Abstract
Background
Currently, physicians are limited in their ability to provide an accurate prognosis for COVID-19 positive patients. Existing scoring systems have been ineffective for identifying patient decompensation. Machine learning (ML) may offer an alternative strategy. A prospectively validated method to predict the need for ventilation in COVID-19 patients is essential to help triage patients, allocate resources, and prevent emergency intubations and their associated risks.
Methods
In a multicenter clinical trial, we evaluated the performance of a machine learning algorithm for prediction of invasive mechanical ventilation of COVID-19 patients within 24 h of an initial encounter. We enrolled patients with a COVID-19 diagnosis who were admitted to five United States health systems between March 24 and May 4, 2020.
Results
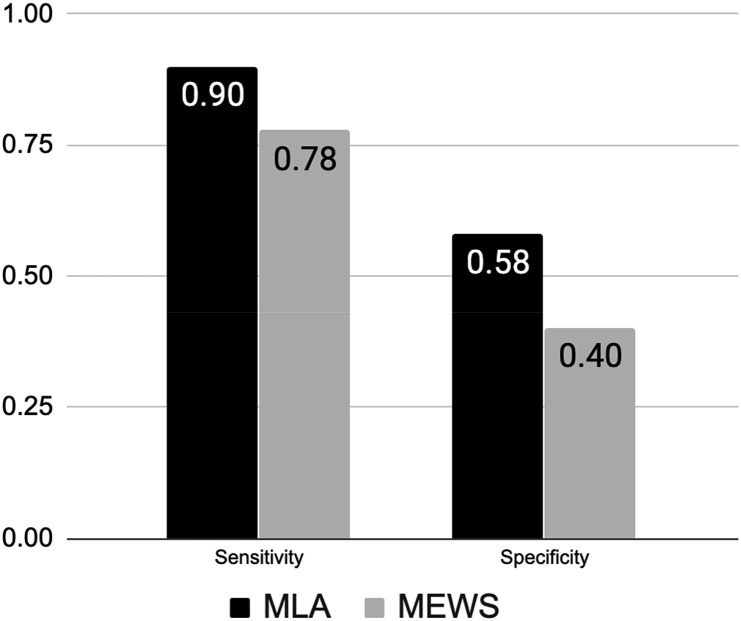
197 patients were enrolled in the REspirAtory Decompensation and model for the triage of covid-19 patients: a prospective studY (READY) clinical trial. The algorithm had a higher diagnostic odds ratio (DOR, 12.58) for predicting ventilation than a comparator early warning system, the Modified Early Warning Score (MEWS). The algorithm also achieved significantly higher sensitivity (0.90) than MEWS, which achieved a sensitivity of 0.78, while maintaining a higher specificity (p < 0.05).
Conclusions
In the first clinical trial of a machine learning algorithm for ventilation needs among COVID-19 patients, the algorithm demonstrated accurate prediction of the need for mechanical ventilation within 24 h. This algorithm may help care teams effectively triage patients and allocate resources. Further, the algorithm is capable of accurately identifying 16% more patients than a widely used scoring system while minimizing false positive results.
Keywords: Machine learning, COVID-19, Mechanical ventilation, Prediction
Highlights
-
•
Validation of prediction algorithm for ventilation requirements in COVID-19 patients.
-
•
Algorithm achieved significantly higher sensitivity than the common scoring system MEWS.
-
•
Algorithm detected 16% more patients who will require invasive ventilation than MEWS.
-
•
Advance warning of ventilation needs can help improve COVID-19 patient outcomes.
1. Introduction
COVID-19, caused by the novel coronavirus SARS-CoV-2, remains a public health emergency in the United States. The rapidly evolving evidence surrounding pharmaceutical treatments and the lack of established preventive resources has made the effective triage of COVID-19 patients challenging. Prognostic scores such as the Modified Early Warning Score (MEWS) [1] guide decision-making for the non-COVID-19 critically ill population [2]. However, literature examining the ability of these scoring systems to predict COVID-19 patient prognosis and mortality is limited, and recent research has suggested that discriminatory ability of such rules-based scores is moderate to poor [3].
