Abstract
Lysosomal enzymes are synthesized in the endoplasmic reticulum (ER) and transferred to the Golgi complex by interaction with the Batten disease protein CLN8 (ceroid lipofuscinosis, neuronal, 8). Here we investigated the relationship of this pathway with CLN6, an ER-associated protein of unknown function that is defective in a different Batten disease subtype. Experiments focused on protein interaction and trafficking identified CLN6 as an obligate component of a CLN6-CLN8 complex (herein referred to as EGRESS: ER-to-Golgi relaying of enzymes of the lysosomal system), which recruits lysosomal enzymes at the ER to promote their Golgi transfer. Mutagenesis experiments showed that the second luminal loop of CLN6 is required for the interaction of CLN6 with the enzymes but dispensable for interaction with CLN8. In vitro and in vivo studies showed that CLN6 deficiency results in inefficient ER export of lysosomal enzymes and diminished levels of the enzymes at the lysosome. Mice lacking both CLN6 and CLN8 did not display aggravated pathology compared with the single deficiencies, indicating that the EGRESS complex works as a functional unit. These results identify CLN6 and the EGRESS complex as key players in lysosome biogenesis and shed light on the molecular etiology of Batten disease caused by defects in CLN6.
Keywords: Cell Biology
Keywords: Genetic diseases, Lysosomes, Molecular pathology
Introduction
Lysosomes contain more than 50 soluble hydrolytic enzymes that mediate the degradation of macromolecules according to various catabolic programs. Lysosomal enzymes are synthesized in the endoplasmic reticulum (ER) and transferred to the endolysosomal system via the secretory route (1, 2). Whereas the post-Golgi trafficking of lysosomal enzymes has been amply characterized (3), the early, pre-Golgi stages of lysosomal enzyme trafficking are only partially understood. We have recently reported that ER-to-Golgi transfer of newly synthesized lysosomal enzymes is mediated by the cargo receptor CLN8 (ceroid lipofuscinosis, neuronal, 8) (4). CLN8 is a ubiquitously expressed multipass membrane protein that forms homodimers and localizes in the ER and the ER-Golgi intermediate compartment (ERGIC) (5, 6). CLN8 interacts with newly synthesized lysosomal enzymes in the ER, transfers them to the Golgi via COPII vesicles, and recycles back to the ER via COPI vesicles (4). CLN8 deficiency results in inefficient ER exit and decreased levels of lysosomal enzymes in mouse tissues and patient-derived cells (4), causing a subtype of Batten disease or neuronal ceroid lipofuscinosis (NCL) (7, 8).
Batten disease is a heterogeneous group of 13 genetically distinct progressive encephalopathies with onset spanning from infancy to adulthood and overall incidence estimated to be 1:12,500 live births in the United States (9). Batten disease is classified as a lysosomal storage disorder due to the observed intralysosomal accumulation of ceroid lipopigment in neurons and other cell types (7). A clear link between Batten disease and lysosomal dysfunction is established by the observation that 4 Batten subtypes are caused by mutations in 1 of 4 lysosomal enzymes (PPT1, TPP1, CTSD, CTSF). In addition, at least 4 other Batten disease genes encode proteins that localize and presumably function in the lysosome (10). Impaired trafficking of lysosomal enzymes due to deficiency of ER- and ERGIC-localized CLN8 protein is another example linking impaired protein function to molecular pathology in Batten disease (4).
The ubiquitously expressed multipass membrane protein CLN6 (ceroid lipofuscinosis, neuronal, 6; OMIM 601780) is the only other Batten protein that resides in the ER (11, 12). Remarkably, the clinical features of Batten patients with mutations in either CLN6 or CLN8 are strikingly similar (13, 14). Commonly associated symptoms include cognitive decline, seizures, retinopathy, and gait difficulties, with patients first reporting to the clinic between 2 and 8 years of age (14–16). Naturally occurring mouse models for CLN6 and CLN8 diseases have been widely characterized and have been shown to mimic many of the disease phenotypes (17). These mouse lines (Cln6nclf and Cln8mnd, henceforth referred to as Cln6–/– and Cln8–/–) carry early frameshift mutations in Cln6 and Cln8 genes, resulting in the complete absence of CLN6 and CLN8 proteins, respectively (11, 18–20). Like human patients, mutant mice die prematurely and exhibit dysfunctional lysosomal metabolism in multiple tissues and organs (19, 21). The underlying defective molecular pathway linking CLN6 deficiency to lysosomal dysfunction is unclear, as the molecular function of CLN6 is yet to be characterized (22). Recent reports show alterations in metal homeostasis pathways — mostly accumulation of zinc and manganese — and aberrant cell signaling related to AKT/GSK3 and ERK/MAPK pathways as characteristic features of CLN6 disease (23). Impaired stability and function of the CLN6 interactor, collapsin response mediator protein 2 (CRMP-2), has also been associated with altered neurite maturation in CLN6 disease, possibly contributing to neuronal dysfunction and encephalopathy (24). While these reports indicate associations between tissue pathology and disease symptoms, however, the function of CLN6 and its relationship with lysosomal pathways has remained elusive.
Based on the similarities between the clinical features associated with CLN6 and CLN8 deficiencies and the partially overlapping subcellular localization of the 2 proteins, we hypothesized that CLN6 also functions in ER-to-Golgi transfer of lysosomal enzymes. Here we investigated whether CLN6 acts as a cargo receptor for lysosomal enzymes and whether CLN6 function is redundant with, or dependent on, the function of CLN8. Our results show that CLN6 and CLN8 are obligate partners for the recruitment of newly synthesized lysosomal enzymes at the ER and that, differently from CLN8, CLN6 is not loaded into COPII vesicles but is retained in the ER, presumably to serve additional cycles of enzyme recruitment. We determined that the second luminal loop of CLN6 is required for the interaction of CLN6 with the enzymes and that CLN6 deficiency results in inefficient ER export of lysosomal enzymes and diminished levels of the enzymes at the lysosome. These results identify CLN6 as a key protein implicated in the biogenesis of lysosomes and shed light on the molecular etiology of Batten disease caused by mutations in CLN6.
Results
CLN6 deficiency results in the depletion of various lysosomal enzymes from the lysosomal compartment.
We recently demonstrated that CLN8 deficiency results in diminished levels of various soluble lysosomal enzymes at the lysosome (4). To investigate whether CLN6 deficiency also results in defective lysosomal composition, we isolated lysosome-enriched fractions from the livers of presymptomatic 6-week-old Cln6–/– mice and age-matched WT mice using a discontinuous Nycodenz-sucrose gradient as described (25). Immunoblot analysis of the lysosomal membrane protein LAMP1 confirmed lysosomal enrichment in the collected fractions and showed no obvious changes in LAMP1 signal between WT and Cln6–/– samples (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI130955DS1). We confirmed lysosomal enrichment by performing an enzyme assay for β-hexosaminidase, a lysosomal enzyme that is not affected by CLN8 deficiency (4) and did not show changes upon deficiency of CLN6 (Supplemental Figure 1B). We then performed immunoblot analysis for a set of enzymes for which antibodies able to recognize the mouse proteins are available. The results showed a general reduction in enzyme levels in the lysosomal fraction from Cln6–/– mice (Figure 1, A and B). Consistent with these results, we observed a decrease in lysosomal enzyme activities using proteins extracted from the lysosome-enriched fraction of Cln6–/– mice (Figure 1C). Real-time qPCR using liver RNAs, however, showed slightly increased transcription of lysosomal enzymes in Cln6–/– mice compared with WT controls (Figure 1D). Consistently, immunoblot analysis of whole liver homogenates showed slightly increased expression of several lysosomal enzymes in Cln6–/– mice compared with WT controls (Supplemental Figure 1, C and D). In addition, confocal microscopy analysis of mouse embryonic fibroblasts (MEFs) from WT and Cln6–/– mice showed decreased overlaps of CTSD and PPT1 with the lysosomal marker LAMP1 in the absence of CLN6, whereas no obvious changes in the overlap of these enzymes with the ER protein KDEL were found (Supplemental Figure 1, E–H). Together, these results show an absence of major defects in the expression of lysosomal enzymes and indicate that the cause of the observed lysosomal enzyme depletion must be posttranslational.
Figure 1. CLN6 deficiency results in the depletion of various lysosomal enzymes from the lysosomal compartment.
(A) Immunoblot analysis of lysosome-enriched fractions confirming depletion of lysosomal enzymes in Cln6–/– mice compared with WT mice. CTSDSC, single-chain processed form; CTSDMH, mature heavy form; CTSDML, mature light form. Blots were run in parallel. (B) Band intensities were quantified and normalized to LAMP1. Data are mean ± SEM (n = 3). (C) Enzymatic assay of TPP1, GBA, GLB1, GAA, and NAGLU in lysosome-enriched fractions from WT and Cln6–/– mice. Activity is expressed as relative fluorescence units compared with WT samples. Data are mean ± SEM (n = 3). (D) Expression analysis of lysosomal genes in the liver of 6-week-old WT and Cln6–/– mice. Shown are expression levels of genes in Cln6–/– mice expressed as fold change of levels in WT mice, normalized to the housekeeping gene Sp16. Data are mean ± SEM (n = 5). Statistical differences between groups were calculated using Student’s t test (B and C). NS, not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001.
CLN6 interacts with CLN8 but does not traffic to the Golgi complex.
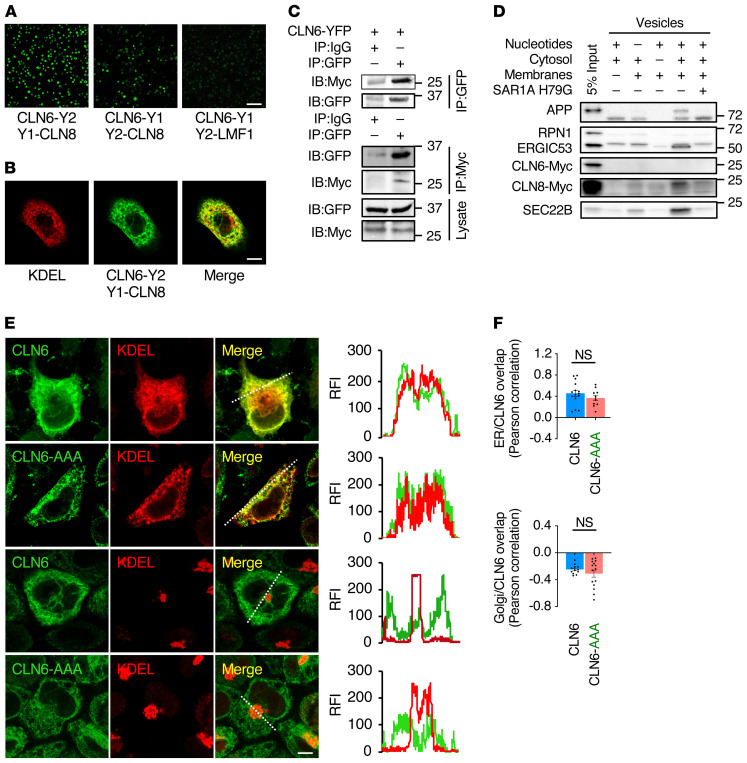
We next investigated whether CLN6 interacts with CLN8 by using bimolecular fluorescence complementation (BiFC), an assay in which proteins are tagged with either the N- or the C-terminus of YFP (Y1 and Y2, respectively), and fluorescence is emitted by reconstituted YFP upon interaction of the 2 tagged proteins (26). BiFC assays performed by cotransfecting either CLN6-Y1 with Y2-CLN8 or CLN6-Y2 with Y1-CLN8 showed interaction between CLN6 and CLN8 under either Y1/Y2 configuration in HeLa cells (Figure 2A and Supplemental Figure 2A) and mouse embryonic fibroblasts (Supplemental Figure 2B). As a control, we verified that CLN6 does not interact with lipase maturation factor 1 (LMF1) (Figure 2A), an ER transmembrane protein that acts as a cargo receptor for lipoprotein lipase (27). Costaining with KDEL showed colocalization with the reconstituted fluorescence, indicating that CLN6 and CLN8 interact at the ER (Figure 2B; R = 0.76 ± 0.08, n = 10 cells). Co-IP experiments using tagged proteins expressed in HEK293T cells confirmed that CLN6 interacts with CLN8 (Supplemental Figure 2C) but not with LMF1 (Supplemental Figure 2D). In addition, we generated a cell line in which endogenous CLN8 is fused to a myc tag by using CRISPR/Cas9 genome editing. Co-IP experiments confirmed the interaction between CLN6 and endogenous CLN8 (Figure 2C).
