Abstract
Coronavirus disease 2019 (COVID-19), an infectious disease caused by the infection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is spreading and causing the global coronavirus pandemic. The viral dynamics of SARS-CoV-2 infection have not been quantitatively investigated. In this paper, we use mathematical models to study the pathogenic features of SARS-CoV-2 infection by examining the interaction between the virus, cells and immune responses. Models are fit to the data of SARS-CoV-2 infection in patients and non-human primates. Data fitting and numerical simulation show that viral dynamics of SARS-CoV-2 infection have a few distinct stages. In the initial stage, viral load increases rapidly and reaches the peak, followed by a plateau phase possibly generated by lymphocytes as a secondary target of infection. In the last stage, viral load declines due to the emergence of adaptive immune responses. When the initiation of seroconversion is late or slow, the model predicts viral rebound and prolonged viral persistence, consistent with the observation in non-human primates. Using the model we also evaluate the effect of several potential therapeutic interventions for SARS-CoV-2 infection. Model simulation shows that anti-inflammatory treatments or antiviral drugs combined with interferon are effective in reducing the duration of the viral plateau phase and diminishing the time to recovery. These results provide insights for understanding the infection dynamics and might help develop treatment strategies against COVID-19.
Highlights
-
•
Mathematical models are developed to study SARS-CoV-2 infection.
-
•
Models are fit to viral load data of patients and non-human primates.
-
•
Viral dynamics of SARS-CoV-2 infection has a few distinct stages.
-
•
Model explains viral rebound and prolonged viral persistence.
-
•
Implication for treatment strategies is discussed.
1. Introduction
In late December 2019, highly contagious pneumonia of unknown etiology was first reported in Wuhan, China [1], [2], [3], [4], [5], [6], [7], [8], [9]. A novel strain of coronavirus was isolated from patients and named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the World Health Organization [10]. Since then, SARS-CoV-2 infection has been detected in about 190 countries and caused more than 600,000 deaths worldwide [9], [11], [12], [13], [14], [15]. Infected individuals develop acute respiratory distress syndrome (ARDS) characterized by lymphocytopenia and hyperactive inflammatory response, which becomes the major cause of lung damage and consequent death [16]. High-throughput sequencing demonstrates that SARS-CoV-2 highly resembles the severe acute respiratory syndrome (SARS-CoV) and the Middle East respiratory syndrome (MERS-CoV), both classified as beta coronaviruses identified in bats [17], [18], [19], [20], [21]. Its genome sequence is 96.2% identical to the bat CoV RaTG13 [22]. It uses the angiotensin-converting enzyme 2 (ACE2), the same receptor used by SARS-CoV [23], [24], [25], [26], [27], to infect humans.
Lymphocytopenia is an indicator of the severity and mortality in patients with coronavirus disease of 2019 (COVI-19) [4], [28], [29], [30], [31], [32], [33]. Critically ill patients have significantly low lymphocyte count, possibly due to the direct infection by coronavirus, leading to cytoplasmic damage and destruction of cellular components [30], [31], [33], [34], [35]. In addition, COVID-19 patients have hyperinflammatory responses that may induce massive recruitment of lymphocytes to the lung and result in the consequent pulmonary injury [36]. These findings suggest that SARS-CoV-2 might rely on “cytokine storm” [37] to develop a vicious cycle to drive lymphocytes to lung for infection to maintain the viral replication and transmissibility.
Currently, there are no approved antiviral drugs for SARS-CoV-2 infection. Many patients were given symptomatic management such as empirical broad-spectrum antibiotics or anti-influenza therapies [38]. Mechanical ventilations were used in patients with respiratory failure. The mortality of COVID-19 patients is primarily caused by severe lung injury induced by hyperactive host inflammation. Corticosteroids were administrated to critically ill patients as immunosuppression agents [4], [29]. The adaptive immunity of a patient may be essential for the recovery from SARS-CoV-2 infection [39], [40]. Convalescent plasma containing neutralizing antibodies donated by recovered patients can improve the clinical status of severely ill patients [41]. However, a fraction of patients recovered from COVID-19 still tested positive or even had long-term viral shedding [29], [42]. The mechanisms underlying these pathogenic features are critically needed to mitigate the COVID-19 pandemic.
Some modeling studies have shed light on the transmission dynamics of COVID-19 at the population level [8], [43], [44], [45], [46], [47], [48], [49]. However, the within-host viral dynamics of SARS-CoV-2 infection have not been quantitatively investigated. Here we use mathematical models to study the pathogenic characteristics of SARS-CoV-2 infection by examining the interaction between viral replication and the host immune responses. We fit models to available data of SARS-CoV-2 infected patients and non-human primates in different countries [14], [50], [51], [52]. We also use model simulations to evaluate some potential therapies against COVID-19. Combination of antiviral drugs and type I interferon (IFN) may control the SARS-CoV-2 replication. This is consistent with the observation that SARS-CoV-2 is sensitive to IFN [53]. These findings may provide new insights into the pathogenesis of SARS-CoV-2 infection and development of new treatment strategies.
2. Methods
2.1. Patient and experiment data
The viral load data in COVID-19 patients with and without treatment we analyzed in this paper were from Germany, South Korea and China. We also studied the viral load data from a SARS-CoV-2 infected adult rhesus macaque in the US. The viral load data of eight patients from Germany were from a larger cohort of COVID-19 cases in Munich [54]. Viral RNA levels of all patients were measured every day after onset of symptoms via reverse transcription polymerase chain reaction (RT-PCR) from specimens of throat swab, sputum, and stool. Seroconversion was detected by IgM and IgG immunofluorescence and virus neutralization assay within cells expressing the spike proteins of SARS-CoV-2 [14]. The data from China were collected by RT-PCR from serial samples (throat swabs, sputum, urine and stool) of two patients after they were hospitalized in Beijing [51]. We fit the models to one patient due to the lack of enough measurements in another. The viral load data from South Korea were from a 35-year-old Chinese woman traveling from Wuhan, China [50]. She was the first case reported in South Korea. The viral load was also measured with RT-PCR every day after diagnosis from samples of nasopharyngeal and oropharyngeal swabs and sputum. This patient was treated with lopinavir/ritonavir on day 5 since symptom’s onset. The rectal viral load data from a rhesus macaque having prolonged viral shedding were collected by RT-PCR every day from its rectal swab samples after inoculation [52].
2.2. A basic viral dynamic model
We use three models, with increasing complexity, to study the viral dynamics of SARS-CoV-2 infection. The first is a very basic model, which only includes target cells, infected cells and the virus. Variations of the model have been used to study the within-host dynamics of many virus infections, such as HIV, hepatitis, and influenza virus [55]. The second model includes an eclipse phase in the basic model and the third one includes a secondary target cell population of SARS-CoV-2 infection. The basic model is given by the following equations.
| (1) |
The exact target of SARS-CoV-2 infection is not fully clear. Based on the findings from SARS-CoV, MERS-CoV [18], [23], [24], [25], [26], [27], [56] and results from SARS-CoV-2 infection [36], [57], [58], SARS-CoV-2 can infect type II pneumocytes [59]. Target cells (T) are assumed to be infected by SARS-CoV-2 (V) to generate infected cells (I) at a mass action rate VT. Infected cells die with a death rate . They produce coronaviruses at a rate p per infected cell. The virus is cleared at a constant rate c.