Epidemiologic predictions indicate that hospitals will continue to see large numbers of COVID-19 patients in the coming months [[4], [5], [6]]. Patient triage will remain important to facilitate the effective allocation of limited resources. Early identification of patients who are at risk of decompensation and who are likely to need mechanical ventilation would enable physicians to more aggressively monitor these patients, which may facilitate a more controlled environment for intubation. Inadequate lead time and subsequent emergency intubation of critically ill patients is associated with known risks, including peri-intubation hypoxia, hypotension, arrhythmia, and cardiac arrest [7].
In an effort to address this growing need, researchers have begun to develop machine learning (ML)-based models for risk prediction critical illness development in COVID-19 patients. Liang et al. [8] developed such a model and achieved strong performance in predicting a composite outcome including admission to the intensive care unit (ICU), invasive ventilation, or death, and reported an area under the curve (AUC) of 0.88. However, the model was only evaluated retrospectively. Although retrospective studies are useful for providing preliminary data and for guiding future research, many of these analyses are subject to threats in internal validity [9,10]. Studies often fail to be replicated in prospective clinical settings, leaving uncertainty regarding the performance and the utility of the intervention in a live clinical setting [11,12].
To assess how ML risk prediction models may assist with caring for COVID-19 patients in a clinical setting, we have performed the first prospective validation of a machine learning algorithm for the prediction of mechanical ventilation requirements in a COVID-19 positive population. In the READY clinical trial, we assessed the performance of a previously developed algorithm at five US health systems. All predictions were made two hours after the start of the patient encounter using patient data obtained within the first two hours of an emergency department (ED) visit. If the patient did not originate in the ED, data as used from the first two hours of hospital admission. The algorithm predicted the need for mechanical ventilation within the next 24 h. Performance was compared to patient evaluation using MEWS, a score commonly used to identify likely patient deterioration and mortality. The primary endpoint of the study was mechanical ventilation within 24 h of the prediction.
The remainder of this paper is organized as follows. Section 2 contains the study methods, including patient enrollment and data processing. Section 3 contains the study results. Section 4 contains the discussion, including study limitations. Section 5 contains the conclusion of the study.
2. Methods
2.1. Patient enrollment
Patients who enrolled in the READY clinical trial visited the emergency department or were admitted to the hospital at five U.S. hospitals between March 24, 2020 and May 4, 2020. Patients were eligible for inclusion in the READY clinical trial if their first set of vital sign and lab measurements were taken within two hours of ED arrival or admission, and if they tested positive for COVID-19 by polymerase chain reaction (PCR) during their visit (Fig. 1 ). In total, 197 patients were eligible for inclusion in our study. We enrolled all eligible patients that were admitted during the study period.
Fig. 1.
Patient inclusion flowchart.
Upon admission of an eligible patient to the ED or hospital, data collection of available vital sign and lab measurements began automatically. The first two hours of data were used to calculate both the machine learning algorithm risk prediction score and the comparison score (MEWS). Additional details of the implementation of the algorithm are provided as a process state diagram in Supplementary Fig. 1. All data were automatically retrieved from the electronic health record (EHR) without requiring clinician intervention. Algorithm predictions were automatically calculated and were made available to clinicians at the hospital to assist with patient care. The outcome of interest was COVID-19 patient decompensation leading to mechanical ventilation, defined as invasive ventilation requiring endotracheal tube or mechanical ventilation not including BIPAP or CPAP. This outcome was assessed 24 h after model predictions were made. Those who were ventilated were considered to be the positive class; all others were considered to be the negative class.
This study is considered to be of minimal risk for human subjects as data collection was passive and did not pose a threat to the subjects involved. All patient data was maintained in compliance with the Health Insurance Portability and Accountability Act (HIPAA). The project was approved by the Pearl Institutional Review Board with a waiver of informed consent under study number 20-DASC-122, and is registered on ClinicalTrials.gov under study number NCT04390516.
2.2. Data processing
The model was created using the XGBoost Classifier method for fitting “boosted” decision trees in Python [13]. Gradient boosting, which XGBoost implements, is an ensemble learning technique that combines results from multiple decision trees to create prediction scores. Each tree splits the patient population into smaller and smaller groups, successively. Each branch splits the patients who enter it into two groups, based on whether their value of some feature is above or below some threshold. For instance, a branch might divide patients according to whether they are male or female, then on the female branch whether their creatinine is above or below 0.97 mg/dl, the average creatinine level for women. If creatinine is above average, then the patient will continue to travel down the higher risk branch; if the creatinine value is absent, the algorithm will choose the default branch that results in more correctly classified patients in the training data; this may default to the low risk or high risk branch depending on the training data. This may default to the low risk or high risk branch depending on the training data. After some number of branches, the tree ends in a set of “leaves.” Each patient falls into exactly one leaf, according to the values of his or her measurements.
