Abstract
There is an urgent need for the identification of effective therapeutics for COVID-19 and we have developed a machine learning drug discovery pipeline to identify several drug candidates. First, we collect assay data for 65 target human proteins known to interact with the SARS-CoV-2 proteins, including the ACE2 receptor. Next, we train machine learning models to predict inhibitory activity and use them to screen FDA registered chemicals and approved drugs (~100,000) and ~14 million purchasable chemicals. We filter predictions according to estimated mammalian toxicity and vapor pressure. Prospective volatile candidates are proposed as novel inhaled therapeutics since the nasal cavity and respiratory tracts are early bottlenecks for infection. We also identify candidates that act across multiple targets as promising for future analyses. We anticipate that this theoretical study can accelerate testing of two categories of therapeutics: repurposed drugs suited for short-term approval, and novel efficacious drugs suitable for a long-term follow up.
Keywords: Microbiology, Virology, Toxicology, Computer-aided drug design, Viruses, Viral disease, Structure activity relationship, SARS-CoV-2, Covid-19, Chemical informatics, Machine learning, Drug discovery, ACE2
Microbiology; Virology; Toxicology; Computer-aided drug design; Viruses; Viral disease; Structure activity relationship; SARS-CoV-2; Covid-19; Chemical informatics; Machine learning; Drug discovery; ACE2
1. Introduction
SARS-CoV-2 is a novel coronavirus that is responsible for the COVID-19 disease which is a rapidly evolving global pandemic. Coronaviruses primarily target the upper respiratory tract and the lungs, with varying degrees of severity. Related coronaviruses such as the SARS-CoV emerging in China in 2002 and the MERS-CoV in the Middle East in 2012 result in severe respiratory conditions. The SARS-CoV-2 also produces similarly severe respiratory conditions, albeit at a lower rate but with a higher contagion factor [1]. Alarmingly, infected individuals may be asymptomatic carriers, presumably harboring the viral infection in the upper airway tract, increasing the likelihood of infecting populations that are most susceptible to severe complications [2, 3].
Although the mechanisms underlying SARS-CoV-2 infection are not completely understood, select human proteins are targets for the virus including ACE2 [4]. The SARS-CoV-2 receptor binding domain (RBD) interacts strongly with the human ACE2 receptor and TMPRSS2 to enter a human cell [5]. In addition to ACE2, a recent systems-level analysis of protein-protein interaction with peptides encoded in the SARS-CoV-2 genome identified ~300 additional human proteins, of which, 66 were considered suitable candidates for identification of therapeutics [6]. Gordon et. al. performed an in vitro assay with human cells expressing 26 SARS-CoV-2 proteins, which was followed by an analysis for high-confidence interactions. Of the 100s of reported interactions 66 were prioritized, and the authors subsequently mined and tested FDA approved drugs that were known or suspected to target these human proteins. Most of the human target proteins are overexpressed in the respiratory tract. Of particular note is the entry receptor ACE2 which is expressed at high levels in a few cell types of the nasal epithelium, as well as elsewhere [6, 7]. This could be an unusual opportunity for volatile inhaled therapeutics and prophylactics that will have direct access to the cells that are infected by the virus.
The Gordon et al study also identified FDA-approved drugs that have known activity against these human protein targets or are structurally related to chemicals with known activity on the targets. While these drugs have not been comprehensively tested on the virus, another study performed high-throughput testing of ~12,000 FDA-approved or clinical stage drugs on viral replication in cell lines [8]. This study identified at least 6 potential leads that include a kinase inhibitor, a CCR1 inhibitor and 4 cysteine protease inhibitors that are candidates for testing in clinical trials.
Since the regulatory process for the approval of new drugs can take several years, the repurposing of FDA approved drugs for COVID-19 offers a potential fast-track to approval. One of the more promising candidates being tested is the antiviral Remdesivir, which has been effective in vitro [9] as well as in non-human primates [10], with human trails currently ongoing. The other drug being tested is the antimalarial, hydroxychloroquine, which showed some promise alongside the antibiotic, azithromycin, in small clinical trials [11, 12]. However, hydroxychloroquine has shown less promise in larger trials for treating COVID-19 [13].
While drug repurposing is expedient, it is possible that drugs designed for other diseases will not be as well suited to respiratory organs, where a large percentage of putative human proteins targeted by the virus are enriched [6], or to the nervous system, implicated by neurological symptoms as well as prior evidence that coronaviruses can cross the blood brain barrier [14, 15]. Drug-development strategies are also often guided by minimizing off-target interactions. Repurposed drugs might have to be used in combination, and the side effects and interactions that this entails are presently not well defined. While there are recent efforts exploring novel, directed therapies from small molecule libraries [16], it is desirable to identify 100–1000s of putative chemicals as the majority may be difficult to synthesize in mass, prove toxic at therapeutic concentrations, or yield inconsistent benefits across patients due to genetic variability. These shortcomings have significantly increased the demand for additional drugs or small molecules that might interfere with viral entry and replication. Additionally, if prophylactics or non-toxic, easy-to-use therapeutics were available even for mild cases that do not require hospitalization and experimental drug treatments, contracting the virus may nevertheless impact long-term health and community transmission [17].
There are subsequently unmet needs in COVID-19 research, including identification of compounds that target the relevant SARS-CoV-2 human proteins from (1) approved drugs, (2) FDA registered chemicals or (3) a large repository of ~14 million purchasable chemicals from the ZINC 15 database [18], which we computed additional properties for such as mammalian toxicity, vapor pressure, and logP. For 65 human protein targets that SARS-CoV-2 interacts with that had publicly available bioassay and chemical data [6], we first generated a database of predictions based on structural similarity to chemicals that interact with the targets and then machine learning models (34). Many chemicals we have identified have little or no known biological activities and are predicted to have low toxicity in addition to a wide range of vapor pressures. These data are a resource to rapidly identify and test novel, safe treatment strategies for COVID-19 and other diseases where the target proteins are relevant.
2. Results
2.1. Identification of important structural features from known inhibitors of human target proteins
In order to test whether there is a structural basis for inhibitors of the target proteins identified previously [5, 6], we used two complementary approaches to evaluate each target's training set of compounds with known activity, compiled from the literature. First, we performed an exhaustive search for maximum common substructures among active chemicals. In some cases, enriched substructures were apparent among known ligands, with slight variation in the substructure based on the sensitivity to the targets, suggesting physicochemical features may be relevant in predicting activity against these targets (Supplementary Table 1). Next, we used a machine learning pipeline for predicting chemicals that interfere with SARS-CoV-2 targets. It involves selection of important physicochemical features for each target, followed by fitting support vector machines (SVM) with these features and then evaluating the predictions using various computational validation methods (Figure 1A). The chemical features that best predicted activity for the different targets included simple 2D information, describing the type and number of bonds, but also more abstract 3D geometries (Tables 1 and 2). Identification of each target-specific feature set provides a foundation to better understand the physicochemical basis of the activity. To that end, Supplementary Tables 2-3 include more comprehensive rank ordered lists of the physicochemical features that optimally predict activity against the targets (details about the feature ranking algorithms in Materials and Methods).
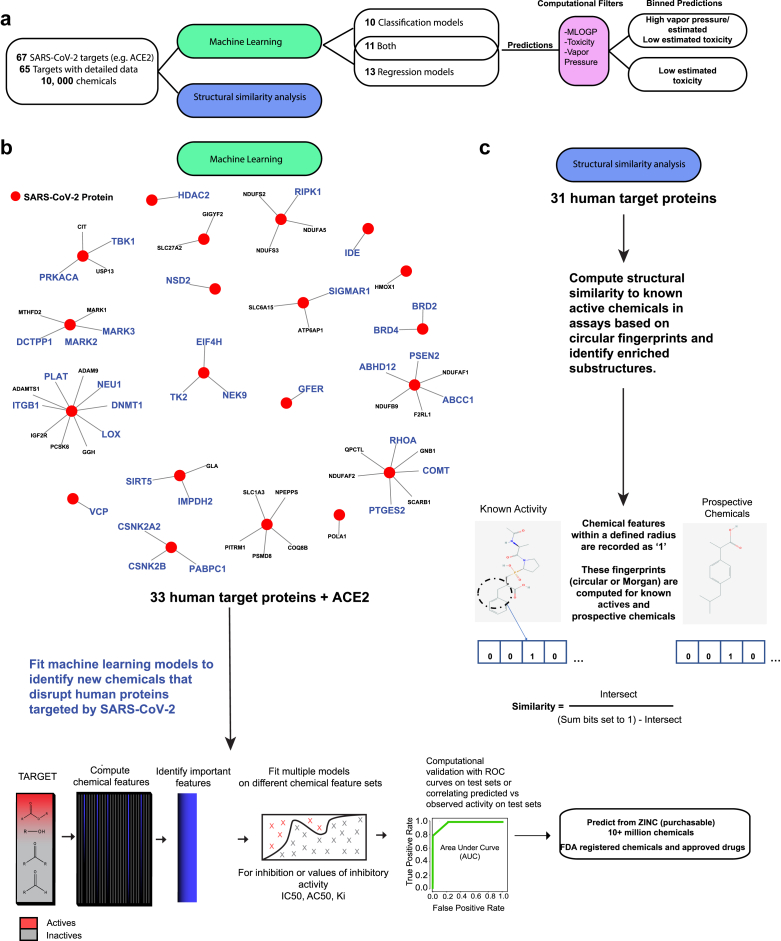
Figure 1.
