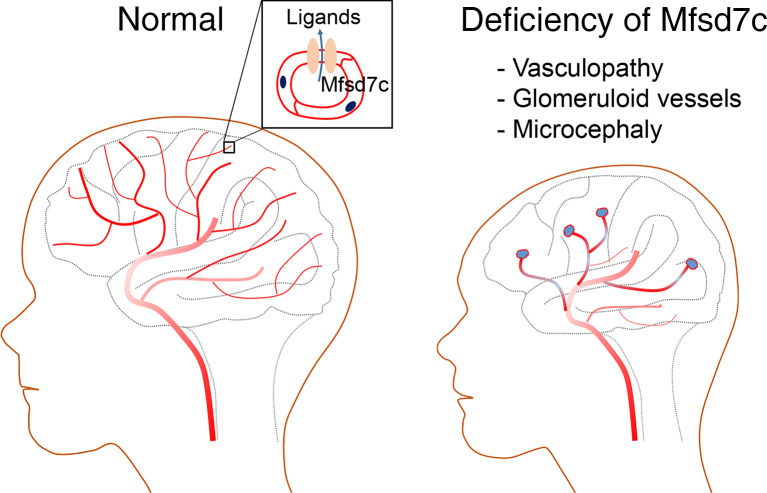
Abstract
Several missense mutations in the orphan transporter FLVCR2 have been reported in Fowler syndrome. Affected subjects exhibit signs of severe neurological defects. We identified the mouse ortholog Mfsd7c as a gene expressed in the blood-brain barrier. Here, we report the characterizations of Mfsd7c-KO mice and compare these characterizations to phenotypic findings in humans with biallelic FLVCR2 mutations. Global KO of Mfsd7c in mice resulted in late-gestation lethality, likely due to CNS phenotypes. We found that the angiogenic growth of CNS blood vessels in the brain of Mfsd7c-KO embryos was inhibited in cortical ventricular zones and ganglionic eminences. Vascular tips were dilated and fused, resulting in glomeruloid vessels. Nonetheless, CNS blood vessels were intact, without hemorrhage. Both embryos and humans with biallelic FLVCR2 mutations exhibited reduced cerebral cortical layers, enlargement of the cerebral ventricles, and microcephaly. Transcriptomic analysis of Mfsd7cK-KO embryonic brains revealed upregulation of genes involved in glycolysis and angiogenesis. The Mfsd7c-KO brain exhibited hypoxia and neuronal cell death. Our results indicate that MFSD7c is required for the normal growth of CNS blood vessels and that ablation of this gene results in microcephaly-associated vasculopathy in mice and humans.
Keywords: Vascular Biology
Keywords: Transport, endothelial cells
Introduction
Proliferative vasculopathy, hydranencephaly-hydrocephaly syndrome (PVHH), also called Fowler syndrome (OMIM 225790), is a severe and often lethal neurological disorder. Hallmarks of this disease are congenital hydrocephalus, hydranencephaly, hypervasculation, and thinning of cortices in the CNS (1). This developmental abnormality is associated with high prenatal lethality (2). Postmortem analysis of 16 PVHH human fetuses showed calcifications, possible vascular pericyte deficiency, and the aforementioned PVHH hallmarks (3). Genetic evidence has linked PVHH to missense mutations of FLVCR2. However, the mechanisms of disease in PVHH are enigmatic, reflecting our lack of understanding of the role or roles of this gene.
Whole-exome sequencing of subjects with Fowler syndrome reveals an association between the disease and biallelic loss-of-function mutations in FLVCR2 (1, 4, 5). Missense point mutations such as R84H, T352R, L398V, G412R, T430M, T430R, N443A, and S203Y are generally located in the transmembrane domain or near the transport cavity of the protein (1, 2, 4–6). Therefore, the mutated residues may affect protein folding or ligand transport activity. Nevertheless, functional assays to confirm the causal effects of these missense mutations in FLVCR2 in Fowler syndrome are lacking. Therefore, it is of interest to characterize the molecular and physiological functions of the protein.
The murine ortholog of FLVCR2 is MFSD7c, which is also conserved in mammals (for simplicity, MFSD7c is used hereafter for both human and mouse FLVCR2). It belongs to the major facilitator superfamily domain proteins, which facilitate the transport of small molecules across membranes. MFSD7c shares high-sequence similarity with MFSD7b, which is the heme exporter in erythroid cells (7, 8). MFSD7c functioned as a heme importer, as assessed by a heme analog zinc mesophorphyrin activity assay, in a Chinese hamster ovary cell line (9). However, physiological evidence is lacking to support this finding. To our knowledge, no previous studies on the physiological roles of MFSD7c have been reported. To understand the physiological as well as the molecular roles of MFSD7c, we generated and studied the phenotypes of the global KO of Mfsd7c in mice. Our results show that MFSD7c is an endothelial transporter in CNS blood vessels, the loss of which causes late-gestation lethality. We observed that deletion of Mfsd7c results in severely reduced angiogenic growth of CNS blood vessels to the subventricular zone (SVZ) and ventricular zone (VZ). As a result, KO brains exhibit severe hypoxia and neuronal cell death likely linked to reduced brain growth. We also report mutations as well as clinical and radiologic findings in a 17-month-old child with Fowler syndrome. Our results provide evidence that lack of MFSD7c strongly suppresses CNS blood vessel growth, which in turn causes a severe delayed growth of brain. Mfsd7c-KO mice are a valuable genetic model for mechanistic investigation of Fowler syndrome.
Results
MFSD7c is a putative membrane transporter that is expressed in endothelial cells of CNS blood vessels.
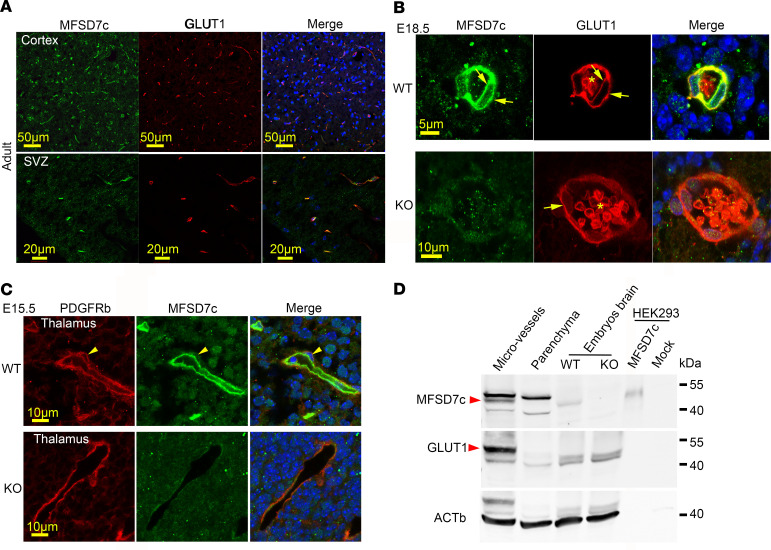
CNS blood vessels comprise tight junction proteins that restrict the diffusion of bloodborne molecules. To facilitate the transport of nutrients, ions, and small molecules, endothelial cells must express membrane transporters, which are used to exchange nutrients and wastes between neuronal cells and blood (10, 11). In search for major facilitator transporters in the brain, we analyzed public transcriptomic data sets and identified Mfsd7c as a potential gene candidate. We were further interested in this gene as it has been reported in Fowler syndrome, a neurological disorder without known molecular mechanism. To study the expression pattern of MFSD7c in vivo, we generated polyclonal antibodies against murine MFSD7c. Using immunohistochemistry with mouse embryos and adult brain sections, we found that MFSD7c is prominently expressed in CNS blood vessels (Figure 1A). The brain blood vessels consist of several cell types, such as pericytes (10). We found that MFSD7c was colocalized with glucose transporter 1 (GLUT1, also known as SLC2A1), but not with pericyte marker PDGFRB, indicating that it is expressed in endothelial cells of CNS blood vessels (Figure 1, B and C). Additionally, MFSD7c was expressed in both luminal and abluminal sides of the blood-brain barrier (BBB); this is similar to the expression pattern of GLUT1 (Figure 1B). MFSD7c protein was enriched in CNS microvessels, but not in parenchymal cell fractions from adult mouse brains (Figure 1D). MFSD7c is predicted to be a membrane protein with molecular weight of 45 to 50 kDa when overexpressed in HEK293 cells (Figure 1D). These results show that MFSD7c is a plasma membrane transporter in CNS blood vessels.
Figure 1. MFSD7c is expressed in CNS blood vessels.
