Abstract
Despite recent advances in understanding chronic inflammation remission, global analyses have not been explored to systematically discover genes or pathways underlying the resolution dynamics of chronic inflammatory diseases. Here, we performed time-course gene expression profiling of mouse synovial tissues along progression and resolution of collagen-induced arthritis (CIA) and identified genes associated with inflammation resolution. Through network analysis of these genes, we predicted 3 key secretory factors responsible for the resolution of CIA: Itgb1, Rps3, and Ywhaz. These factors were predominantly expressed by Tregs and antiinflammatory M2 macrophages, suppressing production of proinflammatory cytokines. In particular, Ywhaz was elevated in the sera of mice with arthritis resolution and in the urine of rheumatoid arthritis (RA) patients with good therapeutic responses. Moreover, adenovirus-mediated transfer of the Ywhaz gene to the affected joints substantially inhibited arthritis progression in mice with CIA and suppressed expression of proinflammatory cytokines in joint tissues, lymph nodes, and spleens, suggesting Ywhaz is an excellent target for RA therapy. Therefore, our comprehensive analysis of dynamic synovial transcriptomes provides previously unidentified antiarthritic genes, Itgb1, Rps3, and Ywhaz, which can serve as molecular markers to predict disease remission, as well as therapeutic targets for chronic inflammatory arthritis.
Keywords: Autoimmunity, Inflammation
Keywords: Arthritis, Rheumatology
Introduction
One of the well-known characteristics of rheumatoid arthritis (RA) is that a large portion of RA patients undergo a clinical course of relapsing-remitting state, rather than experiencing continuous progression of the inflammation (1). During remission periods, RA patients show low disease activity and can be free of symptoms. The maintenance of remission is considered the most effective way to reduce joint damage and, ultimately, prevent long-term disability. In order to sustain remission periods or to induce complete remission, it seems necessary to first understand the factors and pathways underlying the natural resolution of chronic inflammation during remission periods. Proresolving lipid mediators, such as lipoxin A4 (LXA4) and resolvin, are known to be induced along the resolution of inflammation (2). Previous studies have also reported cellular processes contributing to antiinflammation, such as the recognition of resolution-associated molecular patterns (RAMPs), depletion of inflammatory chemokines, inhibition of leukocyte recruitment, activation of suppressive immune cells, and regeneration of tissue (3, 4). Despite these findings, global analyses have not been explored to systematically discover the factors or pathways underlying spontaneous remission mediators of chronic inflammation.
There have been a variety of methods developed toward evaluating RA remission. For instance, musculoskeletal ultrasound or magnetic resonance imaging enables determining the lack of synovitis (5). Remission criteria have been suggested by the European League Against Rheumatism (EULAR)/American College of Rheumatology (ACR) collaborative initiative and have been employed to quantitatively evaluate clinical remission (6, 7). Molecular approaches have also been attempted in order to assess RA remission more accurately, including a multibiomarker disease activity (MBDA) score calculated using concentrations of 12 proteins (7). These molecular approaches have improved the accuracy to some extent, but the development of precise measurements is still necessary because the currently available methods cannot predict whether RA patients will undergo flareup or remission. Therefore, novel molecular candidates for the evaluation of RA remission should be proposed based on an understanding of the dynamic nature of the factors or pathways underlying the aggravation and remission of RA.
The pathology of RA is the result of dynamic changes in the activation of immune cells as well as complex interactions among the immune cells that mediate pro- and antiinflammatory responses. Previous studies have revealed that a balance between proinflammatory cells, such as type 1 macrophages (M1), Th17 cells, and effector B cells, and antiinflammatory cells, such as tolerogenic dendritic cells, type 2 macrophages (M2), Tregs, and regulatory B cells (Bregs), is critical for the development and resolution of RA (8–10). Given the enormous complexity of RA pathology, dynamic analysis of molecular changes in synovial tissues at the global level can effectively summarize a molecular landscape of RA and prioritize key regulators or pathways governing the joint pathology. For comprehensive analyses of diverse types of tissues and/or cells in arthritic conditions, several animal models mimicking RA have been introduced (11–13). Among them, collagen-induced arthritis (CIA) is most widely used to study chronic relapsing-remitting arthritis (14), because it closely mimics the dynamic pathological features of RA during the course of disease progression (13). In CIA, inflammation undergoes dynamic transitions involving induction, peak, and resolution phases following immunization with complete Freund’s adjuvant (CFA) and type II collagen (CII) (15). However, the dynamic expression patterns of the genes involved in the propagation or resolution of inflammation after the initial stage of arthritis have not yet been systematically analyzed.
