Abstract
Dominant mutations in the HSP70 cochaperone DNAJB6 cause a late-onset muscle disease termed limb-girdle muscular dystrophy type D1 (LGMDD1), which is characterized by protein aggregation and vacuolar myopathology. Disease mutations reside within the G/F domain of DNAJB6, but the molecular mechanisms underlying dysfunction are not well understood. Using yeast, cell culture, and mouse models of LGMDD1, we found that the toxicity associated with disease-associated DNAJB6 required its interaction with HSP70 and that abrogating this interaction genetically or with small molecules was protective. In skeletal muscle, DNAJB6 localizes to the Z-disc with HSP70. Whereas HSP70 normally diffused rapidly between the Z-disc and sarcoplasm, the rate of diffusion of HSP70 in LGMDD1 mouse muscle was diminished, probably because it had an unusual affinity for the Z-disc and mutant DNAJB6. Treating LGMDD1 mice with a small-molecule inhibitor of the DNAJ-HSP70 complex remobilized HSP70, improved strength, and corrected myopathology. These data support a model in which LGMDD1 mutations in DNAJB6 are a gain-of-function disease that is, counterintuitively, mediated via HSP70 binding. Thus, therapeutic approaches targeting HSP70-DNAJB6 may be effective in treating this inherited muscular dystrophy.
Keywords: Cell Biology, Muscle Biology
Keywords: Chaperones, Protein misfolding, Skeletal muscle
Introduction
Limb-girdle muscular dystrophies (LGMDs) are a family of hereditary muscle diseases that are inherited in an autosomal recessive or dominant manner (1). Most recessively inherited LGMDs are postulated to be caused by a loss-of-function mechanism, because the protein is absent in patients’ muscle tissue, and many mutations lead to premature stops or frameshifts. However, the mechanisms of dominantly inherited LGMDs are less clear: they may be due to haploinsufficiency, a gain of new toxic function, or a dominant interaction leading to protein dysfunction.
Two common pathologic features seen in some dominantly inherited myopathies, including limb-girdle muscular dystrophy type D1 (LGMDD1), myofibrillar myopathy, and inclusion body myopathy, are protein aggregates and myofiber vacuolation (2). The inclusions in LGMDD1 muscle are heterogeneous and contain myofibrillar proteins, such as desmin and myotilin (3, 4). Additionally, they contain proteins, such as TDP43 and SQSTM1, that aggregate in neurodegenerative disorders (3, 4). Together, these features suggest that a subset of inherited myopathies might involve an impairment in protein quality control. Indeed, 1 family of muscle diseases with myofibrillar disorganization (e.g., myofibrillar myopathies) are caused by missense mutations in Z-disc–associated structural proteins, such as desmin, myotilin, or filamin-C, that lead to their aggregation (2). Likewise, myofibrillar myopathies are also associated with mutations in molecular chaperones, such as BAG3, HSPB5, and DNAJB6 (2, 5, 6). These “chaperonopathies” are autosomally dominantly inherited and lead to the accumulation and aggregation of Z-disc proteins, such as desmin and filamin-C. Moreover, several chaperones are known to be necessary for sarcomere development and maintenance (7). Our aim was to better understand the mechanisms associated with dominantly inherited LGMDD1 caused by mutations in DNAJB6 in order to gain insight into the relationship between protein quality control and myopathy.
DNAJB6 is a ubiquitously expressed HSP40 cochaperone that facilitates HSP70 functionality via its J domain (8). DNAJB6, like other DNAJB family members, has a canonical J domain, a C-terminal dimerization motif, and a glycine- and phenylalanine-rich region (termed a G/F domain) (Figure 1A and ref. 8). Interestingly, all reported LGMDD1 mutations reside within the G/F domain (Figure 1A and refs. 3, 4, 9–11). DNAJB6 has 2 isoforms, DNAJB6a and DNAJB6b, which differ in the length of their C-terminal region and in their cellular localization. DNAJB6a is longer and localizes to the nucleus, whereas DNAJB6b is shorter and cytoplasmic (12). Although both isoforms share the G/F domain containing LGMDD1 disease mutations, it is thought that DNAJB6b is the pathogenic isoform (3, 12). Patients with LGMDD1 have an insidious onset of weakness in the second to sixth decades of life that affects their ability to ambulate (3, 4, 9, 10, 13). The muscle tissue of patients with LGMDD1 is characteristically myopathic, with myofibrillar protein aggregates and rimmed vacuoles (3, 4). Myoblasts lacking DNAJB6 accumulate the Z-disc–associated intermediate filament desmin, suggesting that these phenotypes might involve impaired quality control of Z-disc–associated proteins (14). Consistent with this idea, the skeletal muscle from transgenic mice expressing the F93L mutation in DNAJB6b recapitulated features of LGMDD1 with muscle weakness, atrophy, myofibrillar disorganization, and insoluble desmin aggregates (12).
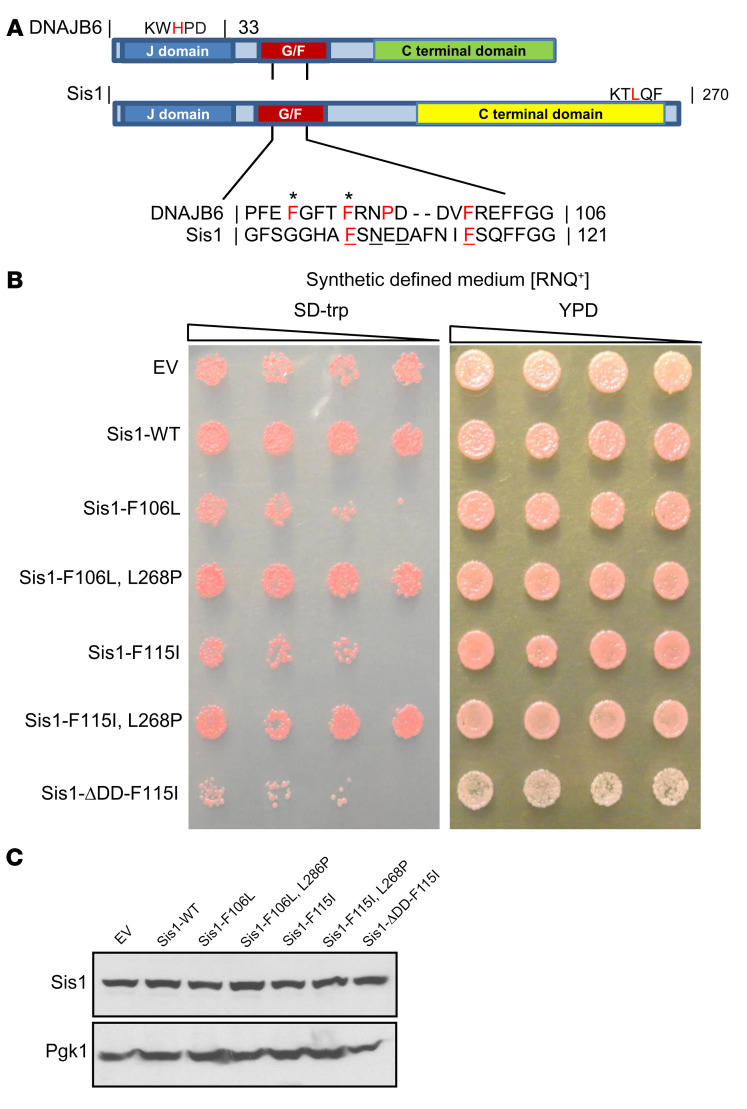
Figure 1. LGMDD1 mutations are toxic in yeast Sis1 and rescued by impairing HSP70 association.
(A) Domain alignment of DNAJB6 and Sis1. Amino acid alignment of the G/F domains denotes sequence homology. Amino acids in bold red represent the mutations used in the study. Amino acids in red represent other disease mutations. Underlined amino acids represent DNAJB6 and Sis1 homologous residues mutated in LGMDD1. The amino acids with asterisks represent residues mutated in our 2 animal models. (B) Yeast cells propagating single-dot medium [RNQ+] were transformed with constructs overexpressing Sis1-WT, Sis1-F106L (DNAJB6-F93L homologous), Sis1-F115I (DNAJB6-F100I homologous), or empty vector control (EV). Yeast were also transformed with Sis1-mutant constructs containing a second mutation that impairs HSP70 association (L268P) or blocks Sis1 dimerization (ΔDD). Cultures were normalized and serially diluted 5-fold and were spotted on medium (SD-trp) to select for the plasmid or medium (yeast extract peptone dextrose [YPD]) that provided no selection. (C) Western blot analysis from lysates of yeast expressing the Sis1 constructs in B. Pgk1 was used as a loading control. Images in A and B are representative of 3 independent experiments.
Yeast has been a powerful model system that has been used to understand the mechanisms of DNAJ proteins, owing to their high conservation. The G/F domain of DNAJB6 is homologous to the specific yeast DNAJ protein Sis1 (15). This is convenient, because, unlike DNAJB6, there are known substrates for Sis1, and the deletion of Sis1 results in strong phenotypes. For example, Sis1 is required for cell viability and is essential for yeast prion propagation (15). Thus, studying the impact of human LGMDD1 mutations in the yeast Sis1 model allows one to form hypotheses about how they might affect DNAJB6 function. Indeed, we previously expressed a chimeric Sis1-DNAJB6, in which the G/F domain of Sis1 was replaced with that of DNAJB6 (15). The yeasts expressing the Sis1-G/F chimera were viable and supported prion propagation, consistent with the homology between these proteins. However, when an LGMDD1 missense mutation within the G/F domain was introduced, prion propagation was impaired, suggesting a partial loss of function (15). This effect on prion propagation appears to be conserved, because DNAJB6b also facilitates the disaggregation of proteins with prion-like, low-complexity sequences such as huntingtin and TDP43 in mammalian cells (15, 16). Moreover, LGMDD1 mutations in DNAJB6 impair this function (15). Together, these observations suggested to us that the Sis1 yeast model might be used to probe the mechanism of LGMDD1 mutations.
Here, we used yeast, mammalian cells, and a new mouse model, combined with chemical probes targeting chaperones, to reveal an unexpected relationship between DNAJB6 mutations and aberrant cycling of HSP70 at the Z-disc. Counterintuitively, we found that inhibiting the interaction of DNAJB6 with HSP70, either genetically or pharmacologically, could partially restore HSP70 mobility and rescue muscle phenotypes in treated mice. These results suggest potential new ways of treating LGMDD1. More broadly, they provide insight into the mechanisms of myopathy-associated chaperonopathies.
