Summary
CD4+ T cells play an important role in the maturation of the antibody responses. Conjugation of identified CD4+ T cell helper epitope to the target antigen has been developed as a strategy to enhance vaccine-induced humoral immunity. In this work, we reported the identification of a novel HLA-IAb helper epitope LS-3 from Aquifex aeolicus. In silico analysis predicted this epitope to have high binding affinity to common human HLA alleles and have complementary binding coverage to the established PADRE epitope. Introduction of HLA-IAb knockout mutations to the LS-3 epitope significantly attenuated humoral responses induced by a vaccine containing this epitope. Finally, engineered fusion of the epitope to a model antigen, influenza hemagglutinin, significantly improved both binding and hemagglutination inhibition antibody responses in mice receiving DNA or protein vaccines. In summary, LS-3 and additional identified CD4+ helper epitopes may be further explored to improve vaccine responses in translational studies.
Subject Areas: Immunology, Cell Biology
Graphical Abstract
Highlights
-
•
Identification of a novel CD4+ helper epitope, LS-3, from Aquifex aeolicus
-
•
In silico analysis predicts high binding affinity of LS-3 to human HLA-DR alleles
-
•
Fusing LS-3 to antigen enhances humoral response by vaccinations
Immunology; Cell Biology
Introduction
Vaccination is an approach where antigenic materials are introduced into the hosts to elicit adaptive immune responses that may confer them with protection from subsequent pathogen exposure (Clem, 2011). Humoral immunity is an important branch of the adaptive immune system, in which antibodies produced by B cells serve either to directly neutralize targets on the pathogens through paratope-epitope interactions (Corti and Lanzavecchia, 2013; Kwong et al., 2013) or to indirectly mediate inactivation of the pathogens by engaging the complement system or effector cells such as macrophages and natural killer cells through Fc-dependent mechanisms (Kurdi et al., 2018; Seidel et al., 2013; van Erp et al., 2019). Antibody responses serve as an important correlate for protection for many emerging and re-emerging infectious diseases, including but not limited to HIV-1 (Burton and Hangartner, 2016), influenza (Laursen et al., 2018), and coronaviruses (Jiang et al., 2020). A strategy to enhance humoral responses induced by vaccination is, therefore, of great significance.
CD4+ T cells, particularly T-follicular helper (Tfh) cells, play a critical role in the maturation of antibody responses (Crotty, 2014). In the germinal center, immunological synapses are formed between Tfh and Germinal Center B (GCB) cells through interactions of pairs of adhesion molecules such as LFA1-ICAM-1 and SAP-Ly108 to enable transfer of soluble cytokines, such as IL-4 and IL-21, from Tfh to GCB cells and promote ligand-receptor interaction, such as CD40L-CD40 binding, to enhance survival, differentiation, somatic hypermutation, and class switching in the GCB cells (Carrasco et al., 2004; Elgueta et al., 2009; Flynn et al., 1998; Kageyama et al., 2012). Provision of T cell help, however, is contingent upon Tfh activation by GCB cells through T cell receptor (TCR) peptide-MHC II interaction (Zhang et al., 2013). As such, robust germinal center B cell responses are dependent on presentation of MHC II-restricted epitope, derived from the antigen, by GCB to Tfh cells. However, different epitopes have varying affinity for binding to MHC-II receptors depending on the hosts' haplotype such that peptide vaccines as well as smaller protein domains may not intrinsically contain a potent CD4+ helper epitope to drive germinal center responses (Elbahnasawy et al., 2018; Falugi et al., 2001; Pichichero, 2013). Such is the rationale for conjugating peptide and carbohydrate vaccines to protein carriers, like Keyhole limpet hemocyanin (KLH) (Ragupathi et al., 2002), tetanus toxin (Diethelm-Okita et al., 2000), or hepatitis B-surface antigen (HbsAg) (Collins et al., 2017). However, these large protein carriers may contain irrelevant immunodominant surfaces that may skew induced antibody responses away from the desired epitopes, creating additional uncertainties and challenges to this approach (Ghosh et al., 2013; Valea et al., 2018; Xu and Kulp, 2019).
Direct incorporation and fusion of a potent CD4+ helper epitope with the target antigen may be a simpler and more effective strategy to enhance the induced humoral immunity. Several important epitopes have been identified in this manner. Incorporation of Pan DR epitope (PADRE), for example, has demonstrated to improve immunogenicity of peptide and protein vaccines in animal studies, and it has also been explored in several clinical studies (Alexander et al., 2000; Ghaffari-Nazari et al., 2015; Snook et al., 2019). Identification of additional potent CD4-helper epitopes can create new tools to be used in conjunction with, or as alternative to, these established CD4-helper epitopes to increase responses induced by various vaccine antigens.
In our prior work, we used synthetic DNA delivery by electroporation to mediate in vivo assembly of nanoparticle vaccines and observed that some nanoparticle scaffolding domains (used to promote self-assembly of scaffolded antigens) could induce CD4+ T cell responses (Xu et al., 2020). In this work, we examined and performed epitope mapping on several bacterial or viral scaffold protein domains and determined that lumazine synthase (LS) from Aquifex aeolicus contained very potent CD4-helper epitopes for both BALB/c and C57BL/6 mice. LS can scaffold the assembly of 60 copies of HIV-priming antigen GT8, eOD-GT8-60mer, as well as other antigens (Jardine et al., 2016; Xu et al., 2020). In silico binding analysis determined that the identified C57BL/6 CD4-helper epitope (LS-3) was predicted also to have high binding affinity (<100 nM) to several common human MHC-II alleles (HLA-DRB1∗07:01, HLA-DRB1∗15:01, and HLA-DRB5∗01:01) and binds to a complementary set of HLA-DR alleles as compared with PADRE. We determined how this epitope might contribute to humoral immunity by engineering mutations that knocked out binding of this epitope to murine HLA I-Ab (LS3KO) and observed that DNA-launched GT8-60mer nanoparticles containing this mutant epitope (DLnano_CD4MutLS_GT8) induced weaker antibody responses than the corresponding DNA-launched wild-type GT8-60mer nanoparticles (DLnano_LS_GT8). Finally, engineered fusion of the identified LS-3 epitope to a different antigen, hemagglutinin (HA) receptor binding domain (RBD) from influenza H1/CA/07/09 (LS3-CA09), improved humoral responses induced to HA by DNA and protein vaccinations. Overall, this study provides a relatively rigorous demonstration that simple fusion of a dominant CD4-helper epitope to a target antigen could improve humoral responses induced by either protein or DNA vaccines in animal models and additionally describes the identification of a novel CD4-helper epitope from a bacterial enzyme, which may help inform the design of additional protein and DNA vaccines and be of translational importance.
