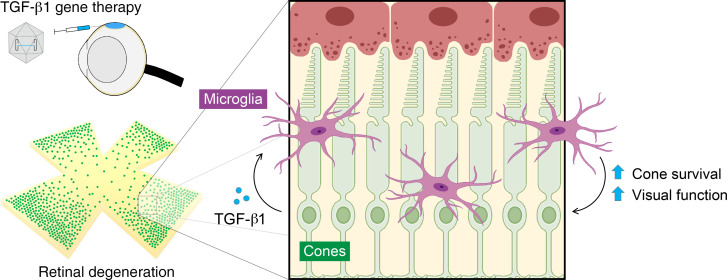
Abstract
Retinitis pigmentosa (RP) is a genetically heterogenous group of eye diseases in which initial degeneration of rods triggers secondary degeneration of cones, leading to significant loss of daylight, color, and high-acuity vision. Gene complementation with adeno-associated viral (AAV) vectors is one strategy to treat RP. Its implementation faces substantial challenges, however; for example, the tremendous number of loci with causal mutations. Gene therapy targeting secondary cone degeneration is an alternative approach that could provide a much-needed generic treatment for many patients with RP. Here, we show that microglia are required for the upregulation of potentially neurotoxic inflammatory factors during cone degeneration in RP, creating conditions that might contribute to cone dysfunction and death. To ameliorate the effects of such factors, we used AAV vectors to express isoforms of the antiinflammatory cytokine transforming growth factor beta (TGF-β). AAV-mediated delivery of TGF-β1 rescued degenerating cones in 3 mouse models of RP carrying different pathogenic mutations. Treatment with TGF-β1 protected vision, as measured by 2 behavioral assays, and could be pharmacologically disrupted by either depleting microglia or blocking the TGF-β receptors. Our results suggest that TGF-β1 may be broadly beneficial for patients with cone degeneration, and potentially other forms of neurodegeneration, through a pathway dependent upon microglia.
Keywords: Inflammation, Ophthalmology
Keywords: Cytokines, Gene therapy, Neurodegeneration
Introduction
Retinitis pigmentosa (RP) is a genetically heterogenous group of eye diseases that causes progressive loss of vision due to the dysfunction and degeneration of photoreceptors. Globally, the condition affects an estimated 2 million people, with thousands of pathogenic mutations identified to date spanning at least 80 different genes (1). In RP, there is early death of rods, the photoreceptors needed for vision in dim light, leading to difficulty with night vision typically by adolescence (2, 3). Rod degeneration is then followed by the dysfunction and death of cones, the cells essential for daylight, color, and high-acuity vision, loss of which can eventually result in blindness (3, 4). The pathogenesis of cone degeneration in RP is not understood, in part due to the fact that causal mutations are often exclusively expressed in rods, suggesting that cone death may be driven by a set of converging mechanisms independent of the genetic lesion (4). Despite ongoing efforts to characterize these mechanisms, there are still no widely accepted interventions to halt primary rod degeneration or secondary cone degeneration in patients with RP (5, 6).
One proposed treatment for RP and other inherited retinal diseases (IRDs) is the use of gene therapy to introduce an allele that can complement the mutation. This strategy recently led to the first commercial gene therapy for an IRD and has tremendous therapeutic promise (7, 8). Nonetheless, its implementation for RP faces several key challenges. Specifically, developing a gene therapy and clinical trial for each disease gene in RP will be logistically difficult considering the large number of genes to target, but the limited number of individuals with any given mutation (1). Because RP may go undiagnosed until the onset of night blindness (3), patients might also not have sufficient rods for correction of the genetic lesion. In addition to these obstacles, RP due to autosomal dominant or unidentified mutations, which together comprise one-third of cases (9), is not amenable to gene complementation and thus requires an alternative approach. To address these challenges, we and others have focused instead on the development of gene therapy targeting secondary cone degeneration (10–12), the process ultimately responsible for loss of quality of life in RP. Such therapies, if successful, would provide a much-needed and broadly applicable treatment option for the many patients with RP for whom gene therapy is otherwise infeasible.
Microglia are the resident immune cells of the retina and central nervous system (CNS). In response to infection or tissue damage, they can become activated, a state characterized by changes in microglial morphology, phagocytosis, and cytokine production (13, 14). Excessive microglial activation has been implicated in virtually every neurodegenerative disorder (13–15), including RP, in which activated microglia in the retina have been shown to phagocytose photoreceptors (16). During primary rod degeneration in RP, activated microglia appear to be harmful, as ablating these cells or suppressing their activation have been reported to enhance rod survival (16, 17). However, how microglia contribute to secondary cone degeneration is less clear. In a previous study of cone degeneration in RP, we overexpressed soluble CX3CL1 (fractalkine), a secreted molecule thought to regulate activation of microglia through a receptor on their surface (12). While soluble fractalkine prolonged cone survival and function in RP mouse models, it surprisingly did not require microglia to do so. In the current study, we further addressed the role of microglia in cone death by overexpressing different isoforms of transforming growth factor beta (TGF-β), an antiinflammatory cytokine known to inhibit microglial activation (18, 19). Using 3 mouse models of RP, we found that TGF-β1 was able to protect degenerating cones and save vision via a mechanism that required both microglia and TGF-β receptor signaling. Our data support the application of TGF-β1 as a generic therapy for patients with RP and highlight the therapeutic potential of modulating microglia to treat neurodegenerative conditions.
Results
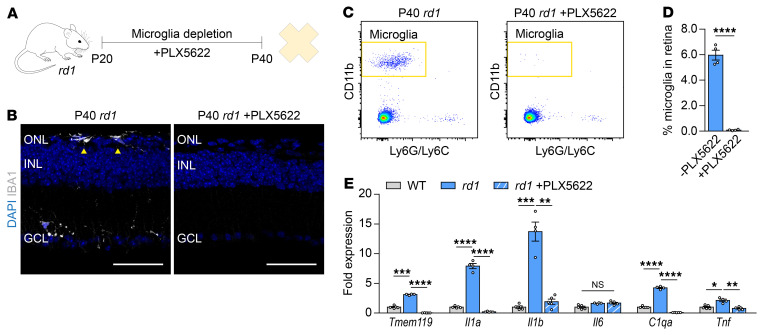
To examine the effects of microglia during secondary cone degeneration, mice were treated with PLX5622, a potent colony-stimulating factor 1 receptor (CSF1R) inhibitor that eliminates microglia (20). In the rd1 mouse line, the most widely used animal model of RP (21), PLX5622 treatment for 20 days depleted approximately 99% of retinal microglia (Figure 1, A–D) but grossly preserved peripheral immune populations, such as circulating monocytes and peritoneal macrophages (Supplemental Figure 1, B–E; supplemental material available online with this article; https://doi.org/10.1172/JCI136160DS1). We previously found that during secondary cone degeneration, there is persistent upregulation in the retina of Tmem119, a marker for microglia (22), as well as Il1a, Il1b, C1qa, and Tnf (12), inflammatory factors that have been shown to induce neurotoxicity both in vitro and in vivo (15, 23, 24). Here, we confirmed these findings (Figure 1E) and sought to determine if microglia were not just correlated with, but responsible for, the upregulation of inflammatory genes. Real-time–polymerase chain reaction (RT-PCR) performed on retinas from rd1 mice with or without PLX5622 treatment demonstrated that increased expression of Il1a, Il1b, C1qa, and Tnf was abolished following microglia depletion (Figure 1E). These data strengthened our hypothesis that microglia play a causal role in retinal inflammation during secondary cone degeneration.