Figure 2. CLN6 interacts with CLN8 in the ER and does not traffic to the Golgi complex.
(A) Shown is a live BiFC assay of CLN6 with CLN8 in the indicated YFP configurations. Green signals (reconstituted YFP) represent CLN6-CLN8 interaction. LMF1 is used as a negative control for interaction with CLN6. Scale bar: 200 μm. (B) Confocal microscopy analysis showing colocalization of reconstituted CLN6-Y2/Y1-CLN8 BiFC signal with the ER marker KDEL. Scale bar: 20 μm. (C) Co-IP analysis of transiently expressed, Y2-tagged CLN6 and endogenous, myc-tagged CLN8. The lysates were immunoprecipitated with both myc and GFP antibodies in separate experiments and analyzed by immunoblotting with the indicated antibodies. IgG antibodies were used as a control. Input represents 10% of the total cell extract used for IP. (D) In vitro COPII vesicle budding assay on digitonin-treated HeLa cell membranes incubated with the indicated combinations of ATP regenerating system, rat liver cytosol, collected donor membranes, and dominant-negative SAR1A H79G; 5% input of donor membranes is included. (E) Confocal microscopy analysis showing that CLN6 resides in the ER upon mutagenesis of a potential retrieval/retention signal (RRR to AAA). Trace outline is used for line-scan analysis of relative fluorescence intensity (RFI) of CLN6, GM130, and KDEL signals. Scale bar: 10 μm. (F) Pearson correlation analysis of the colocalization extent of full-length CLN6 or CLN6-AAA with KDEL or GM130. Data are mean ± SEM; n = 15 (ER/CLN6), n = 10 (ER/CLN6-AAA), n = 12 (Golgi/CLN6), n = 15 (Golgi/CLN6-AAA). Statistical differences between groups were calculated using Student’s t test.
Additional confocal microscopy analyses showed that both CLN6 and CLN8 colocalize with Sec16L, a marker for ER exit sites (Supplemental Figure 2E). To test whether CLN6 is loaded into COPII vesicles, we performed an in vitro COPII vesicle budding assay. We incubated membranes from myc-CLN6–transfected HeLa cells with rat liver cytosol and nucleotides for 1 hour at 30°C. Newly formed vesicles were collected and blotted for CLN6 and CLN8. Three known COPII cargo proteins — APP, ERGIC-53/LMAN1, and SEC22B — were used as positive controls (28, 29), and ribophorin I (RPN1), an ER-resident protein excluded from COPII vesicles (30), was used as a negative control. Unlike CLN8, APP, and ERGIC-53, which were detected in COPII vesicles, CLN6 was excluded from COPII vesicles, similarly to RPN1 (Figure 2D). Addition of SAR1A H79G, a dominant-negative mutant that inhibits COPII vesicle formation (28), blocked the inclusion of APP, SEC22B, ERGIC-53, and CLN8 into newly formed vesicles, demonstrating that the process is specifically dependent on COPII.
In agreement with the observed exclusion of CLN6 from COPII vesicles, mutation of a putative ER retention/retrieval signal (5RRR mutated to AAA) (31) present in the cytosol-facing N-terminus of CLN6 did not change the subcellular localization of CLN6 (Figure 2, E and F), consistent with previous observations (22). Because other unknown protein signals could mediate retrieval of CLN6 from the Golgi to the ER, we also inhibited Golgi-to-ER retrograde trafficking using 1,3-cyclohexanebis(methylamine) (CBM), a chemical inhibitor of COPI-mediated vesicular transport (32). CBM treatment resulted in a change of CLN8 localization to the Golgi (Supplemental Figure 2, F and G) as previously observed (4), whereas it did not result in any discernible overlap of CLN6 with the Golgi marker, confirming exclusive localization of CLN6 in the ER (Supplemental Figure 2, H and I). These results are in agreement with prior work indicating that CLN6 does not localize to the Golgi (22).
CLN8 trafficking to the Golgi is uncoupled from CLN6-CLN8 interaction.
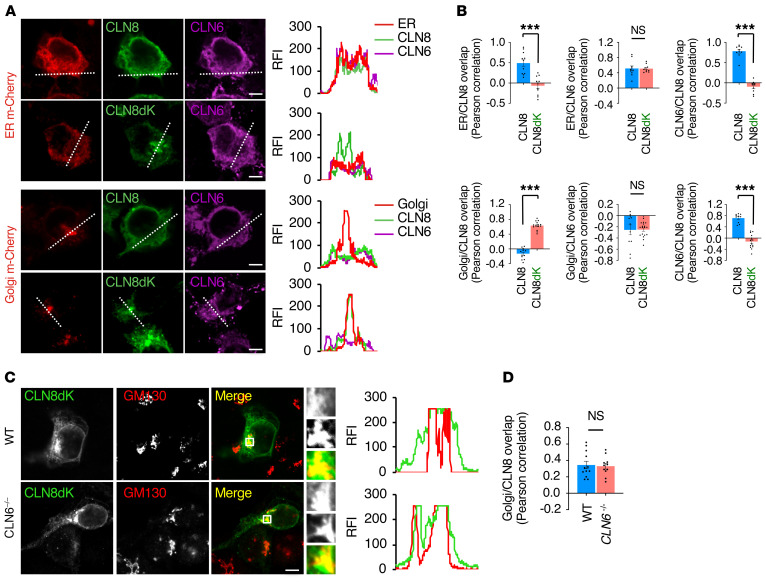
We next investigated whether there is mutual dependence between the subcellular localization of CLN6 and CLN8. We first tested whether abolishing Golgi-to-ER retrieval of CLN8 affects the subcellular localization of CLN6. To this end, we cotransfected myc-tagged CLN6 with either full-length Y2-CLN8 or the retrieval-deficient CLN8 mutant Y2-CLN8dK (4). Confocal microscopy showed that, in either case, CLN6 localized at the ER in HEK293T cells (Figure 3, A and B). To avoid interaction between CLN6 and endogenous CLN8, we repeated the test in a cell line in which CLN8 was knocked out (CLN8–/–) (4) and obtained a similar result (Supplemental Figure 3, A and B). We then tested whether CLN8 can traffic to the Golgi independently of CLN6. To this aim, we generated CLN6–/– cells by CRISPR/Cas9 genome editing (Supplemental Figure 4, A–C); as a control, we confirmed depletion of lysosomal enzymes in these cells, which could be rescued by CLN6 reintroduction (Supplemental Figure 4D). Confocal microscopy showed that retrieval-deficient Y2-CLN8dK localizes to the Golgi in CLN6–/– cells (Figure 3, C and D), indicating that CLN8 trafficking is uncoupled from the interaction of CLN8 with CLN6.
Figure 3. The subcellular localizations of CLN6 and CLN8 are uncoupled from CLN6-CLN8 interaction.
(A) Confocal microscopy analysis showing ER localization of CLN6 and Golgi localization of CLN8 upon cotransfection of full-length CLN6 and retrieval-deficient CLN8. Trace outline is used for RFI line-scan analysis of CLN8, CLN6, Golgi-cherry and ER-cherry signals. Scale bars: 10 μm. (B) Pearson correlation analysis of the colocalization extent of CLN6 and CLN8 constructs with KDEL and GM130. Data are mean ± SEM; n = 11 (ER/CLN8), n = 10 (ER/CLN8dK), n = 10 (ER/CLN6 with CLN8 or CLN8dK), n = 10 (CLN6/CLN8), n = 10 (CLN6/CLN8dK), n = 12 (Golgi/CLN8), n = 15 (Golgi/CLN8dK), n = 11 (Golgi/CLN6 with CLN8), n = 18 (Golgi/CLN6 with CLN8dK), n = 10 (CLN6/CLN8), n = 14 (CLN6/CLN8dK). (C) Confocal microscopy showing that retrieval-deficient CLN8 (CLN8-RRXX, green signal) has partial colocalization with the Golgi marker GM130 (red) both in WT and CLN6–/– cells. Scale bar: 20 μm. Inset magnifications (×5) are reported. (D) Pearson correlation analysis showing partial colocalization of retrieval deficient CLN8dK (green signal) with the Golgi marker GM130 in WT and CLN6–/– cells. Data are mean ± SEM; n = 12 (WT), n = 11 (CLN6–/–). Statistical differences between groups were calculated using Student’s t test (B and D). NS, not statistically significant; ***P < 0.001.
CLN6 interacts with lysosomal enzymes and this interaction requires CLN6’s second luminal loop.
