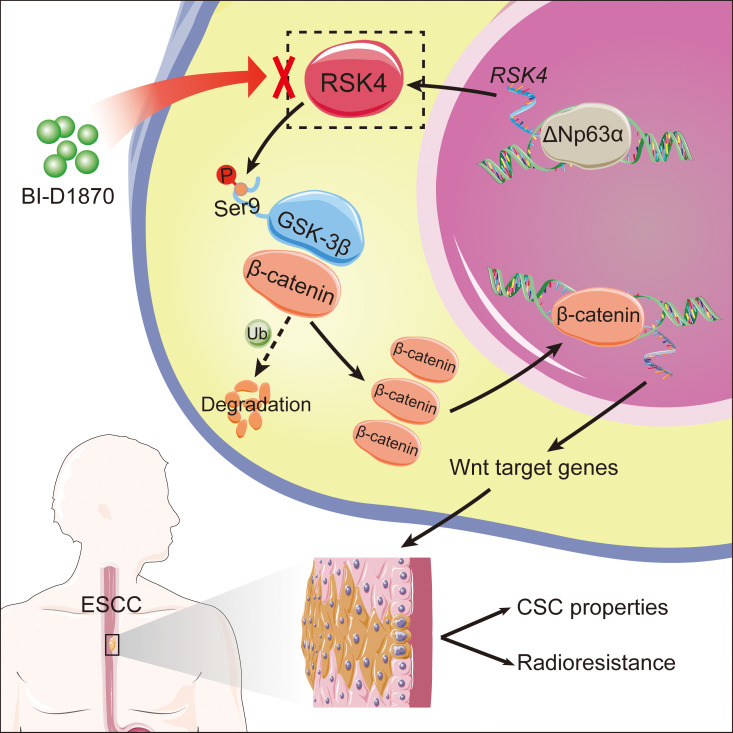
Abstract
Esophageal squamous cell carcinoma (ESCC) is one of the most aggressive cancers and is highly resistant to current treatments. ESCC harbors a subpopulation of cells exhibiting cancer stem-like cell (CSC) properties that contribute to therapeutic resistance including radioresistance, but the molecular mechanisms in ESCC CSCs are currently unknown. Here, we report that ribosomal S6 protein kinase 4 (RSK4) plays a pivotal role in promoting CSC properties and radioresistance in ESCC. RSK4 was highly expressed in ESCC CSCs and associated with radioresistance and poor survival in patients with ESCC. RSK4 was found to be a direct downstream transcriptional target of ΔNp63α, the main p63 isoform, which is frequently amplified in ESCC. RSK4 activated the β-catenin signaling pathway through direct phosphorylation of GSK-3β at Ser9. Pharmacologic inhibition of RSK4 effectively reduced CSC properties and improved radiosensitivity in both nude mouse and patient-derived xenograft models. Collectively, our results strongly suggest that the ΔNp63α/RSK4/GSK-3β axis plays a key role in driving CSC properties and radioresistance in ESCC, indicating that RSK4 is a promising therapeutic target for ESCC treatment.
Keywords: Oncology
Keywords: Cancer
Introduction
Esophageal cancer is one of the most frequently occurring cancers and ranks as the seventh leading cause of cancer-related mortality worldwide. Esophageal squamous cell carcinoma (ESCC) is the most common histological type of esophageal malignancy and has a higher incidence in developing nations (1), with more than half of ESCC cases occurring in China (2). Despite intensive clinical efforts using multiple therapeutic approaches, patients with ESCC still face a poor prognosis, which has not significantly improved in recent decades (3). The treatment of choice for locally advanced ESCC is neoadjuvant chemoradiation followed by radical surgery. Unfortunately, more than 50% of patients with ESCC do not respond adequately, and most die from recurrent cancer (4). ESCC possesses a subpopulation of cells exhibiting cancer stem-like cell (CSC) properties. These CSCs are known to have high tumorigenic potential and resistance to conventional anticancer therapy and are responsible for the intractable features of ESCC (5, 6). Increasing evidence indicates that CSCs contribute to radioresistance, which could result in radiation treatment failure for ESCC (7, 8). Therefore, targeting CSCs may be a promising approach for developing novel combination modalities and overcoming the radiation resistance of ESCC.
p90 ribosomal S6 kinases (RSKs) are a family of serine-threonine kinases involved in the Ras/MAPK pathway. This family consists of 4 members in humans: RSKs 1–4, along with 2 structurally related homologs, mitogen- and stress-activated kinase 1 (MSK1) and MSK2 (9). RSKs phosphorylate many cytosolic and nuclear targets and are integrally linked to a variety of physiological processes, including cell-cycle progression and proliferation, cell growth and protein synthesis, cell migration, and cell survival (10). With regard to the role of different RSKs in cancer, it is generally believed that RSK1 and RSK2 promote cancer cell growth, survival, and motility (11, 12), whereas RSK3 is downregulated in breast tumors (13) and ovarian cancer (14). RSK4, which was first identified as an X-linked gene in patients with mental retardation, is most abundant in the fetal and adult kidney, brain, and thyroid gland (15). Functional characterization analysis shows that RSK4 is constitutively active under serum-starved conditions and independent of 3-phosphoinositide–dependent protein kinase1 (PDK1) for phosphorylation, whereas other RSKs are inactive because of their requirement for growth factors (16), indicating that RSK4 is functionally distinct from the other RSKs. Compared with the more in-depth research on the other RSKs in the field of cancer, there have been limited and conflicting reports describing the role of RSK4 in different cancer types (17–25). Using a tissue microarray (TMA) containing 20 kinds of human tumors and corresponding normal tissues, we found that RSK4 protein was highly expressed in renal cell carcinoma (RCC) and ESCC. We previously reported that RSK4 is overexpressed in RCC and enhances the invasive and metastatic ability of RCC cells by regulating the CSC marker CD44 (23). In this study, we expanded on our previous studies and sought to determine whether RSK4 plays a pathophysiological role in ESCC and CSCs.
Recent genomic analysis has revealed that p63, a p53-related transcriptional factor, is a major oncogenic protein in esophageal cancer; the gene locus is frequently amplified in ESCC, and its expression in ESCC is significantly higher than that in nontumor tissues (26). TP63 contains 2 different promoters that drive 2 distinct isoform classes: with or without the N-terminal transactivation domain, TAp63 and ΔNp63, respectively. In addition, both TAp63 and ΔNp63 have 3 variants with different C-termini (α, β, and γ) generated by alternative splicing (27). ΔNp63 and TAp63 show very different expression patterns, depending on the source of cell lines and tissues (28). ΔNp63α is the main p63 isoform expressed in ESCC (29) and plays an important role in maintaining the properties of CSCs (30), but the relationship between p63 and RSK4 remains to be clarified. In this study, we sought to determine whether the ΔNp63α/RSK4 axis plays a role in establishing CSC properties and radioresistance in ESCC, to define the downstream effector genes and pathways controlled by these factors, and to test the rationale for RSK4 as a therapeutic target in this disease.
Results
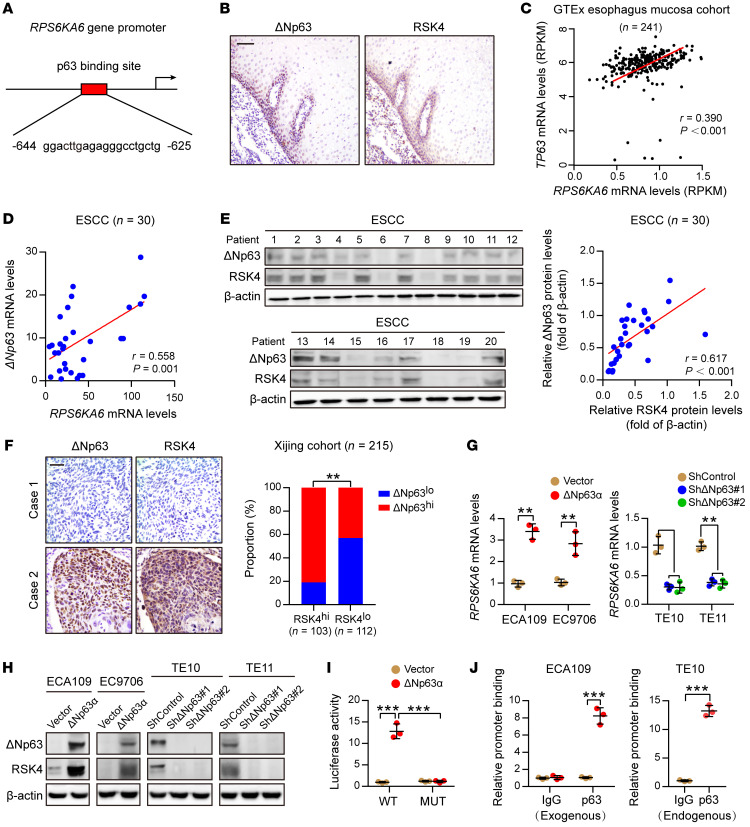
RSK4 is highly expressed in ESCC CSCs and is associated with the radioresistance and poor survival of ESCC patients.
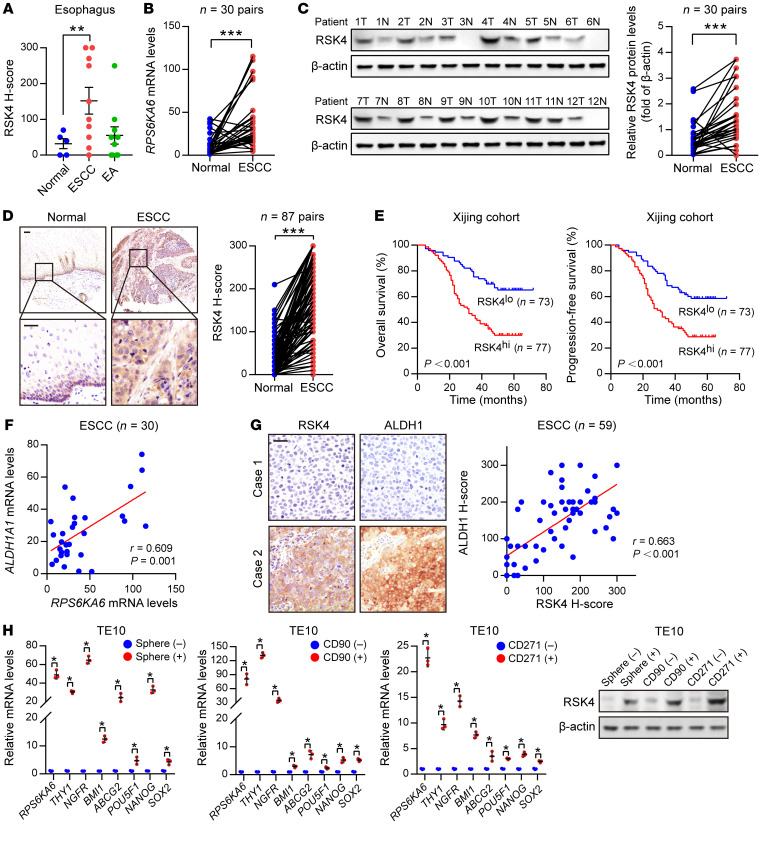
In a TMA containing 20 kinds of human tumors and corresponding normal tissues, IHC showed that RSK4 protein levels were significantly reduced in stomach and testis cancer but highly expressed in kidney and esophageal cancer compared with expression levels in their corresponding nontumor tissues (Supplemental Figure 1A and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI134930DS1). In esophageal cancer, RSK4 protein was highly expressed in ESCC rather than esophageal adenocarcinoma (Figure 1A and Supplemental Figure 1A). In 30 paired ESCC and adjacent nontumor tissues, RPS6KA6 (encoding RSK4) mRNA and RSK4 protein levels were also much higher in ESCC than in normal tissues (Figure 1, B and C, and Supplemental Figure 1B). This result was further confirmed by IHC analyses with 87 paired ESCC and adjacent nontumor tissues (Figure 1D). However, the mRNA levels of the other 2 RSK members, RPS6KA1 (encoding RSK1) and RPS6KA3 (encoding RSK2), showed no significant difference. The mRNA level of RPS6KA2 (encoding RSK3) was much lower in ESCC than in normal tissues (Supplemental Figure 1C). We next used IHC analysis to examine the prognostic significance of RSK4 expression in clinical tumor samples from cohorts of ESCC patients. Importantly, compared with low RSK4 expression, high expression of RSK4 was correlated with poorer overall survival (OS) and progression-free survival (PFS) of patients with ESCC and more aggressive tumor behaviors, including lymph node metastasis and vascular invasion (Figure 1E, Supplemental Figure 1D, and Supplemental Table 2), with similar results found in The Cancer Genome Atlas (TCGA) cohort (Supplemental Figure 1E). In addition, the mRNA levels of RPS6KA6 in patients with grade 2 or grade 3 disease were higher than those in patients with grade 1 ESCC disease (Supplemental Figure 1F). Multivariate Cox regression analysis further indicated RSK4 expression as a potential independent prognostic marker for OS and PFS in patients with ESCC (Supplemental Table 3).