Results
To model LGMDD1-mutant dysfunction, we generated homologous DNAJB6 disease–associated mutations in Sis1 (Figure 1A). We found that yeast expressing Sis1-F106L and Sis1-F115I (corresponding to DNAJB6-F93L and F100I) had reduced viability (Figure 1B). This was similar to previous reports demonstrating that deletion of the Sis1 G/F domain dominantly affects yeast viability in the setting of endogenous Sis1 (17). This dominant effect could be rescued by a second cis mutation within the C-terminus, L268P, which was shown to abrogate Ssa1-HSP70 interaction with Sis1 and was in itself not lethal (Supplemental Figure 1, A and B and ref. 18; supplemental material available online with this article; https://doi.org/10.1172/JCI136167DS1). Similarly, we found that combining the L268P mutation with Sis1-F106L or F115I restored yeast viability (Figure 1B). Notably, generation of a second mutation in Sis1-F115I that impaired Sis1 dimerization, ΔDD, did not rescue viability (Figure 1B). These data indicate that the gain of LGMDD1-mutant toxicity occurred via DNAJ-HSP70 interaction and not DNAJ dimerization. Importantly, we confirmed that these effects were not due to destabilization of Sis1 (Figure 1C).
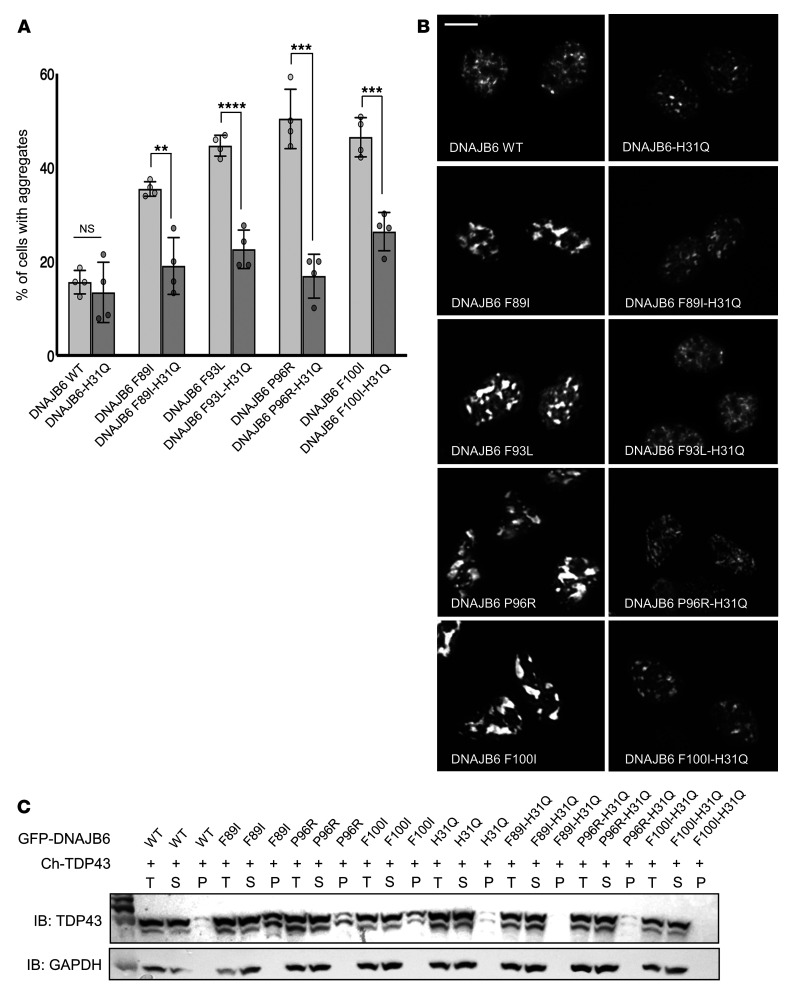
One feature of LGMDD1 muscle pathology is the formation of inclusions composed of RNA-binding proteins, such as TDP43 and hnRNAPA2B1 (4, 11). We have previously demonstrated that expression of LGMDD1-mutant DNAJB6b increases the abundance of TDP43-positive nuclear stress granules following heat shock (15). In light of the results with our LGMDD1 yeast model, we hypothesized that this gain-of-function effect was dependent on HSP70 interaction. To test this, we expressed mCherry-tagged TDP43 with plasmid constructs containing DNAJB6b-WT or LGMDD1-mutant DNAJB6b with or without an additional H31Q mutation that abrogates HSP70 association in HeLa cells (Figure 2, A–C). Although LGMDD1-mutant DNAJB6b expression increased the number of TDP43-positive stress granules after heat shock, addition of the H31Q mutation resulted in TDP43 granules that appeared similar to DNAJB6b-WT expression (Figure 2, A and B). We further tested whether the H31Q mutation ameliorates TDP43 insolubility, as TDP43 stress granules become detergent insoluble in the presence of LGMDD1-mutant DNAJB6b (15). In order to evaluate TDP43 aggregation, we subjected lysates to differential extraction and centrifugation, generating a total lysate fraction (T), a soluble supernatant fraction (S), and an insoluble pelleted fraction (P). Similar to what we observed with fluorescence microscopy, there was an increase in insoluble/pelleted (P) TDP43 following heat shock in cells expressing LGMDD1-mutant DNAJB6b compared with DNAJB6b-WT expression. However, when an H31Q mutation was present with the disease mutation, the increase in pelleted TDP43 was reduced (Figure 2C). In combination with our yeast model, these data provide additional support that the DNAJB6-HSP70 interaction plays a critical role in LGMDD1 pathogenesis.
Figure 2. LGMDD1 mutations in DNAJB6b impair TDP43 disaggregation, which is corrected with a second H31Q mutation.
(A–C) HeLa cells were cotransfected with mCherry-TDP43 and WT or LGMDD1-mutant (F89I, F93L, P96R, or F100I) GFP-DNAJB6b. In some cases, a second mutation in the J domain (H31Q) was also present. Transfected cells were heat shocked at 42°C for 1 hour before analysis. (A) Quantification of the percentage of transfected cells with TDP43 aggregates. **P < 0.005, ***P < 0.0005, and ****P < 0.00005, by paired Student’s t test for comparisons between groups; a 1-way between-conditions ANOVA was performed to compare the effects of mutations. There was a significant effect at P < 0.05 for the 5 conditions [F(4,15) = 54.5, P = 9.3 × 10–9]. n = 300 cells analyzed per condition; the experiment was conducted 3 times. (B) Representative fluorescence images of the mCherry-TDP43–positive nuclei quantified in A. Scale bar: 10 μm. (C) Lysates from cells in A were separated into total (T), soluble (S), and pellet (P) fractions and then subjected to SDS-PAGE and Western blotting using anti-TDP43 and anti-GAPDH antibodies. The immunoblot (IB) is representative of 3 independent experiments. Ch-TDP43, mCherry-tagged TDP43.
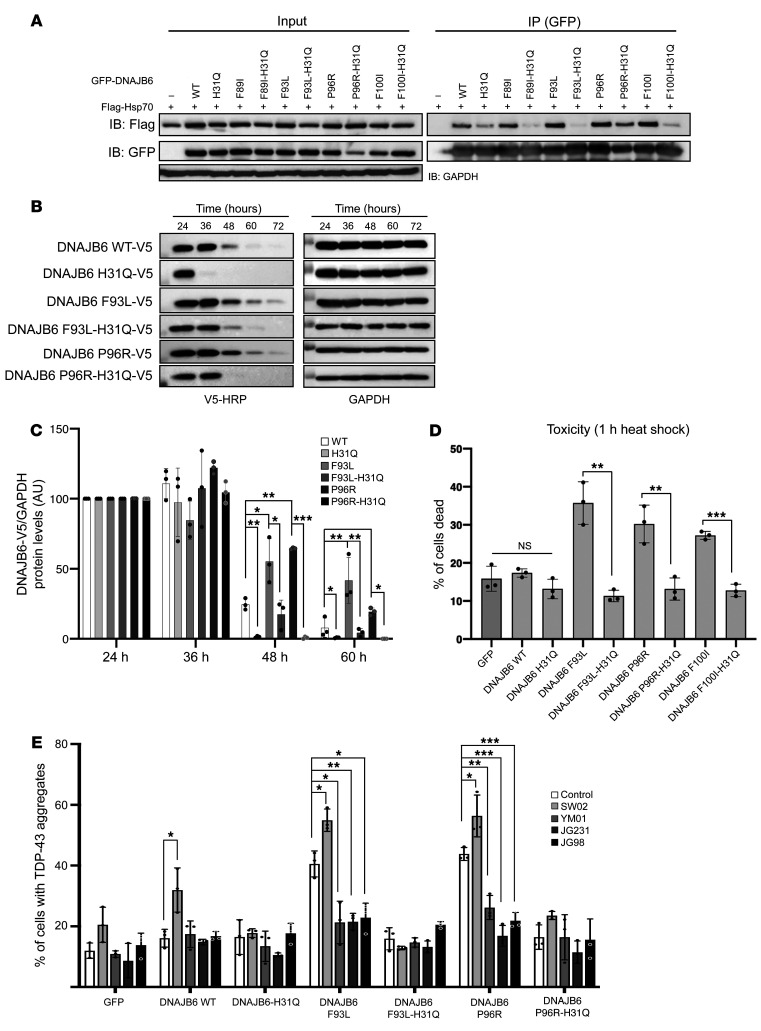
To dissect how the H31Q mutation modulates DNAJB6 function, we specifically examined the binding of DNAJB6 and HSP70. As expected, based on previous results (19), the H31Q mutation decreased the binding of DNAJB6b-WT to coexpressed Flag-HSP70 (Figure 3A and Supplemental Figure 2A). Notably, LGMDD1-mutant DNAJB6b had increased association with Flag-HSP70, and this interaction was weakened by a secondary H31Q mutation (Figure 3A and Supplemental Figure 2A). The degree of reduced interaction was different between LGMDD1 mutations and did not clearly correlate with a reduction in toxicity. Abrogating the HSP70 interaction in LGMDD1-mutant DNAJB6b with the H31Q mutation also corrected other gain-of-function effects, such as an increase in LGMDD1-mutant DNAJB6b stability (Figure 3, B and C), and heat shock–associated cell death (Figure 3D). These data suggest that the gain-of-function effect of LGMDD1-mutant DNAJB6b can be partially corrected by inhibiting the DNAJB6-HSP70 interaction.
Figure 3. LGMDD1-mutant dysfunction is corrected by abrogating HSP70 interaction.