SARS-CoV-2 infection can lead to the diffuse alveolar damage (DAD) [60]. DAD usually has three phases: exudative phase, proliferative phase and fibrotic phase [61]. In the exudative phase, immune cells are recruited to the lung, causing accumulated fluid in the airspace. This prevents alveoli from remaining open for gas exchange. The proliferative phase is characterized by regeneration of alveolar type II epithelial cells or pneumocytes [62]. When replicating, type II epithelial cells transport fluid out of the airspace. SARS-CoV-2 infects pneumocytes and induces hyperactive inflammation that greatly enhances the exudative reaction. Thus, pneumocytes may experience a significant delay in proliferation [63]. In addition, about 30% of COVID-19 patients are asymptomatic [13], [64], [65]. They do not develop DAD so the proliferation of pneumocytes could be at a minimum level. Therefore, when studying the viral dynamics during the early stage of infection, we do not include the proliferation and death of pneumocytes. This also keeps the model in the simplest form for data fitting.
2.3. The model with an eclipse phase
The model with an eclipse phase includes four variables: Target cells (T), infected cells in the eclipse phase (), productively infected cells () and viruses (V). Target cells are infected and enter the eclipse phase, in which they do not produce viruses. They progress to productively infected cells at a transition rate k. The other parameters are the same as those in the basic model.
| (2) |
2.4. The model with a secondary target of infection
Previous studies [30], [31], [33] detected a large number of viral particles within lymphocytes of SARS-CoV and MERS-CoV patients. A new study also found the infection of T lymphocytes by SARS-CoV-2 [35]. We include lymphocytes as a secondary target cell population in the basic model.
| (3) |
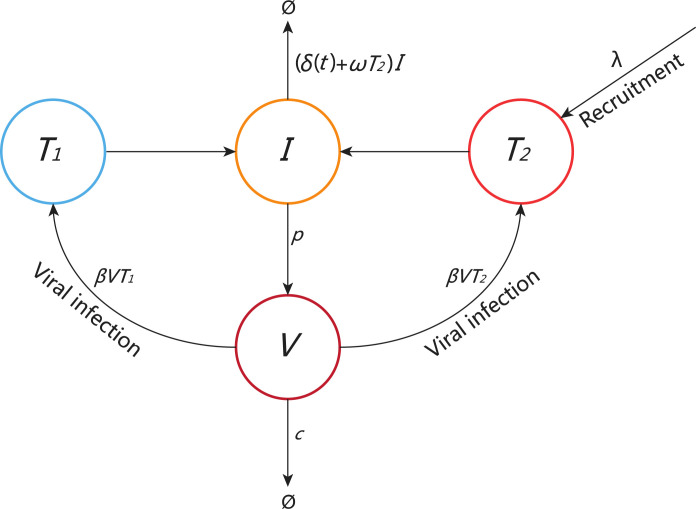
The variable represents the concentration of pneumocytes and represents the concentration of lymphocytes. There is a relatively constant level of lymphocytes in the body of a healthy person. The turnover of uninfected lymphocytes is also slow [66], [67]. Thus, we use a constant rate to represent the recruitment of lymphocytes to the infection site due to the lung inflammatory response. This was also assumed in the viral dynamic models that studied HIV primary infection and influenza virus infection [55], [68]. To minimize the number of parameters for data fitting, we also assume that lymphocytes and pneumocytes are infected by the virus at the same rate . After infection, they are cleared at the same rate (which will be described later) and produce virus at the same rate p. With these assumptions we can use one variable (I) to represent the combined population of infected lymphocytes and pneumocytes. The schematic diagram of model (3) is shown in Fig. 1.
Fig. 1.
Schematic diagram of model (3). The secondary target cells () are recruited to the foci at a constant rate . The two target cell populations are infected by SARS-CoV-2 with the same infection rate . Infected cells can be removed by the innate response with a rate . There is also the death rate of infected cells, (t), due to the adaptive immune response activated on day . The death rate (t) increases from the base rate , following an exponential function , where determines how fast the death rate increases.
Lymphocytes primarily consist of T cells, B cells and natural killer (NK) cells. T cells regulate both innate and adaptive immune responses. B cells produce antibodies and NK cells can recognize and kill stressed cells in the absence of antibodies [70], [71], [72], [73]. Because of the lack of data on the innate and adaptive immune responses for model fitting, we assume that the concentration of lymphocytes is proportional to the innate immune response and use the term T2 I to represent the clearance of infected cells by the innate immune response. The adaptive immune response is assumed to be activated on day and the death rate of infected cells increases from the base rate , following an exponential function , where determines how fast the death rate increases. Thus, is when and it is when . Although we can use a saturating function to set the maximum death rate, as used by Pawelek et al. to study the control of influenza virus infection by the adaptive immune response [68], it will introduce an extra parameter for estimation from data fitting. Because we aim to model the viral elimination in the end of infection, the exponentially increasing function is reasonable for this purpose.
2.5. Model parameters
We fix the base death rate of infected cells () to be 2 cells day−1 [68] and the recruitment rate of lymphocyte () to be 104 cells ml−1 day−1 [69]. The total number of alveoli cells within a human is about 6108 cells [74], [75]. Type II pneumocytes constitute 60% of the total alveolar cells [76] and the volume of lung is about 6000 ml. Thus, the initial value of pneumocytes is chosen to be T(0) (6108 60%)/6000 6104 cells/ml. The initial value of lymphocytes at the infection site is 0. There are no infected cells at the initial time. We fit models to the data of viral load extracted from sputum/saliva. Because much of the early infection occurs in the lung and only a small proportion of the viruses transport to the fluid immediately after infection, we assume V(0) to be an effective initial viral concentration and choose the value of V(0) from 10−6 to 10−3 RNA copies/ml, as used in [77], that can generate the best fits. We estimate the rest of the parameters by fitting models to the data of viral load. All the variables and parameters are summarized in Table 1.
Table 1.
Parameters and variables.
| Variables/parameters | Description | Value | Ref |
|---|---|---|---|
| T | Target cells | – | – |
| Nonproductively infected cells | – | – | |
| Productively infected cells | – | – | |
| Primary target cells/pneumocytes | – | – | |
| Secondary target cells/lymphocytes | – | – | |
| V | Virus | – | – |
| Viral infection rate | Fitted | ||
| Base death rate of productively infected cells | 2 day−1 | [68] | |
| k | Transition rate from exposed to productive stage | Fitted | |
| c | Clearance rate of virus | Fitted | |
| p | Viral production rate | Fitted | |
| Killing rate by innate immune response | Fitted | ||
| Recruitment of lymphocytes to the infection site | 104 day−1 | [69] | |
| Parameter in the killing by adaptive immune response | Fitted | ||
| Time of emergence of adaptive immune response | Fixed |
2.6. Model comparison
We compare the data fits with different models by calculating the Akaike information criterion (AIC), given by the following equation [78], [79]
where m denotes the number of parameters in the model used to fit data and n is the number of observations (data points). The root mean square (RMS) between the prediction of model and data is calculated using the following formula
where V() represents the viral load level at time predicted by the model, is the corresponding data at . Parameter estimates are based on the best fitting that achieves the minimum RMS. Data fitting is performed using the R programming language.
