Abstract
Gasdermin D (GSDMD) induces pyroptosis via the pore-forming activity of its N-terminal domain, cleaved by activated caspases associated with the release of IL-1β. Here, we report a nonpyroptotic role of full-length GSDMD in guiding the release of IL-1β–containing small extracellular vesicles (sEVs) from intestinal epithelial cells (IECs). In response to caspase-8 inflammasome activation, GSDMD, chaperoned by Cdc37/Hsp90, recruits the E3 ligase, NEDD4, to catalyze polyubiquitination of pro–IL-1β, serving as a signal for cargo loading into secretory vesicles. GSDMD and IL-1β colocalize with the exosome markers CD63 and ALIX intracellularly, and GSDMD and NEDD4 are required for release of CD63+ sEVs containing IL-1β, GSDMD, NEDD4, and caspase-8. Importantly, increased expression of epithelial-derived GSDMD is observed both in patients with inflammatory bowel disease (IBD) and those with experimental colitis. While GSDMD-dependent release of IL-1β–containing sEVs is detected in cultured colonic explants from colitic mice, GSDMD deficiency substantially attenuates disease severity, implicating GSDMD-mediated release of IL-1β sEVs in the pathogenesis of intestinal inflammation, such as that observed in IBD.
Keywords: Gastroenterology, Inflammation
Keywords: Cytokines, Inflammatory bowel disease, Innate immunity
Introduction
Inflammasomes are the critical platforms that integrate environmental stimuli to initiate appropriate and necessary innate and adaptive immune responses to maintain homeostasis, particularly at mucosal surfaces in organs that interface with microbial communities, including the gastrointestinal tract (1–5). In this context, increasing evidence also indicates that dysregulated inflammasome activity intimately contributes to chronic intestinal inflammation, such as that observed in inflammatory bowel disease (IBD) (6–8). Myeloid cells, predominantly macrophages, have been the focus of inflammasome research as they are considered to be a major reservoir of IL-1β (9), a classic proinflammatory cytokine known to amplify innate immune responses. More recent studies, however, have demonstrated that inflammasome activation in nonmyeloid compartments can also serve as abundant sources of IL-1β, which is critical for the development of inflammatory pathologies in both humans and mouse models (10–14). In fact, it has long been established that epithelial-derived IL-1 and its family members play a primary role in the pathogenesis of IBD (15, 16). These data collectively suggest temporal and spatial nonredundant roles of different cellular sources of IL-1 during pathogenic inflammatory responses, for which the cellular and molecular mechanisms responsible for specific IL-1β production, especially in nonmyeloid cells such as the intestinal epithelium, are not completely understood.
IL-1β is synthesized as inactive pro-forms with no secretory signal sequence (9–12). The recent discovery of the lipid binding protein, GSDMD, and its requirement for IL-1β release in response to caspase-1/11 (14, 17) and caspase-8 (18, 19) inflammasome activation via pore formation and associated pyroptotic activities in myeloid cells, has introduced paradigm shifting concepts in this area of investigation (20–22). Recent studies have begun to unravel the mechanistic role(s) of GSDMD in mediating IL-1β release, a process that had been regarded as a byproduct of nonselective discharge of cellular contents during pyroptosis (20–22). An increasing number of studies have reported GSDMD-dependent, pyroptosis-independent release of IL-1β (23–25) in a variety of cell types. Notably, it has long been documented that IL-1β can be released in the absence of cell lysis via various mechanisms (23–28), including IL-1β accumulation within membrane vesicles (exosomes and/or secretory autophagy) for export into the extracellular space (29–39). However, whether and how GSDMD participates in these nonpyroptotic pathways of IL-1β release remains unclear.
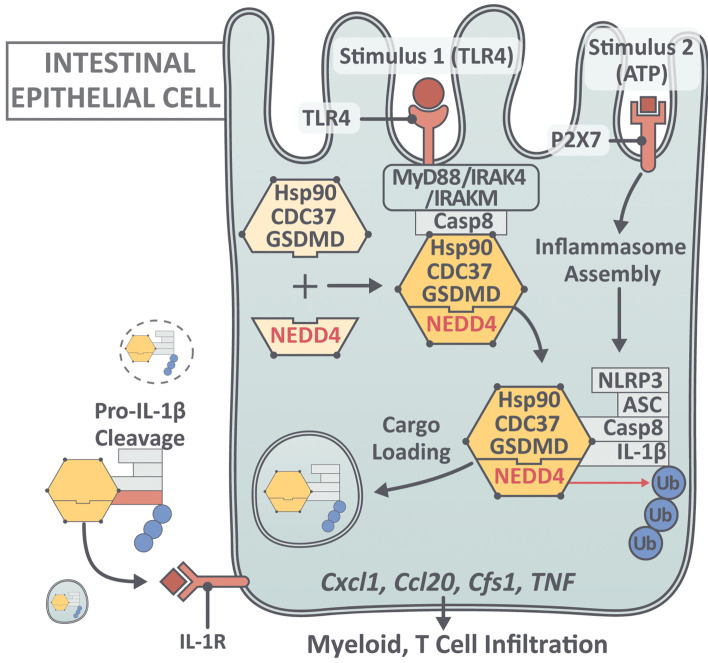
Herein, we report what the seemingly novel observation that the lack of GSDMD effectively reduces the severity of dextran sodium sulfate–induced (DSS-induced) colitis and that the major source of GSDMD within the gut mucosa of colitic mice is IECs. We also report for what we believe is the first time that GSDMD transcripts in mucosal biopsies from patients with ulcerative colitis (UC), and to a lesser extent Crohn’s disease (CD), the 2 main idiopathic forms of IBD, are dramatically increased compared with intestinal tissues from healthy controls. Increased epithelial-specific GSDMD was confirmed in IBD patients versus healthy controls, as well as in UC inflamed versus UC noninflamed tissues, by single-cell (sc)RNA-Seq of gut mucosal biopsies, identifying several different subpopulations of high GSDMD-expressing IECs, particularly secretory progenitor cells and colonocytes/crypt top colonocytes. Importantly, we also discovered a previously undocumented, nonpyroptotic role of GSDMD in guiding release of polyubiquitinated IL-1β via sEVs in response to caspase-8 inflammasome activation in colonic IECs. Through proteomic analysis, we identified a set of novel GSDMD-interacting proteins in IECs, including NEDD4 (an E3 ligase) and the Hsp90 cochaperone Cdc37. Ablation of either GSDMD, NEDD4, or caspase-8 abolished LPS plus ATP-induced IL-1β production from IECs. Moreover, GsdmdD276A (mutation at cleavage site Asp276), with abolished pore-forming and pyroptotic activities, did not lose the ability to guide IL-1β secretion from IECs. Mechanistically, LPS+ATP stimulation induced polyubiquitination of pro–IL-1β proteins, which were secreted and processed into mature forms, along with a complex containing chaperoned-GSDMD (full-length GSDMD/Hsp90/CDC37), NEDD4, and caspase-8, but not caspase-1. NEDD4, an E3 ligase known to interact with LC3 and promote cargo loading into secretory vesicles, catalyzed the polyubiquitination of pro–IL-1β. Whereas GSDMD was associated with LC3-positive vesicles, GSDMD-dependent release of sEVs (<200 nm) was detected by electron microscopy, which contained chaperoned-GSDMD/NEDD4/caspase-8/IL-1β. This GSDMD-guided IL-1β release is crucial for the development of intestinal inflammation. While polyubiquitinated pro–IL-1β, mature IL-1β, and sEVs were induced in a GSDMD-dependent manner in colon explants from DSS-treated mice, GSDMD deficiency attenuated the severity of colitis in these mice. These data collectively demonstrate the association, increased expression, and epithelial localization of GSDMD in patients with IBD, particularly UC, and a novel GSDMD-dependent pathway for IL-1β secretion that is a fundamental process in the pathogenesis of experimental colitis.
Results
GSDMD is increased and localizes to IECs in DSS colitic mice and IBD patients, and its deficiency leads to attenuated intestinal inflammation during experimental colitis.
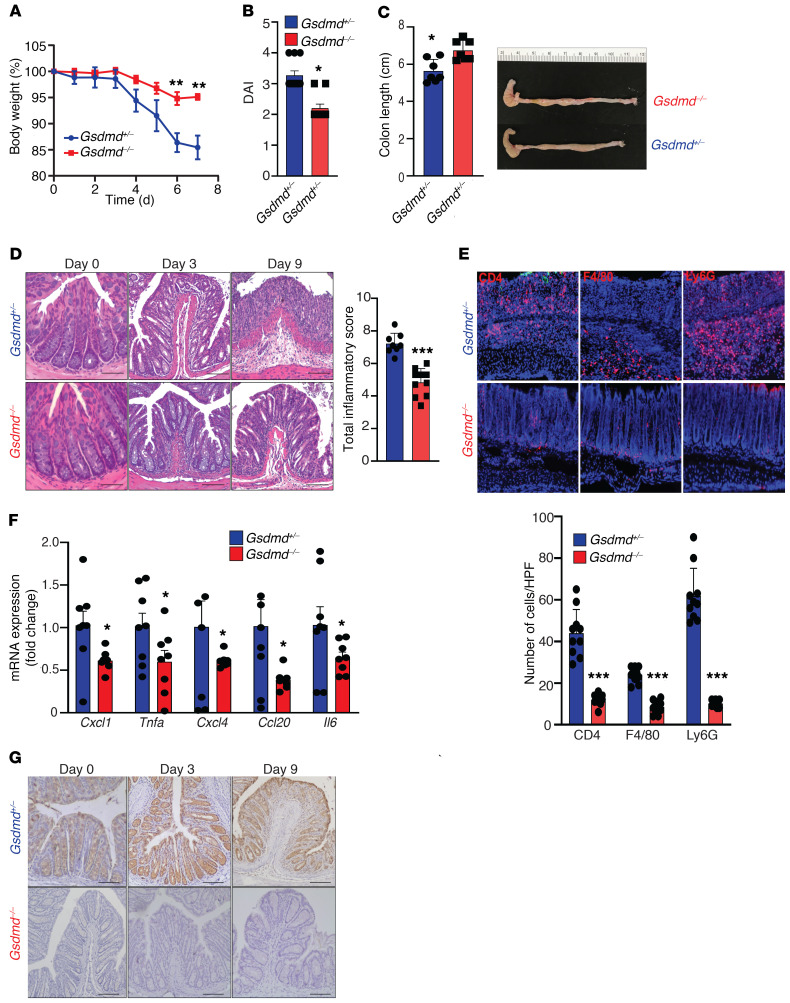
To examine the impact of GSDMD deficiency on acute intestinal inflammation, Gsdmd-KO mice (Gsdmd–/–) and heterozygous control littermates (Gsdmd+/–) were challenged with 3% DSS in drinking water to induce colitis. Gsdmd–/– mice were protected from DSS-induced colitis, as measured by weight loss, disease activity index (DAI), colon length (more severe colitis = decreased colon length), and histologic evaluation of intestinal inflammation (Figure 1, A–D), compared with Gsdmd+/– controls. The reduction in disease severity was accompanied by attenuated infiltration of neutrophils, macrophages, and T cells (Figure 1E), and decreased expression of inflammatory mediators (Figure 1F) in colonic tissues of Gsdmd–/– versus Gsdmd+/– littermate controls. Notably, IHC revealed that GSDMD expression was highly induced in inflamed tissues from colitic mice and localized to the colonic epithelium (Figure 1G).