Machine learning pipeline to identify chemicals that interfere with SARS-CoV-2 targets.a) Overview of the pipeline to predict chemicals for 65 SARS-CoV-2 human targets selected from Gordon et al., 2020 and using bioassay data from publicly available databases. b) Graphically depicts the pipeline details. Available bioassay data on the viral targets were mined for information to use in machine learning or structural analysis. This resulted in 24 targets that could be modeled using values for the most abundant inhibitory assay measure (e.g. Ki or IC50) and 21 targets modeled by classifying broad inhibition or actvity against the proteins (34 unique targets in total). The remaining targets with limited data were funneled into a structural similarity analysis, which aids in developing more bioassay data and helps clarify the chemical features contributing to bioactivity. For targets modeled with supervised machine learning, optimal chemical features were identified on subsets of training data. The top features were sampled by support vector machines (SVM). These models were then aggregated. In certain cases, the Random Forest algorithm was inlcuded to improve the fit. External chemicals were used to verify successful predictions. Models trained for the 34 targets predicted large chemical databases including FDA registered chemicals and approved drugs, as well as 10+ million purchasable chemicals from the ZINC database. Top scoring predicted chemicals were subsequently assigned theoretical toxicity, log vapor pressure, and MLOGP, which estimates membrane permeability.
Table 1.
Important chemical features for regression models. Top three physicochemical features for the viral targets with known bioassay activities.
| Feature | Target | Description |
|---|---|---|
| GATS5s | ABCC1 | Geary autocorrelation of lag 5 weighted by I-state |
| RDF055m | ABCC1 | Radial Distribution Function - 055/weighted by mass |
| SpMax_B(s) | ABCC1 | leading eigenvalue from Burden matrix weighted by I-State |
| CATS2D_08_AA | BRD2 | CATS2D Acceptor-Acceptor at lag 08 |
| RDF035s | BRD2 | Radial Distribution Function - 035/weighted by I-state |
| SpDiam_X | BRD2 | spectral diameter from chi matrix |
| HATS8p | BRD4 | leverage-weighted autocorrelation of lag 8/weighted by polarizability |
| R5i+ | BRD4 | R maximal autocorrelation of lag 5/weighted by ionization potential |
| RDF035m | BRD4 | Radial Distribution Function - 035/weighted by mass |
| Eig02_EA(bo) | CSNK2A2 | eigenvalue n. 2 from edge adjacency mat. weighted by bond order |
| Eig05_EA(bo) | CSNK2A2 | eigenvalue n. 5 from edge adjacency mat. weighted by bond order |
| SpMax2_Bh(m) | CSNK2A2 | largest eigenvalue n. 2 of Burden matrix weighted by mass |
| CATS2D_04_AA | CSNK2B | CATS2D Acceptor-Acceptor at lag 04 |
| SHED_DN | CSNK2B | SHED Donor-Negative |
| SpMin1_Bh(m) | CSNK2B | smallest eigenvalue n. 1 of Burden matrix weighted by mass |
| DISPm | DCTPP1 | displacement value/weighted by mass |
| HATS7u | DCTPP1 | leverage-weighted autocorrelation of lag 7/unweighted |
| Mor31s | DCTPP1 | signal 31/weighted by I-state |
| MATS1e | DNMT1 | Moran autocorrelation of lag 1 weighted by Sanderson electronegativity |
| Mor23m | DNMT1 | signal 23/weighted by mass |
| TDB06u | DNMT1 | 3D Topological distance based descriptors - lag 6 unweighted |
| GATS4m | GFER | Geary autocorrelation of lag 4 weighted by mass |
| Mor14m | GFER | signal 14/weighted by mass |
| R5i | GFER | R autocorrelation of lag 5/weighted by ionization potential |
| DISPp | HDAC2 | displacement value/weighted by polarizability |
| IC2 | HDAC2 | Information Content index (neighborhood symmetry of 2-order) |
| P_VSA_MR_5 | HDAC2 | P_VSA-like on Molar Refractivity, bin 5 |
| F04[C–C] | IMPDH2 | Frequency of C - C at topological distance 4 |
| HOMA | IMPDH2 | Harmonic Oscillator Model of Aromaticity index |
| VE1_B(s) | IMPDH2 | coefficient sum of the last eigenvector (absolute values) from Burden matrix weighted by I-State |
| Eig02_AEA(dm) | ITGB1 | eigenvalue n. 2 from augmented edge adjacency mat. weighted by dipole moment |
| SHED_AA | ITGB1 | SHED Acceptor-Acceptor |
| SpMax2_Bh(s) | ITGB1 | largest eigenvalue n. 2 of Burden matrix weighted by I-state |
| F10[C–N] | MARK2 | Frequency of C - N at topological distance 10 |
| nPyrroles | MARK2 | number of Pyrroles |
| SaaNH | MARK2 | Sum of aaNH E-states |
| max_conj_path | MARK3 | maximum number of atoms that can be in conjugation with each other |
| SaaNH | MARK3 | Sum of aaNH E-states |
| VE1_H2 | MARK3 | coefficient sum of the last eigenvector (absolute values) from reciprocal squared distance matrix |
| GATS3s | NSD2 | Geary autocorrelation of lag 3 weighted by I-state |
| HOMA | NSD2 | Harmonic Oscillator Model of Aromaticity index |
| Mor16s | NSD2 | signal 16/weighted by I-state |
| H7m | PABPC1 | H autocorrelation of lag 7/weighted by mass |
| JGI7 | PABPC1 | mean topological charge index of order 7 |
| P_VSA_MR_2 | PABPC1 | P_VSA-like on Molar Refractivity, bin 2 |
| GATS4m | PLAT | Geary autocorrelation of lag 4 weighted by mass |
| Mor04s | PLAT | signal 04/weighted by I-state |
| R6p+ | PLAT | R maximal autocorrelation of lag 6/weighted by polarizability |
| nPyrroles | PRKACA | number of Pyrroles |
| RDF040v | PRKACA | Radial Distribution Function - 040/weighted by van der Waals volume |
| SpMin3_Bh(m) | PRKACA | smallest eigenvalue n. 3 of Burden matrix weighted by mass |
| Eig02_EA(bo) | PSEN2 | eigenvalue n. 2 from edge adjacency mat. weighted by bond order |
| nArX | PSEN2 | number of X on aromatic ring |
| VE1sign_D/Dt | PSEN2 | coefficient sum of the last eigenvector from distance/detour matrix |
| SHED_DL | PTGES2 | SHED Donor-Lipophilic |
| VE2sign_G | PTGES2 | average coefficient of the last eigenvector from geometrical matrix |
| VE3sign_G | PTGES2 | logarithmic coefficient sum of the last eigenvector from geometrical matrix |
| CATS3D_08_AL | RIPK1 | CATS3D Acceptor-Lipophilic BIN 08 (8.000–9.000 Å) |
| MATS5i | RIPK1 | Moran autocorrelation of lag 5 weighted by ionization potential |
| VE3sign_RG | RIPK1 | logarithmic coefficient sum of the last eigenvector from reciprocal squared geometrical matrix |
| BLTA96 | SIGMAR1 | Verhaar Algae base-line toxicity from MLOGP (mmol/l) |
| F10[C–C] | SIGMAR1 | Frequency of C - C at topological distance 10 |
| TPSA(Tot) | SIGMAR1 | topological polar surface area using N,O,S,P polar contributions |
| Eig01_AEA(dm) | TBK1 | eigenvalue n. 1 from augmented edge adjacency mat. weighted by dipole moment |
| HATS4i | TBK1 | leverage-weighted autocorrelation of lag 4/weighted by ionization potential |
| SdssC | TBK1 | Sum of dssC E-states |
| AROM | VCP | aromaticity index |
| E1m | VCP | 1st component accessibility directional WHIM index/weighted by mass |
| MATS5m | VCP | Moran autocorrelation of lag 5 weighted by mass |
| H5s | ACE2 | H autocorrelation of lag 5/weighted by I-state |
| Mor10m | ACE2 | signal 10/weighted by mass |
| Mor17m | ACE2 | signal 17/weighted by mass |
Table 2.
