Abstract
Next-generation sequencing identified ~60 genes recurrently mutated in chronic lymphocytic leukemia (CLL). We examined the additive prognostic value of the total number of recurrently mutated CLL genes [i.e., tumor mutational load (TML)] or the individually mutated genes beyond the CLL international prognostic index (CLL-IPI) in newly diagnosed CLL and high-count monoclonal B-cell lymphocytosis (HC MBL). We sequenced 59 genes among 557 individuals (112 HC MBL/445 CLL) in a multi-stage design, to estimate hazard ratios (HR) and 95% confidence intervals (CI) for time-to-first treatment (TTT), adjusted for CLL-IPI and sex. TML was associated with shorter TTT in the discovery and validation cohorts, with a combined estimate of continuous HR=1.27 (CI:1.17–1.39, P=2.6×10−8; c-statistic=0.76). When stratified by CLL-IPI, the association of TML with TTT was stronger and validated within low/intermediate risk (combined HR=1.54, CI:1.37–1.72, P=7.0×10−14). Overall, 80% of low/intermediate CLL-IPI cases with 2+ mutated genes progressed to require therapy within 5 years, compared to 24% among those without mutations. TML was also associated with shorter TTT in the HC MBL cohort (HR=1.53, CI:1.12–2.07, P=0.007; c-statistic=0.71). TML is a strong prognostic factor for TTT independent of CLL-IPI, especially among low/intermediate CLL-IPI risk and a better predictor than any single gene. Mutational screening at early stages may improve risk stratification and better predict TTT.
INTRODUCTION
Chronic lymphocytic leukemia (CLL) is a clinically and biologically heterogeneous disease[1] characterized by at least 5×109 clonal lymphocytes/L of characteristic immune-phenotype in the peripheral blood[2]. While most newly diagnosed CLL patients experience an indolent disease course that does not require therapy for years, other patients experience progressive disease that often needs treatment shortly after diagnosis and is characterized by frequent relapses[3, 4]. CLL is always preceded by high-count monoclonal B-cell lymphocytosis (HC MBL), a clonal lymphoproliferative disorder of lymphocytes with the immunophenotype of CLL but an absolute B-cell count (ABC) between 0.5 to 5×109 lymphocytes/L in the peripheral blood in the absence of lymphadenopathy or organomegaly[2, 5, 6]. Individuals with HC MBL progress to CLL requiring therapy at a rate of ~1–5% per year, but it has been difficult to utilize known prognostic biomarkers to consistently predict who will progress[7–10].
CLL is characterized by a heterogeneous genomic landscape[11, 12]. Two large sequencing studies have identified approximately 60 recurrently mutated genes[13, 14], mostly affecting cell cycle, DNA damage response, NOTCH and NF-kB signaling pathways[11, 13–18]. Many of these recurrently mutated genes are already mutated in individuals with HC MBL[13, 19–21] but tend to occur with higher frequency in later stages of CLL, supporting their potential role in disease progression[22–26]. Prior studies have shown the impact of mutations in TP53, NOTCH1, and SF3B1 with progression from HC MBL to CLL[27, 28] and their negative impact on CLL outcome[13, 14, 29–37].
In 2016, a novel CLL International Prognostic Index (CLL-IPI) for overall survival (OS) was developed and validated[38], stratifying CLL patients into 4 risk groups (c-statistic=0.72, 95% confidence interval [CI]: 0.68–0.75) with the following 5-year OS: low risk 93% (CI: 90.5–96.0), intermediate risk 79% (CI: 75.5–83.2), high risk 63% (CI: 57.9–68.8), and very-high risk 23% (CI: 12.5–34.1)[38]. CLL-IPI was also found to predict time-to-first treatment (TTT) in early stage CLL cases (c-statistic=0.72, CI: 0.58–0.81)[39, 40]. It is unknown, however, whether the association between individual recurrently mutated CLL genes (other than TP53) are prognostic for CLL outcome independent of the CLL-IPI. Most sequencing studies that examined individual recurrently mutated genes or total number of mutations either did not include any known CLL prognostic factors in the analyses[14, 26, 32] or did not include all of the components found in CLL-IPI[13, 22, 23, 27, 28, 30, 31, 41–43]. A recent study examined the total number of mutated genes and pathways with TTT adjusting for CLL-IPI[44].
Here, we examined whether individual recurrently mutated genes or aggregate tumor mutational load (TML) (i.e., the total number of recurrently mutated CLL genes) provide prognostic value independent of the well-validated CLL-IPI[38] for time from diagnosis to progression requiring therapy and OS using three CLL cohorts (i.e. discovery and two validation cohorts). We also evaluated, for the first time, the prognostic performance of individual mutated genes, the TML, and CLL-IPI in a cohort of HC MBL individuals.
METHODS
Patients
Based on the 2008 International Workshop CLL (iwCLL) criteria[2], we selected newly diagnosed and treatment naïve CLL and HC MBL individuals whose samples were collected between 2002–2016 from the Mayo Clinic CLL Resource. All individuals had available pre-treatment peripheral blood mononuclear cells (PBMC) collected within 2 years of CLL or HC MBL diagnosis. All individuals provided written informed consent for this research whose protocol was approved by the Mayo Clinic institutional review board.
For this study, we utilized a three-stage study design of CLL cases to minimize false positive findings and improve reproducibility. The three CLL cohorts were formed based on timing of sequencing, with the discovery cohort defined as the first batch of samples sequenced in fall of 2018 and the two validation cohorts as the second and third batch of samples sequenced in spring (i.e., validation cohort 1) and winter (i.e., validation cohort 2) of 2019, respectively. The HC MBL samples were sequenced with the first batch of CLL samples.
DNA sequencing
DNA was extracted from either PBMCs that had a tumor purity >80% or otherwise sorted CD5+/CD19+ clonal cells. We sequenced the entire coding regions of 59 somatically recurring mutated CLL genes using a customized SureSelect panel (Table S1). Samples were paired-end sequenced (150 bp reads), using Illumina HiSeq 4000 sequencer with 24 samples per lane of flow cell. The median coverage depth per sample across the 59 recurrently mutated genes was 2,061 (range 73–5,836) with >98% of the samples having a median coverage depth >1000X per nucleotide, allowing the detection of mutations with variant allelic fraction (VAF) as low as 1%. Somatic mutations were called using MuTect2 in tumor-only mode. After filtering, high impact mutations (frameshift, nonsense, and splicing variants) and missense mutations in previously identified CLL hot spots were used for statistical analyses (Table S2). In secondary analyses we also included all missense mutations that were not considered hot spots or high impact variants. Finally, all TP53 variants were validated as somatic mutations by interrogating the TP53 IARC Database. Additional information can be found in Supplementary Methods.
Statistical analysis
CLL-IPI score was calculated based on clinical stage (Rai 0 vs. Rai I-IV), IGHV-mutation status (mutated vs. unmutated), TP53 status (wild type vs. del17p, TP53 mutations, or both), beta-2 microglobulin level (>3.5 mg/L), and age (>65 years) as previously described[38]. Since HC MBL is not clinically stage, we indicated them as Rai stage 0 when calculating the CLL-IPI score.
TTT was defined as the time from date of sample to date of first treatment or date of last follow-up. We used the guidelines to determine therapy initiation as outlined by the 1996 National Cancer Institute Working Group [45] and reaffirmed in the 2008 International Workshop on CLL [2]. Types of first-line CLL treatments are described in Table S3. OS was defined as time from date of sample to date of death or last follow-up. We used Cox regression models to estimate hazard ratios (HRs) and 95% CIs for TTT and OS associations. The event in the TTT analyses was those individuals who received treatment at the time of treatment otherwise individuals were censored at last follow up. Similarly, the event in OS was those who died otherwise individuals were censored at date of last follow up. TTT models were adjusted for CLL-IPI and sex, while OS models were adjusted for CLL-IPI, sex, and any CLL treatment (time dependent variable). In sensitivity analyses we also evaluated batch effect by adding the batch number as a covariate to the combined cohort analysis. Survival curves were displayed using the Kaplan-Meier method using P-values from the log-rank test.
For the TML analyses, we generated a TML score by counting the number of recurrently mutated CLL genes across all 59 genes, but excluding TP53, which is used in calculating the CLL-IPI. We then categorized the TML either as binary (none or any mutated gene) or as a categorical variable (0, 1, or 2+ mutated genes per patient, with 0 mutated genes serving as the reference category). We also considered TML as a continuous variable. The TML score was evaluated with TTT or OS in the discovery and two validation CLL cohorts separately as well as the three CLL cohorts combined.