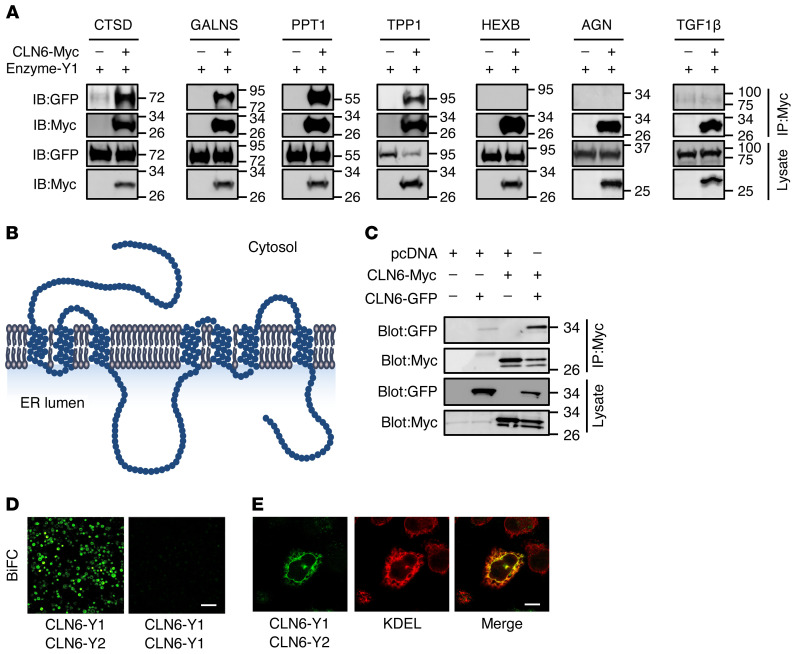
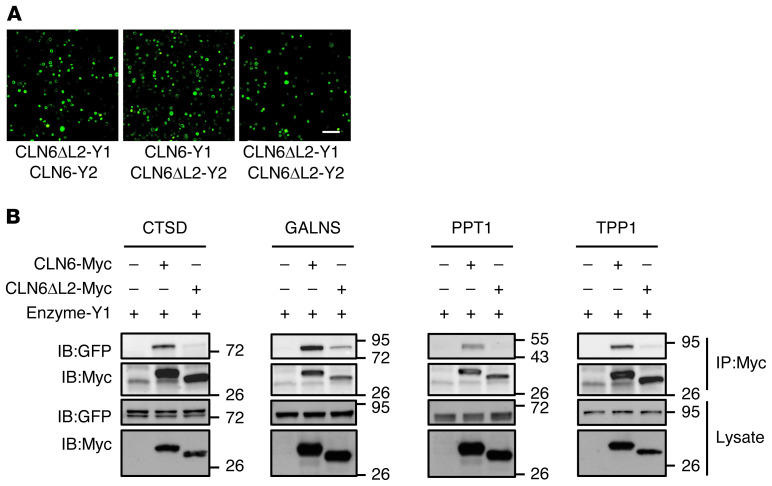
Next, we examined whether CLN6 interacts with lysosomal enzymes. Co-IP assays using myc-tagged CLN6 and a set of YFP-tagged enzymes showed that CLN6 interacts with the precursor forms of enzymes that we previously characterized as CLN8 interactors (CTSD, PPT1, TPP1, and GALNS) but does not interact with HEXB, which also did not interact with CLN8 (ref. 4 and Figure 4A). Co-IP of CLN6 with the nonlysosomal secretory proteins AGN and TGF-1β resulted in the absence of any detectable interaction, thus supporting the notion that CLN6 interactions are specific. To gain insight into the domains of CLN6 involved in the interaction with the enzymes, we analyzed the structure and conservation of the CLN6 protein. CLN6 has a cytosolic N-terminus, 7 transmembrane domains, and a C-terminus in the ER lumen (16, 22, 33). The cytosolic and luminal loops connecting the transmembrane domains are all small (<15 amino acids), with the exception of the second luminal loop, which is 48 amino acids long (Figure 4B). Evolutionary constrained region analysis (34) using 26 vertebrate species showed that CLN6’s second luminal loop (comprised between transmembrane domains 3 and 4) is the most constrained region of the protein (Supplemental Figure 5). Consistently, an analysis of the distribution of disease-causing single-amino acid mutations (missense mutations and deletions; https://www.ucl.ac.uk/ncl-disease) (10) showed that CLN6’s second luminal loop has a higher mutation rate (9 out of 48 positions, 19%) than does the rest of the protein (28 out of 235 positions, 12%). We therefore hypothesized that this loop is involved in the interaction between CLN6 and the lysosomal enzymes. To test this hypothesis, we generated a CLN6 construct lacking the second luminal loop (CLN6ΔL2) by deleting the amino acids from position 135 to position 175 (Supplemental Figure 6, A and B). We first verified that the CLN6ΔL2 protein, like full-length CLN6, localizes at the ER (Supplemental Figure 6C). Given that CLN6 forms homodimers (Figure 4C and ref. 22), we also tested whether CLN6ΔL2 is able to dimerize with full-length and ΔL2 CLN6 proteins. BiFC assays showed that full-length, YFP-tagged CLN6 forms a dimer (CLN6-Y1/CLN6-Y2) that is detectable by BiFC coupled with either confocal microscopy (Figure 4D) or flow cytometry (Supplemental Figure 6, D and E). The CLN6-Y1/CLN6-Y2 dimer correctly localized at the ER as detected by confocal microscopy (Figure 4E). BiFC analysis showed that CLN6ΔL2 retains the ability to form dimers with full-length CLN6 (CLN6ΔL2-Y1/CLN6-Y2 and CLN6ΔL2-Y2/CLN6-Y1) as well as with itself (CLN6ΔL2-Y1/CLN6ΔL2-Y2) (Figure 5A). Thus, the second luminal loop of CLN6 is dispensable for protein stability and self-interaction. We then tested whether CLN6ΔL2 is able to interact with the lysosomal enzymes by performing co-IP assay. Pull-down of myc-tagged CLN6ΔL2 followed by immunoblotting for Y1-tagged lysosomal enzymes (detectable with an anti-GFP antibody) showed that deletion of the second loop of CLN6 disrupted the interaction of CLN6 with the tested enzymes (Figure 5B). Thus, the second loop of CLN6 is required for the interaction of CLN6 with lysosomal enzymes.
Figure 4. CLN6 interacts with lysosomal enzymes.
(A) Co-IP analysis of CLN6 and lysosomal enzymes. Proteins were transiently expressed in HEK293T cells, and immunoprecipitates were analyzed by immunoblotting with the indicated antibodies. Molecular marker analysis indicates that CLN6 interacts with the enzymes’ precursors. Input represents 10% of the total cell extract used for IP. (B) Schematic representation of CLN6 protein. (C) Co-IP analysis of Y2-tagged CLN6 and myc-tagged CLN6. The proteins were transiently expressed in HEK293T cells, and immunoprecipitates were analyzed by immunoblotting with the indicated antibodies. Input represents 10% of the total cell extract used for IP. (D) Shown is a live BiFC assay of CLN6-Y1 with CLN6-Y2 in HeLa cells; expression of CLN6-Y1 is used as a negative control. Scale bar: 200 μm. (E) Confocal microscopy showing colocalization between reconstituted BiFC signal from CLN6-Y1/CLN6-Y2 dimerization (green) and the ER marker KDEL (red). Scale bar: 20 μm.
Figure 5. The second luminal loop of CLN6 is necessary for the interaction of CLN6 with the lysosomal enzymes.
(A) Shown are live BiFC assays of CLN6ΔL2-Y1 with CLN6-Y2, CLN6-Y1 with CLN6ΔL2-Y2, and CLN6ΔL2-Y1 with CLN6ΔL2-Y2 in HeLa cells. Scale bar: 200 μm. (B) Co-IP analysis of CLN6ΔL2 and lysosomal enzymes. Proteins were transiently expressed in HEK293T cells, and immunoprecipitates were analyzed by immunoblotting with the indicated antibodies. Input represents 10% of the total cell extract used for IP.
CLN6 and CLN8 are mutually necessary for their interaction with lysosomal enzymes.
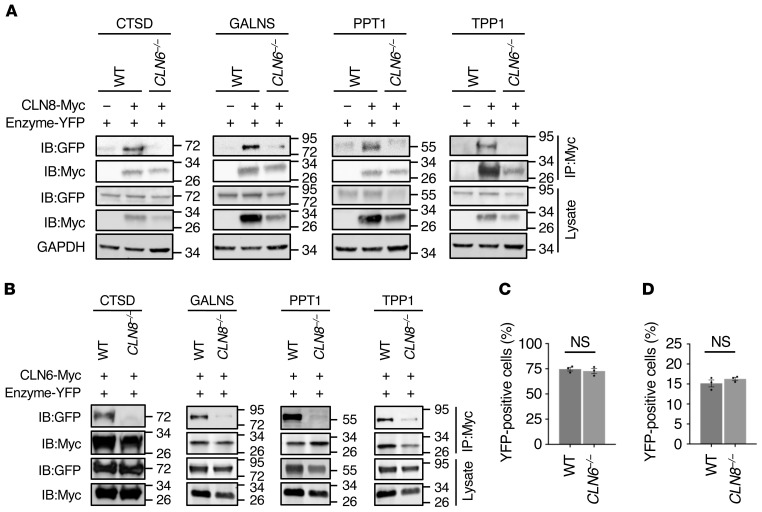
Next, we used CLN6–/– and CLN8–/– cells to investigate whether CLN6 and CLN8 are mutually necessary for their interaction with lysosomal enzymes. Co-IP assays showed that CLN6 is unable to interact with the lysosomal enzymes in CLN8–/– cells (Figure 6A); similarly, the interaction of CLN8 with the lysosomal enzymes is abolished in CLN6–/– cells (Figure 6B). In the absence of CLN6, however, the ability of CLN8 to form homodimers was not abolished as seen by BiFC analysis followed by flow cytometry (Figure 6C). Similarly, CLN6 was able to form homodimers in CLN8–/– cells (Figure 6D).
Figure 6. CLN6 and CLN8 are mutually necessary for their interaction with lysosomal enzymes.
(A) Co-IP analysis of myc-tagged CLN8 and Y2-tagged lysosomal enzymes (TPP1, CTSD, PPT1, and GALNS). Vectors were transiently transfected in WT and CLN6–/– HEK293T cells, and immunoprecipitates were analyzed by immunoblotting with the indicated antibodies. Input represents 10% of the total cell extract used for IP. (B) Co-IP analysis of myc-tagged CLN6 and Y2 tagged lysosomal enzymes (TPP1, CTSD, PPT1, and GALNS). Vectors were transiently transfected in WT and CLN8–/– HEK293T cells, and immunoprecipitates were analyzed by immunoblotting with the indicated antibodies. Input represents 10% of the total cell extract used for IP. (C) Flow cytometry quantification of Y1-CLN8/Y2-CLN8 BiFC signal in WT and CLN6–/– cells. Data are mean ± SEM (n = 3). (D) Flow cytometry quantification of CLN6-Y1/CLN6-Y2 BiFC signal in WT and CLN8–/– cells. Data are mean ± SEM (n = 3). Statistical differences between groups were calculated using Student’s t test (C and D).