3. Results
3.1. Viral dynamics of SARS-CoV-2 infection
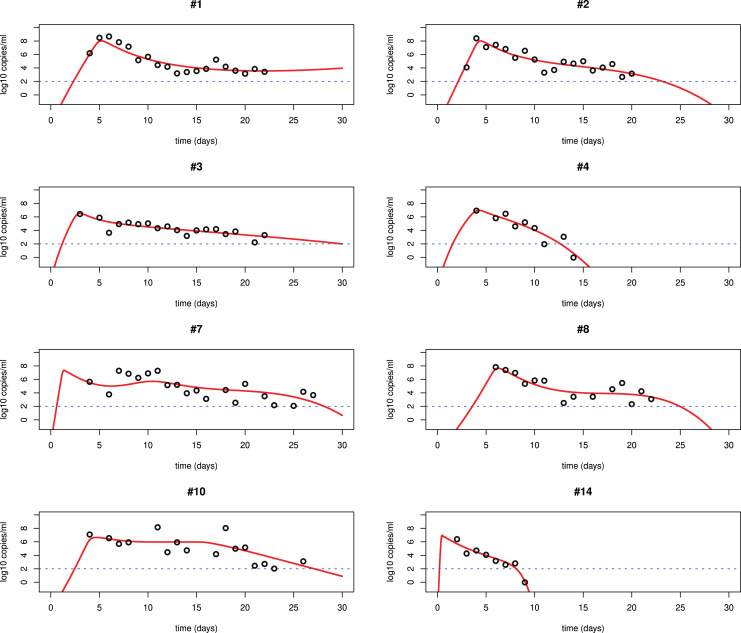
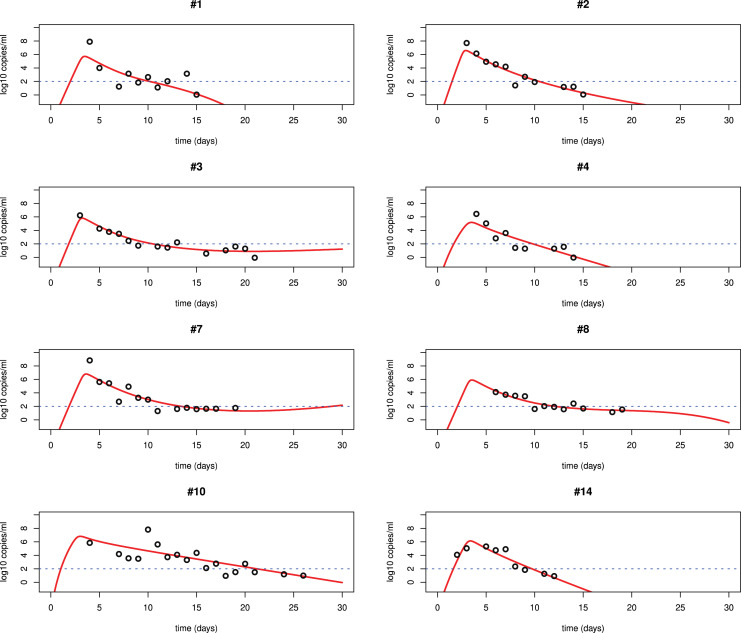
We fit each of the three models Eqs. ((1)–(3)) to the viral load data of SARS-CoV-2 infection in patients from Germany and China [14], [51]. The best fits to the data from lower respiratory tract (LRT) and upper respiratory tract (URT) using model (3) are shown in Fig. 2, Fig. 3, Fig. 4. All the parameters of the best fits are listed in Table 2, Table 3, Table 4. The best fits using models (1) and (2) are shown in the supplemental materials. We find that model (3) significantly improves the fit. The major problem in the fitting with models (1) and (2) is that these models show a single-phase viral decline after the peak. This is not consistent with the viral plateau or even a second peak observed in some patients [14]. Models (1) and (2) also use a constant rate to describe the clearance of infected cells. The killing by the immune responses is complicated. Using a constant rate fails to explain the viral plateau and subsequent decline of the viral load observed in patients [14], [51].
Fig. 2.
Best fits of model (3) to the LRT viral load data in patients from Germany [14]. The patients #1, #2, #3, #4, #7, #8, #10 and #14 are from the cases in [14], [54]. The detection limit is the blue dotted line according to [14]. We fixed according to the emergence time of seroconversion.
Fig. 3.
Best fits of model (3) to the URT viral load data in patients from Germany [14]. The other information is the same as in Fig. 2.
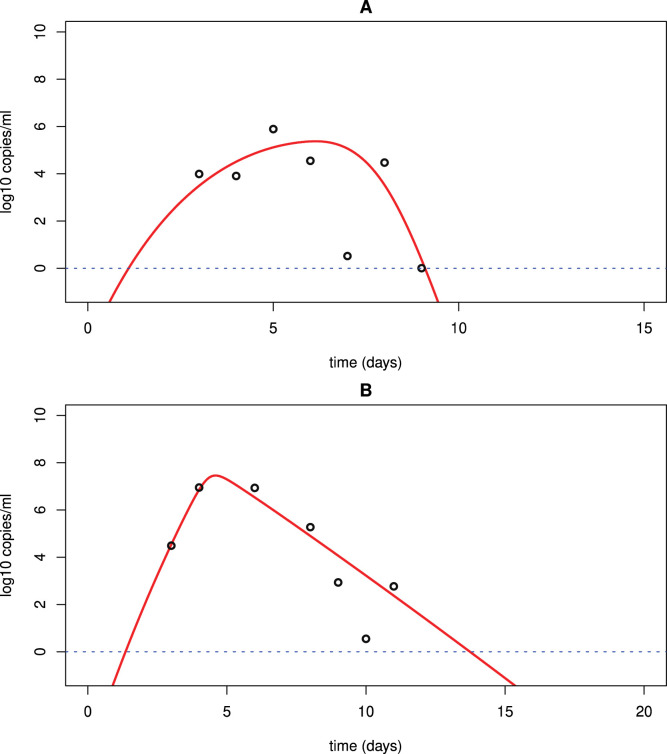
Fig. 4.
Best fits of model (3) to the URT (A) and LRT (B) viral load data in a patient from China [51]. The detection limit is the blue dotted line.
Table 2.