(A) HeLa cells were cotransfected with plasmids expressing Flag-HSP70 and WT or LGMDD1-mutant (F89I, F93L, P96R, or F100I) GFP-DNAJB6b. In some cases, a second mutation in the J domain (H31Q) was also present. Twenty-four hours later, cells were lysed, and GFP-DNAJB6 was immunoprecipitated and subsequently immunoblotted for GFP or Flag. Quantitation of 3 replicates is shown in Supplemental Figure 2A. (B) Tetracycline-inducible isogenic 293 cell lines expressing (WT, H31Q, F93L, F93L-H31Q, P96R, or P96R-H31Q) V5-DNAJB6b were induced with tetracycline for 24 hours and then removed for the indicated durations. Lysates were immunoblotted for V5 and GAPDH. (C) Quantitation of V5-DNAJB6b levels from 3 independent experiments. *P ≤ 0.05, **P < 0.005, and ***P < 0.0005, by paired Student’s t test for comparisons between groups; Bonferroni-corrected P < 0.001 for multiple comparisons. (D) HeLa cells were transfected with the indicated constructs and subjected to heat shock at 42°C for 1 hour, and the percentage of ethidium homodimer-1–positive cells was quantitated. Data are presented as the percentage of cells found positive/dead. **P < 0.005 and ***P < 0.0005, by paired Student’s t test for comparisons between groups. (E) HeLa cells were cotransfected with mCherry-TDP43 and WT or LGMDD1-mutant (F89I, F93L, P96R, or F100I) GFP-DNAJB6b. In some cases, a second mutation in the J domain (H31Q) was also present. After 12 hours of transfection, cells were treated for 16 hours with either YM-01, JG231, JG98 (HSP70 inhibitors), or SW02 (HSP70 activator). The percentage of cells with TDP43-positive nuclear aggregate granules was quantified following 1 hour of heat shock. *P < 0.05, **P < 0.005, and ***P < 0.0005, by paired Student’s t test for comparisons between groups; Bonferroni-adjusted P < 0.0025 for multiple comparisons. n = 300 cells analyzed per condition. The blots and graphs are representative of 3 independent experiments.
Given these results, we wanted to further explore whether HSP70 might be a therapeutic target in LGMDD1. Specifically, we wondered whether inhibiting the association of HSP70 with DNAJB6b with small molecules might partially protect against LGMDD1 phenotypes in mammalian disease models. As a proof-of-concept, we tested 4 different compounds known to modulate HSP70 interactions with cochaperones (20). YM-01, JG98, and JG231 inhibit HSP70 activity by interfering with its interactions with cochaperones, including DNAJ proteins (21). Conversely, SW02 activates HSP70 by enhancing its interaction with DNAJ proteins (22). After 12 hours of transfection with DNAJB6b and TDP43, HeLa cells were treated for 16 hours with vehicle or an HSP70 modulator before heat shock. We quantified the percentage of cells with TDP43 granules (Figure 3E) and found that SW02 increased the gain-of-function effect of LGMDD1-mutant DNAJB6b, whereas YM-01, JG98, and JG231 reduced TDP43 inclusions, specifically in LGMDD1-mutant DNAJB6b–expressing cells. These effects seemed to be dependent on the HSP70 interaction, because there was no change when the HSP70 modulators were applied in the setting of LGMDD1-mutant DNAJB6b harboring an additional H31Q mutation (Figure 3E).
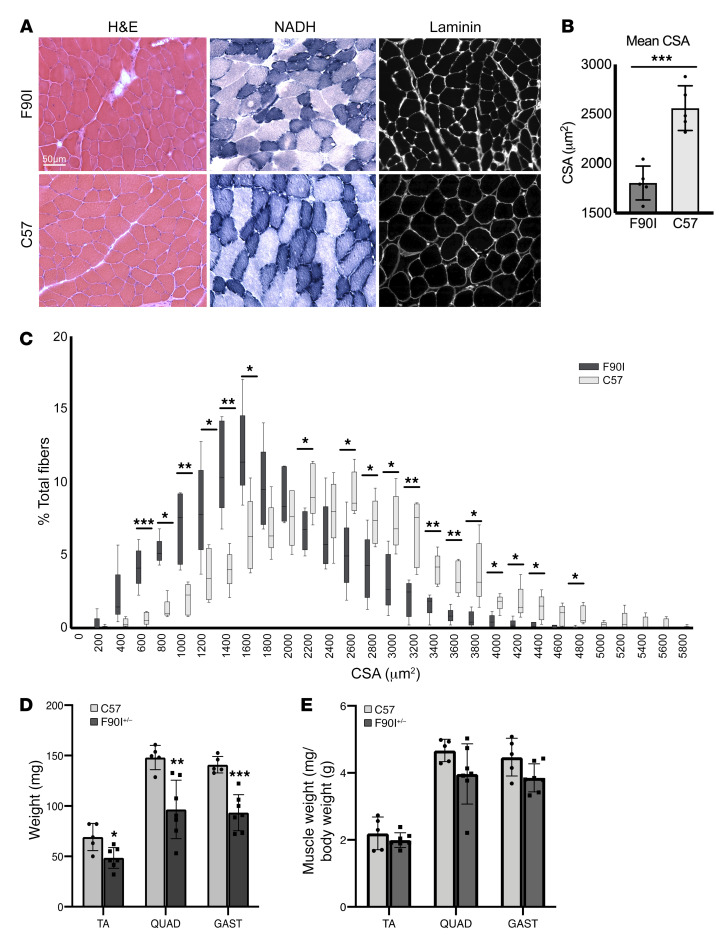
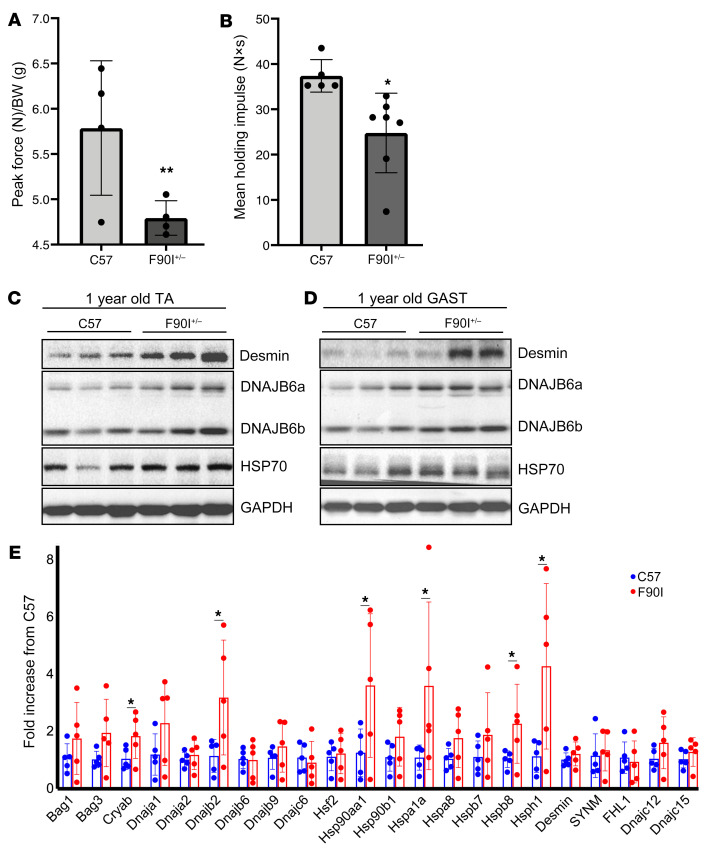
We have previously described a mouse model that overexpresses V5-tagged DNAJB6b-F93L using an MCK promoter in skeletal muscle (12). These mice had premature death, weakness, myopathic muscle, and protein inclusions similar what is seen in patients with LGMDD1 and displayed these phenotypes as early as 2 months of age (12). To generate a model of LGMDD1 that did not rely on overexpression, we used CRISPR/Cas9 to generate the DNAJB6-F90I mutation (homologous to the F89I mutation in humans) in mice. Heterozygous 1-year-old DNAJB6-F90I mice weighed 24.14 ± 2.97 g compared with sex- and age-matched control littermates, which weighed 31.8 ± 3.03 g (P = 0.001) and had smaller-sized muscles (Figure 4D). The DNAJB6-F90I mice developed progressive weakness, muscle atrophy, and scattered fibers with myofibrillar disorganization after 12 months of age (Figure 4, A–E, and Figure 5, A and B). Consistent with the phenotypes seen in the transgenic mouse model, immunoblot analysis showed an increase in both DNAJB6a and DNAJB6b isoforms, as well as in desmin levels (Figure 5, C and D, and Supplemental Figure 2, B and C). In addition, the levels of several chaperone proteins were increased, as assessed by quantitative PCR (qPCR) (Figure 5E and Supplemental Figure 6), suggesting a stress response and impairment of protein quality control. For example, the levels of HSPA1A (also known as HSP70) were elevated, as demonstrated by immunoblotting and qPCR (Figure 5, C–E, and Supplemental Figure 2, B and C). Notably, although DNAJB6 protein accumulated, its mRNA levels were unchanged (Figure 5E). Thus, much like what was previously observed in our transgenic models, LGMDD1 mutations in DNAJB6 seem to lead to an age-associated dysfunction in protein quality control and an aberrant accumulation of Z-disc proteins (12).
Figure 4. DNAJB6-F90I–knockin mice develop myopathy.
(A) Histochemical analysis with H&E and NADH staining of TA muscle from 1-year-old F90I (heterozygous) or control C57 mice. Immunohistochemical analysis with an anti-laminin antibody of the same muscle. Scale bar: 50 μm. (B and C) Quantitation of the muscle fiber CSA from A. (B) Individual data points represent the mean CSA for each mouse, the bar graph represents the mean CSA for all mice ± SD. n = 5 mice per genotype. ***P < 0.001, by paired Student’s t test for comparisons between groups. (C) Frequency distribution of the muscle fiber CSA. Data are presented as the average percentage of total fibers ± SD. n = 5 mice per genotype. *P < 0.05, **P < 0.01, and ***P < 0.001, by 2-way ANOVA for comparisons between groups, followed by Bonferroni’s post hoc multiple comparisons test. (D and E) Graphs represent the average mass (left) (mean ± SD, weight in milligrams) or muscle mass normalized to mouse weight (right) of TA, quadriceps (QUAD), and gastrocnemius (GAST) muscles from 1-year-old C57 (n = 7) and DNAJB6 F90I+/– mice (n = 7). *P < 0.05, **P < 0.005, and ***P < 0.0005, by paired Student’s t test for comparisons between groups.
Figure 5. DNAJB6-F90I–knockin mice show aberrant accumulation of Z-disc protein.
(A) Forelimb grip strength measurements for 1-year-old littermate control (C57) and DNAJB6-F90I heterozygous mice and (B) inverted wire screen holding for the same animals as in A. *P < 0.05 and **P < 0.005, by paired Student’s t test for comparisons between groups. (C and D) Immunoblot of lysates from TA and GAST muscles from 3 different 1-year-old C57 and F90I heterozygous mice using antibodies against DNAJB6, HSP70, desmin, and GAPDH. Quantitation of 3 replicates for each blot is shown in Supplemental Figure 2, B and C. (E) qPCR analysis of TA muscle from five 1-year-old F90I heterozygotic and C57 mice. Data are presented as the fold change versus C57 controls. *P = 0.05–0.01, by Student’s t test; P = 0.0023, Bonferonni-adjusted value for multiple comparisons.