Figure 1. GSDMD localizes to the colonic epithelium and its deficiency protects from DSS-induced colitis.
Colitis was induced in cohoused, sex-matched Gsdmd–/– (n = 10) and control Gdsmd+/– (n = 11) littermate mice by administration of 3% DSS in drinking water. Severity of colitis, assessed by (A) weight loss, (B) disease activity index (DAI), and (C) colon length (decreased length indicates increased inflammation). (D) Representative images of H&E-stained colons from DSS-treated mice (left), with histologic analysis of colitis performed on day 9 (right). Scale bars: 50 µM. (E) Representative images of immunofluorescence staining for CD4, F4/80, and LyG6 on frozen sections of inflamed colons taken at day 9 (left), with number of infiltrating immune cells enumerated and averaged over 10 HPF/section (right). Original magnification, ×40. (F) RT-PCR showing relative differences of indicated proinflammatory gene transcripts in colon tissues harvested on day 9 from experimental mice. Data are presented as fold induction over the mean of Gdsmd+/– group. (G) Representative IHC images showing immunolocalization of GSDMD to IECs in colons of DSS-challenged Gsdmd+/– control mice, with total absence in Gsdmd–/– littermates. Scale bars: 50 µM. Data are presented as mean ± SEM with *P < 0.05, ***P < 0.0001 by Student’s t test. All experiments were repeated twice and yielded consistent results.
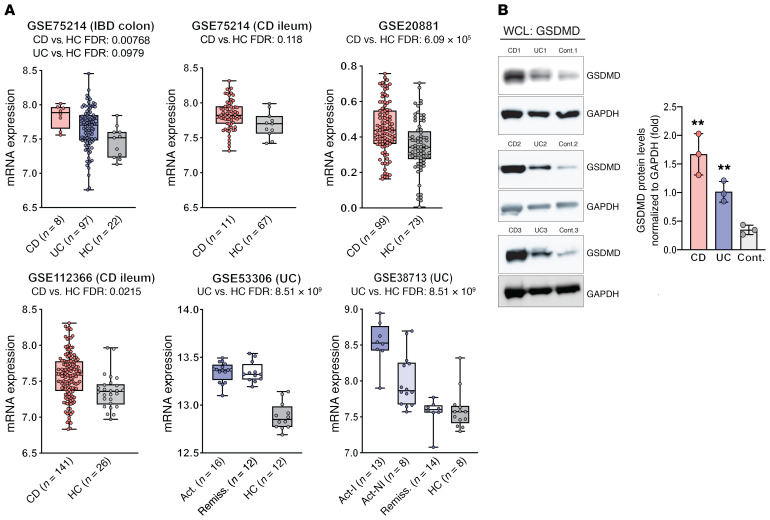
In order to assess the translational relevance of these findings, GSDMD expression was analyzed using the Genomic Spatial Event (GSE) database system in 5 separate expression data sets derived from endoscopically obtained intestinal mucosal biopsies from IBD patients and healthy controls (Supplemental Table 1 and Figures 2 and 3; supplemental material available online with this article; https://doi.org/10.1172/JCI138103DS1). In a cohort that included both CD and UC patients (GSE75214 [ref. 40]), GSDMD was increased in the colons of CD patients compared with healthy controls, but while both colon samples from UC patients and ileal biopsies from CD patients showed an increased trend for GSDMD versus healthy controls, these differences did not reach statistical significance (Figure 2A, upper left and middle). Closer scrutiny of the cohort samples, however, revealed that biopsies were derived from both involved and noninvolved areas of patients with active disease, with the most affected sites taken at a distance from ulcerations in UC patients, which, by inclusion of noninvolved areas, likely normalized GSDMD levels toward that of healthy controls. Therefore, to first confirm whether GSDMD is increased in the gut mucosa of CD patients, 2 larger CD data sets were analyzed: one consisting of gut mucosal biopsies from 1 of 4 anatomic locations (terminal ileum and sigmoid, ascending and descending colon) compared with location-matched healthy controls (GSE20881), and the other from ileal biopsies of CD patients suffering from moderate-to-severe disease (GSE112366 [ref. 41]). Both data sets showed an overall significant increase in GSDMD, comparing CD patients with healthy controls (Figure 2A, upper right and lower left, respectively). Differences in GSDMD, however, comparing IBD phenotype against their respective controls, appear to be more robust in UC versus CD patients. Analyzing 2 UC data sets, one derived from colonic biopsies sampled in consecutive active and quiescent (remission) stages of disease at the same site in individual UC patients (GSE53306 [ref. 42]), and the other comparing involved and noninvolved areas from active UC and inactive UC (remission) (GSE38713 [ref. 43]), GSDMD was found to be markedly increased in active UC compared with healthy controls. Interestingly, while GSDMD appeared to remain elevated in the same sites of individual UC patients, regardless of disease state (Figure 2A, lower middle), GSDMD generally decreased in noninvolved areas in active UC and in UC patients during remission compared with involved (inflamed) areas from active UC patients (Figure 2A, lower right), indicating elevated GSDMD in active IBD.
Figure 2. Increased expression of GSDMD in biopsies and isolated IECs from gut mucosa of IBD patients compared with healthy controls.
(A) Analysis of 5 different IBD gene array expression data sets (Supplemental Table 1) derived from intestinal mucosal biopsies from CD, UC, and healthy controls (HC) displayed as box plots (25th, 50th, and 75th quantiles shown), with differences expressed as FDR between IBD patients and healthy controls. Act, active disease; Act-I, active involved area; Act-NI, active noninvolved area; Remiss., remission from IBD. (B) Representative Western blots of whole-cell lysates (WCLs) showing differences in GSDMD protein levels in freshly isolated IECs from IBD patients versus controls (n = 4),with relative densitometric values calculated as ratio of GSDMD/GAPDH and reported as mean ± SEM; **P < 0.005 versus control (Cont.) by 2-tailed unpaired Student’s t test.
Figure 3. Prevalence of GSDMD expression in specific epithelial subpopulations from inflamed mucosa of IBD patients.
(A) t-SNE plots after scRNa-Seq of colonic IECs depicting expression and distribution of GSDMD, with (B) bar plots showing GSDMD in epithelial subpopulations from UC patients (inflamed and noninflamed) and healthy controls (HC) (n = 3). DESeq2 R package was used to compute library size factors, normalize data, and perform differential expression analysis using negative binomial Wald’s test.
Furthermore, based on the immunolocalization studies showing increased epithelial-derived GSDMD in DSS colitic Gsdmd+/– mice (Figure 1G), we evaluated the relative presence of GSDMD protein in freshly isolated IECs from IBD patients with active disease and healthy controls, and found increased GSDMD protein in IECs from both CD and UC patients (Figure 2B). To confirm IEC localization in both the animal (Figure 1G) and human (Figure 2B) studies and its modulation during active colitis, we performed a biased analysis for GSDMD in a previously published large-scale scRNA-Seq data set compiled from profiled single colonic IECs isolated from inflamed and noninflamed areas of active, immunomodulatory-naive UC patients and from normal, noninflamed healthy controls (44). In this prior study, novel IEC subpopulations were identified, including those that are unique to the pathogenesis of IBD (Supplemental Figure 1). Our analysis revealed that GSDMD displayed both differential distribution and differences in relative expression among IEC subtypes when comparing experimental groups (Figure 3) with the general trend of increased expression in IECs from inflamed compared with noninflamed UC patients and healthy controls. Specifically, GSDMD was predominantly expressed in IEC subpopulations of secretory progenitor cells, as well as in colonocytes, crypt top colonocytes, and absorptive progenitor cells (Figure 3).
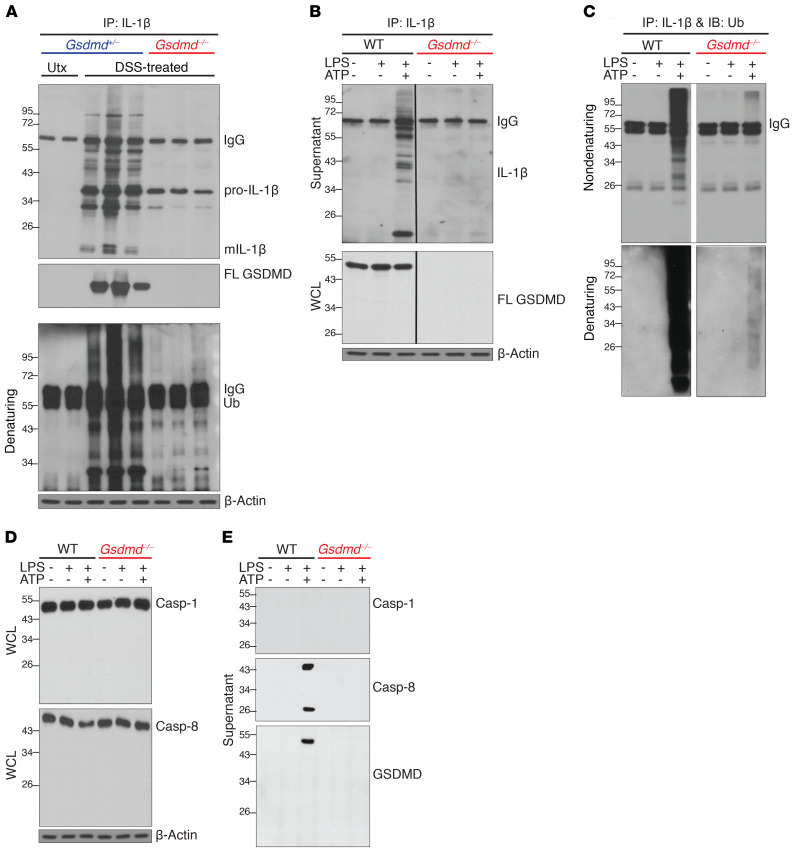
To mechanistically address potential IEC-derived GSDMD function, specifically whether GSDMD deficiency impairs IL-1β release during colitis, we went back to the DSS colitis model and analyzed supernatants from explant cultures of healthy control mice (no DSS) and inflamed colon tissues from both Gsdmd+/– and Gsdmd–/– mice after DSS challenge. Associated with the induction of GSDMD expression in inflamed colons (Figure 1G), mature IL-1β was released from inflamed but not naive colon explants, and IL-1β release was abolished in GSDMD-deficient colons (Figure 4A). Interestingly, IL-1β released from inflamed colon tissues was found in a complex with full-length GSDMD, which was highly modified in a GSDMD-dependent manner (Figure 4A). In fact, immunoprecipitation studies under denaturing conditions indicated that these IL-1β molecules were conjugated with polyubiquitin chains (Figure 4A). Taken together, these data demonstrate a pivotal role for GSDMD in promoting intestinal inflammation and suggest a unique form of GSDMD-dependent IL-1β release in the colon.
Figure 4. GSDMD-dependent release of polyubiquitinated IL-1β from colonic IECs.
Representative Western blots of (A) colon explant cultures from untreated (Utx) and DSS-treated (day 9) Gsdmd+/– and Gsdmd–/– mice, and of (B–E) WT and GSDMD-deficient (Gsdmd–/–) YAMC cells; mature (m)IL-1β, full-length (FL) GSDMD, and ubiquitin (Ub). Supernatants were subjected to IP with anti–IL-1β and probed with anti–IL-1β (B) (WCLs were directly analyzed), or subjected to IP with anti–IL-1β under nondenaturing and denaturing conditions and probed with anti-Ub (C). (D) WCLs were directly analyzed. (E) Total protein was analyzed in supernatants after extraction (WCLs were directly analyzed). All experiments were repeated 3 times and yielded consistent results.