Best fits of model (3) to the LRT viral load data in patients from Germany.
| Patient | (ml/virus/day) | p (day−1) | c (day−1) | (ml/cell/day) | (day−1) | V(0) RNA/ml | AIC value | |
|---|---|---|---|---|---|---|---|---|
| #1 | 4.810−8 | 7.9105 | 17 | 2.110−4 | 10−3 | 10−3 | 10 | −4.042036 |
| #2 | 5.910−8 | 1.3105 | 39 | 1.610−4 | 0.1 | 10−4 | 8 | −1.708228 |
| #3 | 3.610−6 | 1.2103 | 4.4 | 10−3 | 0.1 | 10−3 | 9 | −10.92767 |
| #4 | 7.310−7 | 5.7103 | 4.4 | 1.110−3 | 0.5 | 10−4 | 6 | 5.980355 |
| #7 | 110−6 | 1.1105 | 209 | 4.510−4 | 0.11 | 10−3 | 9 | 17.5142 |
| #8 | 9.110−8 | 1.2105 | 108 | 410−5 | 0.1 | 10−4 | 6 | 6.927418 |
| #10 | 4.610−7 | 1.66102 | 0.89 | 10−9 | 0.93 | 10−4 | 15 | 17.56648 |
| #14 | 410−5 | 6.5103 | 20 | 1.410−2 | 1.8 | 10−5 | 6 | 0.105858 |
Table 3.
Best fits of model (3) to the URT viral load data in patients from Germany.
| Patient | (ml/virus/day) | p (day−1) | c (day−1) | (ml/cell/day) | V(0) RNA/ml | AIC value | ||
|---|---|---|---|---|---|---|---|---|
| #1 | 1.110−5 | 9.4102 | 50 | 1.910−4 | 0.13 | 10−4 | 10 | 15.41636 |
| #2 | 2.210−6 | 4.8103 | 35 | 310−4 | 0.01 | 10−4 | 8 | 1.839616 |
| #3 | 9.810−6 | 1.3103 | 58 | 210−4 | 0.04 | 10−4 | 9 | −7.151683 |
| #4 | 4.510−5 | 8.810 | 5 | 10−3 | 10−3 | 10−4 | 6 | 8.759159 |
| #7 | 9.910−7 | 1.08104 | 48 | 2.110−4 | 10−4 | 10−4 | 9 | 7.385558 |
| #8 | 710−6 | 3.1103 | 118 | 1.510−4 | 0.07 | 10−4 | 6 | −11.03853 |
| #10 | 1.810−6 | 4.9103 | 3.5 | 310−3 | 0 | 10−4 | 15 | 15.05884 |
| #14 | 5.710−6 | 8102 | 10 | 5.310−4 | 0 | 10−4 | 6 | −0.552598 |
Table 4.
Best fits of model (3) to the URT and LRT data of a patient from China.
| (ml/virus/day) | p (day−1) | c (day−1) | (ml/cell/day) | V(0) RNA/ml | AIC value | |||
|---|---|---|---|---|---|---|---|---|
| URT | 7.510−7 | 5103 | 12 | 5.610−4 | 1.5 | 10−3 | 6 | 5.673903 |
| LRT | 1.710−7 | 2.9104 | 19 | 2.910−4 | 0.06 | 10−4 | 6 | 2.669148 |
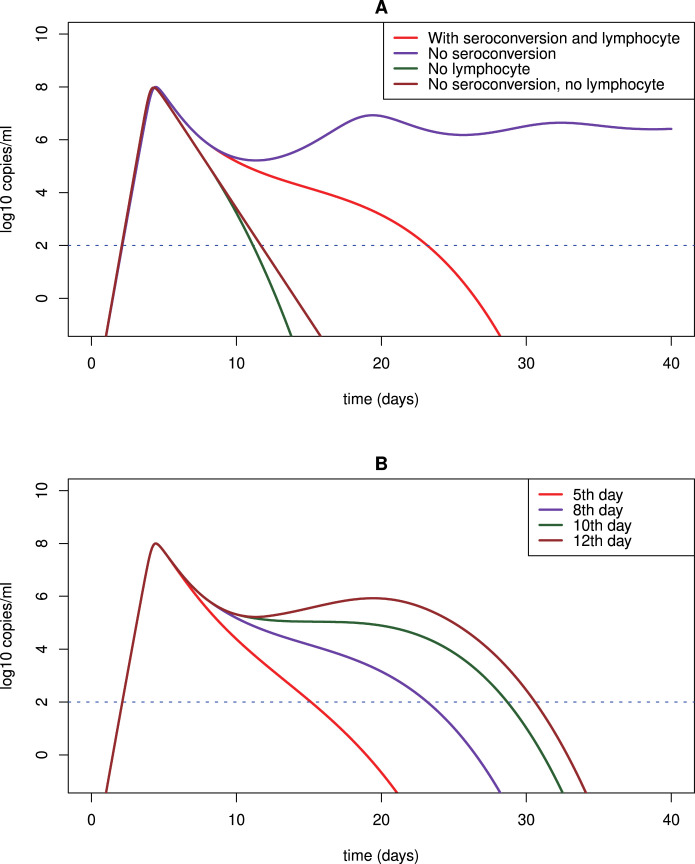
Numerical simulations of model (3) show that the viral dynamics of SARS-CoV-2 infection can exhibit a few distinct phases (Fig. 5A). During the first stage, the primary target cells are infected and consumed rapidly. The viral load experiences a substantial increase to the peak level. This agrees with the clinical data that onsets of ARDS are observed around this time [14]. Following the viral peak, the viral load declines slightly and enters a plateau phase in which the viral load remains approximately unchanged or declines slowly. In our model, the source of this plateau phase is the infection of lymphocytes as a secondary target. Without the infection of lymphocytes, SARS-CoV-2 would continue to infect and deplete pneumocytes. As a consequence, viral load would decline quickly and the infected individual would lose the transmissibility (Fig. 5A). After viral load persists for a period of time (e.g. a week in some patients), the next phase follows as the adaptive immune response emerges. Seroconversion was also detected about a week after the onset of ARDS in COVID-19 patients [14]. During this stage, viral load declines rapidly to an undetectable level. This also agrees with observation in patients [14].
Fig. 5.
Predicted viral dynamics by model (3) under different scenarios. We use the fitting to the LRT viral load data of patient #2 in [14], [54] as an example to study the model prediction under different assumptions. (A) Model prediction with and without lymphocytes (i.e. the secondary target of infection) and seroconversion. The adaptive immune response, if included, is assumed to occur on day 8. (B) The adaptive immune response is assumed to be activated on day 5, 8, 10 and 12, respectively.
Due to the lack of approved antiviral drugs, many patients with severe symptoms are given oxygen treatment, mechanical ventilation, corticosteroids and antibiotics to maintain the respiratory function and ease the inflammatory response. This provides time for the adaptive immune response to emerge to control the virus [4], [29], [37], [38], [39], [40]. In Fig. 5B, we simulate the dynamics of the viral load with adaptive immune response activated at different time. The later the adaptive immune response takes place, the longer the viral load persists. Many senior patients die from COVID-19 because they could not sustain prolonged life support treatment due to multiple significant underlying diseases [80].