Results
Identification of Novel Murine CD4-Helper Epitopes from the LS Domain of Aquifex aeolicus
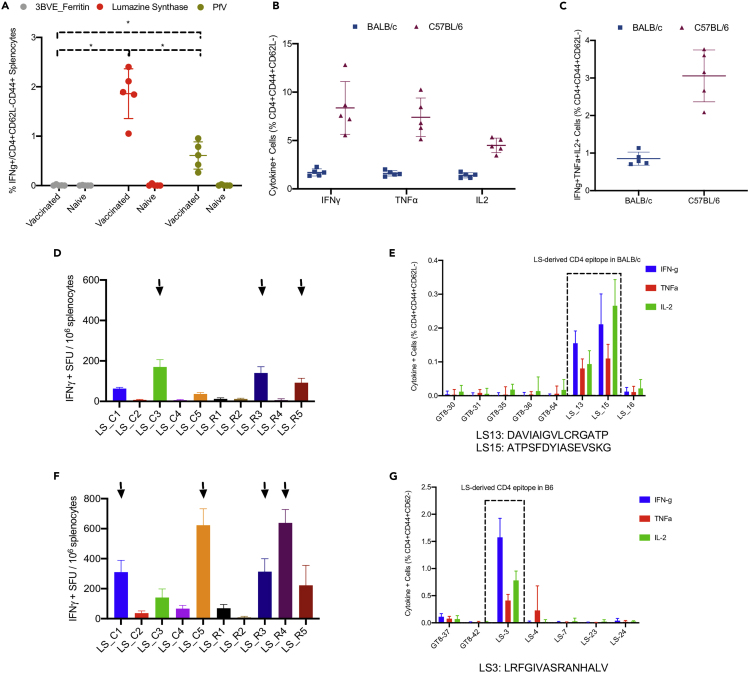
We previously observed that scaffold domains used to drive in vivo assembly of nanoparticle vaccines could sometimes induce CD4+ T cell responses (Xu et al., 2020). Here, we compared CD4+ T cell responses elicited by various nanoparticle scaffolding domains, ferritin from Helicobacter pylori (3BVE), LS from Aquifex aeolicus, and the viral cage of Prototype Foamy Virus (PfV), in BALB/c immunized with DNA-launched GT8 nanoparticle vaccines that incorporate these respective protein domains (DLnano_3BVE_GT8, DLnano_LS_GT8, DLnano_PfV_GT8) (Figure 1A). All mice in the experiments were immunized twice with 25 μg DNA immunogens 3 weeks apart and were euthanized 2 weeks post the second vaccination, at the time point that corresponded to their peak cellular responses. Using intracellular cytokine staining (ICS) to analyze murine splenocytes stimulated with overlapping peptide pools that spanned the respective protein domains, we determined that the LS domain elicited the most potent CD4+ T cell responses (approximately 2% of CD3+CD4+CD62L-CD44+ T cells were observed to IFNγ+ following peptide stimulation), followed by the PfV and the 3BVE domains. Importantly, DLnano_LS_GT8 vaccination elicited even more potent CD4+ T cell responses to the LS domain in the C57BL/6 mice than in the BALB/c mice, as measured by expression of pro-inflammatory cytokines IFNγ, TNF-α, and IL-2 upon peptide stimulation (Figure 1B). LS-specific poly-functional CD4+ T cell responses, as defined by the simultaneous expression of all three cytokines IFNγ, TNF-α, and IL-2, were induced in both the BALB/c and the C57BL/6 mice, accounting for approximately 1% and 3% of all CD3+CD4+CD62L-CD44 + T cells, respectively (Figure 1C). We set out to identify the exact CD4-helper epitope in both the BABL/c and the C57BL/6 mice, using a combination of an IFNγ ELIspot assay for screening and a flow-based ICS assay for confirmation. Two predominant non-overlapping CD4+ epitopes in the LS domain were observed for the BALB/c mice (LS-13: DAVIAIGVLCRGATP and LS-15: ATPSFDYIASEVSKG) (Figures 1D and 1E), whereas a single dominant CD4+ epitope in the LS domain was observed for the C57BL/6 mice (LS-3: LRFGIVASRANHALV) (Figures 1F and 1G). The overall CD4+ T cell responses measured by ICS were lower in the mapping study than in the previous experiment (Figures 1B, 1E and 1G), likely because fresh splenocytes were used for ICS analysis previously (Figure 1B), whereas splenocytes were used 24 h post-harvest in the mapping experiment because of the time required for the preliminary IFNγ ELIspot screen (Figures 1E and 1G). We also characterized additional epitopes identified through the preliminary IFNγ ELIspot screen (Figures S1A and S1B) by ICS and mapped the CD8+ T cell responses to two GT8 peptides in the BALB/c mice (Figure S1C and Table S1) and to one LS peptide in the C57BL/6 mice (Figure S1D and Table S1).
Figure 1.
Identification of LS-3 from Lumazine Synthase as a Potent CD4-Helper Epitope in C57BL/6 Mice
Mice received 25 μg DNA vaccination with EP twice 3 weeks apart and were euthanized 2 weeks post the second vaccination for cellular analysis.
(A) CD4+ T cell IFNγ responses induced to the 3BVE, LS, and PfV domains by DLnano_3BVE_GT8, DLnano_LS_GT8, and DLnano_PfV_GT8 vaccinations in BALB/c mice.
(B) Comparison of CD4+ cytokine responses to the LS domain induced by DLnano_LS_GT8 in BALB/c versus C57BL/6 mice.
(C) Comparison of polyfunctional CD4+ T cell responses to the LS domain induced by DLnano_LS_GT8 in BALB/c versus C57BL/6 mice.
(D and E) Matrix mapping by IFNγ ELISpot assays (D) and ICS (E) to determine HLA I-Ad CD4+ T cell epitopes in the LS domain in BALB/c mice immunized with DLnano_LS_GT8.
(F and G) Matrix mapping by IFNγ ELISpot assays (F) and ICS (G) to determine HLA I-Ab CD4+ T cell epitopes in the LS domain in C57BL/6 mice immunized with DLnano_LS_GT8. Each group includes five mice; each dot represents an animal; error bar represents standard deviation; two-tailed Mann-Whitney rank test used to compare groups; p values were adjusted for multiple comparison where appropriate; ∗, p value<0.05.