Model predictions were generated based only on measurements taken in the first two hours after ED arrival or hospital admission. Information on patient demographics was extracted from the EHR. Diagnosis of acute conditions present during the patient's hospital stay, including acute kidney injury (AKI), sepsis, pneumonia, and acute respiratory distress syndrome (ARDS), were assessed by International Classification of Diseases (ICD)-10 code.
For each patient, exactly 12 values were given to the model. These values were diastolic blood pressure (DBP), systolic blood pressure (SBP), heart rate (HR), temperature, respiratory rate (RR), oxygen saturation (SpO2), white blood cell (WBC), platelet count, lactate, blood urea nitrogen (BUN), creatinine, and bilirubin (Supplementary Table 1). Missing values were left as “Not a Number” or empty placeholders, which are valid inputs to the model. Model prediction scores were therefore able to be calculated in the presence of missing data without imputing missing measurements. Specifically, each node in the decision tree has a default direction that should be traversed in the event that the feature in that node is missing. Imputation of missing measurements was therefore not performed. The model was trained prior to patient enrollment in this study, with training performed on a data set obtained from a separate hospital. Additional details of model training are provided in the supplementary materials and in Supplementary Table 1.
The extracted measurements were also used to generate MEWS comparator scores for each patient. MEWS was calculated at the same time as the algorithm score and were calculated only when all score inputs were available. The algorithm returned prediction scores as probabilities (between 0 and 1) that the patient would require mechanical ventilation within 24 h. MEWS scores were returned as integers, which were converted to probabilities by scaling down by the maximum score in the data set. For example, given a maximum MEWS score of 14, a score of 7 would be scaled down as 7/14 for a probability of 0.50.
Performance metrics were calculated for the algorithm's ability to predict ventilation within 24 h. Reported metrics included sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio. Area under the receiver operating characteristic (AUC) was calculated retrospectively based on the probabilities that were output by the ML algorithm. The ML algorithm was assessed against the MEWS comparator using McNemar's test with an alpha of 0.05.
3. Results
In total, 197 patients who received a positive diagnosis of COVID-19 were included in the study. Of these patients, 10 were placed on mechanical ventilation within 24 h of the algorithm's prediction. Compared to the general patient population, those who tested positive for COVID-19 were likely to be older, more likely to be male, and more likely to receive an in-hospital diagnosis of acute respiratory distress syndrome (ARDS) or pneumonia (Table 1 ). Additional clinical information is presented in Supplementary Table 2.
Table 1.
Demographic characteristics of patients. All characteristics reported as N (%).
| Demographics | All eligible patients (n = 2313) | COVID-19 tested (n = 1286) | COVID-19 positive (n = 197) | |
|---|---|---|---|---|
| Age | Age < 30 | 446 (19.3%) | 151 (11.7%) | 15 (7.6%) |
| 30–49 | 516 (22.3%) | 267 (20.8%) | 30 (15.2%) | |
| 50–59 | 356 (15.4%) | 212 (16.5%) | 32 (16.2%) | |
| 60–69 | 384 (16.6%) | 245 (19.1%) | 41 (20.8%) | |
| 70–79 | 340 (14.7%) | 213 (16.6%) | 44 (22.3%) | |
| Age > 80 | 271 (11.7%) | 195 (15.2%) | 35 (17.8%) | |
| Age unknown |
0 (0.0%) |
3 (0.2%) |
0 (0.0%) |
|
| Gender | Female | 1309 (56.6%) | 683 (53.1%) | 96 (48.7%) |
| Male | 1004 (43.4%) | 603 (46.9%) | 101 (51.3%) | |
| Unknown Sex |
0 (0.0%) |
3 (0.2%) |
0 (0.0%) |
|
| Acute Diagnoses | Sepsis | 19 (0.8%) | 17 (1.3%) | 10 (5.1%) |
| ARDS | 30 (1.3%) | 43 (3.3%) | 19 (9.6%) | |
| Pneumonia | 44 (1.9%) | 52 (4.0%) | 26 (13.2%) | |
| AKI | 97 (4.2%) | 71 (5.5%) | 8 (4.1%) |
ARDS: Acute Respiratory Distress Syndrome. AKI: Acute Kidney Injury.
