Abstract
Advances in genetic and genomic technologies over the last thirty years have greatly enhanced our knowledge concerning the genetic architecture of Alzheimer’s disease (AD). Several genes including APP, PSEN1, PSEN2, and APOE have been shown to exhibit large effects on disease susceptibility, with the remaining risk loci having much smaller effects on AD risk. Notably, common genetic variants impacting AD are not randomly distributed across the genome. Instead, these variants are enriched within regulatory elements active in human myeloid cells, and to a lesser extent liver cells, implicating these cell and tissue types as critical to disease etiology. Integrative approaches are emerging as highly effective for identifying the specific target genes through which AD risk variants act and will likely yield important insights related to potential therapeutic targets in the coming years. In the future, additional consideration of sex- and ethnicity-specific contributions to risk as well as the contribution of complex gene-gene and gene-environment interactions will likely be necessary to further improve our understanding of AD genetic architecture.
1. Introduction to Alzheimer’s disease
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder and the most common cause of dementia in the elderly [1]. Dementia, caused by a variety of disorders, is clinically characterized by a deterioration in memory, learning, spatial orientation, language, comprehension, and judgement severe enough to interfere with daily living [2]. AD usually presents with deficits in short-term memory formation and disturbance of additional cognitive functions including word-finding, spatial orientation, reasoning, judgement, and problem-solving [3, 4]. Of all dementia patients, 70% are diagnosed with AD, which approximates to 44 million individuals worldwide as of 2019 [2]. As age is one of the most significant risk factors for the disease, this number is expected to increase dramatically in the coming years due to the growing population of aged individuals. Given there is still no cure for disease, this presents a major public health concern.
In addition to clinical symptoms, AD is characterized by specific pathological features [5]. First described by Alois Alzheimer in 1906 [6], AD is a neurodegenerative disease defined by the presence of extracellular plaques primarily composed of beta-amyloid (Aβ) protein, a byproduct of sequential cleavage of the amyloid precursor protein (APP) by the enzymatic complexes beta (β) and gamma (γ) secretase [7, 8]. Intracellular aggregates composed of hyperphosphorylated forms of the microtubule-associated protein tau (encoded by the MAPT gene), often referred to as neurofibrillary tangles, also accumulate primarily in neurons, although they have been reported in other cell types of the brain [9, 10]. Beyond these “classical” neuropathologies, several additional cellular and molecular characteristics are associated with AD. For example, an increase in the number of reactive astrocytes and activated microglia (i.e. gliosis) has been described, especially in close proximity to plaques [11–13]. Interestingly, markers of gliosis seem to linearly increase across the course of disease despite a plateau in plaque pathology, suggesting independent mechanisms drive each cellular phenotype [14]. In his original report, Alois Alzheimer also described “adipose inclusions”, now recognized as likely lipid deposits, in glial cells of the brain [15]. Lipid accumulation in AD has been under-studied relative to other pathologies, but alterations in lipid metabolism and homeostasis are now emerging as a hallmark of AD brains [16–18]. A better understanding of these additional AD-associated features will likely shed light on mechanisms driving disease development across the lifespan.
While postmortem identification of these pathologies, particularly the presence of amyloid plaques and tau tangles, is still considered the gold standard for diagnosis, advances in brain imaging now allow clinicians to obtain accurate estimates of these features in living patients [19], which has aided in the clinical diagnosis of AD. Early detection is thought to be a critical aspect of effective therapeutic interventions, and there has been much work in the field aimed at identifying biomarkers that detect high-risk patients or those in early stages of disease [20]. Research has shown that measurements of Aβ and tau in cerebrospinal fluid (CSF) exhibit high diagnostic sensitivity, with low levels of CSF Aβ42 and high levels of total tau or phosphorylated tau (particularly p-tau181) capable of differentiating AD from mild cognitive impairment and other dementias such as frontotemporal dementia and Lewy body dementia [21–23].
AD is typically divided into two categories; early-onset AD (EOAD) where symptoms appear before the age of 65 years, and late-onset AD (LOAD), in which symptoms do not appear until after the age of 65 years. Despite the earlier age of onset and exacerbated clinical phenotypes in some EOAD patients [2], the two subsets share common pathological features and are both considered to be highly heritable. Estimates of heritability for EOAD are as high as 90–100% [24], while heritability estimates for LOAD are slightly lower, in the range of 60–80% [25]. Both forms of the disease can be further classified into sporadic or familial forms. Most sporadic cases present with late-onset symptoms, but instances of sporadic early-onset cases with no previous family history of disease have been documented [26]. Likewise, familial inheritance of AD is typically associated with early-onset forms of the disease, but large families with multiple individuals affected by LOAD are prevalent [27, 28]. While EOAD and LOAD have traditionally been considered distinct [29, 30], this review will consider the underlying genetic architecture of both forms of the disease, which in recent years has come to highlight a significant overlap between the subtypes. As no disease-modifying intervention currently exists, understanding the biological function of AD-associated variants (i.e the functional genetic architecture) across the spectrum of disease is likely to provide insight into potential therapeutic targets. Therefore, this review will also consider insights into the basic biology of disease provided by genetic studies, and discuss likely mechanisms involved in AD pathogenesis.
2. Genetics of AD
a. Monogenic causes of AD
The most well-studied forms of EOAD which have contributed most to our knowledge regarding disease etiology are those that are inherited in an autosomal dominant fashion, often referred to as autosomal dominant Alzheimer’s disease (ADAD). ADAD is very rare, accounting for less than <1% of all cases of AD [2, 31–33], but the clear inheritance pattern facilitates the identification of causal mutations. A majority of ADAD families carry a pathogenic mutation in one of three genes: APP, presenilin 1 (PSEN1), or presenilin 2 (PSEN2) [2, 33]. To date, more than 50 highly penetrant mutations have been identified in APP [2]. These mutations are typically localized either near the sites where cleavage by secretases occurs or within the domain encoding the Aβ peptide. The overall effect of APP mutations is generally increased production and/or aggregation of amyloid, although the precise mechanism differs by mutation type and localization. For example, mutations occurring near the βsecretase cleavage site, such as the Swedish mutation (K670N/M671L), appear to result in increased production of total Aβ [34, 35]. Mutations in the Aβ domain itself alter the hydrophobicity and downstream aggregation propensity of Aβ peptides, leading to an increase in plaque load [36]. Other mutations such as those located near the C-terminal of the Aβ domain (e.g. V717L, V717I) influence the activity of g-secretase altering APP processing so that longer Aβ peptides more prone to aggregation are generated, including species 42–43 amino acids in length (Aβ1–42 and Aβ1–43) [37]. In contrast, a protective mutation in APP (A673T) identified in the Icelandic population [38] results in approximately a 40% reduction in the formation of amyloidogenic peptides. Together, this evidence has led to the development of the “amyloid cascade hypothesis” which posits that amyloid deposition is an initial causative event in a cascade of symptoms resulting in neurodegeneration and cognitive decline [39]. This hypothesis is further supported by the identification of additional APP genomic features associated with AD, including missense mutations inherited in an autosomal recessive fashion that drive amyloidogenic APP processing [40] and APP duplications which result in increased Aβ production [36].
The other EOAD-causative genes are highly homologous members of the presenilin gene family [41]. The first disease-causative mutations in these genes were identified in 1995 [42, 43], although it wasn’t until 2000 that it was discovered these proteins are critical subunits of the γ-secretase complex responsible for processing of APP [44–46] and other membrane-bound proteins such as NOTCH [33]. Similar to some mutations in APP, familial EOAD mutations in PSEN1 and PSEN2 typically result in an increased production of longer and more aggregation-prone derivatives of APP such as Aβ1–42 [47–49]. Mutations in PSEN1 are the most common cause of ADAD; as of April 2020, over 350 mutations (some of unclear pathogenicity) have been identified (www.Alzforum.org). PSEN1 mutations are estimated to contribute to around 80% of monogenic AD. PSEN2 mutations are much rarer, with only around 30 mutations identified in ADAD families [2]. The effect of PSEN2 mutations is more variable than either PSEN1 or APP mutations, with age of onset in PSEN2 mutation carriers ranging from 40 to 85 years of age [50, 51]. This suggests additional modifier variants may be capable of protecting against what are typically thought to be highly penetrant deleterious mutations. This raises the possibility that these “early-onset” mutations may play a significant and under-appreciated role in LOAD, an idea that will be explored later in this review.
b. APOE as a major genetic risk factor for AD
Beyond disease-causative mutations in APP and the presenilins, common polymorphisms in the gene apolipoprotein E (APOE) are the most significant genetic risk factor for AD [52–54]. APOE is a lipoprotein with various biological functions depending on the cell and tissue type in which it is expressed; high expression is observed in the liver, brain, and peripheral immune cells including monocytes and macrophages [55]. Broadly, APOE is responsible for maintaining lipid homeostasis by mediating lipid transport from one cell or tissue type to another [56]. In the central nervous system, APOE is primarily produced by astrocytes under normal conditions and transports cholesterol to neurons via APOE receptors, which are members of the low-density lipoprotein receptor (LDLR) family [57]. Under times of stress, APOE can also be produced by both neurons and microglia [47, 48].
Three different alleles of APOE exist, each encoding a unique isoform differentiated by the identity of the amino acids solely at positions 112 and 158, where either cysteine or arginine is present. These isoforms are referred to as APOEε2 (Cys112/Cys158), APOEε3 (Cys112, Arg158), and APOEε4 (Arg112, Arg158) [57]. Of the three alleles, APOEε3 is the most common in all human populations, present in almost 80% of individuals [57]. APOEε4 is less frequent (~14%) among the general population but drastically increases risk for AD in a dose-dependent manner [54, 57]; Caucasian individuals with two copies of APOEε4 have on average a 15-fold increased risk for LOAD relative to those carrying no APOEε4 alleles, whereas risk for individuals carrying only one APOEε4 allele decreases to ~3 times that of non-carriers [58]. Interestingly, the effect of APOEε4 has been reported to differ across ethnicities [58–61], suggesting additional variants may interact with APOE to mediate its effect on AD risk. For example, up to a 30-fold increase in disease risk was observed among Japanese individuals with two ε4 alleles as compared to a 15-fold increased risk in Caucasians [59, 60], whereas the association between ε4 and disease is reported to be weaker among African-American and Hispanic individuals [58, 61, 62] and nonexistent among Nigerian individuals [63]. Additionally, APOEε4 has been shown to modify the age at onset in both EOAD and LOAD [64–66]. In contrast, APOEε2 appears to reduce risk for AD, with carriers of even one APOEε2 allele in combination with APOEε3 exhibiting only about half the risk of those carrying two APOEε3 alleles [58]. However, APOEε2/APOEε4 individuals are still at increased risk for AD, suggesting the APOEε4 isoform exhibits some dominant effect over the APOEε2 isoform [58]. Homozygous APOEε2 individuals are relatively rare, making the dose-dependent effect of APOEε2 difficult to assess in the general population. It has been suggested that APOEε2 may protect against AD in a dose-dependent manner, with ε2/ε2 individuals exhibiting up to a four-fold decreased risk [67, 68].
Despite the long-standing recognition of the impact of APOEε4 on AD risk, the precise mechanism by which APOEε4 confers elevated risk for AD remains unclear [69]. Although the isoforms differ by only two amino acids, it has been demonstrated these alterations have profound impacts on the structure and function of the APOE protein [70]. For example, the ε2 allele has been demonstrated to exhibit impaired LDLR binding relative to both ε3 and ε4, and structural alterations unique to the ε4 allele shift binding preferences from small, phospholipid-rich high-density lipoproteins (HDL) to large, triglyceride-rich very-low-density lipoproteins (VLDL) in the periphery [70]. In the context of the central nervous system, APOEε4 has been linked to impaired Aβ clearance [71, 72], disruption of lysosomes and neuronal apoptosis [73, 74], as well as mitochondrial dysfunction [75]. Further investigations into these functions and how they may contribute to the development of disease will be critical in the coming years for targeted therapeutic development.
c. Genome-wide association studies (GWAS)
Despite the substantial increase in risk conferred by APOEε4, variation at this locus is estimated to explain only a portion of the heritability of LOAD [76], suggesting additional genetic variants play a significant role in determining an individual’s risk for disease. Following the sequencing of the first human genome, platforms enabling the simultaneous genotyping of millions of single nucleotide polymorphisms (SNPs) across the genome were developed, providing a powerful framework with which to study the contribution of common genetic variation to complex diseases. This is most commonly performed through what is known as a genome-wide association study (GWAS), designed to test the association of genetic variants across the genome with a particular trait, often presence or absence of a disease. GWAS take advantage of the linkage disequilibrium (LD) structure across the human genome, where large blocks of the genome are inherited together [77]. Due to this phenomenon, a single genotyped SNP can tag an entire region of high LD, enabling the surrounding genomic sequence to be imputed, or predicted with high accuracy [78, 79] based on the reference human genome sequence. Genotype imputation allows data from different genotyping platforms to be harmonized and greatly enhances the feasibility of genome-wide studies without the need for expensive sequencing. However, if variants are rare (minor allele occurs in less than 1% of the general population [80]), they are unlikely to be captured by standard SNP genotyping or imputation using the publicly available reference resources [79]. The contribution of rare variants to disease must be assessed by other methods and will be discussed later in this review.
The first association studies covering most of the genome were published in 2007 [81, 82]. As these studies utilized a relatively small number of samples, only a single SNP in APOE reached genome-wide significance (typically considered p < 5.0 × 10−8, based on a Bonferroni correction for testing all independent SNPs in the human genome [83]). In fact, seven of the earliest GWAS, each of which used less than 2,000 cases, identified only APOE as significantly associated with AD [84–90]. This is likely due to small sample numbers, as GWAS requires a large sample size for sufficient statistical power to detect small-effect associations. This is because the tests require simultaneous testing of hundreds of thousands of SNPs across the genome, resulting in a high penalty for multiple testing corrections. The identification of APOE as the sole risk factor in small GWAS demonstrates that genetic variation at this locus is the most significant common genetic risk factor for LOAD, and genetic variation at other loci individually confer only small effects on disease risk.
In 2009, the first large-scale GWAS were published and loci outside of APOE were implicated in LOAD risk for the first time. The larger of these, involving over 16,000 individuals, identified SNPs on chromosomes 8 and 11 as significantly associated with disease risk [91]. The effects of these loci were attributed to nearby genes, clusterin (CLU) and phosphatidylinositol binding clathrin assembly protein (PICALM), respectively. However, due to the underlying LD structure, GWAS cannot identify which variant or gene in the identified region is truly causal. As multiple SNPs are inherited together in any given haplotype block, the GWAS SNP should only be thought of as ‘tagging’ an underlying association in its given genomic region. A second 2009 GWAS replicated the association of SNPs near CLU with LOAD and identified another locus on chromosome 1 as associated with disease, near the gene encoding the complement receptor 1 (CR1) [92]. These studies have since been expanded and many loci replicated across studies and sample sets [93–96], see Figure 1 and Table S1. The most recent meta-analysis of LOAD risk included data from over 95,000 individuals across 46 datasets and validated over 20 loci previously associated with disease, including well-replicated variants located near genes including BIN1, CD2AP, CLU, ABCA7, and INPP5D, among others [97] (Table S1). In addition, this GWAS identified novel variants not previously associated with the disease by standard GWAS, including variants near IQCK [97].
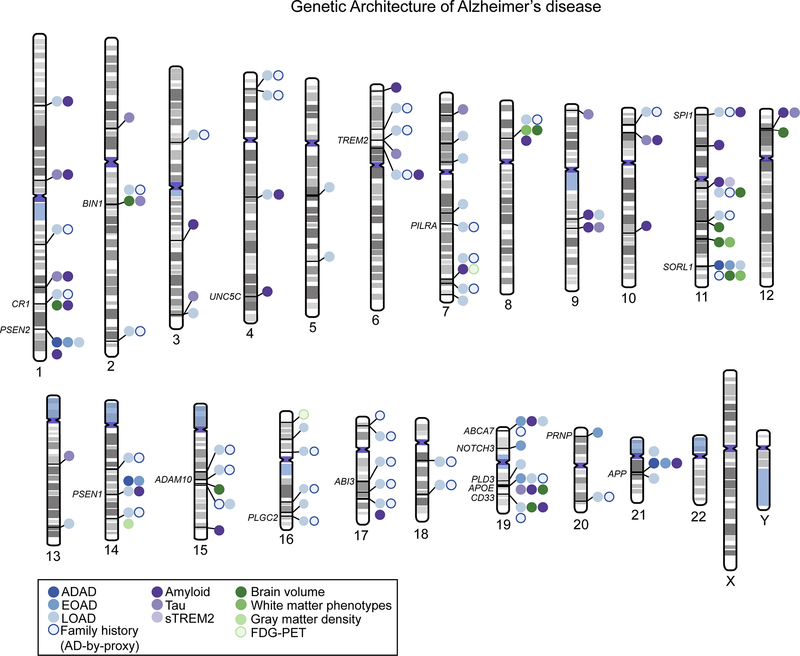
Figure 1: Genetic loci associated with Alzheimer’s disease (AD).