For the individual gene analyses, we evaluated TTT or OS in the discovery and two validation CLL cohorts separately as well as the three CLL cohorts combined for only those recurrent genes (including TP53) that were found to be mutated in more than 15 individuals in the combined CLL cohort (Table S1). Furthermore, in the HC MBL cohort, we evaluated the prognostic implications of TML as well as single genes for those genes with at least 5 individuals with mutations.
Finally, Rossi et al. integrated mutational profiling of 4 genes (TP53, BIRC3, NOTCH1 and SF3B1) and FISH-detected cytogenetic abnormalities together and identified four subgroups with different treatment free survival[36]. We applied these risk groups on our combined CLL cohorts with TTT and OS (Supplementary Methods).
To evaluate model discriminative ability, we computed a c-statistic and 95% CI[46] for the adjusted Cox regression models (Supplementary Methods). Among the CLL analyses, significant findings were those that had a P<0.05 in both the discovery and validation cohorts with HRs in the same direction across the three cohorts. Among the HC MBL analyses, significant results were reported for those with P<0.05.
RESULTS
Baseline characteristics
Among 557 study participants, 445 had CLL (152 in the discovery cohort and 175 and 118 in the two validation cohorts), and 112 had HC-MBL. Collectively, 73% of CLL and 65% of HC MBL were male; median age at diagnosis was 61 years (range 28–87) for CLL and 66 (range 43–87) for HC-MBL. Overall, 301 CLL (68%) and 58 HC MBL (52%) individuals had high-impact mutations (Table S4). We observed fewer mutated genes in HC MBL than CLL (P=0.003; Table S4).
Additional clinical characteristics are described in Table 1. A total of 234 individuals progressed requiring therapy [214 CLL (median follow-up of 1.5 years, range:0–17.3), and 20 HC-MBL (4.7 years, range:0–14.9)]; and 128 individuals died [116 CLL (5.7 years, range:0–17.3), and 12 HC MBL (7.1 years, range:1.0–14.9)].
Table 1:
Clinical characteristics for CLL and HC MBL by cohorts and overall
| Cohorts | CLL discovery cohort | CLL validation Cohort 1 | CLL Validation Cohort 2 | CLL overall | HC MBL | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (N=152) | (N=175) | (N=118) | (N=445) | (N=112) | |||||||
| Clinical characteristic | N | % | N | % | N | % | N | % | N | % | |
| Gender | female | 48 | 31.6% | 41 | 23.4% | 32 | 27.1% | 121 | 27.2% | 39 | 34.8% |
| male | 104 | 68.4% | 134 | 76.6% | 86 | 72.9% | 324 | 72.8% | 73 | 65.2% | |
| Age | Median (range) | 62 | (28–87) | 60 | (31–86) | 62 | (36–87) | 61 | (28–87) | 66 | (43–87) |
| Rai stage risk group | Rai 0 | 111 | 73.0% | 75 | 43.1% | 69 | 59.0% | 255 | 57.5% | - | - |
| Rai I–II | 35 | 23.0% | 80 | 46.0% | 39 | 33.3% | 154 | 34.8% | - | - | |
| Rai III–IV | 6 | 4.0% | 19 | 10.9% | 9 | 7.7% | 34 | 7.7% | - | - | |
| Missing | 1 | 1 | 2 | - | - | ||||||
| FISH* | 13q deletion | 82 | 56.2% | 67 | 39.4% | 53 | 48.7% | 202 | 47.5% | 58 | 51.8% |
| Trisomy 12 | 14 | 9.6% | 30 | 17.6% | 14 | 12.8% | 58 | 13.6% | 11 | 9.8% | |
| 11q deletion | 11 | 7.5% | 30 | 17.6% | 10 | 9.2% | 51 | 12.0% | 9 | 8.0% | |
| 17p deletion | 6 | 4.1% | 12 | 7.1% | 8 | 7.3% | 26 | 6.1% | 4 | 3.6% | |
| Normal | 33 | 22.6% | 31 | 18.2% | 24 | 22.0% | 88 | 20.7% | 30 | 26.8% | |
| Missing | 6 | 5 | 9 | 20 | |||||||
| TP53 | Mutated | 12 | 7.9% | 22 | 7.8% | 13 | 11.0% | 43 | 9.7% | 7 | 6.2% |
| Unmutated | 140 | 92.1% | 261 | 92.2% | 105 | 89.0% | 402 | 90.3% | 105 | 93.8% | |
| IGHV | Mutated | 99 | 65.1% | 61 | 35.7% | 58 | 51.3% | 218 | 50.0% | 85 | 75.9% |
| Unmutated | 53 | 34.9% | 110 | 64.3% | 55 | 48.7% | 218 | 50.0% | 27 | 24.1% | |
| mutation status | Missing | 4 | 5 | 9 | |||||||
| Beta-2 microglobulin | ≤3.5 mg/L | 127 | 83.6% | 123 | 70.3% | 91 | 78.4% | 341 | 77.0% | 99 | 88.4% |
| >3.5 mg/L | 25 | 16.4% | 52 | 29.7% | 25 | 21.6% | 102 | 23.0% | 13 | 11.6% | |
| Missing | 2 | 2 | |||||||||
| CLL-IPI** | Low risk (0–1) | 77 | 50.8% | 47 | 27.0% | 39 | 36.8% | 163 | 37.7% | 68 | 60.7% |
| Intermediate risk (2–3) | 44 | 28.9% | 62 | 35.6% | 39 | 36.8% | 145 | 33.6% | 30 | 26.8% | |
| High risk (4–6) | 25 | 16.4% | 48 | 27.6% | 20 | 18.9% | 93 | 21.5% | 14 | 12.5% | |
| Very high risk (7–10) | 6 | 3.9% | 17 | 9.8% | 8 | 7.5% | 31 | 7.2% | 0 | 0.0% | |
| Missing | 1 | 12 | 13 | ||||||||
| B cell count × 109/L | Median (range) | 11.6 | (2.2–145) | 24.9 | (5.2–235) | 10.9 | (3.4–395) | 15.1 | (2.2–395) | 3.2 | (0.6–5.0) |
| Missing | 5 | 28 | 25 | 58 | 4 | ||||||
| ALC × 109/L | Median (range) | 16.6 | (3.4–183) | 35.2 | (1.9–317) | 17.3 | (2.6–386) | 23.5 | (1.9–386) | 6.12 | (2.35–12.6) |
| Missing | 1 | 0 | 1 | 2 | 0 | ||||||
CLL, chronic lymphocytic leukemia; HC MBL, high-count monoclonal B-cell lymphocytosis; CLL-IPI, CLL international prognostic index; ALC, absolute lymphocyte count
cases with >1 FISH abnormality were counted for the worse abnormality.
HC MBL were scored as 0 points for Rai stage.
Impact of CLL-IPI on TTT and OS
The CLL-IPI (continuous per category increase) was associated with shorter TTT (HR=2.25, CI:1.96–2.57, P=4.5×10−32) and OS (HR=2.28, CI:1.86–2.80, P=2.5×10−15) in the combined cohort of 445 CLL patients. The CLL-IPI model had a c-statistic of 0.74 (CI: 0.71–0.77) to predict TTT. Among the 112 HC-MBL, CLL-IPI was associated with a shorter TTT (HR=1.97, CI:1.14–3.41, P=0.016) but not OS (HR=1.49, CI:0.75–2.94, P=0.26).