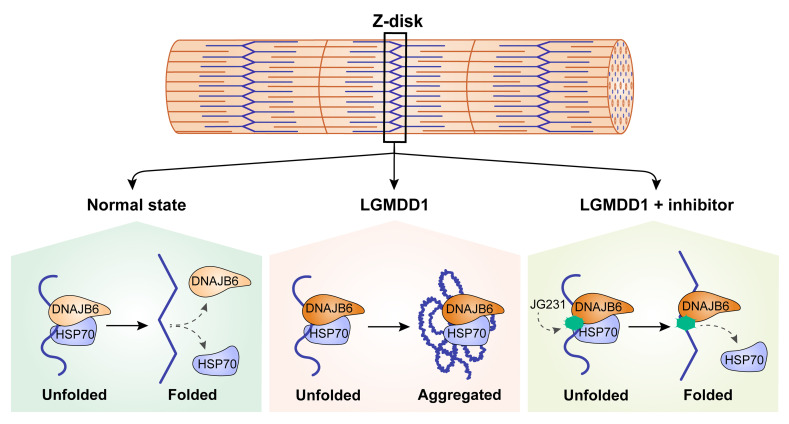
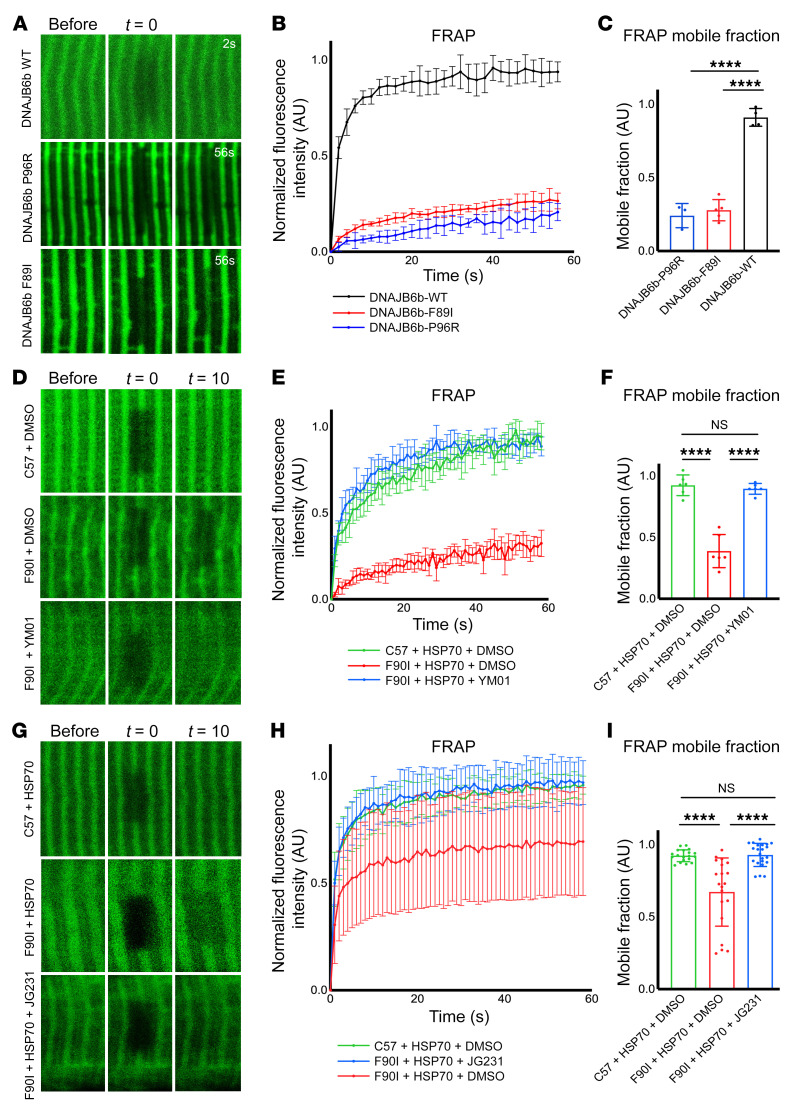
Because DNAJB6 normally collaborates with HSP70 to reversibly cycle onto “client” Z-disc proteins (14), and myofibrillar disorganization is a pathologic feature of LGMDD1, we hypothesized that this process might be damaged by LGMDD1 mutations. To test this idea, we evaluated the localization of DNAJB6b and HSP70 in live mouse muscle by imaging the flexor digitorum brevis (FDB) following electroporation of GFP-tagged DNAJB6b or HSP70. As expected, DNAJB6b-WT protein localized to the Z-disc, whereas HSP70 localized to the Z-disc and, to varying degrees, the M-line (Supplemental Figure 3A). Localizations were confirmed by second harmonic generation imaging, which identifies the A bands, and by expression of desmin-GFP with mCherry–DNAJB6b-WT (Supplemental Figure 3, A and B). To understand chaperone dynamics at the Z-disc, we performed fluorescence recovery after photobleaching (FRAP) of DNAJB6b and HSP70. Fluorescence from DNAJB6b-WT was rapidly recovered at the Z-disc (Figure 6, A and B), indicating that most of the protein was mobile (Figure 6C) and transiently associated with Z-disc clients. By contrast, 2 different LGMDD1 mutations (F89I and P96R) in DNAJB6b showed slower recovery at the Z-disc (Figure 6, A and B).
Figure 6. Imaging of DNAJB6 and HSP70 kinetics at the Z-disc.
(A) Fluorescence recovery after photobleaching was performed with 2PEM footpads of mice following electroporation into the FDB with constructs expressing GFP-DNAJB6b-WT, -P96R, or -F89I. Representative images show before bleaching, after bleaching (t = 0 seconds), and after t = 2 seconds or t = 56 seconds. (B) Graph of the normalized relative fluorescence intensity (RFI) versus the time in seconds for the studies in A. (C) Graph of the percentage of maximum fluorescence recovery corresponding to the mobile versus immobile fraction. n = 3–5 separate myofibers from 1 mouse per condition. (D–I) Experiments similar to those in A following electroporation of HSP70-GFP in C57 control or DNAJB6-F90I heterozygous mice. The footpads of mice were injected with YM01 (D–F), or mice were injected i.p. with JG231 (G–I). (D and G) Representative images show before bleaching, after bleaching (t = 0 seconds), and after t = 10 seconds. (E and H) Graph of the normalized RFI versus the time in seconds for the studies in D and G, respectively. (F and I) Graph of the percentage of maximum fluorescence recovery corresponding to the mobile versus immobile fraction for the studies in D and G, respectively. All individual data points represent the mean of 3 separate FRAP experiments within 1 myofiber. FRAP curves (B, E, and H) and mobile fraction bar graphs (C, F, and I) represent the mean of all myofiber averages ± SD. For YM01 experiments in D–F, n = 6 myofibers from 2 mice per condition. For JG231 experiments in G–I, n = 19–22 myofibers from 3 mice per condition. ****P < 0.0001, by 1-way ANOVA followed by Tukey’s post hoc test for group comparisons. The high variability in the FRAP mobile fraction for the DNAJB6-F90I mice electroporated with HSP70-GFP and treated with i.p. DMSO was largely driven by 4 data points (H and I). These 4 data points were derived from different myofibers within 1 animal. Reanalysis of the data after removing this mouse showed that the results remained statistically significant (***P = 0.0002).
Using this system, we next wanted to test whether the slow recovery of DNAJB6b-F89I might also affect HSP70 in a dominant manner. To test this concept, we performed a similar FRAP experiment using 3-month-old DNAJB6-F90I–knockin mice that express 1 copy of the DNAJB6-F90I (F89I in humans) at endogenous levels. Three months was chosen as the time point because, at this early age, the mice do not show any obvious muscle pathology. Whereas HSP70 had rapid fluorescence recovery in the littermate control mice, its recovery was slowed in the DNAJB6-F90I–knockin mice (Figure 6, D–F). Local delivery of YM-01 to the footpad (Figure 6, D–F), or systemic treatment with JG231 (Figure 6, G–I), increased HSP70 recovery kinetics in DNAJB6-F90I mouse muscle. Importantly, JG231 did not significantly affect the recovery kinetics of a mutant HSP70, Y149W, in which the compound-binding site is occluded, supporting the idea that the compound’s effect was mediated by binding to HSP70 (Supplemental Figure 4, A–C).