A literature search was used to identify studies reporting genomic loci associated with either risk for AD or various endophenotypes including cerebrospinal fluid measurements of amyloid and/or tau, amyloid deposition as measured by positon emission tomography (PET) or postmortem studies, and brain glucose metabolism as measured by fluorodeoxyglucose (FDG)-PET. Genes with sufficient functional evidence to suggest they are the causal gene in their respective loci are annotated on the figure. Unlabeled loci indicate the causal gene in the region is still unknown. Source information for this figure can be found in Supplementary Table 1. Phenogram was constructed at http://visualization.ritchielab.org/phenograms and modified in Adobe Illustrator for clarity.
A majority of these initial GWAS have utilized data from populations of European ancestry due to sample availability and the ease of analyzing and interpreting data originating from a relatively homogenous population. However, the prevalence and incidence of AD is reported to vary widely by ethnic background, suggesting there may be ethnicity-specific contributors to disease risk [98]. To test this hypothesis, significant efforts have been made to expand AD GWAS to multiple diverse populations. As a result, several ethnicity-specific loci associated with disease have been identified, including variants on 18q23 in Caribbean Hispanics [99] and variants near KCHN15 in Chinese [100]. Additionally, these studies have identified several loci that show conserved association with AD across population groups. For example, a large transethnic GWAS utilizing samples from Caucasians, African Americans, Japanese, and Israeli-Arabs identified several loci contributing to disease risk in all populations [101]. Consideration of multiple genetic backgrounds may also provide insight into the mechanisms by which common genetic variation influences disease risk. For example, APOEε4 confers differing risk based on ethnicity, as discussed above. Studies comparing “global” ancestry (i.e. ancestry across the genome) to “local” ancestry (i.e. ancestral background of the APOE region alone) found that APOEε4 risk was modified by local ancestry such that individuals that inherited the APOEε4 allele (and surrounding region) from an African ancestor had a lower risk of developing AD than an individual who inherited APOEε4 from a European ancestor [62, 102]. This finding suggests either 1) APOE genotype interacts with some surrounding genetic variants to dictate disease risk, or 2) protective alleles within the African APOE haplotype reduce the effect of APOEε4 on disease, providing hypotheses to test in future studies.
Utilizing multiple ethnic groups in GWAS also enhances the ability of researchers to identify putative causal variants in associated loci due to variations in LD structure across populations [101]. As the number and identity of specific SNPs inherited together change, the preservation of association signals in the region allows researchers to infer which SNPs may be driving the signal versus those that are non-functional. While differences in LD structure and allele frequencies have traditionally presented a computational barrier to multi-ethnic GWAS, statistical models designed to account for these differences are constantly being developed and improved. Large-scale sequencing efforts targeting under-represented populations are also aiding in rare-variant detection and imputation, all of which will likely lead to improved consideration of ethnicity-specific genetic architecture in the context of AD risk [101]. Indeed, these approaches when applied to the ABCA7 locus identified a deleterious frameshift deletion in ABCA7 present in African Americans with AD at a higher frequency than either control African Americans or Caucasians [103]. While loss-of-function mutations in ABCA7 have been identified as associated with disease risk in European populations [104–106], this study highlighted a novel pathogenic alteration that confers disease risk in an ethnicity-specific manner. Together, these approaches will help identify which therapeutic targets may be leveraged for the development of population-specific treatments or which may be useful across multiple genetic contexts.
d. Variations on traditional GWAS
In recent years, several variations on the traditional case-control and SNP-trait association studies discussed above have been developed. For example, researchers have hypothesized that the large effect of APOE genotype on AD risk may preclude the identification of smaller-effect variants. To test this hypothesis, researchers have re-analyzed data from the International Genomics of Alzheimer’s Project (IGAP) following separation of APOEε4+ and APOEε4− subjects [107]. Among APOEε4− subjects, several novel associations between AD risk and SNPs near the MAPT locus were identified. Researchers then used genome-wide expression data to link these SNPs to expression of nearby genes KANSL1, LRRC37A4P, C17orf69, and MAPT. While these results suggest the identified SNPs may have multiple downstream targets, the identification of MAPT as a candidate target is particularly notable, as MAPT encodes for the tau protein which becomes aberrantly phosphorylated in AD. Specifically, SNPs protective against AD were associated with increased expression of MAPT exon 3, which has previously been associated with decreased aggregation of tau protein [108, 109] and protection from neurodegeneration [110]. As no disease-causative mutations in MAPT have previously been linked to AD, the findings here provide a putative mechanism by which regulation of MAPT splicing and exclusion of exon 3 may promote AD independently of APOEε4 genotype. Additional analyses in stratified GWAS suggest association signals in the MS4A region were also stronger in APOEε4− subjects [107], while variants in the PICALM region exhibit stronger associations in APOEε4+ subjects [111], further suggesting independent mechanisms may partially contribute to AD across APOE genotypes.
Additionally, recent GWAS aimed at identifying genetic variants influencing risk for AD have moved beyond simple case-control designs, in part to reach the large sample sizes needed for sufficient power. In 2017, Liu et al. [112] proposed utilizing family history of AD as a proxy measurement for cases. In this design, individuals with a family history of AD are grouped as “cases” whereas individuals with no family history of disease are considered “controls”. This enables the identification of patients who likely have a high genetic risk for AD but who may not yet present with clinical symptoms. Jansen et al. [113] expanded on this approach, differentially weighting various family history scenarios. For example, the “risk” of individuals with two parents affected with AD was upweighted relative to those with only one affected parent. In addition, the age of parents was taken into consideration. Cognitively normal parents who died at a relatively young age were not treated the same as cognitively normal parents who died at an elderly age, as the younger parents could have gone on to develop dementia later in life.
While proxy analyses themselves are less well-powered than traditional case control GWAS analyses, these approaches require less detailed phenotype data, thus enabling much larger sample sizes than standard case-control analyses (close to 500,000 participants in recent proxy GWAS compared to 94,000 in standard GWAS [97]), which require detailed clinical data for each participant. Proxy risk for AD, defined as a parental history of AD, exhibits strong genetic correlation with AD [113] and can thus be combined with existing GWAS datasets to enhance statistical power to find true genetic associations with disease. Together, these approaches show substantial genetic overlap with previous case-control GWAS and nominate a number of novel loci associated with proxy risk for AD [112–114]. While these results should be interpreted with some caution, as some proxy “cases” may never go on to develop dementia just as some individuals with no family history may go on to develop disease, the loci identified here provide further insight into the genetic architecture of AD and partially confirm family history of AD is a valid proxy measurement for AD risk.
In addition to alterations to study design and subject classification, several variations on the standard SNP-trait association test have been developed. One of the most common variations used in GWAS are gene-based association tests, which assign SNPs to nearby protein coding genes and aggregate the effect of all SNPs in any given gene on disease risk [115]. These approaches provide some advantages over SNP-based tests. First, as individual SNPs are aggregated together, there is a lower multiple testing penalty for gene-based tests. In addition, these tests are less reliant on LD structure present in a population and thus enable GWAS across genetically diverse populations. Several genome-wide significant loci not previously detected at the SNP level have been identified using gene-based approaches, including those assigned to TP53INP [115] and PPARGC1A [116]. At first glance, gene-based tests also seem to ease downstream interpretation as they implicate specific genes rather than individual SNPs in disease risk. However, the assignment of SNPs to genes can be relatively arbitrary and typically fails to consider the impact of SNPs on the function of genes which may not be the closest gene (e.g. SNPs within cell type-specific enhancers may be at some distance from the gene(s) they regulate).
As mentioned above, a limitation of GWAS is that this approach cannot identify which variant or gene in the identified region is truly causal. Further work is needed beyond GWAS to understand the downstream biological impact of genetic variation in the region. Interpretation of GWAS hits is made more straightforward when GWAS SNPs themselves or nearby variants in high LD are located within coding regions of the genome, although this happens relatively infrequently [97, 113]. For example, only 2% of variants identified as associated with LOAD in the latest meta-analysis fell within exons [97]. Missense variants with predicted deleterious effects on protein function have been identified by GWAS in TREM2, CR1, MS4A2, MS4A6A, TOMM40, IQCK, and CD33, among others [97, 113], although many of these variants remain to be functionally validated using experimental systems. Many of these genes containing coding variants are highly expressed in microglia and function within the immune system, highlighting this pathway as critical for disease risk.
e. Polygenic risk scores
Despite the inherent complexities associated with GWAS, the information provided by these studies is valuable in order to better understand the genetic architecture of AD. Given the small effect sizes of loci other than APOE identified by GWAS, it is likely an individual’s risk for AD is determined by the combinatorial effect of multiple variants acting together across the genome. Approaches to better understand how many associated variants may act together to predict risk of disease were first developed by the International Schizophrenia Consortium in 2009 [117]. These “polygenic risk scores” (PRS) have since been expanded upon in the context of AD and used for individualized risk prediction [118], which is important for the identification of at-risk individuals who may benefit from early therapeutic interventions, once developed.
Several diverse approaches can be taken when developing PRS from genetic data. One strategy is to simply identify an individual’s genotype at each of the SNPs identified as associated with AD by GWAS and derive a weighted summary risk score based on the predicted effect of each SNP on disease risk. These scores can then be used to test whether the SNPs associated with disease in the initial GWAS population significantly predict disease in an independent sample. PRS incorporating only genome-wide significant SNPs have showed only a modest improvement in disease prediction over known risk factors such as APOEε4, age, and sex [119–121]. Another strategy designed to identify patients at risk for developing AD is the expansion of the PRS beyond SNPs reaching genome-wide significance in GWAS. This approach likely better captures the underlying genetic architecture of AD, since as discussed above GWAS SNPs simply act as tagging variants for a given risk locus and are often not themselves the causal variants. In addition, as mentioned, standard GWAS often suffer from being under-powered to identify true associations and as such, may miss variants modifying disease risk. By expanding PRS analyses to include variants below the genome-wide statistical cutoff, this approach allows for a more complete view of the genetic contribution to disease risk and seems to provide an improved method for risk prediction over more selective PRS or standard predictors such as APOEε4 alleles, age, and sex [118, 122, 123]. However, evidence suggests the most accurate prediction results from a combined consideration of the PRS as well as these additional predictors (APOE genotype, age, and sex) [123]. In addition to overt disease risk, PRS scores derived from large numbers of common variants have been used to significantly predict age at onset of AD as well as likelihood of longitudinal progression from mild cognitive impairment to AD [118]. This supports the notion that AD is likely influenced by a large number of susceptibility genes, reflecting a complex set of biological pathways related to the disease [123]. In addition, the increased power provided by expansion of the PRS beyond GWAS risk variants suggests that there are still unidentified common variants that could contribute to disease.
One of the benefits of a PRS is that it can be used to compare the genetic architecture of the trait from which it was derived (in this case, LOAD), to the genetic architecture of other traits of interest. For example, Cruchaga et al. [26] compared a PRS generated using LOAD risk alleles identified by GWAS to disease risk in families with high LOAD prevalence (i.e. familial LOAD) as well as sporadic and familial EOAD. It was found that the burden (PRS) of LOAD GWAS variants significantly predicted disease risk and age at onset in both familial LOAD and sporadic EOAD [26]. Although GWAS variants were not associated with risk for familial EOAD inherited in an autosomal dominant fashion, the researchers noted that the sample size was small and that variant burden may have been associated with age at symptom onset in these families [26]. Together, these results suggest shared genetic architecture exists between both EOAD and LOAD, with some of the same loci influencing risk for disease in both cases. Overlap in the genetic architecture between AD and other diseases has also been reported, including type 2 diabetes [124] and immune-mediated diseases such as Crohn’s disease [125, 126], although the precise mechanisms underlying this overlap remain to be elucidated.
f. Rare variants
Interestingly, estimates suggest common variants identified by GWAS explain only 30% of the heritability of AD [76], suggesting the genetic architecture of AD is more complex than that suggested by the “common disease, common variant” hypothesis GWAS is designed to test [127]. As mentioned above, the contribution of rare variants to AD cannot be adequately assessed by GWAS, although improved imputation methods are being developed to address this limitation [128]. The best way to assess the impact of rare variants on disease risk is by direct sequencing of either whole genomes or whole exomes. Fortunately, significant resources have been directed towards this effort in large populations of AD cases and controls through large initiatives such as the Alzheimer’s Disease Sequencing Project (ADSP) [129]. Several additional cost-effective approaches have been developed, including targeted sequencing of selected loci, directed genotyping, and improved imputation of large cohorts [130].
One of the most well-studied rare variants that confers elevated risk for AD is the R47H variant in TREM2 [131, 132]. Since its identification, the R47H variant has been associated not only with AD risk but also neuropathology, brain atrophy, and additional behavioral symptoms such as anxiety in AD patients [133–135]. Several studies have investigated the impact of the R47H variant on the function of TREM2, which is highly expressed on myeloid cells and known to be involved in the clearance of various cellular substrates including lipids, apoptotic cells, and Aβ plaques [136]. Evidence suggests the R47H variant reduces the ability of TREM2 to bind ligands such as lipidated APOE [137] and decreases the stability of the protein, potentially conferring reduced functionality [136]. Since the identification of R47H, additional rare variants in TREM2 have been associated with AD risk [138–140], highlighting the critical role of this protein in the etiology of AD.
Several additional genes beyond TREM2 have been identified as containing rare variants that modify risk for AD, including several others that are highly expressed by microglia in the brain [130] (Figure 1 and Table S1). For example, rare variants in PLCG2 and ABI3 have been associated with disease risk [130, 141]. PLGC2, which harbors a protective variant, encodes a transmembrane signaling enzyme implicated in lipid metabolism, calcium homeostasis, and immune response [130, 142]. ABI3 has similarly been implicated in regulation of the immune response through interferon signaling [143] and also plays a role in actin cytoskeletal organization [144]. Additional genes identified to harbor rare variants associated with AD risk include AKAP9, UNC5, and ADAM10, although the strength of these associations and the role of these genes in disease etiology remains less well-established [145, 146]. Interestingly, several genes harboring rare variants, including ADAM10 [114] and SORL1 (discussed below), also emerge as candidates nominated by GWAS studies due to their proximity to AD-associated SNPs. This provides further evidence these genes are causally involved in disease etiology and facilitates the interpretation of complex GWAS loci.
One insight into the etiology of AD that has emerged due to rare variant sequencing is the importance of APP processing in both early- and late-onset forms of the disease. For example, rare variants in SORL1 and ABCA7 have been identified in family-based studies of LOAD [147–150], each of which have been associated with alterations in APP processing. ABCA7, an ATP-binding cassette transporter protein, is known to be involved in endocytosis and intracellular trafficking of APP [151]. Suppression of ABCA7 led to increased β-secretase-mediated processing of APP and subsequent elevation of Aβ species [151]. SORL1, a sorting receptor located in neurons [152], astrocytes, and microglia [153] is known to direct APP toward non-amyloidogenic processing in the trans-Golgi network [148, 154]. Disease-associated mutations in SORL1 have been linked with altered levels of APP at the cell surface and increased production of toxic Aβ species [147]. Notably, an enrichment for rare disruptive and putatively damaging variants in SORL1 were also identified among EOAD cases [155, 156]. Common variants near SORL1 have also been identified as associated with disease through GWAS [96], suggesting the same loci may contribute to both EOAD and LOAD.
In support, sequencing studies have identified rare variants in APP, PSEN1, and PSEN2 that increase risk for LOAD [157]. Although causal variants in these genes are typically thought to be inherited in an autosomal dominant fashion, de novo variants in APP, PSEN1, and PSEN2 have also been identified by sequencing in sporadic cases of EOAD that occur with no family history of disease [158, 159]. These data suggest APP, PSEN1, and PSEN2 may play a larger role in the development of AD than previously thought and highlight the need to screen both EOAD and LOAD cases by sequencing to identify pathogenic mutations in these genes. Notably, most of the APP variants associated with disease have been evaluated in terms of their effects on Aβ production. However, Aβ-targeting strategies have largely failed to produce disease-modifying therapeutics, suggesting APP plays a role in disease pathogenesis beyond Aβ. Indeed, there is evidence that alternative APP-derived fragments as well as APP itself play important roles in the modulation of immune cell activation [160], protein clearance [161], and lipid homeostasis [162]. Together, this suggests the impact of these variants may have as-yet undescribed impacts on cellular function beyond Aβ generation, an idea which should be further explored in future studies.