Here, we systematically analyzed dynamic transcriptomes in the synovia of mice with CIA. Through network analysis, we identified antiinflammatory and antiarthritic genes — Itgb1, Rps3, and Ywhaz — primarily responsible for the spontaneous resolution of arthritis. The 3 genes were predominantly expressed by Tregs and antiinflammatory macrophages, substantially suppressing proinflammatory cytokine responses in vitro. Particularly, Ywhaz was elevated in the sera of mice with resolution and the urine of RA patients with clinical remission, and its overexpression markedly ameliorated arthritis progression in mice. Collectively, our systems approach to resolution dynamics provides key molecules indicating resolution states of inflammatory arthritis as well as having therapeutic potential for chronic inflammatory arthritis.
Results
Dynamic transcriptome analysis of synovial tissues during progression of CIA.
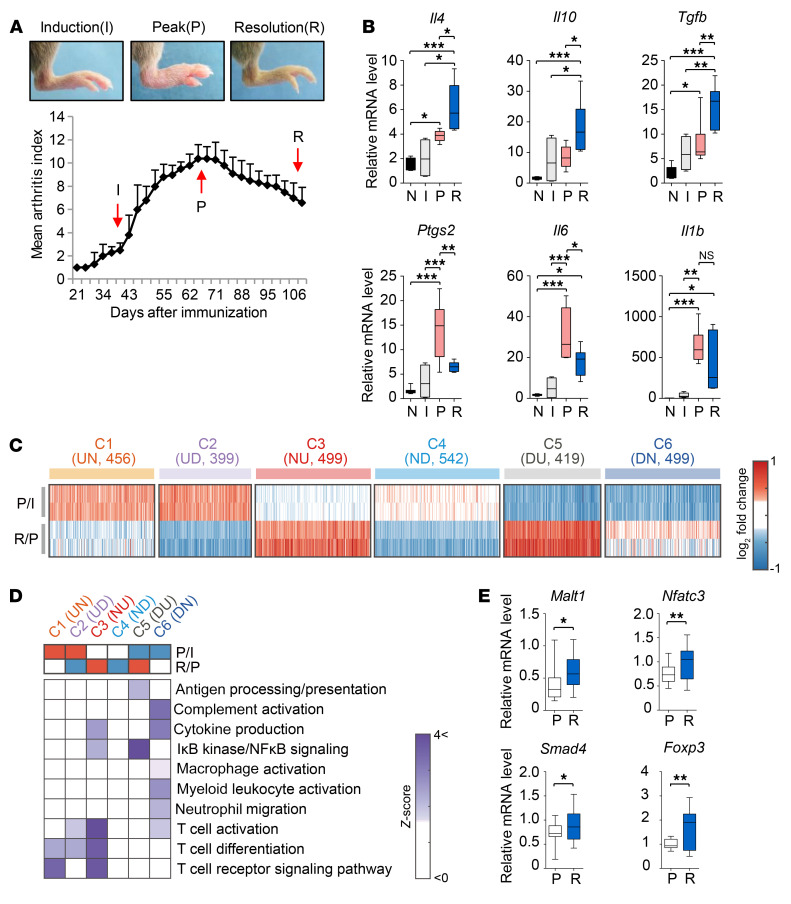
To characterize inflammatory phases in CIA, we first measured temporal changes in the arthritis score (13) to monitor clinical severity over 15 weeks after the injection of CFA and CII. Arthritis started to be induced at 4 weeks and peaked at 9 weeks, but then spontaneously resolved at 15 weeks (Figure 1A), similar to a previous report (15). To confirm the resolution of inflammation, we measured mRNA expression levels of proinflammatory (Ptgs2, Il6, and Il1b) and antiinflammatory cytokines (Il4, Il10, and Tgfb1) in the joint tissues before immunization (nonimmunization) and at the induction (6 weeks), peak (9 weeks), and resolution (15 weeks) phases after immunization (Figure 1B). We found remarkable increases in Ptgs2, Il6, and Il1b mRNA expression levels at the peak phase compared with those at the nonimmunization and induction phases. Conversely, there were significant decreases in mRNA levels of Ptgs2 and Il6 between the peak and resolution phase, suggesting suppression of proinflammatory responses. Meanwhile, antiinflammatory Il4, Il10, and Tgfb1 expression levels showed substantial upregulation at the resolution phase compared with those at the nonimmunization and induction phases, suggesting enhanced antiinflammatory responses; mean expression levels of Il4, Il10, and Tgfb1 at the resolution phase were, respectively, 3.7-, 11.4-, and 7.0-fold higher than those before the immunization and 3.0-, 2.5-, and 2.6-fold higher than those at the induction phase, and remained significantly higher than those at the peak phase. We next performed gene expression profiling of joint tissues from CIA mice at the induction, peak, and resolution phases. Known resolution-related genes involved in inhibition of immune cell infiltration, efferocytosis of apoptotic immune cells, and macrophage activation or tissue repair (refs. 3, 16; see Supplemental Note 1; supplemental material available online with this article; https://doi.org/10.1172/JCI126866DS1) were up- or downregulated at the resolution phase (Supplemental Figure 1A). Moreover, the assessment of dynamic changes in the activities of diverse immune cells during CIA progression using the CIBERSORT method (17) revealed that Tregs and M2 macrophages, known to be involved in resolution (3, 10), were significantly (P < 0.05) increased at the resolution phase, compared with other immune cells, which was confirmed by increased expression levels of the marker genes for the 2 suppressive immune cells (Supplemental Figure 1, B and C, and Supplemental Tables 1 and 2). All these data support the validity of our global analysis.