Important chemical features for classification models. Top three physicochemical features for viral targets where the models classified chemicals as active vs inactive relative to broad inhibition or activition rather than a specific assay value (e.g. Ki, IC50, and AC50).
| Feature | Target | Description |
|---|---|---|
| Mor18s | BRD4 | signal 18/weighted by I-state |
| SpMAD_G/D | BRD4 | spectral mean absolute deviation from distance/distance matrix |
| SpMax3_Bh(p) | BRD4 | largest eigenvalue n. 3 of Burden matrix weighted by polarizability |
| P_VSA_LogP_3 | HDAC2 | P_VSA-like on LogP, bin 3 |
| SHED_DA | HDAC2 | SHED Donor-Acceptor |
| SHED_DL | HDAC2 | SHED Donor-Lipophilic |
| G(N..N) | IDE | sum of geometrical distances between N..N |
| SM1_Dz(i) | IDE | spectral moment of order 1 from Barysz matrix weighted by ionization potential |
| Wap | IDE | all-path Wiener index |
| CATS2D_08_DA | TBK1 | CATS2D Donor-Acceptor at lag 08 |
| F08[N–N] | TBK1 | Frequency of N - N at topological distance 8 |
| P_VSA_e_3 | TBK1 | P_VSA-like on Sanderson electronegativity, bin 3 |
| H7m | PRKACA | H autocorrelation of lag 7/weighted by mass |
| H7s | PRKACA | H autocorrelation of lag 7/weighted by I-state |
| RDF060m | PRKACA | Radial Distribution Function - 060/weighted by mass |
| GATS6e | MARK3 | Geary autocorrelation of lag 6 weighted by Sanderson electronegativity |
| GATS6m | MARK3 | Geary autocorrelation of lag 6 weighted by mass |
| Mor02m | MARK3 | signal 02/weighted by mass |
| CATS2D_02_DL | IMPDH2 | CATS2D Donor-Lipophilic at lag 02 |
| CATS3D_07_DL | IMPDH2 | CATS3D Donor-Lipophilic BIN 07 (7.000–8.000 Å) |
| NaasC | IMPDH2 | Number of atoms of type aasC |
| C-039 | ABCC1 | Ar-C(=X)-R |
| VE2sign_Dz(p) | ABCC1 | average coefficient of the last eigenvector from Barysz matrix weighted by polarizability |
| VE3sign_Dz(v) | ABCC1 | logarithmic coefficient sum of the last eigenvector from Barysz matrix weighted by van der Waals volume |
| Mor31s | ABHD12 | signal 31/weighted by I-state |
| RTi+ | ABHD12 | R maximal index/weighted by ionization potential |
| VE3sign_Dz(p) | ABHD12 | logarithmic coefficient sum of the last eigenvector from Barysz matrix weighted by polarizability |
| E2m | BRD2 | 2nd component accessibility directional WHIM index/weighted by mass |
| GATS2m | BRD2 | Geary autocorrelation of lag 2 weighted by mass |
| TDB03i | BRD2 | 3D Topological distance based descriptors - lag 3 weighted by ionization potential |
| MAXDP | COMT | maximal electrotopological positive variation |
| nDB | COMT | number of double bonds |
| P_VSA_MR_2 | COMT | P_VSA-like on Molar Refractivity, bin 2 |
| CATS2D_02_AL | DNMT1 | CATS2D Acceptor-Lipophilic at lag 02 |
| Mor04s | DNMT1 | signal 04/weighted by I-state |
| VE3sign_Dt | DNMT1 | logarithmic coefficient sum of the last eigenvector from detour matrix |
| ChiA_B(i) | EIF4H | average Randic-like index from Burden matrix weighted by ionization potential |
| F05[C–O] | EIF4H | Frequency of C - O at topological distance 5 |
| NaasC | EIF4H | Number of atoms of type aasC |
| CENT | LOX | centralization |
| EE_G | LOX | Estrada-like index (log function) from geometrical matrix |
| VE2_D/Dt | LOX | average coefficient of the last eigenvector (absolute values) from distance/detour matrix |
| Eta_D_beta | MARK2 | eta measure of electronic features |
| Mor29v | MARK2 | signal 29/weighted by van der Waals volume |
| SpPosA_B(i) | MARK2 | normalized spectral positive sum from Burden matrix weighted by ionization potential |
| CATS2D_07_AL | NEK9 | CATS2D Acceptor-Lipophilic at lag 07 |
| CATS2D_08_AL | NEK9 | CATS2D Acceptor-Lipophilic at lag 08 |
| TDB05p | NEK9 | 3D Topological distance based descriptors - lag 5 weighted by polarizability |
| CATS2D_06_DL | NEU1 | CATS2D Donor-Lipophilic at lag 06 |
| TDB04i | NEU1 | 3D Topological distance based descriptors - lag 4 weighted by ionization potential |
| X3A | NEU1 | average connectivity index of order 3 |
| nR06 | RHOA | number of 6-membered rings |
| R8s+ | RHOA | R maximal autocorrelation of lag 8/weighted by I-state |
| SpMin1_Bh(m) | RHOA | smallest eigenvalue n. 1 of Burden matrix weighted by mass |
| CATS3D_08_NL | SIRT5 | CATS3D Negative-Lipophilic BIN 08 (8.000–9.000 Å) |
| O-057 | SIRT5 | phenol, enol, carboxyl OH |
| SpMax2_Bh(s) | SIRT5 | largest eigenvalue n. 2 of Burden matrix weighted by I-state |
| CATS2D_04_AL | TK2 | CATS2D Acceptor-Lipophilic at lag 04 |
| JGI3 | TK2 | mean topological charge index of order 3 |
| MATS1i | TK2 | Moran autocorrelation of lag 1 weighted by ionization potential |
| P_VSA_e_3 | VCP | P_VSA-like on Sanderson electronegativity, bin 3 |
| RDF020p | VCP | Radial Distribution Function - 020/weighted by polarizability |
| SpMaxA_AEA(dm) | VCP | normalized leading eigenvalue from augmented edge adjacency mat. weighted by dipole moment |
2.2. Machine learning models can successfully predict activity from chemical structure
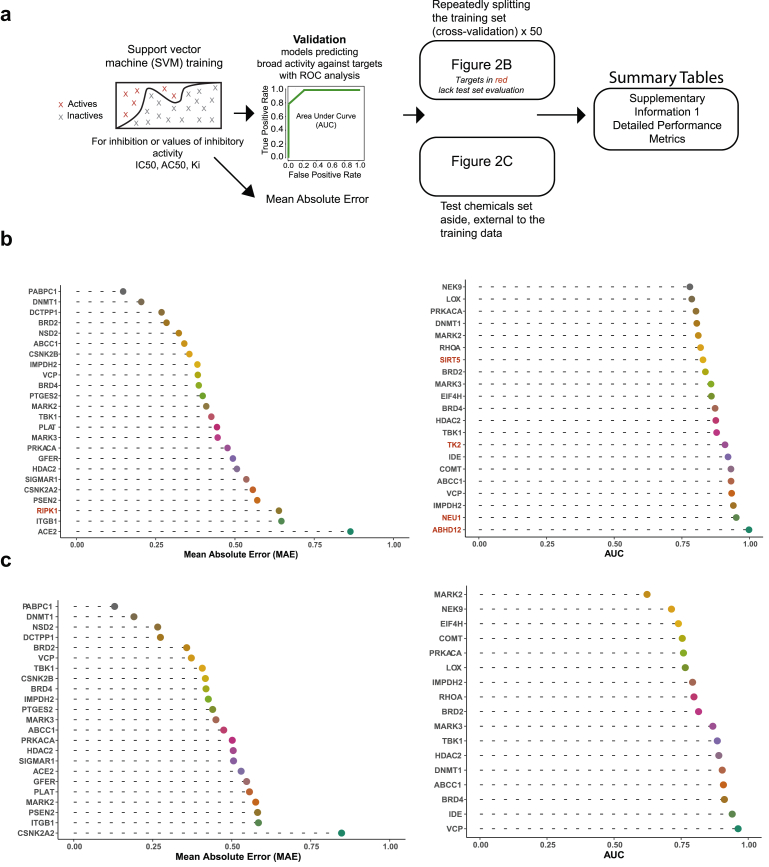
We identified 24 targets with training sets large enough to model the log IC50, Ki, or AC50 (Figure 2A). Rigorous computational validation was performed and the results on training (Figure 2B, left) and test data that had been set aside (Figure 2C, left) indicated good overall performance according to the average mean absolute error (MAE) and the correlation between predicted and observed assay measures (MAE = 0.48; R = 0.62). Predictions of log Ki for the viral entry receptor, ACE2, were also accurate (test set R = 0.92; test set mean absolute error (MAE) = 0.53) (Figure 2C, left; Supplementary Information 1).