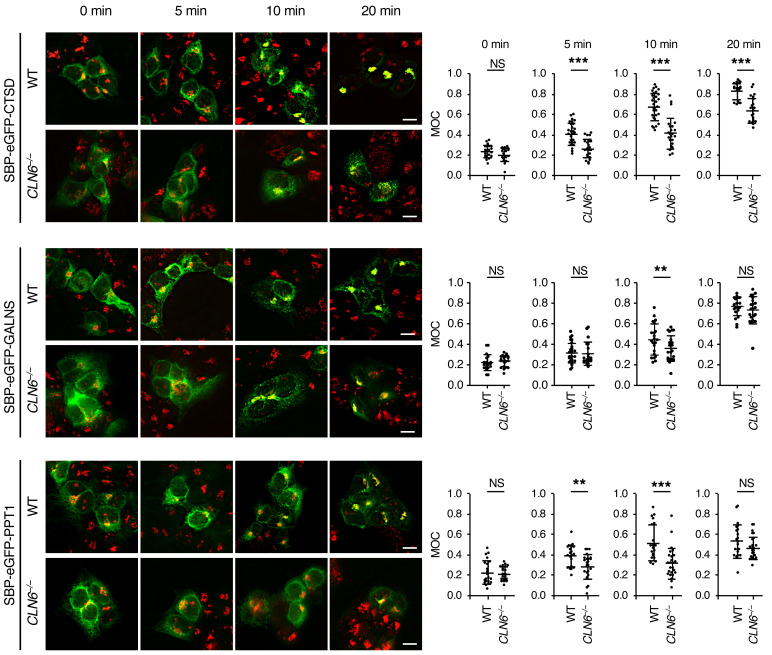
Based on these results, we conclude that CLN6 and CLN8 are obligate partners in the recruitment of newly synthesized lysosomal enzymes in the ER, and that the subsequent transfer of enzymes to the Golgi is mediated by CLN8 only. Although CLN6 is not loaded in COPII vesicles along with CLN8 and lysosomal enzymes, the observed decreased levels of enzymes in the lysosomal compartment upon CLN6 deficiency and the fact that CLN6 is essential for their recruitment in complex with CLN8 predict that, in the absence of CLN6, ER-to-Golgi transfer of the enzymes may be inefficient. To test this hypothesis, we set out to monitor ER-to-Golgi trafficking of CTSD, PPT1, and GALNS by using the RUSH (retention using selective hooks) system (35) based on a recent report that showed that ER-to-Golgi transfer of CTSD occurs in a short time (<30 minutes) and therefore requires a highly synchronized system for careful evaluation (36). In the RUSH system, the test protein is fused with a KDEL-tagged streptavidin binding protein (SBP), which enables retention of the test protein in the subcellular compartment of choice by simultaneous expression of an organelle-targeted hook protein that contains a streptavidin domain. The interaction between the test protein and the hook protein is stable and can be reversed by the addition of biotin to outcompete SBP binding. Biotin supplementation thus results in a synchronous release of the test protein, which can be monitored for relocation by time-course confocal microscopy. For our experiments, we coexpressed an ER-targeted hook protein (35) with SBP- and GFP-fused CTSD, PPT1, and GALNS. We used SBP-, GFP-fused lysozyme C (LyzC/LYZ), a nonlysosomal soluble protein that traffics through the ER and Golgi (36), as a control protein. We monitored the subcellular localization of CTSD, PPT1, GALNS, and LyzC at 0, 5, 10, and 20 minutes from their biotin-induced synchronous release in WT and CLN6–/– cells. As a quantitative measure, we assessed the overlap of the enzyme signal with that of the Golgi marker GM130 at each time point. The results showed that the absence of CLN6 caused a significant delay in ER-to-Golgi trafficking of the tested lysosomal enzymes, but not of LyzC (Figure 7 and Supplemental Figures 7 and 8), thus confirming that the lack of a functional CLN6-CLN8 complex for the recruitment of lysosomal enzymes results in their inefficient exit from the ER.
Figure 7. CLN6 deficiency impairs trafficking of lysosomal enzymes.
Confocal microscopy analysis of WT and CLN6–/– HEK293T cells transfected with plasmids expressing enzymes fused with SBP-EGFP (SBP-EGFP-CTSD, SBP-EGFP-GALNS, and SBP-EGFP-PPT1) and streptavidin-KDEL ‘‘anchor’’ that retains SBP-containing proteins in the ER. Shown are representative images of cells without addition of biotin (0 minutes) and at 5, 10, and 20 minutes from the addition of biotin. Manders’ overlap coefficients (MOC) measuring the degree of colocalization between the test protein (green signal) and the Golgi marker GM130 (red signal) are reported. Data are mean ± SEM; WT cells, n = 21 (CTSD, 0 minutes), n = 27 (CTSD, 5 minutes), n = 30 (CTSD, 10 minutes), n = 20 (CTSD, 20 minutes), n = 20 (GALNS, 0 minutes), n = 30 (GALNS, 5 minutes), n = 22 (GALNS, 10 minutes), n = 20 (GALNS, 20 minutes), n = 21 (PPT1, 0 minutes), n = 21 (PPT1, 5 minutes), n = 20 (PPT1, 10 minutes), n = 21 (PPT1, 20 minutes); CLN6–/– cells, n = 20 (CTSD, 0 minutes), n = 23 (CTSD, 5 minutes), n = 23 (CTSD, 10 minutes), n = 21 (CTSD, 20 minutes), n = 22 (GALNS, 0 minutes), n = 21 (GALNS, 5 minutes), n = 22 (GALNS, 10 minutes), n = 22 (GALNS, 20 minutes), n = 20 (PPT1, 0 minutes), n = 23 (PPT1, 5 minutes), n = 25 (PPT1, 10 minutes), n = 24 (PPT1, 20 minutes). Statistical differences between groups were calculated using Student’s t test. NS, not statistically significant; **P < 0.01; ***P < 0.001. Scale bars: 100 μm.
Loss of CLN6 does not aggravate pathology of CLN8-deficient mice.
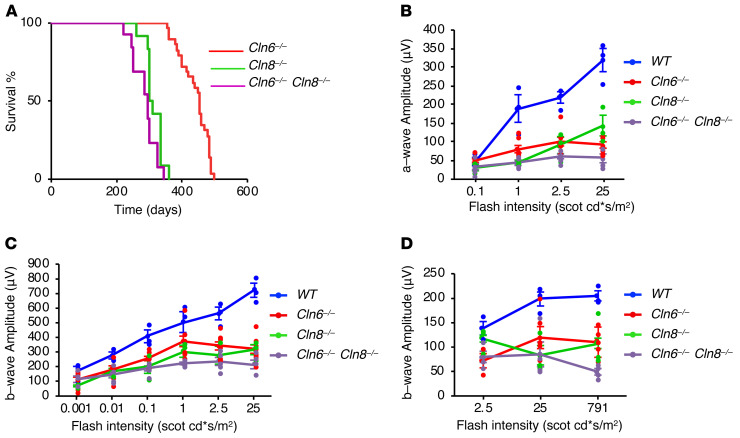
If CLN6 and CLN8 work as a functional unit in vivo, it is expected that simultaneous deletion of both would lead to consequences similar to either single deletion. Life span analyses showed that Cln6–/– and Cln8–/– mice had a slightly different survival rate. The median survival of Cln6–/– mice was 450 days, whereas the median survival of Cln8–/– mice was 302 days (log-rank test; z = 7.07, P < 0.001), in line with previous reports (18, 19, 21). Double KO mice had a median survival of 290 days, which was indistinguishable from that of Cln8–/– mice (log-rank test; z = 2.24, P = 0.025) (Figure 8A). Thus, simultaneous deficiency of CLN6 and CLN8 does not accelerate disease progression compared with CLN8 single deficiency.
Figure 8. Combined deficiency of CLN6 and CLN8 in mice does not accelerate disease progression compared with single deficiencies.
(A) Life span analysis of Cln6–/– (n = 29), Cln8–/– (n = 12), and Cln6–/– Cln8–/– (n = 13) mice. (B) Graph of a-wave amplitudes of scotopic ERG waveforms from WT (n = 3), Cln6–/– (n = 8), Cln8–/– (n = 3), and Cln6–/– Cln8–/– (n = 5) mice in response to a series of light stimuli. (C) Graph of b-wave amplitudes of scotopic ERG waveforms from WT (n = 3), Cln6–/– (n = 8), Cln8–/– (n = 3), and Cln6–/– Cln8–/– (n = 5) mice. (D) Graph of b-wave amplitudes of photopic ERG waveforms from WT (n = 3), Cln6–/– (n = 6), Cln8–/– (n = 3), and Cln6–/– Cln8–/– (n = 4) mice.
Activation of ER stress pathways was previously observed in the brain of Cln8–/– mice (37). We performed a comparative analysis of ER stress pathways in Cln6–/–, Cln8–/–, and double-KO mice by analyzing the expression of several protein markers of ER stress pathways — BiP/HSPA5, CHOP/DDIT3, eIF2α/EIF2S1 (phosphorylated and total protein), JNK1 (phosphorylated and total protein), IRE1/ERN1, and ATF6 — in extracts from whole brain and liver tissue homogenates and from 3 subregions of the brain (cortex, hippocampus, and cerebellum). Overall, the results showed dynamic responses to gene deficiencies for most of the markers analyzed, with individual intragroup mouse variations generally greater than intergroup variations (Supplemental Figure 9, A and B, and Supplemental Figure 10, A–C). Exceptions were BiP, which was upregulated in the hippocampus of single- and double-KO mice and in the liver of double-KO mice, and ATF6, which was upregulated in the brain of Cln8–/– mice. Interestingly, the double-KO mice did not generally recapitulate an additive effect of the variations observed in the single-KO mice, indicating the possible presence of adaptive responses that could differ across genotypes and at the level of individual mice.
A notable phenotype described in most mouse models of Batten disease is retinal degeneration, which eventually leads to complete blindness (38, 39). Both Cln6–/– and Cln8–/– mouse lines display early-onset loss of vision that can be quantified by electroretinogram (ERG) analysis (38, 40). To compare the visual function of single- and double-KO mouse lines, we performed scotopic and photopic ERGs to assess the function of rods and cones, respectively. Scotopic ERG was performed by measuring a-wave (photoreceptor) and b-wave (inner retina) amplitudes. The results from both scotopic (Figure 8, B and C) and photopic (Figure 8D) ERGs showed that, while all mutants displayed significant differences compared with age-matched WT mice, there was no worsening of visual function in the double-KO mice compared with Cln8–/– mice (which had a subtly worse phenotype than Cln6–/– mice; Supplemental Table 1). Together, these analyses support the notion that CLN6 and CLN8 work as a functional unit in vivo.