Beyond the identification of risk factors of LOAD, rare variant sequencing studies have been beneficial in exploring the causal genes responsible for a subset of EOAD not explained by mutations in APP, PSEN1, or PSEN2. While mutations in these genes are the most well-studied mutations associated with EOAD, it is estimated that a significant portion of EOAD remains unexplained genetically [2]. Besides mutations in SORL1 described above, mutations in NOTCH3 were identified in a Turkish family with AD [163]. Targeted sequencing analyses have also provided insight into genetic modifiers of the age at first symptom onset in EOAD families [164, 165]. Given the overlap between EOAD and LOAD as discussed above, these modifiers may provide potential therapeutic targets for a more general population.
In summary, rare variant studies have yielded tremendous insight into mechanisms underlying the development of AD in diverse populations. Rare variants typically have a more deleterious impact on protein structure and/or function than common variants [166, 167] and thus have a larger effect on disease susceptibility than those identified by GWAS. This, combined with the effect that the gene impacted by the identified variant is often immediately apparent, makes it possible to more easily model the effect of the variant in cell-based assays or model organisms. While the variant itself may not contribute to disease across a broad population, the insight gained into disease mechanisms, particularly immune cell function and APP processing, by the study of genes identified by rare variant studies is likely to generate important therapeutic avenues in the coming years.
3. Functional genomic architecture
a. Understanding the regulatory function of non-coding risk variants
The goal of genetic studies of AD is to gain insight into the mechanisms underlying disease risk in order to develop targeted treatments to prevent or delay the onset of disease. However, translating the statistical associations identified by GWAS into biological insights remains a challenge. As mentioned above, GWAS variants often fall within regions of high LD such that multiple non-causal variants are inherited together with putative causal variant(s). A large portion of GWAS variants and even variants in high LD with the lead variant fall within non-coding regions of the genome, making their functional importance difficult to infer [168]. Much of the non-coding genome has regulatory functions, suggesting a majority of GWAS variants act by perturbing gene expression, presumably in a subset of disease-relevant cell or tissue types. However, regulatory elements can act on downstream target genes located a long distance away and have diverse effects depending on the context in which they are activated [169], making the specific genes impacted by GWAS SNPs difficult to identify. Further complicating the identification of downstream target genes is the fact that large numbers of protein-coding genes are often inherited together; for example in the most recent large GWAS of AD, implicated loci contained an average of 16 genes [97].
Due to these challenges, the earliest attempts to assign biological function to identified variants simply assigned SNPs by proximity to their nearest protein-coding gene. This approach has prioritized genes enriched for several major biological pathways including lipid metabolism, immune response, regulation of endocytosis, and protein ubiquitination, implicating these pathways in disease pathogenesis [170, 171]. Interestingly, there is evidence that groups of genes close together in chromosomal space function in similar biological pathways [172], suggesting this approach may be valid to identify overall pathways and functions perturbed by GWAS variants, although the specific genes investigated may not be direct targets of the causal variants in the region. For a number of genes including APOE, ABCA7, BIN1, TREM2, SORL1, ADAM10, and SPI1, there exists a number of functional studies and genetic evidence to suggest they are indeed the causal genes in their respective loci [97, 173], although their identification does not preclude the existence of additional pathogenic genes in these loci.
There has been a growing appreciation in recent years that simply assigning a GWAS-identified SNP to the nearest gene in linear genomic space may lead to false associations [174]. Instead, an increased emphasis has been placed on understanding the regulatory function of identified variants, as well as nearby variants in high LD (i.e. the functional genomic architecture). This has been greatly facilitated by the development of a variety of techniques designed to better understand how regulatory elements function to impact gene expression, which can happen in a variety of ways. For example, promoters proximal to the transcription start site of protein coding genes can recruit the binding of specific transcription factors, activators, or repressors [175]. Enhancers, short genomic sequences distally located from their target genes, confer additional cell- and tissue-type specificity to the regulation of gene expression. Enhancers are difficult to confidently identify from DNA sequence alone, but are instead defined by the presence of several functional marks including specific histone modifications that promote transcription [176] and by accessibility of local chromatin. Histone modifications can reliably be measured across the genome using chromatin immunoprecipitation followed by sequencing (ChIP-seq), while chromatin accessibility is typically measured by either DNase hypersensitivity analysis (DNase-seq) [168] or assay for transposase-accessible chromatin using sequencing (ATAC-seq) [177]. As enhancers are often located at a considerable distance from their target genes, it is believed they function by recruiting specific chromatin remodeling factors that promote the formation of chromatin loops, bringing the enhancer and promoter into close proximity [178]. A variety of assays now exist to capture this three-dimensional architecture, including various adaptations of chromosome conformation capture (3C) [179].
Regulatory elements across the genome exhibit a high degree of cell-type specificity, enabling the existence of diverse cell and tissue types as well as carefully orchestrated transcriptional responses to stimuli from a single genome. Identification of the precise cells and tissues in which regulatory elements containing AD risk variants are active would provide critical insight into where these variants may be acting to perturb gene expression and mediate functional effects that translate into modified risk for disease. This search has been facilitated by large-scale annotation projects such as the Roadmap Epigenomics Project [180], the Blueprint Project [181], and PsychENCODE [182]. These projects aim to generate, and make publicly available, high-quality reference annotation maps across a wide variety of human cell and tissue types. Combined with cell- and tissue-specific gene expression sets, these annotations can provide a comprehensive picture of how and where variants and associated regulatory elements may be impacting gene expression, and presumably, disease risk.
b. Cell-type specificity of AD risk
Several studies have assessed whether AD risk variants (both GWAS-tagged variants and associated variants in high LD) were enriched for localization to functional annotations from specific cell or tissue types [173, 183–185]. These studies largely expanded on the method described by Finucane et al. [186] for partitioning the genome into functional categories based on publicly available annotation data. This method combines annotations including “coding”, “intron”, “promoter”, and “enhancer” with cell-type specific annotations derived from measurements of four histone marks (H3K4me1, H3K4me3, H3K9ac, and H3K27ac) across over 200 different cell types. In combination with additional annotation information including chromatin accessibility as measured by DNase hypersensitivity, Gagliano et al. [183] identified a significant enrichment for AD-associated variants in functional annotations active in the “immune/hematopoietic cell” and “liver” categories, but not in other categories such as brain, cardiovascular, kidney, etc. These associations were observed whether APOE was included or not, confirming the enrichment was not driven by a single moderate-effect variant [183]. Similar enrichment for immune and liver cell categories was replicated by a second group utilizing an expanded set of tissue-specific annotations [185]. The identification of enrichment among “liver” datasets is particularly notable given that the liver is the principal site responsible for maintaining cholesterol homeostasis in the body [187]. Several factors implicate cholesterol metabolism in the pathogenesis of AD; a major role of APOE is to facilitate the transport of cholesterol from astrocytes to neurons in the CNS [188]. Interestingly, several genes expressed in the liver including liver X receptors (LXRs) have been suggested as potential therapeutic targets for AD [189–191]. Together, these findings suggest insights into the localization of genetic risk for AD identifies clinically relevant tissue- and cell-type enrichments.
Using a similar technique and annotation dataset, Huang et al. found a significant enrichment of AD SNPs in functional annotations attributed to hematopoietic cells, specifically monocytes and B-cells [173]. Using measurements of chromatin accessibility and histone modification maps, Tansey et al. [184] also sought to identify whether AD risk variants were preferentially located at DNase hypersensitivity sites across a panel of 38 tissues. Remarkably, immune cell types, specifically hematopoietic stem cells and primary blood monocytes, were again identified as primary cell types containing areas of accessible chromatin in which AD-associated variants were preferentially located [184]. No enrichment was observed for AD variants in brain-specific DNase sites, although the only two datasets included were generated from fetal tissue, which is likely not reflective of the regulatory chromatin landscape observed in adult or diseased brains. A targeted secondary analysis revealed an enrichment for AD variants in regions of open chromatin as measured by ATAC-seq in microglia [184], suggesting a lack of signal in brain tissue is due to the heterogeneity of the bulk tissue from which it was derived. This is supported by a recent identification of the enrichment of AD-associated variants in enhancers active in primary human microglia [192]. Together, these studies suggest the genetic architecture of AD primarily impacts regulatory elements active in cells of the immune system.
Given the critical role of regulatory elements in regulating gene expression, it can be hypothesized based on the above findings that AD variants likely modify risk for AD by having a functional impact on gene expression in immune cells, particularly myeloid cells such as monocytes, macrophages, and microglia. This idea is supported by studies showing that alleles associated with AD show an effect on gene expression specifically in monocytes. Raj et al. [193] performed genome-wide transcriptional profiling of T cells and monocytes, representative cells of the adaptive and innate immune system, respectively. Genetic mapping was then performed in order to identify genomic regions responsible for regulating quantitative gene expression levels, also known as expression quantitative trait loci (eQTLs). AD-associated variants were enriched for colocalization with eQTLs specifically regulating gene expression in monocytes, but not in T-cells, demonstrating AD variants have a cell-type specific effect on gene expression. Huang et al. [173] leveraged this information to identify SPI1 as the most likely candidate causal gene in the genomic region traditionally annotated as the CELF1 locus due to the proximity of the lead GWAS SNP to the gene CELF1. Instead, it was shown that variants in high LD with the lead SNP were significantly associated with regulation of SPI1 levels in myeloid cells, likely causally modifying disease through this mechanism [173].
Exactly how AD-associated variants influence myeloid cell gene expression is still being explored. One plausible mechanism is that these variants disrupt the binding of lineage-specific transcription factors and alter the transcriptional landscape of these cells. In support of this hypothesis, regions of open chromatin in human monocytes, macrophages, and microglia [173, 184] containing AD risk variants were shown to be enriched for binding motifs of the transcription factor SPI1, providing a mechanistic explanation by which alterations in SPI1 levels conferred by variants in the SPI1 locus may have widespread effects on cellular function. SPI1 is particularly important for myeloid cell differentiation and function, and has been linked to the ability of microglia to clear pathogens and cellular debris [173, 194]. AD-associated variants may also alter sites typically targeted for reversible epigenetic modifications that regulate gene expression such as methylation or acetylation. Early clues to the involvement of epigenetic modifications in AD came from observations that altered methylation at the promoters of APP, PSEN1, and APOE could modify risk for disease. Targeted studies have revealed methylation alterations at additional loci associated with disease [195–197], and two independent genome-wide studies recently identified a differentially methylated region in the ANK1 gene as associated with the burden of AD pathology [198, 199]. Additional possible explanations include altered recruitment and/or binding of chromatin remodeling factors such as CTCF or alterations in promoter-enhancer interactions. It is likely that AD risk variants will have diverse effects on gene expression and will need to be investigated individually to gain a complete understanding of how variants function to modify AD risk.
While no enrichment of AD-associated variants were identified in bulk brain datasets, the brain is still the organ most affected in AD, and the regulation of gene expression in this critical tissue is undoubtedly important for disease pathogenesis. Several studies have performed targeted analysis of the impact of identified AD SNPs on brain-specific gene expression [200–202]. While the use of bulk datasets does not provide specific information about the cell types through which AD variants act, they still provide useful information as to which genes may be targeted by GWAS SNPs. Advances in single-cell techniques will enable the further interrogation of these identified eQTLs and enhanced understanding of which cell types are critical to disease etiology. Based on data from peripheral cells and select microglia-specific datasets [184, 192], it is likely the regulation of gene expression in microglia will emerge as critical to disease pathogenesis. Identification of specific target genes and their downstream impact on cellular function will be particularly important, as it is still relatively unclear as to how these cells are involved in promoting or protecting against disease. While microglia exhibit several beneficial functions in the brain such as clearance of cellular debris and orchestration of appropriate immune responses, their chronic activation can lead to toxic neuroinflammation and aberrant pruning of synapses [203, 204]. Insight into how risk or protective variants alter these critical cellular functions should provide insight into how to harness the beneficial properties of these cells for therapeutic development. It should be noted that most of the currently available functional annotation data is derived from healthy donors. Given the dynamic changes in gene expression observed in disease, it may be necessary to investigate the regulatory landscape of cells under a variety of conditions in order to gain a full picture of how the localization of AD risk variants changes with time, cellular stress, and under disease conditions.
4. Genetic architecture of AD endophenotypes
The GWAS discussed above were primarily performed on case/control datasets and provide information regarding the genetic architecture of disease risk. However, this approach does not provide information about other aspects of the disease such as age of onset or disease progression, or as discussed, biological mechanisms involved in disease pathogenesis. In addition, the case/control design can be complicated by pre-symptomatic AD patients who are misclassified as “controls” due to a current absence of clinical symptoms and non-AD dementias misclassified as probable AD. To both better classify cases and controls as well as gain insights into additional mechanisms underlying disease, many groups have turned to the use of endophenotypes. Endophenotype, a term coined in 1966, is defined as a feature of disease that can be objectively and quantitatively measured across individuals, and that which exists between the behavioral symptoms of a disease (e.g. cognitive decline in AD) and its underlying (e.g. genetic) cause [205]. When an endophenotype reliably distinguishes between cases and controls, it can be used as a biomarker enabling the identification of at-risk populations and enhancing diagnostic sensitivity. For example, “core” biomarkers and endophenotypes of AD include low CSF levels of Aβ1–42, shown to indicate reduced clearance of Aβ1–42 from the brain, and elevated CSF tau (both total tau and phosphorylated tau), thought to indicate neuronal degeneration or injury [206].
Using quantitative endophenotypes as the primary endpoint for GWAS instead of a binary classification of presence/absence of disease enables a more precise phenotype measurement, thus increasing the power for identification of genetic variants associated with disease [207] (Figure 1 and Table S1). In addition, this approach greatly facilitates interpretation of the downstream functional consequences of the prioritized loci. In targeted analyses, it has been shown that genetic variants that increase risk for AD also influence CSF Aβ1–42 levels, including pathogenic mutations in APP, PSEN1, and PSEN2, as well as APOE [208–210]. These results have been expanded upon with large GWAS using CSF Aβ1–42 and/or tau levels as the trait of interest. These analyses have identified variants in the TREM2 cluster as associated with these quantitative endophenotypes, as well as variants in the ABCA7 and FERMT2 gene regions [23, 211]. The identification of distinct loci for Aβ1–42 versus tau traits suggest these endophenotypes are regulated, in part, by distinct genetic mechanisms. While the majority of loci overlap with GWAS loci, several novel loci not previously associated with AD risk have also been identified through these analyses, including variants located near the genes GLIS3, GLIS1, GEMC1, and within SERPINB1 [23, 211]. Notably, several of these novel variants, including those near GLIS1 and within SERPINB1, were associated with additional aspects of disease including risk, age at onset, and progression [211], suggesting improved phenotype definition afforded by quantitative measurements is capable of identifying small-effect loci that may be missed by a case/control GWAS.
While Aβ1–42 and tau are considered “core” endophenotypes or biomarkers of AD, additional quantitative measurements have been observed to be related to AD risk. For example, the levels of soluble TREM2 (sTREM2) have been shown to increase five years before symptom onset in autosomal dominant forms of AD [212]. sTREM2 is thought to be produced in one of two ways; either by cleavage of TREM2 by proteases such as ADAM10, ADAM17, and γ-secretase, or by alternative splicing which generates a short isoform lacking a transmembrane domain (~25% of TREM2 transcripts) [134]. sTREM2 is then released into the CSF where it can be measured, and has been shown to correlate with CSF levels of tau [213, 214], suggesting it may be a relevant secondary biomarker of disease progression. Common variants in the MS4A gene cluster were shown to significantly regulate sTREM2 levels in CSF [134], demonstrating regulation of this endophenotype is orchestrated by genetic variants that also modify risk for disease. CSF levels of APOE protein have also been suggested to act as a valuable endophenotype for AD [215], along with levels of various additional proteins, the clinical and biological relevance of which remain to be elucidated [216–218].