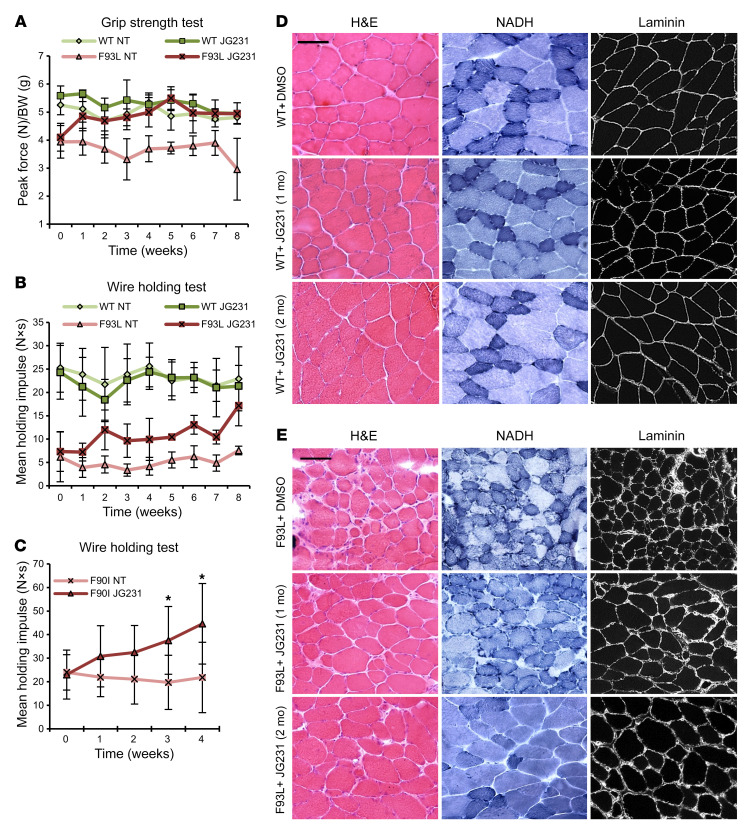
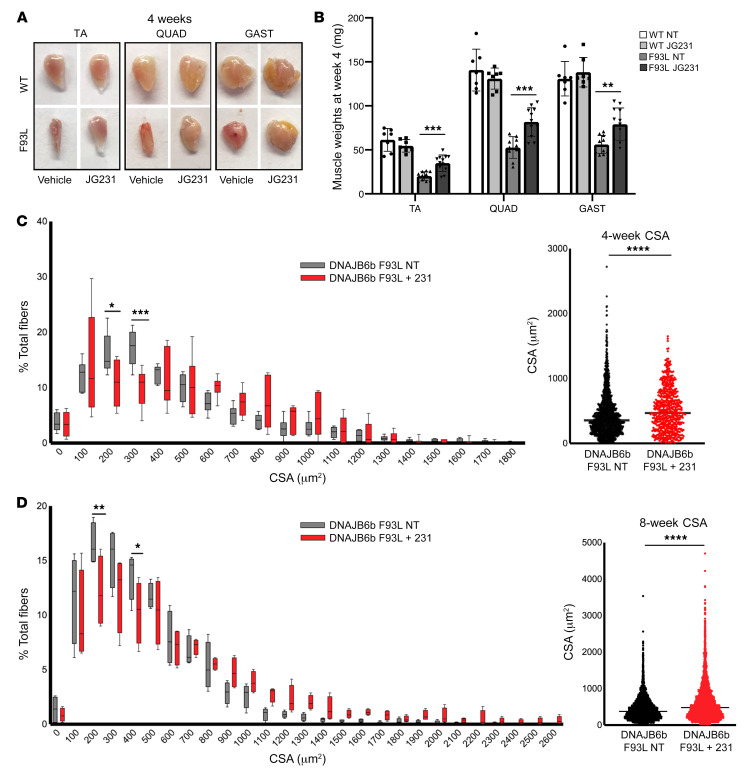
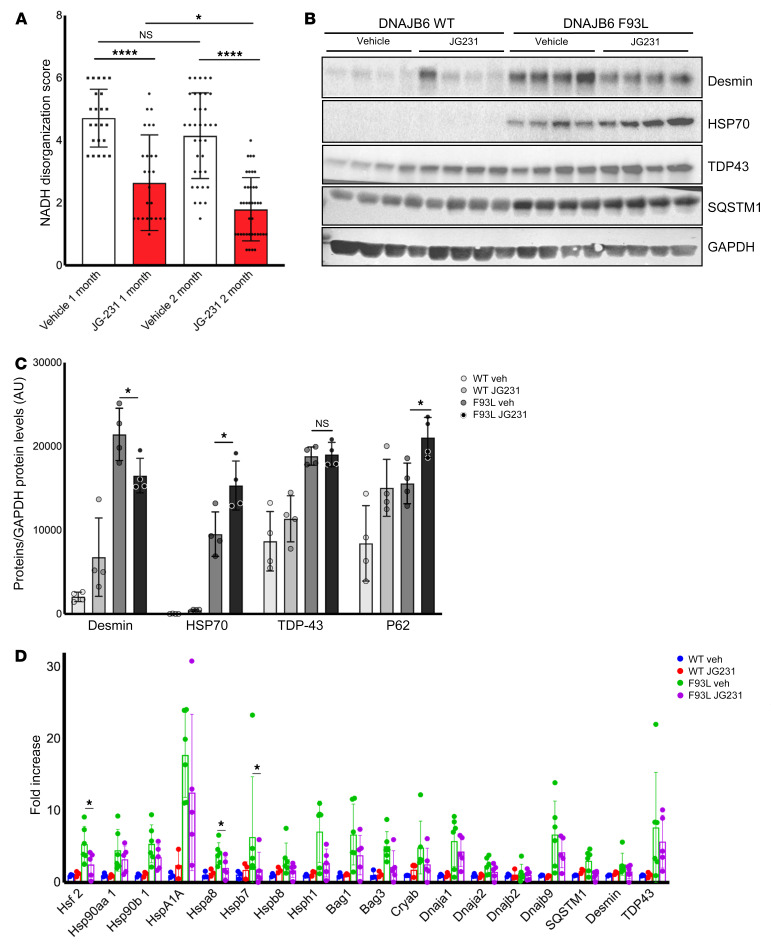
To explore whether abrogation of DNAJB6-HSP70 interactions with JG231 might also restore muscle functions, we treated transgenic DNAJB6b-WT and DNAJB6b-F93L–overexpressing mice with vehicle (DMSO) or 4 mg/kg i.p. JG231 every other day for 8 weeks and measured mouse performance on forelimb grip and hanging grid tests (Figure 7, A and B). Mice were sacrificed at 4 and 8 weeks for muscle histopathology (Figure 7, D and E). This dose and schedule were chosen on the basis of previous safety and pharmacokinetics studies (23). Although DNAJB6b-F93L mice were significantly weaker than DNAJB6b-WT mice, JG231 treatment increased their performance by as early as 1 week (Figure 7, A and B), and at 4 and 8 weeks, these mice also showed an increase in isolated muscle mass (Figure 8, A and B). This increase in performance was also seen when 1-year-old DNAJB6-F90I–knockin mice were treated with JG231 for 1 month and subjected to a hanging grid test (Figure 7C). To further assess the effect of JG231 treatment, we quantitated myofiber size and the degree of myofibrillar disarray in DNAJB6b-WT and DNAJB6b-F93L mice 4 and 8 weeks after treatment with JG231 or vehicle (Figure 8, C and D, and Figure 9A). Abnormal internal architecture was quantitated using an NADH stain and scored from 0–6 in a blinded fashion by 2 neuromuscular physicians with muscle pathology expertise. Examples of scored images are shown in Supplemental Figure 5. Notably, there was a significant shift toward larger myofibers (Figure 8, C and D) and a decrease in the percentage of fibers with abnormal internal architecture, as demonstrated on NADH staining at both 4 and 8 weeks (Figure 9A).
Figure 7. JG231 improves strength and myopathology in LGMDD1 mice.
(A and B) Sex-matched 4-month-old DNAJB6b-WT (n = 8 vehicle; n = 8 treatment) and DNAJB6b-F93L mice (n = 10 vehicle; n = 12 treatment) were treated for 4 or 8 weeks with i.p. vehicle or JG231. (A) Forelimb grip strength and (B) inverted wire screen holding results. (C) Weekly measurements of mean holding impulse for sex-matched 1-year-old DNAJB6-F90I heterozygous mice treated for 4 weeks with i.p. vehicle or JG231.*P < 0.05. (D and E) Histochemical analysis with H&E and NADH staining or immunohistochemical analysis with anti-laminin of TA muscle from DNAJB6b-WT and DNAJB6b-F93L mice after 4 or 8 weeks of treatment. Scale bars: 100 μm. NT, nontreated.
Figure 8. JG231 increases myofiber size in LGMDD1 mice.
(A) Representative mages of TA, QUAD, and GAST muscles from 4-month-old DNAJB6b-WT and DNAJB6b-F93L mice treated with vehicle or JG231 for 4 weeks. (B) Average mass (mean ± SD, weight in milligrams) of the TA, QUAD, and GAST muscles from six DNAJB6b-WT and DNAJB6b-F93L mice treated with vehicle or JG231 for 4 or 8 weeks. **P < 0.005 and ***P < 0.0005, by paired Student’s t test. We used frozen sections of TA muscle from 4-month-old DNAJB6b-WT and DNAJB6b-F93L control mice or DNAJB6b-F93L mice treated with JG231 for 4 or 8 weeks. (C and D) Frequency distributions of muscle fiber CSA in TA muscle from 4-month-old DNAJB6b-F93L mice treated with vehicle or JG231 for 4 weeks (n = 6) (C) or 8 weeks (n = 4) (D). Data are presented as the average percentage of total fibers ± SD. *P < 0.05 and **P < 0.01, by 2-way ANOVA for comparisons between groups, followed by Bonferroni’s post hoc multiple comparisons test. ****P < 0.0001, by paired Student’s t test for comparisons between groups. Scatter plot graphs in C and D represent the mean CSA for all animals, where each point indicates an individual muscle fiber CSA.
Figure 9. JG231 induces a partial recovery of protein quality control in LGMDD1 mice.
(A) Graph of average NADH disorganization, scored from TA muscle from 4-month-old DNAJB6b-F93L mice treated with vehicle or JG231 for 4 weeks (n = 6 mice per cohort with 3–4 images per mouse) or 8 weeks (n = 4 per cohort with 9–10 images per mouse). Error bars indicate the SD. *P < 0.05 and ****P < 0.0001, by 1-way ANOVA followed by Tukey’s post hoc test for comparisons between groups. (B and C) Immunoblot and quantification of TA muscle lysates from 4 different DNAJB6b-WT and DNAJB6b-F93L mice treated with vehicle (Veh) or JG231 for 8 weeks using antibodies against desmin, HSP70, TDP43, SQSTM1, and GAPDH. *P ≤ 0.001, by 2-way ANOVA with Bonferroni’s correction for multiple comparisons. (D) qPCR analysis of TA muscle from 4-month-old DNAJB6b-WT and DNAJB6b-F93L mice treated with vehicle or JG231 for 4 weeks. Data are presented as the fold change compared with C57 controls. *P < 0.05, by paired Student’s t test for comparisons between groups; Bonferroni-adjusted P < 0.0028 for multiple comparisons.
JG231 treatment had a modest effect on the expression of other chaperone proteins, as evaluated by qPCR of tibialis anterior (TA) muscle (Figure 9D and Supplemental Figure 6), suggesting that this effect was independent of a global stress response. To determine whether there were changes in other proteins found to accumulate in LGMDD1 muscle, we immunoblotted for TDP43, SQSTM1, HSP70, and desmin (Figure 9, B and C). Muscle from DNAJB6-F93L–mutant mice treated with JG231 had an increase in HSP70 protein levels (Figure 9, B and C). Whether this was due to a change in its stability in the setting of JG231 remains to be established. In contrast, after 8 weeks of treatment with JG231, we observed a significant decrease in total desmin protein levels (Figure 9, B and C), as might be expected for a partial recovery of quality control. The observed decrease in desmin accumulation also matched the improvement in muscle internal architecture that was seen in the treated mice. Together, these results suggest that DNAJB6-HSP70 interaction is an unexpected drug target for LGMDD1.
Discussion
Protein chaperones are important for the maintenance of sarcomere structure and muscle function (7). Dominant mutations in DNAJB6 cause LGMDD1, a myopathy with myofibrillar disorganization and protein inclusions (3, 4). Our data support the idea of an unexpected mechanism in which the dominant effect of mutations in the G/F domain of DNAJB6 require its interaction with HSP70. Specifically, yeasts expressing homologous LGMDD1 mutations in Sis1 had reduced viability, but this toxicity was corrected by including a second mutation that abrogates binding to HSP70. Moreover, genetically blocking the interaction of DNAJB6 with HSP70 rescued the dominant effect of LGMDD1 mutations on TDP43 disaggregation and post–heat shock toxicity. There are a number of possible mechanisms to link these observations. For example, the interactions of mutant DNAJB6 with HSP70 might reduce the stability of DNAJB6 clients. HSP70 has been shown to prevent proper folding if it is not allowed to cycle off its client protein (24). Indeed, although HSP70 is often associated with its roles in protein folding, its prolonged binding appears to be antagonistic to this process in many settings (25). Alternatively, the prolonged interaction of mutant DNAJB6b with HSP70 might titrate it away from other clients, leading to broader disruption of HSP70-dependent processes (26). These mechanisms are not mutually exclusive, and both could result in a more general impairment of protein quality control.
Several lines of evidence suggest that Z-disc proteins, such as desmin, may be DNAJB6b clients. For example, loss of DNAJB6 in myoblasts leads to the accumulation of desmin (14). We found that normal DNAJB6b and HSP70 transiently localized to the Z-disc in mouse muscle, as measured by FRAP. However, in mice expressing an LGMDD1 mutation, the movement of HSP70 on the Z-disc was slowed, suggesting that it was more avidly associated with mutant DNAJB6. Strikingly, treatment with small molecules known to interrupt DNAJ and HSP70 interactions (21) partially recovered HSP70 kinetics. These results further substantiate the idea that the phenotypes associated with LGMDD1-mutant DNAJB6 require HSP70 interactions.