The ML algorithm demonstrated improved performance over MEWS for predicting ventilation within 24 h (Table 2 ). Note that while the algorithm is able to compute risk scores in the presence of missing patient measurements, MEWS was calculated only when all measurements were available. The sample on which MEWS was calculated is therefore smaller than the full study sample and includes only 183 patients. Given the more dire implications of missing patients over providing false alerts, we chose an operating point that favored sensitivity over specificity [14]. The algorithm achieved higher sensitivity (0.90) than MEWS (0.78) for a higher value of specificity (Fig. 2 ). The concurrent increase in sensitivity and specificity indicates that the algorithm is capable of detecting 16% more patients who will require ventilation as compared to the traditional scoring system, while simultaneously reducing false positive alerts. Similarly, the algorithm demonstrated improved positive and negative likelihood ratios as well as a DOR approximately five times higher than MEWS (12.58 vs 2.36). The difference in performance was statistically significant (p < 0.05). The algorithm additionally demonstrated superior positive and negative predictive values when compared to MEWS (Supplementary Table 3). The algorithm also obtained a higher AUC (0.866) than MEWS when calculated retrospectively (Supplementary Fig. 2). Confusion matrices for the algorithm and for MEWS are presented in the supplement as Supplementary Tables 4 and 5, respectively. Additionally, a detailed breakdown of model inputs and scoring system decisions for two patients is presented in Supplementary Table 6.
Table 2.
Performance metrics of the machine learning algorithm and the Modified Early Warning Score.
| MLA (n = 197) | MEWS (n = 183) | |
|---|---|---|
| AUC | 0.866 | 0.637 |
| Sensitivity | 0.900 | 0.778 |
| Specificity | 0.583 | 0.402 |
| LR+ | 2.158 | 1.301 |
| LR- | 0.172 | 0.552 |
| DOR | 12.577 | 2.356 |
AUC: Area under the receiver operating characteristic. LR+/-: Positive/Negative likelihood ratio. DOR: Diagnostic Odds Ratio. MLA: Machine Learning Algorithm. MEWS: Modified Early Warning Score.
Fig. 2.
Comparison of sensitivity and specificity for the machine learning algorithm and MEWS score. Abbreviations: MLA: Machine Learning Algorithm. MEWS: Modified Early Warning Score.
4. Discussion
The READY study is the first clinical trial of a machine learning algorithm for the prediction of ventilation requirements among COVID-19 patients. We found that the ML algorithm predicted the need for mechanical ventilation within 24 h among COVID-19 patients with high sensitivity and specificity. This work builds upon our prior work developing algorithms to predict patient outcomes including sepsis [15], acute kidney injury [16], mortality [17], and patient stability and decompensation [18]. While machine learning algorithms have been applied to retrospective COVID-19 patient data, no equivalent algorithms have yet been validated in a prospective setting, despite urgent need.
The high sensitivity and specificity achieved by the algorithm demonstrate that it is capable of accurate discrimination between COVID-19 patients at high risk versus low risk of requiring ventilation within 24 h. The high sensitivity, in particular, suggests the algorithm is unlikely to provide false negative classifications and that patients in need of mechanical ventilation are therefore unlikely to be missed by the algorithm. Further, the algorithm's improvements in sensitivity as compared to the traditional scoring system show that the algorithm is capable of detecting 16% more patients who will be in need of mechanical ventilation; this is a meaningful improvement that can allow for effective patient triage and resource allocation. The algorithm also achieved this increase in sensitivity while demonstrating a higher specificity as compared to MEWS. This suggests the algorithm will produce a reduced false positive rate, which may enable more efficient allocation of clinician time and of resources.