GSDMD mediates the release of polyubiquitinated IL-1β via a caspase-8–dependent, nonpyroptotic pathway in colonic IECs.
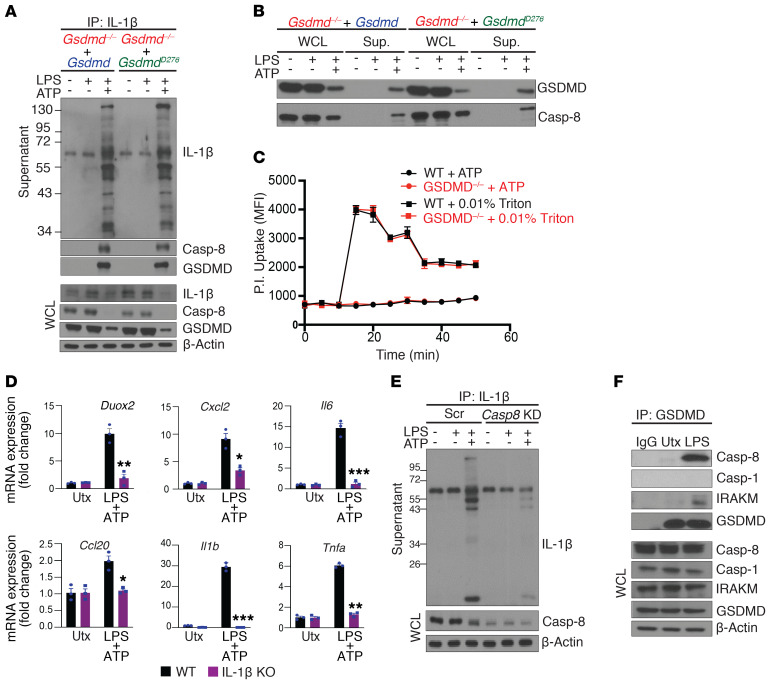
Intrigued by the ex vivo observations, we asked whether GSDMD mediates a previously undocumented mechanism of IL-1β secretion that ultimately promotes intestinal inflammation. While myeloid cells are known to be a critical source of IL-1β (3), a recent study showed that LPS can also induce IL-1β and GSDMD expression in colonic IECs (45). Noting that epithelial-derived GSDMD is highly induced in both IBD patients with active disease (Figures 2 and 3) and in DSS colitic mice (Figure 1G), we generated GSDMD-deficient(Gsdmd–/–) young adult mice colonic (YAMC) epithelial cells as an in vitro model system. In response to activation of the NLRP3 inflammasome (LPS priming followed by ATP stimulation), highly modified pro–IL-1β and mature IL-1β were released by WT, but not Gsdmd–/– YAMC cells (Figure 4B). By performing IP under denaturing conditions, we confirmed that the modified pro–IL-1β proteins from YAMC cells were also conjugated with polyubiquitin chains (Figure 4C), recapitulating our observations in inflamed colon explants (Figure 4A).
Pyroptotic activation of GSDMD is triggered via cleavage at D276 located in its linker region by either caspase-1 or -8 in response to inflammasome assembly (14). Surprisingly, LPS+ATP did not induce the activation of either caspase-1 (p20 fragment), caspase-8 (p18 fragment), or GSDMD cleavage (p30 fragment) in YAMC cells (Figure 4, D and E). Instead, full-length GSDMD was released along with caspase-8 (both full-length and cleaved forms), but not caspase-1, into supernatants upon stimulation (Figure 4, D and E). The secretion of caspase-8, like polyubiquitinated pro–IL-1β, was also dependent on GSDMD (Figure 4, D and E). Consistently, both WT and the D276A-GSDMD mutant that is resistant to proteolytic activation (GsdmdD276A) restored secretion of highly modified IL-1β in a complex with GSDMD from Gsdmd–/– YAMC cells (Figure 5, A and B). Of note, co-IP suggests that caspase-8 is released in a complex with highly modified pro–IL-1β. Although we did not detect any activated, intracellular caspase-8, the enzymatically active caspase-8 (p18) was readily found in supernatants from LPS+ATP treated cells (Figure 4E).
Figure 5. Polyubiquitinated IL-1β from colonic IECs is released via a caspase-8–dependent nonlytic pathway.
(A and B) Gsdmd–/– YAMC cells were stably restored with either WT or D276A GSDMD (GsdmdD276A), and stimulated as indicated. (A) Supernatants from stimulated cells were subjected to IP with anti–IL-1β (WCLs were directly analyzed). (B) Total protein in cell supernatants was extracted and analyzed, alongside WCLs. (C) Pore-forming ability was assessed by kinetic analysis of propidium iodide (PI) uptake in LPS- and ATP-stimulated WT and Gsdmd–/– YAMC cells, with triton treatment used as a positive control. Error bars are shown as SEM of technical replicates (n = 3). (D) IL-1β–deficient (IL-1β–KO) and WT control YAMC cells were left untreated or stimulated with LPS followed by ATP, and analyzed for indicated genes. (E) WT YAMC cells transduced with scramble (Scr) or caspase-8 targeting (Casp8 KD) shRNA were subjected to co-IP with anti–IL-1β. Precipitated supernatant proteins and WCLs were analyzed by Western blot. (F) WT YAMC cells were subjected to co-IP with anti-GSDMD and Western analysis as indicated. All experiments were repeated 3 times and yielded consistent results. All error bars show SEM with *P < 0.05, **P < 0.01, ***P < 0.001 by 2-tailed unpaired Student’s t test.
Furthermore, propidium iodide (PI) uptake indicated that LPS+ATP did not induce GSDMD-dependent pyroptotic pores in YAMC cells (Figure 5C). In fact, nonlytic IL-1β secretion is now appreciated as having distinct physiological significance from pyroptotic IL-1β release because the source of IL-1β remains viable and continues to participate in the inflammatory response (23, 26). Indeed, we found that LPS + ATP treatment induces the production of IL-1β target genes (45) in an IL-1β–dependent manner in YAMC cells (Figure 5D). Taken together, these data suggest that the GSDMD axis mediates the release of polyubiquitinated IL-1β via a nonpyroptotic pathway, specifically in colonic epithelial cells.
Knockdown of Casp8 greatly attenuated LPS+ATP-induced release of modified pro–IL-1β, mature IL-1β (p17), and GSDMD (Figure 5E), suggesting that caspase-8–mediated processing of IL-1β might take place within the extracellular space. We previously reported that LPS stimulation can specifically engage caspase-8 by instigating its recruitment to the receptor proximal kinase, IRAKM (46). In support of this concept, co-IP showed that GSDMD formed a complex with caspase-8 and IRAKM, but not with caspase-1, in response to LPS stimulation of YAMC cells (Figure 5F). These data imply that IRAKM-dependent caspase-8 engagement occurs in colonic epithelial cells and may underlie the selective involvement of caspase-8 inflammasome activity.
A chaperoned pool of GSDMD guides assembly of a secretory complex required for nonpyroptotic IL-1β release.
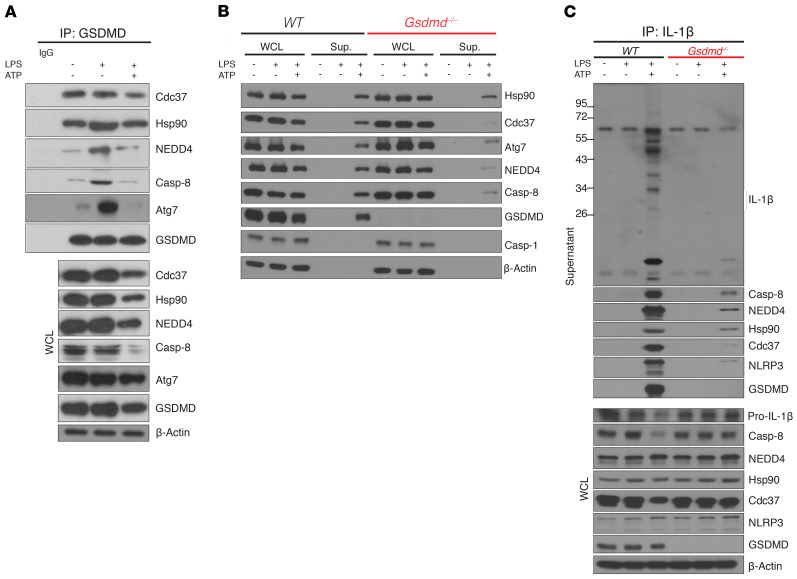
To delineate the pathway of GSDMD-dependent nonpyroptotic release of IL-1β, we performed co-IP of GSDMD from LPS-treated YAMC cell lysates followed by mass spectrometry, which led to the identification of 2 previously unknown GSDMD-interacting proteins, Cdc37 and NEDD4 (Figure 6A and Supplemental Table 2). Cdc37 is a cochaperone of Hsp90 (47–51), a known player in assisting IL-1β loading into membrane vesicles (39), whereas NEDD4 is an E3 ligase that participates in the cargo loading, formation, and trafficking of membrane vesicles, such as exosomes (52–56). By co-IP, we found that full-length GSDMD formed a constitutive complex with Cdc37 and its partner protein, Hsp90 (Figure 6A), whereas NEDD4 was recruited to GSDMD upon LPS stimulation (Figure 6A). Importantly, these newly identified GSDMD-interacting partners were all released into the supernatant upon LPS+ATP stimulation in a GSDMD-dependent manner (Figure 6, A–C). Furthermore, co-IP experiments showed that these GSDMD-interacting partners, including NEDD4, Cdc37, and Hsp90, were present in a secretory complex with modified pro–IL-1β and caspase-8–NLRP3 inflammasome (Figure 6C). Taken together, these data suggest a chaperoned pool of GSDMD that recruits NEDD4 to mediate nonpyroptotic release of IL-1β.
Figure 6. Nonlytic release of IL-1β from IEC requires assembly of a secretory complex guided by chaperoned pool of GSDMD.
Representative Western blots of (A) lysates from unstimulated, LPS-treated, and LPS+ATP-stimulated WT YAMC cells subjected to IP with anti-GSDMD and probed for indicated proteins, (B) WT and Gsdmd–/– YAMC cells either unstimulated or LPS/LPS+ATP-stimulated, with WCL and protein extracts from supernatants probed for indicated proteins, and (C) supernatants from WT and Gsdmd–/– YAMC cells with indicated treatments subjected to IP with anti–IL-1β, and probed for indicated proteins. All experiments were repeated at least 3 times and yielded consistent results.
NEDD4 catalyzes polyubiquitination of pro–IL-1β to facilitate its secretion through a Casp8-dependent GSDMD-guided nonpyroptotic pathway.