Impact of TML on TTT and OS
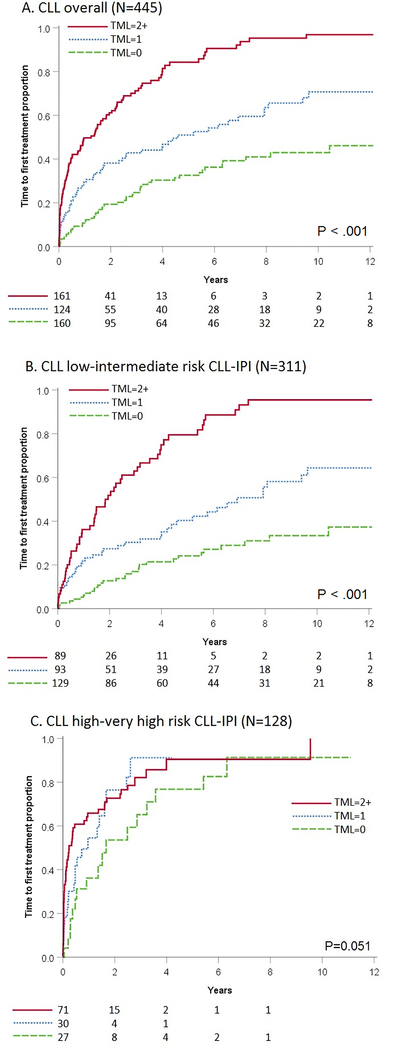
TML (as a continuous variable), was associated with a shorter TTT among CLL patients, after adjusting for CLL-IPI and sex in the discovery cohort (HR=1.41, CI:1.16–1.72, P=1.6×10−5) and subsequently confirmed in the validation cohorts (Table S5). In the full 445 CLL cohort, the TML modeled by itself had a c-statistic of 0.67 (CI:0.64–0.71) to predict TTT. When TML and CLL-IPI were modeled simultaneously (along with sex) for TTT, the c-statistic increased to 0.76 (CI: 0.73–0.79, Table 2). Overall, individuals with 2+ mutated genes showed a shorter TTT (HR=3.03, CI:2.10–4.38, P=2.9×10−9) compared to those without mutations (Table 2, Figure 1A). We did not observe any batch effect in the combined CLL cohort analysis (Table S6). Next, we evaluated the effect of missense mutations that were not part of the CLL described hot spots in the TML calculation and the results were consistent with or without them in the TML (Table S7). TML was associated with worse OS in the CLL discovery cohort, (HR=1.28, CI:1.02–1.61, P=0.03, Table S8), but not in the validation cohorts.
Table 2:
Association between the tumor mutational load^ and time to first treatment – CLL overall and by CLL-IPI
| CLL overall (N=445) | |||||||
|---|---|---|---|---|---|---|---|
| Total | Event | ||||||
| CLL-IPI | Mutated genes | N | % | N | HR | 95% CI | P |
| Overall* | None | 160 | 36 | 48 | 1 | ref | |
| Any | 285 | 64 | 166 | 2.45 | (1.76-3.420 | 1.4×10−7 | |
| 0 | 160 | 36 | 48 | 1 | ref | ||
| 1 | 124 | 27.9 | 60 | 1.94 | (1.31-2.85) | 1.0×10−3 | |
| 2+ | 161 | 36.1 | 106 | 3.03 | (2.10-4.38) | 2.9×10−9 | |
| Cont. | 445 | 214 | 1.27 | (1.17-1.39) | 2.6×10−8 | ||
| c-statistic | 0.76 | (0.73-0.79) | |||||
| Low-intermediate risk CLL-IPI** | None | 129 | 41.5 | 30 | 1 | ref | |
| Any | 182 | 58.5 | 94 | 3.32 | (2.20-5.02) | 1.2×10−8 | |
| 0 | 129 | 41.5 | 30 | 1 | ref | ||
| 1 | 93 | 29.9 | 40 | 2.14 | (1.33-3.44) | 2.0×10−3 | |
| 2+ | 89 | 28.6 | 54 | 5.93 | (3.75-9.36) | 2.4×10−14 | |
| Cont. | 311 | 124 | 1.54 | (1.37-1.72) | 7.0×10−14 | ||
| c-statistic | 0.69 | (0.64-0.73) | |||||
| High-very high risk CLL-IPI** | None | 27 | 21.1 | 17 | 1 | ref | |
| Any | 101 | 78.9 | 72 | 1.92 | (1.11-3.29) | 0.02 | |
| 0 | 27 | 21.1 | 17 | 1 | ref | ||
| 1 | 30 | 23.4 | 20 | 1.67 | (0.86-3.25) | 0.13 | |
| 2+ | 71 | 55.5 | 52 | 2.02 | (1.15-3.53) | 0.01 | |
| Cont. | 128 | 89 | 1.17 | (1.03-1.32) | 0.02 | ||
| c-statistic | 0.60 | (0.54-0.67) | |||||
CLL, chronic lymphocytic leukemia; CLL-IPI, CLL international prognostic index; HR, hazard ratio; CI, confidence interval; Cont., continuous
using 58 genes without counting mutations in TP53
Adjusted for CLL-IPI and sex
Adjusted for sex
Figure 1: Time to first treatment by tumor mutational load – CLL overall and stratified by CLL-IPI.
Kaplan-Meier survival curves with time to first treatment (TTT) by TML of 0 recurrently mutated genes (dashed green), 1 recurrently mutated gene (dotted blue), and 2+ recurrently mutated genes (solid red). (A) TTT Overall with 445 cases; (B) TTT Low to intermediate risk CLL-IPI group with 311 cases; (C) High to very-high risk CLL-IPI group with 128 cases.
CLL, chronic lymphocytic leukemia; CLL-IPI, CLL international prognostic index; TML, tumor mutational load
Similarly to CLL, in HC-MBL TML was associated with shorter TTT after adjusting for CLL-IPI and sex (HR=1.53, CI:1.12–2.07, P=0.007), but not with OS (Table S9).
Incremental effect of TML according to the CLL-IPI risk score
Next, we evaluated the association between TTT and TML stratified by CLL-IPI, with similar findings across the discovery and two validation CLL cohorts (Table S5). In CLL patients with low to intermediate risk CLL-IPI, we found that the TML was associated with shorter TTT, with an overall HR=1.54 (CI:1.37–1.72, P=7.0×10−14, Table 2). CLL patients with 2+ mutated genes had a 5.9-fold increased risk of progression requiring therapy compared to those with no mutations (Table 2). Among CLL patients with low to intermediate risk CLL-IPI, the 5-year risk of needing therapy was 80% for those with 2+ mutated genes, compared to 40% and 24% among those with 1 and 0 mutated genes, respectively (Figure 1B). No association was found between TTT with TML in high and very-high risk CLL-IPI (Table S5, Figure 1C). Similar patterns were observed when evaluating associations separately within each of the four CLL-IPI risk levels (Figure S1). When we evaluated the association between OS and TML stratified by CLL-IPI, we did not find a significant association at the 0.05 level (Table S8).
Among HC-MBL, TML was associated with shorter TTT in individuals with low to intermediate risk CLL-IPI (HR=2.18, CI:1.19–4.01, P=0.01), but not for high to very-high risk CLL-IPI (Table 3). No association was found between TML and OS stratified by CLL-IPI among the HC-MBL (Table S9).