Figure 1. Dynamic expression changes of genes and cellular processes associated with resolution of CIA.
(A) Arthritis score (bottom) evaluated at 2-day intervals during CIA progression. Data are the mean ± SEM (n = 8 mice per time point). Induction (I), peak (P), and resolution (R) phases (6, 9, and 15 weeks, respectively) are indicated by arrows. Representative arthritic joints (top) at the 3 phases are shown. (B) Quantitative real-time polymerase chain reaction (qRT-PCR) assays for proinflammatory (Ptgs2, Il6, and Il1b) and antiinflammatory cytokines (Il4, Il10, and Tgfb1) before immunization (N) and at the I, P, and R phase after the immunization. Expression levels of each gene at the 4 phases (n = 4–8 per phase) are shown using box-and-whisker plots. (C) Resolution-related clusters (C1–C6) of genes (columns in the heat maps). Colors represent up- (red) and downregulation (blue) of the genes in the indicated clusters. Log2-fold changes of the genes from the comparisons of P/I (first and second rows) and R/P (third and fourth rows) are shown for 2 replicates in each condition. U, D, and N in parentheses denote up- and downregulation and no expression changes, respectively; e.g., UN (for C1) indicates upregulation in the comparison of P/I and no expression change in the comparison of R/P. The numbers in parentheses denote the numbers of DEGs in the individual clusters. (D) GOBPs enriched by the genes in C1–C6. Color bars represent the gradients of Z scores (enrichment significance) defined as Z = N–1(1 – P), where N–1(·) is the inverse standard normal distribution and P is the enrichment P value. (E) qRT-PCR assays for the representative genes. For qRT-PCR assays, mRNA expression levels of each gene (n ≥ 8 per group) were normalized to that of Gapdh (internal control) using the 2–ΔΔCt method. In the box-and-whisker plots, the boxes display the lower, median, and upper quartiles and the whiskers represent the minimum and maximum values. *P < 0.05, **P < 0.01, ***P < 0.001 by 1-way ANOVA with a post hoc test (Tukey’s correction; B) or Student’s t test (E).
Cellular processes of immune cells associated with resolution of CIA.
In order to identify and characterize resolution-associated genes, we next selected 1795 (P/I) and 1881 (R/P) differentially expressed genes (DEGs) from the comparisons of peak versus induction phase (P/I) and resolution versus peak phase (R/P) (Supplemental Table 3). We then selected 6 clusters (C1–C6) of the DEGs showing up- (C3 and C5) and downregulation (C2 and C4) at the resolution phase, as well as up- or downregulation at the peak phase and leveling off at the resolution phase (Figure 1C and Supplemental Table 4). The enrichment analysis of gene ontology biological processes (GOBPs) revealed that C3 and C5 most significantly (P < 0.01) represented T cell differentiation/activation and macrophage (NF-κB signaling) functions, respectively, and C6 significantly (P ≤ 0.05) represented macrophage and myeloid leukocyte activation (Figure 1D, Supplemental Figure 2A, and Supplemental Table 5). The network model for the DEGs involved in T cell functions showed the activation of TGF-β, JAK/STAT, and TCR signaling during CIA progression (Supplemental Figure 2B). Quantitative real-time polymerase chain reaction (qRT-PCR) analysis confirmed upregulation of 4 genes at the resolution phase, which were associated with TGF-β (Smad4), JAK/STAT (Foxp3) (18), and TCR (Malt1 and Nfatc3) signaling (Figure 1E). These data suggest that cellular pathways in multiple immune cells, including T cells and macrophages, cooperatively contribute to the resolution of CIA.