While the studies discussed above have largely utilized CSF biomarkers, considered advantageous due to the proximity of this compartment with the brain, collection of CSF samples is quite invasive. Recent advances in imaging technology have enabled high-resolution scanning of the brain in living patients, facilitating non-invasive identification of a number of imaging-based endophenotypes for AD. For example, amyloid burden across the brain can be localized and quantified through the combined use of a radiolabeled ligand such as Pittsburgh B and positron emission tomography (PET) imaging [219]. PET imaging can also be used to measure glucose metabolism in the brain through the utilization of a radiolabeled alternate form of glucose, 18F-fluorodeoxyglucose (FDG) [220]. AD patients exhibit decreased uptake of FDG on PET [220], and this measurement can be used to differentiate AD patients from other clinical diagnoses [221], suggesting FDG-PET provides an additional reliable endophenotype of AD. The final major class of imaging endophenotypes are thought to be indicative of neurodegeneration and include traits such as increased ventricular volume and reduced hippocampal volume. These structure-based endophenotypes are particularly valuable, as they reliably predict cognitive decline [222], are heritable [223, 224], and are readily quantifiable by magnetic resonance imaging (MRI).
Programs such as the Alzheimer’s Disease Neuroimaging Initiative (ADNI) have facilitated efforts to perform standardized imaging across cohorts of AD cases, those diagnosed with MCI, and cognitively normal controls [225]. These efforts have greatly expanded the number of imaging datasets available and have enabled investigation of the genetic architecture of imaging-related AD endophenotypes. However, imaging remains a relatively expensive technique and as such, the sample sizes of well-characterized cohorts are much smaller than those utilized for traditional case-control analyses. The first investigations into the genetic architecture of imaging endophenotypes largely utilized a candidate gene approach, where genes previously implicated in AD risk were tested for association with imaging traits. SNPs in and around candidate genes including APOE, BIN1, SORL1, and CR1 have been associated with traits including medial temporal lobe atrophy and cortical thickness [225–228], suggesting these AD risk variants may act, in part, by regulating neurodegeneration. Despite relatively small sample sizes, GWAS on specific quantitative imaging phenotypes have been conducted by several groups. Interestingly, while several genes previously implicated in AD (including APOE and PICALM [229]) have been associated with imaging endophenotypes, a large number of novel loci and genes have been nominated by these analyses [222, 229–231] (Figure 1 and Table S1). As the quantitative nature of endophenotypes results in improved phenotype definition beyond traditional case-control analyses, the loci identified by imaging studies may contain novel genes contributing to the etiology of AD that may have been missed by traditional approaches. Groups are actively working to integrate multiple imaging traits together, providing an even further improved diagnostic classification [232, 233] which will likely advance our understanding of brain-wide pathologies that occur in AD.
In order to more accurately model the overlap between the genetic architecture of AD risk and regulation of endophenotypes, several groups have turned to using PRS to evaluate overlap between traits. For example, Deming et al. [211] compared a PRS derived from genome-wide significant SNPs associated with AD to levels of CSF pathologies measured in an independent patient cohort. Although in their analysis the individual GWAS SNPs were not significantly associated with CSF levels of Aβ1–42 or tau, together the additive risk conferred by all variants was significantly associated with both Aβ1–42 and tau. Similarly, it has been demonstrated that increased genetic risk for LOAD as defined by a high PRS is associated with several imaging endophenotypes, including decreased cortical thickness in brain regions known to be vulnerable to degeneration in AD, decreased total brain volume, and decreased hippocampal volume [234]. Across studies, the association of GWAS-derived PRS with endophenotypes remained significant even after removal of APOE, suggesting the contribution of small-effect GWAS loci to AD risk is, in part, mediated by the effect of these variants on specific neuropathologies, including Aβ1–42 and tau levels as well as neurodegeneration.
Together, these endophenotype-focused GWAS analyses highlight the overlap in genetic architecture across various aspects of AD and provide important insight into the mechanisms by which variants modifying disease risk may act. The enhanced patient classification scheme afforded by quantitative biomarkers will likely facilitate enhanced power to diagnose AD in general, possibly providing a framework in which to identify subtypes of patients with distinct endophenotype profiles. It has been hypothesized that several subtypes of AD exist [235], and improved identification and diagnosis will likely enable targeted intervention and therapeutic development. As the collection of CSF samples remains invasive and imaging is quite expensive, both approaches are relatively difficult to utilize across very large samples such as those required for well-powered GWAS. As such, there has been an emphasis in recent years to identify blood- or plasma-based biomarkers for disease, the search for which is still ongoing [217]. These approaches will likely provide valuable alternatives to CSF and imaging and yield important insight into how identified risk variants might influence disease onset, severity, and progression.
5. Genetic architecture of AD: beyond SNPs
a. Somatic variation
Typically, an organism is considered to have one DNA sequence across all cells in the body. However, recent advances in single-cell genomic technology have identified a surprising amount of genetic mosaicism, that is, accumulation of non-inherited SNPs, indels, and copy number variants not present in germline cells. Perhaps the most striking example of this in AD is the finding of a sporadic AD patient who harbored a disease-causative mutation in PSEN1 only in approximately 14% of brain cells [236]. The patient was originally classified as negative for PSEN1 mutations after peripheral blood was drawn and blood DNA sequenced, but after DNA was obtained from the cerebral cortex, evidence of a cell-type specific mutation was observed. One offspring of the affected patient inherited the PSEN1 mutation in all cells of her body and exhibited signs of dementia by age 27, indicating the degree to which mutations that are expressed throughout the body have a profound effect on disease severity [236]. Additional somatic mutations in genes known to confer risk for AD such as APP and SORL1 [237] have been identified in the brains of both EOAD and LOAD patients [238, 239]. In general, neurons from AD patients have been observed to have an enriched number of somatic variations not observed in peripheral DNA [239, 240].
Several mechanisms may contribute to the abundance of somatic mutations in the AD brain. Transposable elements (TEs), mobile genetic sequences present in all eukaryotic genomes, account for up to 45% of the human genome. When TEs, especially retrotransposons, are activated, they mobilize through an RNA intermediate and can cause somatic mutations or large genomic rearrangements via their insertion into random segments of the genome [241, 242]. Under normal conditions, surveillance mechanisms including those associated with DNA repair and chromatin remodeling function to repress TE activity and largely maintain genomic stability [243]. However, with aging, the efficacy of these systems is known to be reduced, providing a genomic environment in which TEs may become activated and contribute to somatic mutagenesis [244, 245]. Neuropathologies such as tau have been observed to promote activation of TEs, providing an additional stimulus which may drive TE activation in the AD brain [245]. In addition, aberrant re-entry of neurons into the cell cycle has been hypothesized to be a mechanism underlying development of AD. As these cells do not typically divide, DNA replication stress and insufficient repair mechanisms may contribute to the development of somatic mutations in neurons [246], rendering these cells uniquely susceptible to eventual death as is observed during the neurodegenerative phase of AD. Together, this work suggests the genetic architecture of AD may not be completely accurately represented by studies utilizing peripheral blood DNA, which should be considered when interpreting the results of GWAS and other rare-variant sequencing studies.
b. Copy number variation
GWAS are uniquely designed to identify the impact of common polymorphisms on complex traits. However, SNPs are not the only mechanism by which genetic variation contributes to external traits. Another main source of genetic variation are copy number variants (CNVs) in which regions of the genome have been duplicated, inserted, or deleted [247]. CNVs may alter gene dosage and have a profound impact on both gene and protein function. In addition, CNVs may span multiple genes and have a measurable impact on more than one target [247]. It is estimated about 15% of the human genome is affected by CNVs, suggesting these may have a substantial impact on disease susceptibility. Gene duplications of APP are well-known to cause AD, with as many as 25 duplications of variable length associated with AD in EOAD families [2]. Neuropathological examination of patient brains harboring APP duplications show abundant plaque and tangle pathology as well cerebral amyloid angiopathy (CAA), which is characterized by deposition of Aβ in the cerebrovasculature and by hemorrhagic stroke [248, 249]. In addition, APP gene duplications that result from trisomy of chromosome 21 (Down’s Syndrome) result in near-complete penetrance of AD neuropathology and increased incidence of dementia by age 30–40y [250, 251]. Rare autosomal CNVs in genes besides APP have been implicated in EOAD [252], demonstrating consideration of genetic variation beyond SNPs can improve our understanding of the genetic architecture of AD. CNVs have also been identified as modifying risk for LOAD; a functional CNV in the gene CR1 modifying isoform production was shown to increase risk for LOAD in two populations [253], among others [254–256].
6. Additional Considerations
a. Sex-specific genetic architecture
AD is well-known to be more prevalent among elderly females, although it has been difficult to elucidate whether this is simply attributable to the longer lifespan of women [1, 257]. Notable sex differences in the clinical and pathological presentation between the sexes exist, each of which have been extensively reviewed previously [258–261]. For example, studies have reported more rapid cognitive decline in women, increased susceptibility to neuropathology such that each unit increase of AD pathology confers a 22-fold increase in risk for AD in women compared to a 3-fold increase in males, as well as important differences in clinical and pathological disease presentation [262]. One putative explanation for these differences is the presence of sex-specific genetic factors that influence risk for AD, although these have been difficult to identify in a systematic way. Several studies have demonstrated APOE genotype has an exacerbated effect on disease risk in females [58]. Notably, APOE has been associated with sex-specific alterations in CSF tau levels and disease progression but not robustly with CSF Aβ1–42, suggesting sex-specific effects of APOE genotype emerge downstream of amyloidosis [258]. Given the large sample sizes needed for sufficiently powered GWAS, investigation of sex-specific effects of loci outside of APOE on AD risk are less well-explored. A recent sex-stratified GWAS identified a male-specific protective variant located on chromosome 7 that conferred specific protection from tau pathology and was associated with delayed age at onset [263]. Several additional loci, including APOE [264], have been identified as exhibiting sex-specific association with AD endophenotypes, providing insight into biological mechanisms by which females may be at increased risk for AD [258, 265]. Overall, there is a critical need for a more detailed exploration of the mechanisms by which males and females exhibit differential susceptibility to AD, although current evidence points to important differences in the AD genetic architecture across sexes.
b. Missing heritability of AD
Despite decades of research, our understanding of the genetic architecture of AD is far from complete. Current estimates suggest only about 30–40% of the heritability of AD is explained by identified GWAS variants, leaving over 25% of the variance unexplained by known genetic factors [76]. Some of this “missing heritability” can be attributed to low frequency variants not evaluated in the general population or to variations in the epigenetic profile of patients not captured by standard genetic evaluation, both of which were discussed above. In addition, most GWAS are conducted using a standard additive model [266], where it is hypothesized heterozygous variant carriers exhibit a phenotype approximately intermediate in severity relative to each homozygous genotype. However, it has been estimated that up to 90% of EOAD may be caused by recessive mutations [24]. As such, it can be hypothesized that at least some percentage of LOAD is also caused by recessive mutations, where heterozygous mutation carriers will appear phenotypically similar to homozygotes carrying two copies of the dominant allele. These recessive effects may be missed by standard GWAS. Analyses aimed at genes which influence disease in a non-additive fashion have identified novel candidate genes for obesity and type 2 diabetes [267], suggesting these approaches may be useful in identifying recessive mutations that modify an individual’s risk for AD.
The large variability in the age at onset of AD symptoms even among families who share common pathogenic mutations [31] suggest variants in an individual’s genetic background modify the effect of specific variants on phenotypic penetrance. This highlights the possibility that extensive gene-gene interactions may contribute significantly to the genetic architecture of AD such that the effect of any AD-associated variant may largely depend on surrounding genetic context [268]. Hohman et al. [268] performed the most extensive analysis of gene-gene interactions in AD to date and identified three gene-gene combinations which significantly interact to determine an individual’s risk for AD, suggesting gene-gene interactions do indeed contribute to the heritability of AD. Similarly, an individual’s genetic background has been reported to influence how they respond to environmental exposures or lifestyle factors [269]. Several modifiable risk factors have been associated with AD risk across populations, including educational attainment, obesity, cholesterol levels, social isolation, alcohol consumption, and exposure to pesticides [269, 270]. It has been shown that an individual’s genetics affects the impact of many of these risk factors on AD; for example, APOEε4 carriers with low self-reported exercise levels exhibited an increased risk for disease beyond those with either APOEε4 or low exercise alone [271]. However, genome-wide gene-environment interactions are difficult to identify for various reasons, including the fact that data regarding an individual’s environmental exposures and/or lifestyle choices are unreliable due to incomplete recall or reporting, inconsistent dose, timing, and duration of exposure, and/or additional confounds produced by unreported exposures [272]. More thorough prospective analyses that document an individual’s lifestyle choices and environmental exposures prior to disease onset (such the NIH’s All of Us program) will provide enhanced resources through which to examine the environmental impact on disease risk. Combined with statistical approaches such as Mendelian Randomization [273, 274] that provide estimates of the causal association between environmental exposures (or other lifestyle factors) and disease risk, these approaches will likely enable an improved understanding of how genetics and environment interact to predict disease risk.
Finally, some of the “missing heritability” of AD may be attributable to the heterogeneity of the disease itself. Across the population, patients clinically diagnosed with AD display a wide variation in the onset, duration, and severity of symptoms across multiple functional domains and may experience deficits in cognitive function, speech, mood regulation and psychosis, motor function, and regulation of circadian rhythms [275]. This heterogeneity is observed even in carriers of highly penetrant APP and PSEN mutations [276], suggesting additional factors such as background genetic modifiers or even non-genetic lifestyle factors play a substantial role in dictating disease risk and severity. In addition, although AD is defined pathologically by the presence of Aβ and tau tangles, these pathologies often do not occur in isolation. A significant majority of patients diagnosed with AD also exhibit signs of additional pathologies including vascular lesions, Lewy bodies, TDP43 pathology or cerebral amyloid angiopathy [275]. First, this suggests patients diagnosed with clinical AD and included in many GWAS as “cases” may in fact not be pure AD cases but rather be suffering from a type of dementia due to mixed causes. As distinct genetic variants likely contribute to each aspect of any individual case, including these complex cases in the AD pool will likely reduce statistical power to identify genetic variants associated specifically with AD. Even among patients with neuropathologically confirmed AD, the clinical and pathological heterogeneity of the disease itself has led to the hypothesis that specific genetic variants may be associated only with specific aspects or features of the disease. This has led many to attempt to identify distinct subtypes of AD [235, 277]. In theory, the utilization of distinct subtype classification schema would provide a cleaner trait to test for genome-wide genetic association. Although this has proven difficult in practice, this should be an area of focus in coming years, as subtype-specific analyses may ultimately provide useful information for the development of personalized therapeutics for individual AD patients depending on their symptom spectrum.
c. Use of model systems that reflect the genetic architecture of disease
The substantial increase in our understanding regarding the genetic architecture of AD has resulted in the development of novel model systems in which to experimentally investigate mechanisms driving disease. Since the discovery that genetic variants are enriched within regulatory regions active in myeloid cells [173, 183, 184], a large emphasis in the field has been placed on improving resources to better model the impact of these variants in a disease-relevant context. This has resulted in highly efficient techniques to derive microglia-like cells from human induced pluripotent stem cells (iPSCs) [278, 279]. As these cells are typically difficult to access from human patients, this represents an exciting development in experimental capabilities. The addition of microglia-like cells engineered by genome editing to express disease-relevant mutations to established protocols for neuron/astrocyte co-culture [280] and the development of brain organoids is likely to greatly contribute to our understanding of how these cells contribute to disease [281]. Together, these approaches provide a tractable human model system in which to study AD [282].
As microglia are intrinsically designed to respond to their environment, it is likely disease-relevant insights into the impact of disease-associated variants on their biological function can best be elucidated via in vivo experiments. To this end, innovative transplantation methods have been established to implant hematopoietic stem cells (microglia precursors) or microglia themselves directly into the brains of mouse models, where they have been shown to fully integrate and differentiate into mature microglia [278, 279, 283]. Due to the strong biochemical, developmental, physiological, and genetic similarities between the mouse and human, the mouse represents an incredible resource for neuroscience research and has been used for decades to gain insight into mechanisms underlying the development of AD in vivo [284, 285]. However, traditional models created by incorporating a highly penetrant mutation in APP, PSEN1, or PSEN2 onto a single inbred background strain have largely failed to yield successful results in human clinical trials [286, 287]. Recent attempts to create improved mouse models have benefitted from the insights regarding the genetic architecture of AD described here. Widespread efforts to develop new models based on specific AD-associated variants have begun (e.g., www.model-ad.org), which promise to yield insight into the impact of these variants on gene expression and cellular function. In addition, efforts to expand existing AD mouse models through the inclusion of genetic diversity [288, 289] have resulted in resources that show a more significant overlap with human AD transcriptional profiles than models on a single genetic background [288], suggesting these genetically diverse models may represent improved preclinical resources than traditional approaches. In support, a PRS derived from human LOAD GWAS [96] showed significant association with cognitive outcomes across genetically diverse AD mice at 14 months of age [288], suggesting similar genetic mechanisms underlie AD susceptibility across species. This has important implications for the general use of AD mouse models and demonstrates the importance of considering genetic diversity in preclinical studies.