Myofibrillar proteins at the Z-disc undergo stress and strain with normal muscle contraction, and protein quality control is essential to maintain proper folding of these proteins. Our model suggests that DNAJB6 binds to Z-disc proteins such as desmin and then recruits HSP70. Following the DNAJ:HSP70 catalytic cycle, both were released from the client protein (the Z-disc), allowing them to facilitate protein homeostasis of other clients throughout the myofiber. In the setting of an LGMDD1 disease mutation, DNAJB6 bound its client and recruited HSP70 but failed to release from the Z-disc. Whether this was due to an impairment in the DNAJB6 stimulation of HSP70 ATPase activity or to a failed release from its client due to aberrant interactions with nucleotide exchange factors such as HSP110 remains to be established. Regardless, our data support the idea that abrogating the initial recruitment of HSP70 to DNAJB6 may be beneficial. Unlike a genetic modification of the LGMDD1-mutant DNAJB6 allele (e.g., a second H31Q mutation) that would selectively inactivate the pathogenic DNAJB6 protein, our small-molecule approach should equally impair DNAJB6-WT and LGMDD1-mutant DNAJB6 function. However, our data showed that under conditions of DNAJB6b-WT overexpression, YM-01, JG98, and JG231 had a negligible effect. Whether this was due to a selective interference with a more abundant and/or client-bound LGMDD1-mutant DNAJB6 remains to be established.
A recent study exploring the pathogenic mechanism of mutations in BAG3 associated with another chaperonopathy showed striking similarities (27). Dominant mutations in BAG3 lead to its destabilization and subsequent aggregation (28). Mutant BAG3 aggregates contain additional chaperones, including HSP70 and DNAJB6. Likewise, blocking the interaction of HSP70 with BAG3 can partially rescue phenotypes, suggesting that BAG3-associated myopathy is, like LGMDD1, due to a dominant interaction with HSP70. The similarities between these findings suggest that myopathies might share a common molecular feature of aberrant protein quality control, leading to a “traffic jam” of chaperones on Z-disc proteins. Remobilizing these chaperones might be a generalizable approach to partially restore muscle function in myopathies.
In the present study, we used 2 different LGMDD1 mouse models. We previously described the DNAJB6b-F93L mouse model, in which a MCK promoter is used to selectively overexpress V5-tagged DNAJB6b-F93L in skeletal muscle (12). This model develops an aggressive myopathy, with 40% of mice dying by 2 months of age. The surviving mice develop weakness, myopathic muscle, desmin inclusions, and hnRNPA2B1/A1 aggregates similar to what is seen in patients with LGMDD1 (12). Notably, overexpression of DNAJB6b-WT, DNAJB6a-WT, or the DNAJB6a-F93L isoform did not result in myopathic features at the same time point, suggesting that the DNAJB6b-mutant isoform is the pathogenic driver of myopathy (12). The DNAJB6b-F93L overexpression mouse model is limited, in that only the DNAJB6b isoform is highly expressed exclusively in skeletal muscle and thus may not model other aspects of the disease related to the other isoforms and nonmuscle tissue. The second mouse model, preliminarily described in this study, has a gene-edited LGMDD1 mutation (DNAJB6-F90I, corresponding to the human DNAJB6-F89I mutation) allowing expression of the mutant DNAJB6 allele at endogenous levels, on both isoforms, and in all tissues. At 1 year of age, DNAJB6-F90I mice have muscle hypotrophy, myofiber disorganization, weakness, increased expression of chaperones, and desmin accumulation (all features seen in DNAJB6b-F93L–overexpressing mice and patients with LGMDD1). Future characterization of the DNAJB6-F90I–knockin mouse will be essential to understand the pathogenic mechanism of LGMDD1.
More immediately, this study suggests that the HSP70-DNAJB6 interaction could be a drug target for the treatment of LGMDD1. However, while HSP70 inhibitors have been studied in preclinical safety studies in mice (29, 30), any treatment for human myopathy would require long-term administration, and the safety of chronic treatment is not yet clear. The key to translation of this concept might be to create more selective inhibitors that act exclusively on the DNAJB6-HSP70 interaction. The inhibitors currently available, although useful chemical probes for proof-of-concept studies, have broad effects on HSP70 cochaperone contacts. Thus, a selective inhibitor of mutant DNAJB6-HSP70 interactions may be the (safest and) most effective.
Methods
Yeast studies.
The yeast strains used in this study were derived from Saccharomyces cerevisiae 74-D694 (ade1-14 his3-Δ200 leu2-3, 112 trp1-289 ura 3-52). Yeasts were grown and manipulated using standard techniques. As indicated, cells were grown in rich media YPD (1% yeast extract, 2% peptone, 2% dextrose) or in synthetic defined media (0.67% yeast nitrogen base without amino acids, 2% dextrose) lacking specific nutrients, to select for appropriate plasmids. WT yeast harboring the single-dot medium [RNQ+] variant and the [rnq-] control strain were provided by Susan Liebman (Department of Pharmacology, University of Nevada, Reno, Nevada, USA) (31). Construction of ΔSis1 [rnq-] and single-dot medium [RNQ+] yeast strains were described previously (15). Medium containing 1 mg/mL 5-fluoroorotic acid (5-FOA) that selects against cells maintaining URA3-marked plasmids was used to replace WT Sis1 with the mutant constructs using the plasmid shuffle technique. Plasmid transformations were done using the polyethylene glycol/lithium acetate (PEG/LioAC) technique, and the cells were selected using SD-trp plates. Plasmids pRS316-SIS1 and pRS314-sis1-L268P were gifts from Elizabeth Craig (Department of Biochemistry, University of Wisconsin, Madison, Madison, Wisconsin, USA) (17). Construction of pRS314-SIS1 was described earlier (15). Using pRS314-SIS1 as a template, mutations in SIS1 were generated using bridge PCR with the following oligonucleotides: sis1-F106L (5′-GATTCTCCGGAGGACATGCGCTCAGTAATGAGGATGC-3′ and 5′-GCATCCTCATTACTGAGCGCATGTCCTCCGGAGAATC-3′) and sis1-F115I (5′-GATGCTTTCAATATTATTTCACAATTCTTTGGC-3′ and 5′-GCCAAAGAATTGTGAAATAATATTGAAAGCATC-3′). Using pRS314-sis1-F106L and pRS314-sis1-F115I as templates, double mutations with sis1-L268P were generated using the primer pair 5′-GTTTCTCTAGTTATCCATCTG-3′ and 5′-GTTTCTCTAGTTATCCATCTG-3′. pRS414GPD-sis1-ΔDD-F115I was created using primers (5′-ATACTAGTATGGTCAAGGAGACAAAAC-3′ and 5′-CGCATCGATTTATGGATAGTCCACTTTATATTTTAC-3′), followed by digestion with SpeI/ClaI, and ligation with pRS314GPD that was digested with the same enzymes. Yeast cells were spotted as described previously (32). Briefly, yeast cells grown overnight were pelleted, washed, and normalized with water to an optical density of 1.0. The cells were serially diluted (1:5) using a multichannel pipette into a 96-well plate and were spotted onto agar plates using an ethanol-sterilized 48-pin replicator. For protein analysis, yeast cells were lysed by vortexing with glass beads in buffer containing 100 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA, 5% glycerol, 0.5 mM DTT, 50 mM N-ethylmalemide (NEM), 3 mM PMSF, and cOmplete Mini Protease Inhibitor Cocktail (Roche). Following lysis, an equal volume of RIPA buffer (50 mM Tris-HCl, pH 7, 200 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) was added to the lysate and centrifuged briefly to obtain the total protein fraction. Protein concentrations were normalized and subjected to SDS-PAGE, transferred onto PVDF membranes, and probed with anti-Sis1 and anti-Pgk1 antibodies.
qPCR.
Total RNA was isolated from TA muscle with the SV Total RNA Isolation Kit (Promega; Z3100) according to the manufacturer’s instructions. The concentration and quality of the total RNA isolated were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized using the Transcriptor First-Strand cDNA Synthesis Kit (Roche; 04379012001). Gene expression levels were analyzed by real-time PCR on an Applied Biosystems model 7500 Software (version 2.0.5) using FastStart Universal SYBR Green Master ROX qPCR Mastermix (Roche; 04913850001). qPCR was performed with primers for chaperone proteins (CRYAB, HSP, and BAG families), cytoskeletal proteins (desmin, SYNM), and FHL1, HSF, TDP43, SQSTM1, and hDNAJA/B/C families (Supplemental Figure 6). The values were normalized to GAPDH and are represented as the fold change. The primer sequences are listed in Supplemental Figure 6.
Antibodies.
The antibodies used were as follows: anti–rabbit Sis1 (Cosmo Bio; COP-080051), anti–mouse Pgk1 (Abcam, 113687), anti–rabbit GAPDH (Cell Signaling Technology; 2118), anti–V5 HRP (Invitrogen, Thermo Fisher Scientific; B96125), anti–mouse desmin (Dako; M0760), anti–rabbit TDP43 (Proteintech; 10782-2-AP), anti–rabbit DNAJB6 (Abcam; ab198995), anti–mouse HSP70 (Enzo Life Sciences; ADI-SPA-812), anti–rabbit P62 “SQSTM1” (Proteintech; 18420-1-AP), anti–rabbit laminin (Abcam; ab11575), anti–rabbit GFP (MilliporeSigma; g1544), and anti–mouse Flag (MilliporeSigma; F3165). The following secondary antibodies were used: anti–mouse HRP (Cell Signaling Technology; 7076S), anti–rabbit HRP (Cell Signaling Technology; 7074S), and anti–rabbit Alexa Fluor 555 and 488 (Invitrogen, Thermo Fisher Scientific; A21428 and A11034, respectively).
Plasmids and constructs.
Mammalian constructs of DNAJB6b were cloned using site-directed mutagenesis, digested with HindIII/XhoI, and ligated into the vector pcDNA3.1 containing a GFP tag. DNAJB6b H31Q, F93L, P96R, F100I, F93L-H31Q, P96R-H31Q, and F100I-H31Q mutations were generated with the QuikChange Mutagenesis Kit (Agilent Technologies; 200517). Human TDP43 fused to pCherry was described previously (33). The GFP-HSP70 plasmid was obtained from Addgene (Addgene plasmid 15215).
Western blotting.