Discussion
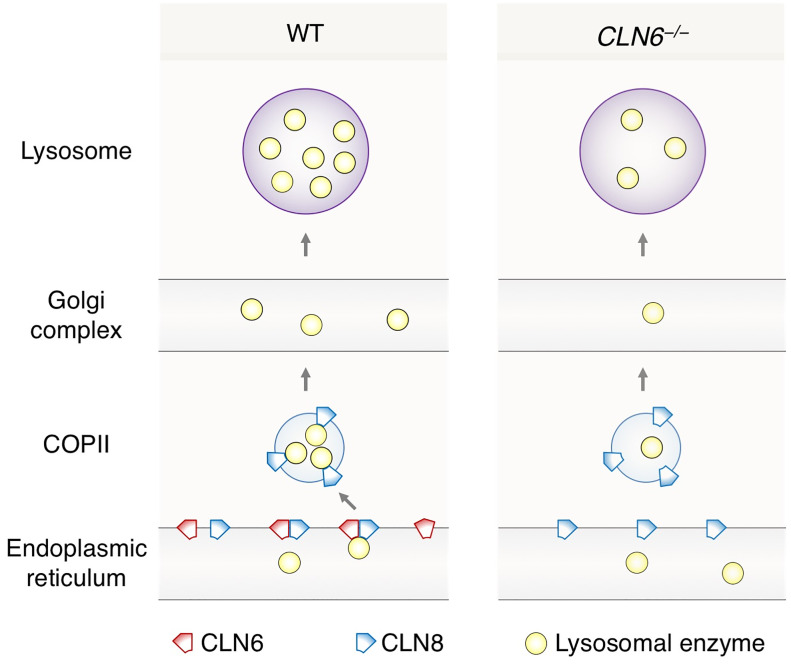
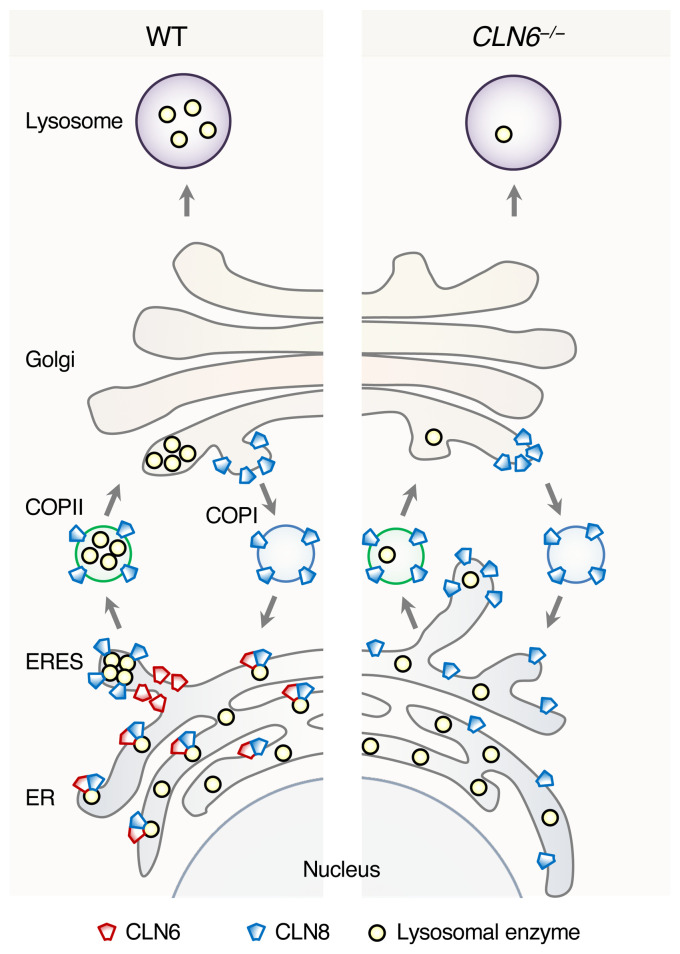
This study identifies CLN6 as a key factor in the biogenesis of lysosomes by uncovering its function as an obligate component of a CLN6-CLN8 complex that recruits lysosomal enzymes at the ER to promote their Golgi transfer. We refer to this complex as EGRESS (ER-to-Golgi relaying of enzymes of the lysosomal system) because of its role in recruiting lysosomal enzymes for Golgi delivery. The emerging model identifies a 2-step process for the engagement of lysosomal enzymes in the ER and their transfer to the Golgi based on the cooperation between CLN6 and CLN8. In the first step, CLN6 and CLN8 form the EGRESS complex, which recruits the enzymes via an interaction that depends on both the large luminal loop of CLN6 (which is dispensable for the interaction with CLN8) and the large luminal loop of CLN8 (4). The absence of either protein prevents the interaction of the other with the enzymes, indicating that the EGRESS complex is the minimal unit required for such interaction. In the second step, CLN8 is loaded into COPII vesicles to escort the enzymes to the cis-Golgi. It is possible that during the loading into COPII vesicles CLN6 is displaced by the interaction between CLN8 and COPII components, thus ending the loading cycle of the EGRESS complex. A dual function of CLN8 as a member of the EGRESS complex and as the cargo receptor that interacts with components of COPII vesicles is consistent with the observed data indicating a more rapid progression of the disease in Cln8–/– mice compared with Cln6–/– mice. Upon Golgi transfer, the enzymes are subsequently trafficked to the lysosomes via selective transport from the trans-Golgi network, whereas the empty receptor is recycled back to the ER. If either component of the EGRESS complex is absent (or functionally defective), ER exit of the enzymes is inefficient, thereby resulting in enzyme depletion at the lysosome. A graphical depiction of inefficient enzyme exit from the ER upon CLN6 deficiency is depicted in Figure 9.
Figure 9. Schematic model of ER-to-Golgi lysosome enzyme trafficking.
Shown is a comparison between WT conditions and deficiency of CLN6.
Inefficient organelle exit and depletion of cargo proteins upon deficiency of components of their receptor systems are commonly observed in mammalian cells. For example, ERGIC-53 promotes ER export of immunoglobulin IgM and coagulation factors V/VII in complex with ERp44 and MCFD2, respectively (41, 42). In cells lacking functional ERGIC-53, its cargo proteins reach their destination at a slower rate and in reduced amounts (43, 44). Similarly, exit of LYZ/LyzC from the Golgi is delayed upon deletion of its sorting protein, CAB45/SDF4 (36). Additional examples from recent studies include a variety of receptor systems and cargo proteins — VTI1A/B for Golgi export of neuropeptide Y (45), SURF4 for the secretion of PCSK9 (46), and p24/TMED2 for ER export of GPI-anchored proteins (47). Bulk flow or lower-affinity binding to other cargo receptors can explain slower cargo transport (48).
Recent studies focused on TFEB, a master regulator of lysosomal biogenesis and function (49, 50), indicate that lysosomal enhancement may counteract disease progression in animal models of lysosomal storage disorders, including a model of Batten disease (51, 52). TFEB increases the expression and activity of most lysosomal enzymes by directly enhancing the transcription rates of their genes and of several additional genes that contribute to lysosomal acidification, lysosomal homeostasis, and Golgi-to-lysosome transport pathways (2). Although CLN6 and CLN8 genes are not among the direct transcriptional targets of TFEB (49, 53), the finding that defects in the EGRESS complex result in reduced amounts of enzymes at the lysosome identifies CLN6 and CLN8 diseases as candidate conditions for testing TFEB-mediated lysosomal enhancement therapy to investigate its capacity to increase the levels and activity of lysosomal enzymes in these conditions (54). In a different approach, CLN6 and CLN8 could be tested in the treatment of lysosomal storage diseases caused by partial enzyme deficiency — conditions that are most typically due to point mutations in enzymes that reduce their stability or trafficking. This strategy could be tested in appropriate models of partial lysosomal enzyme deficiency either with a direct genetic approach — gene therapy of CLN6, CLN8, or both — or pharmacologically by using yet-to-be-discovered drug activators or stabilizers of the EGRESS complex. Future studies will determine whether increased expression or activity of the EGRESS complex is able to enhance the stability or trafficking of mutant enzymes and ameliorate disease phenotypes. If successful, this approach could complement current methods to treat lysosomal storage diseases based on enzyme replacement therapy and gene therapy.
Remarkably, among the enzymes that are depleted upon deficiency of CLN6 or CLN8 are TPP1 and CTSD, which are the defective proteins in 2 other Batten disease subtypes — CLN2 and CLN10, respectively (55–57). Both TPP1 and CTSD are involved in the degradation of subunit c of mitochondrial ATP synthase (SCMAS) (58–60), and studies that have characterized the composition of the storage material in various Batten subtypes have determined that SCMAS accumulates not only in CLN2 and CLN10 but also in CLN6 and CLN8 diseases (61–63). Our results thus establish a framework to interpret the accumulation of SCMAS upon CLN6 and CLN8 deficiency as being caused by the partial depletion of the SCMAS degrading proteins, TPP1 and CTSD, at the lysosome. It should be noticed that the clinical courses of CLN2 and CLN10 cases caused by a complete lack of TPP1 or CTSD activity are much more severe than that of even the most severe cases of CLN6 and CLN8 diseases (7, 8). This indicates that in the absence of functional CLN6 or CLN8, nonnegligible amounts of TPP1 and CTSD (and presumably other enzymes) are still active in the lysosome, helping the cell cope with lysosomal homeostasis and metabolism for a relatively long period of time. As in the case of other lysosomal storage disorders, the reasons for the observed progressive decline in CLN6 and CLN8 diseases could be rooted in the progressive nature of the accumulation of undegraded substrates in the lysosome. Alternatively, the physiological role of CLN6 and CLN8 might vary with organismal development and become more essential at later stages. The complexity of this matter is compounded by the recent observation that different brain cell types respond differently to the lack of CLN6, as recently observed with a gene therapy–mediated approach in mice with a CLN6-defective background (64). These themes could be explored in future work by analyzing the expression of CLN6 and CLN8 in various organs at various developmental ages when reliable antibodies for the mouse proteins become available, or by generating tissue- or cell-specific knockout lines for Cln6 and Cln8. These observations also suggest therapeutic opportunities for CLN6 and CLN8 diseases based on the possibility to provide the depleted enzymes exogenously via enzyme replacement therapy, an option that is being tested for several lysosomal enzymes including CLN2 but that is not available for transmembrane proteins such as CLN6 and CLN8 (65–68). Follow-up studies in preclinical models are needed to explore whether or not this approach would add any benefits to established procedures such as gene therapy for these diseases (69).
In summary, our findings address the molecular mechanism underlying Batten disease caused by loss of functional CLN6 and identify the EGRESS complex as the mediator of the recruitment of lysosomal enzymes in the ER for Golgi transfer. These findings uncover a previously unappreciated complexity of the early steps of lysosomal enzyme trafficking and shed light on the molecular pathogenesis of CLN6 disease, which can henceforth be interpreted as a disorder of impaired lysosomal biogenesis.
Methods
Animal husbandry.
Cln6nclf mice on C57/BL6J background (Cln6–/– mice) were a gift from Susan Cotman (Harvard Medical School, Boston, Massachusetts, USA) and are available from the Jackson Laboratory (catalog 003605). The Cln8mnd mouse line (Cln8–/– line) was purchased from the Jackson Laboratory (catalog 001612). Cln6–/– and Cln8–/– double-KO mice were generated by crossing the single-KO lines. Food and water were provided ad libitum. For subcellular fractionation and ERG experiments we used mice at 6 weeks of age. Only males were used for all analyses. Investigators were blinded to mouse genotype during data acquisition, and no randomization was necessary. No data were excluded from this study.
Molecular biology.
Supplemental Table 2 lists the antibodies used throughout the study. Supplemental Table 3 lists the oligonucleotides used for cloning, genome editing, and PRC. CLN6 and CLN8 were cloned into myc and YFP vectors (backbone: pcDNA3.1 from Invitrogen) by retrotranscription of RNAs from HeLa and HEK293T cells using the QuantiTect Reverse Transcription kit (Qiagen) followed by PCR-mediated amplification and plasmid insertion with the in-Fusion cloning kit (Clontech). The CLN6ΔL2 construct was obtained by removing the codons for amino acids 155-222 using the Q5 Site-Directed Mutagenesis kit (New England Biolabs).
Immunoblotting.
Before harvesting, cells were rinsed in cold PBS and lysed using RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 2 mM EDTA) or NP40 lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Np-40 [vol/vol], 10% glycerol) with protease (Roche) and phosphatase (Roche) inhibitors (1:100). Cells were left in lysis buffer on a nutator for 1 hour at 4°C. After 1 hour of lysis the solution was centrifuged at 15,700g for 20 minutes and the supernatant was collected. Protein concentrations were measured with the bicinchoninic acid protein assay kit (Pierce), using bovine serum albumin as standard. Lysates were separated on a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinyl difluoride (PVDF) membranes. Blots were incubated in blocking buffer (5% dried skimmed milk in Tris-buffered saline, pH 7.4, and 0.2% Tween-20, TBST) followed by overnight incubation with appropriate antibodies, which were diluted in blocking buffer. The following day, membranes were washed 3 times for 10 minutes each in TBST before incubation for 1 hour with secondary HRP antibodies diluted in blocking buffer. Detection was carried out with ECL Western blotting detection reagent (GE Healthcare). Images were detected with ImageQuant LAS 4000 (GE Healthcare) and quantified by Fiji analysis software.