7. Conclusions and future directions
In summary, substantial advances in genetic and genomic technologies over the last thirty years have significantly contributed to our understanding of the genetic architecture of AD (Figure 1). Several genes including APP, PSEN1, PSEN2, and APOE have been shown to exhibit relatively large effects on disease susceptibility, with the remaining implicated loci having a much smaller individual impact on AD risk. It is only in recent decades that the technologies to 1) identify, and 2) effectively link these genetic variants to their downstream biological impact have become available. Specifically, advances in broad -omics technologies now enable genome-wide sampling across thousands of individuals with differing genetic backgrounds, providing unprecedented insight into how genetic variation leads to changes not only in RNA and protein expression, but also epigenetic signatures, chromatin accessibility, and long-range 3D genomic interactions. Integrative approaches are emerging as highly effective for identifying the specific target genes through which AD risk variants act. Indeed, we now understand that common genetic variants impacting AD are not randomly distributed across the genome. Instead, these variants seem to be enriched for localization to regulatory elements active largely in human monocytes, macrophages, and microglia.
In addition, advances in genome editing technologies and model organism development will greatly facilitate the understanding of the functional genetic architecture of AD. As there is currently no cure for AD, together these advances represent a significant and exciting opportunity to explore the therapeutic utility of identified candidate genes and pathways. A better understanding of the genetic architecture of a complex disease such as AD is likely to provide several important benefits to patients and their families as well as clinicians and researchers. First, identifying and understanding the biological function of AD-associated variants (i.e the functional genetic architecture) is likely to provide insight into potential therapeutic targets. Second, genetic variants associated with disease can be used to identify at-risk populations that may benefit most from early interventions or specific therapies. When combined with imaging and/or CSF biomarkers, genetic variants may provide an important tool by which to better understand a given patient’s risk or disease status. Indeed, nearly all clinical trials include stratification by APOEε4, an acknowledgement of the importance of this risk factor. As an alternative to focusing on one or even a few genetic variants, a better understanding of the genetic architecture of AD will also enhance our ability to calculate PRS that broadly encompass genetic risk. At the present time PRS only provide a modest advantage over APOE genotype alone, but improved PRS in combination with profiling of non-genetic risk factors such as various lifestyle factors may further improve our ability to stratify patients based on risk. With this information, we may be able to provide individualized recommendations, including both pharmacological and non-pharmacological approaches (e.g. improved diet and exercise), to those most at risk. Finally, additional characterization of how the architecture of disease differs across ethnicities, sexes, and disease subtypes will provide important insight into the specific populations in which each identified therapeutic may be most effective, a consideration which will likely prove critical in the coming years.
Supplementary Material
8. Acknowledgements
This study was funded by NIA K01AG062683 (JTCW), U01AG058635 (AMG), U01AG052411 (AMG) the JPB foundation (AMG), the Neurodegeneration Consortium (AMG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alzheimer’s Association. (2019). 2019 Alzheimer’s disease facts and figures. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 15(3), 321–387. [Google Scholar]
- 2.Cacace R, Sleegers K, & Van Broeckhoven C (2016). Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 12(6), 733–748. [DOI] [PubMed] [Google Scholar]
- 3.Karantzoulis S, & Galvin JE (2011). Distinguishing Alzheimer’s disease from other major forms of dementia. Expert review of neurotherapeutics, 11(11), 1579–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr., Kawas CH, … Phelps CH (2011). The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 7(3), 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selkoe DJ (1991). The molecular pathology of Alzheimer’s disease. Neuron, 6(4), 487–498. [DOI] [PubMed] [Google Scholar]
- 6.Hippius H, & Neundorfer G (2003). The discovery of Alzheimer’s disease. Dialogues in clinical neuroscience, 5(1), 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selkoe DJ (2008). Biochemistry and molecular biology of amyloid beta-protein and the mechanism of Alzheimer’s disease. Handbook of clinical neurology, 89, 245–260. [DOI] [PubMed] [Google Scholar]
- 8.Hardy J (1997). Amyloid, the presenilins and Alzheimer’s disease. Trends in neurosciences, 20(4), 154–159. [DOI] [PubMed] [Google Scholar]
- 9.Braak H, & Braak E (1998). Evolution of neuronal changes in the course of Alzheimer’s disease. Journal of neural transmission. Supplementum, 53, 127–140. [DOI] [PubMed] [Google Scholar]
- 10.Arnold SE, Hyman BT, Flory J, Damasio AR, & Van Hoesen GW (1991). The topographical and neuroanatomical distribution of neurofibrillary tangles and neuritic plaques in the cerebral cortex of patients with Alzheimer’s disease. Cerebral cortex, 1(1), 103–116. [DOI] [PubMed] [Google Scholar]
- 11.Beach TG, Walker R, & McGeer EG (1989). Patterns of gliosis in Alzheimer’s disease and aging cerebrum. Glia, 2(6), 420–436. [DOI] [PubMed] [Google Scholar]
- 12.Serrano-Pozo A, Frosch MP, Masliah E, & Hyman BT (2011). Neuropathological alterations in Alzheimer disease. Cold Spring Harbor perspectives in medicine, 1(1), a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopperton KE, Mohammad D, Trepanier MO, Giuliano V, & Bazinet RP (2018). Markers of microglia in post-mortem brain samples from patients with Alzheimer’s disease: a systematic review. Molecular psychiatry, 23(2), 177–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingelsson M, Fukumoto H, Newell KL, Growdon JH, Hedley-Whyte ET, Frosch MP, … Irizarry MC (2004). Early Abeta accumulation and progressive synaptic loss, gliosis, and tangle formation in AD brain. Neurology, 62(6), 925–931. [DOI] [PubMed] [Google Scholar]
- 15.Foley P (2010). Lipids in Alzheimer’s disease: A century-old story. Biochimica et biophysica acta, 1801(8), 750–753. [DOI] [PubMed] [Google Scholar]
- 16.Liu Q, & Zhang J (2014). Lipid metabolism in Alzheimer’s disease. Neuroscience bulletin, 30(2), 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farmer BC, Kluemper J, & Johnson LA (2019). Apolipoprotein E4 Alters Astrocyte Fatty Acid Metabolism and Lipid Droplet Formation. Cells, 8(2). 10.3390/cells8020182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong MW, Braidy N, Poljak A, Pickford R, Thambisetty M, & Sachdev PS (2017). Dysregulation of lipids in Alzheimer’s disease and their role as potential biomarkers. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 13(7), 810–827. [DOI] [PubMed] [Google Scholar]
- 19.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, … Beckett L (2005). Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 1(1), 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frisoni GB, Boccardi M, Barkhof F, Blennow K, Cappa S, Chiotis K, … Winblad B (2017). Strategic roadmap for an early diagnosis of Alzheimer’s disease based on biomarkers. Lancet neurology, 16(8), 661–676. [DOI] [PubMed] [Google Scholar]
- 21.Sunderland T, Linker G, Mirza N, Putnam KT, Friedman DL, Kimmel LH, … Cohen RM (2003). Decreased beta-amyloid1–42 and increased tau levels in cerebrospinal fluid of patients with Alzheimer disease. JAMA: the journal of the American Medical Association, 289(16), 2094–2103. [DOI] [PubMed] [Google Scholar]
- 22.Welge V, Fiege O, Lewczuk P, Mollenhauer B, Esselmann H, Klafki HW, … Bibl M (2009). Combined CSF tau, p-tau181 and amyloid-beta 38/40/42 for diagnosing Alzheimer’s disease. Journal of neural transmission, 116(2), 203–212. [DOI] [PubMed] [Google Scholar]
- 23.Cruchaga C, Kauwe JS, Harari O, Jin SC, Cai Y, Karch CM, … Goate AM (2013). GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron, 78(2), 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wingo TS, Lah JJ, Levey AI, & Cutler DJ (2012). Autosomal recessive causes likely in early-onset Alzheimer disease. Archives of neurology, 69(1), 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, Mortimer JA, … Ahlbom A (1997). Heritability for Alzheimer’s disease: the study of dementia in Swedish twins. The journals of gerontology. Series A, Biological sciences and medical sciences, 52(2), M117–25. [DOI] [PubMed] [Google Scholar]
- 26.Cruchaga C, Del-Aguila JL, Saef B, Black K, Fernandez MV, Budde J, … Harari O (2018). Polygenic risk score of sporadic late-onset Alzheimer’s disease reveals a shared architecture with the familial and early-onset forms. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 14(2), 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wijsman EM, Pankratz ND, Choi Y, Rothstein JH, Faber KM, Cheng R, … Nia-Load Ncrad Family Study Group. (2011). Genome-wide association of familial late-onset Alzheimer’s disease replicates BIN1 and CLU and nominates CUGBP2 in interaction with APOE. PLoS genetics, 7(2), e1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vardarajan BN, Faber KM, Bird TD, Bennett DA, Rosenberg R, Boeve BF, … Nia-Load Ncrad Family Study Group. (2014). Age-specific incidence rates for dementia and Alzheimer disease in NIA-LOAD/NCRAD and EFIGA families: National Institute on Aging Genetics Initiative for Late-Onset Alzheimer Disease/National Cell Repository for Alzheimer Disease (NIA-LOAD/NCRAD) and Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA). JAMA neurology, 71(3), 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smits LL, Pijnenburg YA, Koedam EL, van der Vlies AE, Reuling IE, Koene T, … van der Flier WM (2012). Early onset Alzheimer’s disease is associated with a distinct neuropsychological profile. Journal of Alzheimer’s disease: JAD, 30(1), 101–108. [DOI] [PubMed] [Google Scholar]
- 30.Koedam EL, Lauffer V, van der Vlies AE, van der Flier WM, Scheltens P, & Pijnenburg YA (2010). Early-versus late-onset Alzheimer’s disease: more than age alone. Journal of Alzheimer’s disease: JAD, 19(4), 1401–1408. [DOI] [PubMed] [Google Scholar]
- 31.Campion D, Dumanchin C, Hannequin D, Dubois B, Belliard S, Puel M, … Frebourg T (1999). Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. American journal of human genetics, 65(3), 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brickell KL, Steinbart EJ, Rumbaugh M, Payami H, Schellenberg GD, Van Deerlin V, … Bird TD (2006). Early-onset Alzheimer disease in families with late-onset Alzheimer disease: a potential important subtype of familial Alzheimer disease. Archives of neurology, 63(9), 1307–1311. [DOI] [PubMed] [Google Scholar]
- 33.Bekris LM, Yu CE, Bird TD, & Tsuang DW (2010). Genetics of Alzheimer disease. Journal of geriatric psychiatry and neurology, 23(4), 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Busciglio J, Gabuzda DH, Matsudaira P, & Yankner BA (1993). Generation of beta-amyloid in the secretory pathway in neuronal and nonneuronal cells. Proceedings of the National Academy of Sciences of the United States of America, 90(5), 2092–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Citron M, Vigo-Pelfrey C, Teplow DB, Miller C, Schenk D, Johnston J, … Selkoe DJ (1994). Excessive production of amyloid beta-protein by peripheral cells of symptomatic and presymptomatic patients carrying the Swedish familial Alzheimer disease mutation. Proceedings of the National Academy of Sciences of the United States of America, 91(25), 11993–11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tcw J, & Goate AM (2017). Genetics of beta-Amyloid Precursor Protein in Alzheimer’s Disease. Cold Spring Harbor perspectives in medicine, 7(6). 10.1101/cshperspect.a024539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardy J (2017). The discovery of Alzheimer-causing mutations in the APP gene and the formulation of the “amyloid cascade hypothesis.” The FEBS journal, 284(7), 1040–1044. [DOI] [PubMed] [Google Scholar]
- 38.Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, … Stefansson K (2012). A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature, 488(7409), 96–99. [DOI] [PubMed] [Google Scholar]
- 39.Hardy JA, & Higgins GA (1992). Alzheimer’s disease: the amyloid cascade hypothesis. Science, 256(5054), 184–185. [DOI] [PubMed] [Google Scholar]
- 40.Giaccone G, Morbin M, Moda F, Botta M, Mazzoleni G, Uggetti A, … Tagliavini F (2010). Neuropathology of the recessive A673V APP mutation: Alzheimer disease with distinctive features. Acta neuropathologica, 120(6), 803–812. [DOI] [PubMed] [Google Scholar]
- 41.Cruts M, Hendriks L, & Van Broeckhoven C (1996). The presenilin genes: a new gene family involved in Alzheimer disease pathology. Human molecular genetics, 5 Spec No, 1449–1455. [DOI] [PubMed] [Google Scholar]
- 42.Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, … et al. (1995). Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature, 376(6543), 775–778. [DOI] [PubMed] [Google Scholar]
- 43.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, … St George-Hyslop PH (1995). Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature, 375(6534), 754–760. [DOI] [PubMed] [Google Scholar]
- 44.Kopan R, & Goate A (2000). A common enzyme connects notch signaling and Alzheimer’s disease. Genes & development, 14(22), 2799–2806. [DOI] [PubMed] [Google Scholar]
- 45.Esler WP, Kimberly WT, Ostaszewski BL, Diehl TS, Moore CL, Tsai JY, … Wolfe MS (2000). Transition-state analogue inhibitors of gamma-secretase bind directly to presenilin-1. Nature cell biology, 2(7), 428–434. [DOI] [PubMed] [Google Scholar]
- 46.Li YM, Xu M, Lai MT, Huang Q, Castro JL, DiMuzio-Mower J, … Gardell SJ (2000). Photoactivated gamma-secretase inhibitors directed to the active site covalently label presenilin 1. Nature, 405(6787), 689–694. [DOI] [PubMed] [Google Scholar]
- 47.Citron M, Westaway D, Xia W, Carlson G, Diehl T, Levesque G, … Selkoe DJ (1997). Mutant presenilins of Alzheimer’s disease increase production of 42-residue amyloid beta-protein in both transfected cells and transgenic mice. Nature medicine, 3(1), 67–72. [DOI] [PubMed] [Google Scholar]
- 48.Mehta ND, Refolo LM, Eckman C, Sanders S, Yager D, Perez-Tur J, … Hutton M (1998). Increased Abeta42(43) from cell lines expressing presenilin 1 mutations. Annals of neurology, 43(2), 256–258. [DOI] [PubMed] [Google Scholar]
- 49.Cacquevel M, Aeschbach L, Houacine J, & Fraering PC (2012). Alzheimer’s disease-linked mutations in presenilin-1 result in a drastic loss of activity in purified gamma-secretase complexes. PloS one, 7(4), e35133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryan NS, & Rossor MN (2010). Correlating familial Alzheimer’s disease gene mutations with clinical phenotype. Biomarkers in medicine, 4(1), 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pilotto A, Padovani A, & Borroni B (2013). Clinical, biological, and imaging features of monogenic Alzheimer’s Disease. BioMed research international, 2013, 689591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pericak-Vance MA, Bebout JL, Gaskell PC Jr., Yamaoka LH, Hung WY, Alberts MJ, … et al. (1991). Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. American journal of human genetics, 48(6), 1034–1050. [PMC free article] [PubMed] [Google Scholar]
- 53.Roses AD, & Saunders AM (1994). APOE is a major susceptibility gene for Alzheimer’s disease. Current opinion in biotechnology, 5(6), 663–667. [DOI] [PubMed] [Google Scholar]
- 54.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, … Pericak-Vance MA (1993). Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science, 261(5123), 921–923. [DOI] [PubMed] [Google Scholar]
- 55.Van Cauwenberghe C, Van Broeckhoven C, & Sleegers K (2016). The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genetics in medicine: official journal of the American College of Medical Genetics, 18(5), 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahley RW, & Rall SC Jr. (2000). Apolipoprotein E: far more than a lipid transport protein. Annual review of genomics and human genetics, 1, 507–537. [DOI] [PubMed] [Google Scholar]
- 57.Liu CC, Liu CC, Kanekiyo T, Xu H, & Bu G (2013). Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nature reviews. Neurology, 9(2), 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, … van Duijn CM (1997). Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA: the journal of the American Medical Association, 278(16), 1349–1356. [PubMed] [Google Scholar]
- 59.Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, … Kuwano R (2013). SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PloS one, 8(4), e58618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jun GR, Chung J, Mez J, Barber R, Beecham GW, Bennett DA, … Farrer LA (2017). Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 13(7), 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murrell JR, Price B, Lane KA, Baiyewu O, Gureje O, Ogunniyi A, … Hall KS (2006). Association of apolipoprotein E genotype and Alzheimer disease in African Americans. Archives of neurology, 63(3), 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajabli F, Feliciano BE, Celis K, Hamilton-Nelson KL, Whitehead PL, Adams LD, … Pericak-Vance MA (2018). Ancestral origin of ApoE epsilon4 Alzheimer disease risk in Puerto Rican and African American populations. PLoS genetics, 14(12), e1007791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gureje O, Ogunniyi A, Baiyewu O, Price B, Unverzagt FW, Evans RM, … Murrell JR (2006). APOE epsilon4 is not associated with Alzheimer’s disease in elderly Nigerians. Annals of neurology, 59(1), 182–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Houlden H, Crook R, Hardy J, Roques P, Collinge J, & Rossor M (1994). Confirmation that familial clustering and age of onset in late onset Alzheimer’s disease are determined at the apolipoprotein E locus. Neuroscience letters, 174(2), 222–224. [DOI] [PubMed] [Google Scholar]