Network-based selection of key regulators for resolution of CIA.
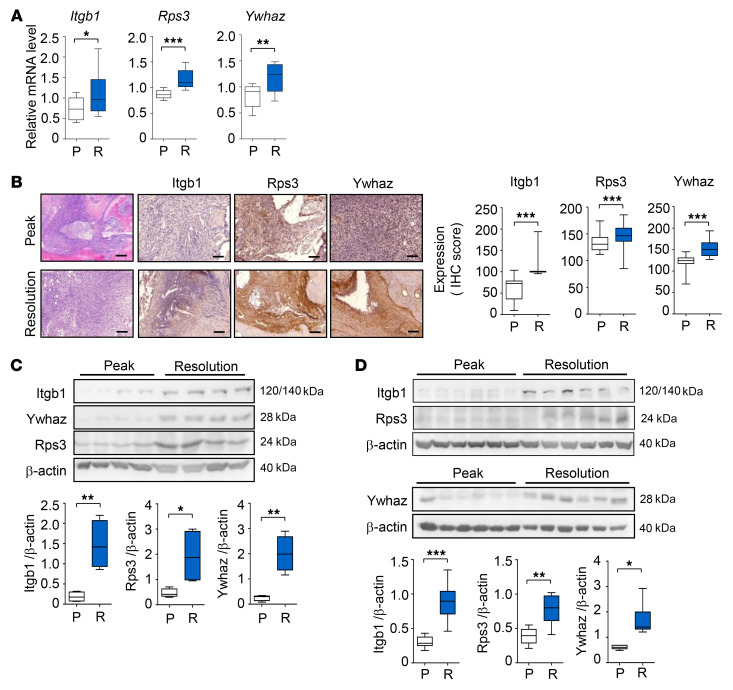
To prioritize key resolution regulators, we constructed a resolution-associated network model for 2836 DEGs from the P/I and R/P comparisons using protein-protein interactions (Supplemental Table 6). We then selected the 3 key regulators — integrin subunit β 1 (Itgb1) and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein ζ (Ywhaz) in C3 and ribosomal protein S3 (Rps3) in C5 — based on the following criteria: (a) hub-like molecules playing significant (P < 0.05) roles in the resolution-associated network and (b) secretory antiinflammatory regulators functionally associated with activation of T cells and/or macrophages (See Supplemental Note 2, Supplemental Figure 3, and Supplemental Tables 7 and 8). Next, through the use of qRT-PCR, we confirmed the upregulation of Itgb1, Rps3, and Ywhaz at the resolution phase (Figure 2A) compared with at the peak phase. We then examined their protein expression in mouse synovial tissues by immunohistochemistry (Figure 2B) and Western blotting (Figure 2C). As expected, both experiments showed significantly higher expression levels of the 3 proteins in synovial tissues at the resolution phase than those at the peak phase. The increased expression of Itgb1, Rps3, and Ywhaz was confirmed in rat models of CIA (Figure 2D), suggesting that their associations with CIA resolution are conserved in other mammalian species.
Figure 2. Key regulators of resolution of CIA.
(A and B) Increased mRNA (A) and protein (B) expression of the 3 key regulators, Itgb1, Ywhaz, and Rps3, measured by qRT-PCR (n = 6) and immunohistochemical (n = 6) assays, respectively, in mouse synovial tissues at the resolution (R) phase, compared with at the peak (P) phase. For qRT-PCR, Gapdh mRNA was used as an internal control. Scale bars: 200 μm. (C) Western blot analysis for Itgb1, Ywhaz, and Rps3 expression in mouse synovia (n = 4 mice per group). (D) Increased protein expression of Itgb1, Rps3, and Ywhaz measured by Western blotting in rat synovial tissues (n = 6 rats per group) at the R phase, compared with that at the P phase. For Western blotting, protein expression levels were normalized to that of β-actin (internal control). In the box-and-whisker plots, the boxes display the lower, median, and upper quartiles and the whiskers represent the minimum and maximum values. Data are the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t test.
Predominant expression of Itgb1, Rps3, and Ywhaz in Tregs and M2 macrophages.
Our next experiment was conducted to determine which types of immune cells are responsible for increased Itgb1, Rps3, and Ywhaz expression at the resolution phase. We postulated that the upregulation of Itgb1, Rps3, and Ywhaz might stem from the activation of immune cells expressing these molecules during the resolution phase. In order to address this, we investigated whether the 3 selected key regulators are expressed and upregulated in B cells, macrophages, or T cells upon their activation. Indeed, Itgb1, Rps3, and Ywhaz were upregulated in CD4+ and CD8+ T cells, B cells, and macrophages when they were activated by treatment with anti-CD3 plus anti-CD28 antibodies, anti-IgM antibodies, and lipopolysaccharide (LPS), respectively, as determined by flow cytometry (Supplemental Figure 4A). Significantly lower expression of the regulators in CD4+ T cells treated with target gene siRNAs compared with in those treated with control siRNAs confirmed the specificity of the antibodies used (Supplemental Figure 4B).