Figure 2.
Models of chemical features accurately predict inhibitors of SARS-CoV-2 targets.a) Pipeline for fitting and validating models that predict IC50, Ki, or AC50 or a classification score, which reflects broad inhibitory activity against the listed viral targets. b) Left, mean absolute error (MAE) in predicting the log transformed endpoints (IC50, Ki, AC50). Right, classification of chemicals for broad inhibition or activity against targets, validating using the area under the receiver operating characteristic (ROC) curve (AUC). Plots are for 10-fold cross validation, repeated 5 times. The model predictions are from an ensemble of three support vector machines (SVM), trained on different chemical feature sets or in some cases SVM and Random Forest. c) Left, external test set performance for regression models, where possible. Right, external test set performance for classification models, where possible. More comprehensive performance data in Supplementary Information 1.
For some of the viral targets, we noticed that assay data included additional inhibitory measurements or descriptions of general activity against the targets. Some of the available data such as % inhibition, for instance, are less quantitative. However, to include as much of the available data as possible, we created models to identify physicochemical features that might broadly contribute to inhibition or activity against the targets. We therefore assigned binary, active and inactive, labels to the chemicals, then trained models as outlined before (Figure 2A; Materials and Methods). The models that were developed using this classification approach similarly proved successful, validating over partitions of the training data (avg. AUC = 0.87, avg. Shuffle AUC = 0.50, p < 10−19) (Figure 2B, right), as well as over sets of external test chemicals (avg. AUC = 0.83, avg. Shuffle AUC = 0.51, p < 10−8) (Figure 2C, right) (Supplementary Information 1). Collectively, these results suggested the models provided accurate predictions and could be used to screen approved drug libraries as well as databases of commercially available chemicals for novel therapeutics.
2.3. Predicting candidates for repurposing of FDA-approved drugs
Repurposing of existing FDA approved drugs offers a path towards rapid deployment of therapeutics against SARS-CoV-2. Approved drugs may have activity that extend beyond the original target protein. Accordingly, we used the machine learning models to predict activities of ~100,000 FDA registered chemicals (UNII database) [19] as well as the DrugBank [20] and Therapeutic Targets [21, 22] databases, which include information on drug interactions, pathways, and approval status. Interestingly, some of the approved drugs are predicted to have high activity against the SARS-CoV-2 targets (Figure 3A). In order to identify more efficacious candidates, we isolated the drugs scoring in the top 25 for multiple targets and found a few of high priority (Figure 3B). The structural analysis suggested that hits visually display 2D similarity to known active chemicals as well. (Supplementary Information 2).
Figure 3.
Approved drugs with putative activity against SARS-CoV-2 targets. a) The best predicted activity against SARS-CoV-2 targets among databases of approved drugs. Viral targets with few promising candidates are omitted. Comprehensive table in Supplementary Information 2. b) Network showing drugs that are among the top 25 for multiple viral targets (drugs: black nodes; viral targets: red nodes).
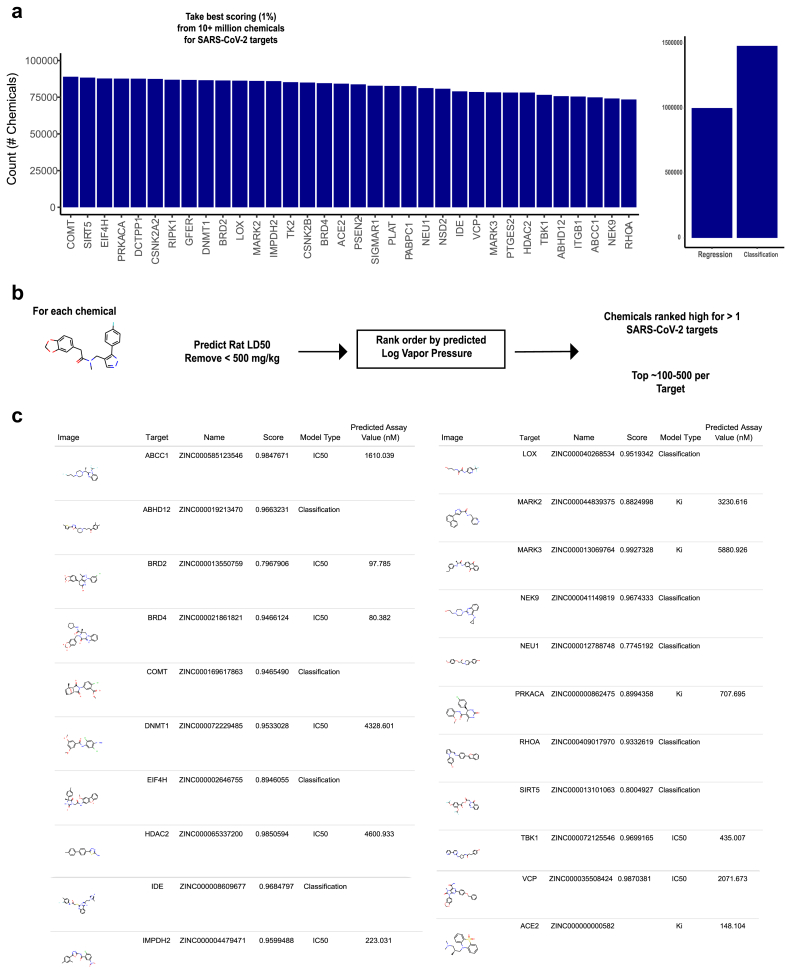
2.4. Predicting volatile drug candidates from a large ~14M chemical space
Given that many of the human target proteins are overexpressed in the respiratory tract, including the entry receptor ACE2 in only a few cells types of the nasal epithelium, the upper airways and lungs [7, 23], we reasoned that volatile chemicals may offer a unique opportunity as inhaled therapeutics that will have direct access to the cells and tissues that are infected by the virus. We used the machine learning models to search a large database of ~14 million commercially available chemicals (ZINC) for volatile candidates. We initially isolated the top 1% of the predicted scoring distribution (Figure 4A, left), which resulted in >1 million chemicals in total (Figure 4A, right). To prioritize the hits for potential human use, we next developed machine learning models to predict volatility (vapor pressure) (Supplementary Figure 1) and mammalian toxicity (LD50) (Supplementary Figure 2). The toxicity and vapor pressure estimates helped identify smaller priority sets (Figure 4B). Although the vapor pressures were not especially high, we rank ordered the top candidates according to the best values (Figure 4C; Supplementary Information 3).
Figure 4.
Predicting activity against SARS-CoV-2 targets among theoretical volatile chemicals. a) Left, count of chemicals per target after initially filtering based on predicted scores. Right, chemical counts across all viral targets for the models predicting general inhibitory or activity against (Classification) and those for specific inhibitory endpoints (Regression) (e.g. IC50). b) Pipeline for further prioritizing chemical sets according to estimated log vapor pressure and low mammalian toxicity (LD50). c) Top ranking predictions of general inhibition or activity against targets (Score) and/or specific inhibitory endpoints (Predicted Assay Value) against SARS-CoV-2 targets from the ZINC database, filtered to the highest estimated log vapor pressures.