Physicians have reported difficulty in predicting the disease course of hospitalized COVID-19 patients, as well as difficulties in the identification of patients at high risk of rapid decompensation [19,20]. Without the benefit of timely warnings, rapid and unexpected deterioration in patient conditions come with the high risks of emergency transfers to the ICU and emergency intubations. Emergency intubations, in particular, have well-documented risks [7,21,22], with at least one complication occurring in 22–54% of all intubations performed in critically ill patients [23]. Cook et al. found that intubations in the ICU are associated with a more than 4-fold higher risk of death or brain damage as compared to intubations performed in the operating room; this may be attributable to a lack of preparedness due to the increased need for emergency intubations in the ICU setting [24]. Complications related to intubation are more likely in patients with limited pulmonary reserve, in patients with poor physiological status, and in patients for whom pre-oxygenation was not possible [22]. Receiving advance notice of patients for whom deterioration is more likely may allow care teams to better prepare for intubation procedures and minimize risk to the patient. Further, early identification of patients for whom ventilation will be required may allow physicians to minimize the risk of patient self-inflicted lung injury (P-SILI). Vigorous breathing and associated high transpulmonary pressures in patients with respiratory distress may contribute to the development of P-SILI [25]. Early intubation of patients requiring mechanical ventilation, when performed with sedation and physician control of mechanical power applied to the lung (determined by transpulmonary pressures and other ventilator-setting determined variables), may minimize the risk of P-SILI due to vigorous spontaneous breathing [26,27].
Accurate and early predictions of risk of patient deterioration may improve patient triage procedures and resource allocation. The model predicted the need for mechanical ventilation using only routinely available labs and vital sign data. Demographic data was not required as in similar work [8]. Of note, the measurements used as inputs to our model were taken during the first two hours after ED arrival or hospital admission. Our model was also able to generate predictions in the absence of certain inputs. Because the algorithm was developed from real world EHR data that contained missing values, we do not anticipate missing values to have significantly affected the output of the model. This is because some data are missing may be the result of clinicians who may have deemed that it was not important to measure that particular vital sign or lab value. This can provide useful information about the patient in the form of “informative missingness” [28]. This model could therefore be used to identify which patients should be considered for direct admission to an area of more intensive monitoring, even if they appear stable at admission, to prevent emergency transfers and minimize patient morbidity. It is possible that patients at a high risk of requiring mechanical ventilation within 24 h have progressed further along their disease course as compared to patients who were at low risk or, alternatively, are experiencing a more intense host response to the virus. High-risk patients may therefore benefit more from supportive or immunologic therapies than their low-risk counterparts, who may need only antiviral medications. Effective discrimination between these two groups may therefore have broad implications for future research into patient care beyond triage and admission decisions [29].
A literature review has shown that there is limited evidence supporting the use of existing non-ML comparators for the COVID-19 population [30,31]. These studies have largely predicted in-hospital mortality rather than short term critical care needs and have generally used data from only a single hospital or health center, thus increasing uncertainty about the generalizability of results. To fill this gap, there have been other attempts to develop ML algorithms to predict patient deterioration and mortality in a COVID-19 population. In addition to the work by Liang et al. [8], Vaid et al. [32] evaluated the performance of an ML algorithm for the prediction of mortality and critical events (defined as any of intubation, discharge to hospice, or death) at three, five, seven, and ten days. Vaid and colleagues utilized retrospective data from a single New York City health system and did not evaluate shorter prediction intervals or examine mechanical ventilation as an individual outcome. There is therefore potential ambiguity about how the algorithm may perform in novel clinical settings or for detecting more rapid patient deterioration. Singh et al. [33] evaluated the Epic Deterioration Index (EDI) prospectively in a population of 174 COVID-19 patients, assessing its performance for a composite outcome of requirement of ICU care, initiation of mechanical ventilation, or in-hospital death. While the EDI showed moderate discrimination for the COVID-19 with a maximum AUC of 0.76, the authors again do not examine ventilation as an individual outcome. This limits the utility of this score for effective patient triage and resource allocation. Further, Singh et al. assessed the performance of the EDI at only a single medical center, leaving uncertainty about its performance at medical centers with different patient demographic characteristics. While much research has been done on the use of ML methods to assist with epidemiologic models and population-level forecasting, there remains a significant need for investigating the potential for ML methods to assist with prediction and decision making on the individual patient level [34]. Several studies have explored the potential utility of machine learning for diagnosing and detecting COVID-19, largely using imaging data [35,36], though the area of patient decompensation prediction remains less explored.