(A) Immunostaining of MFSD7c with polyclonal antibodies against murine MFSD7c in adult mouse brain. Expression of MFSD7c was found in CNS blood vessels, where it colocalized with GLUT1. Top, cortex; bottom, SVZ. Experiments were performed at least 4 times (n = 6). (B) Representative images of immunostaining of E18.5 WT and KO brain sections showing that MFSD7c was expressed in both apical and luminal side (indicated by arrows) of CNS endothelial cells. Asterisk shows erythrocytes, which were also positive with GLUT1 staining. Experiments were performed at least 4 times (n = 6 embryos per genotype). (C) MFSD7c is not expressed in pericytes. Immunostaining of E15.5 WT and KO brain sections with pericyte marker PDGFRb and MFSD7c. Arrowheads indicate pericytes. Experiments were performed at least 3 times (n = 3–4 embryos per genotype). (D) MFSD7c protein was detected in microvessel, but not parenchyma, fraction from WT adult brain. Protein lysates from E16.5 WT and KO embryos as well as MFSD7c protein expression in HEK293 cells were used to interpret the MFSD7c protein band. Note that the total amount of protein from HEK293 cells was 1 μg. Arrowheads show MFSD7c and GLUT1 protein bands. Experiments were performed 3 times.
KO of Mfsd7c results in late-gestation lethality.
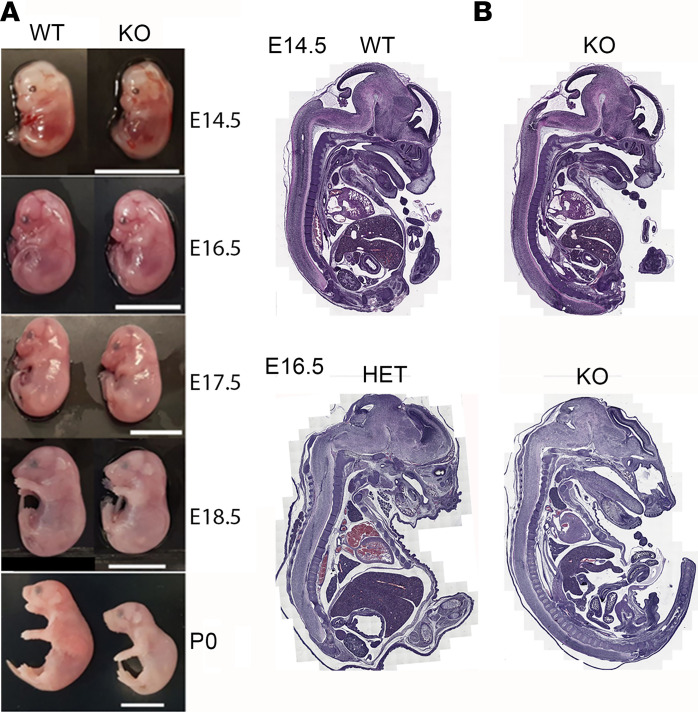
Previously, homozygous human mutations of MFSD7c have been reported in Fowler syndrome. To gain insights into the disease mechanisms of these missense mutations and the physiological roles of MFSD7c, we generated global KO of Mfsd7c in mice (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI136727DS1). We observed that KO mice died before weaning. To assess the lethality phenotype, we performed time mating to obtain embryos at developmental stages and observed that KO embryos had slightly reduced body weight and died in late gestation (Figure 2A). Some of the pups were born with slightly pale color and died within a few hours after birth (Figure 2A). Their milk sacs were empty (Supplemental Figure 2A). The head sizes of KO embryos were significantly smaller than those of WT (Supplemental Figure 2B). To investigate the cause of the lethal phenotype, we performed anatomical analysis of KO and control embryos at E14.5 and E16.5, when the KO embryos were still viable. The gross anatomy of KO embryos appeared normal compared with that of control embryos (Figure 2B). In summary, the anatomical structures of organs from the KO embryos did not differ from those of controls at these time points (Figure 2B). We observed several abnormal brain defects at high magnifications that we were able to elaborate on by histological assessment. These observations suggest neurological defects might be the cause for the death of KO pups and might be related to hypoxia.
Figure 2. Deletion of MFSD7c resulted in late-gestation lethality.
(A) Whole-body deletion of MFSD7c results in late-gestation, perinatal lethality. Appearances of Mfsd7c-KO embryos at indicated time points were comparable with those WT littermates, except that KO embryos and pups exhibited slightly pale color at later times during development. Scale bars: 10 mm. (B) Gross anatomical analysis of E14.5 and E16.5 WT or HET control and KO embryos showing normal organ histology. Tiling scan was used to acquire H&E images. Experiments were repeated twice (n = 2).
Reduced growth of blood vessels in the VZ and SVZ of Mfsd7c-KO embryos.
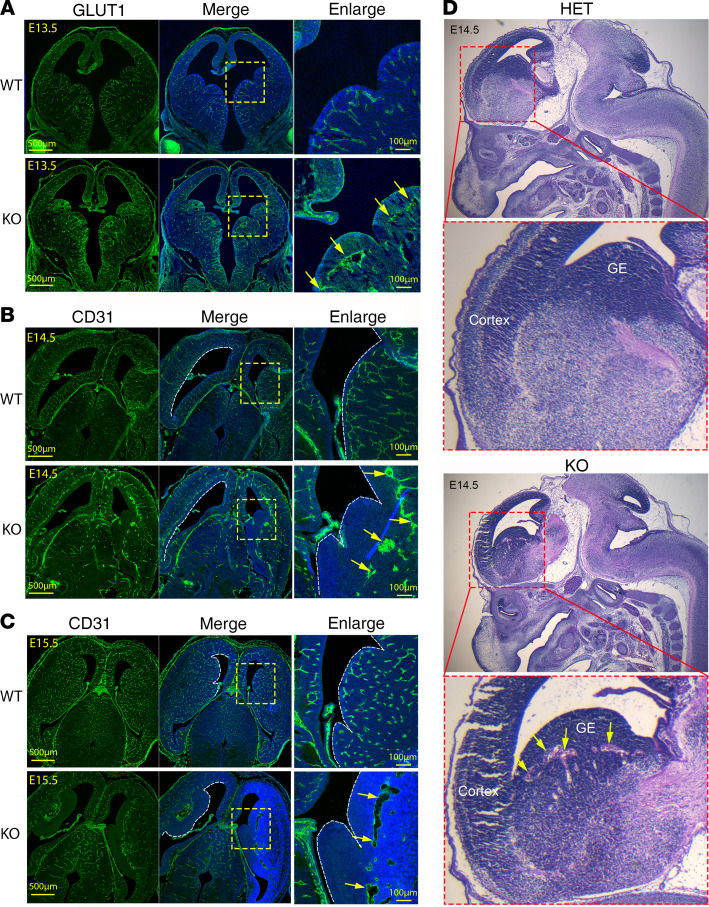
MFSD7c is expressed in endothelial cells of CNS blood vessels, which prompted us to examine the blood vessel phenotypes in the KO embryos. We performed IHC staining with GLUT1 and CD31 antibodies to visualize CNS blood vessels of embryos and examined the formation of blood vessels from E13.5 onwards in coronal sections (Figure 3A). Remarkably, the abnormal growth of vasculature in the brain of KO embryos, especially in the VZ and SVZ regions, was already observed at these time points (Figure 3A). In the cortical VZ regions, there was delayed growth and absence of blood vessels reaching toward the ventricles (Supplemental Figure 3A). Strikingly, the migration of growing blood vessels was stalled near the SVZ regions in the ganglionic eminences (GEs) (Figure 3, A–C). Vessel termini were dilated (Figure 3B). Additionally, vessels were coalesced, creating empty spaces observed in E14.5–E15.5 KO embryos (Figure 3, B and C, and Supplemental Figure 3A). We also noticed a loosened tissue composition in the GE regions using H&E staining (Figure 3D). The vascular density in the midbrain, cortex, and GEs was also significantly reduced in the KO embryos (Supplemental Figure 3, B and C). Our results show that endothelial MFSD7c is required for proper growth of CNS blood vessels in the developing brain. Lack of this protein results in delayed growth of blood vessels in VZ and SVZ regions.
Figure 3. Reduced CNS blood vessel growth in Mfsd7c-KO embryos.
(A–C) Coronal sections of brain from E13.5 (A), E14.5 (B), and E15.5 (C) WT and KO embryos were stained with GLUT1 or CD31 antibody to visualize CNS endothelial cells. The growth of CNS blood vessels of KO embryos was severely reduced, especially in GE regions. Insets show enlarged GE regions. Arrows show VZs, where blood vessel growth was stalled and enlarged blood vessels were observed in the KO embryos. Experiments were repeated twice for A (n = 4 embryos per genotype) and 4 or 5 times for B and C (n = 3–4 embryos per genotype). (D) H&E staining of sagittal sections showed that KO embryos exhibited microaneurism (arrows) in GE regions. Experiments were repeated twice.
MFSD7c is dispensable for BBB integrity.