The best fits with the model show that the viral infection rate is significantly higher in URT, but the viral production rate and the immune response (described by and ) in LRT are much larger (>10 folds) than that in URT (Table 2, Table 3, Table 4). This is consistent with the route of human-to-human spread of SARS-CoV-2. Virus is transmitted from one host to another via exhaled air, respiratory droplets or fine-particle aerosols [81]. The primary infection of SARS-CoV-2 occurs in the URT while the systematic replication takes place within lung. This suggests that wearing face mask is an effective way to prevent transmission of SARS-CoV-2 from infected individuals [82].
3.2. Viral rebound and persistence
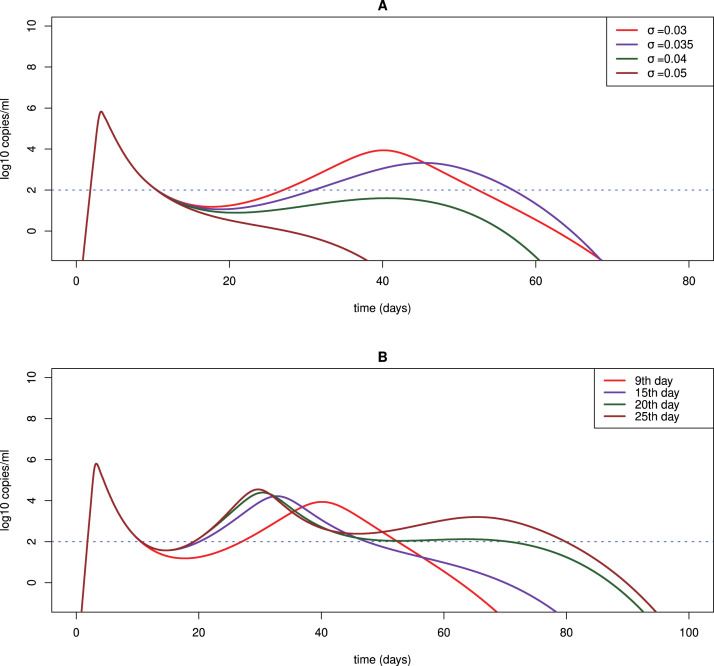
Some patients still tested positive after recovering from COVID-19 [42], [83]. The biological process underlying viral rebound is unknown. Our simulation in Fig. 6A shows that if the value of is small, i.e. the clearance rate of infected cells increases slowly as the adaptive immune response emerges, then a second viral peak is predicted because the adaptive immune response is not sufficient to control the viral replication. Thus, viral rebound that lasts for a period of time can be observed. As (t) increases to above a certain level, the viral load will decline again and eventually be eliminated (Fig. 6A).
Fig. 6.
Predicted viral rebound and persistence under different scenarios. We use the fitting to the URT viral load data of patient #3 in [14], [54] as an example to study viral rebound and persistence with different assumptions. (A) Predicted viral dynamics with different values of , the parameter determining how fast the death rate of infected cells increases as the adaptive immune response emerges. (B) Predicted viral rebound and persistence assuming the adaptive immune response is activated on day 9, 15, 20 and 25, respectively. The parameter is fixed to be 0.03 day−1.
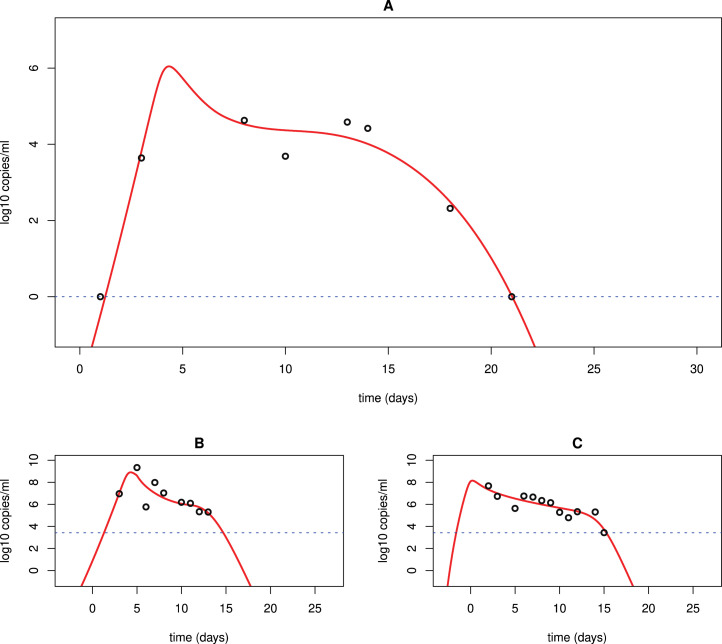
The timing of the emergence of adaptive immune response also affects viral rebound and persistence. If the adaptive immune system is activated later, then the viral load can persist at a higher level for a longer time (Fig. 6B). This is consistent with a report that a SARS-CoV-2 infected non-human primate with the lowest and latest antibody response had prolonged viral shedding from the intestinal tract [52]. We also fit our model to the rectal viral load data of this animal and the fit is excellent (Fig. 7A). The parameter estimates are shown in Table 5. The finding suggests that for infected individuals with compromised or delayed immune responses, the viral load will rebound following the first peak and even persist for a long time [84]. If the virus persists at a low level around the detection limit, the patient may be asymptomatic or only develop very mild symptoms. However, the patient can still test positive and spread virus. This has been reported in a study that suggested the transmission from an asymptomatic contact in Germany [13].
Fig. 7.
Best fit of model (3) to the data of a non-human primate and a patient in South Korea treated with lopinavir/ritonavir. (A) Best fit to long-term viral shedding in an adult rhesus macaque [52]; (B) Best fit to the LRT viral load data of the Korean patient treated with lopinavir/ritonavir [50]; (C) Best fit to the URT data of the same patient in (B).
Table 5.
Best fits of model (3) to the rectal viral load data of a non-human primate.
| (ml/virus/day) | p (day−1) | c (day−1) | (ml/cell/day) | V(0) RNA/ml | AIC value | |||
|---|---|---|---|---|---|---|---|---|
| Rectal | 4.910−6 | 350 | 11 | 5.810−5 | 0.17 | 10−2 | 7 | −5.835853 |
3.3. Potential treatment
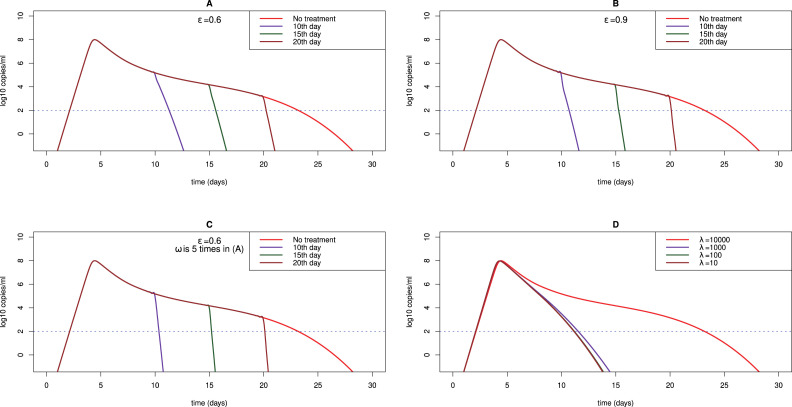
Lopinavir/ritonavir and remdesivir were suggested to be remedy that might inhibit SARS-CoV-2 viral replication [85], [86]. Lopinavir/ritonavir is a combination of protease inhibitors used to treat HIV-1 infection [87], while remdesivir was developed to treat Ebola and Marburg virus infection, acting as an analog of adenosine nucleotide to confuse viral RNA polymerase and evade proofreading by viral exoribonuclease (ExoN) [88]. We assume that they reduce the production of SARS-CoV-2 by a factor (1-) with a constant efficacy . We compare the prediction of model (3) with the LRT and URT viral load data of a patient in South Korea who received lopinavir/ritonavir on day 5 after the onset of symptoms (Fig. 7B, C). The best fits show that the efficacy of lopinavir/ritonavir is 60% effective in reducing viral replication in the LRT while it does not have effect in the URT (Table 6). We also simulate the dynamics with different efficacies (60% in Fig. 8A vs. 90% in Fig. 8B) on 10th, 15th and 20th day, respectively. We find that a higher efficacy of antiviral drugs only makes viral load decline slightly faster. However, early treatment can significantly diminish the time to recovery.