Figure 1. RSK4 is highly expressed in ESCC CSCs.
(A) RSK4 protein was highly expressed in ESCC rather than in esophageal adenocarcinoma (EA) compared with expression in corresponding nontumor tissues. Representative IHC images are shown in Supplemental Figure 1A. (B) mRNA levels of RPS6KA6 in 30 pairs of ESCC samples and adjacent nontumor tissues were determined by real-time PCR. GAPDH was used as a loading control. (C) Western blot analysis and quantification of RSK4 expression in ESCC tumor tissues (T) and adjacent nontumor tissues (N) from 30 patients. The results for the other samples are presented in Supplemental Figure 1B. Protein expression was normalized to β-actin levels. (D) Representative IHC images and H-score of RSK4 protein expression in ESCC tumor tissues and adjacent nontumor tissues. Scale bars: 100 μm. (E) Kaplan-Meier estimation of ESCC OS and PFS based on the RSK4 expression levels in the Xijing cohort. (F) Correlation between RPS6KA6 and ALDH1A1 mRNA expression in 30 ESCC patients. (G) Representative IHC images of RSK4 and ALDH1 protein expression in patients with ESCC from the Xijing cohort. Scale bars: 100 μm. Correlation of IHC data on RSK4 and ALDH1 protein expression in 59 ESCC patients. (H) RSK4 was preferentially expressed in tumor spheres compared with nonspheres, and elevated RSK4 expression was detected in CD90+- or CD271+-enriched cell populations compared with the CD90− or CD271− cell subsets as assessed by real-time PCR (n = 3 independent experiments) and immunoblotting. Data represent the mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001. Differences were tested using a paired (B–D) and unpaired (H) 2-sided Student’s t test, 1-way ANOVA with Tukey’s post hoc test (A), and log-rank test (E). The correlation was determined by Pearson’s correlation test (F and G).
Surprisingly, we found high RSK4 protein levels in the basal layer of esophageal epithelia that decreased progressively in the suprabasal and superficial cell compartments with cellular differentiation (Figure 1D), suggesting that RSK4 may be involved in the stemness properties of esophageal epithelia. In ESCC tissues, RSK4 expression was positively correlated with ALDH1, an ESCC CSC marker that we previously identified (ref. 31 and Figure 1, F and G). In addition, mRNA levels of RPS6KA6 were positively correlated with ALDH1A1 in the TCGA ESCC cohort (Supplemental Figure 1G). Sphere formation has been well established in the enrichment of CSCs on the basis of their self-renewing capacity (32). Like other ESCC CSC markers (6), including CD90, CD271, ABCG2, BMI-1, NANOG, OCT4, and SOX2, we found that RSK4 expression was increased in the spheres of the ESCC cell line TE10 compared with the corresponding adherent cells, and we also observed elevated RSK4 expression in CD90+ or CD271+ cell populations compared with CD90– or CD271– cell subsets (Figure 1H and Supplemental Figure 1H), suggesting that RSK4 is enriched in ESCC CSCs.
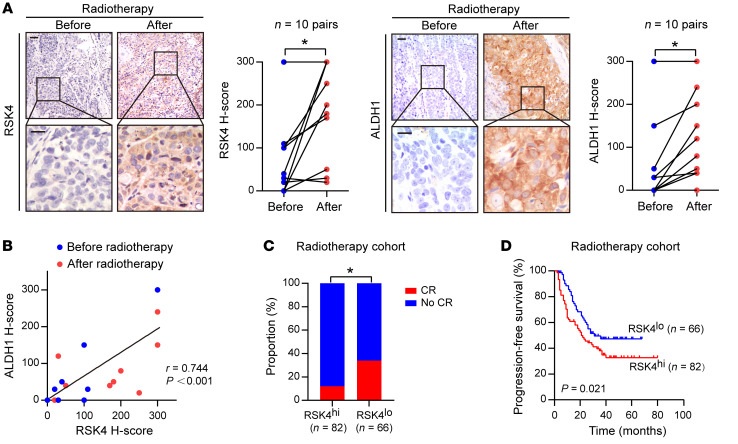
In 10 patients with advanced ESCC treated with definitive radiotherapy, we found that RSK4 expression after radiotherapy was significantly higher than that before treatment, which was consistent with upregulation of the CSC marker ALDH1 after radiotherapy (Figure 2A). We observed a positive correlation between RSK4 and ALDH1 expression (Figure 2B), suggesting that RSK4 activation is induced by irradiation and may be involved in CSC-mediated radioresistance. In an enlarged ESCC radiotherapy cohort of 148 patients, those with high RSK4 expression were significantly associated with radiotherapy resistance; that is, the higher the expression level of RSK4, the lower the complete response rate (Figure 2C). Moreover, patients with high RSK4 expression levels had much worse PFS than did those with low RSK4 expression levels (Figure 2D). In summary, RSK4 was specifically upregulated in ESCC CSCs and was linked with radioresistance and poor prognosis in patients with ESCC, especially those undergoing radiotherapy.
Figure 2. RSK4 is closely linked with the radioresistance and poor survival of ESCC patients.
(A) Representative IHC images and IHC H-scores of RSK4 and ALDH1 protein expression in 10 patients with ESCC before and after receiving radiotherapy. Scale bars: 100 μm. (B) RSK4 protein levels were positively correlated with ALDH1 in these cases. (C) Radiotherapy-treated ESCC patients with higher expression of RSK4 had a lower complete response (CR) ratio. (D) Kaplan-Meier estimation of the PFS curves for 148 patients with ESCC treated with radiotherapy according to RSK4 expression levels in the primary tumor. *P < 0.05. Differences were tested using a paired, 2-sided Student’s t test (A), χ2 test (C), and log-rank test (D). The correlation was determined by Pearson’s correlation test (B).
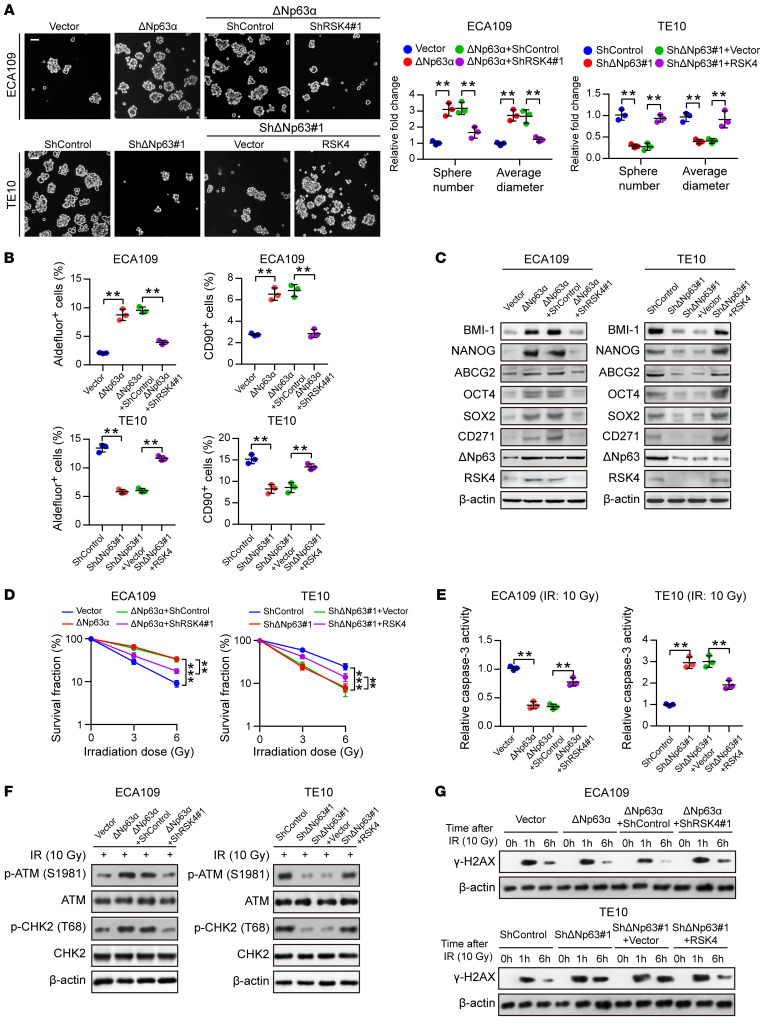
RSK4 promotes the CSC properties and radioresistance of ESCC cells.
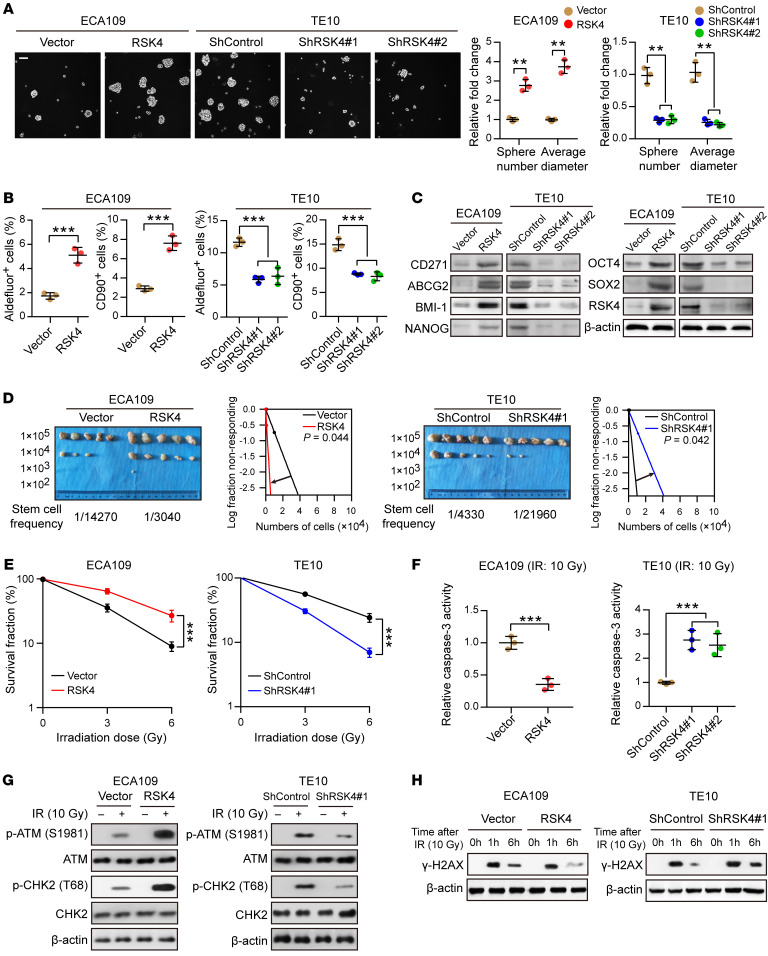
To investigate the biological effects of RSK4 in ESCC, we assessed RPS6KA6 mRNA and RSK4 protein levels in 4 ESCC cell lines (Supplemental Figure 2A) and found that RSK4 expression was higher in the ESCC cell line TE10 and lower in the ECA109 cell line. Therefore, we generated a stable RSK4-overexpressing clone of ECA109 cells along with stable RSK4-knockdown clones of TE10 cells, as confirmed by real-time PCR and Western blotting (Supplemental Figure 2B). Sphere formation ability was increased in RSK4-overexpressing ECA109 cells but decreased in RSK4-knockdown TE10 cells (Figure 3A). According to flow cytometric and Western blot analyses, RSK4 overexpression enhanced, whereas RSK4 knockdown decreased, ALDH activity and the percentage of CD90+ cells as well as protein levels of CD271, ABCG2, BMI-1, NANOG, OCT4, and SOX2 (Figure 3, B and C, and Supplemental Figure 2C). Limiting dilution analysis (33) of xenografted tumors developed from different numbers of injected cells showed that RSK4 overexpression increased the tumorigenic capacity of ESCC cells, whereas RSK4 knockdown had the opposite effect (Figure 3D). To elucidate the role of RSK4 in ESCC CSC maintenance, we examined the effects of RSK4 knockdown on ESCC CSC proliferation. ESCC CSCs (CD90+ subpopulations) transfected with shRSK4 proliferated at a lower rate compared with control cells. In contrast, an identical shRSK4 had little effect on matched non-stem ESCC cells (CD90– subpopulations) (Supplemental Figure 2D). These data suggest that RSK4 exclusively promotes ESCC CSC growth, thereby maintaining the CSC properties of ESCC cells.