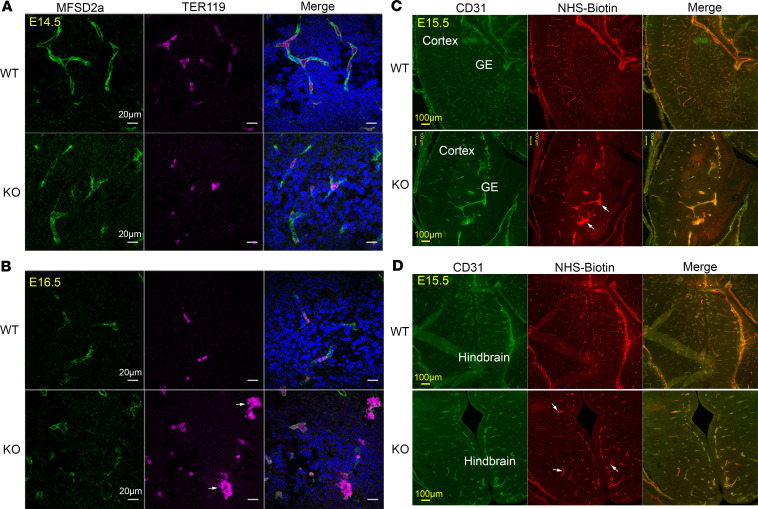
The reduced blood vessel growth to the VZ and SVZ in Mfsd7c-KO embryos particularly resembled the previously published mutants in components of the canonical WNT signaling in CNS (12, 13). Nevertheless, these canonical Wnt mutants exhibit severe cerebral hemorrhage (14, 15), which we did not observe in Mfsd7c-KO embryos. We tested for local disruption of the BBB structure in KO embryos. First, we used MFSD2a as an endothelial cell marker for CNS microvessels and TER119 as a marker for erythrocytes to detect hemorrhage. In WT embryos at E14.5 and E16.5, erythrocytes were found within the lumen of blood vessels (Figure 4, A and B). We did not observe any obvious bleeding phenotype in the brain of KO embryos, although clumps of erythrocytes could be visualized within severely dilated blood vessels (Figure 4B). Thus, lack of MFSD7c does not result in significant bleeding in the brain. Second, we used NHS-biotin as a small molecule to determine whether the KO BBB leaks small molecules. NHS-bi¬otin was found in dilated blood vessels in the brain of KO embryos, but we did not observe any significant extravasation of NHS-biotin into brain parenchyma (Figure 4, C and D). We applied an additional test of whether MFSD7c is required for BBB integrity by staining with PLVAP, a marker for cellular fenestration. Increased expression of PLVAP correlates with leakiness of BBB via a transcellular route. Expression of PLVAP in CNS blood vessels of KO embryos at E16.5 and E18.5 was comparable to that of WT embryos (Supplemental Figure 4, A and B). Expression of genes and proteins in the canonical WNT signaling as well as proteins involved in the TGFB signaling pathway was unchanged in the brains of KO embryos (Supplemental Figure 5, A–C). Together, these results show that MFSD7c is not necessary for maintaining BBB integrity, thus dissociating MFSD7c functions from canonical WNT signaling and linking its unique roles to CNS blood vessel growth.
Figure 4. Deletion of MFSD7c does not result in ruptured CNS blood vessels and increased permeability of BBB.
(A and B) Brain sections of E14.5 (n = 3 embryos per genotype) (A) and E16.5 (n = 4 embryos per genotype) (B). WT and Mfsd7c-KO embryos were stained with MFSD2a (a CNS endothelial cell marker, green) and Ter119 (red blood cell marker, pink). Arrows indicate clumps of erythrocytes surrounded by blood vessels in Mfsd7c-KO embryos. There was no hemorrhage in KO embryonic brains. Shown here are hindbrain regions. Scale bars: 20 μm. Experiments were repeated 3 times. (C and D) Representative coronal brain sections of E15.5 WT and KO showing that NHS-biotin was contained inside KO blood vessels. Arrows show enlarged blood vessels with strong signals of NHS-biotin. Experiments were repeated twice with n = 6 for WT or KO.
Brain blood vessels of Mfsd7c-KO embryos are dilated.
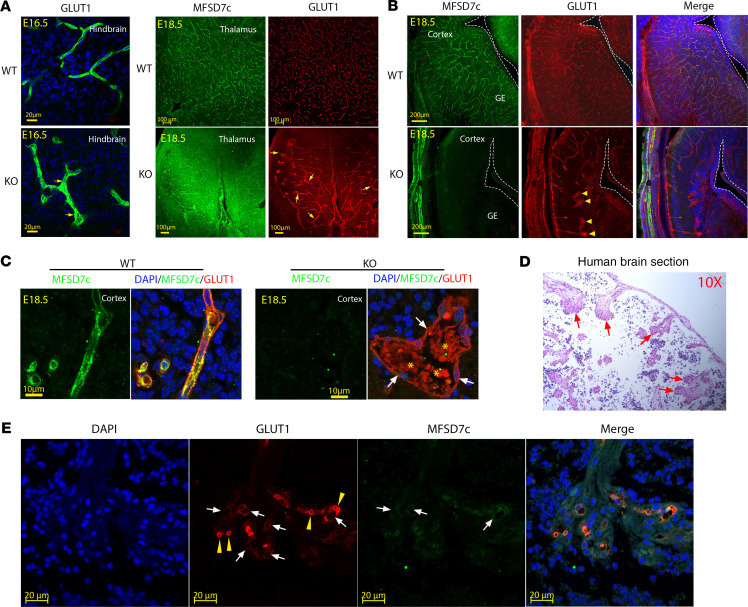
Examining the blood vessel phenotypes, we found that blood vessels were significantly dilated throughout the brain of KO embryos (Figure 5A and Supplemental Figure 6A). During early brain development, the vascular bed is formed by angiogenic sprouting from pial surfaces to VZs to provide oxygen and nutrients for proliferating neurons. The radial growth of blood vessels in the Mfsd7c-KO embryos was severely affected in VZ and SVZ, especially in the GE regions (Figure 3). Additionally, we found glomeruloid structures at the tip of growing vessels stalled near the GE regions in KO (Figure 5B). Interestingly, increased filopodia of endothelial cells from the stalled vessels near ventricles were also observed (Supplemental Figure 6B), suggesting that the growth of blood vessels is inhibited even with the increased expression of angiogenic stimulation by VEGF (Supplemental Figures 10 and 11). The glomeruloid structures contained erythrocytes (Figure 5C and Figure 4), suggesting that the blood vessels of KO embryos are perfused. Furthermore, we obtained brain sections from a Fowler patient who was reported in a Radio et al. study (5). Histological examinations revealed that the patient’s CNS blood vessels exhibited reduced growth and glomeruloid structures (Figure 5D) positive for GLUT1 and MFSD7c, confirming that they were parts of defective CNS blood vessels (Figure 5E). Pericytes are mural cells that enwrap and stabilize CNS blood vessels. Their loss results in dilated blood vessels (16, 17). We stained the KO blood vessels with PDGFRB antibody to examine the coverage of pericytes. However, the dilated blood vessels from KO embryos had normal pericyte staining, suggesting that reduced pericyte coverage is not the cause of blood vessel dilation in the KO brains (Supplemental Figure 7, A and B). Together, our data indicate that deficiency in MFSD7c causes severely dilated CNS blood vessels without affecting pericyte recruitment.
Figure 5. Deficiency of MFSD7c results in glomeruloid vessel structures.
(A) Representative images of GLUT1 staining in hindbrain (left) and GLUT1 and MFSD7c costaining in thalamus regions (right) of E16.5 and E18.5 WT and KO embryos, respectively. Arrows show enlarged blood vessels in KO embryos. Experiments were repeated at least 3 times (n = 6 embryos per genotype). (B) Costaining of MFSD7c and GLUT1 in coronal brain sections of E18.5 WT and KO embryos. In the absence of MFSD7c, glomeruloid blood vessels were observed in brain sections of KO embryos. Arrowheads show the glomeruloid structures at the tips of blood vessels in KO embryos. Experiments were repeated at least 3 times (n = 6 embryos per genotype). (C) Enlarged images of a glomeruloid structure from E18.5 KO brain sections with erythrocytes (GLUT1 is present in both endothelial cells and erythrocytes, red signals). MFSD7c, green; GLUT1, red. Arrows show endothelial cells. Asterisks show erythrocytes. (D) H&E staining of brain sections from a human patient with S203Y homozygous mutation. Glomeruloid blood vessel structures were also observed in the brain sections of the patient. (E) Immunostaining of the human patient brain sections with GLUT1, MFSD7c, and DAPI. Arrows show glomeruliod structures that are positive with GLUT1 and MFSD7c. Arrowheads show the staining of GLUT1 with erythrocytes. n = 1 patient.
Loss of MFSD7c results in microcephaly in Mfsd7c-KO mice and human patient.