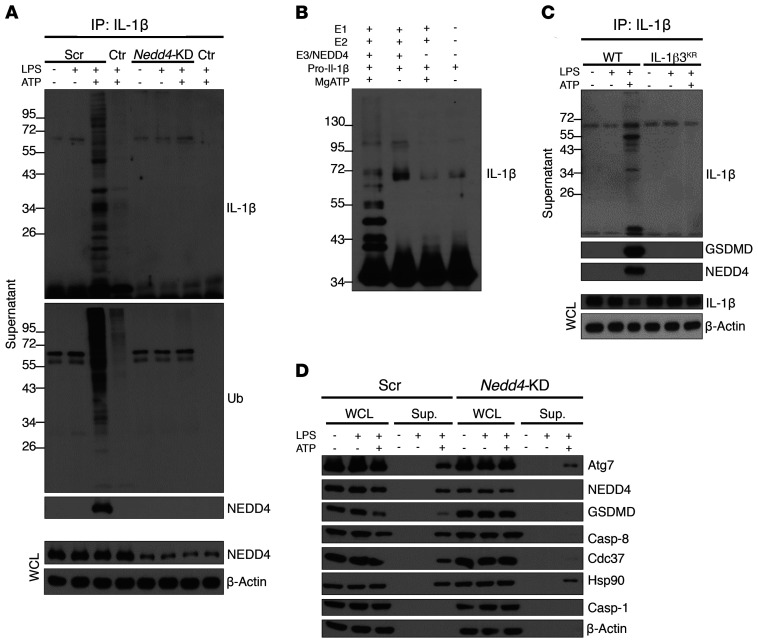
Knockdown of Nedd4 suppressed the release of polyubiquitinated IL-1β in response to LPS+ATP stimulation (Figure 7A), indicating a fundamental role for NEDD4 in LPS+ATP-induced secretion of IL-1β from YAMC cells. Given the reported function of NEDD4/E3-mediated ubiquitination in exosome cargo loading, we hypothesized that NEDD4 may facilitate GSDMD-guided pro–IL-1β secretion by catalyzing the ubiquitination of pro–IL-1β. Indeed, in vitro ubiquitination assays showed that NEDD4/E3 efficiently catalyzed the ubiquitination of pro–IL-1β (Figure 7B). Next, we sought to determine the impact of NEDD4-catalyzed polyubiquitination of pro–IL-1β on its secretion. Using mass spectrometry, we identified 3 lysine residues in pro–IL-1β that were conjugated to polyubiquitin chains by NEDD4 in vitro (Supplemental Table 3). Mutation of these ubiquitination sites (K132/K59/K79 to arginines, IL1b3KR) inhibited the release of the IL-1β–containing secretory complex, comprised of GSDMD and NEDD4, in response to LPS+ATP (Figure 7C). In line with these findings, knockdown of Nedd4 substantially reduced the release of components comprising the GSDMD-containing secretory complex (Figure 7D). Taken together, these data demonstrate that NEDD4-mediated polyubiquitination of pro–IL-1β is required for release of the secretory complex comprised of pro–IL-1β, GSDMD, NEDD4, and caspase-8.
Figure 7. Polyubiquitination of pro–IL-1β by NEDD4 facilitates its secretion via caspase-8–dependent GSDMD-guided nonlytic pathway.
YAMC cells were lentivirally transduced with either scramble (Scr) or NEDD4-targeting shRNA (Nedd4-KD), and stable clones treated as indicated; cells with no guide RNA were used as controls (Ctr). Representative Western blots of (A) total protein from supernatants pulled down with anti–IL-1β, and (B) in vitro ubiquitination assays with recombinant human proteins E1 (Ube1), E2 (UbcH5c/Ube2D3), E3 (NEDD4), and IL-1β as a substrate followed by mass spectometry analysis (Supplemental Table 3). (C) IL-1β–deficient YAMC cells were restored with either WT or mutant (IL-1β3KA) cDNA. Cells were treated as indicated, and total protein from supernatants was directly analyzed or subjected to IL-1β IP followed by Western blotting. (D) WCLs and protein extracts from supernatants were probed for indicated proteins. Except for mass spectrometry analysis, all experiments were repeated 3 times and yielded consistent results.
GSDMD promotes NEDD4-dependent biogenesis and release of extracellular vesicles for IL-1β secretion.
Previous studies have shown that IL-1β can be secreted as cargo within extracellular vesicles (EVs) in response to inflammasome activation (32, 33), and that NEDD4-mediated polyubiquitination can promote the inclusion of cytosolic protein into membrane structures, such as exosomes for extracellular release. Thus, we tested whether GSDMD/NEDD4 mediates IL-1β secretion via biogenesis and release of EVs. We indeed found that LPS+ATP stimulation induced a robust GSDMD-dependent release of sEVs (<200 nm) (Figure 8, A and B), which was confirmed by electron microscopy (Figure 8, C and D). Western blots of the validated sEVs fractions detected modified and mature IL-1β, GSDMD, caspase-8, NEDD4, and CD63 (Figure 8E), confirming these proteins as cargo components of GSDMD-dependent EVs (37, 57). Corroborating these findings, proteomic profiling of IL-1β immunoprecipitates not only validated the association of the GSDMD-complex (GSDMD/CDC37/Hsp90, NEDD4, and Casp8) with IL-1β, but also identified a set of proteins known to participate in IL-1β secretion or exosome formation, including the SNAREprotein, Sec22b (implicated in IL-1β secretion) (37), autophagy proteins (Atg3/7) (58, 59), SNARE-binding proteins (Stxbp2 and Stxbp3) (57), as well as components of ESCRT (60) (endosomal sorting complexes required for transport), ALIX, Vps28, and Vps4b (Supplemental Tables 2 and 4).
Figure 8. Characterization of GSDMD-associated proteins released from isolated IEC-derived sEVs.
WT and Gsdmd–/– YAMC cells were left untreated or stimulated with LPS+ATP. (A) Total concentrations of particles (15-285 nm), and (B) average diameters of particles, in indicated supernatants. Adjusted P values were calculated by Turkey’s multiple comparisons test (n = 3). (C) Representative images of transmission electron microscopic scanning of counterstained EVs from supernatants of untreated and LPS+ATP-treated cells, with (D) examples of EVs of varying sizes. Scale bars: 100 nM. (E) Representative Western blots of EVs from indicated supernatants after enrichment by size exclusion column-based fractionation. All experiments were repeated 3 times and yielded consistent results. Electron microscopy was performed on samples from 2 independent repeats, which generated vesicles of consistent morphology and range of diameters.
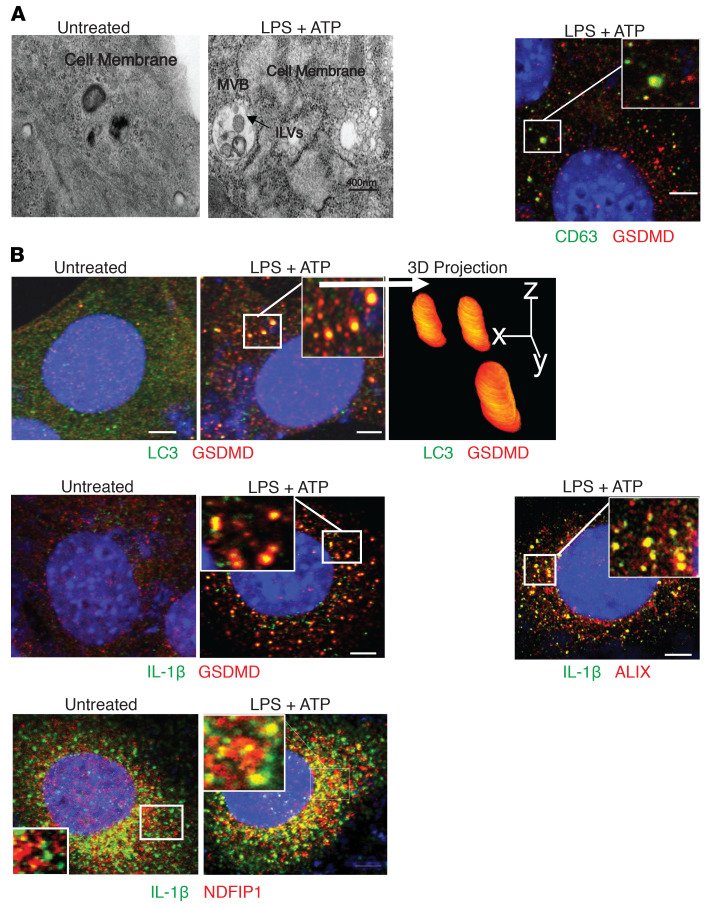
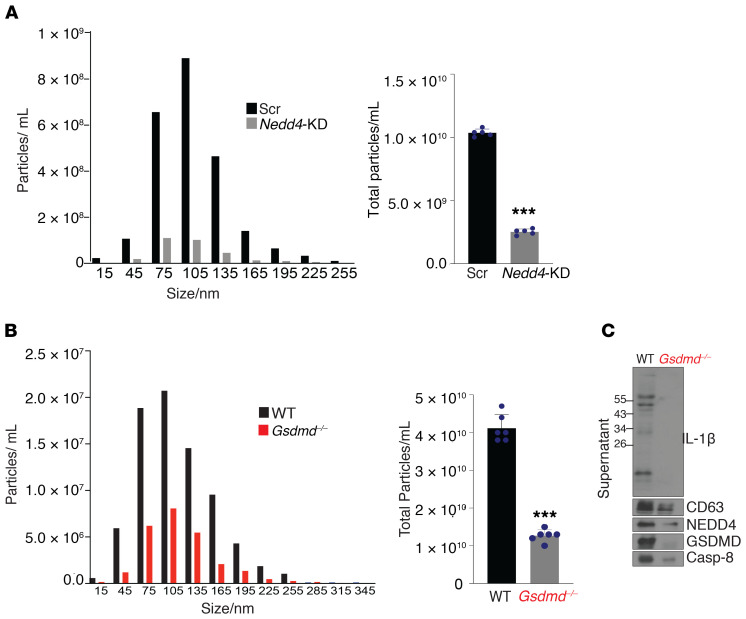
We then performed image analysis to determine the localization of GSDMD, NEDD4, and IL-1β in relation to known subcellular structures involved in the biogenesis of EVs. Transmission electron microscopy suggested that LPS+ATP treatment induced the generation of multivesicular bodies (MVBs) containing intraluminal vesicles (ILVs) (Figure 9A), which are known to give rise to exosomes when released into the extracellular space. Confocal analysis, indeed, revealed a high level of colocalization between GSDMD and CD63, a marker for MVBs, as well as exosomes (Figure 9A). Notably, crystallography studies show that NEDD4 binds to LC3 through a conserved LC3-binding motif (W266/I268/I269) (61, 62), which promotes NEDD4 E3 ligase activity. Interestingly, LPS+ATP stimulation induced the redistribution of GSDMD to the LC3+ membrane structure (Figure 9B). In addition, GSDMD also colocalized with IL-1β, forming punctate structures similar to that observed with GSDMD-LC3 upon LPS+ATP stimulation in YAMC cells (Figure 9B). Furthermore, NEDD4 family interacting protein (NDFIP1), an adaptor that mediates the interaction between NEDD4 and its substrate, was also found in similar structures with IL-1β in response to LPS+ATP (Figure 9B). Importantly, NDFIP1 was shown to promote release of NEDD4 substrates into EVs that contain ALIX, a marker for small EVs, such as exosomes (53, 54). IL-1β also colocalized with ALIX in LPS+ATP-treated cells (Figure 9B). Collectively, these data support a role for GSDMD/NEDD4-mediated biogenesis and release of EVs in mediating IL-1β release from colonic IECs. In further support of this concept, NEDD4 was required for LPS+ATP-induced release of sEVs (<200 nm) (Figure 10A). Remarkably, GSDMD-dependent release of CD63 positive sEVs was also detected from colon explants from DSS-colitic WT and Gsdmd–/– mice (Figure 10, B and C) that contain polyubiquitinated IL-1β, GSDMD, caspase-8, and NEDD4 (Figure 10C). These data suggest that GSDMD-guided secretion of IL-1β likely requires vesicle biogenesis, and results in the release of a GSDMD/NEDD4/IL-1β complex via sEVs.