Table 6.
Best fits of model (3) to the viral data of a patient treated with lopinavir/ritonavir.
| (ml/virus/day) | p (day−1) | c (day−1) | (ml/cell/day) | V(0) RNA/ml | AIC value | ||||
|---|---|---|---|---|---|---|---|---|---|
| URT | 9.610−8 | 7.7104 | 4.4 | 1.810−3 | 1.2 | 10−4 | 11 | 0 | −6.860464 |
| LRT | 910−9 | 8.5104 | 4.3 | 410−5 | 0.69 | 10−4 | 11 | 0.6 | 5.335016 |
Fig. 8.
Predicted viral dynamics under treatment. (A) The efficacy of antiviral drug is assumed to be and (B) . We performed simulation using the fit to the LRT viral load data of patient #2 in [14], [54] as an example. The treatment is assumed to be administrated on day 10, 15 and 20, respectively; (C) For comparison with (A) and (B), the value of is increased by 5 folds to represent IFN intensification. The antiviral drug efficacy is the same as that in (A), i.e. ; (D) Predicted dynamics under anti-inflammation therapy. The value of is fixed to be 104, 103, 102 and 10 cells ml−1 day−1, respectively.
SARS-CoV-2 is shown to be more sensitive to type I interferon (IFN) than SARS-CoV due to the absence of two important IFN antagonists [53]. We examine the impact of IFN as treatment intensification with the above antiviral therapy by increasing the killing due to the innate immune response (i.e. the value of in the model) by 5 folds. We find that increasing the killing by the innate immune response can largely suppress the viral replication (compare Fig. 8A,C). Under IFN treatment, the viral load could be significantly reduced with 60% drug efficacy (lopinavir/ritonavir or remdesivir). The result suggests that early treatment with a combination of IFN and other antiviral drugs might be promising against novel coronavirus infection.
The severe acute respiratory syndrome induced by SARS-CoV-2 infection is characterized by over-exuberant inflammatory response. Baricitinib, fedratinib and ruxolitinib that are known to inhibit JAK-STAT signaling pathway might reduce the levels of cytokines [3], [16]. Very recently, the National Institute of Health has started a clinical trial to test the combination of remdesivir and baricitinib in COVID-19 patients (ClinicalTrials.gov Identifier: NCT04401579). To describe the relieved inflammatory response that may be induced from the anti-inflammation treatment, we decrease the recruitment rate of lymphocytes () in the model. We find that the duration of the plateau phase will be significantly reduced as the recruitment rate decreases (Fig. 8D). The result implies that anti-inflammation drugs can accelerate viral decline because of the limited availability of the secondary target of infection.
4. Discussion
Within-host dynamics of SARS-CoV-2 infection have not been quantitatively investigated. We developed mathematical models to study the viral dynamics and compared modeling prediction with the viral load data in COVID-19 patients and non-human primates from different countries. Our study provides explanations to several pathogenic features of SARS-CoV-2 infection. Firstly, SARS-CoV-2’s very high transmission capacity is likely to come from a viral plateau phase after the peak, which provides a sustainable source of infectious viruses. The duration of the plateau stage is determined by how fast the seroconversion occurs and how strong the immune responses are on the basis of our data fitting and model simulations. Secondly, the model shows that the current best possible control of SARS-CoV-2 infection might be the adaptive immune response of the infected individual. This supports the legitimacy of current treatment with oxygen, mechanical ventilation, corticosteroids and antibiotics that let severely ill patient sustain the life till the activation of adaptive immunity. Thirdly, we explained the scenario of prolonged viral shedding and viral rebound. They originate from the oscillation caused by interplays between viral replication and the adaptive immune response. If seroconversion emerges late, then prolonged viral persistence is generated. As the adaptive immune response is above a threshold, the virus will be eradicated. These results may improve our understanding of the pathogenic features of SARS-CoV-2 infection.
Comparison of the prediction by the three models and the viral load data suggests that SARS-CoV-2 infection might have a secondary target. Without the second target cell population, the model shows a single-phase viral load decline after the peak, which does not agree with the data in many patients. The secondary target of SARS-CoV-2 infection might be lymphocytes. Lymphocytopenia is a predominant feature of COVID-19 patients in which a large number of viral RNAs within lymphocytes were detected [31], [33]. A recent study also found that SARS-CoV-2 can invade T lymphocytes via spike protein mediated membrane fusion [35]. The molecular mechanism underlying the entry of SARS-CoV-2 into lymphocytes is not fully understood. Lymphocytes do not express ACE2, but SARS-CoV-2 has a polybasic furin-type cleavage site at the S1–S2 junction in the spike protein, which can intensify the capacity of virus to penetrate tissues with low ACE2 expression [14], [89]. Lymphocytes may also be infected via the assistance by other co-receptors. The tropism of SARS-CoV could be altered towards human immune cells enhanced by Fc gamma II receptor [90]. The initiation of seroconversion in COVID-19 patients is slightly earlier than in SARS-CoV infected patients [14]. This might facilitate the entry of SARS-CoV-2 into lymphocytes in humans. In addition to lymphocytes, SARS-CoV-2 may also infect some other cells. The virus replicates to a level in the lung where it hits a threshold for which virus spills over into the blood, causing a viremia. This viremia will allow the virus to travel to the gut where it can further replicate, as gut enterocytes have the ACE2 receptor and have been reported to be infected in human COVID-19 infection [91].