Figure 3. RSK4 promotes the CSC properties and radioresistance of ESCC cells.
(A) Tumor sphere formation assay showing that RSK4 overexpression increased the sphere-forming ability of ESCC cells, whereas RSK4 knockdown reduced their sphere-forming ability (n = 3 independent experiments). Scale bar: 100 μm. (B) Flow cytometric analysis showing that RSK4-overexpressing cells had elevated ALDH activity and an increased proportion of CD90+ cells, whereas RSK4-suppressed cells exhibited reduced ALDH activity and a reduced proportion of CD90+ cells (n = 3 independent experiments). (C) Western blot analysis indicating that RSK4 overexpression increased ESCC CSC marker expression, whereas RSK4 knockdown had the opposite effect. (D) Limiting dilution analysis showing higher tumorigenicity of RSK4-overexpressing ECA109 cells in NOD/SCID mice compared with control cells, but RSK4-knockdown TE10 cells had lower tumorigenicity compared with the control group cells (n = 5 mice each). (E) Clonogenic survival assays of ESCC cells with overexpression or knockdown of RSK4 at the indicated irradiation doses (n = 3 independent experiments). (F) Relative caspase-3 activity 24 hours after IR (10 Gy) of ESCC cells with overexpression or knockdown of RSK4 (n = 3 independent experiments). (G) Western blot analysis of phosphorylated and total amounts of the checkpoint proteins ATM and CHK2 from the indicated groups before treatment (–) and 1 hour after 10 Gy IR (+). (H) ESCC cells from the indicated groups were treated with 10 Gy IR and recultured under normal conditions for 1 and 6 hours, and then subjected to Western blot analysis with γ-H2AX antibody. 0h, cells with IR treatment but with no time for DNA repair. Data represent the mean ± SD. **P < 0.01 and ***P < 0.001. Differences were tested using an unpaired, 2-sided Student’s t test (A, B, and D–F).
With regard to the association between RSK4 and radioresistance, RSK4 overexpression in ESCC cells significantly increased their colony-forming ability and decreased caspase-3 activity after irradiation, whereas RSK4 knockdown decreased the colony-forming ability of ESCC cells and increased caspase-3 activity after irradiation (Figure 3, E and F, and Supplemental Figure 2E). Although irradiation damages tumor cells through several mechanisms, irradiation kills cancer cells primarily through DNA damage. Thus, DNA damage checkpoint responses play essential roles in cellular radiosensitivity (34). The activating phosphorylation of checkpoint proteins ATM and CHK2 induced by irradiation was markedly increased in RSK4-overexpressing ECA109 cells but decreased in RSK4-knockdown TE10 cells (Figure 3G), indicating that RSK4 promotes checkpoint activation in response to DNA damage. The primary downstream effect of checkpoint activation is to induce cell-cycle arrest to repair damaged DNA (35). We then used the comet assay to measure the efficiency of DNA repair after inducing DNA damage with irradiation. RSK4-overexpressing ECA109 cells repaired the DNA damage more efficiently than did control cells, as indicated by decreased DNA content in the comet tail 6 hours after irradiation, whereas RSK4 knockdown had the opposite effect (Supplemental Figure 2F). This result was further confirmed by assessing the dynamic changes in γ-H2AX levels after irradiation (Figure 3H). Altogether, these results suggest that RSK4 promotes DNA damage checkpoint responses and DNA damage repair to obtain radioresistance of ESCC cells.
RSK4 is a direct transcriptional target of ΔNp63α in ESCC.
To determine the regulation mechanism of RSK4 in ESCC, we performed a transcription factor prediction analysis and noticed 1 potential binding site of p63 on the RPS6KA6 gene promoter (Figure 4A). Moreover, Rps6ka6 is a leading downregulated gene according to RNA sequencing profiling of Tp63-null squamous epithelia (Supplemental Figure 3A). Considering that RSK4 and ΔNp63 proteins were highly coexpressed in the basal layer and progressively decreased in the suprabasal and superficial cell compartments with cellular differentiation in esophageal epithelia (Figure 4B), that their mRNA levels were positively correlated in esophageal mucosa in the Genotype Tissue Expression (GTEx) cohort (Figure 4C), and that Rps6ka6 mRNA levels were dramatically decreased in squamous epithelium of Tp63-knockout mice (Supplemental Figure 3B) and in p63-knockdown keratinocyte and squamous cell carcinoma (SCC) cell lines (Supplemental Figure 3C), we surmised that ΔNp63 transcriptionally regulated RSK4 expression in ESCC. However, mRNA expression of other RSK family members, including RPS6KA1, RPS6KA3, RPS6KA2, RPS6KA5 (encoding MSK1), and RPS6KA4 (encoding MSK2), did not show any appreciable difference under p63 depletion (Supplemental Figure 3, B and C). As previously reported (36), ΔNp63α was the dominant isoform expressed in ESCC (Supplemental Figure 3D). Real-time PCR analysis of 30 paired ESCC specimens showed a dramatic upregulation of ΔNp63 mRNA expression in tumors compared with expression levels in normal tissues (Supplemental Figure 3E), in accordance with TCGA database (Supplemental Figure 3F). In ESCC tissues, ΔNp63 expression was positively correlated with RSK4 expression at both the mRNA and protein levels (Figure 4, D and E, and Supplemental Figure 3G), and similar results were obtained from IHC of 215 ESCC samples (Figure 4F) and in the Gene Expression Omnibus (GEO) cohort (Supplemental Figure 3H). As previously reported (36, 37), patients with ESCC with high ΔNp63 protein levels had worse OS and PFS than did their low-expression counterparts (Supplemental Figure 3I).
Figure 4. ΔNp63α directly transactivates RSK4 expression in ESCC.
(A) Predicted binding sites for p63 in the promoter regions of RPS6KA6. (B) IHC analysis of ΔNp63 and RSK4 proteins showing an identical coexpression pattern in serial sections of the esophageal epithelial basal and suprabasal layers. Scale bar: 50 μm. (C) Correlation between the RPS6KA6 and TP63 mRNA expression patterns in the GTEx esophagus mucosa data set. (D) mRNA levels of ΔNp63 and RPS6KA6 in 30 ESCC samples were determined by real-time PCR. GAPDH was used as a loading control. (E) Expression of ΔNp63 and RSK4 in 30 ESCC samples was detected by Western blotting. The results of other samples are presented in Supplemental Figure 3G. Protein expression was normalized to β-actin levels. (F) Representative IHC images of ΔNp63 and RSK4 protein expression in patients with ESCC. Scale bar: 100 μm. Histograms show the correlation of the IHC data for high or low RSK4 expression relative to the level of ΔNp63. (G) ΔNp63α overexpression upregulated, whereas ΔNp63 silencing reduced, RPS6KA6 mRNA expression in ESCC cells (n = 3 independent experiments). (H) ΔNp63α overexpression upregulated, whereas ΔNp63 silencing reduced, RSK4 protein expression in ESCC cells. (I) ΔNp63α induced reporter activity of the WT RPS6KA6 promoter rather than the p63 responsive element deletion mutant (MUT) promoter, as determined by a luciferase reporter assay in HEK293T cells (n = 3 independent experiments). (J) Exogenous and endogenous ChIP analysis of the interaction between ΔNp63 protein and the RPS6KA6 promoter in ESCC cells (n = 3 independent experiments). Data represent the mean ± SD. **P < 0.01 and ***P < 0.001. Differences were tested using an unpaired, 2-sided Student’s t test (G, I, and J) and a χ2 test (F). The correlation was determined by Pearson’s correlation test (C–E).
Western blot assays showed that RSK4 expression, but not that of its sibling RSK2, which has been reported to be oncogenic in SCC (12), was positively correlated with ΔNp63 expression in ESCC cell lines (Supplemental Figure 3J). Moreover, ΔNp63α upregulated RSK4 expression when stably transfected into ECA109 and EC9706 cells, whereas knockdown of ΔNp63 decreased the level of RSK4 in TE10 and TE11 cells (Figure 4, G and H). By contrast, RSK2 protein levels and phosphorylation of its known downstream substrates HSP27 and CREB, which have been implicated in SCC (12), were not significantly affected by ΔNp63 overexpression or downregulation (Supplemental Figure 3K). Therefore, it appears that ΔNp63α transcriptionally regulates the expression of RSK4 but not of RSK2 in ESCC. From transcription factor prediction analysis, we identified a putative p63 binding site between –644 and –625 upstream of the transcriptional initiation site in the RPS6KA6 promoter (Figure 4A). The luciferase reporter assays showed that RPS6KA6 promoter activity was greatly enhanced in HEK293T cells transfected with ΔNp63α, whereas a deletion mutation of this p63 binding site nullified this transactivation (Figure 4I). Exogenous and endogenous ChIP assays confirmed the direct binding of ΔNp63α to the RPS6KA6 promoter in both ECA109 and TE10 cells (Figure 4J and Supplemental Figure 3L). Taken together, these results suggest that RSK4 is highly upregulated together with ΔNp63 in ESCC and that RPS6KA6 is a ΔNp63α target gene.
RSK4 mediates the ΔNp63α-enhanced CSC properties and radioresistance of ESCC cells.
To investigate the involvement of RSK4 in the function of ΔNp63α in ESCC, we generated an ECA109 clone with ΔNp63α overexpression and RSK4 knockdown and a TE10 clone with ΔNp63 knockdown and RSK4 overexpression (Supplemental Figure 4A). ΔNp63α overexpression led ECA109 cells to acquire CSC properties, as evidenced by the increased sphere-forming ability, ALDH activity, percentage of CD90+ cells, and protein levels of CD271, ABCG2, BMI-1, NANOG, OCT4, and SOX2. However, the ΔNp63α-induced CSC properties were largely abolished by RSK4 knockdown. We also obtained similar results in TE10 cells, in which the reduction in CSC properties through ΔNp63 knockdown was partially rescued by RSK4 overexpression (Figure 5, A–C, and Supplemental Figure 4B). With respect to radioresistance, ESCC cells with ΔNp63α overexpression had a significantly increased colony-forming ability and decreased caspase-3 activity after irradiation, whereas ΔNp63α-induced radioresistance was largely impaired by RSK4 knockdown. By contrast, RSK4 overexpression greatly restored the radioresistance of ΔNp63-knockdown cells (Figure 5, D and E). The activating phosphorylation of checkpoint proteins ATM and CHK2 induced by irradiation was significantly increased in ΔNp63α-overexpressing ECA109 cells, whereas ΔNp63α-induced DNA damage checkpoint responses were largely impaired by RSK4 knockdown. Similar results were also obtained in TE10 cells, in which the inhibition of DNA damage checkpoint responses through ΔNp63 knockdown was partially rescued by RSK4 overexpression (Figure 5F). The comet assay showed that ESCC cells with ΔNp63α overexpression had a markedly increased DNA damage repair efficiency; however, the ΔNp63α-induced DNA repair efficiency was largely abolished by RSK4 knockdown. By contrast, RSK4 overexpression greatly restored the DNA repair efficiency of ΔNp63-knockdown cells (Supplemental Figure 4C). This result was further confirmed by assessing the dynamic changes in γ-H2AX levels after irradiation (Figure 5G). Altogether, these results suggest that RSK4 is essential for ΔNp63α-mediated CSC properties and the radioresistance of ESCC cells.