- 65.Olarte L, Schupf N, Lee JH, Tang MX, Santana V, Williamson J, … Mayeux R (2006). Apolipoprotein E epsilon4 and age at onset of sporadic and familial Alzheimer disease in Caribbean Hispanics. Archives of neurology, 63(11), 1586–1590. [DOI] [PubMed] [Google Scholar]
- 66.Locke PA, Conneally PM, Tanzi RE, Gusella JF, & Haines JL (1995). Apolipoprotein E4 allele and Alzheimer disease: examination of allelic association and effect on age at onset in both early- and late-onset cases. Genetic epidemiology, 12(1), 83–92. [DOI] [PubMed] [Google Scholar]
- 67.Corneveaux JJ, Myers AJ, Allen AN, Pruzin JJ, Ramirez M, Engel A, … Huentelman MJ (2010). Association of CR1, CLU and PICALM with Alzheimer’s disease in a cohort of clinically characterized and neuropathologically verified individuals. Human molecular genetics, 19(16), 3295–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reiman EM, Arboleda-Velasquez JF, Quiroz YT, Huentelman MJ, Beach TG, Caselli RJ, … Alzheimer’s Disease Genetics Consortium. (2020). Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nature communications, 11(1), 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhong N, & Weisgraber KH (2009). Understanding the association of apolipoprotein E4 with Alzheimer disease: clues from its structure. The Journal of biological chemistry, 284(10), 6027–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mahley RW, Weisgraber KH, & Huang Y (2009). Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. Journal of lipid research, 50 Suppl, S183–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, … Holtzman DM (2011). Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Science translational medicine, 3(89), 89ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wildsmith KR, Holley M, Savage JC, Skerrett R, & Landreth GE (2013). Evidence for impaired amyloid beta clearance in Alzheimer’s disease. Alzheimer’s research & therapy, 5(4), 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ji ZS, Miranda RD, Newhouse YM, Weisgraber KH, Huang Y, & Mahley RW (2002). Apolipoprotein E4 potentiates amyloid beta peptide-induced lysosomal leakage and apoptosis in neuronal cells. The Journal of biological chemistry, 277(24), 21821–21828. [DOI] [PubMed] [Google Scholar]
- 74.Ji ZS, Mullendorff K, Cheng IH, Miranda RD, Huang Y, & Mahley RW (2006). Reactivity of apolipoprotein E4 and amyloid beta peptide: lysosomal stability and neurodegeneration. The Journal of biological chemistry, 281(5), 2683–2692. [DOI] [PubMed] [Google Scholar]
- 75.Chang S, ran Ma T, Miranda RD, Balestra ME, Mahley RW, & Huang Y (2005). Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proceedings of the National Academy of Sciences of the United States of America, 102(51), 18694–18699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ridge PG, Mukherjee S, Crane PK, Kauwe JS, & Alzheimer’s Disease Genetics, Consortium. (2013). Alzheimer’s disease: analyzing the missing heritability. PloS one, 8(11), e79771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ardlie KG, Kruglyak L, & Seielstad M (2002). Patterns of linkage disequilibrium in the human genome. Nature reviews. Genetics, 3(4), 299–309. [DOI] [PubMed] [Google Scholar]
- 78.Bush WS, & Moore JH (2012). Chapter 11: Genome-wide association studies. PLoS computational biology, 8(12), e1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y, Willer C, Sanna S, & Abecasis G (2009). Genotype imputation. Annual review of genomics and human genetics, 10, 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bomba L, Walter K, & Soranzo N (2017). The impact of rare and low-frequency genetic variants in common disease. Genome biology, 18(1), 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, … Stephan DA (2007). A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. The Journal of clinical psychiatry, 68(4), 613–618. [DOI] [PubMed] [Google Scholar]
- 82.Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, … Stephan DA (2007). GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers. Neuron, 54(5), 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu C, Tachmazidou I, Walter K, Ciampi A, Zeggini E, Greenwood CMT, & UK10K Consortium. (2014). Estimating genome-wide significance for whole-genome sequencing studies. Genetic epidemiology, 38(4), 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feulner TM, Laws SM, Friedrich P, Wagenpfeil S, Wurst SH, Riehle C, … Riemenschneider M (2010). Examination of the current top candidate genes for AD in a genome-wide association study. Molecular psychiatry, 15(7), 756–766. [DOI] [PubMed] [Google Scholar]
- 85.Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, … Younkin SG (2009). Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nature genetics, 41(2), 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, Haines JL, & Pericak-Vance MA (2009). Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. American journal of human genetics, 84(1), 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, … Tanzi RE (2008). Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. American journal of human genetics, 83(5), 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, … Roses AD (2008). Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Archives of neurology, 65(1), 45–53. [DOI] [PubMed] [Google Scholar]
- 89.Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, Georgieva L, … Kirov G (2008). A genome-wide association study for late-onset Alzheimer’s disease using DNA pooling. BMC medical genomics, 1, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu F, Arias-Vasquez A, Sleegers K, Aulchenko YS, Kayser M, Sanchez-Juan P, … van Duijn CM (2007). A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. American journal of human genetics, 81(1), 17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, … Williams J (2009). Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nature genetics, 41(10), 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, … Amouyel P (2009). Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nature genetics, 41(10), 1094–1099. [DOI] [PubMed] [Google Scholar]
- 93.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, … Schellenberg GD (2011). Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nature genetics, 43(5), 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, … Williams J (2011). Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nature genetics, 43(5), 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, … Eadi Consortium. (2010). Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA: the journal of the American Medical Association, 303(18), 1832–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, … Amouyel P (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature genetics, 45(12), 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, … Environmental Risk for Alzheimer’s Disease, Consortium. (2019). Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nature genetics, 51(3), 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mayeux Manly J J. (2004). Ethnic differences in dementia and Alzheimer’s disease In Anderson NB, Bulatao RA, Cohen B (Ed.), Critical Perspectives on Racial and Ethnic Differences in Health and DIsease. Washington, DC: National Academies Press. [PubMed] [Google Scholar]
- 99.Lee JH, Cheng R, Barral S, Reitz C, Medrano M, Lantigua R, … Mayeux R (2011). Identification of novel loci for Alzheimer disease and replication of CLU, PICALM, and BIN1 in Caribbean Hispanic individuals. Archives of neurology, 68(3), 320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou X, Chen Y, Mok KY, Zhao Q, Chen K, Chen Y, … Alzheimer’s Disease Neuroimaging, Initiative. (2018). Identification of genetic risk factors in the Chinese population implicates a role of immune system in Alzheimer’s disease pathogenesis. Proceedings of the National Academy of Sciences of the United States of America, 115(8), 1697–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li YR, & Keating BJ (2014). Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome medicine, 6(10), 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blue EE, Horimoto ARVR, Mukherjee S, Wijsman EM, & Thornton TA (2019). Local ancestry at APOE modifies Alzheimer’s disease risk in Caribbean Hispanics. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 15(12), 1524–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cukier HN, Kunkle BW, Vardarajan BN, Rolati S, Hamilton-Nelson KL, Kohli MA, … Alzheimer’s Disease Genetics Consortium. (2016). ABCA7 frameshift deletion associated with Alzheimer disease in African Americans. Neurology. Genetics, 2(3), e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Steinberg S, Stefansson H, Jonsson T, Johannsdottir H, Ingason A, Helgason H, … Stefansson K (2015). Loss-of-function variants in ABCA7 confer risk of Alzheimer’s disease. Nature genetics, 47(5), 445–447. [DOI] [PubMed] [Google Scholar]
- 105.Cuyvers E, De Roeck A, Van den Bossche T, Van Cauwenberghe C, Bettens K, Vermeulen S, … Sleegers K (2015). Mutations in ABCA7 in a Belgian cohort of Alzheimer’s disease patients: a targeted resequencing study. Lancet neurology, 14(8), 814–822. [DOI] [PubMed] [Google Scholar]
- 106.Vardarajan BN, Ghani M, Kahn A, Sheikh S, Sato C, Barral S, … Others. (2015). Rare coding mutations identified by sequencing of A lzheimer disease genome-wide association studies loci. Annals of neurology, 78(3), 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert J-C, Chung J, Naj AC, … Farrer LA (2016). A novel Alzheimer disease locus located near the gene encoding tau protein. Molecular psychiatry, 21(1), 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trabzuni D, Wray S, Vandrovcova J, Ramasamy A, Walker R, Smith C, … Ryten M (2012). MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Human molecular genetics, 21(18), 4094–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhong Q, Congdon EE, Nagaraja HN, & Kuret J (2012). Tau isoform composition influences rate and extent of filament formation. The Journal of biological chemistry, 287(24), 20711–20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Caffrey TM, Joachim C, & Wade-Martins R (2008). Haplotype-specific expression of the N-terminal exons 2 and 3 at the human MAPT locus. Neurobiology of aging, 29(12), 1923–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jun G, Naj AC, Beecham GW, Wang L-S, Buros J, Gallins PJ, … Schellenberg GD (2010). Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Archives of neurology, 67(12), 1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu JZ, Erlich Y, & Pickrell JK (2017). Case-control association mapping by proxy using family history of disease. Nature genetics, 49(3), 325–331. [DOI] [PubMed] [Google Scholar]
- 113.Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, … Posthuma D (2019). Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nature genetics, 51(3), 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marioni RE, Harris SE, Zhang Q, McRae AF, Hagenaars SP, Hill WD, … Visscher PM (2018). GWAS on family history of Alzheimer’s disease. Translational psychiatry, 8(1), 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Escott-Price V, Bellenguez C, Wang L-S, Choi S-H, Harold D, Jones L, … Cardiovascular Health Study (CHS). (2014). Gene-wide analysis detects two new susceptibility genes for Alzheimer’s disease. PloS one, 9(6), e94661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Baker E, Sims R, Leonenko G, Frizzati A, Harwood J, Grozeva D, … EADI Consortium. (n.d.). Gene-Based Analysis in HRC Imputed Genome Wide Association Data Identifies Three Novel Genes For Alzheimer’s Disease. 10.1101/374876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.International Schizophrenia, Consortium, Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, … Sklar P (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature, 460(7256), 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Desikan RS, Fan CC, Wang Y, Schork AJ, Cabral HJ, Cupples LA, … Dale AM (2017). Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLoS medicine, 14(3), e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sleegers K, Bettens K, De Roeck A, Van Cauwenberghe C, Cuyvers E, Verheijen J, … Belneu consortium. (2015). A 22-single nucleotide polymorphism Alzheimer’s disease risk score correlates with family history, onset age, and cerebrospinal fluid Abeta42. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 11(12), 1452–1460. [DOI] [PubMed] [Google Scholar]
- 120.Chouraki V, Reitz C, Maury F, Bis JC, Bellenguez C, Yu L, … International Genomics of Alzheimer’s, Project. (2016). Evaluation of a Genetic Risk Score to Improve Risk Prediction for Alzheimer’s Disease. Journal of Alzheimer’s disease: JAD, 53(3), 921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yokoyama JS, Bonham LW, Sears RL, Klein E, Karydas A, Kramer JH, … Coppola G (2015). Decision tree analysis of genetic risk for clinically heterogeneous Alzheimer’s disease. BMC neurology, 15, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, … Williams J (2015). Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain: a journal of neurology, 138(Pt 12), 3673–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Escott-Price V, Shoai M, Pither R, Williams J, & Hardy J (2017). Polygenic score prediction captures nearly all common genetic risk for Alzheimer’s disease. Neurobiology of aging, 49, 214 e7–214 e11. [DOI] [PubMed] [Google Scholar]
- 124.Hao K, Di Narzo AF, Ho L, Luo W, Li S, Chen R, … Pasinetti GM (2015). Shared genetic etiology underlying Alzheimer’s disease and type 2 diabetes. Molecular aspects of medicine, 43–44, 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yokoyama JS, Wang Y, Schork AJ, Thompson WK, Karch CM, Cruchaga C, … Alzheimer’s Disease Neuroimaging, Initiative. (2016). Association Between Genetic Traits for Immune-Mediated Diseases and Alzheimer Disease. JAMA neurology, 73(6), 691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Potter H (2016). New Genetic Links Between Alzheimer Disease and Immune-Mediated Diseases Revealed by Overlapping Genome-wide Association Study Hits. JAMA neurology, 73(6), 638–640. [DOI] [PubMed] [Google Scholar]
- 127.Guerreiro RJ, Gustafson DR, & Hardy J (2012). The genetic architecture of Alzheimer’s disease: beyond APP, PSENs and APOE. Neurobiology of aging, 33(3), 437–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kunkle BW, Jaworski J, Barral S, Vardarajan B, Beecham GW, Martin ER, … Pericak-Vance MA (2016). Genome-wide linkage analyses of non-Hispanic white families identify novel loci for familial late-onset Alzheimer’s disease. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 12(1), 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Beecham GW, Bis JC, Martin ER, Choi SH, DeStefano AL, van Duijn CM, … Schellenberg G (2017). The Alzheimer’s Disease Sequencing Project: Study design and sample selection. Neurol Genet, 3(5), e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, … Schellenberg GD (2017). Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nature genetics, 49(9), 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, … Stefansson K (2013). Variant of TREM2 associated with the risk of Alzheimer’s disease. The New England journal of medicine, 368(2), 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, … Alzheimer Genetic Analysis, Group. (2013). TREM2 variants in Alzheimer’s disease. The New England journal of medicine, 368(2), 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Luis EO, Ortega-Cubero S, Lamet I, Razquin C, Cruchaga C, Benitez BA, … Pastor P (2014). Frontobasal gray matter loss is associated with the TREM2 p.R47H variant. Neurobiology of aging, 35(12), 2681–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Deming Y, Filipello F, Cignarella F, Cantoni C, Hsu S, Mikesell R, … Cruchaga C (2019). The MS4A gene cluster is a key modulator of soluble TREM2 and Alzheimer’s disease risk. Science translational medicine, 11(505). 10.1126/scitranslmed.aau2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Guerreiro R, Bilgic B, Guven G, Bras J, Rohrer J, Lohmann E, … Emre M (2013). Novel compound heterozygous mutation in TREM2 found in a Turkish frontotemporal dementia-like family. Neurobiology of aging, 34(12), 2890 e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sudom A, Talreja S, Danao J, Bragg E, Kegel R, Min X, … Wang Z (2018). Molecular basis for the loss-of-function effects of the Alzheimer’s disease-associated R47H variant of the immune receptor TREM2. The Journal of biological chemistry, 293(32), 12634–12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yeh FL, Wang Y, Tom I, Gonzalez LC, & Sheng M (2016). TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron, 91(2), 328–340. [DOI] [PubMed] [Google Scholar]