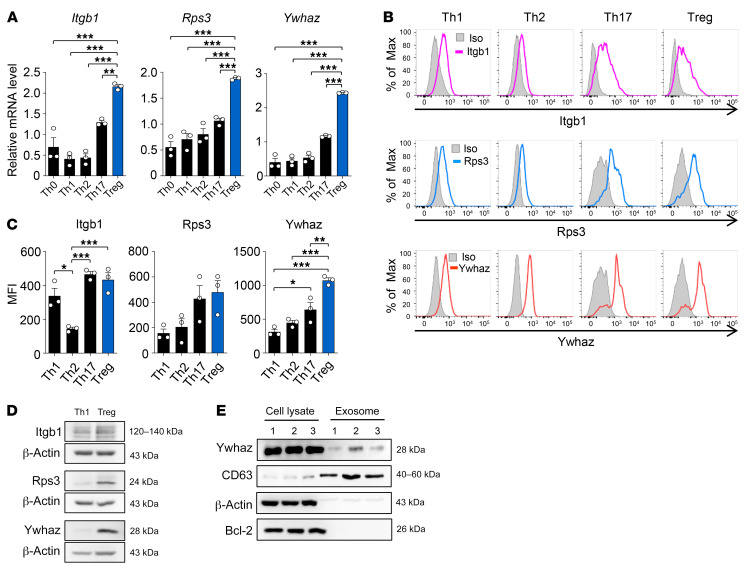
Gene expression analysis suggested that subpopulations of T cells and macrophages, particularly Treg cells and M2 macrophages, are associated with the resolution of CIA. Thus, we first investigated subpopulations of T cells and their association with the resolution using the 3 key regulators. Among the diverse subtypes of CD4+ T helper (Th) cells, Tregs play major roles in the suppression of chronic inflammation. We thus examined expression levels of the 3 regulators in 4 subtypes of CD4+ T helper cells (Th1, Th2, Th17, and Treg; Supplemental Figure 5) and found that they were expressed in all 4 subtypes (Figure 3, A–C). However, the highest expression of Itgb1, Rps3, and Ywhaz was observed in Tregs as compared with the other types of Th cells, as determined by flow cytometry and qRT-PCR; Th17 cells showed the second highest expression of them (Figure 3, A–C). Notably, Ywhaz expression was strikingly high in Tregs, and its expression levels were greater than those in Th17 cells (Figure 3, A–C). A higher expression of Itgb1, Rps3, and Ywhaz protein in Tregs than in Th1 cells was also confirmed by Western blot analysis (Figure 3D). Intriguingly, Ywhaz was recently shown to be one of the top 10 proteins present in exosomes (19), and exosomal Ywhaz derived from tumor cells can be transmitted to tumor-infiltrating T cells, impairing their antitumor activity (20). To assess exosomal Ywhaz, we isolated exosomes derived from Tregs and demonstrated the coexpression of the Ywhaz protein with CD63, an exosome marker (Figure 3E), suggesting the possibility of exosomal transmission of Ywhaz from Tregs to adjacent immune cells. Together, these data demonstrate that the 3 regulators may be associated with Tregs, thereby contributing to inflammation resolution in CIA.
Figure 3. Expression of the 3 key regulators, Itgb1, Rps3, and Ywhaz, in T cells.
(A) Gene expression levels of the 3 regulators in undifferentiated and differentiated subtypes of CD4+ T helper cells isolated from mice. Th cell subsets were defined according to the expression pattern of transcription factors and/or cytokines: Tbx21 and IFN-γ for Th1, Gata3 for Th2, Rorc and IL-17 for Th17, and Foxp3 for Treg (see Methods). mRNA expression levels of the target genes were normalized to that of Gapdh (internal control). (B and C) Expression of Itgb1, Rps3, and Ywhaz proteins in the subtypes of CD4+ T helper cells determined by flow cytometry are presented as overlay histogram (B) and mean fluorescence intensity (MFI) (C). The differentiated Th cells were restimulated with PMA/ionomycin for 4 hours before intracellular cytokine staining. A gray-colored histogram indicates isotype control (Iso). Data are representative (B) or the mean ± SEM of 3 independent experiments (A and C). *P < 0.05, **P < 0.01, ***P < 0.001 as determined by 1-way ANOVA with a post hoc test (Tukey’s correction). (D) Western blot assay for Itgb1, Rps3, and Ywhaz in Th1 cells and Tregs. Data are representative of 3 independent experiments with similar results. (E) Western blot analysis showing coexpression of Ywhaz and Bcl2 in cell lysates or coexpression of Ywhaz and CD63 in exosomes isolated from Treg cells (n = 3).