Murine HLA-IAb Epitope Was Predicted to Have High Binding Affinity for Several Human MHC-II Alleles by In Silico Analysis
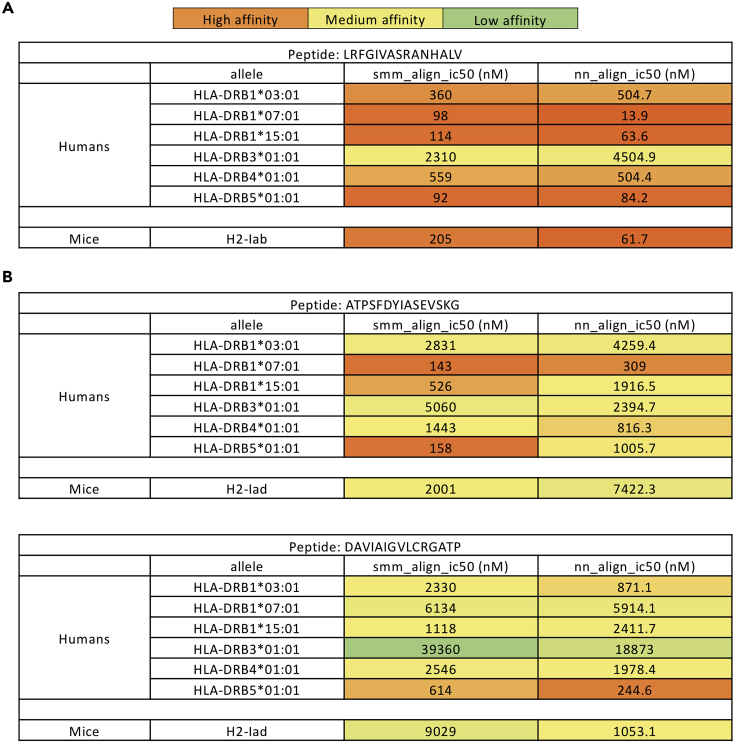
As the identified murine LS CD4-helper epitopes may or may not be conserved in humans, we used in silico analysis to predict the binding affinities of the identified LS-3, LS-13, and LS-15 epitopes to common human MHC-II alleles. Using a stabilization matrix method (SMM-align) and an artificial neural network-based method (NN-align) for alignment (Nielsen and Lund, 2009; Nielsen et al., 2007), the mapped murine C57BL/6 HLA-IAb epitope LS-3 demonstrated high binding affinity (<100 nM) for HLA-DRB1∗07:01, HLA-DRB1∗15:01, and HLA-DRB5∗01:01, which correspond to human allele frequencies of 6.98%, 7.86%, and 14.6% respectively (Louthrenoo et al., 2013; Solberg et al., 2008), and moderate binding affinity (<1,000 nM) for HLA-DRB1∗03:01 and HLA-DRB4∗01:01, which correspond to human allele frequencies of 6.76% and 35% respectively (Geng et al., 1995; Solberg et al., 2008). Low to moderate binding affinity (<5,000 nM) was observed for LS-3 to the human allele HLA-DRB3∗01:01 (Figure 2A). Of note, both the NN-align and the SMM-align correctly predicted high binding affinities of the LS-3 epitope to murine HLA-IAb. In contrast, the identified murine BALB/c HLA-IAd epitopes LS-13 and LS-15 were predicted to have lower binding affinities to either human or murine HLA alleles than the LS-3 epitope (Figure 2B). As such, since the LS-3 epitope was more likely to be conserved in humans, we decided to further characterize the HLA-IAb LS-3 epitope rather than the HLA-IAd LS-13/LS-15 epitopes in our downstream experiments.
Figure 2.
The LS-3 Epitope Was Observed to Have Potent Binding Affinities to Several Human HLA-DR Alleles with In Silico Analysis
Predicted binding affinity, in terms of IC50 value (nM), of the identified LS-3 (A), LS-13 and LS-15 (B) epitopes to common human and murine HLA alleles by NN- and SMM-align.
Identified Murine LS-3 CD4+ Helper Epitope Supported the Induction of Potent Immune Responses by DLnano_LS_GT8
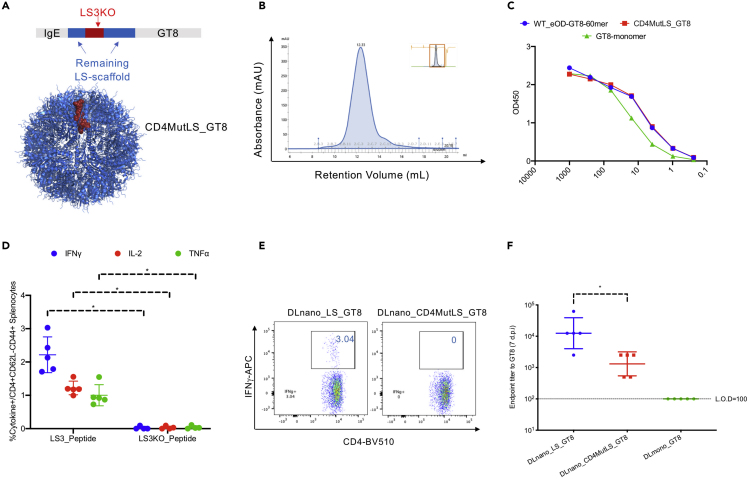
To determine whether CD4+ T cell help provided by the identified LS-3 epitope can contribute to the induction of humoral immunity by DLnano_LS_GT8, we engineered a GT8 nanoparticle variant (DLnano_CD4MutLS_GT8) through a structure-guided design process in which the LS-3 epitope was selectively mutated to ablate its binding to HLA-IAb (as informed by the NN-align and the SMM-align-based binding analysis). Care was taken, simultaneously, to avoid mutations that may disrupt nanoparticle assembly. We mutated 27% residues in the LS-3 epitope (4/15 residues) and generated the corresponding knockout epitope LS3-KO (Figure 3A and Table S1), resulting in reduction of HLA-IAb binding affinity from 205 to 4,261 nM by the SMM-align and 61.7 to 7,668 nM by the NN-align. We verified the engineered variant DLnano_CD4MutLS_GT8 incorporating the LS3-KO epitope could still assemble homogenously by expressing this new construct in vitro and performing size exclusion chromatography (SEC) of the lectin-column-purified DLnano_CD4MutLS_GT8 transfection supernatant. SEC showed CD4MutLS_GT8 assembled homogenously into 60-mer (single peak observed on the SEC trace centering at 12.33 mL retention volume) similar to what we previously observed for the wildtype eOD-GT8-60mer (Figure 3B) (Xu et al., 2020). Additionally, Size Exclusion Chromatography Multi Angle Light Scattering (SEC-MALs) analysis determined the molecular weight of CD4MutLS_GT8 to be around 2 MDa, close to the observed molecular weight of eOD-GT8-60mer (Figure S2A) (Xu et al., 2020). We examined the antigenic profiles of the engineered immunogens, and equivalent binding to VRC01, an HIV-1 broadly neutralizing antibody, was observed for eOD-GT8_60mer and CD4MutLS_GT8_60mer (Figure 3C). ICS analyses of mice immunized with respective DNA-encoded constructs confirmed complete knockout of the LS-3 CD4+ helper epitope in the CD4MutLS_GT8 construct (Figures 3D and 3E). Sera from animals seven d.p.i demonstrated significantly attenuated responses to GT8 in animals immunized with DLnano_CD4MutLS_GT8, although they still had stronger responses than those immunized with DNA-encoded GT8-monomer (Figure 3F). Differences in humoral immunity induced by DLnano_LS_GT8 and DLnano_CD4MutLS_GT8 waned overtime; however, repeat vaccination of DLnano_LS_GT8 but not DLnano_CD4MutLS_GT8 at 21 d.p.i boosted the humoral immunity in mice (Figure S2B). Taken together, this experiment suggests that the identified LS-3 CD4-helper epitope contributes to the overall antibody responses induced, as partial attenuation was observed when binding of this epitope to HLA IAb was knocked out.