This study builds upon existing evidence about the ability of algorithms to successfully provide clinical decision support [[15], [16], [17], [18]]. However, there are several limitations to this study. First, while we included patients from several medical centers in our sample, the total sample remained relatively small and the outcome of mechanical ventilation within 24 h of model prediction was rare in our sample. Building models in the emerging stages of a pandemic is difficult due to data limitations and uncertainty in the data. This is true of both patient-level and population-level prediction [37,38]. These constraints motivate the use of flexible, sparse data tolerant algorithms and careful model evaluation. Because of the prospective nature of this study, we did not directly assess the potential for imbalance in the dataset or consider the use of data balancing techniques. It is therefore possible that our dataset is unbalanced, which in turn may have impacted the observed sensitivity and specificity of the algorithm. Additionally, we cannot make any inference about the generalizability of algorithm performance in new settings based on the present study and this study design may be impacted by multiple hypothesis testing bias. Additionally, because we restricted our study population to patients with a confirmed COVID-19 diagnosis, we cannot infer the performance of this algorithm for predicting respiratory decompensation in prospective settings for non-COVID-19 patients, nor can we make inference about the performance of the algorithm on patients suspected of, but not ultimately diagnosed with COVID-19. The focus of this study was to validate the performance of the predictive algorithm and our study protocol therefore did not directly examine physician response to algorithm alerts. Therefore, we cannot draw conclusions on the effect of patient alerts to influence clinician actions, or on patient outcomes.
5. Conclusion
A machine learning algorithm for prediction of mechanical ventilation of COVID-19 patients within 24 h of their initial hospital encounter demonstrated a high sensitivity (0.90) and specificity (0.58) and outperformed a commonly used early warning scoring system; the algorithm is capable of detecting 16% more patients than the Modified Early Warning Score (p < 0.05) while simultaneously reducing false positive alerts. Given the substantial concerns regarding limited resources, including mechanical ventilators, during the COVID-19 crisis, accurate prediction of patients likely to require mechanical ventilation may help to provide significant guidance with respect to triaging patients and allocating resources to hospitalized individuals. Further, early identification of such individuals may allow for planned ventilation procedures, mitigating some of the known risks associated with emergency intubation. This algorithm may therefore help to improve patient care, minimize clinician burden, and minimize morbidity and mortality during the COVID-19 pandemic.
Summary
In the READY multicenter clinical trial, we evaluated the performance of a machine learning (ML) algorithm for the prediction of invasive mechanical ventilation of COVID-19 patients within 24 h of their initial hospital encounter. We found that our algorithm achieved significantly higher sensitivity (0.90) than MEWS, a scoring system commonly used to assess patient status and assign levels of care while maintaining a higher specificity (p < 0.05). This accurate advance warning of the need for mechanical ventilation of COVID-19 patients is important, as physicians have reported difficulty with predicting which patients are at high risk of rapid respiratory decompensation. Inadequate lead time and subsequent emergency intubation of critically ill patients is associated with significant known risks, including peri-intubation hypoxia, hypotension, arrhythmia, and cardiac arrest. Accurate advance warning can help improve COVID-19 patient outcomes and our algorithm is capable of detecting 16% more patients who will require invasive mechanical ventilation than MEWS while also reducing false positive alerts.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2020.103949.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Subbe C.P., Slater A., Menon D. Validation of physiological scoring systems in the accident and emergency department. Emerg. Med. J. 2006 Nov;23(11):841–845. doi: 10.1136/emj.2006.035816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith M.E.B., Chiovaro J.C., O'Neil M. Early warning system scores for clinical deterioration in hospitalized patients: a systematic review. Ann Am Thorac Soc. 2014 Oct 8;11(9):1454–1465. doi: 10.1513/AnnalsATS.201403-102OC. [DOI] [PubMed] [Google Scholar]