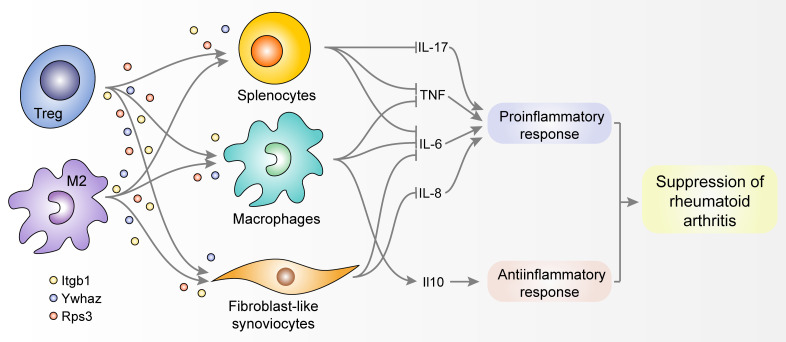
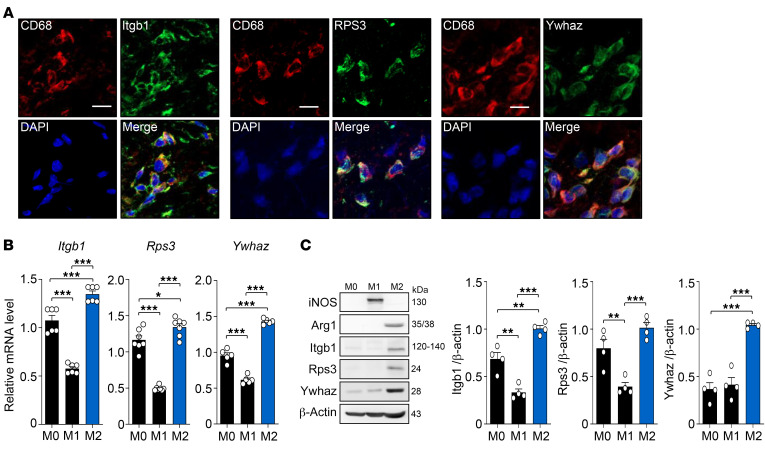
Macrophages are another type of important immune cell involved in the development or resolution of inflammation, and they can differentiate into proinflammatory (M1) or antiinflammatory macrophages (M2), depending on their surrounding microenvironments (10). In order to determine whether Itgb1, Rps3, and Ywhaz are expressed in macrophages, we first examined their expression in synovial macrophages by double immunofluorescence staining of CD68 and each regulator. Marked colocalization of cells expressing CD68 and each regulator was observed (Figure 4A), indicating that synovial macrophages in the joints with CIA express Itgb1, Rps3, and Ywhaz. We also compared the distribution of M1 and M2 macrophage subtypes in synovial tissues between the peak and resolution phases by immunofluorescence staining. CD68-expressing synovial macrophages were universally present at both phases, but Arg1-expressing M2 cells were more frequently colocalized with CD68-expressing synovial macrophages at the resolution phase than at the peak phase (data not shown), confirming the association of M2 macrophages with arthritis resolution (21). We next examined whether the 3 regulators were differentially expressed between M1 and M2 macrophages, which were differentiated from M0 macrophages in vitro (Supplemental Figure 6). Both the mRNA and protein levels of Itgb1, Rps3, and Ywhaz were significantly (P < 0.05) higher in the M2 macrophages than they were in the M0 or M1 macrophages, as determined by qRT-PCR and Western blotting (Figure 4, B and C). Additionally, their mRNA levels were significantly (P < 0.01) lower in the M1 macrophages than in the M0 macrophages (Figure 4B), although such a differential expression pattern was less apparent in protein levels. Collectively, these data suggest that the 3 potential regulators — Itgb1, Rps3, and Ywhaz — are preferentially expressed in antiinflammatory M2 macrophages, rather than in proinflammatory M1 macrophages, and thereby M2 macrophages may contribute to the resolution of CIA.
Figure 4. Expression of the 3 key regulators in macrophages.