Figure 3.
Knockout of the LS-3 Epitope from DLnano_LS_GT8 was Observed to Attenuate Vaccine-Induced Humoral Immunity
Mice received 25 μg DNA vaccination with EP twice 3 weeks apart and were euthanized 2 weeks post the second vaccination for cellular analysis.
(A) Engineering of CD4MutLS_GT8 mutants by selected mutations of the LS-3 epitope (in red) that knocked out C57BL/6 HLA-IAb binding but still preserve assembly of the nanoparticle using structure-guided design, the remaining LS domain is shown in blue, the GT8 domain is not shown.
(B) SEC-trace of lectin-column purified transfection supernatant of CD4MutLS_GT8 to determine the assembly status of designed CD4MutLS_GT8.
(C) Characterization of binding of recombinantly produced CD4MutLS_GT8, eOD-GT8-60mer, and GT8-mono to VRC01 by ELISA.
(D and E) Cytokine expression by the ICS assay in C57BL/6 mice immunized with either DLnano_LS_GT8 or DLnano_CD4MutLS_GT8 to confirm knockout of the dominant LS-3 CD4+ helper epitope in CD4MutLS_GT8.
(F) Humoral responses to GT8 for mice immunized with DLnano_CD4MutLS_GT8, DLnano_LS_GT8, or DLmono_GT8 7 d.p.i. Each group includes five mice; each dot represents a mouse; error bar represents standard deviation; two-tailed Mann-Whitney rank test used to compare groups; p values were adjusted for multiple comparison where appropriate; ∗, p value<0.05.
See also Figure S2.
Engineered Fusion of LS-3 CD4+ Helper Epitope to CA09 HA-RBD Enhanced Anti-HA Antibody Responses Induced by DNA or Protein Vaccines
As we have determined that CD4+ T cell help provided by the LS-3 epitope could contribute to the overall humoral responses, we set out to determine if it can serve as an incorporated CD4-helper epitope to enhance induced antibody responses by engineering fusion of the epitope with a different model antigen, CA09 HA-RBD.
We first set out to compare LS-3 and PADRE epitopes in terms of their biophysical properties and predicted binding affinities to HLA-DR alleles by in silico analysis. LS-3 and PADRE epitopes are predicted to have similar biophysical properties, in that they both have low water solubility, contain several basic residues, and are expected to be slightly positively charged at pH 7.0 (Figure S3A). We used the PADRE epitope to help validate the accuracy of HLA-binding affinity prediction, by comparing in silico predicted IC50 values with experimentally measured values (Figure S3B) (Alexander et al., 1994). Experimentally measured IC50 values were observed to trend in a very similar fashion to in silico predicted values for the binding of PADRE to HLA-DRB1∗07:01, HLA-DRB5∗01:01, HLA-DRB3∗01:01, and murine H2-Iab, particularly with the nn-align prediction algorithm. We next compared LS-3 and PADRE in terms of their binding affinities to various human HLA-DR alleles by computationally applying the MixMHCIIpred model, which has been recently reported (Figure S3C) (Racle et al., 2019). The MixMHCIIpred model incorporates features of smm- and nn-align and also uses a large dataset of empirically determined peptide-HLA-DR binding affinities to train a machine learning model, which was observed to have better performance, in terms of its receiver operating characteristic (ROC) curve in peptide HLA-DR binding prediction, than the classical NetMHCIIPan model. The LS-3 and PADRE epitopes were both predicted to have high binding affinities to several HLA-DR alleles, in terms of percentile rank (0–100, lower rank indicates stronger binding). Nevertheless, as compared with PADRE, LS-3 was observed to bind to several HLA-DR alleles more potently, particularly HLA-DRB1∗01:01, HLA-DRB1∗01:02, HLA-DRB1∗03:01, HLA-DRB1∗04:04, HLA-DRB1∗07:01, HLA-DRB3∗02:02, and HLA-DRB5∗02:02. In contrast, PADRE was observed to have higher binding affinity than LS-3 to other HLA-DR alleles, specifically HLA-DRB1∗04:01, HLA-DRB1∗04:08, and HLA-DRB1∗11:01. A negative control epitope, the first 15 residues of the human IgG1 Fc CH1 domain, was predicted to have low binding affinity (greater than 10% percentile rank) for all HLA-DR alleles, as expected.
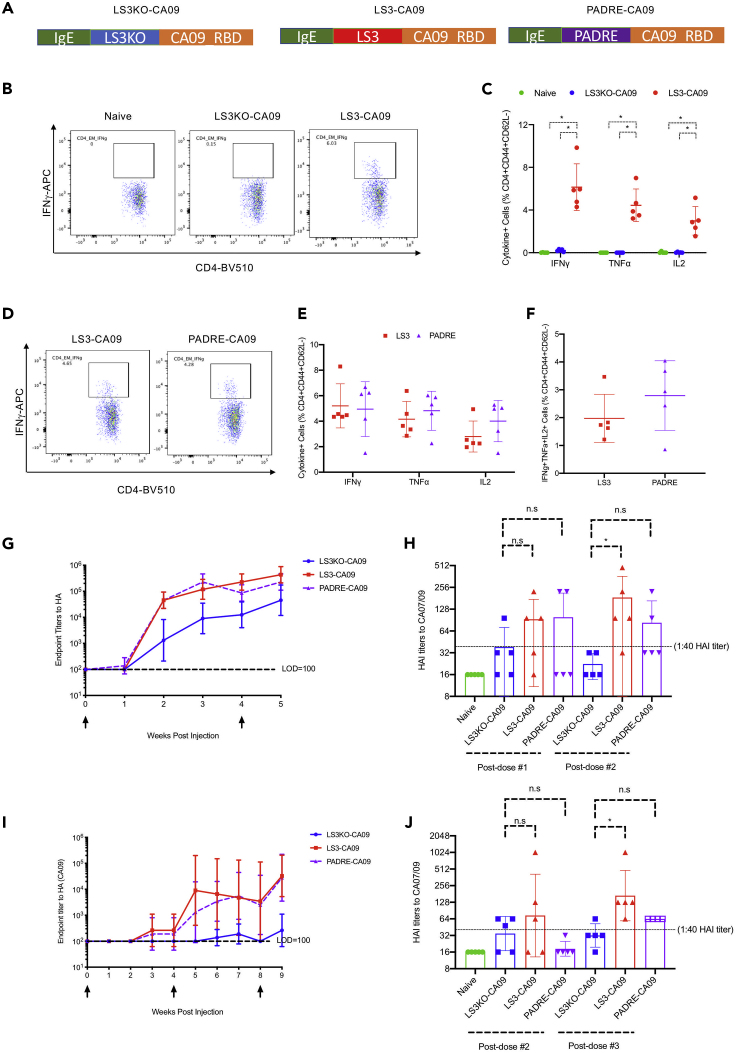
We incorporated either an LS3KO epitope, an LS3 epitope, or a PADRE epitope (AKFVAAWTLKAAA) on the N terminus of CA09 HA-RBD, downstream of the IgE leader sequence, which served as a secretion tag for the antigen (Figure 4A). LS3KO-CA09 served as a better control to which responses induced by LS3-CA09 and PADRE-CA09 would be compared, as the impact of N-terminal peptide fusion on the immunogenicity of an antigen would be considered (protein sequences of LS3KO-CA09 and LS3-CA09 only differed at four residues). First, we confirmed that DNA-encoded LS3-CA09 could induce CD4+ T cell responses to the incorporated LS-3 epitope. Indeed, by ICS analysis, C57BL/6 mice immunized with DNA-encoded LS3-CA09 but not those immunized with LS3KO-CA09 were capable of mounting CD4+ T cell responses to their respective incorporated epitope (Figures 4B and 4C). The finding was similarly validated by IFNγ ELIspot analysis (Figures S4A and S4B). Next, we compared CD4+ T cell responses induced by DNA-encoded LS3-CA09 and PADRE-CA09 to the LS3 and PADRE epitopes, respectively. Both LS-3 and PADRE elicited potent CD4+ T cell responses upon vaccination of DNA-encoded LS3-CA09 and PADRE-CA09 in C57BL/6 mice, with similar levels of cytokine responses induced as determined by ICS (Figures 4D and 4E) and IFNγ ELIspot assays (Figure S4c and S4d). Additionally, by ICS analysis, epitope-specific polyfunctional T cell responses were also similar between the LS-3 and PADRE epitopes (Figure 4F).