Type I IFN was used to treat MERS-CoV, SARS-CoV, and SARS-CoV-2 infection [53], [92], [93]. The early treatment of MERS-CoV infection protected mice from lethal infection but late induction could result in serious pneumonia [92]. SARS-CoV-2 was shown to be more sensitive to type-I IFN treatment than SARS-CoV [53], [93]. Our simulation shows that the antiviral drug efficacy needs to be close to 90% to suppress viral replication effectively. However, intensification with interferon can more effectively reduce the duration of viral plateau with a 60% efficacy of the antiviral drug. Comparison of the genomic sequences of SARS-CoV-2 and SARS-CoV shows that some changes, such as the absence of open reading frame (ORF) 3b and a short truncation of ORF6 in SARS-CoV-2, can reduce the virus’ capacity to interfere with type I IFN [53], [93]. ORF3b is a 154 amino acid (AA) long protein that blocks IRF3 phosphorylation [94]. SARS-CoV-2 ORF3b has a premature terminal codon that produces a truncated protein of 22 AA. The N-terminal domain of ORF6 can disrupt karyopherin transportation of transcriptions factors like STAT1 [53], [95]. SARS-CoV-2 has a two amino acid truncation in its ORF6 [53]. These alterations might make SARS-CoV-2 infection more sensitive to IFN. IFN was also administrated with other drugs to treat COVID-19 patients but the outcomes have not been available [96]. Because IFN might activate immunity to attract more immune cells, further studies are needed to determine if IFN can exacerbate the cytokine storm and evaluate the effectiveness of using IFN with and without other antiviral drugs to treat SARS-CoV-2 infection in humans.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
HM is supported by the NSF grant DMS-1620957. ANB is supported by the University of Florida Clinical and Translational Science Institute , which is supported in part by the NIH National Center for Advancing Translational Sciences under award number UL1TR001427. LR is supported by the NSF grant DMS-1758290. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.mbs.2020.108438.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
S1 Text. Supporting information for “Fits of models (1) and (2) to patient data”.
Fits of models (1) and (2) to patient data.
References
- 1.Hui D.S., E. I.A., Madani T.A., Ntoumi F., Kock R., Dar O. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J. Med. Virol. 2020;92(4):401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X. Clinical characteristics of Coronavirus disease 2019 in China. New Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A Novel Coronavirus from patients with Pneumonia in China, 2019. New Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/s0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of Novel Coronavirus–infected Pneumonia. New Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H. First case of 2019 Novel Coronavirus in the United States. New Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naming the coronavirus disease (COVID-19) and the virus that causes it 2020. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it.
- 11.Kamel Boulos M.N., Geraghty E.M. Geographical tracking and mapping of coronavirus disease COVID-19/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic and associated events around the world: how 21st century GIS technologies are supporting the global fight against outbreaks and epidemics. Int. J. Health Geogr. 2020;19(1):8. doi: 10.1186/s12942-020-00202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phan L.T., Nguyen T.V., Luong Q.C., Nguyen T.V., Nguyen H.T., Le H.Q. Importation and human-to-human transmission of a Novel Coronavirus in Vietnam. New Engl. J. Med. 2020;382(9):872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. New Engl. J. Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 15.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–697. doi: 10.1016/s0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Justin S., Anne P., Ivan G., Catherine T., Olly O., Dan S. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet. 2020 doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting wuhan. Emerg Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS And MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23(2):130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500(7461):227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian Z., Travanty E.A., Oko L., Edeen K., Berglund A., Wang J. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am. J. Respir. Cell. Mol. Biol. 2013;48(6):742–748. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2013;110(40):16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from wuhan, China. Intensive Care Med. 2020 doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020 doi: 10.1016/s2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu H., Zhou J., Wong B.H., Li C., Chan J.F., Cheng Z.S. Middle east respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J. Infect. Dis. 2016;213(6):904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202(3):415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu W.J., Zhao M., Liu K., Xu K., Wong G., Tan W. T-cell immunity of SARS-CoV: Implications for vaccine development against MERS-CoV. Antiviral. Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Mao Y., Ju L., Zhang J., Liu Z., Zhou X. Detection and monitoring of SARS coronavirus in the plasma and peripheral blood lymphocytes of patients with severe acute respiratory syndrome. Clin. Chem. 2004;50(7):1237–1240. doi: 10.1373/clinchem.2004.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perlman S., Dandekar A.A. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5(12):917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Xu W., Hu G., Xia S., Sun Z., Liu Z. SARS-CoV-2 infects T lymphocytes through its spike protein-mediated membrane fusion. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020 doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phua J., Weng L., Ling L., Egi M., Lim C.M., Divatia J.V. Intensive care management of coronavirus disease 2019 (COVID-19): challenges and recommendations. Lancet Respir. Med. 2020 doi: 10.1016/s2213-2600(20)30161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., Sandt C.E.van.de. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nature Med. 2020 doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao X. COVID-19: immunopathology and its implications for therapy. Nature Rev. Immunol. 2020 doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020 doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lan L., Xu D., Ye G., Xia C., Wang S., Li Y. Positive RT-PCR test results in patients recovered from COVID-19. Jama. 2020 doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu T., Hu J., Kang M., Lin L., Zhong H., Xiao J. 2020. Transmission dynamics of 2019 novel coronavirus (2019-nCoV) bioRxiv. 2020.01.25.919787. [DOI] [Google Scholar]
- 44.Shen M., Peng Z., Xiao Y., Zhang L. 2020. Modelling the epidemic trend of the 2019 novel coronavirus outbreak in China. bioRxiv. 2020.01.23.916726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kucharski A.J., Russell T.W., Diamond C., Liu Y., Edmunds J., Funk S. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis. 2020 doi: 10.1016/s1473-3099(20)30144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang Y., Nie Y., Penny M. Transmission dynamics of the COVID-19 outbreak and effectiveness of government interventions: A data-driven analysis. J. Med. Virol. 2020 doi: 10.1002/jmv.25750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu J.T., Leung K., Bushman M., Kishore N., Niehus R., de Salazar P.M. Estimating clinical severity of COVID-19 from the transmission dynamics in wuhan, China. Nature Med. 2020 doi: 10.1038/s41591-020-0822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shim E., Tariq A., Choi W., Lee Y., Chowell G. Transmission potential and severity of COVID-19 in South Korea. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen T.M., Rui J., Wang Q.P., Zhao Z.Y., Cui J.A., Yin L. A mathematical model for simulating the phase-based transmissibility of a novel coronavirus. Infect. Dis. Poverty. 