(A) Colocalization of CD68 with the 3 key regulators. Double immunofluorescence staining was performed in mouse synovial tissues at the resolution phase using Abs against CD68, and Itgb1 (left), Rps3 (middle), or Ywhaz (right). Scale bars: 10 μm. Data are representative of at least 3 independent experiments with similar results. (B and C) Determination of mRNA (B) and protein (C) expression levels of the 3 regulators in undifferentiated (M0) and differentiated M1 and M2 macrophages using qRT-PCR and Western blotting, respectively. Target gene and protein expression levels were normalized by those of Gapdh and β-actin, respectively. Data are the mean ± SEM (n = 4–7). *P < 0.05, **P < 0.01, ***P < 0.001 as determined by 1-way ANOVA with a post hoc test (Tukey’s correction).
Regulation of proinflammatory cytokine production by Itgb1, Rps3, and Ywhaz.
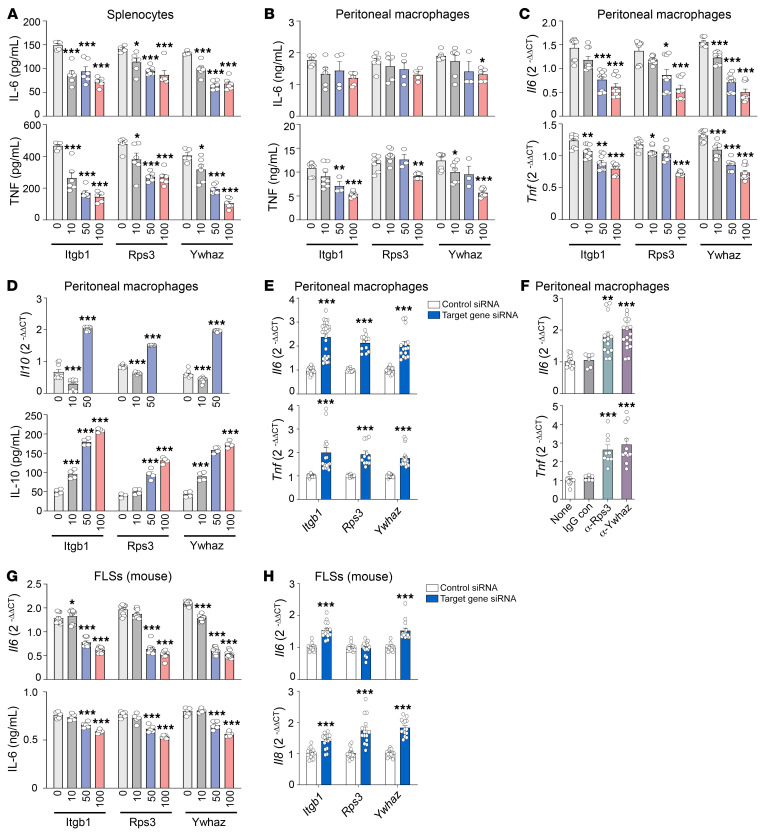
Based on our findings that Itgb1, Rps3, and Ywhaz were predominantly expressed in immune suppressor cells, including Tregs and M2 macrophages, we next attempted to validate the antiinflammatory activities of Itgb1, Rps3, and Ywhaz by performing in vitro functional tests using effector cells of RA, including macrophages and fibroblast-like synoviocytes (FLSs). In mouse splenocytes stimulated with LPS, the addition of recombinant Itgb1, Rps3, and Ywhaz dose-dependently inhibited the production of IL-6 and/or TNF, which are representative proinflammatory cytokines (Figure 5A). In cultured peritoneal macrophages, recombinant Itgb1, Rps3, and Ywhaz were also able to dose-dependently suppress LPS-induced increases in mRNA and/or protein expression of IL-6 and TNF (Figure 5, B and C). In contrast, recombinant Itgb1, Rps3, and Ywhaz promoted mRNA and protein expression of antiinflammatory IL-10 in peritoneal macrophages stimulated with LPS (Figure 5D). We also confirmed that cell proliferation was not significantly affected by the addition of those recombinant proteins (Supplemental Figure 7). In contrast to its effect on macrophages, recombinant Ywhaz had no direct effect on the production of IFN-γ, IL-2, IL-4, IL-10, and IL-17 by CD4+ T cells stimulated with anti-CD3 plus anti-CD28 antibodies (data not shown).
Figure 5. Promotion of antiinflammatory responses by the 3 regulators.