Table 3:
Association between the tumor mutational load^ and time to first treatment –HC MBL
| HC MBL (N=112) | |||||||
|---|---|---|---|---|---|---|---|
| Total | Event | ||||||
| CLL-IPI | Mutated genes | N | col % | N | HR | 95% CI | P |
| Overall* | None | 59 | 52.7 | 5 | 1 | reference | |
| Any | 53 | 47.3 | 15 | 2.88 | (1.01–8.21) | 0.048 | |
| 0 | 59 | 52.6 | 5 | 1 | reference | ||
| 1 | 35 | 31.3 | 7 | 2.07 | (0.63–6.86) | 0.23 | |
| 2+ | 18 | 16.1 | 8 | 4.09 | (1.29–13.0) | 0.017 | |
| Cont. | 112 | 20 | 1.53 | (1.12–2.07) | 0.007 | ||
| c-statistic | 0.71 | (0.59–0.83) | |||||
| Low-intermediate risk CLL-IPI** | None | 51 | 52 | 3 | 1 | reference | |
| Any | 47 | 48 | 13 | 2.74 | (0.77–9.81) | 0.12 | |
| 0 | 51 | 52 | 3 | 1 | reference | ||
| 1 | 33 | 33.7 | 6 | 1.93 | (0.48–7.76) | 0.36 | |
| 2+ | 14 | 14.3 | 7 | 5.21 | (1.20–22.6) | 0.028 | |
| Cont. | 98 | 16 | 2.18 | (1.19–4.01) | 0.01 | ||
| c-statistic | 0.70 | (0.57–0.83) | |||||
| High-very high risk CLL-IPI** | None | 8 | 57.1 | 2 | 1 | reference | |
| Any | 6 | 42.9 | 2 | 2.18 | (0.29–16.6) | 0.45 | |
| 0 | 8 | 57.1 | 2 | 1 | reference | ||
| 1 | 2 | 14.3 | 1 | 4.3 | (0.24–75.8) | 0.32 | |
| 2+ | 4 | 28.6 | 1 | 1.41 | (0.11–17.8) | 0.79 | |
| Cont. | 14 | 4 | 1.13 | (0.71–1.81) | 0.60 | ||
| c-statistic | 0.61 | (0.26–0.95) | |||||
HC MBL, monoclonal B-cell lymphocytosis; CLL-IP, CLL international prognostic index; HR, hazard ratio; CI, confidence interval; Cont., continuous
using 58 genes without counting mutations in TP53
Adjusted for CLL-IPI and sex
Adjusted for sex;
Individual gene mutations and impact on TTT and OS
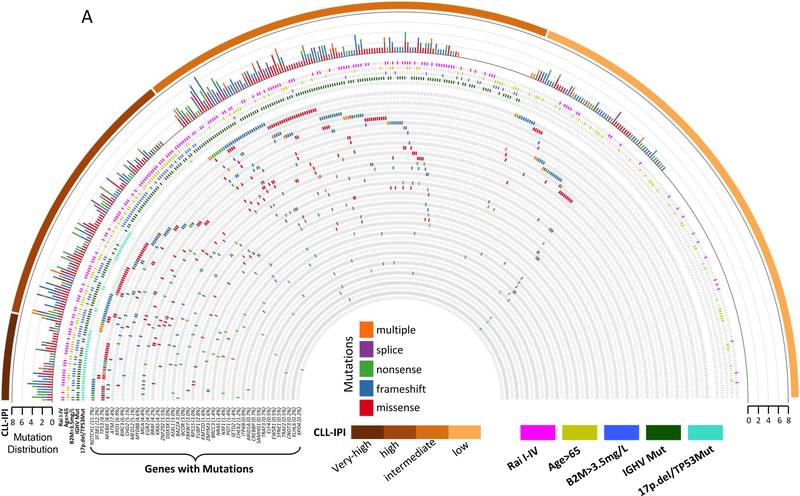
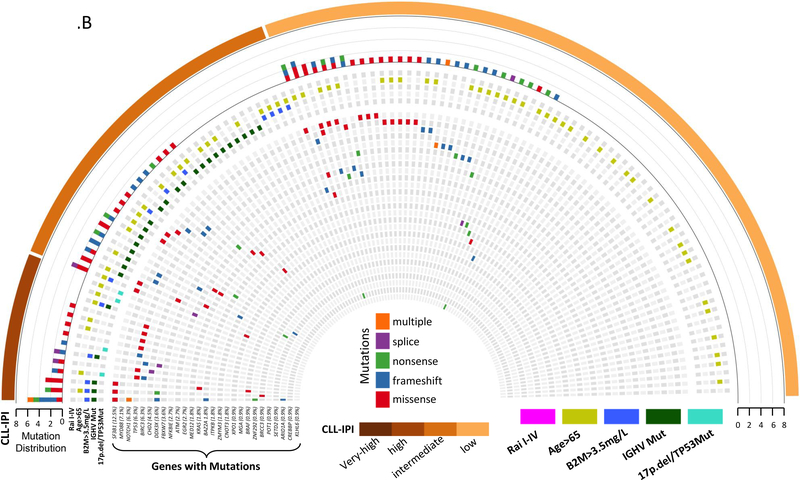
The most commonly mutated genes were NOTCH1 (16%), SF3B1 (13%), TP53 (10%), NFKBIE (9%), ATM (8%), XPO1 (7%), and BIRC3 (7%) in CLL, and SF3B1 (12%) and MYD88 (8%) in HC-MBL (Figure 2.A–B, Table S1). For the single gene association analyses among CLL, we analyzed the 14 genes (including TP53) that had more than 15 individuals with mutations. In the discovery cohort, we found shorter TTT for NOTCH1, SF3B1, and ATM mutations (Table S10). After adjusting for CLL-IPI category and sex, mutations in SF3B1 remained significant, which was confirmed in validation cohort 1 but not in validation cohort 2 (Table S10). In the combined CLL cohort, SF3B1 was associated with shorter TTT (HR=2.05; CI:1.42–2.96, P=4.3×10−4, c-statistic=0.75, Table 4) after adjusting for CLL-IPI and sex.
Figure 2: Heat map of recurrently mutated CLL genes by CLL clinical characteristics for CLL and HC MBL.
(A) Heat map representing recurrently mutated CLL genes that were found to be mutated in 432 newly diagnosed CLL patients with available CLL-IPI score only. The heat map is stratified by CLL-IPI as indicated in the external bar. The top histogram represents the number of mutated genes per patient, while the type of mutation is color-coded for missense (red), frame shift (blue), nonsense (green), splice (purple), or multiple mutations (orange). Mutated genes are indicated regardless of the number of mutations in each gene. The five CLL-IPI components are below the histogram row and color-coded by Rai stage I-IV (pink), age>65 (light green), B2M>3.5mg/L (blue), IGHV-unmutated (green), FISH (del17p or del11q) or TP53 (light blue).
(B) Heat map representing recurrently mutated CLL genes that were found to be mutated in 112 newly diagnosed HC MBL individuals. The heat map is stratified by CLL-IPI as indicated in the external bar. The top histogram represents the number of mutated genes per individual, while the type of mutation is color-coded for missense (red), frame shift (blue), nonsense (green), splice (purple), or multiple mutations (orange). Mutated genes are indicated regardless of the number of mutations in each gene. The five CLL-IPI components are below the histogram row and color-coded by, age>65 (light green), B2M>3.5mg/L (blue), IGHV-unmutated (green), FISH (del17p or del11q) or TP53 (light blue), since HC MBL is not clinically staged there are no Rai stage I-IV.
CLL, chronic lymphocytic leukemia; HC MBL, high-count monoclonal B-cell lymphocytosis; CLL-IPI, CLL international prognostic index; IGHV, immunoglobulin heavy-chain variable region; B2M, Beta-2 microglobulin
Table 4:
Association between recurrently mutated CLL genes and time to first treatment – CLL overall and stratified by CLL-IPI
| CLL combined (N=445) | Low-intermediate risk (N=311) | High-very high risk (N=128) | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Event | Unadjusted | Multivariate | Total | Event | Multivariate | Total | Event | Multivariate | ||||||||||||
| Gene | N | % | N | HR | 95% CI | P | HR* | 95% CI | P | N | % | N | HR** | 95% CI | P | N | % | N | HR** | 95% CI | P |
| NOTCH1 | 69 | 15.5% | 40 | 2.04 | (1.44–2.89) | 5.8×10−5 | 1.40 | (0.98–2.00) | 0.06 | 41 | 13.2% | 20 | 2.35 | (1.45–3.82) | 1.0×10−3 | 28 | 21.9% | 20 | 1.31 | (0.79–2.17) | 0.30 |
| c-statistic | 0.55 | (0.52–0.57) | 0.75 | (0.72–0.78) | |||||||||||||||||
| SF3B1 | 56 | 12.6% | 36 | 2.35 | (1.63–3.38) | 4.0×10−6 | 2.05 | (1.42–2.96) | 1.4×10−4 | 35 | 11.3% | 22 | 3.38 | (2.09–5.45) | 6.2×10−7 | 20 | 15.6% | 14 | 1.49 | (0.83–2.67) | 0.18 |
| c-statistic | 0.55 | (0.52–0.57) | 0.75 | (0.72–0.78) | |||||||||||||||||
| TP53 | 43 | 9.7% | 28 | 1.84 | (1.23–2.74) | 3.0×10−3 | 1.74§ | (1.16–2.61) | 7.0×10−3 | ||||||||||||
| c-statistic | 0.52 | (0.50–0.55) | 0.58 | (0.54–0.62) | |||||||||||||||||
| NFKBIE | 39 | 8.8% | 25 | 2.14 | (1.41–3.24) | 3.9×10−4 | 1.52 | (1.00–2.31) | 0.053 | 21 | 6.8% | 12 | 1.80 | (0.99–3.27) | 0.056 | 18 | 14.1% | 13 | 1.95 | (1.07–3.55) | 0.03 |