Figure 5. RSK4 is essential for ΔNp63α-mediated CSC properties and radioresistance of ESCC cells.
(A) Knockdown of RSK4 abolished ΔNp63α-enhanced sphere formation ability, whereas RSK4 overexpression partially restored the effects of ΔNp63-suppressed sphere formation (n = 3 independent experiments). Scale bars: 100 μm. (B) Flow cytometric analysis of ALDH activity and the proportion of CD90+ cells in ESCC cells from the indicated groups (n = 3 independent experiments). (C) Western blot analysis of ESCC CSC markers in the indicated groups. (D) Clonogenic survival assays of ESCC cells in the indicated groups at IR doses of 0, 3, and 6 Gy (n = 3 independent experiments). (E) Relative caspase-3 activity 24 hours after IR (10 Gy) of ESCC cells in the indicated groups (n = 3 independent experiments). (F) Western blot analysis of phosphorylated and total amounts of the checkpoint proteins ATM and CHK2 from the indicated groups 1 hour after 10 Gy IR. (G) ESCC cells from the indicated groups were treated with 10 Gy IR and recultured under normal conditions for 1 and 6 hours, and then subjected to Western blot analysis with γ-H2AX antibody. 0 h, cells with IR treatment but with no time for DNA repair. Data represent the mean ± SD. **P < 0.01 and ***P < 0.001. Differences were tested using an unpaired, 2-sided Student’s t test (A, B, D, and E).
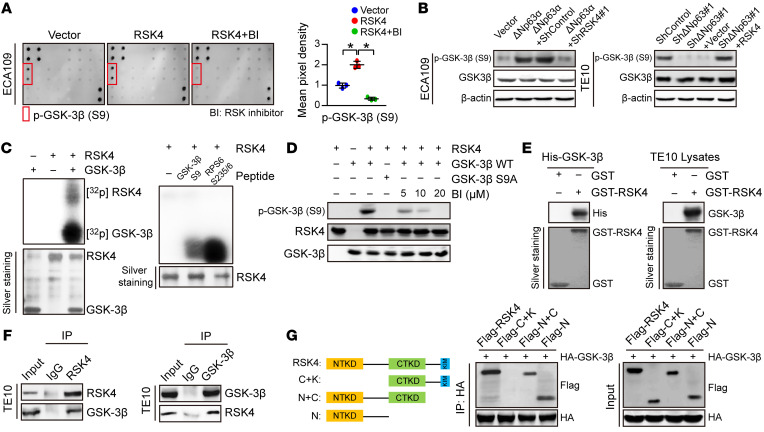
RSK4 directly phosphorylates GSK-3β (Ser9) in ESCC.
To further investigate the mechanism underlying the role of RSK4 in ESCC, we performed a MAPK pathway phosphorylated antibody array analysis (Supplemental Table 4) of ESCC cell lines with exogenous RSK4 expression after treatment with the RSK inhibitor BI-D1870. With respect to the identified phosphorylated proteins, we found GSK-3β, a major serine/threonine kinase that is aberrantly activated in various cancer types and is involved in CSC properties and therapy resistance (38), to be highly phosphorylated at serine residue 9 (Ser9) in ECA109 cells expressing RSK4, whereas treatment with the RSK inhibitor BI-D1870 decreased the level of GSK-3β (Ser9) phosphorylation (Figure 6A). Moreover, Western blotting showed that RSK4 knockdown reduced the ΔNp63α-induced phosphorylation of GSK-3β (Ser9), whereas the reduction in phosphorylated GSK-3β (p–GSK-3β) (Ser9) through ΔNp63 knockdown was rescued by RSK4 overexpression (Figure 6B). Sequence alignment analysis showed that Ser9 of GSK-3β is highly conserved from fruit fly to human, suggesting that this phosphorylation site may have important biological functions (Supplemental Figure 5A). On the basis of these results, we propose that RSK4 may directly phosphorylate GSK-3β (Ser9). The results of an in vitro kinase assay showed that active RSK4 protein directly phosphorylated GSK-3β (Ser9), with phosphorylation of RPS6 at Ser235/236 used as a positive control (Figure 6C). We found that the phosphorylation level of GSK-3β was inhibited in vitro when treated with BI-D1870 (Supplemental Figure 5B), which we confirmed using an antibody that specifically detected the level of p–GSK-3β (Ser9) (Figure 6D). Moreover, RSK4 and GSK-3β colocalized in ESCC cells (Supplemental Figure 5C), and the results of glutathione S-transferase–pulldown (GST- pulldown) assays as well as exogenous and endogenous co-IP assays confirmed the direct binding of RSK4 to GSK-3β (Figure 6, E and F, and Supplemental Figure 5D). Molecular mapping using truncated RSK4 revealed that the N-terminal kinase domain (NTKD) was responsible for the interaction with GSK-3β (Figure 6G).
Figure 6. RSK4 directly phosphorylates GSK-3β (Ser9).
(A) MAPK pathway phosphorylated antibody microarray analysis shows that p–GSK-3β (Ser9) was significantly increased when RSK4 was stably overexpressed and decreased when treated with 10 μM BI-D1870 (an inhibitor of RSK) for 12 hours (n = 3 independent experiments). (B) Western blot analysis showing that downregulation of RSK4 resulted in a reduced level of ΔNp63α-induced p–GSK-3β (Ser9) (S9) (left), whereas RSK4 overexpression partially reversed the reduction in phosphorylation levels through ΔNp63 knockdown (right). (C) Active RSK4 phosphorylated GSK-3β at Ser9 in vitro in the presence of [γ-32P] ATP as visualized by an autoradiograph. The RPS6 (Ser235/236) peptide was used as a positive control. The input was confirmed by silver staining. (D) Validation of p–GSK-3β (Ser9) phosphorylation levels in an in vitro kinase assay by Western blotting. The levels of p–GSK-3β were inhibited when treated with BI-D1870 for 2 hours. WT GSK-3β and mutant GSK-3β S9A were used as substrates for active RSK4. (E) An in vitro GST-pulldown assay was performed to verify the interaction of RSK4 with purified His–GSK-3β protein (left) or GSK-3β from TE10 cell lysates (right). Retrieved proteins were evaluated by immunoblotting. GST-only protein was used as a negative control. GST fusion proteins were confirmed by silver staining. (F) The interaction of RSK4 and GSK-3β was confirmed by an endogenous co-IP assay in TE10 cells. IgG served as a negative control. (G) Mapping analyses of full-length and truncated RSK4 with representative co-IP assays in HEK293T cells showing that the NTKD of RSK4 was responsible for the interaction with GSK-3β. C, CTKD; K, kinase interaction motif (KIM); N, NTKD. Data represent the mean ± SD. *P < 0.05. Differences were tested using 1-way ANOVA with Tukey’s post hoc test (A).
The correlation between RSK4 and p–GSK-3β (Ser9) was also found in ESCC samples by IHC (Supplemental Figure 5E). Clinical association studies found that upregulation of the p–GSK-3β (Ser9) protein was associated with lymph node metastasis and vascular invasion (Supplemental Table 5), similar to that seen with RSK4. Kaplan-Meier survival analysis showed that patients with ESCC patients who had higher p–GSK-3β (Ser9) protein levels had worse OS and PFS than did their counterparts with low expression levels (Supplemental Figure 5F). In summary, RSK4 phosphorylates GSK-3β (Ser9) through direct interactions in ESCC cells, and coexpression of RSK4 and p–GSK-3β has prognostic value for clinical ESCC.
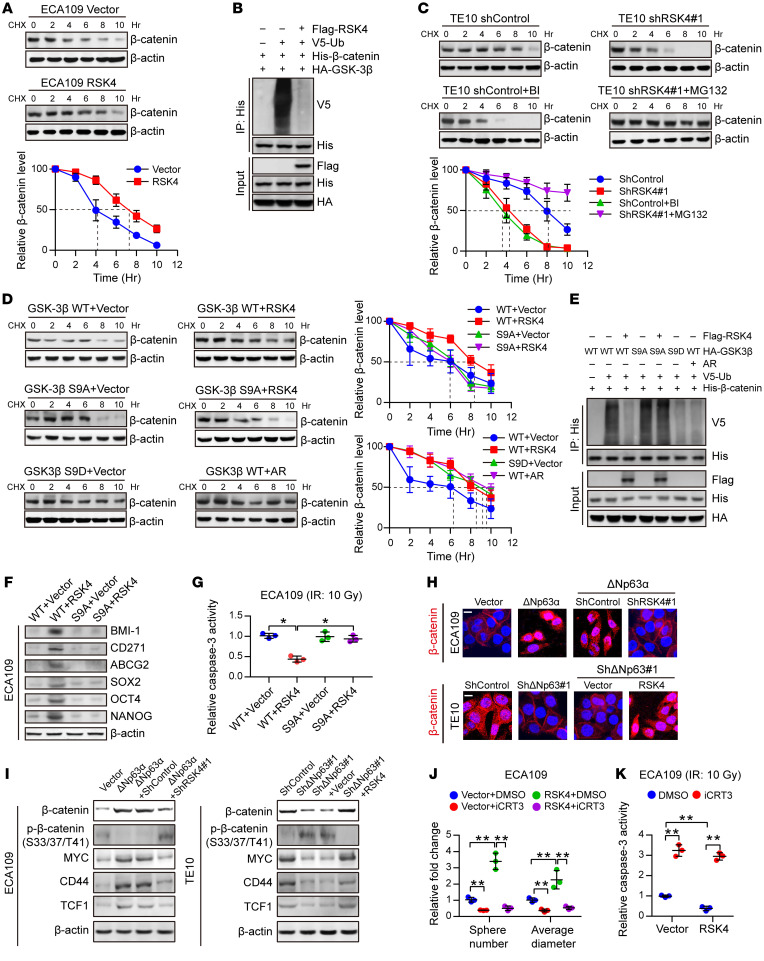
RSK4 stabilizes β-catenin through phosphorylation of GSK-3β (Ser9) in ESCC cells.
It has been reported that the Wnt/β-catenin signaling pathway is closely involved in the maintenance and associated radioresistance of CSCs (39). The protein expression and nuclear localization of β-catenin are tightly regulated by GSK-3β, with phosphorylation of GSK-3β at Ser9 leading to reduced degradation of β-catenin, followed by its subsequent nuclear translocation (40). Here, we examined whether this pathway is altered in response to the modulation of RSK4 in ESCC cells. Western blotting showed that higher RSK4 levels in ESCC cells were associated with a longer half-life and reduced ubiquitination of β-catenin (Figure 7, A and B). By contrast, RSK4 depletion or treatment with BI-D1870 accelerated β-catenin degradation, which was reversed by the addition of MG132, a proteasome inhibitor (Figure 7C). Moreover, RSK4 stabilized β-catenin in the presence of GSK-3β WT but not GSK-3β S9A in ESCC cells, which was similar to overexpression of GSK3β S9D (constitutively inactivated mutation) or addition of the GSK3β inhibitor AR-A014418 (Figure 7, D and E), further highlighting the importance of GSK-3β (Ser9) phosphorylation by RSK4 for β-catenin stabilization. In addition, RSK4 promoted CSC properties and radioresistance in the presence of GSK-3β WT but not GSK-3β S9A in ESCC cells (Figure 7, F and G).
Figure 7. RSK4 activates the Wnt/β-catenin pathway.
(A) Control or RSK4-overexpressing ECA109 cells were incubated with CHX (10 μg/mL) for the indicated durations. High RSK4 expression prolonged the half-life of β-catenin degradation (n = 3 independent experiments). (B) HEK293T cells were transfected with the indicated plasmids, and cell extracts were immunoprecipitated with an anti-His antibody. Ubiquitinated β-catenin was detected by immunoblotting. (C) Control or RSK4-silenced TE10 cells were incubated with CHX (10 μg/mL), CHX plus BI-D1870 (10 μM), or MG132 (10 μM) for the indicated durations. The indicated proteins were detected by immunoblotting (n = 3 independent experiments). (D) HEK293T cells were transfected with the indicated plasmids and incubated with CHX (10 μg/mL) for the indicated durations. The indicated proteins were detected by immunoblotting (n = 3 independent experiments). (E) HEK293T cells were transfected with the indicated plasmids, and cell extracts were immunoprecipitated with an anti-His antibody. Ubiquitinated β-catenin was detected by immunoblotting. (F and G) ECA109 cells were transfected with the indicated plasmids, and then CSC markers (F) and caspase-3 activity after IR (10 Gy) (G) of the indicated groups were detected (n = 3 independent experiments). (H) Representative images of the nuclear localization of β-catenin in ESCC cells from the indicated groups detected using immunofluorescence microscopy. Scale bars: 100 μm. (I) Effects of knockdown or overexpression of RSK4 on the indicated proteins in ΔNp63α-overexpressing or ΔNp63-suppressing ESCC cells. (J and K) Treatment of RSK4-overexpressing cells with 50 μM iCRT3 (an inhibitor of β-catenin signaling) for 24 hours greatly reduced their sphere-forming ability (J) and increased caspase-3 activity after IR (K) (n = 3 independent experiments). BI, BI-D1870; AR, AR-A014418. Data represent the mean ± SD. *P < 0.05 and **P < 0.01. Differences were tested using 1-way ANOVA with Tukey’s post hoc test (G, J, and K).