- 3.Hu H., Yao N., Qiu Y. Comparing rapid scoring systems in mortality prediction of critically ill patients with novel coronavirus disease. Acad. Emerg. Med. 2020;27(6):461–468. doi: 10.1111/acem.13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moghadas S.M., Shoukat A., Fitzpatrick M.C. Projecting hospital utilization during the COVID-19 outbreaks in the United States. Proc. Natl. Acad. Sci. U. S. A. 2020;117(16):9122–9126. doi: 10.1073/pnas.2004064117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC . Centers for Disease Control and Prevention; 2020. Coronavirus Disease 2019 (COVID-19) [Internet]https://www.cdc.gov/coronavirus/2019-ncov/covid-data/forecasting-us.html [cited 2020 May 15]. Available from: [Google Scholar]
- 6.Kissler S.M., Tedijanto C., Goldstein E. 2020 Apr 14. Projecting the Transmission Dynamics of SARS-CoV-2 through the Postpandemic Period. Science [Internet]https://science.sciencemag.org/content/early/2020/05/11/science.abb5793 [cited 2020 May 15]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgs A., McGrath B.A., Goddard C. Guidelines for the management of tracheal intubation in critically ill adults. Br. J. Anaesth. 2018;120(2):323–352. doi: 10.1016/j.bja.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 8.Liang W., Liang H., Ou L. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Internal. Med. 2020 May 12 doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trochim W. Mason Thomson; Ohio: 2005. Research Methods: the Concise Knowledge Base. [Google Scholar]
- 10.Tofthagen C. Threats to validity in retrospective studies. J Adv. Pract. Oncol. 2012 May;3(3):181–183. PMID: 25031944; PMCID: PMC4093311. [PMC free article] [PubMed] [Google Scholar]
- 11.Leviton L.C. Generalizing about public health interventions: a mixed-methods approach to external validity. Annu. Rev. Publ. Health. 2017;38:371–391. doi: 10.1146/annurev-publhealth-031816-044509. [DOI] [PubMed] [Google Scholar]
- 12.Avellar S.A., Thomas J., Kleinman R. External validity: the next step for systematic reviews? Eval. Rev. 2017;41(4):283–325. doi: 10.1177/0193841X16665199. [DOI] [PubMed] [Google Scholar]
- 13.Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining [Internet] ACM; San Francisco California USA: 2016. XGBoost: a scalable tree boosting system.https://dl.acm.org/doi/10.1145/2939672.2939785 [cited 2020 Jun 11]. pp. 785–94. Available from: [Google Scholar]
- 14.Lalkhen A.G., McCluskey A. Clinical tests: sensitivity and specificity. Cont. Educ. Anaesth. Crit. Care Pain. 2008 Dec 1;8(6):221–223. [Google Scholar]
- 15.Shimabukuro D.W., Barton C.W., Feldman M.D. Effect of a machine learning-based severe sepsis prediction algorithm on patient survival and hospital length of stay: a randomised clinical trial. BMJ Open. Resp. Res. 2017;4 doi: 10.1136/bmjresp-2017-000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohamadlou H., Lynn-Palevsky A., Barton C. Prediction of acute kidney injury with a machine learning algorithm using electronic health record data. Can. J. Kidney. Health. Dis. 2018 Jan 1;5 doi: 10.1177/2054358118776326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohamadlou H., Panchavati S., Calvert J. Multicenter validation of a machine-learning algorithm for 48-h all-cause mortality prediction. Health Inf. J. 2019 Dec 30 doi: 10.1177/1460458219894494. [DOI] [PubMed] [Google Scholar]
- 18.Calvert J.S., Price D.A., Barton C.W. Discharge recommendation based on a novel technique of homeostatic analysis. J Am. Med. Inform. Assoc JAMIA. 2017;24(1):24–29. doi: 10.1093/jamia/ocw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartman M.E., Hernandez R.A., Patel K. COVID-19 respiratory failure: targeting inflammation on VV-ECMO support. Am. Soc. Artif. Intern. Organs J. 2020 doi: 10.1097/MAT.0000000000001177. [published online ahead of print, 2020 Apr 16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs J.P., Stammers A.H., St Louis J. Extracorporeal membrane oxygenation in the treatment of severe pulmonary and cardiac compromise in COVID-19: experience with 32 patients. Am. Soc. Artif. Intern. Organs J. 2020 doi: 10.1097/MAT.0000000000001185. [published online ahead of print, 2020 Apr 17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Divatia J.V., Khan P.U., Myatra S.N. Tracheal intubation in the ICU: life saving or life threatening? Indian J. Anaesth. 2011;55(5):470–475. doi: 10.4103/0019-5049.89872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon U., Mojica J., Wiltshire M. Emergent airway management outside of the operating room - a retrospective review of patient characteristics, complications and ICU stay. BMC Anesthesiol. 2019;19(1):220. doi: 10.1186/s12871-019-0894-4. Published 2019 Dec 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natt B.S., Malo J., Hypes C.D. Strategies to improve first attempt success at intubation in critically ill patients. Br. J. Anaesth. 2016;117(Suppl 1):60–68. doi: 10.1093/bja/aew061. [DOI] [PubMed] [Google Scholar]