Figure 4.
Incorporation of Either PADRE or LS-3 Epitopes to a Model CA09 Influenza HA Immunogen (HA-RBD) Induced Epitope-Specific CD4+ T cell Responses and Improved Antigen-Specific Binding and HAI Antibody Responses
C57BL/6 mice received either 25 μg DNA vaccination with EP twice 4 weeks apart and were euthanized 1 week post the second vaccination or 10 μg RIBI-adjuvanted protein vaccinations three times 4 weeks apart and were euthanized 1 week post the third vaccination.
(A) Layouts of the engineered LS3-CA09, LS3KO-CA09, and PADRE-CA09 fusion constructs.
(B and C) Flow plots (B) and groups statistics (C) to compare CD4+ T cell cytokine responses induced by either DNA-encoded LS3-CA09 or LS3KO-CA09 immunizations in mice to LS3 and LS3KO peptides, respectively.
(D and E) Flow plots (D) and groups statistics (E) to compare CD4+ T cell cytokine responses induced by either DNA-encoded LS3-CA09 or PADRE-CA09 immunizations in mice to LS3 and PADRE peptides, respectively.
(F) Comparison of poly-functional IFNγ+TNFα+IL-2+ CD4+ T cell responses to either LS3 or PADRE peptides in mice immunized as described in (D) and (E).
(G and H) Comparison of anti-HA binding antibody responses (G) and HAI titers (H) in mice immunized with DNA-encoded LS3KO-CA09, LS3-CA09, or PADRE CA09.
(I and J) Comparison of anti-HA binding antibody responses (I) and HAI titers (J) in mice immunized with RIBI-adjuvanted protein LS3KO-CA09, LS3-CA09, or PADRE CA09. Each group includes five mice; each dot represents a mouse; error bar represents standard deviation; two-tailed Mann-Whitney rank test used to compare groups; p values were adjusted for multiple comparison where appropriate; ∗, p value<0.05.
See also Figures S3 and S4.
Next, we compared the humoral responses induced by two vaccinations of DNA-encoded LS3KO-CA09, LS3-CA09, and PADRE-CA09 to CA09 HA over time. By ELISA analysis, both DNA-encoded LS3-CA09 and PADRE-CA09 improved induced binding antibody responses to HA as compared with DNA-encoded LS3KO-CA09 prior to and after the boost, with approximately 9.5-fold and 5-fold improvements observed for DNA-encoded LS3-CA09 and PADRE-CA09, respectively (Figure 4G). Most importantly, we observed that functional antibody responses, as measured by the hemagglutination inhibition (HAI) titers, were significantly improved for DNA-encoded LS3-CA09 relative to LS3KO-CA09 after the second vaccination (with a mean titer of 126 versus 21, respectively) (Figure 4H). DNA-encoded PADRE-CA09, on the other hand, did not significantly improve the HAI titers relative to DNA-encoded LS3KO-CA09 after the first or the second vaccination (Figure 4H).
We further determined whether the observed phenomenon can be generalized to other routes of vaccination, such as protein vaccines. C-terminal his-tagged LS3KO-CA09, LS3-CA09, and PADRE-CA09 were expressed in vitro and purified from Expi293F cell transfection supernatant with nickel column. C57BL/6 mice were subsequently immunized with 10 μg recombinant protein LS3KO-CA09, LS3-CA09, or PADRE-CA09 co-formulated with RIBI each time. RIBI adjuvant, or Sigma Adjuvant System, is a stable water-in-oil emulsion with significantly lower local reactogenicity and can be used as an alternative to classical Freund's adjuvant (Lipman et al., 1992); it has also been demonstrated to induce higher IgG titers and a Th1-biased immune response in comparison with incomplete Freund's adjuvant (Chaitra et al., 2007), the latter of which has been used in several clinical trials (Apostolico Jde et al., 2016; Rosenberg et al., 2010). Humoral and cellular responses induced by protein vaccination were observed to be considerably lower than those induced by DNA vaccinations (Figures 4G and 4I), such that three protein vaccinations at weeks 0, 4, and 8 were required for us to observe robust humoral responses. Epitope-specific CD4+ T cell responses induced by protein vaccinations were considerably lower than that by DNA vaccines. However, CD4+ T cell responses directed at the LS-3 epitope could still be observed by ICS (Figure S4E) and by IFNγ ELIspot (Figure S4F and S4G). CD4+ T cell responses to PADRE were not observed (Figures S4E–S4G), likely as a result of the sensitivity of detection of the assays. Regardless, it was observed that HA-binding antibody titers induced by both protein LS3-CA09 and PADRE-CA09 vaccinations were significantly higher (100% sero-conversion in both groups) relative to protein LS3KO-CA09 vaccination, for which 20% sero-conversion (1/5 mice) was observed (Figure 4I). Lastly, similar to what was observed for DNA vaccinations, protein LS3-CA09 vaccination induced significantly improved HAI titers post-dose 3 relative to protein LS3KO-CA09 vaccination (with a mean titer of 169 versus 32, respectively). Protein PADRE-CA09 vaccination, on the other hand, was not observed to induce significantly improved HAI titers (Figure 4J). Taken together, the data suggest that the engineered fusion of the identified LS-3 CD4+ helper epitope to a model antigen can significantly enhance humoral responses to that antigen. Additionally, the incorporation of the LS-3 epitope performed as well as, if not better than, the incorporation of the PADRE epitope in terms of adjuvating humoral responses.