Muscle tissues and cultured cells were homogenized using RIPA lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% Na-deoxycholate, and 1 mM EDTA) supplemented with protease inhibitor cocktail (MilliporeSigma), and lysates were centrifuged at 16,000 g for 10 minutes. Protein concentrations were determined using a BCA Protein Assay Kit (Thermo Fisher Scientific). Aliquots of lysates were solubilized in Laemmli sample buffer, and equal amounts of proteins were separated on 12% SDS-PAGE gels. Proteins were transferred onto nitrocellulose membranes and then blocked with 5% nonfat dry milk in PBS with 0.1% Tween-20 for 1 hour. The membrane was then incubated with primary antibodies, specific to the protein of interest, in 5% nonfat dry milk in PBS with 0.1% Tween overnight at 4°C. After incubation with the appropriate secondary antibody conjugated with HRP, ECL (GH Healthcare) was used for protein detection. Immunoblots were obtained using the G:Box Chemi XT4, Genesys, version 1.1.2.0 (Syngene). Densitometry was measured with ImageJ software (NIH).
Solubility assay.
HeLa cells were collected and homogenized in 250 μL of 2% SDS-radioimmunoprecipitation assay buffer (50 mg Tris–Cl [pH 8.0], 150 mg NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 2% SDS) and protease inhibitor cocktail (MilliporeSigma). Homogenates were precleared with a 30-second low-speed spin, and an aliquot of the supernatant was collected and labeled the total fraction. The additional supernatant was centrifuged at 100,000 g for 30 minutes at 4°C, and this supernatant was collected and named the soluble fraction. The pellet was then sonicated on ice after the addition of 150 μL of 5 M guanidine-HCl and recentrifuged at 100,000 g for 30 minutes at 4°C. This supernatant was removed and labeled the insoluble fraction. The insoluble fraction was precipitated by adding an equal volume of 20% trichloroacetic acid (MilliporeSigma) and incubated on ice for 20 minutes. Samples were then centrifuged at 10,000 g for 15 minutes at 4°C, and the resulting pellet was washed twice with ice-cold acetone. Residual acetone was removed by drying tubes at 95°C, and the samples were resuspended in 50 μL of 5% SDS in 0.1N NaOH. The protein concentrations of all samples were determined using a BCA Protein Assay Kit (Pierce, Thermo Fisher Scientific). Each sample (30 μg) was analyzed by Western blotting for each fraction.
Immunoprecipitation.
For immunoprecipitation (IP) of GFP-tagged DNAJB6, HeLa cells were cotransfected with Flag-HSP70 and different DNAJB6 plasmids for 24 hours. Cell pellets were collected, washed twice with cold PBS, lysed in co-IP buffer (1× tris buffered saline, 1 mM NaCl, 1% Triton X-100, 10% glycerol supplemented with phosphatase and protease inhibitors) for 15 minutes, and spun at 12,000 g for 25 minutes. Lysate was collected and the protein concentration determined using a Micro BCA Protein Assay Kit. Total proteins (1000 mg) from whole-cell extracts were immunoprecipitated overnight at 4°C using GFP-conjugated magnetic beads (MilliporeSigma), and then the beads were washed 3 times with IP wash buffer (20 mM Tris, pH 7.5, 1 mM ethylenediaminetetraacetic acid, 150 mM NaCl, 10% glycerol). GFP-conjugated magnetic beads were resuspended in 40 μL 2× SDS loading buffer with β-mercaptoethanol and heated at 90°C for 5 minutes to elute the samples. Equal amounts of total proteins (35 μg/sample) and 20 mL eluted IP samples were resolved in 10% SDS-PAGE and analyzed by Western blotting according to standard procedures.
Wire screen–holding and grip tests.
Grip strength testing consisted of 5 separate measurements using a trapeze bar attached to a force transducer that recorded peak-generated force (Stoelting Co.). Mice have the tendency to grab the bar with their forepaws and continue to hold while being pulled backwards by the tail, releasing only when unable to maintain their grip. The resulting measurement was recorded, and the average of the highest 3 measurements was determined to give the strength score. For every time point and strain, at least 5 animals were used. P values were determined by a paired Student’s t test. To validate our results, another quantitative strength measurement was performed by a wire screen–holding test. Mice were placed on a grid on which it stood using all 4 limbs. Subsequently, the grid was turned upside down 15 cm above a cage. Latency for the mouse to release the mesh was recorded, and the average hanging time for 3 trials was used as an outcome measure.
Animal and experimental protocols.
Human DNAJB6b-WT plasmids were obtained from Addgene, and the F93L point mutation was generated using QuikChange Site-directed Mutagenesis by changing the cDNA position 277T>C. Both cDNAs were subcloned into a 1256MCKCAT-transgenic targeting vector obtained from Stephen Hauschka (University of Washington, Seattle, Washington, USA). The promoter and coding sequences were confirmed by DNA sequence analysis. A linear fragment containing the MCK-V5-hDNAJB6 sequence was isolated by digesting the targeting vector with HindIII and KpnI and subsequent gel purification. This fragment was sent to the mouse genetics core facility at Washington University for transgenic animal production. Animals were screened for transgene insertion using PCR amplification of tail DNA. For the CRISPR/Cas9-mediated knockin F90I mice, the primers PC484.DS.Dnajb6.F: 5′-GGCAAATAAGACTCTTGGGCT-3′, PC484.DS.Dnajb6.R: 5′-ACGGACCAGGCAGGTACTTA-3′ were used, and the sgRNA (SM721.Dnajb6.g4) TCATTGGGCAGTGTTATCATNGG was used. The generation of these mice was carried out by the mouse genetics core facility for transgenic animal production at Washington University. Both animal lines were bred on a C57/B6 background (The Jackson Laboratory) to at least the F5 generation for phenotypic analysis. The control animals were nontransgenic F90I–/– littermates. Mice were housed in a temperature-controlled environment with 12-hour light/12-hour dark cycles, in which they received food and water ad libitum. Mice were euthanized, and skeletal muscle was dissected.
In vivo electroporation.
Mice were anesthetized using 2.5% isoflurane. The footpad was wiped with ethanol, and 10 μL hyaluronidase at a concentration of 2 mg/mL was injected into the FDB from the base of the pad toward the digits. The mouse was then monitored in a recovery cage for 1–2 hours. Again, the animal was anesthetized with 2.5% isoflurane, and the footpad was wiped with ethanol. The plasmid was then injected into the FDB with endotoxin-free plasmid diluted in sterile PBS to a volume of 50 μL using a 29-gauge × 1/2 needle. Immediately after that, 2-needle array electrodes were inserted longitudinally relative to the muscle fibers. The in vivo electroporation parameters for FDB were as follows: 20 pulses for a 20-ms duration at 1 Hz at 100 V/cm. The procedure was repeated on the contralateral FDB. The needles were removed, and the foot was wiped again with ethanol. The mouse was then return to the recovery cage for monitoring.
Cell culture, transfection, and heat shock.
Flp-In T-REX 293 Cells (Invitrogen, Thermo Fisher Scientific) expressing the pcDNA5/FRT/TO constructs V5-DNAJB6b-WT, V5-DNAJB6b-F93L, V5-DNAJB6b-P96R, V5-DNAJB6b-H31Q, and V5-DNAJB6b-F93L-H31Q were cultured in DMEM containing 4 mM l-glutamine (Invitrogen, Thermo Fisher Scientific; 11965-084), 10% FBS (Atlanta Biologicals; S10350), penicillin (50 IU)/streptomycin (50 μg/mL) (Invitrogen, Thermo Fisher Scientific; 15 140), 50 μg/mL hygromycin B (Invitrogen; 10687-010), and 50 μg/mL blasticidin (Life Technologies, Thermo Fisher Scientific; R21001), and induced with 1 μg/mL tetracycline hydrochloride (MilliporeSigma; T76600) 48 hours before experimentation. Cells were maintained in 5% CO2 at 37°C in 60-mm tissue culture–treated plates until the cells reached 80%–85% confluence. HeLa cells were cultured in a high-glucose formulation of DMEM (MilliporeSigma) supplemented with 10% (vol/vol) FBS, 2 mM l-glutamine, and penicillin/streptomycin. Transfection was performed with Lipofectamine 2000 (Life Technologies, Thermo Fisher Scientific; catalog 11668019) according to the manufacturer’s instructions. For heat shock experiments, HeLa cells were either transfected with GFP-DNAJB6 plasmids or cotransfected with Ch-TDP43 constructs for 24 hours. After transfection, cells were subjected to heat shock (42°C, 5% CO2) for 1 hour. For the drug treatment, HeLa cells were treated for 16 hours with either DMSO, SW02 (20 μM), YM01 (1 μM), JG231 (10 μM), or JG98 (10 μM).
Histochemistry, immunohistochemistry, and microscopy.
Isolated muscle was mounted using tragacanth gum (MilliporeSigma; G1128) and quick-frozen in liquid nitrogen–cooled 2-methylbutane. Samples were stored at –80°C until sectioning into 10-μm sections. H&E, nicotinamide adenine dinucleotide diaphorase (NADH), and laminin staining was performed as previously described (12). Sections were blocked in blocking reagent (PerkinElmer; FP1012), and incubated with a primary antibody followed by the appropriate secondary antibody. Briefly, muscle sections were affixed to slides incubated for 10 minutes in ice-cold acetone, mounted with Mowiol 4-88 (MilliporeSigma) plus DAPI, and examined using a fluorescence microscope (Nikon 80i upright+ and a Roper Scientific EZ monochrome CCD camera with Nikon NIS Elements deconvolution software). Nonfluorescent images were taken with a 5-megapixel color CCD (Nikon). Image processing and analysis were done with Nikon NIS Elements 4.0 software and Adobe Photoshop. Fluorescence Images of random fields were taken with a ×20 objective using a Nikon Eclipse 80i fluorescence microscope. The muscle fiber cross-sectional area (CSA) was measured using ImageJ software. Two separate individuals, blinded to the genotype and treatment status of mice, took photographs and measured the CSA.
FRAP and second harmonic generation.