- 24.Cook T.M., Woodall N., Harper J. Fourth national audit project. Major complications of airway management in the UK: results of the fourth national audit project of the royal college of anaesthetists and the difficult airway society. Part 2: intensive care and emergency departments. Br. J. Anaesth. 2011;106(5):632–642. doi: 10.1093/bja/aer059. [DOI] [PubMed] [Google Scholar]
- 25.Grieco D.L., Menga L.S., Eleuteri D. Patient self-inflicted lung injury: implications for acute hypoxemic respiratory failure and ARDS patients on non-invasive support. Minerva Anestesiol. 2019;85(9):1014–1023. doi: 10.23736/S0375-9393.19.13418-9. [DOI] [PubMed] [Google Scholar]
- 26.Brochard L., Slutsky A., Pesenti A. Mechanical ventilation to minimize progression of lung injury in acute respiratory failure. Am. J. Respir. Crit. Care Med. 2017;195(4):438–442. doi: 10.1164/rccm.201605-1081CP. [DOI] [PubMed] [Google Scholar]
- 27.Marini J.J., Gattinoni L. Management of COVID-19 respiratory distress. JAMA. 2020;24 doi: 10.1001/jama.2020.6825. Published online April. [DOI] [PubMed] [Google Scholar]
- 28.Che Z., Purushotham S., Cho K. Recurrent neural networks for multivariate time series with missing values. Sci. Rep. 2018;8:6085. doi: 10.1038/s41598-018-24271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahsan W., Javed S., Bratty M.A. Treatment of SARS-CoV-2: how far have we reached? Drug Discov Ther. 2020 Apr 30;14(2):67–72. doi: 10.5582/ddt.2020.03008. [DOI] [PubMed] [Google Scholar]
- 30.Su Y., Tu G.W., Ju M.J. Comparison of CRB-65 and quick sepsis-related organ failure assessment for predicting the need for intensive respiratory or vasopressor support in patients with COVID-19. J. Infect. 2020;S0163–4453(20):30281–30284. doi: 10.1016/j.jinf.2020.05.007. [published online ahead of print, 2020 May 7] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou X., Li S., Fang M. Acute physiology and chronic health evaluation II score as a predictor of hospital mortality in patients of coronavirus disease 2019. Crit. Care Med. 2020 doi: 10.1097/CCM.0000000000004411. [published online ahead of print, 2020 May 1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaid A., Somani S., Russak A.J. Machine learning to predict mortality and critical events in COVID-19 positive New York city patients. medRxiv. 2020 Apr 28 doi: 10.1101/2020.04.26.20073411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh K., Valley T.S., Tang S. Validating a widely implemented deterioration Index model among hospitalized COVID-19 patients. medRxiv. 2020 Jan 1 doi: 10.1513/AnnalsATS.202006-698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shinde G.R., Kalamkar A.B., Mahalle P.N. Forecasting models for coronavirus disease (COVID-19): a survey of the state-of-the-art. SN Comput. Sci. 2020;1:197. doi: 10.1007/s42979-020-00209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S., Shukla P.K. IRBM; 2020. Deep Transfer Learning Based Classification Model for COVID-19 Disease.http://www.sciencedirect.com/science/article/pii/S1959031820300993 2020 May 20 [cited 2020 Jul 16]; Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh D., Kumar V., Vaishali, Kaur M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution-based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39(7):1379–1389. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fong S.J., Li G., Dey N., Crespo R.G., Herrera-Viedma E. 2019. Finding an Accurate Early Forecasting Model from Small Dataset: A Case of 2019-ncov Novel Coronavirus Outbreak. arXiv preprint arXiv:2003.10776. 2020 Mar 24. [Google Scholar]
- 38.Fong S.J., Li G., Dey N., Crespo R.G., Herrera-Viedma E. Composite Monte Carlo decision making under high uncertainty of novel coronavirus epidemic using hybridized deep learning and fuzzy rule induction. Appl. Soft Comput. 2020 Apr 9:106282. doi: 10.1016/j.asoc.2020.106282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.