Discussion
The importance of CD4+ T cell help in facilitating antibody maturation and class switching is well established (Crum-Cianflone and Wallace, 2014). Patients with AIDS with low CD4+ T cell count cannot mount effective antibody responses with vaccination. Similarly, laboratory animals that receive transient CD4+ T cell depletion also cannot develop strong antibody responses to a foreign gene or an antigen (Duperret et al., 2018; Wise et al., 2020; Xu et al., 2018). Both secreted soluble cytokine factors as well as surface-displayed ligands from Tfh cells are indispensable to the survival, AID-dependent somatic hypermutation, and proliferation of GCB cells and are necessary for the generation of antibody-secreting plasma cells and long-lived memory B cells (Crotty, 2015).
Although larger antigenic protein domains likely harbor CD4+ T cell epitopes that can be restricted by the host HLA alleles for the induction of Tfh responses, carbohydrate and peptide vaccines are intrinsically minimalistic and unlikely to contain potent CD4+ T cell help epitopes (Astronomo and Burton, 2010). Additionally, domain minimization has now become an increasingly important approach in protein engineering and vaccinology, as researchers begin to appreciate the importance of focusing elicited B cell responses to certain target epitopes by designing protein mini-domain devoid of distracting immunodominant surfaces (van der Lubbe et al., 2018; Yassine et al., 2015). However, these mini-proteins contain fewer overlapping peptides and therefore statistically will be less likely to harbor potent HLA-restricted CD4+ helper epitopes. As such, several studies have explored conjugation of these carbohydrate, peptide, or mini-protein vaccines to carrier proteins, including but not limited to KLH, tetanus toxin, and HbsAg. Induction of more potent antibody responses was observed in many cases (Jin et al., 2017; Marini et al., 2019). However, this approach may undermine the core motivations behind domain minimization by introducing a host of immunodominant distracting surfaces that may skew induced humoral responses.
Conjugation of the antigen with a shorter conserved CD4+ T cell epitope may offer a promising alternative to conjugation with a whole protein carrier. As the CD4+ T cell epitope is intrinsically shorter (12–16 amino acid long), it will less represent a distracting immunodominant surface (Hemmer et al., 2000). Additionally, they may alternatively be used as short linker to connect different protein domains, such as to cross-link a nanoparticle protein scaffold with a target antigen to promote vaccine antigen self-assembly (He et al., 2018). Fusing antigen with the PADRE epitope has been demonstrated to improve antibody responses in several animal studies and has also been explored in the clinic (clinical trials NCT01972737 and NCT02264236) (Rosa et al., 2004). Additional CD4+ helper epitopes have also been mapped and explored. For example, a recent study reported that co-delivery of MPER antigen with a Leishmania major-derived HLA I-Ad helper CD4+ T cell epitope (LACK) in liposomes can improve induced anti-MPER antibody responses (Elbahnasawy et al., 2018).
In this study, we reported the identification and characterization of a novel HLA I-Ab epitope LS-3 from the LS domain of Aquifex aeolicus, which is also the protein domain that can be used to scaffold the assembly of a 60-mer nanoparticle. In silico analysis predicted the LS-3 epitope to have high binding affinity to several common human HLA alleles. Epitope knockout experiment demonstrated that CD4+ T cell help provided by this epitope could indeed contribute to the overall antibody responses. Additionally, with in silico analysis, the LS-3 and PADRE epitopes were predicted to bind to a complementary set of HLA-DR alleles, such that it may be envisioned for them to be used in conjunction to further increase breadth of HLA-DR coverage in diverse human populations. Finally, engineered genetic fusion of the LS-3 epitope with a different target antigen CA09 HA-RBD (LS3-CA09) significantly increased binding and HAI antibody titers elicited by protein and DNA vaccinations to HA as compared with the control antigen, LS3KO-CA09. The study demonstrates the potential utility in this epitope to increase vaccine-induced antibody responses, in both preclinical murine studies as well as possibly in translational vaccine trials.
Limitations of the Study
The main limitation in this study is that the binding affinity of the LS-3 epitope to human HLA alleles was predicted through in silico analysis, the accuracy of which was determined to be around 65% in one report (Andreatta et al., 2015), since the empirical in vitro human HLA binding assays were cost inhibitive. It should be noted, however, even with in vitro binding analysis, a human vaccine trial would be required to validate whether an identified epitope can indeed elicit CD4+ T cell responses in humans. Notably, the LS-domain scaffolded protein eOD-GT8-60mer is currently undergoing a phase I clinical study (NCT03547245), and additional insights may be obtained in the near future as to whether the LS domain, and particularly the LS-3 epitope, can elicit CD4+ T cell responses in the human population with varying HLA-allele distribution and be used to increase vaccine-induced humoral responses more broadly as described in this paper.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David B. Weiner, Email: dweiner@wistar.org. Address: 3601 Spruce St, Room 630, Philadelphia, PA 19104.
Materials Availability
All unique reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
No new code, software, algorithm, and large dataset were generated in this research.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This research is supported by NIH IPCAVD Grant U19 Al109646-04, NIH/NIAID Collaborative Influenza Vaccine Innovation Centers (CIVIC) contract 75N93019C00051, and Inovio Pharmaceuticals Vaccine Grant 5182101374 awarded to D.B.W, W. W. Smith Charitable Trust 68112-01-383 awarded to D.W.K, and by Wistar Monica H.M. Shander Memorial Fellowship awarded to Z.X.
Author Contributions
Z.X., D.B.W., and D.W.K. conceptualized the project. Z.X., D.B.W., and D.W.K. planned the experiments. Z.X., N.C., E.T.-R., and S.W. conducted the experiments. Z.X. analyzed the data. Z.X., N.C., D.B.W., and and D.W.K. wrote the paper.
Declaration of Interests
Z.X., D.W.K., and D.B.W. have a pending US patent on genetic fusion constructs with the identified LS-3 epitope. D.B.W. has received grant funding, participates in industry collaborations, has received speaking honoraria, and has received fees for consulting, including serving on scientific review committees and board series. Remuneration received by D.B.W. includes direct payments, stock or stock options, and in the interest of disclosure he notes potential conflicts associated with his work with Inovio and possible others.
Published: August 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101399.
Contributor Information
Daniel W. Kulp, Email: dwkulp@wistar.org.
David B. Weiner, Email: dweiner@wistar.org.