| c-statistic | 0.53 | (0.51–0.55) | 0.74 | (0.71–0.78) | |||||||||||||||||
| ATM | 35 | 7.9% | 23 | 2.02 | (1.30–3.13) | 2.0×10−3 | 1.66 | (1.06–2.59) | 0.03 | 19 | 6.1% | 13 | 2.97 | (1.65–5.36) | 2.9×10−4 | 15 | 11.7% | 10 | 0.88 | (0.45–1.70) | 0.70 |
| c-statistic | 0.52 | (0.50–0.54) | 0.75 | (0.71–0.78) | |||||||||||||||||
| XPO1 | 31 | 7.0% | 23 | 2.11 | (1.37–3.25) | 1.0×10−3 | 1.53 | (0.99–2.37) | 0.055 | 20 | 6.4% | 14 | 2.26 | (1.29–3.95) | 4.0×10−3 | 11 | 8.6% | 9 | 1.73 | (0.84–3.56) | 0.13 |
| c-statistic | 0.53 | (0.51–0.55) | 0.75 | (0.71–0.78) | |||||||||||||||||
| BIRC3 | 30 | 6.7% | 20 | 2.06 | (1.30–3.27) | 2.0×10−3 | 1.89 | (1.19–3.00) | 7.0×10−3 | 21 | 6.8% | 14 | 2.44 | (1.39–4.28) | 2.0×10−3 | 9 | 7.0% | 6 | 1.33 | (0.58–3.06) | 0.50 |
| c-statistic | 0.53 | (0.50–0.55) | 0.75 | (0.72–0.78) | |||||||||||||||||
| CHD2 | 22 | 4.9% | 7 | 0.51 | (0.24–1.09) | 0.08 | 0.86 | (0.40–1.85) | 0.70 | 19 | 6.1% | 6 | 0.66 | (0.29–1.49) | 0.31 | 3 | 2.3% | 1 | 1.22 | (0.17–8.85) | 0.84 |
| c-statistic | 0.52 | (0.50–0.53) | 0.75 | (0.72–0.78) | |||||||||||||||||
| MED12 | 22 | 4.9% | 14 | 1.98 | (1.15–3.40) | 0.01 | 1.64 | (0.95–2.85) | 0.08 | 11 | 3.5% | 4 | 1.75 | (0.63–4.82) | 0.28 | 11 | 8.6% | 10 | 1.41 | (0.72–2.76) | 0.32 |
| c-statistic | 0.52 | (0.50–0.54) | 0.75 | (0.71–0.78) | |||||||||||||||||
| MYD88 | 20 | 4.5% | 10 | 0.92 | (0.49–1.74) | 0.81 | 1.09 | (0.58–2.06) | 0.79 | 13 | 4.2% | 6 | 1.02 | (0.45–2.31) | 0.97 | 7 | 5.5% | 4 | 0.74 | (0.27–2.02) | 0.55 |
| c-statistic | 0.50 | (0.49–0.52) | 0.75 | (0.72–0.78) | |||||||||||||||||
| MGA | 19 | 4.3% | 14 | 2.53 | (1.47–4.35) | 1.0×10−3 | 1.48 | (0.85–2.59) | 0.17 | 5 | 1.6% | 2 | 1.14 | (0.36–3.59) | 0.83 | 14 | 10.9% | 9 | 2.00 | (1.04–3.85) | 0.04 |
| c-statistic | 0.52 | (0.50–0.54) | 0.75 | (0.71–0.78) | |||||||||||||||||
| EGR2 | 18 | 4.0% | 13 | 2.69 | (1.53–4.73) | 1.0×10−3 | 1.81 | (1.03–3.18) | 0.04 | 12 | 3.9% | 8 | 3.54 | (1.71–7.30) | 1.0×10−3 | 6 | 4.7% | 5 | 1.26 | (0.51–3.14) | 0.61 |
| c-statistic | 0.52 | (0.50–0.53) | 0.75 | (0.72–0.78) | |||||||||||||||||
| KRAS | 18 | 4.0% | 14 | 2.76 | (1.60–4.77) | 2.6×10−4 | 2.66 | (1.54–4.61) | 4.7×10−4 | 12 | 3.9% | 9 | 3.13 | (1.58–6.19) | 1.0×10−3 | 6 | 4.7% | 5 | 2.73 | (1.08–6.89) | 0.03 |
| c-statistic | 0.52 | (0.51–0.54) | 0.75 | (0.72–0.78) | |||||||||||||||||
| BRAF | 18 | 4.0% | 13 | 3.02 | (1.71–5.31) | 1.3×10−4 | 2.24 | (1.27–3.95) | 5.0×10−3 | 9 | 2.9% | 5 | 2.62 | (1.06–6.44) | 0.04 | 9 | 7.0% | 8 | 2.64 | (1.27–5.52) | 0.01 |
| c-statistic | 0.52 | (0.51–0.54) | 0.75 | (0.72–0.78) | |||||||||||||||||
Adj for CLL-IPI, and sex
Adjusted for sex
Adj age, and sex
CLL, chronic lymphocytic leukemia; CLL-IPI, CLL international prognostic index; HR, hazard ratio; CI, confidence interval
When patients were stratified by CLL-IPI status, 8 genes (excluding TP53 since is part of CLL-IPI) remained significantly associated with a shorter TTT in multivariate analyses in the low to intermediate risk CLL-IPI group, with greater effect sizes when compared to the overall CLL cohort (Table 4). In addition, 4 genes were associated with a shorter TTT in the high to very-high risk CLL-IPI group (Table 4).
When evaluating the associations of the 13 individual genes with OS, we found inconsistent evidence (Table S11–12). In stratified analyses by CLL-IPI status, only BRAF was associated in the low to intermediate risk CLL-IPI group (HR=3.05, CI:1.38–6.75, P=0.006), and MGA in the high-very high risk CLL-IPI group (HR=2.50, CI:1.08–5.79, P=0.03; Table S12).
For the single gene association analyses among HC MBL, we observed no evidence of associations for TTT or OS (Table S13).
Applying risk groups
Finally, we applied the risk groups from Rossi et al.[36], predicting both, TTT (HR=1.68, CI:1.49–1.89, P=3.9×10−17) with a c-statistic of 0.67 (CI:0.64–0.71), and OS (HR=1.46, CI:1.23–1.73, P=1.6×10−5) with a c-statistic of 0.62 (CI:0.56–0.77) (Figure S2). However, when we adjusted for other clinical characteristics (age, sex, Rai stage, IGHV mutational status, and Beta-2 microglobulin) these associations attenuated for TTT (HR=1.22, CI:1.06–1.41, P=0.005) and were not significant for OS (HR=1.09, CI:0.90–1.33, P=0.40).
DISCUSSION
The vast majority of newly diagnosed CLL patients (~75%) have early stage disease, and do not meet the iwCLL indications for therapy. Current recommendations are to follow these patients in the clinic every 6–12 months to look for evidence of disease progression (“watch and wait” strategy)[47]. Although this approach is evidence based, it can lead to significant anxiety and distress in a significant number of patients[48]. Since the first description of Rai and Binet staging systems more than 4 decades ago, several prognostic markers have been developed to counsel patients – not only to predict OS, but more importantly time to initial therapy in newly diagnosed CLL patients.
The CLL-IPI, the most widely used prognostic tool to predict time to initial therapy, has a c-statistic of 0.72[38]. Although this is clinically relevant (a c-statistic of >0.7 is considered helpful at an individual patient level) and represents a significant advance, efforts are underway to improve its prognostic ability. With the advent of next generation sequencing, and the identification of recurrently mutated genes in CLL, our study in addition to several other groups have evaluated the utility of individual gene mutations or cumulative number of gene mutations in predicting TTT.