2020;9(1):24. doi: 10.1186/s40249-020-00640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J.Y., Ko J.H., Kim Y., Kim Y.J., Kim J.M., Chung Y.S. Viral load kinetics of SARS-CoV-2 infection in first two patients in Korea. J. Korean Med. Sci. 2020;35(7) doi: 10.3346/jkms.2020.35.e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lokugamage K.G., Hage A., Schindewolf C., Rajsbaum R., Menachery V.D. 2020. SARS-CoV-2 is sensitive to type I interferon pretreatment. bioRxiv. 2020.03.07.982264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Böhmer M.M., Buchholz U., Corman V.M., Hoch M., Katz K., Marosevic D.V. Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: a case series. Lancet Infect. Dis. 2020 doi: 10.1016/s1473-3099(20)30314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perelson A.S., Ribeiro R.M. Modeling the within-host dynamics of HIV infection. BMC Biol. 2013;11(1):96. doi: 10.1186/1741-7007-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuhn J.H., Li W., Choe H., Farzan M. Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus. Cell Mol Life Sci. 2004;61(21):2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuster G.M., Pfister O., Burkard T., Zhou Q., Twerenbold R., Haaf P. SARS-CoV2: should inhibitors of the renin–angiotensin system be withdrawn in patients with COVID-19? Eur. Heart J. 2020 doi: 10.1093/eurheartj/ehaa235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baig A.M., Khaleeq A., Ali U., Syeda H. Evidence of the COVID-19 virus targeting the CNS: Tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem Neurosci. 2020 doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- 59.Haagmans B.L., Kuiken T., Martina B.E., Fouchier R.A., Rimmelzwaan G.F., van Amerongen G. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat Med. 2004;10(3):290–293. doi: 10.1038/nm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/s2213-2600(20)30076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sweeney R.M., McAuley D.F. Acute respiratory distress syndrome. Lancet. 2016;388(10058):2416–2430. doi: 10.1016/s0140-6736(16)00578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manicone A.M. Role of the pulmonary epithelium and inflammatory signals in acute lung injury. Expert. Rev. Clin. Immunol. 2009;5(1):63–75. doi: 10.1586/177666x.5.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mason R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur. Respir. J. 2020;55(4) doi: 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 2020;25(10) doi: 10.2807/1560-7917.Es.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao Z., Xu Y., Sun C., Wang X., Guo Y., Qiu S. A systematic review of asymptomatic infections with COVID-19. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vukmanovic-Stejic M., Zhang Y., Cook J.E., Fletcher J.M., McQuaid A., Masters J.E. Human CD4+ CD25hi Foxp3+ regulatory T cells are derived by rapid turnover of memory populations in vivo. J. Clin. Invest. 2006;116(9):2423–2433. doi: 10.1172/jci28941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaur A., Mascio M.Di., Barabasz A., Rosenzweig M., McClure H.M., Perelson A.S. Dynamics of T- and B-Lymphocyte Turnover in a natural host of simian immunodeficiency virus. J. Virol. 2008;82(3):1084. doi: 10.1128/JVI.02197-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pawelek K.A., Huynh G.T., Quinlivan M., Cullinane A., Rong L., Perelson A.S. Modeling within-host dynamics of influenza virus infection including immune responses. PLoS Comput. Biol. 2012;8(6) doi: 10.1371/journal.pcbi.1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang S., Hottz P., Schechter M., Rong L. Modeling the slow CD4+ T cell decline in HIV-infected individuals. PLoS Comput. Biol. 2015;11(12) doi: 10.1371/journal.pcbi.1004665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Janeway Jr. C.A., Travers P., Walport M., Shlomchik M.J. 5th ed. Garland Science; New York: 2001. Immunobiology: The Immune System in Health and Disease. [Google Scholar]
- 71.Berg R.E., Forman J. The role of CD8 T cells in innate immunity and in antigen non-specific protection. Curr. Opin. Immunol. 2006;18(3):338–343. doi: 10.1016/j.coi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 72.Zhu J., Paul W.E. Paul WE CD4 T cells: fates, functions, and faults. Blood. 2008;112(5):1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gutcher I., Becher B. APC-Derived cytokines and T cell polarization in autoimmune inflammation. J. Clin. Invest. 2007;117(5):1119–1127. doi: 10.1172/JCI31720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ochs M., Nyengaard J.R., Jung A., Knudsen L., Voigt M., Wahlers T. The number of alveoli in the human lung. Am. J. Respir. Crit. Care Med. 2004;169(1):120–124. doi: 10.1164/rccm.200308-1107OC. [DOI] [PubMed] [Google Scholar]
- 75.KidsHealth, Your Lungs & Respiratory System. Available from: https://kidshealth.org/en/kids/lungs.html.
- 76.Castranova V., Rabovsky J., Tucker J.H., Miles P.R. The alveolar type II epithelial cell: a multifunctional pneumocyte. Toxicol. Appl. Pharmacol. 1988;93(3):472–483. doi: 10.1016/0041-008x(88)90051-8. [DOI] [PubMed] [Google Scholar]
- 77.Best K., Guedj J., Madelain V., de Lamballerie X., Lim S.-Y., Osuna C.E. Zika plasma viral dynamics in nonhuman primates provides insights into early infection and antiviral strategies. Proc. Natl. Acad. Sci. USA. 2017;114(33):8847–8852. doi: 10.1073/pnas.1704011114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McElreath R. CRC Press; 2018. Statistical Rethinking: A Bayesian Course with Examples in R and Stan. [Google Scholar]
- 79.Taddy M. McGraw-Hill Education; 2019. Business Data Science: Combining Machine Learning and Economics To Optimize, Automate, and Accelerate Business Decisions. [Google Scholar]
- 80.Du R.-H., Liang L.-R., Yang C.-Q., Wang W., Cao T.-Z., Li M. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: A prospective cohort study. Eur. Respir. J. 2020 doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID- 19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leung N.H.L., Chu D.K.W., Shiu E.Y.C., Chan K.-H., McDevitt J.J., Hau B.J.P. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat. Med. 2020 doi: 10.1038/s41591-020-0843-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yuan J., Kou S., Liang Y., Zeng J., Pan Y., Liu L. 2020. Clinical characteristics on 25 discharged patients with COVID-19 virus returning. medRxiv. 2020.03.06.20031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tan L., Kang X., Zhang B., Zheng S., Liu B., Yu T. 2020. A special case of COVID-19 with long duration of viral shedding for 49 days. medRxiv. 2020.03.22.20040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.lei l, Jian-ya G. Clinical characteristics of 51 patients discharged from hospital with COVID-19 in chongqing, China. medRxiv. 2020 doi: 10.1101/2020.02.20.20025536. 2020.02.20.20025536. [DOI] [Google Scholar]
- 86.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sham H.L., Kempf D.J., Molla A., Marsh K.C., Kumar G.N., Chen C.M. ABT-378, a highly potent inhibitor of the human immunodeficiency virus protease. Antimicrob .Agents Chemother. 1998;42(12):3218–3224. doi: 10.1128/aac.42.12.3218. Epub 1998/12/03. PubMed PMID: 9835517; PubMed Central PMCID: PMCPMC106025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11(4) doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jaume M., Yip M.S., Cheung C.Y., Leung H.L., Li P.H., Kien F. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcR pathway. J. Virol. 2011;85(20):10582–10597. doi: 10.1128/JVI.00671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lamers M.M., Beumer J., Vaart J.van.der., Knoops K., Puschhof J., Breugem T.I. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020 doi: 10.1126/science.abc1669. eabc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M. IFN-I Response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;130(9):3625–3639. doi: 10.1172/jci126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Totura A.L., Baric R.S. SARS Coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2(3):264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81(2):548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81(18):9812–9824. doi: 10.1128/jvi.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pang J., Wang M.X., Ang I.Y.H., Tan S.H.X., Lewis R.F., Chen J.I. Potential rapid diagnostics, vaccine and therapeutics for 2019 Novel Coronavirus (2019-nCoV): A systematic review. J. Clin. Med. 2020;9(3) doi: 10.3390/jcm9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fits of models (1) and (2) to patient data.