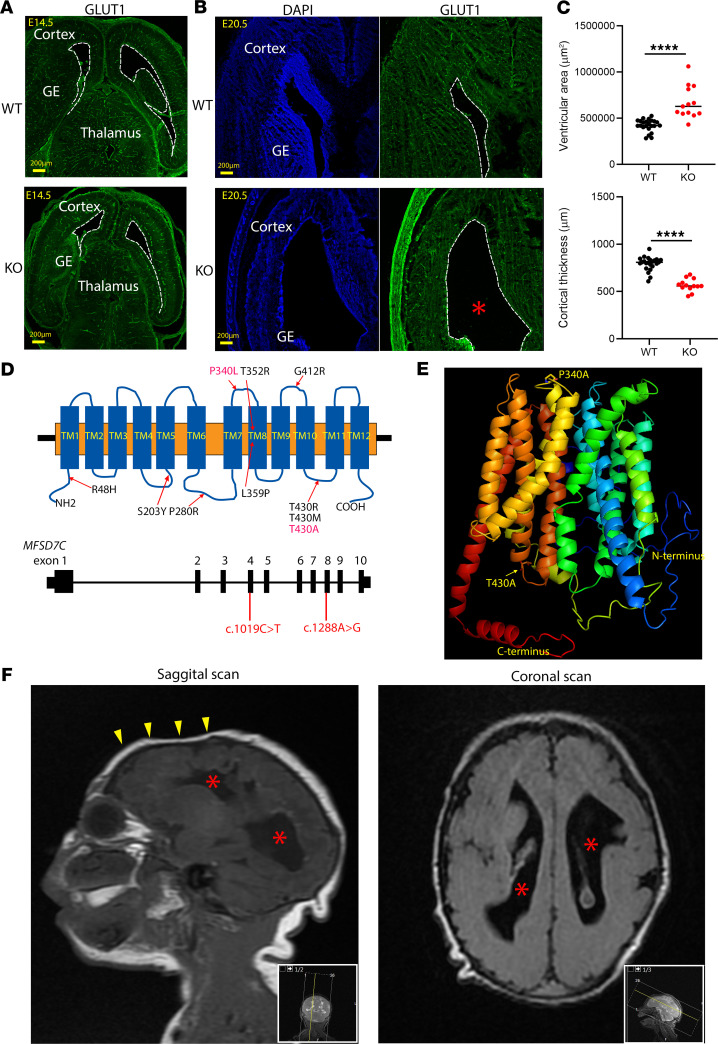
The head size of Mfsd7c-KO embryos was significantly reduced at P0 (Supplemental Figure 2). The brains of KO embryos exhibited enlarged ventricles and thinned cortices at E20.5 (Figure 6, A–C). Additionally, we did not observe hydrocephalus in the KO embryos (Figure 6, A and B). Thus, MFSD7c deficiency, at least in mice, does not result in hydrocephalus as reported in Fowler patients.
Figure 6. Deficiency in MFSD7c results in microcephaly.
(A and B) Representative images of coronal sections of E14.5 and E20.5 WT and KO embryos were stained with DAPI and GLUT1. Thinning of cortices were visible at E20.5. (C) Quantification of VZ and cortex thickness in E20.5 WT and KO embryos. Each dot represents 1 section. Data are represented as mean ± SD. n = 4–5 per genotype. ****P < 0.0001, 2-tailed t test. (D and E) Illustration of human MFSD7c gene and the modeled structure of human MFSD7c. Shown are missense mutations that have been reported for MFSD7c and the 2 novel mutations that occurred in the patient shown in D. (F) MRI images of the patient brain showing that the patient has microcephaly and noticeable thinning of the cortices. The enlarged ventricles in mouse embryos at E20.5 or the human patient are possibly the results of thinning of cortices. Asterisks denote enlarged ventricles in both KO embryos and the human patient. Arrowheads show the reduced frontal brain in the human patient. n = 1 patient.
In line with the brain-growth phenotypes of KO embryos, we identified a young child with a postnatal diagnosis of Fowler syndrome. His phenotype included severe neurological defects, albeit milder than previously reported (2). Whole-exome sequencing of the patient and his parents revealed that the proband carried compound heterozygous mutations in 2 conserved residues of MFSD7c (Supplemental Figure 8A). The variants from the proband and parents were confirmed by Sanger sequencing (Supplemental Figure 8B). We found that the c.1019C>T p.(Pro340Leu) variant was inherited from the mother and the c.1288A>G p.(Thr430Ala) variant was inherited from the father (Figure 6, D and E, and Supplemental Figure 8B). According to the American College of Medical Genetics and Genomics (ACMG) guidelines, the variants are classified as likely pathogenic (18). On physical examination, the patient had severe microcephaly (Figure 6F). Brain MRI scanning also revealed that the patient’s brain has enlarged ventricles and thinning of the cortices (Figure 6F). On his last evaluation (at 17 months of age), the proband had severe global developmental delay and impaired vision and hearing as well as increased muscle tone (see detailed description in Supplemental Methods). Both detected variants are rare and located in highly conserved amino acids predicted to be near the transport cavity (Figure 6, D and E). Other substitutions of Thr430 have been reported in other cases of Fowler syndrome, suggesting that this residue is critical for MFSD7c function (Figure 6D). The mutation Pro340Leu is a novel variant. As MFSD7c is a plasma membrane protein, a reduced expression level or mislocalization to the plasma membrane might result in impaired function. We generated the gene mutants and tested their expression and localization in HEK293 cells. We found that mutations in these 2 conserved residues did not affect the expression and cellular localization of MFSD7c (Supplemental Figure 9, A and B). The mutation S203Y that was reported in the Radio study also did not affect the expression and localization of the protein (5) (Supplemental Figure 9, A and B). In contrast, the previously reported mutation of T430M did affect localization of the protein (Supplemental Figure 9B). These results imply that reduced transport activity rather than defective expression and/or localization of the mutated proteins is responsible for the dysfunctions of MFSD7c in the patient. Our data may indicate that detected mutations are milder and explain the patient’s survival and milder findings upon brain imaging.
Mfsd7c-KO brain exhibits severe hypoxia.
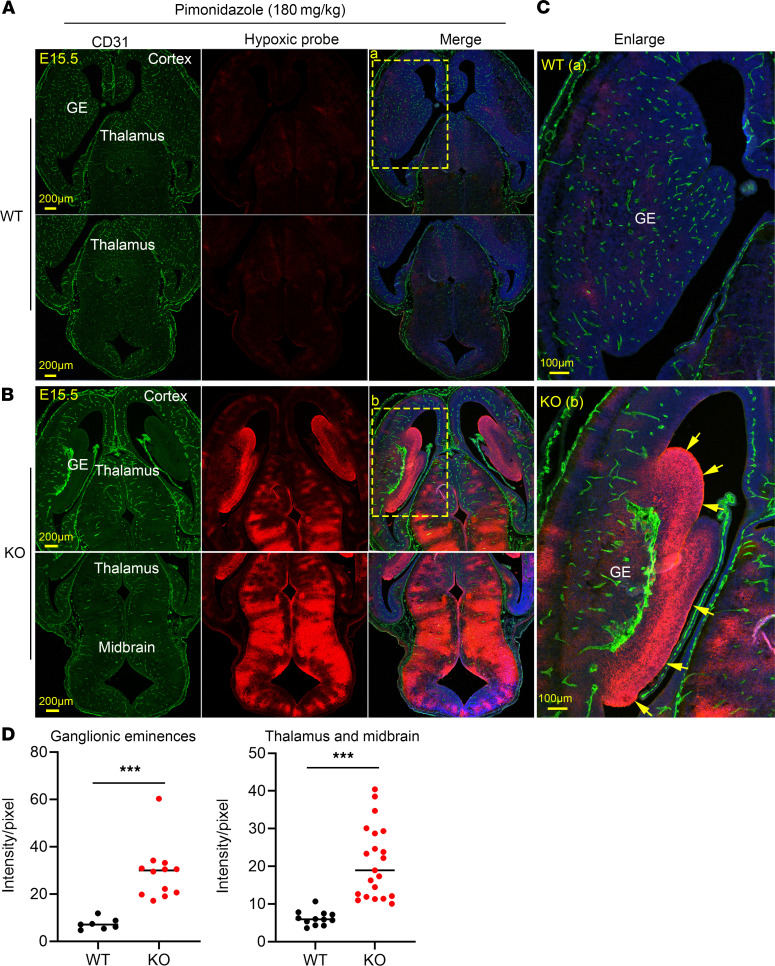
We observed that the KO embryos exhibited slightly pale appearances, especially near birth (Figure 2A). Thus, we performed transcriptomic analysis for gene expression in the whole-brain transcriptome at E14.5 (Supplemental Table 1). We found that genes involved in glycolysis, hypertrophy, angiogenesis, cell-cell adhesion, and inflammatory responses were upregulated by at least 2-fold in the KO embryo brains compared with those of WT controls (Supplemental Figure 10). Most interestingly, many genes involved in glycolysis were upregulated in KO embryos. The upregulation of these genes, including Slc2a1 (Glut1), Hk2 (hexose kinase 2), Ldha (lactate dehydrogenase A), and Tpi (triosephosphate isomerase 1), at E14.5 was confirmed by quantitative reverse-transcriptase PCR (RT-PCR) (Supplemental Figure 11A). Interestingly, expression of several angiogenic genes such as Vegfa and angiopoietin 2 (Angpt2) was also significantly upregulated in the KO brain (Supplemental Figure 11, A and B). The upregulation of glycolytic and angiogenetic genes suggested that neuronal cells of KO embryos experience hypoxia. Thus, we used pimonidazole hydrochloride to detect the hypoxic response in the brain of KO embryos. Under hypoxic conditions, pimonidazole is metabolized to form stable adduct with thiol groups in proteins, which are detected by a monoclonal antibody by immunostaining. We found that in the reduced vascular areas shown by CD31 staining in the KO brain regions, such as cortical hem and the VZs in the cerebral cortex, hypoxia was not detected (Figure 7, A and B). Interestingly, we did observe a strong hypoxic signal in the GE regions, where the growth of blood vessels was severely reduced or absent in KO embryos (Figure 7, B–D). Severe hypoxia was also found in the thalamus and midbrain regions (Figure 7, B and D). Our findings indicate that MFSD7c is required for angiogenic growth of CNS blood vessels, especially in the GE regions. Lack of MFSD7c brings about hypoxia, which alters the expression of several important metabolic genes in the brain.