Generation of CLN6 knockout and CLN8 knockin cells.
We used CRISPR/Cas9 genome editing to introduce a deletion in exon 2 of the CLN6 gene in HEK293T cells. In brief, we designed 2 complementary oligonucleotide couples (Supplemental Table 3) using the online CRISPR design tool (http://crispr.mit.edu), coding for a guide RNA upstream of a protospacer adjacent motif (PAM) site in exon 2 of CLN6. The 2 oligonucleotides were annealed and subsequently cloned into the pX458 plasmid (70), followed by Sanger sequencing of the insert to confirm the correct sequence. HEK293T cells were then transfected with 2 μg plasmid and split into a 96-well plate by single-cell deposition. DNA was isolated from the expanded single colonies when confluent enough and used in PCRs using oligonucleotides to amplify exon 2 of CLN6 with CloneAmp HiFi PCR Premix (Clontech) according to the manufacturer’s instructions; clones that were unable to produce an amplicon were subsequently Sanger sequenced to confirm deletion of exon 2. We inserted a Myc tag (5′-GAACAAAAACTCATCTCAGAAGAGGATCTG-3′) just before the stop codon of endogenous CLN8 in HEK293T cells by combining CRISPR/Cas9 with single-stranded oligodeoxynucleotides (ssODNs) as described (71). We selected recombinant clones by single colony expansion followed by gDNA extraction and PCR using oligonucleotides to amplify exon 3 of CLN8. Sanger sequencing confirmed the correct insertion of the Myc tag in a clone that was subsequently used for the described experiments.
Quantitative real-time PCR.
Total RNA was extracted from control and Cln6nclf mouse embryonic fibroblasts, and from control and CLN6–/– HEK293T cells using the RNEasy kit (Qiagen) according to the manufacturer’s instructions. RNA was quantified using the Nano-Drop 8000 (Thermo Fisher) followed by cDNA synthesis using the QuantiTect Reverse Transcription kit (Qiagen). The primers for PCR with reverse transcription reactions are listed in Supplemental Table 3. Quantitative real-time PCR was performed by using iQ SYBR Green Supermix on the CFX96 Touch Real-Time Detection System (Bio-Rad Laboratories) with the following conditions: 95°C, 5 minutes; (95°C, 10 seconds; 60°C, 10 seconds; 72°C, 15 seconds) × 40. Analyses were conducted using CFX manager software (Bio-Rad) and the threshold cycle (CT) was extracted from the PCR amplification plot. Relative gene expression was determined using methods described previously, normalizing to GAPDH (for human genes) and Sp16 (for mouse genes). The change in messenger RNA level of the genes was expressed as fold change.
RUSH cargo sorting assay.
Plasmids pIRESneo3-LyzC-SBP-EGFP and pIRESneo3-SS-SBP-EGFP-CTSD were a gift from J. Von Blume (Yale School of Medicine, New Haven, Connecticut, USA). WT and CLN6–/– cells were cultured in a 96-well plate precoated for 2 hours with poly-D-Lysine to aid in cell adherence. Cells were transfected for 24 hours using plasmids expressing test (CTSD, PPT1, GALNS) and control (LyzC/LYZ) proteins fused with EGFP and a SBP (pIRESneo3-LyzC-SBP-EGFP, pIRESneo3-SS-SBP-EGFP-CTSD, pIRESneo3-SS-SBP-EGFP-PPT1, and pIRESneo3-SS-SBP-EGFP-GALNS). Cells were incubated with 40 μM d-Biotin (SUPELCO) in DMEM for 10 minutes or without d-Biotin as a control to confirm the retention of the reporter. Cells were then washed once in PBS and fixed in 4% PFA in PBS for 15 minutes and used in confocal microscopy.
Confocal microscopy analysis.
Cells were grown on glass coverslips in 24-well plates overnight before treatment. Cells were transfected using Polyplus reagent, incubated for 6 hours, and left to incubate for an additional 36 hours after changing the culture media. At the end of the treatment, cells were rinsed with PBS and fixed with 4% paraformaldehyde (PFA) in PBS at room temperature (RT) for 15 minutes. Cells were then rinsed 3 times with PBS and blocked with blocking reagent (0.1% saponin, 10% goat serum in PBS) for 60 minutes at RT. After blocking, cells were washed twice with PBS, followed by incubation with appropriate primary antibody in blocking reagent overnight at 4 °C. Cells were then washed 3 times with PBS and later incubated with appropriate Alexa Fluor–conjugated secondary antibodies against primary host animal for 1 hour at RT in the dark. Cells were then washed 5 times with PBS and coverslips were mounted with VECTASHIELD mounting media containing DAPI (Vector Laboratories). Images were acquired with Zeiss 710 and Zeiss 880 confocal microscopes (Carl Zeiss, Inc.). For the RUSH analysis of protein subcellular trafficking, imaging was performed on a GE Healthcare DVLive epifluorescence image restoration microscope using an Olympus PlanApo ×60/1.42 NA objective and a 1.9k × 1.9k pco.EDGE sCMOS_5.5 camera with a 1900 × 1900 FOV. The filter sets used were DAPI, FITC, TRITC, and CY5. For colocalization analysis, quantification was done with organellar markers, pictures were analyzed using Coloc2 and plot profile plugin using ROI manager in Fiji software.
Immunoprecipitation assay.
HEK293T cells after 24 or 48 hours of transfection were scraped off from Petri dishes in cold PBS. Cells were centrifuged at 400g for 5 minutes and the pellet was resuspended in NP40 lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Np-40 [vol/vol], 10% glycerol) with protease (Roche) and phosphatase (Roche) inhibitors (1:100). Cells were lysed for 30 minutes at 4°C on a nutator. Upon lysis, equal amounts of protein lysates (500 μg) were incubated with the indicated primary antibodies for 3 hours at 4°C. After antibody incubation, samples were further incubated for 2 hours in 40 μL precleared protein-G agarose beads (Roche). Beads with immunocomplexes were centrifuged at 3000g and washed 4 times in lysis buffer with intermittent incubations, during each wash, for 6 minutes on the nutator at 4°C. After the fourth wash, beads were resuspended in Laemmli SDS sample buffer with β-Mercaptoethanol heated at 95°C for 10 minutes and then analyzed by immunoblotting. For experiments involving CLN8, samples were incubated at 37°C to avoid formation of CLN8 dimers and smears observed at higher temperatures which are hard to resolve on a SDS gel.
Flow cytometry analysis.
HeLa cells were transfected with 200 ng YFP1-tagged and 200 ng YFP2-tagged constructs, in combination with 200 ng Ruby plasmid that was used as a reference for transfection efficiency. Cells were collected 48 hours after transfection in PBS with 10% FBS. Fluorescence of 10,000 cells per sample was determined by flow cytometry using the BD LSRFortessa Cell Analyzer (BD Biosciences) with the HTS auto-sampler device. Only cells positive for YFP and Ruby were considered for the subsequent analysis of reconstituted YFP signal quantification.
Subcellular fractionation.
Three mouse livers per sample were pooled and centrifuged in a discontinuous Nycodenz (Progen Biotechnik) density gradient as previously described (72), with modifications. Briefly, we homogenized tissues in an assay buffer (0.25 M sucrose, pH 7.2) and centrifuged at 4800g for 5 minutes, and then at 17,000g for 10 minutes. The sediment of the second centrifugation was washed at 17,000g for 10 minutes. Consequently, it was resuspended 1:1 vol/vol in 84.5% Nycodenz and placed on the bottom of an Ultraclear (Beckman) tube. Above this, a discontinuous gradient of Nycodenz was constructed using the following percentages from bottom to top: 32.8%, 26.3%, and 19.8% Nycodenz. Samples were then centrifuged for 1 hour in an SW 40 Ti rotor (Beckman) at 141,000g. Lysosome-enriched fractions were collected from the 26.3/19.8 interface and diluted in 5 to 10 volumes of assay buffer. Finally, they were centrifuged at 37,000g for 15 minutes. Pellets were then resuspended in 500 μL assay buffer.
Enzyme assays.
Enzyme activity assays were conducted by using fluorophore analogs of enzymes substrates, as previously described: TPP1 (73), GAA (74), GBA (75), GLB1 (76), and NAGLU (77).
Electroretinography (ERG).
Mice were dark-adapted overnight and then anesthetized by a single i.p. injection of 22 mg/kg ketamine, 4.4 mg/kg xylazine, and 0.37 mg/kg acepromazine. Pupils were dilated with a drop of tropicamide (1.0%) and phenylephrine (2.5%) and then corneas were anesthetized with a drop of proparacaine (1.0%). After 1 minute, excess fluid was removed and a drop of hypromellose (2%) was placed on each cornea to keep it moistened and provide a good contact between the cornea and the ERG electrode (N1530NNC, LKC Technologies). All tests were performed under a dim red light and a feedback-controlled heating pad was used to keep treated mice at a constant body temperature of 37°C. ERG recordings were performed using UTAS Visual Diagnostic System and EMWIN software (LKC Technologies). Scotopic ERGs were performed using 6 flash intensities (–34, –24, –14, –4, 0, and 10 dB). Photopic ERGs were subsequently recorded. Mice were light-adapted to a 30 cd*s/m2 white background for 2 minutes after scotopic ERG recordings, and then photopic ERGs were recorded with flash intensities of 0, 10, and 25 dB. ERG data was plotted using GraphPad Prism5 software. All mice were analyzed at 4 weeks of age.
CBM assay.
HeLa cells were plated on coverslips, 24 hours before the experiment, in 24-well plates in DMEM at 37°C in the presence of 5% CO2. Cells were transfected the next day with either myc-tagged CLN6 or myc-tagged CLN8. At 24 hours after transfection, the media was removed, cells were washed with PBS, and CBM was added to each well at a final concentration of 2 mM for a 90-minute period.
In vitro COPII vesicle budding reaction.
Rat liver cytosol was prepared from fresh livers as previously described (28). HeLa cells were cotransfected with CLN6-myc and CLN8-myc, permeabilized as previously described (78), and used as donor membranes. Vesicle formation and purification was performed as described previously (78). In brief, donor membranes were incubated with rat liver cytosol (4 mg/mL), ATP regeneration system, 0.3 mM GTP, and SAR1A H79G (10 ng/μl) in a final volume of 100 μL. The reaction was carried out at 30°C for 1 hour and terminated by incubation on ice for 5 minutes. Donor membranes were removed by centrifugation at 14,000g for 12 minutes at 4°C. The resulting supernatant was centrifuged at 115,000g for 25 minutes at 4°C to collect COPII vesicles. Vesicle pellets were then resuspended in 15 μL buffer C (10 mM Tris-HCl, pH 7.6, 100 mM NaCl, 1% Triton X-100) and Laemmli sample buffer and heated at 55°C for 15 minutes. Vesicle samples and original donor membrane samples were resolved by SDS-PAGE, transferred onto PVDF membranes, and subjected to immunoblotting to detect various ER-resident proteins and COPII cargo proteins.
Sequence analysis.