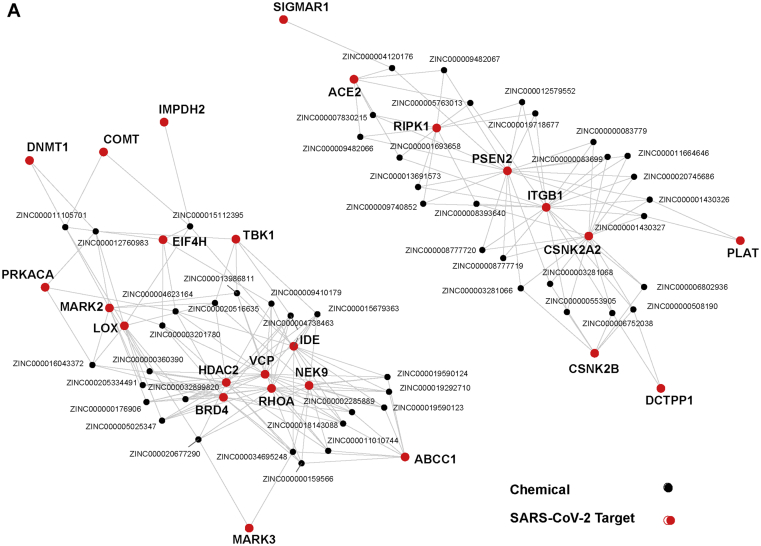
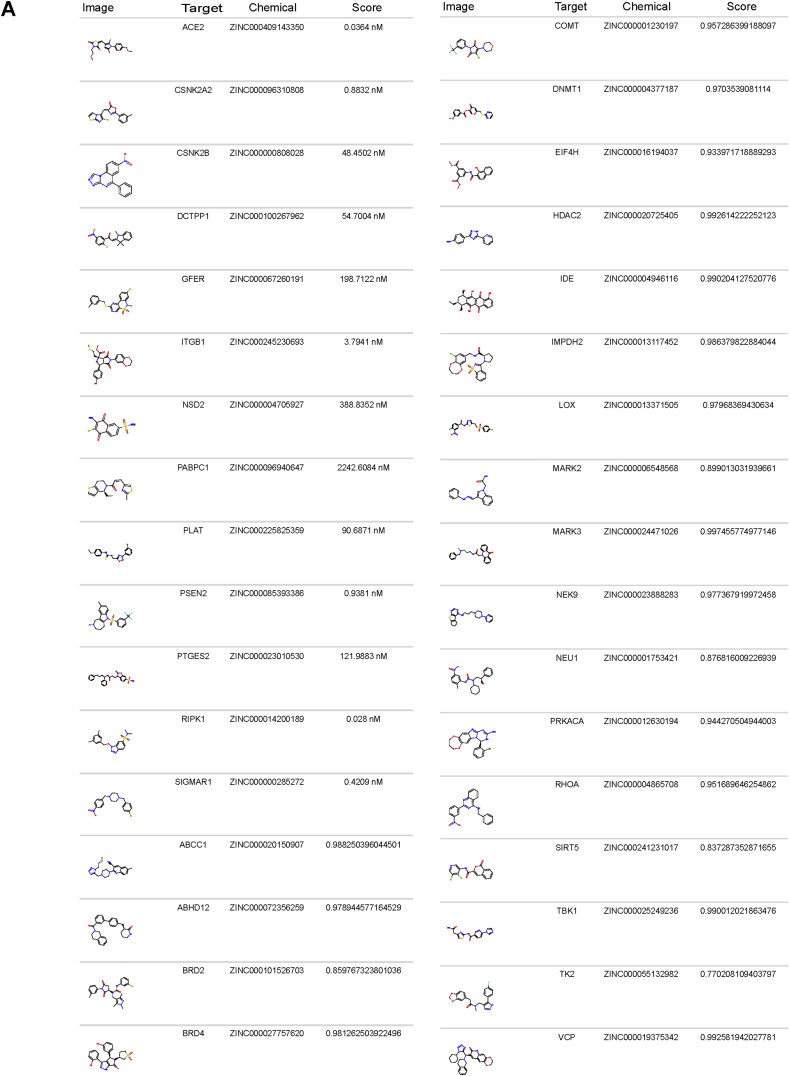
Chemicals with suspected odorant properties, however, represent only a fraction of the chemical space, and these chemicals may not have the activity levels suited for COVID-19 cases. Volatile compounds, for instance, may be biased towards structurally simple chemicals that do not resemble drugs. We therefore also focused on additional chemicals with the high predicted activities for their targets and low estimated toxicities regardless of vapor pressure. We identified numerous candidates with potential activity against multiple viral targets (Figure 5A) and many other others with significant activity against a single target (Figure 6A; Supplementary Information 4).
Figure 5.
Predicted chemicals rank highly for multiple SARS-CoV-2 targets. a) Network of chemicals predicted to have low toxicity that are ranked highly for >1 viral targets. Chemicals were considered if for multiple viral targets they had >0.75 activity/class scores or predictions of specific assay measures (Ki, IC50, and AC50) < 100 nM.
Figure 6.
Predictions of SARS-CoV-2 targets among chemicals lacking odorant properties.a) Sample of ZINC chemicals scoring highly for activity against the viral targets (classification or regression models, Score). Comprehensive tables in Supplementary Information 4, detailing the model type and predicted assay endpoint.
3. Discussion
SARS-CoV-2 is a significant world health crisis. The full scope of COVID-19 disease and any long-term health complications following infection remain unclear. Although vaccines are the best long-term solution, treatments will be necessary to mitigate disease severity in the short term. What is concerning is that several repurposed drugs have already been tested in some form of clinical trial, and only one drug Remdesivir has shown a clear benefit in randomized clinical trials. Additionally, there is no guarantee that an effective vaccine can be found for the SARS-CoV-2 virus, and therefore drug candidate pipelines are extremely important to pursue for the long-term research effort against COVID-19. A vaccine against SARS-CoV-2 would likely need to stimulate local immunity, since the infection is limited to mucosal surfaces, and these could be short-lived immunities.
We have therefore taken a comprehensive approach to try and provide a pipeline for short and long-term use, and for a potentially local application route via inhalation. Existing FDA approved drugs that target a single protein important for viral replication and host entry are currently the highest priority for repurposing as new COVID-19 drugs. However, we think that there are compelling reasons to create pipelines to explore many putative targets, and chemical spaces that are far larger and more diverse than the known approved drugs. We have therefore screened ~10+ million potentially purchasable compounds from the ZINC database and also predicted toxicity values for the numerous candidates. In addition, we have identified chemicals that are predicted to affect more than one of the host proteins, suggesting these may have more efficacy. One unusual category we have emphasized is volatiles, as these compounds may be biologically sourced, and therefore microbes could be genetically engineered to produce them in mass [24]. This would subsequently reduce the strain on global supply chains for chemicals that are necessary in synthesizing certain pharmaceuticals. These chemicals are also intriguing options for drug cocktails. If present in metabolic pathways, they possibly already interact in vivo. Therefore, short-term therapeutic concentrations may be better tolerated in humans.
It is nevertheless important to note that machine learning depends on available data. Because the size and diversity of publicly available bioassay data are limited, caution is required in interpreting the predictions. It is common to find past bioassays focused on similarly shaped chemicals, limiting the scope of the machine learning approach to find new chemistries. Importantly, apart from ACE2, the other human proteins that were identified to interact with SARS-CoV-2 are yet to be tested in vivo for efficacy. And although some of the candidate chemicals we identified may be biologically sourced, the concentrations are not well defined or unknown, nor is there any understanding of a therapeutic concentration in this scenario. These data are presented as a forward-looking resource and a pipeline to evaluate chemical data with additional research. While our motivation was the evolving COVID-19 pandemic, the 65 SARS-CoV-2 targets including ACE2 are relevant to a range of other diseases and conditions. We therefore anticipate that the AI-based predictions of purchasable compounds from 10+ million chemicals will accelerate drug discovery in general and facilitate research on these chemicals in the future for a number of diseases. In general, the use of AI-driven tools could provide additional valuable solutions for tackling Covid-19 [25].
4. Materials and methods
4.1. Data sources for machine learning
4.1.1. ZINC
ZINC is a free database comprised of 230 million chemicals for in silico analyses. It was developed as a resource for non-commercial research. Chemicals predicted here are from a purchasable subset; however, availability is subject to change and pricing may vary widely [18, 26].