Figure 1. Retinal expression of inflammatory genes after microglia depletion.
(A) Timeline of microglia depletion. Microglia from FVB (rd1) mice were pharmacologically depleted with PLX5622 beginning at P20 with harvesting of retinas at P40. (B) Retinal cross-sections from P40 rd1 mice (n = 2) with or without depletion. Arrowheads depict IBA1-positive microglia in the ONL by immunostaining. Scale bar: 50 μm. Representative gating (C) and quantification (D) by flow cytometry of microglia as a percentage of all retinal cells in P40 rd1 mice (n = 4) with or without depletion. Microglia were defined as CD11b-positive Ly6G/Ly6C-negative cells. For full gating strategy, see Supplemental Figure 1A. (E) mRNA expression of indicated genes in retinas (n = 4–5) from 6- to 8-week-old WT (sighted FVB) or P40 rd1 mice with or without 20 days of PLX5622. Fold changes are relative to WT retinas. Data shown are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by 2-tailed Student’s t test for D, 2-tailed Student’s t test with Bonferroni’s correction for E. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
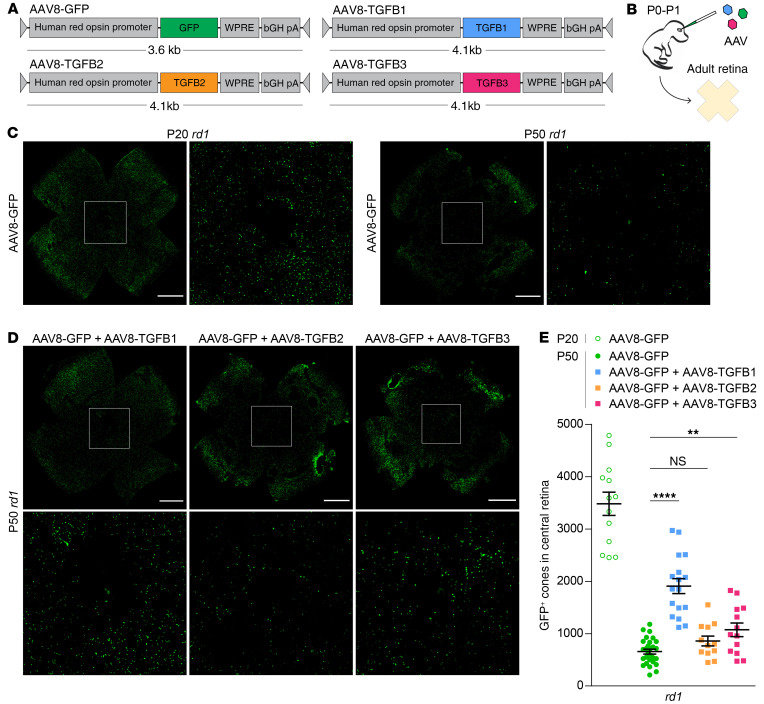
TGF-β is a major antiinflammatory cytokine that signals through the TGF-β type I (TGFBR1) and type II (TGFBR2) receptors to trigger downstream expression of target genes (25). Exogenous TGF-β can inhibit microglial production of inflammatory cytokines such as Tnf and Il6 (18, 19), whereas ablation of TGF-β signaling in microglia via genetic deletion of TGFBR2 leads to activation of these cells (26) and, notably, degenerative changes in the retina highly reminiscent of RP (27). We reasoned that suppressing microglial activation and its resulting inflammation with TGF-β might be beneficial for degenerating cones in RP. To test this idea, adeno-associated viral (AAV) vectors encoding each of the TGF-β isoforms — TGF-β1 (AAV8-TGFB1), TGF-β2 (AAV8-TGFB2), and TGF-β3 (AAV8-TGFB3) — were generated and subretinally injected into rd1 mice at postnatal day 0 to 1 (P0–P1), a time point enabling infection of photoreceptors throughout the entire retina (Figure 2, A and B). These vectors used the human red opsin promoter to drive expression in cones (28) and were coadministered with a previously described GFP vector (AAV8-GFP) employing the same promoter to facilitate cone quantification (11, 12). GFP driven by the human red opsin promoter could first be detected in cones around 7 days after injection, with strong expression by day 14 (Supplemental Figure 2A). AAV vectors with this same promoter resulted in significant upregulation of TGF-β isoforms in infected retinas at both the mRNA and protein levels (Supplemental Figure 2, B and C).
Figure 2. Effect of overexpressing TGF-β isoforms on cone survival.
Schematics of AAV vector design (A) and delivery (B). (C) Representative flat-mounts of FVB (rd1) retinas treated with AAV8-GFP and harvested at P20 or P50. Paired images depict low and high magnifications (boxed areas). Scale bar: 1 mm. (D) Representative flat-mounts of rd1 retinas treated with indicated AAV vectors and harvested at P50. Scale bar: 1 mm. (E) Quantification of GFP-positive cones in central retinas of rd1 mice (n = 12–28) treated with indicated AAV vectors. Data shown are mean ± SEM. **P < 0.01, ****P < 0.0001 by 2-tailed Student’s t test with Bonferroni’s correction.