ΔNp63α overexpression dramatically increased the ratio of nuclear β-catenin and β-catenin protein levels in both the cytoplasm and nucleus, whereas the increase was largely abolished by RSK4 knockdown in ECA109 cells. By contrast, TE10 cells with ΔNp63 downregulation exhibited significant restoration of β-catenin activity when RSK4 was overexpressed (Figure 7H and Supplemental Figure 5G). The importance of these effects was strengthened by the assessment of the total and phosphorylated (inactive state) protein levels of β-catenin and expression of the Wnt/β-catenin pathway downstream targets MYC, CD44, and TCF1 (Figure 7I). In addition, RPS6KA6 mRNA levels were positively correlated with HNF1A (encoding TCF1) and CD44 in ESCC specimens from the GEO cohorts (Supplemental Figure 5H). To demonstrate that RSK4-dependent ESCC CSC properties and radioresistance are mediated through Wnt/β-catenin signaling, we treated RSK4-overexpressing ECA109 cells with the Wnt/β-catenin inhibitor iCRT3. Notably, inhibition of Wnt/β-catenin signaling greatly reduced the sphere-forming ability of CSCs and increased caspase-3 activity after irradiation in RSK4-overexpressing cells (Figure 7, J and K, and Supplemental Figure 5I), showing that this pathway mediates the upregulation of RSK4-dependent CSC properties and radioresistance in ESCC cells. Altogether, these results indicate that RSK4 phosphorylation of GSK-3β (Ser9) is essential for activating the Wnt/β-catenin pathway to promote ESCC CSC properties and radioresistance.
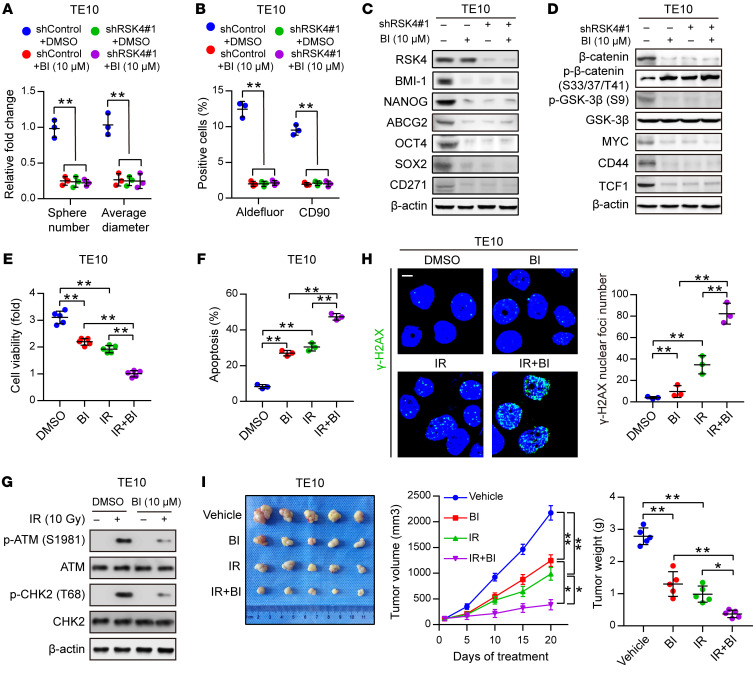
Disruption of the RSK4 pathway reduces CSC properties and improves the radiosensitivity of ESCC.
On the basis of a computational simulation, we constructed a model of the 3D structure of RSK4 NTKD by homology modeling and in silico docking of BI-D1870, a highly specific and potent inhibitor of the NTKD of RSKs (41), to the ATP-binding site of RSK4. BI-D1870 had strong intermolecular interactions with the NTKD of RSK4 by forming 2 hydrogen bonds with the hinge region and another 2 hydrogen bonds with residues Lys105 and Asp216 (Supplemental Figure 6A). BI-D1870 inhibited GSK-3β (Ser9) phosphorylation in ESCC cells in a dose-dependent manner, with nearly 100% inhibition at a concentration of 10 μM (Supplemental Figure 6B). At this concentration, BI-D1870 greatly suppressed CSC properties as well as β-catenin activity and expression of the Wnt/β-catenin pathway downstream targets of ESCC cells. However, RSK4-knockdown cells were not sensitive to BI-D1870 treatment compared with control cells, indicating that RSK4 is a key target of BI-D1870 for the inhibition of CSC properties and the Wnt/β-catenin pathway in ESCC cells (Figure 8, A–D, and Supplemental Figure 6, C–F). Furthermore, BI-D1870 inhibited ESCC CSC growth but had little effect on matched non-stem ESCC cells (Supplemental Figure 6G), indicating that BI-D1870 specifically disrupted ESCC CSC growth and maintenance. On the other hand, treatment of ESCC cells with BI-D1870, with RSK4 knockdown, or with both did not significantly change the expression levels of RSK1–3 proteins or the phosphorylation of their downstream substrates, indicating that RSK1–3 may not be involved with BI-D1870 in inhibiting the malignant phenotypes of ESCC (Supplemental Figure 6H).
Figure 8. Disruption of the RSK4 pathway reduces CSC properties and improves the radiosensitivity of ESCC.
(A and B) Sphere formation assay (A) and flow cytometric analysis (B) of TE10 cells treated with BI-D1870 (10 μM), RSK4 knockdown, or both (n = 3 independent experiments). (C) Immunoblot analyses of BMI-1, NANOG, ABCG2, OCT4, SOX2, and CD271 in TE10 cells with the indicated treatments. (D) Immunoblot analyses of β-catenin, p–β-catenin (Ser33/37/Thr41, inactive state), p–GSK-3β (Ser9), GSK-3β, MYC, CD44, and TCF1 in TE10 cells with the indicated treatments. (E) Cell viability assay of TE10 cells treated with BI-D1870 (10 μM) for 12 hours, IR (10 Gy), or both (n = 3 independent experiments). (F) FACS analyses of apoptosis of TE10 cells treated with BI-D1870 (10 μM) for 12 hours, IR (10 Gy), or both (n = 3 independent experiments). (G) Western blot analysis of phosphorylated and total amounts of the checkpoint proteins ATM and CHK2 from TE10 cells treated with or without BI-D1870 (10 μM) before treatment (–) and 1 hour after 10 Gy IR (+). (H) Immunofluorescence staining of γ-H2AX in TE10 cells with the indicated treatments (n = 3 independent experiments). Scale bar: 100 μm. (I) ESCC-derived xenografts in mice treated with vehicle control, BI-D1870 (50 mg/kg/day, i.p. injection), and/or IR (5 Gy, twice). The growth curve of tumor size and average tumor weight are presented (n = 5 mice each). Data represent the mean ± SD. *P < 0.05 and **P < 0.01. Differences were tested using 1-way ANOVA with Tukey’s post hoc test (A, B, E, F, H, and I).
We next examined whether RSK4 inhibition by BI-D1870 could improve the therapeutic efficacy of radiotherapy for treating ESCC. BI-D1870 or irradiation treatment each suppressed ESCC cell proliferation, whereas the combined treatment achieved the strongest inhibition of proliferation (Figure 8E). BI-D1870 or irradiation treatment each induced cell apoptosis, and the combined treatment again exhibited the strongest effect (Figure 8F and Supplemental Figure 6I). These observations were supported by an assessment of the protein levels of the apoptosis markers cleaved caspase-3 and cleaved PARP (Supplemental Figure 6J). Furthermore, the activating phosphorylation of checkpoint proteins ATM and CHK2 induced by irradiation was markedly decreased with BI-D1870 treatment (Figure 8G), demonstrating that disruption of the RSK4 pathway reduced checkpoint activation in response to DNA damage. In addition, as assessed by immunofluorescence staining for γ-H2AX, combined treatment with BI-D1870 and irradiation induced more DNA damage than did treatment with irradiation alone (Figure 8H). We further confirmed this result by the comet assay after irradiation (Supplemental Figure 6K), which indicated that DNA damage repair was impaired by disruption of the RSK4 pathway. Concomitant treatment of tumor-bearing mice with BI-D1870 and irradiation also reduced the growth and weight of ESCC xenograft tumors more significantly than did treatment with each alone (Figure 8I). Altogether, these results indicate that disruption of the RSK4 pathway by BI-D1870 suppressed the CSC properties and enhanced the radiosensitivity of ESCC cells.
It has been reported that BI-D1870 has other targets such as PLK1 and Aurora B, whose IC50 values are 7-fold and 23-fold higher, respectively, than that of RSK4 in vitro (41). To verify whether BI-D1870 had an off-target effect on PLK1 and Aurora B in ESCC cells, we performed a dose-response analysis to determine the BI-D1870 concentration that would not inhibit PLK1 and Aurora B in TE10 cells. We found that the expression levels of PLK1 and Aurora B proteins and the phosphorylation of their downstream substrates were not affected by BI-D1870 at a concentration of 5 μM (Supplemental Figure 6L). At this concentration, GSK-3β (Ser9) phosphorylation was strongly inhibited in TE10 cells (Supplemental Figure 6B). Next, we tested whether BI-D1870 enhanced the radiosensitivity of ESCC cells at the 5-μM concentration. Combined treatment of BI-D1870 and irradiation achieved the strongest proliferation inhibition and highest caspase-3 activity compared with each treatment alone (Supplemental Figure 6, M and N). Moreover, combined treatment with BI-D1870 and irradiation resulted in higher DNA content in the comet tail and higher γ-H2AX levels 6 hours after irradiation than did treatment with irradiation alone (Supplemental Figure 6, O and P). These results suggest that BI-D1870 can improve the radiosensitivity of ESCC cells at a concentration that does not inhibit PLK1 and Aurora B activity.
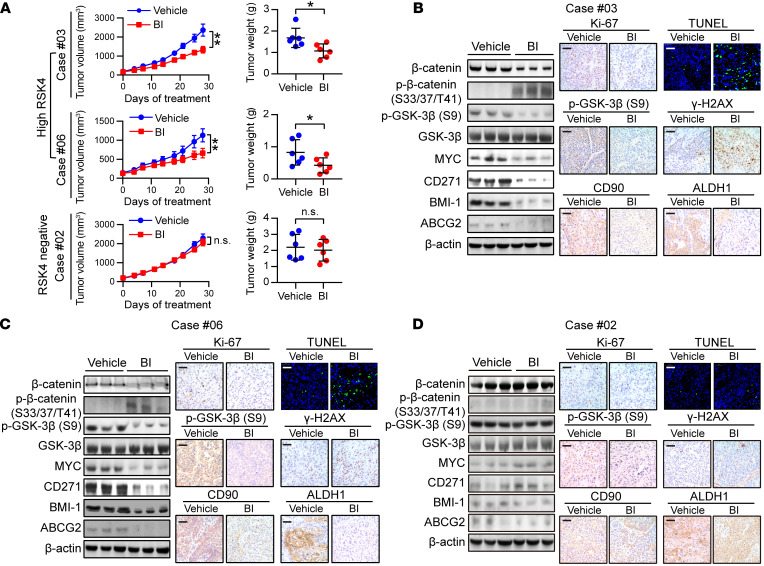
RSK4 inhibition with BI-D1870 sensitizes radiotherapy in ESCC patient–derived xenografts.