4.1.2. Bioassay data
Bioassay data was retrieved from ChEMBL 25 using the associated Python module, which enables access to the API services via Python [27, 28]. The various inhibitory measures/endpoints, wherever possible, are standardized to nM units; the logarithm of the standardized values was used for machine learning. Regression models were fit for a single endpoint. For classification machine learning models, however, ‘active’ class chemicals were defined using the deposited activity comments such as for assays of general activity against proteins, and added active labels for endpoints with values up to 10,000 nM (Ki and IC50) and for the semi-quantitative % inhibition, greater than 10%. The majority class was downsampled during the training and model tuning phases to adjust for possible class imbalances. Because the class labels were assigned using arbitrary cutoffs and the predicted activities for classification models from various assay endpoints are not clearly defined, we also compared each model fit to shuffled labels. Training for the regression and classification approaches was done on 85% of the total data. Notably, in a small number of cases the remaining 15% was insufficient to effectively estimate performance using an external test set. To reduce bias, feature selection (recursive feature elimination (RFE) algorithm) was always run on 85% of the data over 250–300 different partitions (iteratively running the 10-fold cross validation 25–30 times). However, for these cases, the held-out portion (15%) was then incorporated back into the dataset to better estimate performance of the trained model by 10-fold cross-validation (repeated 5 times) and obtain a better fit. We also fit 3 different radial basis function (RBF) support vector machine (SVM) models, wherein the chemical features (predictors) were randomly sampled (50%) from the top 70. This makes the performance estimates more conservative (see Key Resources Table for machine algorithm source files). However, the structural diversity and size of the datasets imply some bias in the performance estimates.
4.1.3. Toxicity data
Training and testing data are curated by various government agencies and provided freely to the general public as databases (see Key Resources Table) [29, 30, 31].
4.1.4. Vapor pressure data
Training and testing data are from EPI Suite [32], which is developed and maintained by the Environmental Protection Agency (EPA) (see Key Resources Table). Methods for fitting these models are as outlined in the Figure 1 pipeline. To compare the vapor pressure model predictions with respect to different machine learning methods as well as EPI suite, data were split into train/test partitions as defined in a previous study [33].
4.2. Selecting optimally predictive chemical features
4.2.1. Optimizing chemical structures
Chemical features were computed with ~5300 AlvaDesc descriptors, from the developers of DRAGON software, and 3D coordinates and optimization performed using RDKit in Python [34].
4.2.2. Chemical feature ranking and importance
4.2.2.1. Cross-validated recursive feature elimination (CV-RFE)
Recursive feature elimination iteratively selects subsets of features to identify optimal sets. The algorithm is a “wrapper” and therefore relies on an additional algorithm to supply predictions and quantify importance. We used two different algorithms, depending on the size and composition of data: (1) Random Forest and (2) Support Vector Machine (SVM). Random forest determines the importance in relation to the % increase in error when permuting a feature or predictor. There is no equivalent method for computing importance with the SVM. Accordingly, the importance is based on fitting a model between the response and each predictor or feature as compared to null. If the response is numeric, importance is derived from the pseudo R2 (non-linear regression). If, however, the response is binary, the AUC is instead computed for each predictor or feature (see Key Resources Table for algorithm source files).
Including cross-validation with the recursive feature elimination (RFE) partitions the training data into multiple folds. This step avoids biasing performance estimates but results in lists of top predictors over the cross-validation folds such that importance of a predictor is based on a selection rate.
4.2.2.2. Selection bias
Selecting features or predictors on the same dataset used for cross validation results in models that have already “seen” possible partitions of the data and therefore performance metrics will be biased. Selection bias [35] was addressed by bootstrapping and cross validation, which ensure some separation between predictor/feature selection and model-fitting/validation. In addition to these methods, we used hidden test sets or more generally performed the feature selection on a portion of the data.
4.3. Selecting optimal machine learning algorithms
The support vector machine (SVM) with the radial basis function kernel (RBF) outperformed regularized Random Forest (regRF) or performed comparably. Rather than utilize many different approaches, we aggregated multiple SVM models to improve generalizability. However, in the case of the classification model for EIF4H, we included the regularized random forest algorithm, as the aggregated prediction (SVM and regRF) was clearly optimal on the test data. Algorithm selection and training was done using the classification and regression training package in R [36], caret [37], and the implementation of the Support Vector Machine (SVM) algorithm in Kernlab [38].
4.4. Enriched substructures/cores
Enriched cores were analyzed using RDKit through Python [34]. The algorithm performs an exhaustive search for maximum a common substructure among a set of chemicals. In practice, larger sets often yield fewer substantive cores. To remedy this, the algorithm includes a threshold parameter that relaxes the proportion of chemicals containing the core. We used a threshold of 0.55, which ensures that the majority of the chemicals contained the core.
4.5. Chemical fingerprinting
Extended Connectivity Fingerprints (ECFP) are a class of cheminformatic algorithms that iteratively combine chemical features that are present within a predefined radius/diameter, representing them by a set of integer values. Typically, the fingerprint is converted into a binary string of fixed length using a hash function. Here, the bit length was set at 1024 and a radius of 2 (diameter = 4 or ECFP4). This structural representation was preferred as it is strongly associated with activity [39]. Accordingly, it is a suitable alternative to identify drug candidates in the absence of machine learning models. We used the ECFP algorithm in RDKit (Morgan or circular fingerprint) [34]. The similarity between the fingerprints of chemicals with known activity against the SARS-CoV-2 targets and prospective chemicals was computed using the Tanimoto index. This index is a similarity coefficient (0–1; 1 = max similarity). It is the overlap of the “on-bits” divided by the sum of the unique “on-bits”. Notably, coefficients of 1 need not imply identical chemicals.
where c = overlapping “on-bits”; a = “on bits” in A; b = “on-bits” in B.
4.6. Support vector machine (SVM)
Training the support vector machine (SVM) involves identifying a set of parameters that optimize a cost function, where cost 1 and cost 0 correspond to training chemicals labeled as “Active” and “Inactive,” respectively. θT is the scoring function or output of the support vector machine. If the output is ≥0, the prediction is “Active.” The function (ƒ) is a kernel function.
The kernel determines the shape of the decision boundary between the active and inactive chemicals from the training set. The radial basis function (RBF) or Gaussian kernel enables the learning of more complex, non-linear boundaries. It is therefore well suited for problems in which the biologically active chemicals cannot be properly classified as a linear function of physicochemical properties. This kernel computes the similarity for each chemical (x) and a set of landmarks (l), where σ2 is a tunable parameter determined by the problem and data. The similarity with respect to these landmarks is used to predict new chemicals (“Active” vs. “Inactive”).
4.6.1. Model performance metrics
The Area under the ROC Curve (AUC) assesses the true positive rate (TPR or sensitivity) as a function of the false positive rate (FPR or 1-specificity) while varying the probability threshold (T) for a label (Active/Inactive). If the computed probability score (x) is greater than the threshold (T), the observation is assigned to the active class. Integrating the curve provides an estimate of classifier performance, with the top left corner giving an AUC of 1.0 denoting maximum sensitivity to detect all targets or actives in the data without any false positives. The theoretical random classifier is reported at AUC = 0.5.
where T is a variable threshold and x is a probability score.
However, we generated classifiers that are more authentic than theoretical random classification, shuffling the chemical feature values in the models and statistically comparing the mean AUCs across multiple partitions of the data. This controls against optimally tuned algorithms predicting well simply because of specific predictor attributes (e.g. range, mean, median, and variance) or models that are of a specific size (number of predictors) performing well even with shuffled values. Additionally, biological data sets are often small, with stimuli or chemicals that—rather than random selection—reflect research biases, possibly leading to optimistic validation estimates without the proper controls.
We used the AUC for evaluating classification models. For the classification-based training, we initially converted the inhibitory data into a binary label (Active/Inactive). For predictions of quantitative bioassay measures (e.g. Ki, IC50, AC50, Log LD50), we computed the mean absolute error (MAE), the correlation coefficient (R) and the squared correlation coefficient (R2). MAE: Mean absolute error is the mean of the absolute difference between predicted and observed (% usage). It therefore assigns equal weight to all prediction errors, whether large or small.
where, = predicted and = observed
where, TP = True Positive and FN = False Negative
where, TN = True Negative and FP = False Positive.