Secondary cone degeneration begins around P20 in rd1 mice, with massive loss of cones by P50, particularly within the central retina (Figure 2C). To measure the effect of TGF-β isoforms on retinal degeneration, the number of GFP-positive cones in the central retina was quantified. Compared with AAV8-GFP alone, there was no significant difference in the number of cones at P50 with the addition of AAV8-TGFB2, and only a modest increase with AAV8-TGFB3 (Figure 2, D and E). In contrast, infection with AAV8-TGFB1 nearly tripled the number of cones in the central retina at P50. To determine whether greater cone numbers with TGF-β1 were a result of cone preservation or rather a perturbation in retinal development, rd1 retinas treated with AAV8-GFP or AAV8-GFP plus AAV8-TGFB1 were examined at P20, before secondary cone degeneration. AAV8-TGFB1 did not alter the number of cones at this time point (Supplemental Figure 3, A and B), suggesting that the difference in cones at P50 was indeed due to prolonged survival. As increased cone counts with TGF-β1 could also be explained by a rearrangement of peripheral cones to the central retina, whole rd1 retinas were analyzed at P30 by flow cytometry, which showed significantly more GFP-positive cones in eyes treated with AAV8-GFP plus AAV8-TGFB1 compared with AAV8-GFP only (Supplemental Figure 3, C and D). Finally, to verify that TGF-β1 was improving the survival of GFP-positive cones and not just upregulating GFP expression, rd1 retinas at P50 were immunostained for cone arrestin, a marker of all cones, which again demonstrated significantly more cones in the central retina with the addition of AAV8-TGFB1 (Supplemental Figure 3, E and F). Together, these data indicated that AAV8-TGFB1 could rescue degenerating cones in the rd1 model of RP.
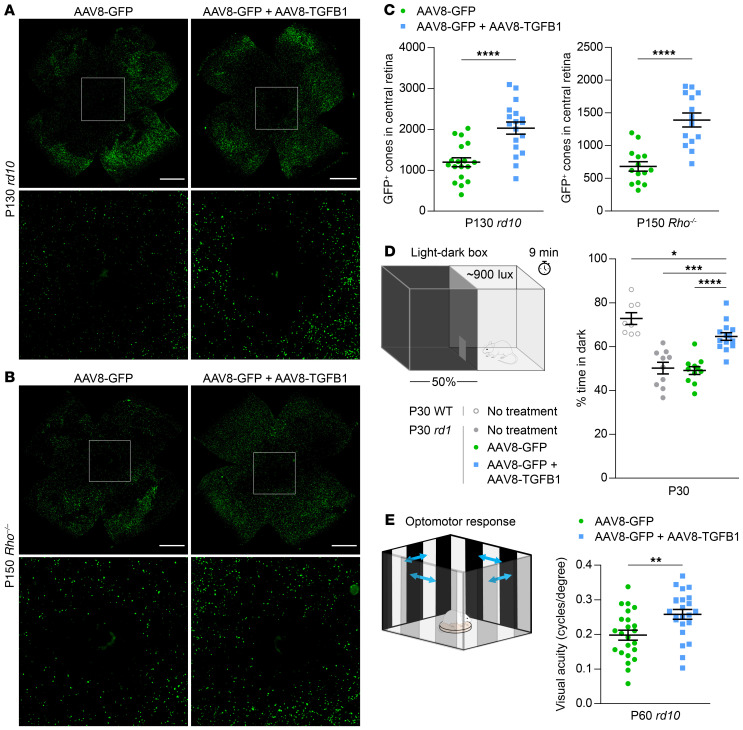
AAV8-TGFB1 was next studied in 2 more slowly degenerating mouse models of RP: rd10, which harbors a mutation in Pde6b, a common cause of autosomal recessive RP (21), and Rho–/–, which lacks rhodopsin, the most frequently mutated gene in autosomal dominant RP (29). Upregulation of Tgfb1 with AAV8-TGFB1 persisted in these older mice (Supplemental Figure 4A). In both strains, AAV8-TGFB1 again significantly improved cone survival (Figure 3, A–C), implying that TGF-β1 might be generically beneficial for cones in RP. The impact of TGF-β1 on rod survival was additionally investigated in rd10 mice by measuring the thickness of the outer nuclear layer (ONL), which normally consists primarily of rods. Despite preserving cones in the same model, AAV8-TGFB1 did not prevent rod death and the reduction of ONL thickness in rd10 retinas (Supplemental Figure 4, B and C). Thus, the therapeutic effect of AAV8-TGFB1 in RP appears to be selective for cones.
Figure 3. Effect of AAV8-TGFB1 on long-term cone survival and cone-mediated vision.
Representative flat-mounts of rd10 (A) and Rho–/– (B) retinas treated with AAV8-GFP or AAV8-GFP plus AAV8-TGFB1. Paired images depict low and high magnifications (boxed areas). Scale bar: 1 mm. (C) Quantification of GFP-positive cones in central retinas of rd10 (n = 18) and Rho–/– (n = 14) mice. (D) Percentage of time spent in dark in a 50:50 light-dark box for untreated animals (n = 8–10) and C3H (rd1) mice (n = 11–14) treated with AAV8-GFP or AAV8-GFP plus AAV8-TGFB1. (E) Visual acuity in eyes from P60 rd10 mice (n = 23) as measured by optomotor after treatment with AAV8-GFP or AAV8-GFP plus AAV8-TGFB1. Data shown are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by 2-tailed Student’s t test for C and E, 2-tailed Student’s t test with Bonferroni’s correction for D.
Encouraged by our histological findings, we assessed the potential clinical relevance of TGF-β1 gene therapy by subjectingtreated mice to a light-dark discrimination test. Sighted mice spend less time in well-illuminated spaces, as demonstrated by the strong preference of WT animals for the dark half of a 50:50 light-dark box (Figure 3D and Supplemental Figure 4D). Conversely, rd1 mice, which can no longer distinguish light from dark by P30 due to loss of functional photoreceptors, equally split their time between the 2 compartments. Compared with animals without treatment or receiving AAV8-GFP only, rd1 mice treated with AAV8-GFP plus AAV8-TGFB1 spent significantly more time in the dark, consistent with an improvement in visual function allowing for light-dark discrimination. As a complementary measure of vision, the optomotor assay was performed on rd10 mice treated with AAV8-GFP in one eye and AAV8-GFP plus AAV8-TGFB1 in the other. In this experiment, moving stripes are used to elicit the visually dependent optomotor response. By adjusting the stripes until the animal can no longer track them, the visual acuity in each eye can be estimated (30). At P60, rd10 eyes treated with AAV8-GFP plus AAV8-TGFB1 exhibited significantly better visual acuity than those only receiving AAV8-GFP (Figure 3E). From these data, we concluded that TGF-β1 in mouse RP not only helps preserve cones, but also importantly protects from vision loss.