Figure 7. Mfsd7c-KO embryos exhibited severe hypoxia.
(A and B) Representative images of brain sections of WT (A) and KO (B) embryos were stained with CD31 to visualize blood vessels and the monoclonal antibody to detect hypoxic neuronal cells after injection of pimonidazole to the pregnant mice. Top panels in A and B are images from ventral sides of the WT and KO embryo brains, and the bottom panels are dorsal sides, including thalamus and hindbrain regions. In the ventral images in B, hypoxia was detected in the GE of KO embryos. The hypoxia was also present in the brainstem from these KO sections. (C) Enlarged images from GEs of WT and KO embryos. Experiment was repeated three times. n = 3–4 embryos per genotype. (D) Quantification of hypoxic signals in the GEs and thalamus/midbrain regions. ***P < 0.001, 2-tailed t test. Each dot represents 1 section from 3 WT and 4 KO embryos.
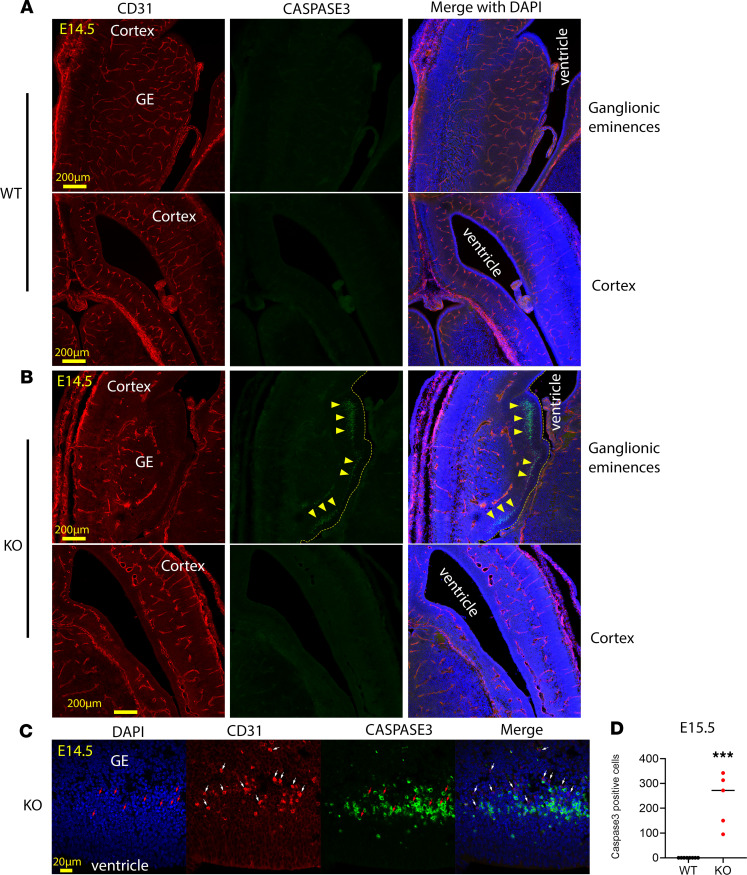
Lack of MFSD7c causes neuronal cell death.
Neuronal progenitor cells are glycolytic during proliferation. Indeed, we observed that the lateral and median GEs were formed even in the absence of blood vessels (see Figure 7, B and C). To gain more insight into the hypoxic response, we examined neuronal cell viability. We costained WT and KO embryo sections with activated caspase-3 and CD31. Interestingly, we found that caspase-3–positive cells were observed in Mfsd7c-KO embryos at least from E14.5 onwards (Figure 8 and Supplemental Figure 12). These activated caspase-3+ cells were mainly found in VZ in the GEs, but not in the cortical VZs of KO embryos (Figure 8, A and B, and quantification in D). The nuclei of these caspase-3+ cells were condensed by DAPI staining (Figure 8C). Remarkably, although there was a clearly delayed growth of blood vessels to the cortical VZ, cell death was not observed in this region. Perhaps oxygen could diffuse through the abnormally grown blood vessels to this region in the KO embryos (Figure 8B). We also noted that not all neuronal cells undergoing hypoxia died because there was only a small population of neuronal cells in the VZs in GE positive for caspase-3 (Figure 8B). Neuronal cell death was more prominent in the E16.5 KO embryos and appeared in several regions in the brain of E18.5 KO embryos (Supplemental Figure 12). As expected, activated caspase-3 staining was negative in all examined brain regions of WT controls at all tested time points (Figure 8 and Supplemental Figure 12). Interestingly, we found that a nonendothelial population of CD31+ cells appeared in the same area where cells with activated caspase-3 were observed. These CD31+ cells were not caspase-3+. These cells appeared near blood vessel sites, suggesting that they might be infiltrated immune cells. CD31 is also present in platelets, lymphocytes, and macrophages. Nevertheless, they were negative for CD3 and B220 antibodies, which are the markers for T and B cells, respectively (Supplemental Figure 13). The cells were also negative for IBA1, a marker for microglia cells (Supplemental Figure 13). Our results show that lack of MFSD7c induces neuronal cell death. The death of neuronal cells might impede the formation of cortical layers reflected in the thinning of cortices and microcephaly in the KO embryos and the human patient.
Figure 8. Mfsd7c-KO embryos exhibited cell death in SVZs.
(A and B) Representative images of coronal sections of E14.5 WT and KO embryos that were immunostained to detect activated caspase-3, a marker for cell death. The activated caspase-3+ cells were only observed in the SVZs in GEs of KO embryos as shown by arrowheads (top), but not in the cortical hem and neocortex (bottom). Dead cells were clearly localized in avascular regions in GEs. The vascular network was visualized by CD31. (C) Enlarged images in the GEs. The activated caspase-3+ cells were found in close proximity to nonendothelial cells that were also positive for CD31 in the VZs of GE. Activated caspase-3+ cells had condensed nuclei (red arrows), and the CD31-positive cells are adjacent to them (white arrows). n = 5–8 per genotype. (D) Quantification of caspase-3+ cells in the GEs. Each dot represents 1 section from 8 WT and 5 KO embryos. ***P < 0.001, 2-tailed t test.
Discussion
The physiological roles of MFSD7c in blood vessel development.
Mutations in MFSD7c have been identified in humans and associated with Fowler syndrome. These patients commonly exhibit neurological defects. However, the mechanistic links between the missense mutations in MFSD7c and the brain defects observed in Fowler syndrome are unknown. Our results here show that MFSD7c is expressed in the endothelial cells of CNS blood vessels. The expression of MFSD7c is specific to CNS endothelial cells, expression in other neuronal cells and endothelial cells in peripheral tissues being low or undetectable (19–21). Therefore, we hypothesized the dysfunctions of MFSD7c in the CNS blood vessels were directly related to the phenotypes of Fowler syndrome patients and that of the KO embryos. We have demonstrated that the defective growth of blood vessels to SVZ and VZ causes the neurological defects in Mfsd7c-KO embryos. The growth of blood vessels to VZ and SVZ regions is blunted, leading to hypoxia and neuronal cell death. We observed that hypoxia occurred in several regions with reduced vascular density and avascular regions in the KO brain. However, the neuronal cell death was initiated in the VZ regions in the GEs, suggesting that neural progenitors in this region are more sensitive to hypoxia (22, 23). Of note, GEs are part of the germinal matrix. Defects in blood vessels in germinal matrix account for a number of neurological disorders (24, 25). Our results show that lack of MFSD7c brings about the blunted growth of blood vessels to SVZ and VZ regions, with consequent hypoxia and neuronal cell death. Thus, we propose that MFSD7c is important for both CNS blood vessel functions and for brain growth.