KEY RESOURCES TABLE
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Deposited Data | ||
| ZINC 15 | Sterling and Irwin, 2015 | https://zinc.docking.org/substances/home/ |
| chEMBL 25 | EMBL-EBI, 2011; Mendez et al., 2019 | https://www.ebi.ac.uk/chembl/ |
| EPI Suite Data | EPA, 2015 | http://esc.syrres.com/interkow/EPiSuiteData.htm |
| DrugBank | Wishart et al., 2018 | https://www.drugbank.ca/ |
| Therapeutic Targets Database (TTD) | Chen, 2002; Zhu et al., 2009 | http://db.idrblab.net/ttd/ |
| FDA: Substance Registration Database (FDA UNII) | FDA, 2020 | https://fdasis.nlm.nih.gov/srs/ |
| Hazardous Substances Data Bank (HSDB) | Fonger et al., 2014 | https://www.nlm.nih.gov/databases/download/hsdb.html |
| Viral Targets | Gordon et al. 2020 | https://www.nature.com/articles/s41586-020-2286-9 |
| Acutoxbase | Kinsner-Ovaskainen et al., 2009 | https://www.acutetox.eu/ |
| DSSTox | Richard and Williams, 2002 | https://www.epa.gov/chemical-research/distributed-structure-searchable-toxicity-dsstox-database |
| Top 50 physicochemical features to predict inhibitory assay activity for each SARS-CoV-2 target | This paper | Supplementary Table 2 |
| Top 50 physicochemical features to predict broadly inhibiting activity for each SARS-CoV-2 target | This paper | Supplementary Table 3 |
| Top predicted drug and FDA registered chemicals. Structural similarity between drugs and chemicals with bioassay activities for SARS-CoV-2 targets |
This paper | Supplementary Information 2 |
| Top predicted chemicals from ZINC, rank ordered by estimated vapor pressure | This paper | Supplementary Information 3 |
| Top predicted chemicals from ZINC, filtered for toxicity | This paper | Supplementary Information 4 |
| Software and Algorithms | ||
| Classification and regression training (caret) | Kuhn, 2008 | https://github.com/topepo/caret |
| Kernlab | Karatzoglou et al., 2004 | https://github.com/cran/kernlab |
| Regularized Random Forest (RRF) | Deng and Runger, 2013 | https://github.com/softwaredeng/RRF |
| RDKit | Landrum, 2006 Python wrapper |
https://github.com/rdkit/rdkit |
| ggplot2 | Wickham, 2016 | https://github.com/tidyverse/ggplot2 |
Declarations
Author contribution statement
Joel Kowalewski: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Anandasankar Ray: Conceived and designed the experiments; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
J.K. and A.R. are listed as inventors in patents submitted by the University of California Riverside. A.R. is also founder of Sensorygen Inc.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Supplementary_Figure_1a) Ensemble model for predicting log vapor pressure is validated on 676 test chemicals. Test set predictions are bootstrapped 500 times, averaged over 100 bins (5 bootstrap samples per bin). Predictive success is quantified as the mean absolute error (MAE), with the test set value reported in the plot area.b) The test chemical predictions are assessed using the R2 value, bootstrapped 500 times and averaged over 100 bins (5 bootstrap samples per bin). Overall R2 value reported in the plot area. Individual models are trained on different chemical feature sets and predictions are aggregated.
a) Ensemble model prediction of rat log LD50 for 2895 test chemicals. Relationship between predicted and observed log LD50 is quantified as the correlation. Value reported in plot area.
Validation statistics using classification and regression-based support vector machine (SVM) models to predict inhibitory activity against SARS-CoV-2 targets. Sheet 1: Test set performance for classification-based models. Models are compared to an otherwise identical model where the training was performed on shuffled or permuted classification labels. P-values are based on a One-tailed Independent Samples T-test over 500 bootstrap iterations. Where possible, exact p-values are reported. Sheet 2: raw data for the Sheet 1 analysis. Sheet 3: Contains test set performance details for regression-based models. Sheet 4: Predictions of drugs provided in the supplementary information of Gordon et. al. (2020). These are expert curated and approved/investigational compounds with reported activities against select SARS-CoV-2 targets. Explanations and formulae for model performance metrics are in Materials and Methods.
Machine learning predictions of SARS-CoV-2 targets for the DrugBank and Therapeutic Targets databases (Sheet 1; Sheet 2). Machine learning predictions of SARS-CoV-2 targets for the FDA UNII database (Sheet 3; Sheet 4). Structural similarity analysis (Sheet 5), which applies a fingerprint (circular or Morgan) approach to identify basic structural overlap between chemicals with known activity against the SARS-CoV-2 targets and drugs as well as other chemicals in the UNII database such as food additives. The similarity coefficient (Tanimoto) is on the scale 0–1 (1 = max similarity). The “DB” column is the database ID or name of the chemical that is compared to the >10, 000 chemicals in assays for the SARS-CoV-2 targets. Data are filtered to reflect the highest similarities.
The best candidates included in the ZINC database with the largest predicted log vapor pressure values.
Top machine learning predictions for SARS-CoV-2 targets, filtered with respect to theoretical LD50 values and without regard for vapor pressure.
Enriched substructures/cores among assay chemicals for different measures, standardized to nanomolar units (nM). Three broad concentration ranges are used to isolate more interesting enriched features with respect to different sensitivities for the viral targets. Images of representative chemicals are shown for each target. Bonds and atoms appear in black. The enriched substructure is in red. GT = greater than; LT = less than; LTE = less than or equal to.
Top 50 physicochemical features for predict raw assay activity for the protein targets (regression). The SVM models in Figure 2 sample these features.
Top 50 physicochemical features to predict classification labels for the protein targets (classification). The classification labels here reflect broad inhibition. SVM models in Figure 2 sample these features.
References
- 1.Sanche S., Lin Y.T., Xu C., Romero-Severson E., Hengartner N., Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. J. 2020;26 doi: 10.3201/eid2607.200282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. JAMA - J. Am. Med. Assoc. 2020;323(14):1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Day M. Covid-19: four fifths of cases are asymptomatic, China figures indicate. BMJ. 2020;369:m1375. doi: 10.1136/bmj.m1375. [DOI] [PubMed] [Google Scholar]
- 4.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y.F., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L., Banovich N.E., Barbry P., Brazma A., Collin J., Desai T.J., Duong T.E., Eickelberg O., Falk C., Farzan M., Glass I., Gupta R.K., Haniffa M., Horvath P., Hubner N., Hung D., Kaminski N., Krasnow M., Kropski J.A., Kuhnemund M., Lako M., Lee H., Leroy S., Linnarson S., Lundeberg J., Meyer K.B., Miao Z., V Misharin A., Nawijn M.C., Nikolic M.Z., Noseda M., Ordovas-Montanes J., Oudit G.Y., Pe’er D., Powell J., Quake S., Rajagopal J., Tata P.R., Rawlins E.L., Regev A., Reyfman P.A., Rozenblatt-Rosen O., Saeb-Parsy K., Samakovlis C., Schiller H.B., Schultze J.L., Seibold M.A., Seidman C.E., Seidman J.G., Shalek A.K., Shepherd D., Spence J., Spira A., Sun X., Teichmann S.A., Theis F.J., Tsankov A.M., Vallier L., van den Berge M., Whitsett J., Xavier R., Xu Y., Zaragosi L.-E., Zerti D., Zhang H., Zhang K., Rojas M., Figueiredo F., Network H.C.A.L.B. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Burgstaller-Muehlbacher S., Pache L., De Jesus P.P., V Hull M., Chang M., Chan J.F.-W., Cao J., Poon V.K.-M., Herbert K., Nguyen T.-T., Pu Y., Nguyen C., Rubanov A., Martinez-Sobrido L., Liu W.-C., Miorin L., White K.M., Johnson J.R., Benner C., Sun R., Schultz P.G., Su A., Garcia-Sastre A., Chatterjee A.K., Yuen K.-Y., Chanda S.K. A large-scale drug repositioning survey for SARS-CoV-2 antivirals. BioRxiv. 2020 04.16.044016. [Google Scholar]
- 9.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williamson B., Feldmann F., Schwarz B., Meade-White K., Porter D., Schulz J., van Doremalen N., Leighton I., Yinda C.K., Perez-Perez L., Okumura A., Lovaglio J., Hanley P., Saturday G., Bosio C., Anzick S., Barbian K., Chilar T., Martens C., Scott D., Munster V., de Wit E. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. BioRxiv. 2020 doi: 10.1038/s41586-020-2423-5. 04.15.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.-M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;56(1):105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Chen Z., Hu J., Zhang Z., Jiang S., Han S., Yan D., Zhuang R., Hu B., Zhang Z. Efficacy of hydroxychloroquine in patients with COVID-19: results of a randomized clinical trial. MedRxiv. 2020 [Google Scholar]
- 13.Mahevas M., Tran V.-T., Roumier M., Chabrol A., Paule R., Guillaud C., Gallien S., Lepeule R., Szwebel T.-A., Lescure X., Schlemmer F., Matignon M., Khellaf M., Crickx E., Terrier B., Morbieu C., Legendre P., Dang J., Schoindre Y., Pawlotski J.-M., Michel M., Perrodeau E., Carlier N., Roche N., De Lastours V., Mouthon L., Audureau E., Ravaud P., Godeau B., Costedoat N. No evidence of clinical efficacy of hydroxychloroquine in patients hospitalized for COVID-19 infection with oxygen requirement: results of a study using routinely collected data to emulate a target trial. MedRxiv. 2020;2020 04.10.20060699. [Google Scholar]