Although we found AAV8-TGFB1 to be beneficial for cones, TGF-β signaling in the eye has also been reported to mediate cataract formation (31, 32), ocular hypertension leading to loss of retinal ganglion cells (RGCs) (32), and epithelial-mesenchymal transition (EMT) in the retinal pigment epithelium (RPE), a process implicated in proliferative vitreoretinopathy (33, 34). Mice treated with AAV8-GFP or AAV8-GFP plus AAV8-TGFB1 were thus examined for these possibilities. At P30, no obvious difference in the opacity of the lens was seen in animals receiving AAV8-GFP plus AAV8-TGFB1 compared with AAV8-GFP only (Supplemental Figure 5A). Moreover, treatment with AAV8-TGFB1 did not impact the number of RGCs at this time point (Supplemental Figure 5, B and C). To assess for EMT in the RPE, immunostaining was performed for ZO-1, a component of epithelial tight junctions (35), and α-smooth muscle actin (α-SMA), which labels RPE cells undergoing EMT (33, 34). Neither of these proteins were qualitatively changed in the RPE with the addition of AAV8-TGFB1 (Supplemental Figure 5, D and E), suggesting that at least up to 1 month after treatment, AAV8-TGFB1 is not noticeably disruptive in the eye.
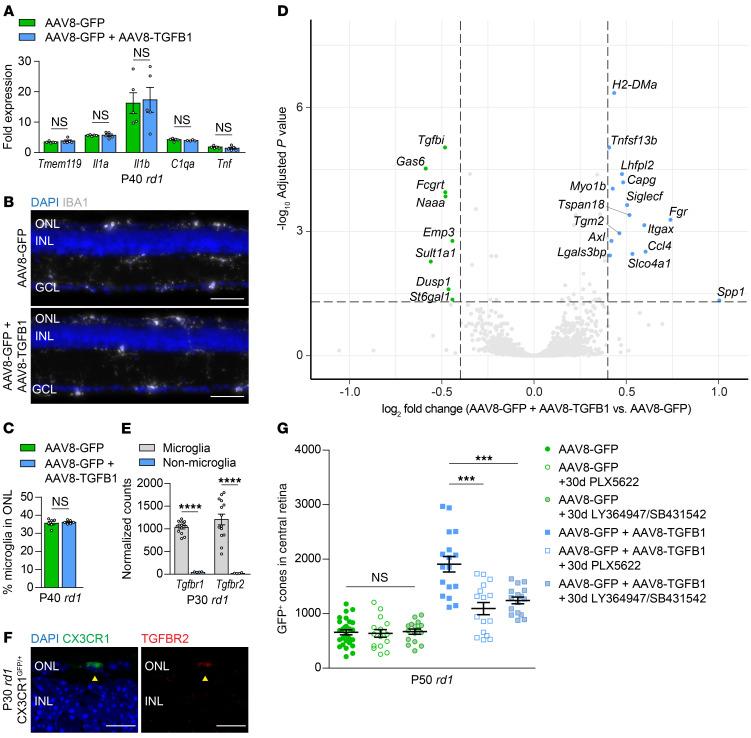
How does AAV8-TGFB1 combat secondary cone degeneration? Given the antiinflammatory properties of TGF-β, mRNA levels of Tmem119, Il1a, Il1b, C1qa, and Tnf in P40 rd1 retinas were quantified and, surprisingly, were found to be unchanged with AAV8-TGFB1 (Figure 4A). AAV8-TGFB1 likewise did not affect the number of microglia in the retina as assayed by flow cytometry (Supplemental Figure 6, A and B), and treatment did not alter the percentage of microglia in the ONL (Figure 4, B and C), the retinal layer in which microglia preferentially localize during degeneration (12). To better understand the microglial response to AAV8-TGFB1, microglia from P30 rd1 retinas treated with AAV8-GFP or AAV8-GFP plus AAV8-TGFB1 were isolated by cell sorting and subjected to RNA sequencing (RNA-seq). Sorted microglia were highly pure, expressing microglia markers such as Tmem119 and P2ry12, but not those of other cell types (Supplemental Figure 6C). Only 23 genes were significantly altered (adjusted P < 0.05, log2 fold change >0.4) in microglia treated with AAV8-TGFB1 (Figure 4D). These included Spp1 and Gas6, the most upregulated and downregulated of the 23 genes, respectively, which were validated by RT-PCR in microglia from both P30 rd1 and P200 rd10 retinas (Supplemental Figure 6D).
Figure 4. Role of retinal microglia in AAV8-TGFB1-mediated cone survival.
(A) mRNA expression of indicated genes in FVB (rd1) retinas (n = 4–5) treated with AAV8-GFP or AAV8-GFP plus AAV8-TGFB1. Fold changes are relative to untreated WT (sighted FVB) retinas. Representative images (B) and quantification (C) of IBA1-positive microglia in the ONL of P40 rd1 retinas (n = 6–7) treated with AAV8-GFP or AAV8-GFP plus AAV8-TGFB1. Scale bar: 50 μm. (D) Volcano plot of up- and downregulated genes in microglia sorted from P30 rd1 retinas (n = 7) after treatment with AAV8-GFP plus AAV8-TGFB1 relative to AAV8-GFP only. Dotted lines indicate adjusted P < 0.05 and log2 fold change >0.4. (E) Normalized RNA-seq counts for expression of Tgfbr1 and Tgfbr2 in microglia versus nonmicroglia cells from P30 rd1 retinas (n = 4–14). (F) Immunostaining for TGFBR2 in rd1 CX3CR1GFP/+ retinas (n = 2). Arrowheads indicate colocalization of TGFBR2 with a CX3CR1-positive microglia in the ONL. Scale bar: 10 μm. (G) Quantification of GFP-positive cones in central retinas of rd1 mice after 30 days of microglia depletion with PLX5622 (n = 16) or inhibition of TGFBR1/2 with LY364947 and SB431542 (n = 16). Data for untreated groups are taken from Figure 2E. Data shown are mean ± SEM. ***P < 0.001, ****P < 0.0001 by 2-tailed Student’s t test with Bonferroni’s correction for A and G, 2-tailed Student’s t test for C and E. INL, inner nuclear layer.