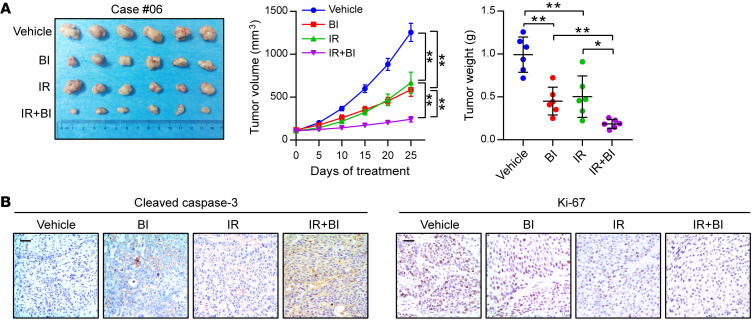
Patient-derived xenograft (PDX) tumors are known to closely resemble primary tumors, and their use has transformed anticancer drug research, enabling the study of therapeutic responses and accelerating the transition from the bench to the clinic (42). We obtained ESCC tumor specimens from 28 patients to establish PDX models, and nearly 80% of these tumors expressed the RSK4 protein at a high level, indicating that RSK4 overexpression is a common event in ESCC. However, we detected no significant differences in RSK4 expression between successful PDX models and unsuccessful ones (Supplemental Figure 7A). Two PDX tumors (cases 03 and 06) with high RSK4 expression and 1 PDX tumor (case 02) with negative RSK4 expression were selected for further analysis by IHC staining of grafted tumors collected from the animals. These tumors were all RSK2 protein negative (Supplemental Figure 7B). BI-D1870 strongly attenuated the growth and weight of high RSK4-expressing PDX tumors; in sharp contrast, BI-D1870 failed to reduce the growth and weight of RSK4-negative PDX tumors (Figure 9A and Supplemental Figure 7C). The levels of Wnt/β-catenin pathway–associated and CSC-associated proteins and the Ki-67 index were greatly reduced, whereas apoptosis and DNA damage were significantly increased in RSK4-high, but not RSK4-negative, PDX tumors after BI-D1870 treatment (Figure 9, B–D). Moreover, the growth curves of body weight; the histology of the liver, lung, and kidney; and the results of functional tests of the liver and kidney from mice treated with BI-D1870 or vehicle suggest that BI-D1870 is a well-tolerated agent without severe toxicity to these organs (Supplemental Figure 7, D–F). Finally, the combination of BI-D1870 and irradiation was even more effective at reducing tumor growth and weight in ESCC PDX models than treatment with either alone (Figure 10A). Furthermore, IHC staining of cleaved caspase-3 and Ki-67 demonstrated that, whereas BI-D1870 or irradiation treatment alone induced apoptosis and reduced the proliferation of tumor cells in ESCC PDX models, combined treatment resulted in substantially more apoptosis and greater proliferation inhibition (Figure 10B). Taken together, these results confirm that BI-D1870 inhibited tumor growth and improved the therapeutic efficacy of radiotherapy in ESCC PDX models and that RSK4 expression levels positively correlated with the therapeutic response.
Figure 9. BI-D1870 inhibits tumor growth in ESCC PDXs.
(A) Tumor volume growth curve and tumor weight of PDX mice with different RSK4 expression levels treated with BI-D1870 (50 mg/kg/day, i.p. injection) or vehicle for 28 days (n = 6 mice each). (B–D) Immunoblotting, IHC, and terminal deoxynucleotidyl transferase–mediated dUTP labeling (TUNEL) analyses of the indicated markers in PDX tumor numbers 3 (B), 6 (C) and 2 (D) treated with BI-D1870 (50 mg/kg/day, i.p. injection) or vehicle. Scale bars: 100 μm. Data represent the mean ± SD. *P < 0.05 and **P < 0.01. Differences were tested using an unpaired, 2-sided Student’s t test (A).
Figure 10. BI-D1870 improves the therapeutic efficacy of radiotherapy in ESCC PDXs.
(A) PDX tumor number 6 treated with the vehicle control, BI-D1870 (50 mg/kg/day, i.p. injection), and/or IR (5 Gy, twice). The growth curve of the tumor size and average tumor weight are presented (n = 6 mice each). (B) IHC analyses of cleaved caspase-3 and Ki-67 in the indicated groups. Scale bars: 100 μm. Data represent the mean ± SD. *P < 0.05 and **P < 0.01. Differences were tested using 1-way ANOVA with Tukey’s post hoc test (A).
Discussion
Here, we revealed RSK4 to be an oncogenic driver in ESCC that promotes CSC properties and radioresistance. First, we systematically identified RSK4 as a direct target of ΔNp63α that was frequently amplified in ESCC. RSK4 phosphorylates GSK-3β at Ser9, activating the Wnt/β-catenin pathway, enhancing CSC properties, and increasing ESCC resistance to radiotherapy. Most important, pharmacologic inhibition of RSK4 using BI-D1870 markedly attenuated CSC properties and enhanced the antitumor activity of radiotherapy in both nude mice and PDX models.
Our findings provide further evidence of the role of p63 as a key regulator in ESCC. ΔNp63α is the predominant p63 isoform expressed in basal epithelial cells and is essential for squamous epithelial development (43). Furthermore, ΔNp63α is frequently overexpressed in ESCC and has been implicated in CSC properties (44, 45). Meera et al. reported that ΔNp63α is associated with the B56α regulatory subunit of protein phosphatase 2A (PP2A), leading to inhibition of PP2A-mediated GSK-3β reactivation, which induces nuclear accumulation of β-catenin and activates β-catenin–dependent transcription in SCC (46). PP2A is a phosphatase that can mediate GSK-3β dephosphorylation, thereby increasing its activity (47). However, how ΔNp63α inhibits the activity of PP2A is not clear. Here, we identified the functional link between p63 and the new target gene RSK4 and demonstrated the transcriptional regulation of RSK4 by ΔNp63α to activate the Wnt/β-catenin signaling pathway in ESCC, which could be another potential mechanism for ΔNp63α to promote CSC properties and radioresistance.
RSK4 has previously been reported to play different roles in multiple cancers. Dewdney et al. reported that RSK4 is frequently hypermethylated in endometrial cancer compared with normal endometrial tissues (17). It was also found that RSK4 expression limits the oncogenic, invasive, and metastatic potential of breast cancer cells (18). These findings suggest that RSK4 may have an antitumor effect, which is further supported by the downregulation of RSK4 expression in ovarian cancer (19) and acute myeloid leukemia (20). By contrast, Thakur et al. (25) observed that the expression of RSK4 mRNA was higher in transgenic mouse mammary tumors and human breast cancer tissues than in normal mammary tissues. Furthermore, RSK4 overexpression is associated with sunitinib resistance in kidney carcinoma and melanoma cell lines (21) and can mediate resistance to PI3K pathway inhibitors in breast cancer (22), indicating that RSK4 may regulate treatment resistance and can promote tumor progression. Our laboratory previously discovered that RSK4 is overexpressed in RCC as well as promotes cell-cycle progression and enhances the invasive and metastatic capability of RCC cell lines (23). This study advanced our knowledge of the association of RSK4 in ESCC. Our data not only reveal roles for RSK4 in promoting ESCC CSC properties and radioresistance but also identify RSK4 as a potential target for treating ESCC through pharmacologic inhibition. Using a panel of antibodies, we have previously reported that RSK4 protein may exist as several isoforms besides the known 84-kDa form (24). Distinct RSK4 isoforms may explain, to some extent, why RSK4 acts so differently in multiple cancers, as different isoforms may have unique or even opposite functions, such as the roles of STAT3 α and β in ESCC (48). In the present study, we used an antibody developed by The Human Protein Atlas project to study the role of RSK4 in ESCC, which, we believe, makes our study more meaningful and consistent.
With regard to other members of the RSK family, RSK2 was reported to be involved in SCC progression (12). In the present study, we found no significant difference in RSK2 mRNA levels between ESCC and adjacent nontumor tissues. Moreover, the protein levels of RSK2 and phosphorylation of its downstream targets were not significantly affected by ΔNp63 overexpression or downregulation in ESCC cells. In addition, RNA-Seq data generated under conditions of p63 depletion in human keratinocytes and SCC cell lines showed that only RSK4, rather than the other members of the RSK family, was downregulated by p63 knockdown. Furthermore, ESCC cells treated with BI-D1870 did not evidently have altered levels of the RSK1–3 proteins or phosphorylation of their downstream substrates. Altogether, these findings suggest that RSK4, but not other RSK family members, plays a key role in ESCC CSC properties and radioresistance.
Canonical Wnt/β-catenin signaling supports the formation and maintenance of CSCs and radioresistance (39). Consistent with the physiological roles of GSK-3β in negatively regulating canonical Wnt/β-catenin signaling, inhibition of GSK-3β is a prerequisite for the maintenance of CSC properties and radioresistance (40). Although it has been reported that RSK2 can phosphorylate GSK-3β at Ser9 (49), we found that GSK-3β was a binding partner and substrate of RSK4. To our knowledge, GSK-3β is the first direct substrate of RSK4 to our date to be identified with direct evidence. RSK4 phosphorylated GSK-3β at Ser9 and reduced its activity, resulting in the enhancement of ESCC CSC properties and radioresistance by stabilizing β-catenin. Surprisingly, using an in vitro kinase assay, we also discovered that GSK-3β may directly phosphorylate RSK4 in turn (Figure 5C). It is well known that a number of proteins phosphorylated by GSK-3β are targeted by E3-ubiquitin ligases, leading to subsequent proteasomal degradation, such as of β-catenin (50), Snail (51), and Mcl-1 (52). Therefore, we speculate that RSK4 phosphorylated by GSK-3β may also be degraded in a proteasome-dependent manner, thus leading to more stabilized RSK4 when GSK-3β is inactivated by RSK4 phosphorylation, which is likely a previously unknown RSK4–GSK-3β feedback loop in ESCC.
Radiotherapy is a primary treatment option for locally advanced or unresectable ESCC; however, there have been no significant improvements in OS of patients with advanced ESCC (53). The current concept to account for the increased mortality and therapy failure is that CSCs are mainly responsible for radioresistance and are the principal cause of cancer relapse (6, 54), suggesting that elimination of CSCs is crucial for improving ESCC treatment and overcoming its therapeutic resistance. However, CSC-targeting drugs are still unavailable in ESCC clinical practice. Thus, the development of anti-CSC therapeutics based on ESCC CSC–specific targets is promising for curing this type of cancer. In the search for CSC-specific functional targets in ESCC, we identified RSK4 as a molecular target and therapeutic candidate, as evidenced by its strong sphere-forming ability and in vivo tumor-initiating potential. These results are in line with evidence that RSK4 is constitutively activated under serum-starved conditions (16), which partly contributes to tumor sphere formation for acquiring stemness under serum-free conditions. There are several potential advantages of targeting RSK4 therapeutically: first, the preferential RSK4 upregulation in ESCC cells compared with normal esophageal epithelium suggested a favorable therapeutic index for RSK4 inhibition in ESCC; second, RSK4 was critical for ESCC CSC proliferation and maintenance of CSC self-renewal and tumorigenic potential, and thus its pharmacological disruption would eradicate CSCs; third, inhibition of RSK4 significantly disrupted DNA damage checkpoint responses and DNA damage repair and increased the radiosensitivity of ESCC cells, suggesting a synergistic potential of RSK4 inhibition with radiotherapy in ESCC; and fourth, RSK4 is a kinase with ATP-binding sites in its kinase domain, which makes it amenable for the development of irreversible and specific small-molecule inhibitors (9). Our current study demonstrates that the combination of BI-D1870 and irradiation markedly attenuated tumor growth in ESCC xenografts and PDX models, suggesting that combinatorial RSK4-targeted therapy may enhance the efficacy of radiotherapy in ESCC.
Several small-molecule inhibitors have been reported that specifically target the NTKD or C-terminal kinase domain (CTKD) of RSKs. However, current RSK inhibitors target more than just 1 RSK isoform, which may limit their efficacy as therapeutic agents (55). BI-D1870 is a dihydropteridinone that reversibly binds the ATP pocket of the NTKD and is a more potent inhibitor of RSK4 than other RSK family members, given its lower IC50 for RSK4 than for other RSKs (41). However, the disadvantages of BI-D1870 include its poor pharmacokinetic profile due to its poor stability, its high clearance, and its short plasma half-life (56, 57). It is suggested that the structural divergence between the NTKDs of the RSK isoforms could be exploited for the design of isoform-selective RSK inhibitors (55). Considering that RSK4 plays a significant role in ESCC progression, high-efficiency and specific RSK4 isoform–selective inhibitors urgently need to be developed.