Supplemental Information
References
- Alexander J., del Guercio M.F., Maewal A., Qiao L., Fikes J., Chesnut R.W., Paulson J., Bundle D.R., DeFrees S., Sette A. Linear PADRE T helper epitope and carbohydrate B cell epitope conjugates induce specific high titer IgG antibody responses. J. Immunol. 2000;164:1625–1633. doi: 10.4049/jimmunol.164.3.1625. [DOI] [PubMed] [Google Scholar]
- Alexander J., Sidney J., Southwood S., Ruppert J., Oseroff C., Maewal A., Snoke K., Serra H.M., Kubo R.T., Sette A. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994;1:751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- Andreatta M., Karosiene E., Rasmussen M., Stryhn A., Buus S., Nielsen M. Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics. 2015;67:641–650. doi: 10.1007/s00251-015-0873-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolico Jde S., Lunardelli V.A., Coirada F.C., Boscardin S.B., Rosa D.S. Adjuvants: classification, modus operandi, and licensing. J. Immunol. Res. 2016;2016:1459394. doi: 10.1155/2016/1459394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astronomo R.D., Burton D.R. Carbohydrate vaccines: developing sweet solutions to sticky situations? Nat. Rev. Drug Discov. 2010;9:308–324. doi: 10.1038/nrd3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Hangartner L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco Y.R., Fleire S.J., Cameron T., Dustin M.L., Batista F.D. LFA-1/ICAM-1 interaction lowers the threshold of B cell activation by facilitating B cell adhesion and synapse formation. Immunity. 2004;20:589–599. doi: 10.1016/s1074-7613(04)00105-0. [DOI] [PubMed] [Google Scholar]
- Chaitra M.G., Nayak R., Shaila M.S. Modulation of immune responses in mice to recombinant antigens from PE and PPE families of proteins of Mycobacterium tuberculosis by the Ribi adjuvant. Vaccine. 2007;25:7168–7176. doi: 10.1016/j.vaccine.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Clem A.S. Fundamentals of vaccine immunology. J. Glob. Infect. Dis. 2011;3:73–78. doi: 10.4103/0974-777X.77299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K.A., Snaith R., Cottingham M.G., Gilbert S.C., Hill A.V.S. Enhancing protective immunity to malaria with a highly immunogenic virus-like particle vaccine. Sci. Rep. 2017;7:46621. doi: 10.1038/srep46621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu. Rev. Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. A brief history of T cell help to B cells. Nat. Rev. Immunol. 2015;15:185–189. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crum-Cianflone N.F., Wallace M.R. Vaccination in HIV-infected adults. AIDS Patient Care STDS. 2014;28:397–410. doi: 10.1089/apc.2014.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diethelm-Okita B.M., Okita D.K., Banaszak L., Conti-Fine B.M. Universal epitopes for human CD4+ cells on tetanus and diphtheria toxins. J. Infect. Dis. 2000;181:1001–1009. doi: 10.1086/315324. [DOI] [PubMed] [Google Scholar]
- Duperret E.K., Trautz A., Stoltz R., Patel A., Wise M.C., Perales-Puchalt A., Smith T., Broderick K.E., Masteller E., Kim J.J. Synthetic DNA-encoded monoclonal antibody delivery of anti-CTLA-4 antibodies induces tumor shrinkage in vivo. Cancer Res. 2018;78:6363–6370. doi: 10.1158/0008-5472.CAN-18-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbahnasawy M.A., Donius L.R., Reinherz E.L., Kim M. Co-delivery of a CD4 T cell helper epitope via covalent liposome attachment with a surface-arrayed B cell target antigen fosters higher affinity antibody responses. Vaccine. 2018;36:6191–6201. doi: 10.1016/j.vaccine.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgueta R., Benson M.J., de Vries V.C., Wasiuk A., Guo Y., Noelle R.J. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol. Rev. 2009;229:152–172. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falugi F., Petracca R., Mariani M., Luzzi E., Mancianti S., Carinci V., Melli M.L., Finco O., Wack A., Di Tommaso A. Rationally designed strings of promiscuous CD4(+) T cell epitopes provide help to Haemophilus influenzae type b oligosaccharide: a model for new conjugate vaccines. Eur. J. Immunol. 2001;31:3816–3824. doi: 10.1002/1521-4141(200112)31:12<3816::AID-IMMU3816>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Flynn S., Toellner K.M., Raykundalia C., Goodall M., Lane P. CD4 T cell cytokine differentiation: the B cell activation molecule, OX40 ligand, instructs CD4 T cells to express interleukin 4 and upregulates expression of the chemokine receptor, Blr-1. J. Exp. Med. 1998;188:297–304. doi: 10.1084/jem.188.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L., Imanishi T., Tokunaga K., Zhu D., Mizuki N., Xu S., Geng Z., Gojobori T., Tsuji K., Inoko H. Determination of HLA class II alleles by genotyping in a Manchu population in the northern part of China and its relationship with Han and Japanese populations. Tissue Antigens. 1995;46:111–116. doi: 10.1111/j.1399-0039.1995.tb02486.x. [DOI] [PubMed] [Google Scholar]
- Ghaffari-Nazari H., Tavakkol-Afshari J., Jaafari M.R., Tahaghoghi-Hajghorbani S., Masoumi E., Jalali S.A. Improving multi-epitope long peptide vaccine potency by using a strategy that enhances CD4+ T help in BALB/c mice. PLoS One. 2015;10:e0142563. doi: 10.1371/journal.pone.0142563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Solanki A.K., Roy K., Dhoke R.R., Ashish, Roy S. Carrier protein influences immunodominance of a known epitope: implication in peptide vaccine design. Vaccine. 2013;31:4682–4688. doi: 10.1016/j.vaccine.2013.06.110. [DOI] [PubMed] [Google Scholar]
- He L., Kumar S., Allen J.D., Huang D., Lin X., Mann C.J., Saye-Francisco K.L., Copps J., Sarkar A., Blizard G.S. HIV-1 vaccine design through minimizing envelope metastability. Sci. Adv. 2018;4:eaau6769. doi: 10.1126/sciadv.aau6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmer B., Kondo T., Gran B., Pinilla C., Cortese I., Pascal J., Tzou A., McFarland H.F., Houghten R., Martin R. Minimal peptide length requirements for CD4(+) T cell clones--implications for molecular mimicry and T cell survival. Int. Immunol. 2000;12:375–383. doi: 10.1093/intimm/12.3.375. [DOI] [PubMed] [Google Scholar]