The importance of these gene expression changes in microglia was subsequently evaluated by depleting microglia from mice treated with AAV8-TGFB1 during secondary cone degeneration. Beginning at P20, rd1 mice were administered PLX5622, which eliminated approximately 99% of retinal microglia even in eyes infected with AAV vectors (Supplemental Figure 6E). While microglia depletion had no significant effect on cone survival in rd1 retinas treated with AAV8-GFP only (Figure 4G), consistent with our prior observations (12), it significantly abrogated cone rescue by AAV8-TGFB1. These findings indicate that microglia are not inherently helpful or harmful for degenerating cones, but are necessary for the cone survival mediated by TGF-β1 gene therapy. In the retina, microglia are among the only cells that highly express TGFBR1 and TGFBR2 (Figure 4, E and F, and ref. 27), both of which are required for TGF-β signaling (25). We therefore hypothesized that AAV8-TGFB1 might act via TGF-β receptors on microglia in order to promote cone survival. To test this, rd1 mice treated with AAV8-GFP or AAV8-GFP plus AAV8-TGFB1 were administered a combination of LY364947 and SB431542, potent TGFBR1/2 inhibitors capable of blocking these receptors in vivo (36). As with microglia depletion, TGFBR1/2 inhibition had no discernable effect on retinas treated with AAV8-GFP only (Figure 4G), suggesting that any endogenous signaling through these receptors during cone degeneration did not dramatically affect cone survival. On the other hand, treatment with LY364947 and SB431542 significantly disrupted the ability of AAV8-TGFB1 to preserve cones (Figure 4G). Collectively, these results demonstrate that both microglia and TGF-β signaling through TGFBR1 and TGFBR2 are needed for AAV8-TGFB1 to function therapeutically.
Discussion
AAV8-TGFB1 provides a novel gene therapy that protects cones and vision in multiple mouse models of RP, supporting its potential translation to different genetic forms of retinal degeneration in patients. Interestingly, although depletion of microglia itself does not help or hinder cone survival, cone rescue by AAV8-TGFB1 requires microglia. Together, these data suggest that microglia do not play a significantly negative role during cone degeneration in RP, and under certain conditions, can be induced to benefit cones. Our study further shows a dependence of TGF-β1 gene therapy on TGFBR1 and TGFBR2, which likely mediate signaling directly within microglia. We favor a model in which this signaling induces microglia to create a retinal environment favorable to cone survival. Our findings thus highlight a new immunomodulatory strategy centered around microglia for treating patients with RP, an approach that may also be relevant for other degenerative diseases of the visual system and CNS.
Of note, dependence of TGF-β1 gene therapy on microglia in this study was determined using PLX5622, a CSF1R inhibitor which depleted up to approximately 99% of retinal microglia. However, CSF1R is likewise present on monocytes and other macrophages in the body, and although the majority of these populations are not depleted with PLX5622 (37), their functions could theoretically be affected by CSF1R inhibition. While we cannot exclude possible contributions from monocytes or macrophages residing in the choroid, we argue that retinal microglia are the most likely effector cells of AAV8-TGFB1 given their high expression of the TGF-β receptors and proximity to degenerating cones. It should further be mentioned that in both our microglia depletion and TGFBR1/2 inhibition experiments, cone rescue with AAV8-TGFB1 was not fully eliminated. This could have been due to therapeutic activity from TGF-β1 before P20, as PLX5622, LY364947, and SB431542 were not administered until this age. Alternatively, blocking of the relevant receptors by these drugs may have been incomplete, leading to ablation of most, but not all, of the treatment effect.
One question that remains is how exactly microglia preserve cones in response to TGF-β1. Surprisingly, RNA-seq of retinal microglia from P30 rd1 mice only identified 23 genes that were significantly altered with AAV8-TGFB1. This list included Spp1, which is upregulated in microglia associated with RPE protection (38), but did not contain any genes already known to aid in cone survival. In treated eyes, it is conceivable that microglia may become less sensitive to elevated TGF-β1 levels over time, resulting in fewer and less pronounced transcriptional changes. RNA-seq of these microglia at a time point earlier than P30 may therefore uncover additional differences in gene expression that were not captured in our analysis. Alternatively, the therapeutic effects of AAV8-TGFB1 may occur via changes not detectable by RNA-seq, such as through posttranslational modifications of the proteome.
Clinically, a major appeal of AAV-mediated gene therapy is the prospect of sustained or even lifelong treatment following a single dose of vector. Nonetheless, receiving a long-term treatment also carries risks, which must be carefully weighed against the benefits of therapy. With AAV8-TGFB1, we were particularly concerned about the possibilities of cataract formation, RGC death, and EMT in the RPE, any of which could be detrimental to vision. Reassuringly, none of these complications were observed at 1 month after vector delivery, supporting the safety of TGF-β1 gene therapy in the eye. Notable differences between the methodologies of our paper and prior studies may explain why we found AAV8-TGFB1 to be well tolerated. Using an adenoviral vector, Robertson et al. showed that overexpression of TGF-β1 in rats could cause fibrosis in the lens and severe RGC loss as early as 2 weeks after injection (32). However, this vector was administered in the anterior segment of the eye rather than the subretinal space, employed a promoter with much broader expression, and, being an adenoviral vector, was substantially more inflammatory than the AAV vectors tested here (39). In the RPE, TGF-β1 has been widely used to study EMT in vitro as it initiates loss of epithelial markers and upregulates α-SMA in cultured RPE cells (33, 34). Even so, it is unclear whether this effect of TGF-β1 can be extended in vivo, as experiments conducted on sheets of RPE suggest that normal cell-cell contact, which is absent from cell culture models, is sufficient to prevent induction of EMT by TGF-β signaling (40). Finally, while we did not detect any changes in the lens, RGCs, or RPE with AAV8-TGFB1 after 1 month, the same assessment at a later time point could yield different results. An important future step will be to reevaluate the safety of TGF-β1 in the eye following a longer duration of therapy, ideally in large animals.
What might treatment with TGF-β1 look like in patients? Compared with mouse models of RP, which undergo cone degeneration over the span of months, humans with the disease typically begin losing their cones as young adults, with progression over multiple decades (3, 41). Based on these kinetics, we speculate that prolonged cone survival for several months with AAV8-TGFB1 as demonstrated in this study may potentially translate to years of meaningful vision for patients. With the addition of TGF-β1, there is now a growing list of promising molecules and mechanisms capable of alleviating cone death in RP (4, 10–12). Although not individually curative, a combination of these and other treatments may ultimately slow the disease enough to provide lifelong preservation of sight.
Methods
Animals.
CD-1 (catalog 022) and FVB (rd1) (catalog 207) mice were purchased from Charles River Laboratories. Sighted FVB (catalog 004828), rd10 (catalog 004297), C3H (rd1) (catalog 000659), sighted C3H (catalog 003648), and CX3CR1GFP/+ (catalog 005582) mice were purchased from The Jackson Laboratory. Rhodopsin null (Rho–/–) mice were a gift from Janis Lem (Tufts University, Boston, Massachusetts, USA) (29). FVB and CX3CR1GFP/+ mice were crossed for at least 4 generations to obtain rd1 CX3CR1GFP/+ animals. Mice were subsequently bred and maintained at Harvard Medical School on a 12-hour alternating light and dark cycle. Both male and female mice were used in all experiments.
Histology.