The migratory defects of blood vessels to the SVZ and VZ regions in Mfsd7c-KO brain are remarkable. In these regions, the radial growth of blood vessels is inhibited even though there is increased signaling from VEGF throughout the KO brain. We found that filopodia of stalled vessels were elongated to the ventricular surfaces, but the blood vessels failed to migrate. These blood vessels then appeared to grow perpendicularly and fuse together. The fusion of these blood vessels generated the glomeruloid structures with large intravascular lumenal spaces. The increased expression of several angiogenic factors such as VEGF and ANGPT2 has been associated with the glumeruloid vessels (26, 27). However, the molecular mechanisms underpinning the glomeruloid phenotype of MFSD7c absence remain to be determined. Interestingly, we noticed that the phenotypes of blood vessels in Mfsd7c-KO embryos are similar to the phenotypes from canonical WNT signaling components in CNS and TGFB signaling pathway (13, 27–31). In contrast, MFSD7c is apparently dispensable for blood vessel integrity, as the global KO embryos show no cerebral hemorrhage, as observed in deficiency of the canonical WNT signaling components or TGFB signaling deficient models. Expression of MFSD7c is reduced in Reck and Norrin knockout mice, the 2 components of canonical WNT signaling in CNS and retina, respectively, and is induced in β-catenin expression (32–34). Nevertheless, our data show that WNT and TGFB signaling pathways are not disrupted in the brain of KO embryos, suggesting that MFSD7c is only required for proper CNS blood vessel development. It is interesting to note that MFSD7c is a predicted plasma membrane transporter for metabolites. Thus, we speculate that the ligand or ligands of this protein are required for proper growth of CNS blood vessels. Several brain disorders, such as vascular dementia and stroke, are associated with defects in CNS blood vessels. Therefore, the identification of MFSD7c ligands may offer insight into the roles of metabolites in the physiological functions of blood vessels in brain.
The molecular functions of MFSD7c.
Little is known about the molecular functions of MFSD7c. MFSD7c is similar to the known heme exporter MFSD7a (also called FLVCR1a, an isoform for the plasma membrane exporter of heme in erythroid cells; and FLVCR1b is the mitochondrial isoform). Deletion of these heme exporters results in heme accumulation and toxicity in erythroid cells. A prior study showed that MFSD7c/FLVCR2 is a heme importer in vitro (9). We performed the heme-uptake and heme-release assays using HEK293 cells overexpressed with mouse or human MFSD7c and did not observe the results as reported for MFSD7c (Supplemental Figure 14). Additionally, heme levels were comparable in the embryonic brains of Mfsd7c-KO mice and their WT littermates (data not shown). Thus, it seems that the heme-transporting roles for MFSD7c need further investigation. Our results here show that MFSD7c is a plasma membrane protein that is expressed in both sides of endothelial cells in the brain, suggesting its ligand or ligands are transported into or out of the brain directly. The current study did not elucidate the molecular roles of MFSD7c. Nonetheless, our findings here have enabled us to anticipate that the ligand or ligands of MFSD7c should be essential for CNS blood vessel structure and function as well as brain growth.
The roles of MFSD7c in neuronal cell migration to cortical sites for brain formation.
The patient described here exhibited ventriculomegaly and intracranial calcifications without hydrocephalus. The absence of the hydrocephaly symptom in the patient may be linked to the partial loss of MFSD7c functions. However, we did not observe the hydrocephalus phenotype (e.g., enlargement of brain) in the KO embryos’ brains. Thus, hydrocephalus might not be the major consequence of MFSD7c deficiency. Interestingly, the KO embryos and humans with mutations of MFSD7c experience microcephaly with thinning of the cerebral cortices. What drives these phenotypes? In growing brains, the development of vasculature and neuronal networks needs to be well coordinated. Angiogenic sprouting of CNS blood vessels occurs from the pial surface to the neuronal layers to provide cells with oxygen and nutrients. Our findings suggest that the delayed growth of blood vessels in the Mfsd7c-KO embryos’ brains, especially in the GEs, impacts negatively on the survival and migration of the neural progenitor cells that are required for forming the cortical layers. It has been established that GEs are enriched with neural progenitors. These cells migrate tangentially via the axons established by radial glial cells (35). They can also migrate in axon-free regions to the cerebral cortex, likely via blood vessel routes (36, 37). The reduced growth and absence of blood vessels in the GEs are the most striking phenotypes that we observed in the Mfsd7c-KO embryos. The vasculature network is necessary to provide routes for the migration of the neural progenitor cells to cerebral cortex regions in the growing brain. The blunted growth of the blood vessels to these regions will impair oxygen availability and the delivery of nutrients to these proliferating cells (38–41). Therefore, the lack of blood vessels penetrating into these inner brain regions brings about hypoxic environment, affecting the metabolism, survival, and migration of neural stem cells to build cerebral cortex during brain development. We propose that the thinning of cortices and microcephaly observed both in Mfsd7c-KO embryos and the human patient are in part due to the reduced blood vessel growth during the critically developing stages of brain. It is also possible that MFSD7c transports essential ligands for neuronal cell growth, such as the ligands of MFSD2a and GLUT1 (42, 43). In summary, we report here that defects in MFSD7c functions result in a microcephaly-associated vasculopathy in mice and humans.
Methods
Mice.
Mice with whole-body KO of Mfsd7c were obtained from the International Mouse Program Consortium (IMPC). Mice were maintained on normal chow diets and housed with regular light/dark cycles. To generate the KO embryos, mice heterozygous for Mfsd7c were time mated. Embryos were collected from time-mated female mice and genotyped using the following primers: Mfsd7c WT: 5′-CACTACAGAAACTCTGGCTGTTGC-3′; Mfsd7c common primer: 5′-CATCCCAGCATTGGAAGGAC-3′; Mfsd7c KO: 5′-TCGTGGTATCGTTATGCGCC-3′.
Antibodies.
In-house polyclonal antibodies for human and mouse MFSD7c proteins were generated in rabbit. These purified polyclonal antibodies were raised in rabbits with antigens comprising the 29–amino acid sequence (APETKVQEEEEEEEGSNTSKVPVVSEAHL) at the C-terminus of mouse MFSD7c protein or the 25 amino acids (ENKLQEEEEESNTSKVPTAVSEDHL) at the C-terminus of human MFSD7c protein. Commercial primary and secondary antibodies used are detailed in Supplemental Table 2.
Western blot.
Whole-brain tissue from E16.5 WT and KO embryos or microvessels isolated from adult mouse brain as described below were lysed in RIPA buffer (25 mM Tris pH 7–8, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate 0.5% Triton X-100), then supplemented with protease inhibitor cocktail (Roche, 11836170001) and phosphatase inhibitor cocktail 1 (MilliporeSigma, P2850) for detection of phosphorylated proteins. For detection of TGFB signaling components, 100 μg of total protein from whole-brain lysates of E16.5 WT and KO embryos was resolved in 10% to 12% SDS-PAGE and transferred to membrane before probing with phospho-SMAD1/5 antibody (Cell Signaling Technology, catalog 9516) and SMAD4 antibody (Cell Signaling Technology, catalog 38454). The membranes were also probed with MFSD7c antibody (1:500) and reprobed with β-actin antibody (MilliporeSigma, catalog A5441). For detection of WNT signaling components, whole-brain lysates of E16.5 WT and KO embryos were similarly analyzed. The membrane was probed with β-catenin antibody (Cell Signaling Technology, catalog 19807) and phospho–β-catenin antibody (Cell Signaling Technology, 9561). For microvessels, 100 μg of total proteins from brain fractions and 1 μg of total proteins from HEK293 cells were used for Western blot. The membrane was probed with MFSD7c antibody and GLUT1 antibody (Abcam, catalog ab40084) and reprobed with β-actin antibody. Images were acquired with the ChemiDoc MP Imaging System (Bio-Rad).
Isolation of microvessels from adult mouse brains.
Brain microvessels were obtained by dextran gradient centrifugation. Briefly, 4 to 6 adult mouse brains were collected after PBS perfusion. The brainstem, cerebellum, and thalamus were removed. The cortical gray matter was rolled on filter paper to remove the meninges, then briefly homogenized by 3 strokes with a Dounce homogenizer in PBS, transferred into a Falcon tube, and centrifuged at 1500 g for 20 minutes at 4°C. The pellet was resuspended in 4 volumes of 18% dextran in PBS and centrifuged at 1500 g for 20 minutes at 4°C. The pellet containing microvessels was saved and the remaining tissue reprocessed twice in similar fashion. All 3 pellets were pooled and washed again with PBS. The parenchyma fraction (mainly composed of neurons and glial cells) was also collected for analysis. The pellet was lysed with 100 μL RIPA containing proteinase and phosphatase inhibitors.
Immunostaining of embryo sections.