Figure 9. GSDMD-guided secretion of IL-1β requires vesicle biogenesis.
Representative photomicrographs of WT YAMC cells untreated or treated with LPS+ATP imaged by (A) transmission electron microscopy (left), and by confocal microscopy after staining for (B) CD63 (green) and GSDMD (red) (right), and (B) for indicated proteins, with 3D projections based on Z stack scanning of 100 planes of boxed region. All immunofluorescence imaging was repeated over 3 biological replicates in 3 independent experiments, which yielded consistent pattern of staining. Electron microscopy was performed on samples from 2 independent repeats. Confocal microscopy was performed 3 times and revealed consistent morphology. Scale bars: 10 µM (A), 15 µM (B).
Figure 10. GSDMD and NEDD4 mediate the release of a GSDMD/NEDD4/IL-1β complex via sEVs.
(A) YAMC cells transduced with either scramble shRNA (Scr) or shRNA targeting Nedd4 (Nedd4-KD) were stimulated with LPS, and supernatants were concentrated and subjected to size-exclusion columns to isolate exosomes, followed by nanoparticle tracking analysis with ZetaView for size distribution and total particle concentration (n = 5). (B) Supernatants from cultured colon explants from DSS-treated (day 9) WT and Gsdmd–/– mice were fractioned by size-exclusion columns to enrich for EVs followed by nanoparticle tracking analysis for size distribution and total particle concentration (n = 6), and evaluated for (C) indicated proteins, as shown by representative Western blots. ***P < 0.0001 by 2-tailed unpaired Student’s t test. All experiments were repeated 3 times and yielded consistent results.
Discussion
Herein, we report a novel nonpyroptotic role of GSDMD in guiding release of IL-1β–containing vesicles from IECs in response to caspase-8 inflammasome activation. Interestingly, while this GSDMD-guided secretory pathway led to pro–IL-1β polyubiquitination, NEDD4, known for promoting cargo loading into secretory vesicles (52–56, 63), also catalyzed the polyubiquitination of IL-1β. Previous studies, in fact, have described NEDD4’s role in EV generation for exporting ubiquitinated target proteins (52–56, 63), and NEDD4 itself has been detected in EVs (53). In line with these findings, we found that an IL-1β ubiquitination mutant failed to be released from activated IECs, but detected GSDMD-dependent release of sEVs that contained chaperoned-GSDMD/NEDD4/caspase-8/IL-1β, together indicating a novel, nonpyroptotic role for GSDMD in IL-1β secretion from nonmyeloid cells.
The impact of GSDMD-guided IL-1β secretion from colonic IECs is reminiscent of the autocrine IL-1β function of TH17 and microglia cells that we previously reported (46, 64). In all cases, a caspase-8–dependent pathway is initiated, in lieu of caspase-1 inflammasome activation, with resulting IL-1β serving as a positive feedback signal to promote cell survival/proliferation and inflammation (46, 64). In fact, we found that GSDMD-guided IL-1β secretion also operates in TH17 cells (Supplemental Figure 2A). Moreover, while GSDMD-LC3 colocalization was observed in ATP-treated TH17 cells (Supplemental Figure 2C), CD3+ATP induced robust exosome production from polarized TH17 cells, which was substantially impaired by Gsdmd-deficiency (Supplemental Figure 2B). Importantly, Gsdmd-deficiency reduced CD3+ATP-induced expression of Ifng, Csf2, and Tnfa, which collectively signals the transition to inflammatory TH17 cells that drive intestinal inflammation (Supplemental Figure 2D). Using a T cell–dependent mouse model of colitis, we found that donor T cells derived from either Asc- or Gsdmd-deficient versus WT mice exhibited reduced colitigenicity when transferred into recipients (Supplemental Figure 2, E–H), which was associated with a reduction in inflammatory CD4+ T cells (Supplemental Figure 2, I and J). Together, these data suggest that GSDMD-guided IL-1β secretion from TH17 cells, similar to IECs, can also enable autocrine IL-1β signaling, in this case for T cell–mediated colitis.
Of note, unlike pyroptosis-mediated IL-1β release, we did not detect generation of the pyroptotic-inducing GSDMD fragment in YAMC cells. Instead, chaperoned full-length GSDMD formed a caspase-8–containing secretory complex with IL-1β to guide its export. Although caspase-8 was not initially identified as a pyroptotic caspase, recent studies have shown that it is also competent at cleaving GSDMD (18, 19); thus, a key question is how GSDMD evades cleavage by caspase-8. Interestingly, GSDMD was constitutively chaperoned by Hsp90-Cdc37 complexes in IECs. Previous studies have reported that GSGSFG, a glycine-rich motif, is required for the stable client protein’s interaction with Cdc37 (65), followed by interaction with Hsp90 to form a ternary complex (47, 48, 50, 51). There is, indeed, a GSGSFG-like glycine-rich motif in the N-terminal region of GSDMD. Based on structural modeling, we posit that cleavage at D276 might be masked by the GSDMD/Cdc73/Hsp90 complex in IECs, as well as TH17 cells, preventing caspases from accessing this site. Future mutagenesis studies of these glycine-rich motifs will be required to test this possibility. Notably, fully activated caspase-8 (p18) was only detected in supernatants, but not in cell lysates, in response to stimulation. These data suggest that activation of caspase-8 occurs in a compartmentalized fashion, simultaneously with the export of polyubiquitinated pro–IL-1β. GSDMD-guided secretion of sEVs containing polyubiquitinated pro–IL-1β complexed with caspase-8 may allow processing of IL-1β in the extracellular space. We detected mature IL-1β in the sEVs.
We also identified NEDD4 as a novel GSDMD-interacting protein. Crystal structure revealed that NEDD4 can be activated by interacting with LC3 (62). Thus, assembly of chaperoned GSDMD with NEDD4 may direct its localization to LC3-II+ membranes, leading to its activation. In fact, our imaging studies showed that GSDMD, NEDD4, and IL-1β were associated with LC3-II+ vesicles upon stimulation, which is consistent with previous studies of IL-1β release by secretory autophagy via LC3+ IL-1β–carrying vesicles (37, 39). Biogenesis of LC3-II+ membranes is dependent on Atg3/7-mediated lipidation of LC3, which is traditionally used as a marker for degradative autophagy. One key question is how GSDMD/NEDD4-IL-1β/IL-18–containing LC3-II+ vesicles are directed away from degradative autophagy. Recent studies have shown that IL-1β is recognized by TRIM16, which interacts with the SNARE protein Sec22b, and fused with LC3-II+ vesicles (Sec22b+LC3+). Sec22b (in LC3-II+IL-1β+ membranes), in turn, engages plasma membrane syntaxin-3 and syntaxin-4 for IL-1β secretion, but not syntaxin-17, a SNARE-promoting autophagosome–lysosome fusion (for degradation) (37, 57). We found Sec22b, several SNARE-interacting proteins (stxbp2/3), and Atg3/7 in our mass spectometry of IL-1β pull downs. These data collectively point to a critical role for GSDMD and NEDD4 in loading IL-1β as cargo proteins in vesicles destined for extracellular release (Supplemental Figure 3).
Alternatively, NEDD4 has previously been implicated in EV biogenesis (exosome generation) for exporting its ubiquitinated target proteins (52–55). IL-1β–containing exosomes have been reported to be released via MVB fusion with the plasma membrane (32). Our data show that, while activation of the NLPR3 inflammasome induced formation of MVBs with ILVs, GSDMD and NEDD4 mediated release of EVs (<200 nm) that contained polyubiquitinated IL-1β from IECs and TH17 cells. Notably, polyubiquitin chains function as a marker that triggers the ESCRT machinery to sort and load ubiquitinated proteins into ILVs through invagination of CD63+ MVBs (60). We found that mutagenesis abolishing NEDD4-mediated polyubiquitination of pro–IL-1β impaired the release of IL-1β and GSDMD-containing secretory complexes, suggesting that NEDD4 may facilitate the loading of pro–IL-1β into ILVs through the ESCRT system. In support of this, we detected several ESCRT proteins, including ALIX, Vps28, and Vps4b, by proteomic profiling of protein coimmunoprecipitates of IL-1β. Collectively, these data suggested that GSDMD mediates NEDD4-dependent biogenesis of sEVs for IL-1β export using select autophagy components, including LC3+ vesicles, supporting the interplay between autophagy and exosome biogenesis.
It has been well-documented that IL-1β can be released in the absence of cell lysis via various mechanisms (23–28), including IL-1β accumulation within membrane vesicles (exosomes and/or secretory autophagy) for export to the extracellular space (29–39). A growing body of literature has demonstrated GSDMD-dependent pyroptosis-independent release of IL-1β in response to caspase-1 inflammasome activation in a variety of cell types, including neutrophils, hyperactivated macrophages, dendritic cells, microglia, and human monocytes (23–25). Based on our data, we propose a two-step activation model for GSDMD-guided IL-1β release from IECs: LPS stimulation or TCR activation (stimulus 1) engages chaperoned GSDMD and recruits NEDD4 to caspase-8/receptor complexes. In response to ATP stimulation (stimulus 2), the GSDMD-NEDD4 complex is assembled onto the NLRP3/Casp8 inflammasome and pro–IL-1β is subsequently ubiquitinated by NEDD4, resulting in loading of the whole complex into LC3-II+ vesicles and release into the extracellular space for pro–IL-1β processing. With emerging studies also highlighting the potential involvement of GSDMD in intracellular vesicle-sorting machineries, such as ESCRT (25) and Golgi reassembly stacking proteins (GRASPs) (66), future studies are required to examine whether this pathway is operative in other cell types in response to various inflammasomes.
GSDMD-guided IL-1β secretion through exosomes has distinct physiological significance from pyroptotic IL-1β release because producing cells remain viable and continue to participate in the inflammatory response. As a result, NLRP3 inflammasome activation leads to autocrine IL-1β signaling to propagate and amplify the inflammatory response, promoting acquisition of an inflammatory phenotype. Since producing cells can survive inflammasome activation with nonlytic IL-1β release, we posit that the duration of inflammatory responses may be sustained over an extended period of time, which might underlie the pathogenesis of chronic and recurring inflammation in many disease settings, including IBD.
In fact, it has long been established that increased and persistent IL-1β is present in intestinal lesions and mucosal cells from IBD patients (67–69). However, to date, no clinical trials have shown definitive efficacy of IL-1β inhibition in patients with IBD, and no drug specifically targeting IL-1 has been FDA-approved for the treatment of this condition. Although considered a classic proinflammatory cytokine, the role of IL-1β in gut injury and acute colitis is not without controversy, as mice deficient in IL-1β are reported to display more aggressive intestinal inflammation compared with controls, suggesting a potential protective role of IL-1β in this process (70). However, how epithelial-derived and/or pyroptotic versus nonlytic IL-1β release specifically contributes to chronic intestinal inflammation, such as that observed in IBD, is unclear. Our present study indicates that GSDMD-guided IL-1β secretion from IEC may represent an important mechanism by which intestinal inflammation is sustained, as increased GSDMD transcripts are observed in locally inflamed gut mucosal tissues from IBD patients. Single-cell profiling enabled us to determine cell-type specific expression of GSDMD, revealing its upregulation in specific clusters of inflamed IECs, including colonocytes and crypt-top colonocytes. Upregulation of inflammatory pathways, including cytokine production, across these clusters has been previously shown (44) and indicates how GSDMD may actively promote these processes during chronic intestinal inflammation. Interestingly, increased GSDMD was detected in absorptive and secretory progenitor cells characterized by specific upregulation of differentiation and cell-migration pathways in inflamed tissues, suggesting that GSDMD may also modulate the potential role of these subpopulations to repair inflammation-induced damage. However, whether the effects of GSDMD promote or interfere with these particular events remains to be determined. Further studies are also required to dissect the contribution of IL-1β from lytic versus nonlytic release during chronic intestinal inflammation, such as in IBD, as well as in other GI-related inflammatory pathologies. Importantly, our study has identified a set of IL-1β mutations that specifically ablates GSDMD-guided IL-1β secretion, affording genetic tools to evaluate the mechanistic, pathological role of this pathway using knockin mouse models in the future.