To prepare retinal cross-sections, enucleated eyes were dissected in phosphate-buffered saline (PBS) to remove the cornea, iris, lens, and ciliary body. The remaining eye cup was fixed in 4% paraformaldehyde for 1 hour at room temperature, cryoprotected in a sucrose gradient, and embedded in a 1:1 solution of 30% in PBS and optimal cutting temperature compound (Tissue-Tek) on dry ice. Frozen eye cups were cut on a Leica CM3050S cryostat (Leica Microsystems) into 20- to 30-μm sections. If applicable, sections were then blocked for 1 hour at room temperature in PBS containing 5% goat serum and 0.1% Triton X-100, stained with primary antibodies (Supplemental Table 1) in blocking buffer overnight at 4°C, and incubated with the appropriate secondary antibody in PBS for 2 hours at room temperature. All sections were incubated for 5 minutes at room temperature with PBS containing 0.5 μg/mL of 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific) and mounted using Fluoromount-G (SouthernBiotech). To prepare retinal flat-mounts for quantifying GFP-positive cones, whole retinas were fixed in 4% paraformaldehyde for 30 minutes at room temperature. After PBS washes, retinas were relaxed with 4 radial incisions, flattened onto a microscope slide, and mounted with the ganglion cell layer facing up. Descriptions of additional histology procedures can be found in the Supplemental Methods.
Microglia depletion.
Microglia were depleted using PLX5622 (a gift from Plexxikon, Berkeley, California, USA), an orally available CSF1R inhibitor, formulated into AIN-76A rodent chow (Research Diets) at 1200 mg/kg and provided ad libitum.
Flow cytometry and cell sorting.
All flow cytometry and cell sorting were performed on a BD FACSAria II and analyzed using FlowJo 10 (Tree Star). For retinal cells, freshly dissected retinas were dissociated as previously described (12) using cysteine-activated papain followed by gentle trituration with a micropipette. If applicable, harvested cells were then blocked for 5 minutes with 1:100 rat anti-mouse CD16/32 (BD Pharmingen) and incubated for 20 minutes on ice with the antibodies listed in Supplemental Table 1. Before analysis, all samples were passed through a 40-μm filter and stained with 0.5 μg/mL of DAPI (Thermo Fisher Scientific) in FACS buffer (PBS containing 2% fetal bovine serum and 2 mM ethylenediaminetetraacetic acid [EDTA]) to exclude nonviable cells. Descriptions of additional flow cytometry procedures can be found in the Supplemental Methods.
RT-PCR.
mRNA was isolated from whole retinas or sorted microglia using an RNeasy Micro Kit (Qiagen) as previously described (12), with the exception of mRNA from Rho–/– retinas, which was isolated from fixed tissues using the RecoverAll Total Nucleic Acid Isolation Kit for FFPE (Thermo Fisher Scientific). One whole retina or 1000–2000 sorted microglia were collected per sample. cDNA was synthesized using the SuperScript III First-Strand Synthesis System (Invitrogen) with oligo(dT) primers, followed by RT-PCR using the Power SYBR Green PCR Master Mix (Applied Biosystems) on a CFX96 real-time PCR detection system (Bio-Rad). Reactions were performed in triplicate with expression normalized to the housekeeping gene Gapdh. Sequences for RT-PCR primers were designed using PrimerBank (42) and are listed in Supplemental Table 2. For Rho–/– samples, primers targeting shorter amplicons (Gapdh-s and Tgfb1-s) were used to account for potential fragmentation of mRNA following fixation.
AAV vector design and production.
The AAV–human red opsin–GFP-WPRE-bGH (AAV8-GFP) plasmid was a gift from Botond Roska (Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland) (43). To generate plasmids for TGF-β isoforms, the GFP coding sequence from AAV8-GFP was replaced with the full-length mouse cDNA for TGF-β1 (NM_011577.2), TGF-β2 (NM_009367.4), or TGF-β3 (NM_009368.3) flanked by NotI and AgeI restriction sites. Recombinant AAV serotype 8 (AAV8) vectors were produced as previously described (11, 12, 44). Briefly, HEK293T cells were transfected using polyethylenimine with a mixture of the vector plasmid, adenovirus helper plasmid, and rep2/cap8 packaging plasmid. Supernatant was harvested 72 hours after transfection, and viral particles were PEGylated overnight and precipitated by centrifugation. Viral particles were subsequently centrifuged through an iodixanol gradient to remove cellular debris, and the recovered vectors were washed 3 times with PBS and collected in a final volume of 100–200 μL. AAV vectors were semiquantitatively titered by SYPRO Ruby (Molecular Probes) staining for viral capsid proteins (VP1, VP2, and VP3) in comparison with a reference vector titered by RT-PCR.
Subretinal injections.
All subretinal injections were performed on neonatal mice at P0–P1. After anesthetization of the mouse on ice, the palpebral fissure was carefully opened with a 30-gauge needle and the eye exposed. Using a glass needle controlled by a FemtoJet microinjector (Eppendorf), approximately 0.25 μL AAV vectors was then injected into the subretinal space. For each eye, 5 × 108 vector genomes (vg) per eye of AAV8-GFP were administered. All other vectors were administered at 1 × 109 vg per eye.
Image acquisition and analysis.
Images of retinal cross-sections and GFP-positive cones in retinal flat-mounts were acquired using a Zeiss LSM710 scanning confocal microscope (×20 air objective or ×40 oil objective) and Nikon Ti inverted widefield microscope (×10 air objective), respectively. All image analysis was performed using ImageJ. Quantification of GFP-positive cones was performed as previously described (12) using a custom ImageJ module (available at https://sites.imagej.net/Seankuwang/). For each flat-mount, the user indicated the location of the optic nerve head and each of the 4 retinal leaflets. The module then automatically defined the region corresponding to the central retina and counted the number of GFP-positive objects within the region. This value was used to represent the number of GFP-positive cones in the central retina for each sample. Quantification of microglia in the ONL of retinal cross-sections was performed by dividing the number of microglia in the ONL across 5 random fields by the total number of microglia in those fields. Microglia were defined as residing in the ONL if 50% or more of the cell body was located in that layer. Descriptions of additional image analysis procedures can be found in the Supplemental Methods.
Light-dark discrimination.
Innate light-avoidance behavior in mice was assessed as previously described (45) with minor modifications. A 28 cm (length) by 28 cm (width) by 21 cm (height) plastic chamber (Med Associates) was divided into 2 equally sized compartments: one dark and one brightly illuminated (~900 lux). Temperatures in the 2 compartments differed by less than 1°C. A small opening connected the 2 compartments, allowing subjects to freely travel throughout the chamber. At the beginning of each trial, a mouse was placed in the illuminated compartment and its activity recorded for 10 minutes. If after one minute, the animal had not yet entered the dark compartment, it was gently directed there, removed from the chamber, and the trial restarted. The location and movement of each mouse were determined by infrared sensors and analyzed with Activity Monitor (Med Associates). Percentage of time spent in dark was calculated based on activity during the final 9 minutes of each trial.