Embryos from time-matched heterozygous female mice were collected. Whole embryos were fixed with 4% paraformaldehyde at 4°C for between 1 and 5 days depending on embryo size. They were washed with PBS and dehydrated with 15% and then 30% sucrose for 1 to 2 days. Samples were then embedded in OCT compound and snap-frozen with dry ice. A Leica CM3060S Cryostat was used to prepare sagittal and coronal sections, 15 μm to 50 μm thick, that were then mounted onto Fisherbrand Superfrost Plus Microscope Slides. The specific antibodies and the concentrations used for immunostaining embryo sections are detailed in Supplemental Table 2. For hypoxia staining, the Hypoxyprobe-Red549 Kit (Dylight 549-Mab) was used. Heterozygous pregnant females at gestation day 15.5 were injected with 180 mg/kg body weight for 3 hours before embryo collection. The embryos were harvested, genotyped, and processed as described above. The brain sections were then stained with Dylight 549-Mab to visualize hypoxic cells and costained with CD31 to visualize blood vessels. Images were taken with a Zeiss LSM710 confocal microscope.
NHS-biotin injections.
NHS-biotin, a small molecular weight dye, was used to assess blood vessel leakage. A volume of 5 μL of 4 mg/mL NHS-biotin (Thermo Fisher Scientific, 21335) in PBS was injected into the liver of E16.5 WT and KO embryos using a Hamilton syringe. Briefly, as previously described, embryos were exposed by removing the yolk sac while retaining umbilical cord connection to the maternal circulation. Circulation of NHS-biotin dye was permitted for 10 minutes before the embryos were collected for fixation and dissection. The NHS-biotin signal was visualized with Texas red streptavidin, as described previously (44, 45).
Vascular measurements.
To quantify the vascular density and diameter, CNS blood vessels were stained with CD31. The tiling scan of whole coronal sections was acquired using a Zeiss LSM710 confocal microscope. Vascular density measurements in midbrain, GEs, and cerebral cortex were quantified by ImageJ (NIH) and expressed as percentages of measured areas. The diameters of CNS blood vessels in the same regions were also quantified by ImageJ.
H&E staining from paraffin sections.
Embryos at various stages were collected and fixed as described above. Tissue-mounted slides were treated with xylene to remove paraffin. Slides were rehydrated progressively with 100% to 70% ethanol with 10-minute intervals and under running water for 5 minutes. The sections were stained with hematoxylin (MilliporeSigma) for 1 minute and then washed under running water to remove excess stain. Slides were treated with 95% ethanol to dehydrate and then stained for 1.5 minutes with eosin (MilliporeSigma). Excess dye was removed by washing in 50% to 100% ethanol for 5 minutes, and slides were then cleared with xylene for 1 minute. Slides were air-dried overnight, and slide covers were mounted onto the samples with DPX mountant (Merck). Similar procedures were performed with human brain sections.
Expression of MFSD7c in HEK293 cells.
The mouse, human, and human mutant plasmids were constructed in pcDNA3.1 plasmid and transfected into HEK293 cells (2–3 μg plasmid per well in 24-well plates) using Lipofectamine 2000. At 24 hours after transfection, the HEK293 cells were used for protein isolation, localization, or heme transport assay as described below.
Heme assay.
Heme assays were performed using the Hemin Colorimetric Assay Kit (catalog K672, Biovision) and tritium-labeled [3H]hemin (American Radiochemicals). In these experiments, human or mouse MFSD7c was expressed in HEK293 cells. Empty pcDNA3.1 plasmid was used as mock control, and pcDNA3.1 containing human or mouse MFSD7c was used for overexpression. The transfected HEK293 cells were incubated with 1 μM to 50 μM hemin (prepared in DMSO) in growth medium. Various time points ranging from 30 minutes to overnight incubation with hemin were tested. The HEK293 cells were then lysed with RIPA buffer. Clear lysates were used in the hemin assay. For [3H]hemin assays, the HEK293 cells were prepared as described above and incubated with 1 μM to 50 μM [3H]hemin. The cells were washed with PBS and collected in RIPA buffer. Radioactive signals from the RIPA lysates were quantified by scintillation counter.
Transcriptomic analysis.
Total RNA from whole brain of WT and KO embryos at E14.5 was extracted using the QIAGEN Mini RNA Extraction Kit per the manufacturer’s protocol. For RNA-Seq, whole-brain tissue from individual E14.5 KO and WT embryos was extracted with TRIzol reagent for total RNA isolation. RNA-Seq was performed with Novogene. Over 99% of RNA-Seq samples were of appropriate quality for use in downstream analysis. The RNA-Seq data were aligned to the mm10 mouse genome using the Ensembl Release 98 version annotation file STAR package. After alignment, all the samples achieved more than 95% unique mapped reads. Reads were quantified using the featureCount package. The DESeq2 package was used for differential gene analysis. All original RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE148854).
Quantitative real-time PCR.
To quantify mRNA from mouse brain, total RNA samples were prepared as described above for microarray analysis. A total cDNA library was synthesized using the Super-Script First-Strand Synthesis System (Invitrogen, Thermo Fisher Scientific). Quantitative RT-PCR was performed with specific primers (Supplemental Table 3) for each gene of interest using SYBR Green mix. Gene expression level was normalized to β-actin and expressed as fold change.
Patient recruitment.
The family was evaluated at the medical genetics clinic at the Rambam Health Care campus.
Genetic evaluation.
Exome sequencing was performed on a NovaSeq 6000 platform (Illumina) using the Twist Human Core Exome Kit. Mapping of the obtained reads to the reference genome (build GRCh37/hg19), variant calling, annotation, and data analysis were done using the Genoox Data Analysis Platform. We filtered sequencing data on a trio-based paradigm to identify recessive (homozygous and compound heterozygous), X-linked, and potential de novo variants in the proband. Variants were prioritized based on their effect on the protein and minor allele frequency below 1% in general population databases, such as gnomAD (http://gnomad.broadinstitute.org) and the Rambam Genetics Institute internal database. We identified compound heterozygous variants in FLVCR2 (NM_017791.2): c.1019C>T;p.Pro340Leu and c.1288A>G;p.Thr430Ala. The variants were confirmed by Sanger sequencing.
Statistics.
Data were analyzed using GraphPad Prism, version 7.2, software for Windows. Statistical significance was calculated using 2-tailed t test, as indicated in the figure legends. A P value of less than 0.05 was considered statistically significant.
Study approval.
All experimental protocols were approved by the IACUC committee of the National University of Singapore. Parents of the patients gave informed consent to clinical exome sequencing and to the publication of clinical data and photographs.
Author contributions
PK, KQW, and XRT performed immunostaining of embryo sections. LNN performed confocal imaging. TQN and XRT performed Western blot analyses and generated the human mutant plasmids. MC and XRT provided H&E staining data and data for Figure 3A. PK performed qPCR analysis. KW, CH, and AS provided the data from the patient with P340L and T430A mutations. FCR and BD provided human brain sections from the S203Y patient. ZH performed IHC staining for human brain sections. LNN conceived and designed the study and experiments, analyzed the data, composed all figures, and wrote the paper with assistance from KW. The order of shared first authors were determined by their respective contributions to the data in this paper.
Supplementary Material
Acknowledgments
This study was supported in part by Singapore Ministry of Health’s National Research Council NMRC grant OFIRG/0066/20; Ministry of Education grants MOE2018-T2-1-126, MOE-Tier-1, and MOE PDF fellowships; NUS Young Investigator Award grant NUSYIA_ FY16_P19; NUH bridging fund grant NUHSRO/2017/066/BRIDGING/04; and NUSMED-FOS Joint Research Programme grant on Healthy Brain Aging (to LNN). We thank Tan Tze King for assistance with the analysis of RNA-Seq samples. We thank Barry Halliwell, Roger Foo, and Mark Richards for proofreading the manuscript.
Version 1. 05/05/2020
In-Press Preview
Version 2. 06/22/2020
Electronic publication
Version 3. 08/03/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(8):4081–4093.https://doi.org/10.1172/JCI136727.
Contributor Information
Pazhanichamy Kalailingam, Email: bchpaz@nus.edu.sg.
Kai Qi Wang, Email: e0035940@u.nus.edu.
Xiu Ru Toh, Email: xiuru95@gmail.com.
Toan Q. Nguyen, Email: bchnqt@nus.edu.sg.
Madhuvanthi Chandrakanthan, Email: madhuvanthi.jc@gmail.com.
Zafrul Hasan, Email: bchmzh@nus.edu.sg.
Clair Habib, Email: c_habib@rambam.health.gov.il.
Aharon Schif, Email: ngnlong@gmail.com.
Francesca Clementina Radio, Email: fclementina.radio@opbg.net.
Bruno Dallapiccola, Email: Bruno.dallapiccola@opbg.net.
Karin Weiss, Email: K_WEISS@rambam.health.gov.il.
Long N. Nguyen, Email: bchnnl@nus.edu.sg.