CLN6 sequence analysis was performed by retrieving CLN6 sequences from the NCBI database (79) or from the UCSC Genome Browser (80) by BLAT searches using available CLN6 protein or DNA sequences. Local evolutionary rates of CLN6 protein sequence were calculated using the webtool Aminode (34) by feeding the indicated sequences as inputs for the custom analysis. Downloaded data were graphically combined with a multiple protein alignment (81) to generate the graph of the local evolutionary rates.
Statistics.
All experiments were performed at least in biological triplicate. Experimental analysis was performed in a blinded fashion for quantification of the Pearson correlation in immunofluorescence analysis. Statistical significance was tested using 2-sided t test for simple comparisons, whereas false discovery rate (FDR) was used for multiple comparisons. A value of P less than 0.05 (FDR < 0.1 for multiple comparisons) was considered significant.
Study approval.
All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Author contributions
MS conceived and supervised the study. LB and MS designed the experiments with the contributions of JS, ADR, PZ, AE, RP, DR, JRC, CB, KTC, RNS, SYJ, JMW, RC, and RWS. LB performed most cell biology, molecular biology, and mouse analyses. ADR, RP, and JS performed molecular analyses. LB and PZ performed in vitro budding assay under the supervision of RWS. AE, DR, and LB performed ERG analysis under the supervision of RC. CB and KTC participated in mouse tissue collection under the supervision of JMW and MS. LB and MS wrote the manuscript with input from all authors.
Supplementary Material
Acknowledgments
We thank H. Bellen, H. Zoghbi, M. Wang, and T. Eissa for helpful discussion and K. Venkatachalam and H. Jafar-Nejad for critical reading of the manuscript. This work was supported by NIH grants NS079618 and GM127492 (to MS) and grants from the Gwenyth Gray Foundation, Beyond Batten Disease Foundation, and NCL-Stiftung (to MS). This project was supported in part by IDDRC grant number 1U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (Core: Integrated Microscopy). This project was supported by the Integrated Microscopy Core and the Proteomics Core at Baylor College of Medicine with funding from NIH (DK56338, CA125123), CPRIT (RP150578, RP170719), the Dan L. Duncan Comprehensive Cancer Center, and the John S. Dunn Gulf Coast Consortium for Chemical Genomics.
Version 1. 06/29/2020
Electronic publication
Version 2. 08/03/2020
Print issue publication
Footnotes
PZ’s present address is: Department of Neurology, Weill Institute for Neurosciences, UCSF, San Francisco, California, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(8)4118–4132.https://doi.org/10.1172/JCI130955.
Contributor Information
Lakshya Bajaj, Email: Lakshya_Bajaj@hms.harvard.edu.
Jaiprakash Sharma, Email: jsharma@bcm.edu.
Alberto di Ronza, Email: dironzalberto@gmail.com.
Aiden Eblimit, Email: aeblimit@central.uh.edu.
Rituraj Pal, Email: riturajpalster@gmail.com.
Dany Roman, Email: dany.roman@bcm.edu.
John R. Collette, Email: john.collette@bcm.edu.
Rui Chen, Email: ruichen@bcm.edu.
Marco Sardiello, Email: sardiell@bcm.edu.
References
- 1.Hu YB, Dammer EB, Ren RJ, Wang G. The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl Neurodegener. 2015;4:18. doi: 10.1186/s40035-015-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajaj L, Lotfi P, Pal R, Ronza AD, Sharma J, Sardiello M. Lysosome biogenesis in health and disease. J Neurochem. 2019;148(5):573–589. doi: 10.1111/jnc.14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rohrer J, Kornfeld R. Lysosomal hydrolase mannose 6-phosphate uncovering enzyme resides in the trans-Golgi network. Mol Biol Cell. 2001;12(6):1623–1631. doi: 10.1091/mbc.12.6.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.di Ronza A, et al. CLN8 is an endoplasmic reticulum cargo receptor that regulates lysosome biogenesis. Nat Cell Biol. 2018;20(12):1370–1377. doi: 10.1038/s41556-018-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lonka L, Kyttälä A, Ranta S, Jalanko A, Lehesjoki AE. The neuronal ceroid lipofuscinosis CLN8 membrane protein is a resident of the endoplasmic reticulum. Hum Mol Genet. 2000;9(11):1691–1697. doi: 10.1093/hmg/9.11.1691. [DOI] [PubMed] [Google Scholar]
- 6.Passantino R, et al. Identifying protein partners of CLN8, an ER-resident protein involved in neuronal ceroid lipofuscinosis. Biochim Biophys Acta. 2013;1833(3):529–540. doi: 10.1016/j.bbamcr.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 7.Williams RE, Mole SE. New nomenclature and classification scheme for the neuronal ceroid lipofuscinoses. Neurology. 2012;79(2):183–191. doi: 10.1212/WNL.0b013e31825f0547. [DOI] [PubMed] [Google Scholar]
- 8.Dyken PR. Reconsideration of the classification of the neuronal ceroid-lipofuscinoses. Am J Med Genet Suppl. 1988;5:69–84. doi: 10.1002/ajmg.1320310610. [DOI] [PubMed] [Google Scholar]
- 9.Haltia M. The neuronal ceroid-lipofuscinoses: from past to present. Biochim Biophys Acta. 2006;1762(10):850–856. doi: 10.1016/j.bbadis.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 10.Mole SE, Cotman SL. Genetics of the neuronal ceroid lipofuscinoses (Batten disease) Biochim Biophys Acta. 2015;1852(10 Pt B):2237–2241. doi: 10.1016/j.bbadis.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao H, et al. Mutations in a novel CLN6-encoded transmembrane protein cause variant neuronal ceroid lipofuscinosis in man and mouse. Am J Hum Genet. 2002;70(2):324–335. doi: 10.1086/338190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mole SE, Michaux G, Codlin S, Wheeler RB, Sharp JD, Cutler DF. CLN6, which is associated with a lysosomal storage disease, is an endoplasmic reticulum protein. Exp Cell Res. 2004;298(2):399–406. doi: 10.1016/j.yexcr.2004.04.042. [DOI] [PubMed] [Google Scholar]
- 13.Williams RE, Aberg L, Autti T, Goebel HH, Kohlschütter A, Lönnqvist T. Diagnosis of the neuronal ceroid lipofuscinoses: an update. Biochim Biophys Acta. 2006;1762(10):865–872. doi: 10.1016/j.bbadis.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Mole SE, Williams RE, Goebel HH. Correlations between genotype, ultrastructural morphology and clinical phenotype in the neuronal ceroid lipofuscinoses. Neurogenetics. 2005;6(3):107–126. doi: 10.1007/s10048-005-0218-3. [DOI] [PubMed] [Google Scholar]
- 15.Cannelli N, et al. Variant late infantile ceroid lipofuscinoses associated with novel mutations in CLN6. Biochem Biophys Res Commun. 2009;379(4):892–897. doi: 10.1016/j.bbrc.2008.12.159. [DOI] [PubMed] [Google Scholar]
- 16.Wheeler RB, Sharp JD, Schultz RA, Joslin JM, Williams RE, Mole SE. The gene mutated in variant late-infantile neuronal ceroid lipofuscinosis (CLN6) and in nclf mutant mice encodes a novel predicted transmembrane protein. Am J Hum Genet. 2002;70(2):537–542. doi: 10.1086/338708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan JP, et al. A murine model of variant late infantile ceroid lipofuscinosis recapitulates behavioral and pathological phenotypes of human disease. PLoS ONE. 2013;8(11):e78694. doi: 10.1371/journal.pone.0078694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bronson RT, Donahue LR, Johnson KR, Tanner A, Lane PW, Faust JR. Neuronal ceroid lipofuscinosis (nclf), a new disorder of the mouse linked to chromosome 9. Am J Med Genet. 1998;77(4):289–297. doi: 10.1002/(SICI)1096-8628(19980526)77:4<289::AID-AJMG8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 19.Bronson RT, Lake BD, Cook S, Taylor S, Davisson MT. Motor neuron degeneration of mice is a model of neuronal ceroid lipofuscinosis (Batten’s disease) Ann Neurol. 1993;33(4):381–385. doi: 10.1002/ana.410330408. [DOI] [PubMed] [Google Scholar]
- 20.Ranta S, et al. The neuronal ceroid lipofuscinoses in human EPMR and mnd mutant mice are associated with mutations in CLN8. Nat Genet. 1999;23(2):233–236. doi: 10.1038/13868. [DOI] [PubMed] [Google Scholar]
- 21.Bolivar VJ, Scott Ganus J, Messer A. The development of behavioral abnormalities in the motor neuron degeneration (mnd) mouse. Brain Res. 2002;937(1-2):74–82. doi: 10.1016/S0006-8993(02)02470-8. [DOI] [PubMed] [Google Scholar]
- 22.Heine C, et al. Topology and endoplasmic reticulum retention signals of the lysosomal storage disease-related membrane protein CLN6. Mol Membr Biol. 2007;24(1):74–87. doi: 10.1080/09687860600967317. [DOI] [PubMed] [Google Scholar]
- 23.Kanninen KM, et al. Increased zinc and manganese in parallel with neurodegeneration, synaptic protein changes and activation of Akt/GSK3 signaling in ovine CLN6 neuronal ceroid lipofuscinosis. PLoS ONE. 2013;8(3):e58644. doi: 10.1371/journal.pone.0058644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benedict JW, Getty AL, Wishart TM, Gillingwater TH, Pearce DA. Protein product of CLN6 gene responsible for variant late-onset infantile neuronal ceroid lipofuscinosis interacts with CRMP-2. J Neurosci Res. 2009;87(9):2157–2166. doi: 10.1002/jnr.22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt H, Gelhaus C, Lucius R, Nebendahl M, Leippe M, Janssen O. Enrichment and analysis of secretory lysosomes from lymphocyte populations. BMC Immunol. 2009;10:41. doi: 10.1186/1471-2172-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc. 2006;1(3):1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Péterfy M, et al. Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nat Genet. 2007;39(12):1483–1487. doi: 10.1038/ng.2007.24. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Hamamoto S, Ravazzola M, Orci L, Schekman R. Uncoupled packaging of amyloid precursor protein and presenilin 1 into coat protein complex II vesicles. J Biol Chem. 2005;280(9):7758–7768. doi: 10.1074/jbc.M411091200. [DOI] [PubMed] [Google Scholar]
- 29.Nufer O, Kappeler F, Guldbrandsen S, Hauri HP. ER export of ERGIC-53 is controlled by cooperation of targeting determinants in all three of its domains. J Cell Sci. 2003;116(Pt 21):4429–4440. doi: 10.1242/jcs.00759. [DOI] [PubMed] [Google Scholar]
- 30.Schindler AJ, Schekman R. In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proc Natl Acad Sci USA. 2009;106(42):17775–17780. doi: 10.1073/pnas.0910342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang ZN, Li Q, Liu C, Wang HB, Wang Q, Bao L. The voltage-gated Na+ channel Nav1.8 contains an ER-retention/retrieval signal antagonized by the beta3 subunit. J Cell Sci. 2008;121(Pt 19):3243–3252. doi: 10.1242/jcs.026856. [DOI] [PubMed] [Google Scholar]