- 138.Benitez BA, Cooper B, Pastor P, Jin S-C, Lorenzo E, Cervantes S, & Cruchaga C (2013). TREM2 is associated with the risk of Alzheimer’s disease in Spanish population. Neurobiology of aging, 34(6), 1711.e15–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jin SC, Benitez BA, Karch CM, Cooper B, Skorupa T, Carrell D, … Cruchaga C (2014). Coding variants in TREM2 increase risk for Alzheimer’s disease. Human molecular genetics, 23(21), 5838–5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jin SC, Carrasquillo MM, Benitez BA, Skorupa T, Carrell D, Patel D, … Ertekin-Taner N (2015). TREM2 is associated with increased risk for Alzheimer’s disease in African Americans. Molecular neurodegeneration, 10, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dalmasso MC, Brusco LI, Olivar N, Muchnik C, Hanses C, Milz E, … Ramirez A (2019). Transethnic meta-analysis of rare coding variants in PLCG2, ABI3, and TREM2 supports their general contribution to Alzheimer’s disease. Translational psychiatry, 9(1), 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Milner JD (2015). PLAID: a Syndrome of Complex Patterns of Disease and Unique Phenotypes. Journal of clinical immunology, 35(6), 527–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, … Knight JC (2014). Innate Immune Activity Conditions the Effect of Regulatory Variants upon Monocyte Gene Expression. Science, 343(6175), 1118–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sekino S, Kashiwagi Y, Kanazawa H, Takada K, Baba T, Sato S, … Tani K (2015). The NESH/Abi-3-based WAVE2 complex is functionally distinct from the Abi-1-based WAVE2 complex. Cell communication and signaling: CCS, 13, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lord J, Lu AJ, & Cruchaga C (2014). Identification of rare variants in Alzheimer’s disease. Frontiers in genetics, 5, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Del-Aguila JL, Koboldt DC, Black K, Chasse R, Norton J, Wilson RK, & Cruchaga C (2015). Alzheimer’s disease: rare variants with large effect sizes. Current opinion in genetics & development, 33, 49–55. [DOI] [PubMed] [Google Scholar]
- 147.Vardarajan BN, Zhang Y, Lee JH, Cheng R, Bohm C, Ghani M, … Mayeux R (2015). Coding mutations in SORL1 and Alzheimer disease. Annals of neurology, 77(2), 215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Raghavan NS, Brickman AM, Andrews H, Manly JJ, Schupf N, Lantigua R, … Alzheimer’s Disease Sequencing, Project. (2018). Whole-exome sequencing in 20,197 persons for rare variants in Alzheimer’s disease. Ann Clin Transl Neurol, 5(7), 832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, … Goate AM (2014). Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature, 505(7484), 550–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Le Guennec K, Nicolas G, Quenez O, Charbonnier C, Wallon D, Bellenguez C, … Cnr-Maj collaborators. (2016). ABCA7 rare variants and Alzheimer disease risk. Neurology, 86(23), 2134–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Satoh K, Abe-Dohmae S, Yokoyama S, St George-Hyslop P, & Fraser PE (2015). ATP-binding cassette transporter A7 (ABCA7) loss of function alters Alzheimer amyloid processing. The Journal of biological chemistry, 290(40), 24152–24165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Grear KE, Ling IF, Simpson JF, Furman JL, Simmons CR, Peterson SL, … Estus S (2009). Expression of SORL1 and a novel SORL1 splice variant in normal and Alzheimers disease brain. Molecular neurodegeneration, 4, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, … Barres BA (2016). Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron, 89(1), 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, … St George-Hyslop P (2007). The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nature genetics, 39(2), 168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pottier C, Hannequin D, Coutant S, Rovelet-Lecrux A, Wallon D, Rousseau S, … Phrc Gmaj Collaborators. (2012). High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Molecular psychiatry, 17(9), 875–879. [DOI] [PubMed] [Google Scholar]
- 156.Nicolas G, Charbonnier C, Wallon D, Quenez O, Bellenguez C, Grenier-Boley B, … Cnr-Maj collaborators. (2016). SORL1 rare variants: a major risk factor for familial early-onset Alzheimer’s disease. Molecular psychiatry, 21(6), 831–836. [DOI] [PubMed] [Google Scholar]
- 157.Cruchaga C, Haller G, Chakraverty S, Mayo K, Vallania FL, Mitra RD, … Nia-Load Ncrad Family Study Consortium. (2012). Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s disease families. PloS one, 7(2), e31039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lanoiselee HM, Nicolas G, Wallon D, Rovelet-Lecrux A, Lacour M, Rousseau S, … collaborators of the, C. N. R. M. A. J. project. (2017). APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: A genetic screening study of familial and sporadic cases. PLoS medicine, 14(3), e1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rovelet-Lecrux A, CNR-MAJ collaborators, Charbonnier C, Wallon D, Nicolas G, Seaman MNJ, … Campion D (2015). De novo deleterious genetic variations target a biological network centered on Aβ peptide in early-onset Alzheimer disease. Molecular Psychiatry. 10.1038/mp.2015.100 [DOI] [PubMed] [Google Scholar]
- 160.Sondag CM, & Combs CK (2004). Amyloid precursor protein mediates proinflammatory activation of monocytic lineage cells. The Journal of biological chemistry, 279(14), 14456–14463. [DOI] [PubMed] [Google Scholar]
- 161.Lauritzen I, Pardossi-Piquard R, Bourgeois A, Pagnotta S, Biferi MG, Barkats M, … Checler F (2016). Intraneuronal aggregation of the beta-CTF fragment of APP (C99) induces Abeta-independent lysosomal-autophagic pathology. Acta neuropathologica, 132(2), 257–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Grimm MO, Mett J, Grimm HS, & Hartmann T (2017). APP Function and Lipids: A Bidirectional Link. Frontiers in molecular neuroscience, 10, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Guerreiro RJ, Lohmann E, Kinsella E, Bras JM, Luu N, Gurunlian N, … Singleton A (2012). Exome sequencing reveals an unexpected genetic cause of disease: NOTCH3 mutation in a Turkish family with Alzheimer’s disease. Neurobiology of aging, 33(5), 1008 e17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Velez JI, Rivera D, Mastronardi CA, Patel HR, Tobon C, Villegas A, … Arcos-Burgos M (2016). A Mutation in DAOA Modifies the Age of Onset in PSEN1 E280A Alzheimer’s Disease. Neural plasticity, 2016, 9760314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lee JH, Cheng R, Vardarajan B, Lantigua R, Reyes-Dumeyer D, Ortmann W, … Mayeux R (2015). Genetic Modifiers of Age at Onset in Carriers of the G206A Mutation in PSEN1 With Familial Alzheimer Disease Among Caribbean Hispanics. JAMA neurology, 72(9), 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Park JH, Gail MH, Weinberg CR, Carroll RJ, Chung CC, Wang Z, … Chatterjee N (2011). Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proceedings of the National Academy of Sciences of the United States of America, 108(44), 18026–18031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Eyre-Walker A (2010). Evolution in health and medicine Sackler colloquium: Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proceedings of the National Academy of Sciences of the United States of America, 107 Suppl 1, 1752–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, … Stamatoyannopoulos JA (2012). Systematic localization of common disease-associated variation in regulatory DNA. Science, 337(6099), 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jowett JB, Curran JE, Johnson MP, Carless MA, Goring HH, Dyer TD, … Blangero J (2010). Genetic variation at the FTO locus influences RBL2 gene expression. Diabetes, 59(3), 726–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Karch CM, Cruchaga C, & Goate AM (2014). Alzheimer’s disease genetics: from the bench to the clinic. Neuron, 83(1), 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Karch CM, & Goate AM (2015). Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biological psychiatry, 77(1), 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Thevenin A, Ein-Dor L, Ozery-Flato M, & Shamir R (2014). Functional gene groups are concentrated within chromosomes, among chromosomes and in the nuclear space of the human genome. Nucleic acids research, 42(15), 9854–9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Huang KL, Marcora E, Pimenova AA, Di Narzo AF, Kapoor M, Jin SC, … Goate AM (2017). A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nature neuroscience, 20(8), 1052–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Brodie A, Azaria JR, & Ofran Y (2016). How far from the SNP may the causative genes be? Nucleic acids research, 44(13), 6046–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fuda NJ, Ardehali MB, & Lis JT (2009). Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature, 461(7261), 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Catarino RR, & Stark A (2018). Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes & development, 32(3–4), 202–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Buenrostro JD, Wu B, Chang HY, & Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Current protocols in molecular biology / edited by Frederick M. Ausubel ... [et al.], 109, 21 29 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Krivega I, & Dean A (2012). Enhancer and promoter interactions-long distance calls. Current opinion in genetics & development, 22(2), 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gavrilov A, Eivazova E, Priozhkova I, Lipinski M, Razin S, & Vassetzky Y (2009). Chromosome conformation capture (from 3C to 5C) and its ChIP-based modification. Methods in molecular biology, 567, 171–188. [DOI] [PubMed] [Google Scholar]
- 180.Roadmap Epigenomics, Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, … Kellis M (2015). Integrative analysis of 111 reference human epigenomes. Nature, 518(7539), 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fernandez JM, de la Torre V, Richardson D, Royo R, Puiggros M, Moncunill V, … Valencia A (2016). The BLUEPRINT Data Analysis Portal. Cell Syst, 3(5), 491–495 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Psych, Encode Consortium, Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, … Sestan N (2015). The PsychENCODE project. Nature neuroscience, 18(12), 1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gagliano SA, Pouget JG, Hardy J, Knight J, Barnes MR, Ryten M, & Weale ME (2016). Genomics implicates adaptive and innate immunity in Alzheimer’s and Parkinson’s diseases. Ann Clin Transl Neurol, 3(12), 924–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tansey KE, Cameron D, & Hill MJ (2018). Genetic risk for Alzheimer’s disease is concentrated in specific macrophage and microglial transcriptional networks. Genome medicine, 10(1), 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lu Q, Powles RL, Abdallah S, Ou D, Wang Q, Hu Y, … Zhao H (2017). Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease. PLoS genetics, 13(7), e1006933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, … Price AL (2015). Partitioning heritability by functional annotation using genome-wide association summary statistics. Nature genetics, 47(11), 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Trapani L, Segatto M, & Pallottini V (2012). Regulation and deregulation of cholesterol homeostasis: The liver as a metabolic “power station.” World journal of hepatology, 4(6), 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mahley RW (2016). Central Nervous System Lipoproteins: ApoE and Regulation of Cholesterol Metabolism. Arteriosclerosis, thrombosis, and vascular biology, 36(7), 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sodhi RK, & Singh N (2013). Liver X receptors: emerging therapeutic targets for Alzheimer’s disease. Pharmacological research: the official journal of the Italian Pharmacological Society, 72, 45–51. [DOI] [PubMed] [Google Scholar]
- 190.Zelcer N, Khanlou N, Clare R, Jiang Q, Reed-Geaghan EG, Landreth GE, … Tontonoz P (2007). Attenuation of neuroinflammation and Alzheimer’s disease pathology by liver x receptors. Proceedings of the National Academy of Sciences of the United States of America, 104(25), 10601–10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Koldamova RP, Lefterov IM, Staufenbiel M, Wolfe D, Huang S, Glorioso JC, … Lazo JS (2005). The liver X receptor ligand T0901317 decreases amyloid beta production in vitro and in a mouse model of Alzheimer’s disease. The Journal of biological chemistry, 280(6), 4079–4088. [DOI] [PubMed] [Google Scholar]
- 192.Nott A, Holtman IR, Coufal NG, Schlachetzki JCM, Yu M, Hu R, … Glass CK (2019). Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science, 366(6469), 1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, … De Jager PL (2014). Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science, 344(6183), 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Smith AM, Gibbons HM, Oldfield RL, Bergin PM, Mee EW, Faull RL, & Dragunow M (2013). The transcription factor PU.1 is critical for viability and function of human brain microglia. Glia, 61(6), 929–942. [DOI] [PubMed] [Google Scholar]
- 195.Lunnon K, & Mill J (2013). Epigenetic studies in Alzheimer’s disease: current findings, caveats, and considerations for future studies. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics, 162B(8), 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yu L, Chibnik LB, Srivastava GP, Pochet N, Yang J, Xu J, … Bennett DA (2015). Association of Brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA neurology, 72(1), 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chibnik LB, Yu L, Eaton ML, Srivastava G, Schneider JA, Kellis M, … De Jager PL (2015). Alzheimer’s loci: epigenetic associations and interaction with genetic factors. Ann Clin Transl Neurol, 2(6), 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M, … Mill J (2014). Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nature neuroscience, 17(9), 1164–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, … Bennett DA (2014). Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nature neuroscience, 17(9), 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Karch CM, Ezerskiy LA, Bertelsen S, Alzheimer’s Disease Genetics, Consortium, & Goate AM (2016). Alzheimer’s Disease Risk Polymorphisms Regulate Gene Expression in the ZCWPW1 and the CELF1 Loci. PloS one, 11(2), e0148717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Katsumata Y, Nelson PT, Estus S, Fardo DW, Weiner MW, Aisen P, … Initia ADN (2019). Translating Alzheimer’s disease-associated polymorphisms into functional candidates: a survey of IGAP genes and SNPs. Neurobiology of aging, 74, 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Allen M, Kachadoorian M, Carrasquillo MM, Karhade A, Manly L, Burgess JD, … Ertekin-Taner N (2015). Late-onset Alzheimer disease risk variants mark brain regulatory loci. Neurol Genet, 1(2), e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Salter MW, & Stevens B (2017). Microglia emerge as central players in brain disease. Nature medicine, 23(9), 1018–1027. [DOI] [PubMed] [Google Scholar]
- 204.Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, … Stevens B (2016). Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science, 352(6286), 712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Flint J, & Munafo MR (2007). The endophenotype concept in psychiatric genetics. Psychological medicine, 37(2), 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ferreira D, Perestelo-Perez L, Westman E, Wahlund LO, Sarria A, & Serrano-Aguilar P (2014). Meta-Review of CSF Core Biomarkers in Alzheimer’s Disease: The State-of-the-Art after the New Revised Diagnostic Criteria. Frontiers in aging neuroscience, 6, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hall MH, & Smoller JW (2010). A new role for endophenotypes in the GWAS era: functional characterization of risk variants. Harvard review of psychiatry, 18(1), 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ringman JM, Younkin SG, Pratico D, Seltzer W, Cole GM, Geschwind DH, … Cummings JL (2008). Biochemical markers in persons with preclinical familial Alzheimer disease. Neurology, 71(2), 85–92. [DOI] [PubMed] [Google Scholar]
- 209.Kauwe JS, Wang J, Mayo K, Morris JC, Fagan AM, Holtzman DM, & Goate AM (2009). Alzheimer’s disease risk variants show association with cerebrospinal fluid amyloid beta. Neurogenetics, 10(1), 13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kauwe JS, Jacquart S, Chakraverty S, Wang J, Mayo K, Fagan AM, … Goate AM (2007). Extreme cerebrospinal fluid amyloid beta levels identify family with late-onset Alzheimer’s disease presenilin 1 mutation. Annals of neurology, 61(5), 446–453. [DOI] [PubMed] [Google Scholar]
- 211.Deming Y, Li Z, Kapoor M, Harari O, Del-Aguila JL, Black K, … Cruchaga C (2017). Genome-wide association study identifies four novel loci associated with Alzheimer’s endophenotypes and disease modifiers. Acta neuropathologica, 133(5), 839–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Suarez-Calvet M, Araque Caballero MA, Kleinberger G, Bateman RJ, Fagan AM, Morris JC, … Dominantly Inherited Alzheimer, Network. (2016). Early changes in CSF sTREM2 in dominantly inherited Alzheimer’s disease occur after amyloid deposition and neuronal injury. Science translational medicine, 8(369), 369ra178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Henjum K, Almdahl IS, Årskog V, Minthon L, Hansson O, Fladby T, & Nilsson LNG (2016). Cerebrospinal fluid soluble TREM2 in aging and Alzheimer’s disease. Alzheimer’s research & therapy, 8(1), 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Suárez-Calvet M, Kleinberger G, Araque Caballero MÁ, Brendel M, Rominger A, Alcolea D, … Haass C (2016). sTREM2 cerebrospinal fluid levels are a potential biomarker for microglia activity in early-stage Alzheimer’s disease and associate with neuronal injury markers. EMBO molecular medicine, 8(5), 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Cruchaga C, Kauwe JS, Nowotny P, Bales K, Pickering EH, Mayo K, … Goate AM (2012). Cerebrospinal fluid APOE levels: an endophenotype for genetic studies for Alzheimer’s disease. Human molecular genetics, 21(20), 4558–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hampel H, Toschi N, Baldacci F, Zetterberg H, Blennow K, Kilimann I, … Alzheimer Precision Medicine, Initiative. (2018). Alzheimer’s disease biomarker-guided diagnostic workflow using the added value of six combined cerebrospinal fluid candidates: Abeta1–42, total-tau, phosphorylated-tau, NFL, neurogranin, and YKL-40. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 14(4), 492–501. [DOI] [PubMed] [Google Scholar]