Methods
Animals.
Gasdermin D-KO mice were a gift from Vishva M. Dixit (Genetech, South San Francisco, California, USA) and have been described in previous studies (17). Asc–/– mice were generated as described previously (64).
Generation of knockdown, KO, and restored cell lines.
The immortalized mouse colonic epithelial cell line, YAMC, was a gift from Robert H. Whitehead at Vanderbilt University (71). To generate GsdmD- and IL-1β–KO cells, sgRNA sequences from the GeCKO (v2) library were used. Specifically, 5′-AGCATCCTGGCATTCCGAG-3′ was used to knock out GsdmD and 5′-TTATCCCTATATGACTGGGC-3′ was used to knock out IL-1β. These sgRNA sequences were cloned into the pLenti-Crispr v2 vector and transduced into YAMC cells by lentiviral infection, which were dispersed into single-cell suspensions to generate independent clones, validated by Western blot. Control cells were generated by transducing YAMC cells with the same pLenti-Crispr that carried no guide RNA. To generate Nedd4, Atg7, and Casp8 knockdown (KD) cells, control and targeting shRNAs (Nedd4 - ATATTCTGCTACGGATAATTA, Atg7- CCAGCTCTGAACTCAATAATA, Casp8 - TCCTGACTGGCGTGAACTATG, control - CAACAAGATGAAGAGCACCAA) were delivered to YAMC cells by lentiviral infection. Infected cells were cultured in selective medium for 48 hours (Casp8-KD) or sorted for EGFP (Nedd4-KD) or mCherry (Atg7-KD) to enrich for transduced cells before subjected to treatment and analysis. For restoration experiments, HA-tagged WT and 276mutant of GSDMD or WT and ubiquitination mutant of IL-1β were transduced into YAMC cells using lentiviral infection.
Microarray analysis.
R package GEOquery (72) was used to download gene array expression data from the following GSE database accessions: GSE20881, GSE39713 (43), GSE53306 (42), GSE75214 (40), and GSE112366 (41). Ambiguous and control probes were filtered out from each data set (presented as log2M). limma R package (73) was used to fit generalized linear models for each cohort and the empirical Bayes variance moderation method was used to estimate moderated t statistics and P values. Benjamini-Hochberg correction was used to estimate FDR, and q values were estimated using R package qvalue (74, 75).
Single-cell data analysis.
10x Genomics single-cell data were processed as previously described (44). Normalized single-cell expression values were visualized using Seurat (76) package, with data embeddings generated as previously described (44). Pseudo-bulk analyses were carried out as follows: epithelial cell populations were isolated in silico, discarding contaminating immune types, and a pseudo-bulk for each sample was generated by aggregating raw unique molecular identifier counts from selected cellular barcodes for each subpopulation. DESeq2 R package (77) was used to compute library size factors, normalize data, and perform differential expression analysis using negative binomial Wald’s test. Benjamini-Hochberg correction was used to estimate FDR.
Antibodies.
Antibodies were purchased from the following companies: NEDD4 (catalog 2740), caspase-8 (catalog 4790), Atg7 (catalog 8558), Cdc37 (catalog 4793), NLRP3 (catalog 15101), β-actin (catalog 3700) (all from Cell Signaling Technology), GSDMD (ab209845 and ab219800), CD63 (ab217345), ubiquitin (ab7780) (all from Abcam), Hsp90 (catalog sc-13119; Santa Cruz Biotechnology, Inc.), caspase-1 (catalog AG-20B-0042-C100; AdipoGen Life Sciences), IL-1β (catalog AF-401, R&D Systems; catalog 503505, BioLegend), and IL-18 (catalog 210-401-323; Rockland Immunochemicals, Inc.).
Western blotting.
Freshly isolated human IECs were provided by the Biorepository Core of the NIH-funded Cleveland Digestive Diseases Research Core Center (P30 DK097948). In brief, surgically resected colon tissues were obtained from age- and sex-matched CD and UC patients, as well as noninflammatory controls who underwent therapeutic bowel resection for malignant and nonmalignant noninflammatory conditions. IECs from the latter group were derived from healthy margins of diseased/malignant tissues. All specimens were processed as previously described (78) to obtain IEC preparations with less than 1% contaminating lamina propria mononuclear cells, as assessed by staining with a mAb directed against a leukocyte common antigen (CD45).
IEC cell lines were stimulated as indicated and harvested, washed once with cold PBS, and lysed for 30 minutes at 4°C in 1% Triton X-100, 20 mM HEPES (pH 7.4), 150 mM NaCl, 1.5 mM MgCl2, 2 mM EGTA, protease and phosphatase inhibitors (Roche Life Science). Cellular debris were removed by centrifugation at 16,000g for 10 minutes. For immunoblotting, cell extracts were fractionated by SDS-PAGE and transferred to Immobilon-P transfer membranes (Millipore) using a wet transfer apparatus (Bio-Rad Laboratories). IB analysis was performed, and bands were visualized with HRP-coupled goat anti-rabbit, goat anti-mouse, or donkey anti-goat IgG, as appropriate (Rockland Immunochemicals, Inc.), using the ECL Western blotting detection system (GE Healthcare). Semiquantitative analysis was performed by densitometry, calculating the ratio of target protein(s) to GAPDH (Image J version 1.52, NIH software). Protein levels were equilibrated with the Protein Assay Reagent (Bio-Rad Laboratories).
Coimmunoprecipitation.
For co-IPs, cells were harvested, washed once with cold PBS, and lysed in Triton X-100–containing buffer (0.5% Triton X-100, 20 mM HEPES [pH 7.4], 150 mM NaCl, 1.5 mM MgCl2, 2 mM EGTA, protease and phosphatase inhibitors) (Roche Life Science). Cell extracts were incubated with 1 μg of indicated Ab for 2 hours, followed by incubation for 12 hours with 30 μL protein G–Sepharose beads (prewashed and resuspended in lysis buffer at a 1:1 ratio). After incubations, beads were washed 4 times with lysis buffer, separated by SDS-PAGE, and analyzed by immunoblotting. IL-1β IP was performed as previously described (79). In brief, 5-mL supernatants were incubated overnight at 4°C with 5 μg biotinylated hamster monoclonal IgG1, clone B122 (BioLegend). The next day, 25 μL neutravidin agarose (Pierce Biotechnology) was added into each lysate. After 1 hour of incubation at 4°C, agarose resins were centrifuged, washed 3 times in lysis buffer, and heated in 30–40 μL 2X Laemmli buffer before SDS-PAGE.
Protein extracts.
Cell-free supernatants were mixed in a 1:1 ratio with methanol and then mixed with chloroform at a 1:4 ratio (chloroform/starting supernatant volume). Precipitates were collected by centrifugation and washed with methanol followed by centrifugation. Precipitates were dissolved in 4X Laemmli buffer for Western blot analysis.
LC-MS/MS method.
IP and control IgG samples were fractionated on an SDS-PAGE gel and the entire lane was cut into 2 bands, which were subjected to in-gel digestion. Gel bands were divided into small pieces, washed with water, and dehydrated in acetonitrile. Bands were reduced with DTT and alkylated with iodoacetamide before in-gel digestion, achieved by adding 5 μL of 10 ng/μL trypsin in 50 mM ammonium bicarbonate, and incubating overnight at room temperature. Formed peptides were extracted from polyacrylamide into 2 aliquots of 30 μL 50% acetonitrile with 5% formic acid. Extracts were combined and evaporated to less than 10 μL by Speedvac and resuspended in 1% acetic acid to a final volume of approximately 30 μL for LC-MS analysis.
Digested peptides were analyzed on a Thermo Fisher Scientific UltiMate 3000 HPLC system (Thermo Fisher Scientific) interfaced with either a Thermo Fisher Orbitrap Elite or Thermo Fisher Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific). Liquid chromatography was performed before MS/MS analysis for peptide separation. The HPLC column used was a Dionex 15 cm × 75 μm Acclaim Pepmap C18, 2 μm, 100 Å reversed-phase capillary chromatography column. A 5 μL volume of peptide extracts was injected and eluted from the column by a 90-minute acetonitrile/0.1% formic acid gradient at a flow rate of 0.30 μL per minute and introduced to the mass spectrometer source online. Digests were analyzed using data-dependent multitask capability of the instrument acquiring full scan mass spectra on a Fourier Transform (FT) orbitrap analyzer to determine peptide molecular weights and collision-induced dissociation (CID) MS/MS product ion spectra with an ion-trap analyzer to determine the aa sequence in successive instrument scans.
In order to do label-free quantitative and qualitative proteomic analysis, data was searched using X! Tandem (The GPM) and Sequest (bundled into Proteome Discoverer 2.2, Thermo Fisher Scientific). The database used in these searches corresponds to the mouse SwissProtKB database. The parameters used for these searches include enzyme-specific trypsin with a maximum of 2 missed cleavages, carbamidomethyl (C) as a fixed modification, oxidation of methionine and protein N-terminal acetylation as variable modifications, peptide mass tolerance of 10 ppm and fragment ion mass tolerance of 0.6 Da.
Scaffold (version 4.8.9, Proteome Software Inc.) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 0.0% probability by the Peptide Prophet algorithm (80) with Scaffold delta-mass correction. Protein identifications were accepted if they could be established at greater than 99.9% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (81). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Label-free spectral counting was used to determine relative differences in the IgG control and IP samples.
Transmission electron microscopy.
For ultrastructural analysis, isolated exosomes or cells were fixed for 1 hour at room temperature in 2% paraformaldehyde, 2.5% glutaraldehyde (Polysciences), and 0.05% malachite green (MilliporeSigma) in 100 mM sodium cacodylate buffer, pH 7.2. Malachite green was incorporated into the fixative for stabilization of lipid constituents soluble in aqueous glutaraldehyde. Samples were washed in cacodylate buffer and postfixed for 1 hour in 1% osmium tetroxide (Polysciences). Samples were then rinsed extensively in distilled water before en bloc staining for 1 hour with 1% aqueous uranyl acetate (Ted Pella). Following several rinses in distilled water, samples were dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella). Sections 95 nm in thickness were cut with a Leica Ultracut UC7 ultramicrotome (Leica Microsystems), stained with uranyl acetate and lead citrate, and viewed on a Tecnai G2 Spirit BioTWIN Transmission Electron Microscope (FEI Co.) at 60 kV.
In vitro ubiquitination assay.