In conclusion, we established RSK4 as a key oncogenic factor in ESCC (Supplemental Figure 8). This study not only describes a ΔNp63α/RSK4/GSK-3β axis to promote CSC properties and radioresistance in ESCC, but also reveals a clinical opportunity involving combined RSK4 inhibitor and radiotherapy for treating patients with ESCC. These findings establish a working model encompassing the function of RSK4 action on malignant progression and prognostic prediction in ESCC and reveal a promising drug target for the treatment of these aggressive malignancies.
Methods
Further details on methods can be found in the Supplemental Methods.
Cell culture, reagents, and irradiation.
HEK293T cells and the human ESCC cell lines ECA109, EC9706, TE10, and TE11 were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cell lines were verified through short tandem repeat DNA profiling. Mycoplasma contamination was checked, and the test results were negative. Cells were cultured in DMEM (Thermo Fisher Scientific) with 10% FBS (Gibco, Thermo Fisher Scientific) at 37°C in an incubator with 5% CO2. BI-D1870, AR-A014418, iCRT3, and MG-132 were purchased from Selleckchem Inc., and cycloheximide (CHX) was purchased from MedChemExpress (MCE). The cells and mice were irradiated by x-rays using the MBR-1520R-3 system (Hitachi Medico Technology) with the indicated dosages.
Tissue microarray.
Multiple human normal and tumor organ tissue arrays were purchased from US Biomax Inc. (MC5003c). This array contained 20 types of normal human tissues (n = 5 specimens for each tissue type) and corresponding tumor tissues (n = 20 specimens for each tissue type), including stomach, skin, prostate, brain, ovary, breast, testis, colon, bladder, uterus, thyroid, lung, head and neck, lymph node, soft tissue, liver, pancreas, kidney, cervix, and esophagus (Supplemental Table 1). ESCC specimens and matched adjacent normal tissues were used to construct a tissue microarray (HEso-Squ180Sur-03, Shanghai Biochip).
IHC.
Paraffin-embedded tissues were sectioned at 4-μm thickness. Slides were baked at 60°C for 1 hour and then deparaffinized, rehydrated, and treated with 3% hydrogen peroxide for 10 minutes. Antigen retrieval was performed in citrate buffer, pH 6.0, in a steamer for 2 minutes or in Tris-EDTA buffer, pH 9.0, at 100°C for 20 minutes. After the slides were blocked with 5% BSA in PBS for 30 minutes, tissue sections were incubated overnight at 4°C with the indicated primary antibodies. Antibody information is shown in Supplemental Table 6. Subsequently, a standard rapid EnVision (Dako) technique was used to detect the protein conjugates and develop the color. Finally, the sections were visualized after counterstaining with hematoxylin. Serial sections of ESCC were run in parallel with the primary antibody replaced by PBS and mouse IgG1 (Santa Cruz Biotechnology) as blank and negative controls.
Evaluation of IHC staining.
The sections were photographed under an optical microscope (BX51, Olympus), and photos were captured with the software DP2-BSW (Olympus). IHC staining was evaluated simultaneously by 2 observers who had no knowledge of the clinicopathological features of the patients. The H-score was calculated by adding the multiplication of the different staining intensities in 4 gradations (0, 1+, 2+, 3+), with each percentage of positive cells: H-score = 1× (% cells 1+) + 2× (% cells 2+) + 3× (% cells 3+). Finally, a score from 0–300 points was obtained (58). We used the median H-score in the cohort as a cutoff to distinguish between high and low protein expression levels.
Clonogenic assay.
Equal numbers of ESCC cells were plated in 6-cm tissue culture dishes at a clonogenic density (100 cells/dish) and allowed to adhere overnight. Subsequently, cells were irradiated with a single dose of 0, 3, or 6 Gy. Clonogenic assay procedures were performed as described previously (59). The survival fraction was calculated as follows: (number of colonies/number of cells plated)irradiated/(number of colonies/number of cells plated)nonirradiated.
In vitro kinase assay.
RSK4 active kinase and 10× kinase buffer were purchased from MilliporeSigma. His-GSK-3β WT protein was purchased from Sino Biological (10044-H07B). GSK-3β S9 peptide was synthesized by GL Biochem, and RPS6 peptide was purchased from Abcam (ab204879). His–GSK-3β S9A protein was expressed in E. coli BL21 bacteria. Bacteria were grown at 37°C to an absorbance of 0.6–0.8 at 600 nm and induced with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG) at 30°C for 4 hours. All proteins were purified using nickel-nitrilotriacetic acid agarose (QIAGEN) overnight at 4°C and eluted with 200 mM imidazole. After protein quantitation, the samples were separated by 10% SDS-PAGE and visualized by Coomassie brilliant blue staining. The GSK-3β WT and GSK-3β S9A substrate (1 μg) and the active kinase (0.2 μg) in a 30-μL reaction were incubated at 37°C for 40 minutes with 1× kinase buffer containing 100 μmol/L unlabeled ATP (9804, Cell Signaling Technology) or 1 μCi [γ-32P] ATP (NEG502A, Thermo Fisher Scientific). The samples were added to 5× SDS buffer and then resolved by SDS-PAGE and visualized by autoradiography or Western blotting. The input was confirmed using an identical experimental set with a Silver Stain Kit (CW2012, CW Biotech).
PDX tumors and drug sensitivity assay.
PDX tumors were generated as described previously (60). Fresh tumor specimens were procured from previously established PDX models (passages 2–3) and cut into small tissue blocks (~50 mm3) before being engrafted subcutaneously into male BALB/c nude mice. Xenografts were allowed to grow until they reached a size of 100–200 mm3, and then mice were randomized into 2 groups (6 animals/group) for treatment. BI-D1870 (50 mg/kg) dissolved in DMSO and sterile saline was administered daily by i.p. injection for 28 consecutive days. Drug vehicle–treated mice received daily injections of an identical solution without BI-D1870. Tumor size was measured every 3 days with a digital caliper using the following formula: length × width2 × 0.5. Investigators were blinded to the case number and the correspondent RSK4 level during the experiment. At the end of treatment, all mice were euthanized, and tumors were excised and weighed. The lungs, liver, and kidneys of mice were removed for pathological examination, and serological examination of alanine transaminase (ALT), aspartate amino transferase (AST), blood urea nitrogen (BUN), and creatinine (Cr) was performed with a Chemray 240 Automatic Biochemical Analyzer (Rayto). To evaluate changes in tumor volume after each therapeutic regimen in PDX models, when tumors had grown to a volume of 100–200 mm3, mice were randomized into 4 groups (6 mice/group): vehicle; vehicle plus ionizing radiation (IR); BI-D1870; and BI-D1870 plus IR. A total of 10 Gy (5 Gy, 2 times) was delivered to animals restrained in custom lead jigs for localized IR treatment at the seventh and 14th day after dividing the mice into groups. BI-D1870 was administered at 50 mg/kg by daily i.p. injection. Animals were raised for 25 days, and tumor volume was measured every 5 days and calculated as follows: length × width2 × 0.5. All animals were housed in a virus-free facility and maintained in a temperature- and light-controlled (12-hour light/12-dark cycle) animal facility.
Data mining.
The ESCC cohort of TCGA database (https://portal.gdc.cancer.gov/) was used to analyze mRNA expression of RSK4 in ESCC and its association with clinical data and the CSC marker ALDH1. The Genotype-Tissue Expression (GTEx) database (https://www.gtexportal.org/) was used to detect the correlation between TP63 and RSK4 mRNA in esophageal mucosa. The JASPAR database (jaspar.genereg.net/) was used to predict the potential transcription factor binding site on the RSK4 promoter. The GEO data set (https://www.ncbi.nlm.nih.gov/geo/) (GSE 20347 [ref. 61]) was used to analyze the correlation between TP63 and RSK4 mRNA in ESCC. GEO data sets (GSE 3108 [ref. 62] and GSE 4975 [ref. 63]) were used to show the effect of p63 depletion on RSK4 mRNA expression. GEO data sets (GSE 23400 [ref. 64] and GSE 32701 [ref. 65]) were used to analyze the correlation between RSK4 mRNA and the Wnt/β-catenin pathway targets TCF1 and CD44 in ESCC. The raw data were downloaded from these data sets and manually graphed in GraphPad Prism 7.0 (GraphPad Software) with relative mRNA alterations.
Statistics.
Data are expressed as the mean ± SD. Comparisons between 2 groups were performed using an unpaired or paired, 2-tailed Student’s t test, and comparisons of more than 2 groups were conducted with a 1-way ANOVA with Tukey’s post hoc test. Differences among variables were assessed using the χ2 test. The correlation between groups was determined by Pearson’s correlation test. The survival rates were analyzed by the Kaplan-Meier method. The log-rank test was used to compare the survival of patients between subgroups. Multivariate analyses were performed using the multivariate Cox proportional hazard regression model. Sample numbers indicate the number of independent biological samples in each experiment. Generally, all experiments were replicated at least 3 independent times. A P value of less than 0.05 was considered statistically significant. Analyses were performed using GraphPad Prism 7.0.
Study approval.
Ethics approval was granted by the Xijing Hospital Research Ethics Committee, and written informed consent was obtained from each patient. All experimental procedures were performed in accordance with relevant institutional and national guidelines and approved by the IACUC of the Fourth Military Medical University.
Author contributions
MYL designed and performed the experiments, analyzed data, and wrote the manuscript. LNF and DHH performed animal experiments and analyzed data; ZY, JM, and YXL performed the bioinformatics analysis. PFL, DHZ, and JC performed Western blot experiments and histopathological analysis. LJ and SLL performed homology modeling. JJX and QHD performed the in vitro kinase assay. JY, MS, YZN, KCW, DJL, YS, YW, QGY, SPG, and XWB wrote the manuscript and provided important advice for this study. FZ, JZ, and ZW designed experiments, wrote the manuscript, and supervised the study. All authors read the manuscript and approved the study.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81272651, 81570180, 81421003, 81770523, and 81472402) and the State Key Laboratory of Cancer Biology (CBSKL2014Z07). We thank Deng-Xu Tan (Laboratory Animal Center, Fourth Military Medical University) for assistance in establishing the PDX models. We thank Pei-Pei Xue and Ling Zou (Department of Biochemistry and Molecular Biology, Huazhong University of Science and Technology) for help with in vitro kinase analyses. We thank Hui-Ping Liu (Department of Pharmacology, Feinberg School of Medicine, Northwestern University) for the important suggestions for this study.
Version 1. 05/12/2020
In-Press Preview
Version 2. 07/13/2020
Electronic publication
Version 3. 08/03/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(8):4301–4319.https://doi.org/10.1172/JCI134930.
Contributor Information
Zhou Yu, Email: yz20080512@163.com.
Jing Ma, Email: 854700623@qq.com.
Jia Chai, Email: 18292866925@163.com.
Lei Jiang, Email: 18463285147@163.com.
Jing Ye, Email: yejing@fmmu.edu.cn.
Mei Shi, Email: mshi23@163.com.
Kai-Chun Wu, Email: kaicwu@fmmu.edu.cn.
Dezhong Joshua Liao, Email: djliao@gmc.edu.cn.
Yu Shi, Email: drshiyu@126.com.
Yan Wang, Email: wang_yan1977@hotmail.com.
Xiu-Wu Bian, Email: bianxiuwu@263.net.
Feng Zhu, Email: fengzhu@hust.edu.cn.
Jian Zhang, Email: biozhangj@fmmu.edu.cn.
Zhe Wang, Email: zhwang@fmmu.edu.cn.