- 217.Blennow K (2017). A Review of Fluid Biomarkers for Alzheimer’s Disease: Moving from CSF to Blood. Neurol Ther, 6(Suppl 1), 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Olsson B, Lautner R, Andreasson U, Ohrfelt A, Portelius E, Bjerke M, … Zetterberg H (2016). CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: a systematic review and meta-analysis. Lancet neurology, 15(7), 673–684. [DOI] [PubMed] [Google Scholar]
- 219.Suppiah S, Didier MA, & Vinjamuri S (2019). The Who, When, Why, and How of PET Amyloid Imaging in Management of Alzheimer’s Disease-Review of Literature and Interesting Images. Diagnostics (Basel), 9(2). 10.3390/diagnostics9020065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ou YN, Xu W, Li JQ, Guo Y, Cui M, Chen KL, … Alzheimer’s Disease Neuroimaging, Initiative. (2019). FDG-PET as an independent biomarker for Alzheimer’s biological diagnosis: a longitudinal study. Alzheimer’s research & therapy, 11(1), 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Dukart J, Mueller K, Horstmann A, Vogt B, Frisch S, Barthel H, … Schroeter ML (2010). Differential effects of global and cerebellar normalization on detection and differentiation of dementia in FDG-PET studies. NeuroImage, 49(2), 1490–1495. [DOI] [PubMed] [Google Scholar]
- 222.Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC, & Alzheimer’s Disease Neuroimaging, Initiative. (2009). Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Current Alzheimer research, 6(4), 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kremen WS, Panizzon MS, Neale MC, Fennema-Notestine C, Prom-Wormley E, Eyler LT, … Dale AM (2012). Heritability of brain ventricle volume: converging evidence from inconsistent results. Neurobiology of aging, 33(1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lunetta KL, Erlich PM, Cuenco KT, Cupples LA, Green RC, Farrer LA, … Mirage Study Group. (2007). Heritability of magnetic resonance imaging (MRI) traits in Alzheimer disease cases and their siblings in the MIRAGE study. Alzheimer disease and associated disorders, 21(2), 85–91. [DOI] [PubMed] [Google Scholar]
- 225.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Cedarbaum J, … Alzheimer’s Disease Neuroimaging, Initiative. (2015). 2014 Update of the Alzheimer’s Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 11(6), e1–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, … Alzheimer’s Disease Neuroimaging, Initiative. (2010). Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 6(3), 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, … Alzheimer’s Disease Neuroimaging, Initiative. (2010). Genetic variation and neuroimaging measures in Alzheimer disease. Archives of neurology, 67(6), 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Cuenco K T, Lunetta KL, Baldwin CT, McKee AC, Guo J, Cupples LA, … Mirage Study Group. (2008). Association of distinct variants in SORL1 with cerebrovascular and neurodegenerative changes related to Alzheimer disease. Archives of neurology, 65(12), 1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Furney SJ, Simmons A, Breen G, Pedroso I, Lunnon K, Proitsi P, … AddNeuroMed, Consortium. (2011). Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Molecular psychiatry, 16(11), 1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, … Alzheimer’s Disease Neuroimaging, Initiative. (2009). Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PloS one, 4(8), e6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Stein JL, Hua X, Morra JH, Lee S, Hibar DP, Ho AJ, … Alzheimer’s Disease Neuroimaging, Initiative. (2010). Genome-wide analysis reveals novel genes influencing temporal lobe structure with relevance to neurodegeneration in Alzheimer’s disease. NeuroImage, 51(2), 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Xu Z, Wu C, Pan W, & Alzheimer’s Disease Neuroimaging, Initiative. (2017). Imaging-wide association study: Integrating imaging endophenotypes in GWAS. NeuroImage, 159, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Scelsi MA, Khan RR, Lorenzi M, Christopher L, Greicius MD, Schott JM, … Altmann A (2018). Genetic study of multimodal imaging Alzheimer’s disease progression score implicates novel loci. Brain: a journal of neurology, 141(7), 2167–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Harrison TM, & Bookheimer SY (2016). Neuroimaging genetic risk for Alzheimer’s disease in preclinical individuals: From candidate genes to polygenic approaches. Biological psychiatry. Cognitive neuroscience and neuroimaging, 1(1), 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Murray ME, Graff-Radford NR, Ross OA, Petersen RC, Duara R, & Dickson DW (2011). Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: a retrospective study. Lancet neurology, 10(9), 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Beck JA, Poulter M, Campbell TA, Uphill JB, Adamson G, Geddes JF, … Tabrizi SJ (2004). Somatic and germline mosaicism in sporadic early-onset Alzheimer’s disease. Human molecular genetics, 13(12), 1219–1224. [DOI] [PubMed] [Google Scholar]
- 237.Nicolas G, Acuna-Hidalgo R, Keogh MJ, Quenez O, Steehouwer M, Lelieveld S, …Hoischen A (2018). Somatic variants in autosomal dominant genes are a rare cause of sporadic Alzheimer’s disease. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 14(12), 1632–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Bushman DM, Kaeser GE, Siddoway B, Westra JW, Rivera RR, Rehen SK, … Chun J (2015). Genomic mosaicism with increased amyloid precursor protein (APP) gene copy number in single neurons from sporadic Alzheimer’s disease brains. eLife, 4 10.7554/eLife.05116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sala Frigerio C, Lau P, Troakes C, Deramecourt V, Gele P, Van Loo P, … De Strooper B (2015). On the identification of low allele frequency mosaic mutations in the brains of Alzheimer’s disease patients. Alzheimer’s & dementia: the journal of the Alzheimer’s Association, 11(11), 1265–1276. [DOI] [PubMed] [Google Scholar]
- 240.Parcerisas A, Rubio SE, Muhaisen A, Gomez-Ramos A, Pujadas L, Puiggros M, …Soriano E (2014). Somatic signature of brain-specific single nucleotide variations in sporadic Alzheimer’s disease. Journal of Alzheimer’s disease: JAD, 42(4), 1357–1382. [DOI] [PubMed] [Google Scholar]
- 241.Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, … Faulkner GJ (2011). Somatic retrotransposition alters the genetic landscape of the human brain. Nature, 479(7374), 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sanchez-Luque FJ, Bodea GO, … Faulkner GJ (2015). Ubiquitous L1 mosaicism in hippocampal neurons. Cell, 161(2), 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Levin HL, & Moran JV (2011). Dynamic interactions between transposable elements and their hosts. Nature reviews. Genetics, 12(9), 615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Maxwell PH, Burhans WC, & Curcio MJ (2011). Retrotransposition is associated with genome instability during chronological aging. Proceedings of the National Academy of Sciences of the United States of America, 108(51), 20376–20381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Li W, Prazak L, Chatterjee N, Gruninger S, Krug L, Theodorou D, & Dubnau J (2013). Activation of transposable elements during aging and neuronal decline in Drosophila. Nature neuroscience, 16(5), 529–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yurov YB, Vorsanova SG, & Iourov IY (2011). The DNA replication stress hypothesis of Alzheimer’s disease. TheScientificWorldJournal, 11, 2602–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Li W, & Olivier M (2013). Current analysis platforms and methods for detecting copy number variation. Physiological genomics, 45(1), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Planton M, Raposo N, Albucher JF, & Pariente J (2017). Cerebral amyloid angiopathy-related cognitive impairment: The search for a specific neuropsychological pattern. Revue neurologique, 173(9), 562–565. [DOI] [PubMed] [Google Scholar]
- 249.Ellis RJ, Olichney JM, Thal LJ, Mirra SS, Morris JC, Beekly D, & Heyman A (1996). Cerebral amyloid angiopathy in the brains of patients with Alzheimer’s disease: the CERAD experience, Part XV. Neurology, 46(6), 1592–1596. [DOI] [PubMed] [Google Scholar]
- 250.Wisniewski KE, Wisniewski HM, & Wen GY (1985). Occurrence of neuropathological changes and dementia of Alzheimer’s disease in Down’s syndrome. Annals of neurology, 17(3), 278–282. [DOI] [PubMed] [Google Scholar]
- 251.Menendez M (2005). Down syndrome, Alzheimer’s disease and seizures. Brain & development, 27(4), 246–252. [DOI] [PubMed] [Google Scholar]
- 252.Hooli BV, Kovacs-Vajna ZM, Mullin K, Blumenthal MA, Mattheisen M, Zhang C, … Tanzi RE (2014). Rare autosomal copy number variations in early-onset familial Alzheimer’s disease. Molecular psychiatry, 19(6), 676–681. [DOI] [PubMed] [Google Scholar]
- 253.Brouwers N, Van Cauwenberghe C, Engelborghs S, Lambert JC, Bettens K, Le Bastard N, … an Broeckhoven C (2012). Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing C3b/C4b binding sites. Molecular psychiatry, 17(2), 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Swaminathan S, Huentelman MJ, Corneveaux JJ, Myers AJ, Faber KM, Foroud T, … Nia-Load Ncrad Family Study Group. (2012). Analysis of copy number variation in Alzheimer’s disease in a cohort of clinically characterized and neuropathologically verified individuals. PloS one, 7(12), e50640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Szigeti K, Lal D, Li Y, Doody RS, Wilhelmsen K, Yan L, … Care, Consortium. (2013). Genome-wide scan for copy number variation association with age at onset of Alzheimer’s disease. Journal of Alzheimer’s disease: JAD, 33(2), 517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Heinzen EL, Need AC, Hayden KM, Chiba-Falek O, Roses AD, Strittmatter WJ, … Goldstein DB (2010). Genome-wide scan of copy number variation in late-onset Alzheimer’s disease. Journal of Alzheimer’s disease: JAD, 19(1), 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Arias E, Xu J, & Kochanek KD (2019). United States Life Tables, 2016. National vital statistics reports: from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System, 68(4), 1–66. [PubMed] [Google Scholar]
- 258.Dumitrescu L, Mayeda ER, Sharman K, Moore AM, & Hohman TJ (2019). Sex Differences in the Genetic Architecture of Alzheimer’s Disease. Current genetic medicine reports, 7(1), 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Mielke MM, Vemuri P, & Rocca WA (2014). Clinical epidemiology of Alzheimer’s disease: assessing sex and gender differences. Clinical epidemiology, 6, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Laws KR, Irvine K, & Gale TM (2018). Sex differences in Alzheimer’s disease. Current opinion in psychiatry, 31(2), 133–139. [DOI] [PubMed] [Google Scholar]
- 261.Laws KR, Irvine K, & Gale TM (2016). Sex differences in cognitive impairment in Alzheimer’s disease. World J Psychiatry, 6(1), 54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Koran MEI, Wagener M, Hohman TJ, & Alzheimer’s Neuroimaging, Initiative. (2017). Sex differences in the association between AD biomarkers and cognitive decline. Brain imaging and behavior, 11(1), 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Dumitrescu L, Barnes LL, Thambisetty M, Beecham G, Kunkle B, Bush WS, … Hohman TJ (2019). Sex differences in the genetic predictors of Alzheimer’s pathology. Brain: a journal of neurology, 142(9), 2581–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hohman TJ, Dumitrescu L, Barnes LL, Thambisetty M, Beecham G, Kunkle B, … the Alzheimer’s Disease Neuroimaging, Initiative. (2018). Sex-Specific Association of Apolipoprotein E With Cerebrospinal Fluid Levels of Tau. JAMA neurology, 75(8), 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Deming Y, Dumitrescu L, Barnes LL, Thambisetty M, Kunkle B, Gifford KA, … Hohman TJ (2018). Sex-specific genetic predictors of Alzheimer’s disease biomarkers. Acta neuropathologica, 136(6), 857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Sebastiani P, Timofeev N, Dworkis DA, Perls TT, & Steinberg MH (2009). Genome-wide association studies and the genetic dissection of complex traits. American Journal of Hematology. 10.1002/ajh.21440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Wood AR, Tyrrell J, Beaumont R, Jones SE, Tuke MA, Ruth KS, … Weedon MN (2016). Variants in the FTO and CDKAL1 loci have recessive effects on risk of obesity and type 2 diabetes, respectively. Diabetologia, 59(6), 1214–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hohman TJ, Bush WS, Jiang L, Brown-Gentry KD, Torstenson ES, Dudek SM, … Alzheimer’s Disease Genetics, Consortium. (2016). Discovery of gene-gene interactions across multiple independent data sets of late onset Alzheimer disease from the Alzheimer Disease Genetics Consortium. Neurobiology of aging, 38, 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Eid A, Mhatre I, & Richardson JR (2019). Gene-environment interactions in Alzheimer’s disease: A potential path to precision medicine. Pharmacology & therapeutics, 199, 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Livingston G, Sommerlad A, Orgeta V, Costafreda SG, Huntley J, Ames D, … Mukadam N (2017). Dementia prevention, intervention, and care. The Lancet, 390(10113), 2673–2734. [DOI] [PubMed] [Google Scholar]
- 271.Luck T, Riedel-Heller SG, Luppa M, Wiese B, Kohler M, Jessen F, … Maier W (2014). Apolipoprotein E epsilon 4 genotype and a physically active lifestyle in late life: analysis of gene-environment interaction for the risk of dementia and Alzheimer’s disease dementia. Psychological medicine, 44(6), 1319–1329. [DOI] [PubMed] [Google Scholar]
- 272.Dunn AR, O’Connell KMS, & Kaczorowski CC (2019). Gene-by-environment interactions in Alzheimer’s disease and Parkinson’s disease. Neuroscience and biobehavioral reviews, 103, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Andrews SJ, Goate A, & Anstey KJ (n.d.). Association between alcohol consumption and Alzheimer’s disease: A Mendelian randomization Study. 10.1101/190165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Burgess S, & Thompson SG (2015). Mendelian Randomization: Methods for Using Genetic Variants in Causal Estimation. CRC Press. [Google Scholar]
- 275.DeTure MA, & Dickson DW (2019). The neuropathological diagnosis of Alzheimer’s disease. Molecular neurodegeneration, 14(1), 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Ryman DC, Acosta-Baena N, Aisen PS, Bird T, Danek A, Fox NC, … Dominantly Inherited Alzheimer Network. (2014). Symptom onset in autosomal dominant Alzheimer disease: a systematic review and meta-analysis. Neurology, 83(3), 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Petersen RC (1998). Clinical subtypes of Alzheimer’s disease. Dementia and geriatric cognitive disorders, 9 Suppl 3, 16–24. [DOI] [PubMed] [Google Scholar]
- 278.McQuade A, Coburn M, Tu CH, Hasselmann J, Davtyan H, & Blurton-Jones M (2018). Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Molecular neurodegeneration, 13(1), 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, … Blurton-Jones M (2017). iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron, 94(2), 278–293 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Park J, Wetzel I, Marriott I, Dreau D, D’Avanzo C, Kim DY, … Cho H (2018). A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nature neuroscience, 21(7), 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Chen HI, Song H, & Ming GL (2019). Applications of Human Brain Organoids to Clinical Problems. Developmental dynamics: an official publication of the American Association of Anatomists, 248(1), 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Tcw J (2019). Human iPSC application in Alzheimer’s disease and Tau-related neurodegenerative diseases. Neuroscience letters, 699, 31–40. [DOI] [PubMed] [Google Scholar]
- 283.Hasselmann J, Coburn MA, England W, Figueroa Velez DX, Kiani Shabestari S, Tu CH, … Blurton-Jones M (2019). Development of a Chimeric Model to Study and Manipulate Human Microglia In Vivo. Neuron, 103(6), 1016–1033 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Esquerda-Canals G, Montoliu-Gaya L, Guell-Bosch J, & Villegas S (2017). Mouse Models of Alzheimer’s Disease. Journal of Alzheimer’s disease: JAD, 57(4), 1171–1183. [DOI] [PubMed] [Google Scholar]
- 285.Jankowsky JL, & Zheng H (2017). Practical considerations for choosing a mouse model of Alzheimer’s disease. Molecular neurodegeneration, 12(1), 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Onos KD, Sukoff Rizzo SJ, Howell GR, & Sasner M (2016). Toward more predictive genetic mouse models of Alzheimer’s disease. Brain research bulletin, 122, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Hargis KE, & Blalock EM (2017). Transcriptional signatures of brain aging and Alzheimer’s disease: What are our rodent models telling us? Behavioural brain research, 322(Pt B), 311–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Neuner SM, Heuer SE, Huentelman MJ, O’Connell KMS, & Kaczorowski CC (2019). Harnessing Genetic Complexity to Enhance Translatability of Alzheimer’s Disease Mouse Models: A Path toward Precision Medicine. Neuron, 101(3), 399–411 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Onos KD, Uyar A, Keezer KJ, Jackson HM, Preuss C, Acklin CJ, … Howell GR (2019). Enhancing face validity of mouse models of Alzheimer’s disease with natural genetic variation. PLoS genetics, 15(5), e1008155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.