E1- Ube1 (100 nM), E2- UbcH5c/Ube2D3 (1 μM), E3-NEDD4 (1 μM), 1× E3 ligase reaction buffer, ubiquitin (100 μM), MgATP solution (10 mM) (Boston Biochem, Inc.), and substrate IL-1β (5 μM, Sino Biological, Inc.) were mixed and incubated at 37°C for 1 hour. For negative controls, MgATP solution was replaced with dH2O. The reaction was terminated by addition of an equal volume of 2× SDS-PAGE sample buffer.
Exosomes analysis.
EVs were isolated from culture medium using Size Exclusion Chromatography (qEV, Izon Science) and subjected to ZetaView analysis for quantity and size.
DSS colitis model.
Eight-week-old sex-matched Gsdmd+/– and Gsdmd–/– littermate mice were given 3% DSS (MP Biomedicals, LLC) in drinking water for up to 9 days. Body weight was recorded daily until sacrifice. Different parts of the colon (proximal, middle, and distal) were collected, fixed in 10% formalin overnight, and paraffin embedded. For frozen sections, colon tissues were fixed in 4% paraformaldehyde (PFA) for 4 hours, followed by 15% and 30% sucrose overnight, or until tissue sank, respectively. After sucrose dehydration, tissues were embedded in OCT and snap-frozen in liquid nitrogen for further histology analysis. Histologic scores were evaluated based on H&E sections of colons taken on day 9 of DSS treatment following a previously published scoring system (82).
RNA isolation, purification, and qPCR analysis.
Total RNA was isolated using TRIzol reagent (Invitrogen), according to the manufacturer’s instruction. In brief, YAMC cells or colon tissues were first lysed or homogenized, respectively, in 1 mL TRIzol, followed by addition of 200 μL chloroform. Samples were vortexed and spun down at 12,000g for 10 minutes. Upper aqueous phase was transferred to a new tube and an equal amount of isopropanol was added to precipitate RNA. Samples were then centrifuged at 12,000g for 10 minutes. Supernatants were discarded and resulting pellets washed with 70% ethanol twice and dissolved in RNase-free water. RNA purification was done using lithium chloride as previously described (83, 84). Briefly, RNA solution was mixed with 0.1 volume of 8M lithium chloride (LiCl) and incubated on ice for 2 hours. Samples were centrifuged at 14,000g for 30 minutes. Pellets were dissolved in 200 μL RNase-free water. LiCl purification process was repeated again. A 0.1 volume of 3M sodium acetate (pH 5.2) and 2.0 volume of −20°C prechilled 100% ethanol was added to 1 volume of RNA solution and incubated at −20°C for 30 minutes and spun down. Pellet was washed with 70% ethanol and RNA was dissolved in 50 μL RNase-free water for RT-PCR. All gene expression data were normalized to actin.
T cell transfer colitis.
T cells were collected from WT, Gsdmd–/–, and Asc–/– mice. Lymph nodes and spleens were collected and gently smashed to prepare single-cell suspensions; RBCs were lysed with ACK lysis buffer. T cells were then stained with anti-CD4 and anti-CD45RB antibodies. CD4+ CD45RBhi/+ cells were sorted and injected i.p. at 0.5 million/mouse into Rag1–/– recipient mice. Body weights were recorded once a week until endpoint. Colon weights and lengths were measured. Histologic scores were based on evaluation of H&E staining of colon sections following a previously reported system (85).
Statistics.
Data are expressed as mean ± SEM. Two-tailed unpaired Student’s t test was used to compare the difference of means between 2 groups throughout the manuscript. Turkey’s multiple comparisons test was used to adjust for analysis involving multiple groups. P values were calculated in GraphPad Prism (version 8.1.1), and a value of less than 0.05 was considered significant for all statistical tests.
Study approval.
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the Cleveland Clinic (protocol 2017-1814). Experiments involving patient samples were approved by the Institutional Review Board at Case Western Reserve University and the University Hospitals (protocol 02-10-07). All patients provided written informed consent.
Author contributions
KB conceptualized the study, curated data, engaged in formal analysis, acquired funding, validated, investigated, visualized, developed methodology, and reviewed and edited the finished draft. JZ conceptualized the study, curated data, engaged in formal analysis, validated, investigated, visualized, developed methodology, wrote the original draft, and reviewed and edited the final draft. YL conceptualized the study, ran software, engaged in formal analysis, validated, investigated, visualized, developed methodology, and reviewed and edited the final draft. NR curated data, engaged in formal analysis, and reviewed and edited the final draft. DC curated data, visualized, and reviewed and edited the final draft. AA was responsible for computational analyses. XC, HW, WQ, SS, and FT curated data, engaged in formal analysis, validated, and investigated. WAML curated data, validated, and investigated. BBW curated data, engaged in formal analysis, acquired funding, and developed methodology. ZK provided resources and supervision. GRD conceptualized the study and curated data. KM and FC reviewed and edited the final draft. AS curated data, visualized, and reviewed and edited the final draft. TTP conceptualized the study, curated data, engaged in formal analysis, acquired funding, wrote the original draft, and reviewed and edited the final draft. XL conceptualized the study, curated data, engaged in formal analysis, acquired funding, investigated, developed methodology, wrote the original draft, administered the project, and reviewed and edited the final draft. The authorship order for the co–first authors was assigned randomly.
Supplementary Material
Acknowledgments
This work is supported by grants from the NIH (P01 HL029582 to XL, P01 DK091222 and R01 DK042191 to TTP) and NMSS (RG-1707-28180 to XL), and by grant R01 NS104164 (to ZK). KB is supported by the National Science Centre, Poland (2015/19/B/NZ6/01578). Selected studies were supported by the Cleveland Digestive Diseases Research Core Center (NIH P30 DK097948). The Fusion Lumos and Orbitrap Elite instruments were purchased via NIH shared instrument grants (S10 OD023436 and S10 RR031537, respectively). The funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Version 1. 06/29/2020
Electronic publication
Version 2. 08/03/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(8):4218–4234.https://doi.org/10.1172/JCI138103.
Contributor Information
Katarzyna Bulek, Email: kjbulek@gmail.com.
Junjie Zhao, Email: zhaoj2@ccf.org.
Yun Liao, Email: liaoy@ccf.org.
Nitish Rana, Email: nxr186@case.edu.
Daniele Corridoni, Email: daniele.corridoni@ndm.ox.ac.uk.
Agne Antanaviciute, Email: agne.antanaviciute@ndm.ox.ac.uk.
Xing Chen, Email: chenx@ccf.org.
Han Wang, Email: wangh3@ccf.org.
Wen Qian, Email: qianw@ccf.org.
William A. Miller-Little, Email: millerw@ccf.org.
Shadi Swaidani, Email: swaidas@ccf.org.
Fangqiang Tang, Email: fangqiang.tang@gmail.com.
Zizhen Kang, Email: kangz@ccf.org.
George R. Dubyak, Email: gxd3@case.edu.
Fabio Cominelli, Email: fabio.cominelli@uhhospitals.org.
Alison Simmons, Email: alison.simmons@ndm.ox.ac.uk.
Theresa T. Pizarro, Email: theresa.pizarro@case.edu.
Xiaoxia Li, Email: lix@ccf.org.
References
- 1.de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13(1):13–27. doi: 10.1038/nrgastro.2015.186. [DOI] [PubMed] [Google Scholar]
- 2.Levy M, et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015;163(6):1428–1443. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seo SU, et al. Distinct commensals induce interleukin-1β via NLRP3 inflammasome in inflammatory monocytes to promote intestinal inflammation in response to injury. Immunity. 2015;42(4):744–755. doi: 10.1016/j.immuni.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorbara MT, Girardin SE. Mitochondrial ROS fuel the inflammasome. Cell Res. 2011;21(4):558–560. doi: 10.1038/cr.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477(7366):596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 6.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145(5):745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Opipari A, Franchi L. Role of inflammasomes in intestinal inflammation and Crohn’s disease. Inflamm Bowel Dis. 2015;21(1):173–181. doi: 10.1097/MIB.0000000000000230. [DOI] [PubMed] [Google Scholar]
- 8.Zhu S, et al. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature. 2017;546(7660):667–670. doi: 10.1038/nature22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubartelli A, Cozzolino F, Talio M, Sitia R. A novel secretory pathway for interleukin-1 beta, a protein lacking a signal sequence. EMBO J. 1990;9(5):1503–1510. doi: 10.1002/j.1460-2075.1990.tb08268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black RA, Kronheim SR, Sleath PR. Activation of interleukin-1 beta by a co-induced protease. FEBS Lett. 1989;247(2):386–390. doi: 10.1016/0014-5793(89)81376-6. [DOI] [PubMed] [Google Scholar]
- 11.Dubyak GR. P2X7 receptor regulation of non-classical secretion from immune effector cells. Cell Microbiol. 2012;14(11):1697–1706. doi: 10.1111/cmi.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostura MJ, et al. Identification of a monocyte specific pre-interleukin 1 beta convertase activity. Proc Natl Acad Sci USA. 1989;86(14):5227–5231. doi: 10.1073/pnas.86.14.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowarski R, et al. Epithelial IL-18 equilibrium controls barrier function in colitis. Cell. 2015;163(6):1444–1456. doi: 10.1016/j.cell.2015.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 15.Lopetuso LR, Chowdhry S, Pizarro TT. Opposing functions of classic and novel IL-1 family members in gut health and disease. Front Immunol. 2013;4:181. doi: 10.3389/fimmu.2013.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastorelli L, De Salvo C, Mercado JR, Vecchi M, Pizarro TT. Central role of the gut epithelial barrier in the pathogenesis of chronic intestinal inflammation: lessons learned from animal models and human genetics. Front Immunol. 2013;4:280. doi: 10.3389/fimmu.2013.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kayagaki N, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526(7575):666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 18.Sarhan J, et al. Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc Natl Acad Sci USA. 2018;115(46):E10888–E10897. doi: 10.1073/pnas.1809548115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orning P, et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science. 2018;362(6418):1064–1069. doi: 10.1126/science.aau2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding J, et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535(7610):111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 21.Aglietti RA, et al. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc Natl Acad Sci USA. 2016;113(28):7858–7863. doi: 10.1073/pnas.1607769113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535(7610):153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evavold CL, Ruan J, Tan Y, Xia S, Wu H, Kagan JC. The pore-forming protein gasdermin D regulates interleukin-1 secretion from living macrophages. Immunity. 2018;48(1):35–44.e6. doi: 10.1016/j.immuni.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carty M, et al. Cell survival and cytokine release after inflammasome activation is regulated by the Toll-IL-1R protein SARM. Immunity. 2019;50(6):1412–1424.e6. doi: 10.1016/j.immuni.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Rühl S, Shkarina K, Demarco B, Heilig R, Santos JC, Broz P. ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science. 2018;362(6417):956–960. doi: 10.1126/science.aar7607. [DOI] [PubMed] [Google Scholar]