References
- 1.Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154(2):360–373. doi: 10.1053/j.gastro.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Liang H, Fan JH, Qiao YL. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol Med. 2017;14(1):33–41. doi: 10.20892/j.issn.2095-3941.2016.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Hagen P, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366(22):2074–2084. doi: 10.1056/NEJMoa1112088. [DOI] [PubMed] [Google Scholar]
- 5.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16(3):225–238. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D, Plukker JTM, Coppes RP. Cancer stem cells with increased metastatic potential as a therapeutic target for esophageal cancer. Semin Cancer Biol. 2017;44:60–66. doi: 10.1016/j.semcancer.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Krause M, Dubrovska A, Linge A, Baumann M. Cancer stem cells: Radioresistance, prediction of radiotherapy outcome and specific targets for combined treatments. Adv Drug Deliv Rev. 2017;109:63–73. doi: 10.1016/j.addr.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Chen GZ, Zhu HC, Dai WS, Zeng XN, Luo JH, Sun XC. The mechanisms of radioresistance in esophageal squamous cell carcinoma and current strategies in radiosensitivity. J Thorac Dis. 2017;9(3):849–859. doi: 10.21037/jtd.2017.03.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9(10):747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 10.Romeo Y, Zhang X, Roux PP. Regulation and function of the RSK family of protein kinases. Biochem J. 2012;441(2):553–569. doi: 10.1042/BJ20110289. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y, et al. Crucial roles of RSK in cell motility by catalysing serine phosphorylation of EphA2. Nat Commun. 2015;6:7679. doi: 10.1038/ncomms8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang S, et al. p90 ribosomal S6 kinase 2 promotes invasion and metastasis of human head and neck squamous cell carcinoma cells. J Clin Invest. 2010;120(4):1165–1177. doi: 10.1172/JCI40582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao H, et al. The clinical implications of RSK1-3 in human breast cancer. Anticancer Res. 2016;36(3):1267–1274. [PubMed] [Google Scholar]
- 14.Bignone PA, et al. RPS6KA2, a putative tumour suppressor gene at 6q27 in sporadic epithelial ovarian cancer. Oncogene. 2007;26(5):683–700. doi: 10.1038/sj.onc.1209827. [DOI] [PubMed] [Google Scholar]
- 15.Yntema HG, et al. A novel ribosomal S6-kinase (RSK4; RPS6KA6) is commonly deleted in patients with complex X-linked mental retardation. Genomics. 1999;62(3):332–343. doi: 10.1006/geno.1999.6004. [DOI] [PubMed] [Google Scholar]
- 16.Dümmler BA, et al. Functional characterization of human RSK4, a new 90-kDa ribosomal S6 kinase, reveals constitutive activation in most cell types. J Biol Chem. 2005;280(14):13304–13314. doi: 10.1074/jbc.M408194200. [DOI] [PubMed] [Google Scholar]
- 17.Dewdney SB, et al. Aberrant methylation of the X-linked ribosomal S6 kinase RPS6KA6 (RSK4) in endometrial cancers. Clin Cancer Res. 2011;17(8):2120–2129. doi: 10.1158/1078-0432.CCR-10-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thakur A, et al. Anti-invasive and antimetastatic activities of ribosomal protein S6 kinase 4 in breast cancer cells. Clin Cancer Res. 2008;14(14):4427–4436. doi: 10.1158/1078-0432.CCR-08-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arechavaleta-Velasco F, et al. Ribosomal S6 kinase 4 (RSK4) expression in ovarian tumors and its regulation by antineoplastic drugs in ovarian cancer cell lines. Med Oncol. 2016;33(2):11. doi: 10.1007/s12032-015-0724-6. [DOI] [PubMed] [Google Scholar]
- 20.Rafiee M, et al. Down-regulation of ribosomal S6 kinase RPS6KA6 in acute myeloid leukemia patients. Cell J. 2016;18(2):159–164. doi: 10.22074/cellj.2016.4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bender C, Ullrich A. PRKX, TTBK2 and RSK4 expression causes Sunitinib resistance in kidney carcinoma- and melanoma-cell lines. Int J Cancer. 2012;131(2):E45–E55. doi: 10.1002/ijc.26486. [DOI] [PubMed] [Google Scholar]
- 22.Serra V, et al. RSK3/4 mediate resistance to PI3K pathway inhibitors in breast cancer. J Clin Invest. 2013;123(6):2551–2563. doi: 10.1172/JCI66343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan L, et al. Ribosomal s6 protein kinase 4: a prognostic factor for renal cell carcinoma. Br J Cancer. 2013;109(5):1137–1146. doi: 10.1038/bjc.2013.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, et al. Basic anatomy and tumor biology of the RPS6KA6 gene that encodes the p90 ribosomal S6 kinase-4. Oncogene. 2013;32(14):1794–1810. doi: 10.1038/onc.2012.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thakur A, et al. Aberrant expression of X-linked genes RbAp46, Rsk4, and Cldn2 in breast cancer. Mol Cancer Res. 2007;5(2):171–181. doi: 10.1158/1541-7786.MCR-06-0071. [DOI] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research Network, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su X, Chakravarti D, Flores ER. p63 steps into the limelight: crucial roles in the suppression of tumorigenesis and metastasis. Nat Rev Cancer. 2013;13(2):136–143. doi: 10.1038/nrc3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sethi I, et al. A global analysis of the complex landscape of isoforms and regulatory networks of p63 in human cells and tissues. BMC Genomics. 2015;16:584. doi: 10.1186/s12864-015-1793-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thépot A, et al. Intraepithelial p63-dependent expression of distinct components of cell adhesion complexes in normal esophageal mucosa and squamous cell carcinoma. Int J Cancer. 2010;127(9):2051–2062. doi: 10.1002/ijc.25221. [DOI] [PubMed] [Google Scholar]
- 30.Saladi SV, et al. ACTL6A is co-amplified with p63 in squamous cell carcinoma to drive YAP activation, regenerative proliferation, and poor prognosis. Cancer Cell. 2017;31(1):35–49. doi: 10.1016/j.ccell.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, et al. ALDH1A1 defines invasive cancer stem-like cells and predicts poor prognosis in patients with esophageal squamous cell carcinoma. Mod Pathol. 2014;27(5):775–783. doi: 10.1038/modpathol.2013.189. [DOI] [PubMed] [Google Scholar]
- 32.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1-2):70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanz MC, Dibitetto D, Smolka MB. DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J. 2019;38(18):e101801. doi: 10.15252/embj.2019101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hibi K, et al. AIS overexpression in advanced esophageal cancer. Clin Cancer Res. 2001;7(3):469–472. [PubMed] [Google Scholar]
- 37.Lo Muzio L, et al. p63 overexpression associates with poor prognosis in head and neck squamous cell carcinoma. Hum Pathol. 2005;36(2):187–194. doi: 10.1016/j.humpath.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Domoto T, et al. Glycogen synthase kinase-3β is a pivotal mediator of cancer invasion and resistance to therapy. Cancer Sci. 2016;107(10):1363–1372. doi: 10.1111/cas.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19(2):150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Trowbridge JJ, Xenocostas A, Moon RT, Bhatia M. Glycogen synthase kinase-3 is an in vivo regulator of hematopoietic stem cell repopulation. Nat Med. 2006;12(1):89–98. doi: 10.1038/nm1339. [DOI] [PubMed] [Google Scholar]
- 41.Sapkota GP, et al. BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem J. 2007;401(1):29–38. doi: 10.1042/BJ20061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer. 2015;15(5):311–316. doi: 10.1038/nrc3944. [DOI] [PubMed] [Google Scholar]
- 43.Daniely Y, et al. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am J Physiol, Cell Physiol. 2004;287(1):C171–C181. doi: 10.1152/ajpcell.00226.2003. [DOI] [PubMed] [Google Scholar]
- 44.Melino G, Memmi EM, Pelicci PG, Bernassola F. Maintaining epithelial stemness with p63. Sci Signal. 2015;8(387):re9. doi: 10.1126/scisignal.aaa1033. [DOI] [PubMed] [Google Scholar]
- 45.Nekulova M, Holcakova J, Coates P, Vojtesek B. The role of p63 in cancer, stem cells and cancer stem cells. Cell Mol Biol Lett. 2011;16(2):296–327. doi: 10.2478/s11658-011-0009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patturajan M, et al. DeltaNp63 induces beta-catenin nuclear accumulation and signaling. Cancer Cell. 2002;1(4):369–379. doi: 10.1016/S1535-6108(02)00057-0. [DOI] [PubMed] [Google Scholar]
- 47.Seeling JM, Miller JR, Gil R, Moon RT, White R, Virshup DM. Regulation of beta-catenin signaling by the B56 subunit of protein phosphatase 2A. Science. 1999;283(5410):2089–2091. doi: 10.1126/science.283.5410.2089. [DOI] [PubMed] [Google Scholar]
- 48.Zhang HF, et al. The opposing function of STAT3 as an oncoprotein and tumor suppressor is dictated by the expression status of STAT3β in esophageal squamous cell carcinoma. Clin Cancer Res. 2016;22(3):691–703. doi: 10.1158/1078-0432.CCR-15-1253. [DOI] [PubMed] [Google Scholar]
- 49.Lee CJ, Lee MH, Lee JY, Song JH, Lee HS, Cho YY. RSK2-induced stress tolerance enhances cell survival signals mediated by inhibition of GSK3β activity. Biochem Biophys Res Commun. 2013;440(1):112–118. doi: 10.1016/j.bbrc.2013.09.042. [DOI] [PubMed] [Google Scholar]
- 50.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16(13):3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou BP, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6(10):931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 52.Ding Q, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27(11):4006–4017. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He L, et al. Re-evaluating the optimal radiation dose for definitive chemoradiotherapy for esophageal squamous cell carcinoma. J Thorac Oncol. 2014;9(9):1398–1405. doi: 10.1097/JTO.0000000000000267. [DOI] [PubMed] [Google Scholar]
- 54.Islam F, Gopalan V, Wahab R, Smith RA, Lam AK. Cancer stem cells in oesophageal squamous cell carcinoma: identification, prognostic and treatment perspectives. Crit Rev Oncol Hematol. 2015;96(1):9–19. doi: 10.1016/j.critrevonc.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 55.Casalvieri KA, Matheson CJ, Backos DS, Reigan P. Selective targeting of RSK isoforms in cancer. Trends Cancer. 2017;3(4):302–312. doi: 10.1016/j.trecan.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 56.Pambid MR, et al. Overcoming resistance to sonic hedgehog inhibition by targeting p90 ribosomal S6 kinase in pediatric medulloblastoma. Pediatr Blood Cancer. 2014;61(1):107–115. doi: 10.1002/pbc.24675. [DOI] [PubMed] [Google Scholar]
- 57.Hammoud L, et al. Identification of RSK and TTK as modulators of blood vessel morphogenesis using an embryonic stem cell-based vascular differentiation assay. Stem Cell Reports. 2016;7(4):787–801. doi: 10.1016/j.stemcr.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Specht E, Kaemmerer D, Sänger J, Wirtz RM, Schulz S, Lupp A. Comparison of immunoreactive score, HER2/neu score and H score for the immunohistochemical evaluation of somatostatin receptors in bronchopulmonary neuroendocrine neoplasms. Histopathology. 2015;67(3):368–377. doi: 10.1111/his.12662. [DOI] [PubMed] [Google Scholar]
- 59.Frankle RT. Nutrition education in the medical school curriculum: a proposal for action: a curriculum design. Am J Clin Nutr. 1976;29(1):105–109. doi: 10.1093/ajcn/29.1.105. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, et al. PP2AC level determines differential programming of p38-TSC-mTOR signaling and therapeutic response to p38-targeted therapy in colorectal cancer. EBioMedicine. 2015;2(12):1944–1956. doi: 10.1016/j.ebiom.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu N, et al. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics. 2010;11:576. doi: 10.1186/1471-2164-11-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koster MI, Kim S, Huang J, Williams T, Roop DR. TAp63alpha induces AP-2gamma as an early event in epidermal morphogenesis. Dev Biol. 2006;289(1):253–261. doi: 10.1016/j.ydbio.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 63.Barbieri CE, Tang LJ, Brown KA, Pietenpol JA. Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 2006;66(15):7589–7597. doi: 10.1158/0008-5472.CAN-06-2020. [DOI] [PubMed] [Google Scholar]
- 64.Su H, et al. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res. 2011;17(9):2955–2966. doi: 10.1158/1078-0432.CCR-10-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aoyagi K, et al. Artificially induced epithelial-mesenchymal transition in surgical subjects: its implications in clinical and basic cancer research. PLoS ONE. 2011;6(4):e18196. doi: 10.1371/journal.pone.0018196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.