In the present analysis, we demonstrated and validated in three independent CLL cohorts that the total number of genes with high impact or hotspot mutations provides important prognostic information in newly diagnosed CLL patients, independently of CLL-IPI[38]. Both, the presence and the total number of mutated genes identified those individuals who were more likely to progress requiring therapy. Based on our study, those individuals within the low to intermediate CLL-IPI category who had TML of 2+ had a ~6-fold increased risk of progression requiring therapy. Interestingly, the incorporation of TML, in addition to other important prognostic factors, improved the discriminatory power of the CLL-IPI in this cohort with c-statistic of 0.76 for CLL, compared to the CLL-IPI alone (c=0.74 in our CLL data versus c=0.72 in a previously published paper)[39] or Rossi et al integrated subgroups applied on our data (c=0.67). Our results evaluating the ability of TML to provide prognostic value independent of the CLL-IPI also complement recent work assessing the cumulative number of mutations in 314 CLL patients and found an association with shorter TTT[44]. In the context of stratification systems previous to CLL-IPI, Puente et al.[13] assessed the cumulative number of recurrently mutated CLL genes per case (based on the presence of mutations in 29 genes) and found a progressively worse effect on TTT, after adjusting for IGHV status and Binet stage[13]; and Nadeu et al.[42] (based on 28 genes) adjusted for Binet stage, age, and IGHV mutation status. Furthermore, we replicated the findings of Rossi et al. [36] regarding an integrated mutational and cytogenetic risk model and TTT or OS; however, the TML was a better prognostic factor in our cohort. As new and more effective targeted therapies emerge for CLL, there is a renewed interest in consideration of early intervention studies. The TML may be used to identify patients predicted to have a shorter TTT for such early intervention studies.
For the first time, our study also demonstrates that the CLL-IPI score is associated with TTT among individuals with HC-MBL. Moreover, we found that the TML was also associated with TTT among individuals with HC MBL, after controlling for CLL-IPI category and sex. Small clonal B-cell populations are present in the peripheral blood of 3–5% of all U.S. adults over the age of 40. Although many individuals with these small clones, defined as low-count MBL, never come to clinical attention, those with HC-MBL (making up ~2% of MBLs) have been shown to progress to CLL requiring therapy about 1–5%/year. The conundrum is how to identify HC MBLs who remain asymptomatic versus individuals that will progress to active leukemia. Our results provide, for the first time, evidence that both CLL-IPI and TML are useful prognostic factors for TTT in the pre-malignant, HC MBL phase. Studies are needed to evaluate the prognostic ability of TML in individuals with low-count MBL.
Since CLL is a genetically heterogeneous disease, characterized by the presence of ~60 genes mutated in 1–15% of patients each, efforts are ongoing in determining the biological and clinical effect of individually mutated genes[13, 34, 35]. Although some prior studies controlled for prognostic factors (e.g. TP53, IGHV mutation status, FISH, clinical stage) known to be associated with CLL outcome, none considered these collectively or adjusted for CLL-IPI risk category. The only CLL driver gene previously found to be associated with TTT or OS after adjusting for some prognostic factors was SF3B1[13, 34, 35]. Results from our study also indicate that in the combined cohort of CLL patients, mutations in SF3B1 were associated with a shorter TTT, after adjusting for CLL-IPI and sex. Only 10–15% of CLL patients have impactful mutations in SF3B1. Sequencing only this gene in clinical practice would misclassify those CLL patients who have mutations in other recurrently mutated CLL genes. Herein, we found that having one or more recurrently mutated CLL gene is an important prognostic marker. Our study provides strong evidence of an association for TTT independent of CLL-IPI risk category and sex. We found an interesting association with single genes and TTT, suggesting power may be an issue for individual genes. In addition, we found interesting associations between single genes and TTT in the low to intermediate risk CLL-IPI, with larger effect sizes than the overall cohort including high to very-high risk patients. These results need further validation but provide strong evidence that single genes are important prognostic factors for TTT in CLL with low to intermediate risk CLL-IPI.
Strengths of this study include using newly diagnosed, treatment naïve CLL and HC MBL cases and employing deep tumor sequencing data of over 1000X on a comprehensive set of recurrently mutated CLL genes. All of our samples had robustly annotated CLL clinical data as well as all the components necessary to calculate the CLL-IPI. Finally, we used a three-stage study design among the CLL cases with a discovery and two validation cohorts to minimize the chance of false positives and demonstrate reproducibility. A limitation of this study is the potential of limited power for evaluating associations in the high to very-high risk CLL-IPI group. With 128 CLL individuals, we had 90% power to detect a minimal HR of 2.5-fold assuming type I error rate of 0.01. Another limitation is that we did not consider other genetic factors that may be driving CLL progression requiring therapy in these individuals, including copy number variation, DNA methylation status, or chromosomal complexity[41, 42, 49].
In summary, our analyses emphasize that the TML has prognostic value independent of the CLL-IPI and that incorporating this with the CLL-IPI increases its prognostic accuracy, especially for CLL or HC MBL individuals in the low to intermediate risk CLL-IPI groups. Performing a focused mutation panel evaluating recurrently mutated CLL genes among individuals with HC MBL and CLL at time of diagnosis may improve risk stratification and the ability to predict time to progression requiring therapy.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grants, R25 CA92049 (Mayo Cancer Genetic Epidemiology Training Program), R01 CA235026, R01 CA197120, and P50 CA097274.
Footnotes
Disclosure of Conflicts of Interest
Research funding has been provided to the institution from Pharmacyclics, MorphoSys, Janssen, AstraZeneca, and Ascentage Pharma for clinical studies in which Sameer A. Parikh is a principal investigator.
Sameer A. Parikh has also participated in Advisory Board meetings of Pharmacyclics, AstraZeneca, Genentech, Gilead, and AbbVie (he was not personally compensated for his participation).
Neil E. Kay has Research funding from: Acerta Pharm, Pharmacyclics, MEI Pharma, and Tolero. He is on a data safety Monitoring committee for: Agios Pharm, Celgene, Sunesis, Cytomx Therapeutics, Morpho-Sys, Rigel Pharm and Juno Therapeutics. He is on an advisory board: Astra Zeneca, Cytomx Therapeutics, Pharmacyclics Dava Oncology, Acerta Pharma BV, and Juno Therapeutics.
Wei Ding has a research funding from Merck. Advisory board: Merck and Octapharma (no personal compensation).
Esteban Braggio is a consultant of DASA.
The authors declare no competing financial interests
REFERENCES
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic Lymphocytic Leukemia. New England Journal of Medicine, 2005. 352: 804–815. [DOI] [PubMed] [Google Scholar]
- 2.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, Hillmen P, Keating MJ, Montserrat E, Rai KR, Kipps TJ, International Workshop on Chronic Lymphocytic Leukemia. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood, 2008. 111: 5446–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parikh SA, Shanafelt TD. Prognostic factors and risk stratification in chronic lymphocytic leukemia. Seminars in Oncology, 2016. 43: 233–240. [DOI] [PubMed] [Google Scholar]
- 4.Scarfò L, Ferreri AJM, Ghia P. Chronic lymphocytic leukaemia. Critical Reviews in Oncology/Hematology, 2016. 104: 169–182. [DOI] [PubMed] [Google Scholar]