- 26.Zanoni I, et al. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science. 2016;352(6290):1232–1236. doi: 10.1126/science.aaf3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaidt MM, et al. Human monocytes engage an alternative inflammasome pathway. Immunity. 2016;44(4):833–846. doi: 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Chen KW, et al. The neutrophil NLRC4 inflammasome selectively promotes IL-1β maturation without pyroptosis during acute Salmonella challenge. Cell Rep. 2014;8(2):570–582. doi: 10.1016/j.celrep.2014.06.028. [DOI] [PubMed] [Google Scholar]
- 29.Pizzirani C, et al. Stimulation of P2 receptors causes release of IL-1beta-loaded microvesicles from human dendritic cells. Blood. 2007;109(9):3856–3864. doi: 10.1182/blood-2005-06-031377. [DOI] [PubMed] [Google Scholar]
- 30.Bianco F, et al. Astrocyte-derived ATP induces vesicle shedding and IL-1 beta release from microglia. J Immunol. 2005;174(11):7268–7277. doi: 10.4049/jimmunol.174.11.7268. [DOI] [PubMed] [Google Scholar]
- 31.MacKenzie A, Wilson HL, Kiss-Toth E, Dower SK, North RA, Surprenant A. Rapid secretion of interleukin-1beta by microvesicle shedding. Immunity. 2001;15(5):825–835. doi: 10.1016/S1074-7613(01)00229-1. [DOI] [PubMed] [Google Scholar]
- 32.Qu Y, Franchi L, Nunez G, Dubyak GR. Nonclassical IL-1 beta secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J Immunol. 2007;179(3):1913–1925. doi: 10.4049/jimmunol.179.3.1913. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. Inflammasome-derived exosomes activate NF-κB signaling in macrophages. J Proteome Res. 2017;16(1):170–178. doi: 10.1021/acs.jproteome.6b00599. [DOI] [PubMed] [Google Scholar]
- 34.Carta S, et al. Histone deacetylase inhibitors prevent exocytosis of interleukin-1beta-containing secretory lysosomes: role of microtubules. Blood. 2006;108(5):1618–1626. doi: 10.1182/blood-2006-03-014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrei C, Margiocco P, Poggi A, Lotti LV, Torrisi MR, Rubartelli A. Phospholipases C and A2 control lysosome-mediated IL-1 beta secretion: implications for inflammatory processes. Proc Natl Acad Sci USA. 2004;101(26):9745–9750. doi: 10.1073/pnas.0308558101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Claude-Taupin A, Bissa B, Jia J, Gu Y, Deretic V. Role of autophagy in IL-1β export and release from cells. Semin Cell Dev Biol. 2018;83:36–41. doi: 10.1016/j.semcdb.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 2011;30(23):4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iula L, et al. Autophagy mediates interleukin-1β secretion in human neutrophils. Front Immunol. 2018;9:269. doi: 10.3389/fimmu.2018.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang M, Kenny SJ, Ge L, Xu K, Schekman R. Translocation of interleukin-1β into a vesicle intermediate in autophagy-mediated secretion. Elife. 2015;4:e11205. doi: 10.7554/eLife.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vancamelbeke M, et al. Genetic and transcriptomic bases of intestinal epithelial barrier dysfunction in inflammatory bowel disease. Inflamm Bowel Dis. 2017;23(10):1718–1729. doi: 10.1097/MIB.0000000000001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.VanDussen KL, et al. Abnormal small intestinal epithelial microvilli in patients with Crohn’s disease. Gastroenterology. 2018;155(3):815–828. doi: 10.1053/j.gastro.2018.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao X, et al. Mobilization of epithelial mesenchymal transition genes distinguishes active from inactive lesional tissue in patients with ulcerative colitis. Hum Mol Genet. 2015;24(16):4615–4624. doi: 10.1093/hmg/ddv192. [DOI] [PubMed] [Google Scholar]
- 43.Planell N, et al. Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut. 2013;62(7):967–976. doi: 10.1136/gutjnl-2012-303333. [DOI] [PubMed] [Google Scholar]
- 44.Parikh K, et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019;567(7746):49–55. doi: 10.1038/s41586-019-0992-y. [DOI] [PubMed] [Google Scholar]
- 45.Price AE, et al. A map of Toll-like receptor expression in the intestinal epithelium reveals distinct spatial, cell type-specific, and temporal patterns. Immunity. 2018;49(3):560–575.e6. doi: 10.1016/j.immuni.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang CJ, et al. TLR-stimulated IRAKM activates caspase-8 inflammasome in microglia and promotes neuroinflammation. J Clin Invest. 2018;128(12):5399–5412. doi: 10.1172/JCI121901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eckl JM, Scherr MJ, Freiburger L, Daake MA, Sattler M, Richter K. Hsp90·Cdc37 complexes with protein kinases form cooperatively with multiple distinct interaction sites. J Biol Chem. 2015;290(52):30843–30854. doi: 10.1074/jbc.M115.693150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keramisanou D, et al. Molecular mechanism of protein kinase recognition and sorting by the Hsp90 kinome-specific cochaperone Cdc37. Mol Cell. 2016;62(2):260–271. doi: 10.1016/j.molcel.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taipale M, et al. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150(5):987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaughan CK, et al. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23(5):697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaughan CK, et al. Hsp90-dependent activation of protein kinases is regulated by chaperone-targeted dephosphorylation of Cdc37. Mol Cell. 2008;31(6):886–895. doi: 10.1016/j.molcel.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Putz U, et al. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Sci Signal. 2012;5(243):ra70. doi: 10.1126/scisignal.2003084. [DOI] [PubMed] [Google Scholar]
- 53.Putz U, et al. Nedd4 family-interacting protein 1 (Ndfip1) is required for the exosomal secretion of Nedd4 family proteins. J Biol Chem. 2008;283(47):32621–32627. doi: 10.1074/jbc.M804120200. [DOI] [PubMed] [Google Scholar]
- 54.Segura-Morales C, Pescia C, Chatellard-Causse C, Sadoul R, Bertrand E, Basyuk E. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J Biol Chem. 2005;280(29):27004–27012. doi: 10.1074/jbc.M413735200. [DOI] [PubMed] [Google Scholar]
- 55.Blot V, et al. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J Cell Sci. 2004;117(Pt 11):2357–2367. doi: 10.1242/jcs.01095. [DOI] [PubMed] [Google Scholar]
- 56.Rotin D, Staub O, Haguenauer-Tsapis R. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J Membr Biol. 2000;176(1):1–17. doi: 10.1007/s00232001079. [DOI] [PubMed] [Google Scholar]
- 57.Kimura T, et al. Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 2017;36(1):42–60. doi: 10.15252/embj.201695081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408(6811):488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 59.Frudd K, Burgoyne T, Burgoyne JR. Oxidation of Atg3 and Atg7 mediates inhibition of autophagy. Nat Commun. 2018;9(1):95. doi: 10.1038/s41467-017-02352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Christ L, Raiborg C, Wenzel EM, Campsteijn C, Stenmark H. Cellular functions and molecular mechanisms of the ESCRT Membrane-scission machinery. Trends Biochem Sci. 2017;42(1):42–56. doi: 10.1016/j.tibs.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 61.Sun A, et al. The E3 ubiquitin ligase NEDD4 is an LC3-interactive protein and regulates autophagy. Autophagy. 2017;13(3):522–537. doi: 10.1080/15548627.2016.1268301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qiu Y, Zheng Y, Wu KP, Schulman BA. Insights into links between autophagy and the ubiquitin system from the structure of LC3B bound to the LIR motif from the E3 ligase NEDD4. Protein Sci. 2017;26(8):1674–1680. doi: 10.1002/pro.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sterzenbach U, Putz U, Low LH, Silke J, Tan SS, Howitt J. Engineered exosomes as vehicles for biologically active proteins. Mol Ther. 2017;25(6):1269–1278. doi: 10.1016/j.ymthe.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin BN, et al. T cell-intrinsic ASC critically promotes T(H)17-mediated experimental autoimmune encephalomyelitis. Nat Immunol. 2016;17(5):583–592. doi: 10.1038/ni.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Terasawa K, Yoshimatsu K, Iemura S, Natsume T, Tanaka K, Minami Y. Cdc37 interacts with the glycine-rich loop of Hsp90 client kinases. Mol Cell Biol. 2006;26(9):3378–3389. doi: 10.1128/MCB.26.9.3378-3389.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chiritoiu M, Brouwers N, Turacchio G, Pirozzi M, Malhotra V. GRASP55 and UPR control interleukin-1β aggregation and secretion. Dev Cell. 2019;49(1):145–155.e4. doi: 10.1016/j.devcel.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 67.Casini-Raggi V, Kam L, Chong YJ, Fiocchi C, Pizarro TT, Cominelli F. Mucosal imbalance of IL-1 and IL-1 receptor antagonist in inflammatory bowel disease. A novel mechanism of chronic intestinal inflammation. J Immunol. 1995;154(5):2434–2440. [PubMed] [Google Scholar]
- 68.Ligumsky M, Simon PL, Karmeli F, Rachmilewitz D. Role of interleukin 1 in inflammatory bowel disease--enhanced production during active disease. Gut. 1990;31(6):686–689. doi: 10.1136/gut.31.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Youngman KR, et al. Localization of intestinal interleukin 1 activity and protein and gene expression to lamina propria cells. Gastroenterology. 1993;104(3):749–758. doi: 10.1016/0016-5085(93)91010-F. [DOI] [PubMed] [Google Scholar]
- 70.Bersudsky M, et al. Non-redundant properties of IL-1α and IL-1β during acute colon inflammation in mice. Gut. 2014;63(4):598–609. doi: 10.1136/gutjnl-2012-303329. [DOI] [PubMed] [Google Scholar]
- 71.Whitehead RH, VanEeden PE, Noble MD, Ataliotis P, Jat PS. Establishment of conditionally immortalized epithelial cell lines from both colon and small intestine of adult H-2Kb-tsA58 transgenic mice. Proc Natl Acad Sci USA. 1993;90(2):587–591. doi: 10.1073/pnas.90.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 73.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Storey JD. The positive false discovery rate: a Bayesian interpretation and the q -value. Annals of Statistics. 2003;31(6):2013–2035. doi: 10.1214/aos/1074290335. [DOI] [Google Scholar]
- 75. Storey JD BA, Dabney A, Robinson D. qvalue: Q-value estimation for false discovery rate control. R package version 2.18.0. https://www.bioconductor.org/packages/release/bioc/html/qvalue.html Accessed June 2, 2020.
- 76.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pastorelli L, et al. Epithelial-derived IL-33 and its receptor ST2 are dysregulated in ulcerative colitis and in experimental Th1/Th2 driven enteritis. Proc Natl Acad Sci USA. 2010;107(17):8017–8022. doi: 10.1073/pnas.0912678107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duong BH, et al. A20 restricts ubiquitination of pro-interleukin-1β protein complexes and suppresses NLRP3 inflammasome activity. Immunity. 2015;42(1):55–67. doi: 10.1016/j.immuni.2014.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 81.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 82.Mähler M, et al. Differential susceptibility of inbred mouse strains to dextran sulfate sodium-induced colitis. Am J Physiol. 1998;274(3):G544–G551. doi: 10.1152/ajpgi.1998.274.3.G544. [DOI] [PubMed] [Google Scholar]
- 83.Barlow JJ, Mathias AP, Williamson R, Gammack DB. A simple method for the quantitative isolation of undegraded high molecular weight ribonucleic acid. Biochem Biophys Res Commun. 1963;13:61–66. doi: 10.1016/0006-291X(63)90163-3. [DOI] [PubMed] [Google Scholar]
- 84.Cathala G, et al. A method for isolation of intact, translationally active ribonucleic acid. DNA. 1983;2(4):329–335. doi: 10.1089/dna.1983.2.329. [DOI] [PubMed] [Google Scholar]
- 85.Ostanin DV, et al. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am J Physiol Gastrointest Liver Physiol. 2009;296(2):G135–G146. doi: 10.1152/ajpgi.90462.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.