- 14.Li Y.C., Bai W.Z., Hashikawa T. The neuroinvasive potential of SARS-CoV2 may be at least partially responsible for the respiratory failure of COVID-19 patients. J. Med. Virol. 2020;92(6):552–555. doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao L., Wang M., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D., Li Y., Jin H., Hu B. Neurological Manifestations of Hospitalized Patients with COVID-19 in Wuhan, China: a retrospective case series study. MedRxiv. 2020 [Google Scholar]
- 16.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12(541):eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagheri S.H.R., Asghari A.M., Farhadi M., Shamshiri A.R., Kabir A., Kamrava S.K., Jalessi M., Mohebbi A., Alizadeh R., Honarmand A.A., Ghalehbaghi B., Salimi A. Coincidence of COVID-19 epidemic and olfactory dysfunction outbreak. MedRxiv. 2020 doi: 10.34171/mjiri.34.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sterling T., Irwin J.J. ZINC 15 - Ligand discovery for everyone. J. Chem. Inf. Model. 2015;55(11):2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.F.D.S.C.V.S.W. Group Food and drug administration substance registration system standard operating procedure. Language (Baltim) 2007 [Google Scholar]
- 20.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., MacIejewski A., Gale N., Wilson A., Chin L., Cummings R., Le Di., Pon A., Knox C., Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(Database issue):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X. TTD: therapeutic target database. Nucleic Acids Res. 2002;30(1):412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu F., Han B.C., Kumar P., Liu X.H., Ma X.H., Wei X.N., Huang L., Guo Y.F., Han L.Y., Zheng C.J., Chen Y.Z. Update of TTD: therapeutic target database. Nucleic Acids Res. 2009;38(Database issue):D787–D791. doi: 10.1093/nar/gkp1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., O’Meara M.J., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Naing Z.Z.C., Zhou Y., Peng S., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Shen W., Shi Y., Zhang Z., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Ramachandran R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Wankowicz S.A., Bohn M., Sharp P.P., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Roesch F., Vallet T., Meyer B., White K.M., Miorin L., Rosenberg O.S., Verba K.A., Agard D., Ott M., Emerman M., Ruggero D., García-Sastre A., Jura N., von Zastrow M., Taunton J., Schwartz O., Vignuzzi M., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S., Fraser J.S., Gross J., Sali A., Kortemme T., Beltrao P., Shokat K., Shoichet B.K., Krogan N.J. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. BioRxiv. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hug J.J., Krug D., Müller R. Bacteria as genetically programmable producers of bioactive natural products. Nat. Rev. Chem. 2020;4:172–193. doi: 10.1038/s41570-020-0176-1. [DOI] [PubMed] [Google Scholar]
- 25.Santosh K.C. AI-driven tools for coronavirus outbreak: need of active learning and cross-population train/test models on multitudinal/multimodal data. J. Med. Syst. 2020;44 doi: 10.1007/s10916-020-01562-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irwin J.J., Sterling T., Mysinger M.M., Bolstad E.S., Coleman R.G. ZINC: a free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012;52(7):1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., Magariños M.P., Mosquera J.F., Mutowo P., Nowotka M., Gordillo-Marañón M., Hunter F., Junco L., Mugumbate G., Rodriguez-Lopez M., Atkinson F., Bosc N., Radoux C.J., Segura-Cabrera A., Hersey A., Leach A.R. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019;47(D1):D930–D940. doi: 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.EMBL-EBI, ChEMBL . 2011. ChEMBL. [Google Scholar]
- 29.Kinsner-Ovaskainen A., Rzepka R., Rudowski R., Coecke S., Cole T., Prieto P. Acutoxbase, an innovative database for in vitro acute toxicity studies. Toxicol. Vitr. 2009;23(3):476–485. doi: 10.1016/j.tiv.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Richard A.M., Williams C.L.R. Distributed structure-searchable toxicity (DSSTox) public database network: a proposal. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2002;499(1):27–52. doi: 10.1016/s0027-5107(01)00289-5. [DOI] [PubMed] [Google Scholar]
- 31.Fonger G.C., Hakkinen P., Jordan S., Publicker S. The national library of medicine’s (NLM) hazardous substances data bank (HSDB): background, recent enhancements and future plans. Toxicology. 2014;0:209–216. doi: 10.1016/j.tox.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.U.S. EPA . United States Environ. Prot. Agency; Washington, DC, USA: 2015. Estimation Programs Interface SuiteTM for Microsoft® Windows. [Google Scholar]
- 33.Zang Q., Mansouri K., Williams A.J., Judson R.S., Allen D.G., Casey W.M., Kleinstreuer N.C. In Silico prediction of physicochemical properties of environmental chemicals using molecular fingerprints and machine learning. J. Chem. Inf. Model. 2017;57(1):36–49. doi: 10.1021/acs.jcim.6b00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landrum G. 2006. RDKit: Open-Source Cheminformatics, Online)http://Www.Rdkit.Org [Google Scholar]
- 35.Ambroise C., McLachlan G.J. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6562–6566. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R Development Core Team R: a language and environment for statistical computing. R Found. Stat. Comput. Vienna Austria. 2016 [Google Scholar]
- 37.Kuhn M. Caret Package. J. Stat. Softw. 2008;28:1–26. [Google Scholar]
- 38.Karatzoglou A., Smola A., Hornik K., Zeileis A. Kernlab – an S4 package for kernel methods in R. J. Stat. Softw. 2004;11:1–20. [Google Scholar]
- 39.Rogers D., Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50(5):742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_Figure_1a) Ensemble model for predicting log vapor pressure is validated on 676 test chemicals. Test set predictions are bootstrapped 500 times, averaged over 100 bins (5 bootstrap samples per bin). Predictive success is quantified as the mean absolute error (MAE), with the test set value reported in the plot area.b) The test chemical predictions are assessed using the R2 value, bootstrapped 500 times and averaged over 100 bins (5 bootstrap samples per bin). Overall R2 value reported in the plot area. Individual models are trained on different chemical feature sets and predictions are aggregated.
a) Ensemble model prediction of rat log LD50 for 2895 test chemicals. Relationship between predicted and observed log LD50 is quantified as the correlation. Value reported in plot area.
Validation statistics using classification and regression-based support vector machine (SVM) models to predict inhibitory activity against SARS-CoV-2 targets. Sheet 1: Test set performance for classification-based models. Models are compared to an otherwise identical model where the training was performed on shuffled or permuted classification labels. P-values are based on a One-tailed Independent Samples T-test over 500 bootstrap iterations. Where possible, exact p-values are reported. Sheet 2: raw data for the Sheet 1 analysis. Sheet 3: Contains test set performance details for regression-based models. Sheet 4: Predictions of drugs provided in the supplementary information of Gordon et. al. (2020). These are expert curated and approved/investigational compounds with reported activities against select SARS-CoV-2 targets. Explanations and formulae for model performance metrics are in Materials and Methods.
Machine learning predictions of SARS-CoV-2 targets for the DrugBank and Therapeutic Targets databases (Sheet 1; Sheet 2). Machine learning predictions of SARS-CoV-2 targets for the FDA UNII database (Sheet 3; Sheet 4). Structural similarity analysis (Sheet 5), which applies a fingerprint (circular or Morgan) approach to identify basic structural overlap between chemicals with known activity against the SARS-CoV-2 targets and drugs as well as other chemicals in the UNII database such as food additives. The similarity coefficient (Tanimoto) is on the scale 0–1 (1 = max similarity). The “DB” column is the database ID or name of the chemical that is compared to the >10, 000 chemicals in assays for the SARS-CoV-2 targets. Data are filtered to reflect the highest similarities.
The best candidates included in the ZINC database with the largest predicted log vapor pressure values.
Top machine learning predictions for SARS-CoV-2 targets, filtered with respect to theoretical LD50 values and without regard for vapor pressure.
Enriched substructures/cores among assay chemicals for different measures, standardized to nanomolar units (nM). Three broad concentration ranges are used to isolate more interesting enriched features with respect to different sensitivities for the viral targets. Images of representative chemicals are shown for each target. Bonds and atoms appear in black. The enriched substructure is in red. GT = greater than; LT = less than; LTE = less than or equal to.
Top 50 physicochemical features for predict raw assay activity for the protein targets (regression). The SVM models in Figure 2 sample these features.
Top 50 physicochemical features to predict classification labels for the protein targets (classification). The classification labels here reflect broad inhibition. SVM models in Figure 2 sample these features.