- 5.Marti GE, Rawstron AC, Ghia P, Hillmen P, Houlston RS, Kay N, Schleinitz TA, Caporaso N, International Familial CLL Consortium. Diagnostic criteria for monoclonal B-cell lymphocytosis. British Journal of Haematology, 2005. 130: 325–332. [DOI] [PubMed] [Google Scholar]
- 6.Shanafelt TD, Ghia P, Lanasa MC, Landgren O, Rawstron AC. Monoclonal B-cell lymphocytosis (MBL): biology, natural history and clinical management. Leukemia, 2010. 24: 512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strati P, Shanafelt TD. Monoclonal B-cell lymphocytosis and early-stage chronic lymphocytic leukemia: diagnosis, natural history, and risk stratification. Blood, 2015. 126: 454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shanafelt TD, Kay NE, Rabe KG, Call TG, Zent CS, Maddocks K, Jenkins G, Jelinek DF, Morice WG, Boysen J, Schwager S, Bowen D, Slager SL, Hanson CA. Brief report: natural history of individuals with clinically recognized monoclonal B-cell lymphocytosis compared with patients with Rai 0 chronic lymphocytic leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology, 2009. 27: 3959–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawstron AC, Bennett FL, O’Connor SJM, Kwok M, Fenton JAL, Plummer M, de Tute R, Owen RG, Richards SJ, Jack AS, Hillmen P. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. The New England journal of medicine, 2008. 359: 575–83. [DOI] [PubMed] [Google Scholar]
- 10.Parikh SA, Chaffee KG, Larson MC, Hampel PJ, Call TG, Ding W, Kenderian SS, Leis JF, Chanan-Khan AA, Conte MJ, Bowen D, Schwager SM, Slager SL, Hanson CA, Kay NE, Shanafelt TD. Outcomes of a large cohort of individuals with clinically ascertained high-count monoclonal B-cell lymphocytosis. Haematologica, 2018. 103: e237–e240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, Zhang W, Vartanov AR, Fernandes SM, Goldstein NR, Folco EG, Cibulskis K, Tesar B, Sievers QL, Shefler E, Gabriel S, Hacohen N, Reed R, Meyerson M, Golub TR, Lander ES, Neuberg D, Brown JR, Getz G, Wu CJ. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. The New England journal of medicine, 2011. 365: 2497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns A, Alsolami R, Becq J, Stamatopoulos B, Timbs A, Bruce D, Robbe P, Vavoulis D, Clifford R, Cabes M, Dreau H, Taylor J, Knight SJL, Mansson R, Bentley D, Beekman R, Martín-Subero JI, Campo E, Houlston RS, Ridout KE, Schuh A. Whole-genome sequencing of chronic lymphocytic leukaemia reveals distinct differences in the mutational landscape between IgHVmut and IgHVunmut subgroups. Leukemia, 2018. 32: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puente XS, Beà S, Valdés-Mas R, Villamor N, Gutiérrez-Abril J, Martín-Subero JI, Munar M, Rubio-Pérez C, Jares P, Aymerich M, Baumann T, Beekman R, Belver L, Carrio A, Castellano G, Clot G, Colado E, Colomer D, Costa D, Delgado J, Enjuanes A, Estivill X, Ferrando AA, Gelpí JL, González B, González S, González M, Gut M, Hernández-Rivas JM, López-Guerra M, Martín-García D, Navarro A, Nicolás P, Orozco M, Payer ÁR, Pinyol M, Pisano DG, Puente DA, Queirós AC, Quesada V, Romeo-Casabona CM, Royo C, Royo R, Rozman M, Russiñol N, Salaverría I, Stamatopoulos K, Stunnenberg HG, Tamborero D, Terol MJ, Valencia A, López-Bigas N, Torrents D, Gut I, López-Guillermo A, López-Otín C, Campo E. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature, 2015. 526: 519–524. [DOI] [PubMed] [Google Scholar]
- 14.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Böttcher S, Carter SL, Cibulskis K, Mertens D, Sougnez CL, Rosenberg M, Hess JM, Edelmann J, Kless S, Kneba M, Ritgen M, Fink A, Fischer K, Gabriel S, Lander ES, Nowak MA, Döhner H, Hallek M, Neuberg D, Getz G, Stilgenbauer S, Wu CJ. Mutations driving CLL and their evolution in progression and relapse. Nature, 2015. 526: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, Wan Y, Zhang W, Shukla SA, Vartanov A, Fernandes SM, Saksena G, Cibulskis K, Tesar B, Gabriel S, Hacohen N, Meyerson M, Lander ES, Neuberg D, Brown JR, Getz G, Wu CJ. Evolution and Impact of Subclonal Mutations in Chronic Lymphocytic Leukemia. Cell, 2013. 152: 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez-Trillos A, López-Guerra M, Colomer D, Navarro A, Baumann T, Aymerich M, Rozman M, Delgado J, Giné E, Hernández JM, González-Díaz M, Puente DA, Velasco G, Freije JMP, Tubío JMC, Royo R, Gelpí JL, Orozco M, Pisano DG, Zamora J, Vázquez M, Valencia A, Himmelbauer H, Bayés M, Heath S, Gut M, Gut I, Estivill X, López-Guillermo A, Puente XS, Campo E, López-Otín C. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nature genetics, 2011. 44: 47–52. [DOI] [PubMed] [Google Scholar]
- 17.Schuh A, Becq J, Humphray S, Alexa A, Burns A, Clifford R, Feller SM, Grocock R, Henderson S, Khrebtukova I, Kingsbury Z, Luo S, McBride D, Murray L, Menju T, Timbs A, Ross M, Taylor J, Bentley D. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood, 2012. 120: 4191–6. [DOI] [PubMed] [Google Scholar]
- 18.Ojha J, Secreto C, Rabe K, Ayres-Silva J, Tschumper R, Dyke D V, Slager S, Fonseca R, Shanafelt T, Kay N, Braggio E. Monoclonal B-cell lymphocytosis is characterized by mutations in CLL putative driver genes and clonal heterogeneity many years before disease progression. Leukemia, 2014. 28: 2395–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasi S, Monti S, Spina V, Foa R, Gaidano G, Rossi D. Analysis of NOTCH1 mutations in monoclonal B-cell lymphocytosis. Haematologica, 2012. 97: 153–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imielinski M, Guo G, Meyerson M. Insertions and Deletions Target Lineage-Defining Genes in Human Cancers. Cell, 2017. 168: 460–472.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ojha J, Secreto CR, Rabe KG, Van Dyke DL, Kortum KM, Slager SL, Shanafelt TD, Fonseca R, Kay NE, Braggio E. Identification of recurrent truncated DDX3X mutations in chronic lymphocytic leukaemia. British journal of haematology, 2015. 169: 445–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M, Fangazio M, Vaisitti T, Monti S, Chiaretti S, Guarini A, Del Giudice I, Cerri M, Cresta S, Deambrogi C, Gargiulo E, Gattei V, Forconi F, Bertoni F, Deaglio S, Rabadan R, Pasqualucci L, Foa R, Dalla-Favera R, Gaidano G. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood, 2011. 118: 6904–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossi D, Fangazio M, Rasi S, Vaisitti T, Monti S, Cresta S, Chiaretti S, Del Giudice I, Fabbri G, Bruscaggin A, Spina V, Deambrogi C, Marinelli M, Famà R, Greco M, Daniele G, Forconi F, Gattei V, Bertoni F, Deaglio S, Pasqualucci L, Guarini A, Dalla-Favera R, Foà R, Gaidano G. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia. Blood, 2012. 119: 2854–62. [DOI] [PubMed] [Google Scholar]
- 24.Schnaiter A, Paschka P, Rossi M, Zenz T, Bühler A, Winkler D, Cazzola M, Döhner K, Edelmann J, Mertens D, Kless S, Mack S, Busch R, Hallek M, Döhner H, Stilgenbauer S. NOTCH1, SF3B1, and TP53 mutations in fludarabine-refractory CLL patients treated with alemtuzumab: results from the CLL2H trial of the GCLLSG. Blood, 2013. 122: 1266–70. [DOI] [PubMed] [Google Scholar]
- 25.Agathangelidis A, Ljungström V, Scarfò L, Fazi C, Gounari M, Pandzic T, Sutton L-A, Stamatopoulos K, Tonon G, Rosenquist R, Ghia P. Highly similar genomic landscapes in monoclonal B-cell lymphocytosis and ultra-stable chronic lymphocytic leukemia with low frequency of driver mutations. Haematologica, 2018. 103: 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrio S, Shanafelt TD, Ojha J, Chaffee KG, Secreto C, Kortüm KM, Pathangey S, Van-Dyke DL, Slager SL, Fonseca R, Kay NE, Braggio E. Genomic characterization of high-count MBL cases indicates that early detection of driver mutations and subclonal expansion are predictors of adverse clinical outcome. Leukemia, 2017. 31: 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lionetti M, Fabris S, Cutrona G, Agnelli L, Ciardullo C, Matis S, Ciceri G, Colombo M, Maura F, Mosca L, Gentile M, Recchia AG, Ilariucci F, Musolino C, Molica S, Di Raimondo F, Cortelezzi A, Rossi D, Gaidano G, Morabito F, Ferrarini M, Neri A. High-throughput sequencing for the identification of NOTCH1 mutations in early stage chronic lymphocytic leukaemia: biological and clinical implications. British journal of haematology, 2014. 165: 629–39. [DOI] [PubMed] [Google Scholar]