The mice were treated as follows: YM01 was diluted in DMSO to 24 mg/mL to mimic the conditions used in cell culture. YM01 (10 μL) or vehicle was injected into the footpad of mice 24–48 hours before imaging. JG2321 was diluted in DMSO to 8 mg/mL to mimic the conditions used in cell culture. JG231 (50 μL) or vehicle was injected i.p. every other day for 2 weeks before imaging. For imaging, the mice were anesthetized with isoflurane with 5% used for induction and 1%–1.5% for maintenance during surgery at the level of reflex suppression. Skin overlying the FDB was removed. The mice were placed onto a customized viewing pane to facilitate imaging with the inverted 2-photon microscope. Electroporated skeletal muscle in live mice was observed with a Zeiss LSM 880 Airyscan confocal microscope with a motorized inverted Zeiss Axio Observer Z1 microscope frame. We used a ×40 1.2 NA water immersion objective, an excitation wavelength of 950 nm, and an emission filter of 499–571 nm for GFP probes. Regions of interest (2.4 M × 8.49 μM) were bleached with the Zeiss software. Laser settings were adjusted depending on myofiber depth to achieve at least a 60% reduction in GFP signal in myofibers with variable tissue depths. This was achieved with an excitation wavelength of 950 nm at 2%–10% laser power for 2–5 iterations for a duration of 0.24 μs. One image was captured immediately before and after photobleaching, followed by a series of post-bleaching images every 1–2 seconds, depending on the experiment. At least 3 different myofibers per animal were used for FRAP experiments for each condition. Each myofiber was tested at 3 different locations along its length. A single excitation wavelength of 800 nm was used for 2-photon electron microscopy (2PEM) and recorded using a ×40 1.2 NA water immersion objective. Second harmonic generation was detected with a nondescanned detector through a bandpass filter of 380–430 nm. As described previously, second harmonic generation occurs mostly from 2PEM of noncentrosymetric structures and is restricted to collagen fibers and striated muscle myosin rod domains (34). Data were analyzed using ImageJ. Measurements were made of the average fluorescence intensity of the bleached Z-disc, a nonbleached Z-disc as a control, and a background region for each time point. The background intensity was subtracted from the bleached and unbleached intensity values. A bleached/nonbleached ratio was calculated between the corrected intensities to account for photobleaching from image capture at each time point. The value from the initial bleach image (t = 0) was then subtracted from each time point, and the data were normalized so that the pre-bleach intensity was set to 1, and the intensity of the initial bleach image (t = 0) was set to 0. The normalized recovery data from at least 3 Z-discs per myofiber were used to generate a myofiber average. At least 3 different myofibers per animal were then averaged for further quantitative analysis. Grouped data are presented as the mean, and error bars represent the SD. The mobile fraction was calculated from the FRAP curve as previously described (35) with some modifications. GraphPad Prism 8 (GraphPad Software) was used for nonlinear regression analysis. FRAP data were fit to an exponential model with 1-phase association: Y = Y0 + (plateau – Y0) × (1 – exp[–K × X]). Y0 is the Y value when X (time) is zero. Plateau is the Y value at infinite time and represents the mobile fraction. K is the rate constant. Tau is the time constant. Half-time is computed as ln(2)/K. A mobile fraction myofiber average was calculated from 3 separate FRAP experiments within each myofiber. At least 3 different myofibers per animal were then averaged for further quantitative analysis. Grouped data are presented as the mean, and error bars represent the SD. Statistical significance was assessed using 1-way ANOVA with Turkey’s post hoc test.
Statistics.
Comparisons between 2 groups were made using a 2-tailed, paired Student’s t test. Comparisons between several groups were made using a 1-way ANOVA with Tukey’s post hoc test or Bonferroni’s correction to adjust for multiple comparisons. All analyses were performed with GraphPad Prism 8 (GraphPad Software). Data are presented as the mean ± SD, and results were considered statistically significant if P was less than 0.05. In some cases, the Student’s t test P value was calculated, and a Bonferroni-adjusted P value was determined for multiple comparisons.
Study approval.
All animal experimental protocols were approved by the Animal Studies Committee of the Washington University School of Medicine.
Author contributions
CCW, RB, ARF, and HLT conceived and designed the studies, analyzed data, and wrote the manuscript. RB, ARF, and AKB performed all experiments. RB performed cell culture and mouse functional studies, and ARF performed all histopathologic analyses and in vivo imaging studies. SKP, JAWD, HS and JEG provided key reagents and essential technical assistance. All authors reviewed the manuscript. Shared first authorship was agreed upon by all authors. RB performed the first set of experiments and was thus placed before ARF, whose studies were performed later.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AG031867 (to CCW), AG042095 (to CCW), AR068797 (to CCW and HLT), and AR075894 (to ARF); an American Academy of Neurology Neuroscience Research Training Scholarship 1350 (to ARF); the Muscular Dystrophy Association (to CCW); and by a gift from the LGMD-1D DNAJB6 Foundation.
Version 1. 05/19/2020
In-Press Preview
Version 2. 07/20/2020
Electronic publication
Version 3. 08/03/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(8):4470–4485.https://doi.org/10.1172/JCI136167.
Contributor Information
Rocio Bengoechea, Email: bengoecheaibacetar@wustl.edu.
Andrew R. Findlay, Email: arfindlay@wustl.edu.
Ankan K. Bhadra, Email: ankan@wustl.edu.
Hao Shao, Email: hao.shao@ucsf.edu.
Kevin C. Stein, Email: kstein14@gmail.com.
Sara K. Pittman, Email: pittmans@wustl.edu.
Jason E. Gestwicki, Email: jason.gestwicki@ucsf.edu.
Heather L. True, Email: heather.true@wustl.edu.
Conrad C. Weihl, Email: weihlc@WUSTL.EDU.
References
- 1.Iyadurai SJ, Kissel JT. The Limb-Girdle Muscular Dystrophies and the Dystrophinopathies. Continuum (Minneap Minn) 2016;22(6, Muscle and Neuromuscular Junction Disorders):1954–1977. doi: 10.1212/CON.0000000000000406. [DOI] [PubMed] [Google Scholar]
- 2.Kley RA, Olivé M, Schröder R. New aspects of myofibrillar myopathies. Curr Opin Neurol. 2016;29(5):628–634. doi: 10.1097/WCO.0000000000000357. [DOI] [PubMed] [Google Scholar]
- 3.Sarparanta J, et al. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat Genet. 2012;44(4):450–455. doi: 10.1038/ng.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harms MB, et al. Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann Neurol. 2012;71(3):407–416. doi: 10.1002/ana.22683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Homma S, Iwasaki M, Shelton GD, Engvall E, Reed JC, Takayama S. BAG3 deficiency results in fulminant myopathy and early lethality. Am J Pathol. 2006;169(3):761–773. doi: 10.2353/ajpath.2006.060250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wójtowicz I, et al. Drosophila small heat shock protein CryAB ensures structural integrity of developing muscles, and proper muscle and heart performance. Development. 2015;142(5):994–1005. doi: 10.1242/dev.115352. [DOI] [PubMed] [Google Scholar]
- 7.Smith DA, Carland CR, Guo Y, Bernstein SI. Getting folded: chaperone proteins in muscle development, maintenance and disease. Anat Rec (Hoboken) 2014;297(9):1637–1649. doi: 10.1002/ar.22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11(8):579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruggieri A, et al. Complete loss of the DNAJB6 G/F domain and novel missense mutations cause distal-onset DNAJB6 myopathy. Acta Neuropathol Commun. 2015;3:44. doi: 10.1186/s40478-015-0224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmio J, et al. Novel mutations in DNAJB6 gene cause a very severe early-onset limb-girdle muscular dystrophy 1D disease. Neuromuscul Disord. 2015;25(11):835–842. doi: 10.1016/j.nmd.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Sato T, et al. DNAJB6 myopathy in an Asian cohort and cytoplasmic/nuclear inclusions. Neuromuscul Disord. 2013;23(3):269–276. doi: 10.1016/j.nmd.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Bengoechea R, Pittman SK, Tuck EP, True HL, Weihl CC. Myofibrillar disruption and RNA-binding protein aggregation in a mouse model of limb-girdle muscular dystrophy 1D. Hum Mol Genet. 2015;24(23):6588–6602. doi: 10.1093/hmg/ddv363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandell S, et al. The enigma of 7q36 linked autosomal dominant limb girdle muscular dystrophy. J Neurol Neurosurg Psychiatry. 2010;81(8):834–839. doi: 10.1136/jnnp.2009.192351. [DOI] [PubMed] [Google Scholar]
- 14.Findlay AR, Bengoechea R, Pittman SK, Chou TF, True HL, Weihl CC. Lithium chloride corrects weakness and myopathology in a preclinical model of LGMD1D. Neurol Genet. 2019;5(2):e318. doi: 10.1212/NXG.0000000000000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein KC, Bengoechea R, Harms MB, Weihl CC, True HL. Myopathy-causing mutations in an HSP40 chaperone disrupt processing of specific client conformers. J Biol Chem. 2014;289(30):21120–21130. doi: 10.1074/jbc.M114.572461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hageman J, et al. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol Cell. 2010;37(3):355–369. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+] Mol Biol Cell. 2003;14(3):1172–1181. doi: 10.1091/mbc.e02-09-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aron R, Lopez N, Walter W, Craig EA, Johnson J. In vivo bipartite interaction between the Hsp40 Sis1 and Hsp70 in Saccharomyces cerevisiae. Genetics. 2005;169(4):1873–1882. doi: 10.1534/genetics.104.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3(1):28–36. doi: 10.1379/1466-1268(1998)003<0028:SFAEOD>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gestwicki JE, Shao H. Inhibitors and chemical probes for molecular chaperone networks. J Biol Chem. 2019;294(6):2151–2161. doi: 10.1074/jbc.TM118.002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young ZT, et al. Stabilizing the Hsp70-tau complex promotes turnover in models of tauopathy. Cell Chem Biol. 2016;23(8):992–1001. doi: 10.1016/j.chembiol.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wisén S, et al. Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS Chem Biol. 2010;5(6):611–622. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moses MA, et al. Targeting the Hsp40/Hsp70 chaperone axis as a novel strategy to treat castration-resistant prostate cancer. Cancer Res. 2018;78(14):4022–4035. doi: 10.1158/0008-5472.CAN-17-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirschke E, Goswami D, Southworth D, Griftfin PR, Agard DA. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014;157(7):1685–1697. doi: 10.1016/j.cell.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekhar A, Santiago M, Lam HN, Lee JH, Cavagnero S. Transient interactions of a slow-folding protein with the Hsp70 chaperone machinery. Protein Sci. 2012;21(7):1042–1055. doi: 10.1002/pro.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SH, et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154(1):134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Meister-Broekema M, et al. Myopathy associated BAG3 mutations lead to protein aggregation by stalling Hsp70 networks. Nat Commun. 2018;9(1):5342. doi: 10.1038/s41467-018-07718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selcen D, et al. Mutation in BAG3 causes severe dominant childhood muscular dystrophy. Ann Neurol. 2009;65(1):83–89. doi: 10.1002/ana.21553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eftekharzadeh B, et al. Hsp70 and Hsp40 inhibit an inter-domain interaction necessary for transcriptional activity in the androgen receptor. Nat Commun. 2019;10(1):3562. doi: 10.1038/s41467-019-11594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao H, et al. Exploration of benzothiazole rhodacyanines as allosteric inhibitors of protein-protein interactions with heat shock protein 70 (Hsp70) J Med Chem. 2018;61(14):6163–6177. doi: 10.1021/acs.jmedchem.8b00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA. 2002;99 Suppl 4:16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keefer KM, True HL. A toxic imbalance of Hsp70s in Saccharomyces cerevisiae is caused by competition for cofactors. Mol Microbiol. 2017;105(6):860–868. doi: 10.1111/mmi.13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuentealba RA, et al. Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J Biol Chem. 2010;285(34):26304–26314. doi: 10.1074/jbc.M110.125039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plotnikov SV, Millard AC, Campagnola PJ, Mohler WA. Characterization of the myosin-based source for second-harmonic generation from muscle sarcomeres. Biophys J. 2006;90(2):693–703. doi: 10.1529/biophysj.105.071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, et al. Dynamics of Z-band based proteins in developing skeletal muscle cells. Cell Motil Cytoskeleton. 2005;61(1):34–48. doi: 10.1002/cm.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.