References
- 1.Kvarnung M, et al. Mutations in FLVCR2 associated with Fowler syndrome and survival beyond infancy. Clin Genet. 2016;89(1):99–103. doi: 10.1111/cge.12565. [DOI] [PubMed] [Google Scholar]
- 2.Meyer E, et al. Mutations in FLVCR2 are associated with proliferative vasculopathy and hydranencephaly-hydrocephaly syndrome (Fowler syndrome) Am J Hum Genet. 2010;86(3):471–478. doi: 10.1016/j.ajhg.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessières-Grattagliano B, et al. Refining the clinicopathological pattern of cerebral proliferative glomeruloid vasculopathy (Fowler syndrome): report of 16 fetal cases. Eur J Med Genet. 2009;52(6):386–392. doi: 10.1016/j.ejmg.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Lalonde E, et al. Unexpected allelic heterogeneity and spectrum of mutations in Fowler syndrome revealed by next-generation exome sequencing. Hum Mutat. 2010;31(8):918–923. doi: 10.1002/humu.21293. [DOI] [PubMed] [Google Scholar]
- 5.Radio FC, et al. Proliferative vasculopathy and hydranencephaly-hydrocephaly syndrome or Fowler syndrome: Report of a family and insight into the disease’s mechanism. Mol Genet Genomic Med. 2018;6(3):446–451. doi: 10.1002/mgg3.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas S, et al. High-throughput sequencing of a 4.1 Mb linkage interval reveals FLVCR2 deletions and mutations in lethal cerebral vasculopathy. Hum Mutat. 2010;31(10):1134–1141. doi: 10.1002/humu.21329. [DOI] [PubMed] [Google Scholar]
- 7.Keel SB, et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science. 2008;319(5864):825–828. doi: 10.1126/science.1151133. [DOI] [PubMed] [Google Scholar]
- 8.Doty RT, et al. Single-cell analyses demonstrate that a heme-GATA1 feedback loop regulatesred cell differentiation. Blood. 2019;133(5):457–469. doi: 10.1182/blood-2018-05-850412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy SP, et al. The Fowler syndrome-associated protein FLVCR2 is an importer of heme. Mol Cell Biol. 2010;30(22):5318–5324. doi: 10.1128/MCB.00690-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Z, Nelson AR, Betsholtz C, Zlokovic BV. Establishment and dysfunction of the blood-brain barrier. Cell. 2015;163(5):1064–1078. doi: 10.1016/j.cell.2015.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sweeney MD, Zhao Z, Montagne A, Nelson AR, Zlokovic BV. Blood-brain barrier: from physiology to disease and back. Physiol Rev. 2019;99(1):21–78. doi: 10.1152/physrev.00050.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stenman JM, Rajagopal J, Carroll TJ, Ishibashi M, McMahon J, McMahon AP. Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science. 2008;322(5905):1247–1250. doi: 10.1126/science.1164594. [DOI] [PubMed] [Google Scholar]
- 13.Daneman R, Agalliu D, Zhou L, Kuhnert F, Kuo CJ, Barres BA. Wnt/beta-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc Natl Acad Sci U S A. 2009;106(2):641–646. doi: 10.1073/pnas.0805165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho C, Smallwood PM, Nathans J. Reck and Gpr124 are essential receptor cofactors for Wnt7a/Wnt7b-specific signaling in mammalian CNS angiogenesis and blood-brain barrier regulation. Neuron. 2017;95(5):1056–1073.e5. doi: 10.1016/j.neuron.2017.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, et al. Canonical WNT signaling components in vascular development and barrier formation. J Clin Invest. 2014;124(9):3825–3846. doi: 10.1172/JCI76431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armulik A, et al. Pericytes regulate the blood-brain barrier. Nature. 2010;468(7323):557–561. doi: 10.1038/nature09522. [DOI] [PubMed] [Google Scholar]
- 17.Daneman R, Zhou L, Kebede AA, Barres BA. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 2010;468(7323):562–566. doi: 10.1038/nature09513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanlandewijck M, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554(7693):475–480. doi: 10.1038/nature25739. [DOI] [PubMed] [Google Scholar]
- 21.Sabbagh MF, et al. Transcriptional and epigenomic landscapes of CNS and non-CNS vascular endothelial cells. Elife. 2018;7:e36187. doi: 10.7554/eLife.36187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson CB, Momma S, Clarke DL, Risling M, Lendahl U, Frisén J. Identification of a neural stem cell in the adult mammalian central nervous system. Cell. 1999;96(1):25–34. doi: 10.1016/S0092-8674(00)80956-3. [DOI] [PubMed] [Google Scholar]
- 23.Gage FH. Mammalian neural stem cells. Science. 2000;287(5457):1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 24.Flores JJ, Klebe D, Tang J, Zhang JH. A comprehensive review of therapeutic targets that induce microglia/macrophage-mediated hematoma resolution after germinal matrix hemorrhage. J Neurosci Res. 2020;98(1):121–128. doi: 10.1002/jnr.24388. [DOI] [PubMed] [Google Scholar]
- 25.Luo J, Luo Y, Zeng H, Reis C, Chen S. Research advances of germinal matrix hemorrhage: an update review. Cell Mol Neurobiol. 2019;39(1):1–10. doi: 10.1007/s10571-018-0630-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundberg C, et al. Glomeruloid microvascular proliferation follows adenoviral vascular permeability factor/vascular endothelial growth factor-164 gene delivery. Am J Pathol. 2001;158(3):1145–1160. doi: 10.1016/S0002-9440(10)64062-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crist AM, et al. Angiopoietin-2 Inhibition Rescues Arteriovenous Malformation in a Smad4 Hereditary Hemorrhagic Telangiectasia Mouse Model. Circulation. 2019;139(17):2049–2063. doi: 10.1161/CIRCULATIONAHA.118.036952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, et al. Interplay of the Norrin and Wnt7a/Wnt7b signaling systems in blood-brain barrier and blood-retina barrier development and maintenance. Proc Natl Acad Sci U S A. 2018;115(50):E11827–E11836. doi: 10.1073/pnas.1813217115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F, et al. Endothelial Smad4 maintains cerebrovascular integrity by activating N-cadherin through cooperation with Notch. Dev Cell. 2011;20(3):291–302. doi: 10.1016/j.devcel.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Arnold TD, et al. Excessive vascular sprouting underlies cerebral hemorrhage in mice lacking αVβ8-TGFβ signaling in the brain. Development. 2014;141(23):4489–4499. doi: 10.1242/dev.107193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dave JM, Mirabella T, Weatherbee SD, Greif DM. Pericyte ALK5/TIMP3 axis contributes to endothelial morphogenesis in the developing brain. Dev Cell. 2018;47(3):388–389. doi: 10.1016/j.devcel.2018.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho C, Wang Y, Smallwood PM, Williams J, Nathans J. Molecular determinants in Frizzled, Reck, and Wnt7a for ligand-specific signaling in neurovascular development. Elife. 2019;8:e47300. doi: 10.7554/eLife.47300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heng JS, et al. Hypoxia tolerance in the Norrin-deficient retina and the chronically hypoxic brain studied at single-cell resolution. Proc Natl Acad Sci U S A. 2019;116(18):9103–9114. doi: 10.1073/pnas.1821122116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Sabbagh MF, Gu X, Rattner A, Williams J, Nathans J. Beta-catenin signaling regulates barrier-specific gene expression in circumventricular organ and ocular vasculatures. Elife. 2019;8:e43257. doi: 10.7554/eLife.43257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marín O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 36.Licht T, Dor-Wollman T, Ben-Zvi A, Rothe G, Keshet E. Vessel maturation schedule determines vulnerability to neuronal injuries of prematurity. J Clin Invest. 2015;125(3):1319–1328. doi: 10.1172/JCI79401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Won C, et al. Autonomous vascular networks synchronize GABA neuron migration in the embryonic forebrain. Nat Commun. 2013;4:2149. doi: 10.1038/ncomms3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kojima T, et al. Subventricular zone-derived neural progenitor cells migrate along a blood vessel scaffold toward the post-stroke striatum. Stem Cells. 2010;28(3):545–554. doi: 10.1002/stem.306. [DOI] [PubMed] [Google Scholar]
- 39.Lange C, et al. Relief of hypoxia by angiogenesis promotes neural stem cell differentiation by targeting glycolysis. EMBO J. 2016;35(9):924–941. doi: 10.15252/embj.201592372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen Q, et al. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304(5675):1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- 41.Tavazoie M, et al. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3(3):279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen LN, et al. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature. 2014;509(7501):503–506. doi: 10.1038/nature13241. [DOI] [PubMed] [Google Scholar]
- 43.Seidner G, et al. GLUT-1 deficiency syndromecaused by haploinsufficiency of the blood-brain barrier hexose carrier. Nat Genet. 1998;18(2):188–191. doi: 10.1038/ng0298-188. [DOI] [PubMed] [Google Scholar]
- 44.Ben-Zvi A, et al. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature. 2014;509(7501):507–511. doi: 10.1038/nature13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong BH, et al. Mfsd2a is a transporter for the essential ω-3 fatty acid docosahexaenoic acid (DHA) in eye and is important for photoreceptor cell development. J Biol Chem. 2016;291(20):10501–10514. doi: 10.1074/jbc.M116.721340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.