Optomotor assay.
Visual acuity was measured by an observer blinded to the treatment assignment using the OptoMotry System (CerebralMechanics) as previously described (11, 12, 46). Mice were placed inside a virtual-reality chamber with bright background luminance to saturate rods and presented with sine wave gratings of varying spatial frequencies. During each test, the observer assessed reflexive head-tracking movements of the animal in response to the sine wave grating, and for each eye, the highest spatial frequency at which the animal tracked the grating was determined to be the visual acuity. Left and right eyes were tested independently using clockwise and counterclockwise gratings, respectively, as only motion in the temporal-to-nasal direction is known to evoke the optomotor response in mice (4).
RNA sequencing.
Transcriptional profiling of microglia (7 biological replicates per experimental condition) or nonmicroglia (4 biological replicates total) was performed as previously described (12). One thousand microglia (CD11b+ Ly6G/Ly6C–) or nonmicroglia cells (CD11b–) from each retina were sorted into 10 μL of Buffer TCL (Qiagen) containing 1% β-mercaptoethanol and immediately frozen on dry ice. Samples were subsequently sent to the Broad Institute Genomics Platform for cDNA library synthesis and sequencing using a modified Smart-Seq2 protocol (47) with an expected coverage of approximately 6 million reads per sample. Before analysis, reads were subjected to quality control measures and mapped to the GRCm38.p6 reference genome. Reads assigned to each gene were then quantified using featureCounts (48) and normalized and analyzed for differential expression using DESeq2 (49).
TGFBR1/2 inhibition.
Pharmacological inhibition of the TGF-β type I and II receptors in vivo was performed using a combination of SB431542 (SelleckChem) and LY364947 (SelleckChem) as previously described (36). Both compounds were dissolved in PBS containing 5% dimethyl sulfoxide (DMSO) and 30% polyethylene glycol 300 and dosed at 10 mg/kg daily via intraperitoneal injections.
Data availability.
All RNA-seq data generated in this work have been deposited in the Gene Expression Omnibus (GEO) repository (accession number GSE145601).
Statistics.
All group data are shown as mean ± SEM. Two-tailed Student’s t tests were used to compare experimental groups, with the addition of a Bonferroni’s correction if 3 or more hypotheses were tested. Differences between groups were considered significant when the P value was less than 0.05.
Study approval.
All animal experiments were approved by the IACUC of Harvard University in accordance with institutional guidelines.
Author contributions
SKW and CLC designed the study. SKW and YX performed the experiments and analyzed the data. SKW and CLC wrote the manuscript with input from YX.
Supplementary Material
Acknowledgments
The authors thank Sophia Zhao, Christin Hong, Ryoji Amamoto, the Flow Cytometry Core at Joslin Diabetes Center, the Mouse Behavior Core at Harvard Medical School, and Microscopy Resources on the North Quad at Harvard Medical School for discussions and technical support. This work was supported by the Howard Hughes Medical Institute (SKW, CLC) and Foundation Fighting Blindness (SKW).
Version 1. 04/30/2020
In-Press Preview
Version 2. 07/13/2020
Electronic publication
Version 3. 08/03/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(8):4360–4369.https://doi.org/10.1172/JCI136160.
See the related Commentary at Precision metabolome reprogramming for imprecision therapeutics in retinitis pigmentosa.
Contributor Information
Sean K. Wang, Email: sean_wang@hms.harvard.edu.
Yunlu Xue, Email: yunlu_xue@hms.harvard.edu.
Constance L. Cepko, Email: cepko@genetics.med.harvard.edu.
References
- 1.Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin Genet. 2013;84(2):132–141. doi: 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merin S, Auerbach E. Retinitis pigmentosa. Surv Ophthalmol. 1976;20(5):303–346. doi: 10.1016/S0039-6257(96)90001-6. [DOI] [PubMed] [Google Scholar]
- 3.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368(9549):1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 4.Narayan DS, Wood JP, Chidlow G, Casson RJ. A review of the mechanisms of cone degeneration in retinitis pigmentosa. Acta Ophthalmol. 2016;94(8):748–754. doi: 10.1111/aos.13141. [DOI] [PubMed] [Google Scholar]
- 5.Sahni JN, Angi M, Irigoyen C, Semeraro F, Romano MR, Parmeggiani F. Therapeutic challenges to retinitis pigmentosa: from neuroprotection to gene therapy. Curr Genomics. 2011;12(4):276–284. doi: 10.2174/138920211795860062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang XF. Current pharmacological concepts in the treatment of the retinitis pigmentosa. Adv Exp Med Biol. 2018;1074:439–445. doi: 10.1007/978-3-319-75402-4_54. [DOI] [PubMed] [Google Scholar]
- 7.Russell S, et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390(10097):849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trapani I, Banfi S, Simonelli F, Surace EM, Auricchio A. Gene therapy of inherited retinal degenerations: prospects and challenges. Hum Gene Ther. 2015;26(4):193–200. doi: 10.1089/hum.2015.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daiger SP, Bowne SJ, Sullivan LS. Perspective on genes and mutations causing retinitis pigmentosa. Arch Ophthalmol. 2007;125(2):151–158. doi: 10.1001/archopht.125.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrne LC, et al. Viral-mediated RdCVF and RdCVFL expression protects cone and rod photoreceptors in retinal degeneration. J Clin Invest. 2015;125(1):105–116. doi: 10.1172/JCI65654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong W, MacColl Garfinkel AE, Li Y, Benowitz LI, Cepko CL. NRF2 promotes neuronal survival in neurodegeneration and acute nerve damage. J Clin Invest. 2015;125(4):1433–1445. doi: 10.1172/JCI79735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang SK, Xue Y, Rana P, Hong CM, Cepko CL. Soluble CX3CL1 gene therapy improves cone survival and function in mouse models of retinitis pigmentosa. Proc Natl Acad Sci USA. 2019;116(20):10140–10149. doi: 10.1073/pnas.1901787116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Block ML, Zecca L, Hong JS. Microglia-mediated neurotoxicity: uncovering the molecular mechanisms. Nat Rev Neurosci. 2007;8(1):57–69. doi: 10.1038/nrn2038. [DOI] [PubMed] [Google Scholar]