- Jardine J.G., Kulp D.W., Havenar-Daughton C., Sarkar A., Briney B., Sok D., Sesterhenn F., Ereno-Orbea J., Kalyuzhniy O., Deresa I. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Gibani M.M., Moore M., Juel H.B., Jones E., Meiring J., Harris V., Gardner J., Nebykova A., Kerridge S.A. Efficacy and immunogenicity of a Vi-tetanus toxoid conjugate vaccine in the prevention of typhoid fever using a controlled human infection model of Salmonella Typhi: a randomised controlled, phase 2b trial. Lancet. 2017;390:2472–2480. doi: 10.1016/S0140-6736(17)32149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R., Cannons J.L., Zhao F., Yusuf I., Lao C., Locci M., Schwartzberg P.L., Crotty S. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity. 2012;36:986–1002. doi: 10.1016/j.immuni.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdi A.T., Glavey S.V., Bezman N.A., Jhatakia A., Guerriero J.L., Manier S., Moschetta M., Mishima Y., Roccaro A., Detappe A. Antibody-dependent cellular Phagocytosis by macrophages is a novel mechanism of action of elotuzumab. Mol. Cancer Ther. 2018;17:1454–1463. doi: 10.1158/1535-7163.MCT-17-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Mascola J.R., Nabel G.J. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat. Rev. Immunol. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- Laursen N.S., Friesen R.H.E., Zhu X., Jongeneelen M., Blokland S., Vermond J., van Eijgen A., Tang C., van Diepen H., Obmolova G. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science. 2018;362:598–602. doi: 10.1126/science.aaq0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman N.S., Trudel L.J., Murphy J.C., Sahali Y. Comparison of immune response potentiation and in vivo inflammatory effects of Freund's and RIBI adjuvants in mice. Lab. Anim. Sci. 1992;42:193–197. [PubMed] [Google Scholar]
- Louthrenoo W., Kasitanon N., Wichainun R., Wangkaew S., Sukitawut W., Ohnogi Y., Hong G.H., Kuwata S., Takeuchi F. The genetic contribution of HLA-DRB5∗01:01 to systemic lupus erythematosus in Thailand. Int. J. Immunogenet. 2013;40:126–130. doi: 10.1111/j.1744-313X.2012.01145.x. [DOI] [PubMed] [Google Scholar]
- Marini A., Zhou Y., Li Y., Taylor I.J., Leneghan D.B., Jin J., Zaric M., Mekhaiel D., Long C.A., Miura K. A universal plug-and-display vaccine carrier based on HBsAg VLP to maximize effective antibody response. Front. Immunol. 2019;10:2931. doi: 10.3389/fimmu.2019.02931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Lundegaard C., Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007;8:238. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichichero M.E. Protein carriers of conjugate vaccines: characteristics, development, and clinical trials. Hum. Vaccin. Immunother. 2013;9:2505–2523. doi: 10.4161/hv.26109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racle J., Michaux J., Rockinger G.A., Arnaud M., Bobisse S., Chong C., Guillaume P., Coukos G., Harari A., Jandus C. Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat. Biotechnol. 2019;37:1283–1286. doi: 10.1038/s41587-019-0289-6. [DOI] [PubMed] [Google Scholar]
- Ragupathi G., Cappello S., Yi S.S., Canter D., Spassova M., Bornmann W.G., Danishefsky S.J., Livingston P.O. Comparison of antibody titers after immunization with monovalent or tetravalent KLH conjugate vaccines. Vaccine. 2002;20:1030–1038. doi: 10.1016/s0264-410x(01)00451-0. [DOI] [PubMed] [Google Scholar]
- Rosa D.S., Tzelepis F., Cunha M.G., Soares I.S., Rodrigues M.M. The pan HLA DR-binding epitope improves adjuvant-assisted immunization with a recombinant protein containing a malaria vaccine candidate. Immunol. Lett. 2004;92:259–268. doi: 10.1016/j.imlet.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Rosenberg S.A., Yang J.C., Kammula U.S., Hughes M.S., Restifo N.P., Schwarz S.L., Morton K.E., Laurencot C.M., Sherry R.M. Different adjuvanticity of incomplete freund's adjuvant derived from beef or vegetable components in melanoma patients immunized with a peptide vaccine. J. Immunother. 2010;33:626–629. doi: 10.1097/CJI.0b013e3181dac9de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel U.J., Schlegel P., Lang P. Natural killer cell mediated antibody-dependent cellular cytotoxicity in tumor immunotherapy with therapeutic antibodies. Front. Immunol. 2013;4:76. doi: 10.3389/fimmu.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook A.E., Baybutt T.R., Xiang B., Abraham T.S., Flickinger J.C., Jr., Hyslop T., Zhan T., Kraft W.K., Sato T., Waldman S.A. Split tolerance permits safe Ad5-GUCY2C-PADRE vaccine-induced T-cell responses in colon cancer patients. J. Immunother. Cancer. 2019;7:104. doi: 10.1186/s40425-019-0576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg O.D., Mack S.J., Lancaster A.K., Single R.M., Tsai Y., Sanchez-Mazas A., Thomson G. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum. Immunol. 2008;69:443–464. doi: 10.1016/j.humimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valea I., Adjei S., Usuf E., Traore O., Ansong D., Tinto H., Owusu Boateng H., Leach A., Mwinessobaonfou Some A., Buabeng P. Immune response to the hepatitis B antigen in the RTS,S/AS01 malaria vaccine, and co-administration with pneumococcal conjugate and rotavirus vaccines in African children: a randomized controlled trial. Hum. Vaccin. Immunother. 2018;14:1489–1500. doi: 10.1080/21645515.2018.1442996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lubbe J.E.M., Verspuij J.W.A., Huizingh J., Schmit-Tillemans S.P.R., Tolboom J., Dekking L., Kwaks T., Brandenburg B., Meijberg W., Zahn R.C. Mini-HA is superior to full length hemagglutinin immunization in inducing stem-specific antibodies and protection against group 1 influenza virus challenges in mice. Front. Immunol. 2018;9:2350. doi: 10.3389/fimmu.2018.02350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp E.A., Luytjes W., Ferwerda G., van Kasteren P.B. Fc-Mediated antibody effector functions during respiratory syncytial virus infection and disease. Front. Immunol. 2019;10:548. doi: 10.3389/fimmu.2019.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise M.C., Xu Z., Tello-Ruiz E., Beck C., Trautz A., Patel A., Elliott S.T., Chokkalingam N., Kim S., Kerkau M.G. In vivo delivery of synthetic DNA-encoded antibodies induces broad HIV-1-neutralizing activity. J. Clin. Invest. 2020;130:827–837. doi: 10.1172/JCI132779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Kulp D.W. Protein engineering and particulate display of B-cell epitopes to facilitate development of novel vaccines. Curr. Opin. Immunol. 2019;59:49–56. doi: 10.1016/j.coi.2019.03.003. [DOI] [PubMed] [Google Scholar]
- Xu Z., Wise M.C., Choi H., Perales-Puchalt A., Patel A., Tello-Ruiz E., Chu J.D., Muthumani K., Weiner D.B. Synthetic DNA delivery by electroporation promotes robust in vivo sulfation of broadly neutralizing anti-HIV immunoadhesin eCD4-Ig. EBioMedicine. 2018;35:97–105. doi: 10.1016/j.ebiom.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wise M.C., Chokkalingam N., Walker S., Tello-Ruiz E., Elliott S.T.C., Perales-Puchalt A., Xiao P., Zhu X., Pumroy R.A. In vivo assembly of nanoparticles achieved through synergy of structure-based protein engineering and synthetic DNA generates enhanced adaptive immunity. Adv. Sci. (Weinh) 2020;7:1902802. doi: 10.1002/advs.201902802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine H.M., Boyington J.C., McTamney P.M., Wei C.J., Kanekiyo M., Kong W.P., Gallagher J.R., Wang L., Zhang Y., Joyce M.G. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- Zhang X., Ing S., Fraser A., Chen M., Khan O., Zakem J., Davis W., Quinet R. Follicular helper T cells: new insights into mechanisms of autoimmune diseases. Ochsner J. 2013;13:131–139. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new code, software, algorithm, and large dataset were generated in this research.