- 32.Hu T, Kao CY, Hudson RT, Chen A, Draper RK. Inhibition of secretion by 1,3-cyclohexanebis(methylamine), a dibasic compound that interferes with coatomer function. Mol Biol Cell. 1999;10(4):921–933. doi: 10.1091/mbc.10.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 34.Chang KT, Guo J, di Ronza A, Sardiello M. Aminode: identification of evolutionary constraints in the human proteome. Sci Rep. 2018;8(1):1357. doi: 10.1038/s41598-018-19744-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boncompain G, et al. Synchronization of secretory protein traffic in populations of cells. Nat Methods. 2012;9(5):493–498. doi: 10.1038/nmeth.1928. [DOI] [PubMed] [Google Scholar]
- 36.Deng Y, Pakdel M, Blank B, Sundberg EL, Burd CG, von Blume J. Activity of the SPCA1 calcium pump couples sphingomyelin synthesis to sorting of secretory proteins in the trans-Golgi network. Dev Cell. 2018;47(4):464–478.e8. doi: 10.1016/j.devcel.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galizzi G, et al. Different early ER-stress responses in the CLN8(mnd) mouse model of neuronal ceroid lipofuscinosis. Neurosci Lett. 2011;488(3):258–262. doi: 10.1016/j.neulet.2010.11.041. [DOI] [PubMed] [Google Scholar]
- 38.Mirza M, et al. Progressive retinal degeneration and glial activation in the CLN6 (nclf) mouse model of neuronal ceroid lipofuscinosis: a beneficial effect of DHA and curcumin supplementation. PLoS ONE. 2013;8(10):e75963. doi: 10.1371/journal.pone.0075963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei B, Tullis GE, Kirk MD, Zhang K, Katz ML. Ocular phenotype in a mouse gene knockout model for infantile neuronal ceroid lipofuscinosis. J Neurosci Res. 2006;84(5):1139–1149. doi: 10.1002/jnr.21008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang B, et al. Retinal degeneration in motor neuron degeneration: a mouse model of ceroid lipofuscinosis. Invest Ophthalmol Vis Sci. 1994;35(3):1071–1076. [PubMed] [Google Scholar]
- 41.Zhang B, et al. Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nat Genet. 2003;34(2):220–225. doi: 10.1038/ng1153. [DOI] [PubMed] [Google Scholar]
- 42.Cortini M, Sitia R. ERp44 and ERGIC-53 synergize in coupling efficiency and fidelity of IgM polymerization and secretion. Traffic. 2010;11(5):651–659. doi: 10.1111/j.1600-0854.2010.01043.x. [DOI] [PubMed] [Google Scholar]
- 43.Nyfeler B, et al. Identification of ERGIC-53 as an intracellular transport receptor of alpha1-antitrypsin. J Cell Biol. 2008;180(4):705–712. doi: 10.1083/jcb.200709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang B. Recent developments in the understanding of the combined deficiency of FV and FVIII. Br J Haematol. 2009;145(1):15–23. doi: 10.1111/j.1365-2141.2008.07559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emperador-Melero J, et al. Vti1a/b regulate synaptic vesicle and dense core vesicle secretion via protein sorting at the Golgi. Nat Commun. 2018;9(1):3421. doi: 10.1038/s41467-018-05699-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emmer BT, et al. The cargo receptor SURF4 promotes the efficient cellular secretion of PCSK9. Elife. 2018;7:e38839. doi: 10.7554/eLife.38839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takida S, Maeda Y, Kinoshita T. Mammalian GPI-anchored proteins require p24 proteins for their efficient transport from the ER to the plasma membrane. Biochem J. 2008;409(2):555–562. doi: 10.1042/BJ20070234. [DOI] [PubMed] [Google Scholar]
- 48.Barlowe C, Helenius A. Cargo capture and bulk flow in the early secretory pathway. Annu Rev Cell Dev Biol. 2016;32:197–222. doi: 10.1146/annurev-cellbio-111315-125016. [DOI] [PubMed] [Google Scholar]
- 49.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 50.Sardiello M. Transcription factor EB: from master coordinator of lysosomal pathways to candidate therapeutic target in degenerative storage diseases. Ann N Y Acad Sci. 2016;1371(1):3–14. doi: 10.1111/nyas.13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmieri M, et al. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun. 2017;8:14338. doi: 10.1038/ncomms14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lotfi P, et al. Trehalose reduces retinal degeneration, neuroinflammation and storage burden caused by a lysosomal hydrolase deficiency. Autophagy. 2018;14(8):1419–1434. doi: 10.1080/15548627.2018.1474313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palmieri M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20(19):3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 54.Sharma J, di Ronza A, Lotfi P, Sardiello M. Lysosomes and Brain Health. Annu Rev Neurosci. 2018;41:255–276. doi: 10.1146/annurev-neuro-080317-061804. [DOI] [PubMed] [Google Scholar]
- 55.Sleat DE, et al. Association of mutations in a lysosomal protein with classical late-infantile neuronal ceroid lipofuscinosis. Science. 1997;277(5333):1802–1805. doi: 10.1126/science.277.5333.1802. [DOI] [PubMed] [Google Scholar]
- 56.Siintola E, et al. Cathepsin D deficiency underlies congenital human neuronal ceroid-lipofuscinosis. Brain. 2006;129(Pt 6):1438–1445. doi: 10.1093/brain/awl107. [DOI] [PubMed] [Google Scholar]
- 57.Steinfeld R, et al. Cathepsin D deficiency is associated with a human neurodegenerative disorder. Am J Hum Genet. 2006;78(6):988–998. doi: 10.1086/504159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ezaki J, Tanida I, Kanehagi N, Kominami E. A lysosomal proteinase, the late infantile neuronal ceroid lipofuscinosis gene (CLN2) product, is essential for degradation of a hydrophobic protein, the subunit c of ATP synthase. J Neurochem. 1999;72(6):2573–2582. doi: 10.1046/j.1471-4159.1999.0722573.x. [DOI] [PubMed] [Google Scholar]
- 59.Ezaki J, Wolfe LS, Kominami E. Specific delay in the degradation of mitochondrial ATP synthase subunit c in late infantile neuronal ceroid lipofuscinosis is derived from cellular proteolytic dysfunction rather than structural alteration of subunit c. J Neurochem. 1996;67(4):1677–1687. doi: 10.1046/j.1471-4159.1996.67041677.x. [DOI] [PubMed] [Google Scholar]
- 60.Ezaki J, Takeda-Ezaki M, Kominami E. Tripeptidyl peptidase I, the late infantile neuronal ceroid lipofuscinosis gene product, initiates the lysosomal degradation of subunit c of ATP synthase. J Biochem. 2000;128(3):509–516. doi: 10.1093/oxfordjournals.jbchem.a022781. [DOI] [PubMed] [Google Scholar]
- 61.Koike M, et al. Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci. 2000;20(18):6898–6906. doi: 10.1523/JNEUROSCI.20-18-06898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pardo CA, Rabin BA, Palmer DN, Price DL. Accumulation of the adenosine triphosphate synthase subunit C in the mnd mutant mouse. A model for neuronal ceroid lipofuscinosis. Am J Pathol. 1994;144(4):829–835. [PMC free article] [PubMed] [Google Scholar]
- 63.Palmer DN, et al. Mitochondrial ATP synthase subunit c storage in the ceroid-lipofuscinoses (Batten disease) Am J Med Genet. 1992;42(4):561–567. doi: 10.1002/ajmg.1320420428. [DOI] [PubMed] [Google Scholar]
- 64.Kleine Holthaus SM, et al. Neonatal brain-directed gene therapy rescues a mouse model of neurodegenerative CLN6 Batten disease. Hum Mol Genet. 2019;28(23):3867–3879. doi: 10.1093/hmg/ddz210. [DOI] [PubMed] [Google Scholar]
- 65.Ohashi T. Enzyme replacement therapy for lysosomal storage diseases. Pediatr Endocrinol Rev. 2012;10 Suppl 1:26–34. [PubMed] [Google Scholar]
- 66.Geraets RD, Koh Sy, Hastings ML, Kielian T, Pearce DA, Weimer JM. Moving towards effective therapeutic strategies for Neuronal Ceroid Lipofuscinosis. Orphanet J Rare Dis. 2016;11:40. doi: 10.1186/s13023-016-0414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kohlschütter A, Schulz A, Bartsch U, Storch S. Current and emerging treatment strategies for neuronal ceroid lipofuscinoses. CNS Drugs. 2019;33(4):315–325. doi: 10.1007/s40263-019-00620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neverman NJ, Best HL, Hofmann SL, Hughes SM. Experimental therapies in the neuronal ceroid lipofuscinoses. Biochim Biophys Acta. 2015;1852(10 Pt B):2292–2300. doi: 10.1016/j.bbadis.2015.04.026. [DOI] [PubMed] [Google Scholar]
- 69.Kleine Holthaus SM, Smith AJ, Mole SE, Ali RR. Gene therapy approaches to treat the neurodegeneration and visual failure in neuronal ceroid lipofuscinoses. Adv Exp Med Biol. 2018;1074:91–99. doi: 10.1007/978-3-319-75402-4_12. [DOI] [PubMed] [Google Scholar]
- 70.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshimi K, Kunihiro Y, Kaneko T, Nagahora H, Voigt B, Mashimo T. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun. 2016;7:10431. doi: 10.1038/ncomms10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wattiaux R, Wattiaux-De Coninck S, Ronveaux-dupal MF, Dubois F. Isolation of rat liver lysosomes by isopycnic centrifugation in a metrizamide gradient. J Cell Biol. 1978;78(2):349–368. doi: 10.1083/jcb.78.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ezaki J, Takeda-Ezaki M, Oda K, Kominami E. Characterization of endopeptidase activity of tripeptidyl peptidase-I/CLN2 protein which is deficient in classical late infantile neuronal ceroid lipofuscinosis. Biochem Biophys Res Commun. 2000;268(3):904–908. doi: 10.1006/bbrc.2000.2207. [DOI] [PubMed] [Google Scholar]
- 74.Kishnani PS, et al. Chinese hamster ovary cell-derived recombinant human acid alpha-glucosidase in infantile-onset Pompe disease. J Pediatr. 2006;149(1):89–97. doi: 10.1016/j.jpeds.2006.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schueler UH, Kolter T, Kaneski CR, Zirzow GC, Sandhoff K, Brady RO. Correlation between enzyme activity and substrate storage in a cell culture model system for Gaucher disease. J Inherit Metab Dis. 2004;27(5):649–658. doi: 10.1023/B:BOLI.0000042959.44318.7c. [DOI] [PubMed] [Google Scholar]
- 76.Berg JD, Fiksdal L. Rapid detection of total and fecal coliforms in water by enzymatic hydrolysis of 4-methylumbelliferone-beta-D-galactoside. Appl Environ Microbiol. 1988;54(8):2118–2122. doi: 10.1128/AEM.54.8.2118-2122.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marsh J, Fensom AH. 4-Methylumbelliferyl alpha-N-acetylglucosaminidase activity for diagnosis of Sanfilippo B disease. Clin Genet. 1985;27(3):258–262. doi: 10.1111/j.1399-0004.1985.tb00217.x. [DOI] [PubMed] [Google Scholar]
- 78.Zhang P, Schekman R. Distinct stages in the recognition, sorting, and packaging of proTGFα into COPII-coated transport vesicles. Mol Biol Cell. 2016;27(12):1938–1947. doi: 10.1091/mbc.e16-02-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46(D1):D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.