- 28.Winkelmann N, Rose-Zerilli M, Forster J, Parry M, Parker A, Gardiner A, Davies Z, Steele AJ, Parker H, Cross NCP, Oscier DG, Strefford JC. Low frequency mutations independently predict poor treatment-free survival in early stage chronic lymphocytic leukemia and monoclonal B-cell lymphocytosis. Haematologica, 2015. 100: e237–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Döhner K, Bühler A, Böttcher S, Ritgen M, Kneba M, Winkler D, Tausch E, Hoth P, Edelmann J, Mertens D, Bullinger L, Bergmann M, Kless S, Mack S, Jäger U, Patten N, Wu L, Wenger MK, Fingerle-Rowson G, Lichter P, Cazzola M, Wendtner CM, Fink AM, Fischer K, Busch R, Hallek M, Döhner H. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood, 2014. 123: 3247–54. [DOI] [PubMed] [Google Scholar]
- 30.Zhou W, Goldin L, Wang M, McMaster ML, Jones K, Burdett L, Chanock SJ, Yeager M, Dean M, Caporaso NE. Combined somatic mutation and copy number analysis in the survival of familial CLL. British Journal of Haematology, 2018. 181: 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young E, Noerenberg D, Mansouri L, Ljungström V, Frick M, Sutton L-A, Blakemore SJ, Galan-Sousa J, Plevova K, Baliakas P, Rossi D, Clifford R, Roos-Weil D, Navrkalova V, Dörken B, Schmitt CA, Smedby KE, Juliusson G, Giacopelli B, Blachly JS, Belessi C, Panagiotidis P, Chiorazzi N, Davi F, Langerak AW, Oscier D, Schuh A, Gaidano G, Ghia P, Xu W, Fan L, Bernard OA, Nguyen-Khac F, Rassenti L, Li J, Kipps TJ, Stamatopoulos K, Pospisilova S, Zenz T, Oakes CC, Strefford JC, Rosenquist R, Damm F. EGR2 mutations define a new clinically aggressive subgroup of chronic lymphocytic leukemia. Leukemia, 2017. 31: 1547–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leeksma AC, Taylor J, Wu B, Gardner JR, He J, Nahas M, Gonen M, Alemayehu WG, te Raa D, Walther T, Hüllein J, Dietrich S, Claus R, de Boer F, de Heer K, Dubois J, Dampmann M, Dürig J, van Oers MHJ, Geisler CH, Eldering E, Levine RL, Miller V, Mughal T, Lamanna N, Frattini MG, Heaney ML, Zelenetz A, Zenz T, Abdel-Wahab O, Kater AP. Clonal diversity predicts adverse outcome in chronic lymphocytic leukemia. Leukemia, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amin NA, Malek SN. Gene mutations in chronic lymphocytic leukemia. Seminars in Oncology, 2016. 43: 215–221. [DOI] [PubMed] [Google Scholar]
- 34.Jeromin S, Weissmann S, Haferlach C, Dicker F, Bayer K, Grossmann V, Alpermann T, Roller A, Kohlmann A, Haferlach T, Kern W, Schnittger S. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia, 2014. 28: 108–117. [DOI] [PubMed] [Google Scholar]
- 35.Baliakas P, Hadzidimitriou A, Sutton L-A, Rossi D, Minga E, Villamor N, Larrayoz M, Kminkova J, Agathangelidis A, Davis Z, Tausch E, Stalika E, Kantorova B, Mansouri L, Scarfò L, Cortese D, Navrkalova V, Rose-Zerilli MJJ, Smedby KE, Juliusson G, Anagnostopoulos A, Makris AM, Navarro A, Delgado J, Oscier D, Belessi C, Stilgenbauer S, Ghia P, Pospisilova S, Gaidano G, Campo E, Strefford JC, Stamatopoulos K, Rosenquist R, European Research Initiative on CLL (ERIC). Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia, 2015. 29: 329–36. [DOI] [PubMed] [Google Scholar]
- 36.Rossi D, Rasi S, Spina V, Bruscaggin A, Monti S, Ciardullo C, Deambrogi C, Khiabanian H, Serra R, Bertoni F, Forconi F, Laurenti L, Marasca R, Dal-Bo M, Rossi FM, Bulian P, Nomdedeu J, Del Poeta G, Gattei V, Pasqualucci L, Rabadan R, Foà R, Dalla-Favera R, Gaidano G. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood, 2013. 121: 1403–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oscier DG, Rose-Zerilli MJJ, Winkelmann N, Gonzalez de Castro D, Gomez B, Forster J, Parker H, Parker A, Gardiner A, Collins A, Else M, Cross NCP, Catovsky D, Strefford JC. The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood, 2013. 121: 468–75. [DOI] [PubMed] [Google Scholar]
- 38.International CLL-IPI working group. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. The Lancet Oncology, 2016. 17: 779–790. [DOI] [PubMed] [Google Scholar]
- 39.Molica S, Giannarelli D, Levato L, Mirabelli R, Gentile M, Morabito F. Assessing time to first treatment in early chronic lymphocytic leukemia (CLL): a comparative performance analysis of five prognostic models with inclusion of CLL-international prognostic index (CLL-IPI). Leukemia & Lymphoma, 2017. 58: 1736–1739. [DOI] [PubMed] [Google Scholar]
- 40.Molica S, Shanafelt TD, Giannarelli D, Gentile M, Mirabelli R, Cutrona G, Levato L, Di Renzo N, Di Raimondo F, Musolino C, Angrilli F, Famà A, Recchia AG, Chaffee KG, Neri A, Kay NE, Ferrarini M, Morabito F. The chronic lymphocytic leukemia international prognostic index predicts time to first treatment in early CLL: Independent validation in a prospective cohort of early stage patients. American journal of hematology, 2016. 91: 1090–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu L, Kim HT, Kasar SN, Benien P, Du W, Hoang K, Aw A, Tesar B, Improgo R, Fernandes SM, Radhakrishnan S, Klitgaard JL, Lee C, Getz G, Setlur SR, Brown JR. Survival of Del17p CLL Depends on Genomic Complexity and Somatic Mutation. Clinical Cancer Research, 2017. 23: 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nadeu F, Clot G, Delgado J, Martín-García D, Baumann T, Salaverria I, Beà S, Pinyol M, Jares P, Navarro A, Suárez-Cisneros H, Aymerich M, Rozman M, Villamor N, Colomer D, González M, Alcoceba M, Terol MJ, Navarro B, Colado E, Payer Á, Puente XS, López-Otín C, López-Guillermo A, Enjuanes A, Campo E. Clinical impact of the subclonal architecture and mutational complexity in chronic lymphocytic leukemia. Leukemia, 2018. 32: 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strati P, Wang F, Tambaro FP, Thompson PA, Burger JA, Jain N, Ferrajoli A, Bose P, Estrov Z, Keating MJ, Futreal A, Takahashi K, Wierda WG. The landscape of genetic mutations in patients with chronic lymphocytic leukaemia and complex karyotype. British Journal of Haematology, 2019. bjh.16117. [DOI] [PubMed] [Google Scholar]
- 44.Brieghel C, da Cunha-Bang C, Yde CW, Schmidt AY, Kinalis S, Nadeu F, Andersen MA, Jacobsen LO, Andersen MK, Pedersen LB, Delgado J, Baumann T, Mattsson M, Mansouri L, Rosenquist R, Campo E, Nielsen FC, Niemann CU. The Number of Signaling Pathways Altered by Driver Mutations in Chronic Lymphocytic Leukemia Impacts Disease Outcome. Clinical cancer research : an official journal of the American Association for Cancer Research, 2020. clincanres.4158.2018. [DOI] [PubMed] [Google Scholar]
- 45.Cheson B, Bennett J, Grever M, Kay N, Keating M, O’Brien S, Rai K. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood, 1996. 87: 4990–4997. [PubMed] [Google Scholar]
- 46.Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, Pencina MJ, Kattan MW. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology (Cambridge, Mass), 2010. 21: 128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parikh SA. Chronic lymphocytic leukemia treatment algorithm 2018. Blood Cancer Journal, 2018. 8: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shanafelt TD, Bowen D, Venkat C, Slager SL, Zent CS, Kay NE, Reinalda M, Sloan JA, Call TG. Quality of life in chronic lymphocytic leukemia: an international survey of 1482 patients [Internet]. British Journal of Haematology, 2007. 139: 255–264. [cited 2020 Jan 17] Available from: http://doi.wiley.com/10.1111/j.1365-2141.2007.06791.x [DOI] [PubMed] [Google Scholar]
- 49.Oakes CC, Claus R, Gu L, Assenov Y, Hullein J, Zucknick M, Bieg M, Brocks D, Bogatyrova O, Schmidt CR, Rassenti L, Kipps TJ, Mertens D, Lichter P, Dohner H, Stilgenbauer S, Byrd JC, Zenz T, Plass C. Evolution of DNA Methylation Is Linked to Genetic Aberrations in Chronic Lymphocytic Leukemia. Cancer Discovery, 2014. 4: 348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.