- 79.Smith A.D., Cheung L., Botero S. Long-term selenium deficiency increases the pathogenicity of a Citrobacter rodentium infection in mice. Biol. Trace Elem. Res. 2011;144(1-3):965–982. doi: 10.1007/s12011-011-9071-4. [DOI] [PubMed] [Google Scholar]