- 14.Subhramanyam CS, Wang C, Hu Q, Dheen ST. Microglia-mediated neuroinflammation in neurodegenerative diseases. Semin Cell Dev Biol. 2019;94:112–120. doi: 10.1016/j.semcdb.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Smith JA, Das A, Ray SK, Banik NL. Role of pro-inflammatory cytokines released from microglia in neurodegenerative diseases. Brain Res Bull. 2012;87(1):10–20. doi: 10.1016/j.brainresbull.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao L, et al. Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol Med. 2015;7(9):1179–1197. doi: 10.15252/emmm.201505298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng B, Xiao J, Wang K, So KF, Tipoe GL, Lin B. Suppression of microglial activation is neuroprotective in a mouse model of human retinitis pigmentosa. J Neurosci. 2014;34(24):8139–8150. doi: 10.1523/JNEUROSCI.5200-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim WK, Hwang SY, Oh ES, Piao HZ, Kim KW, Han IO. TGF-beta1 represses activation and resultant death of microglia via inhibition of phosphatidylinositol 3-kinase activity. J Immunol. 2004;172(11):7015–7023. doi: 10.4049/jimmunol.172.11.7015. [DOI] [PubMed] [Google Scholar]
- 19.Taylor RA, et al. TGF-β1 modulates microglialphenotype and promotes recovery after intra-cerebral hemorrhage. J Clin Invest. 2017;127(1):280–292. doi: 10.1172/JCI88647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elmore MR, et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82(2):380–397. doi: 10.1016/j.neuron.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang B, Hawes NL, Hurd RE, Davisson MT, Nusinowitz S, Heckenlively JR. Retinal degeneration mutants in the mouse. Vision Res. 2002;42(4):517–525. doi: 10.1016/S0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- 22.Bennett ML, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA. 2016;113(12):E1738–E1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541(7638):481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chitnis T, Weiner HL. CNS inflammation and neurodegeneration. J Clin Invest. 2017;127(10):3577–3587. doi: 10.1172/JCI90609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370(6488):341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 26.Zöller T, et al. Silencing of TGFβ signalling in microglia results in impaired homeostasis. Nat Commun. 2018;9(1):4011. doi: 10.1038/s41467-018-06224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma W, et al. Absence of TGFβ signaling in retinal microglia induces retinal degeneration and exacerbates choroidal neovascularization. Elife. 2019;8:e42049. doi: 10.7554/eLife.42049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Timmers AM, Guy J, Pang J, Hauswirth WW. Cone-specific expression using a human red opsin promoter in recombinant AAV. Vision Res. 2008;48(3):332–338. doi: 10.1016/j.visres.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 29.Lem J, et al. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc Natl Acad Sci USA. 1999;96(2):736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Douglas RM, Alam NM, Silver BD, McGill TJ, Tschetter WW, Prusky GT. Independent visual threshold measurements in the two eyes of freely moving rats and mice using a virtual-reality optokinetic system. Vis Neurosci. 2005;22(5):677–684. doi: 10.1017/S0952523805225166. [DOI] [PubMed] [Google Scholar]
- 31.Srinivasan Y, Lovicu FJ, Overbeek PA. Lens-specific expression of transforming growth factor beta1 in transgenic mice causes anterior subcapsular cataracts. J Clin Invest. 1998;101(3):625–634. doi: 10.1172/JCI1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson JV, Golesic E, Gauldie J, West-Mays JA. Ocular gene transfer of active TGF-beta induces changes in anterior segment morphology and elevated IOP in rats. Invest Ophthalmol Vis Sci. 2010;51(1):308–318. doi: 10.1167/iovs.09-3380. [DOI] [PubMed] [Google Scholar]
- 33.Dvashi Z, Goldberg M, Adir O, Shapira M, Pollack A. TGF-β1 induced transdifferentiation of rpe cells is mediated by TAK1. PLoS ONE. 2015;10(4):e0122229. doi: 10.1371/journal.pone.0122229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang S, Yao H, Li M, Li H, Wang F. Long non-coding RNA MALAT1 mediates transforming growth factor beta1-induced epithelial-mesenchymal transition of retinal pigment epithelial cells. PLoS ONE. 2016;11(3):e0152687. doi: 10.1371/journal.pone.0152687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgiadis A, et al. The tight junction associated signalling proteins ZO-1 and ZONAB regulate retinal pigment epithelium homeostasis in mice. PLoS ONE. 2010;5(12):e15730. doi: 10.1371/journal.pone.0015730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maddaluno L, et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 2013;498(7455):492–496. doi: 10.1038/nature12207. [DOI] [PubMed] [Google Scholar]
- 37.Feng X, Valdearcos M, Uchida Y, Lutrin D, Maze M, Koliwad SK. Microglia mediate postoperative hippocampal inflammation and cognitive decline in mice. JCI Insight. 2017;2(7):e91229. doi: 10.1172/jci.insight.91229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Koren EG, et al. Microglial function is distinct in different anatomical locations during retinal homeostasis and degeneration. Immunity. 2019;50(3):723–737.e7. doi: 10.1016/j.immuni.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaiss AK, Liu Q, Bowen GP, Wong NC, Bartlett JS, Muruve DA. Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J Virol. 2002;76(9):4580–4590. doi: 10.1128/JVI.76.9.4580-4590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamiya S, Liu L, Kaplan HJ. Epithelial-mesenchymal transition and proliferation of retinal pigment epithelial cells initiated upon loss of cell-cell contact. Invest Ophthalmol Vis Sci. 2010;51(5):2755–2763. doi: 10.1167/iovs.09-4725. [DOI] [PubMed] [Google Scholar]
- 41.Milam AH, Li ZY, Fariss RN. Histopathology of the human retina in retinitis pigmentosa. Prog Retin Eye Res. 1998;17(2):175–205. doi: 10.1016/S1350-9462(97)00012-8. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40(Database issue):D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Busskamp V, et al. Genetic reactivation of cone photoreceptors restores visual responses in retinitis pigmentosa. Science. 2010;329(5990):413–417. doi: 10.1126/science.1190897. [DOI] [PubMed] [Google Scholar]
- 44.Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nat Protoc. 2006;1(3):1412–1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- 45.Gaub BM, et al. Restoration of visual function by expression of a light-gated mammalian ion channel in retinal ganglion cells or ON-bipolar cells. Proc Natl Acad Sci USA. 2014;111(51):E5574–E5583. doi: 10.1073/pnas.1414162111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiong W, et al. AAV cis-regulatory sequences are correlated with ocular toxicity. Proc Natl Acad Sci USA. 2019;116(12):5785–5794. doi: 10.1073/pnas.1821000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10(11):1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 48.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 49.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data generated in this work have been deposited in the Gene Expression Omnibus (GEO) repository (accession number GSE145601).