(A–C) Decrease in mRNA and protein expression of IL-6 and TNF induced by Itgb1, Rps3, or Ywhaz. Mouse splenocytes (A) and peritoneal macrophages (B and C) were cultured with multiple concentrations (0, 10, 50, or 100 ng/mL) of recombinant Itgb1, Rps3, or Ywhaz in the absence or presence of 10 ng/mL LPS for 6 hours (for peritoneal macrophages) or 24 hours (for splenocytes). IL-6 and TNF protein concentrations in the culture supernatants were measured by ELISA (A and B), and their mRNA levels in the cells were determined by qRT-PCR (C). (D) Increase in IL-10 mRNA (top) and protein (bottom) expression induced by the 3 regulators. (E) Increase in Il6 and Tnf expression induced by the knockdown of Itgb1, Rps3, or Ywhaz transcripts. Peritoneal macrophages were transfected with siRNAs for Itgb1, Rps3, or Ywhaz for 24 hours. (F) Promotion of Il6 and Tnf production by anti-Rps3 or anti-Ywhaz Abs for 6 or 12 hours. (G) Decrease in IL-6 mRNA (top) and protein (bottom) expression in mouse fibroblast-like synoviocytes (FLSs) after treatment with recombinant Itgb1, Rps3, or Ywhaz for 6 hours. (H) Increase in mRNA levels of Il6 and Il8 in FLSs transfected with Itgb1, Rps3, or Ywhaz siRNAs. For qRT-PCR assays (C–H), mRNA levels of the target gene were first normalized to those of Gapdh, and then further normalized to the mean mRNA expression levels in the cells treated with LPS only (C, D, and G), those without treatment (none; F), or those transfected with control siRNA (E and H). Data are the mean ± SEM of more than 2 (F) or 3 independent experiments (A–E, G, and H). *P < 0.05, **P < 0.01, ***P < 0.001 by 1-way ANOVA with a post hoc test (Dunnett’s correction; A–D, F, and G) or Student’s t test (E and H).
We next tested whether the blockade of Itgb1, Rps3, and Ywhaz, in contrast to treatment with their recombinant proteins, increases the production of proinflammatory cytokines. As expected, knockdown of Itgb1, Rps3, and Ywhaz transcripts using siRNAs increased the mRNA expression levels of Il6 and Tnf in peritoneal macrophages, compared with the control siRNA–treated peritoneal macrophages (Figure 5E and Supplemental Figure 8). Moreover, treatment with anti-Rps3 or anti-Ywhaz antibodies resulted in an increase in Il6 and Tnf production by peritoneal macrophages (Figure 5F), indicating that the reduction of endogenous Rps3 and Ywhaz promotes the production of proinflammatory cytokines.
FLSs are the representative effector cells in RA that actively participate in joint destruction as a major component of invasive pannus. We therefore sought to test whether or not Itgb1, Rps3, and Ywhaz also show antiinflammatory activities in these cells. As a result, recombinant Itgb1, Rps3, and Ywhaz dose-dependently suppressed mRNA and protein expression of IL-6 in mouse FLSs stimulated with LPS (Figure 5G); the proliferation of FLSs was not significantly altered (Supplemental Figure 7). In contrast, as in peritoneal macrophages, knockdown of Itgb1, Rps3, and Ywhaz genes promoted the expression of Il6 and/or Il8 in the FLSs (Figure 5H and Supplemental Figure 8), suggesting that the antiinflammatory functions of Itgb1, Rps3, and Ywhaz are not limited to macrophages but are also apparent in FLSs.
Taken together, these results demonstrate that Itgb1, Rps3, and Ywhaz may have antiinflammatory activities by repressing proinflammatory cytokine production and/or promoting antiinflammatory cytokine production in activated effector cells in RA, including macrophages and FLSs.
Ywhaz as a potential diagnostic target for arthritis resolution.
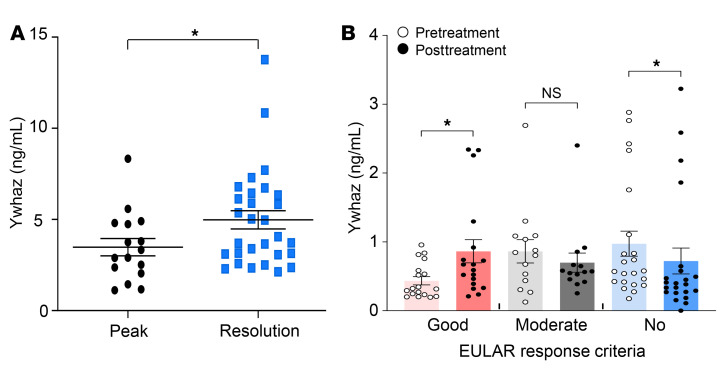
Our final goal was to determine whether or not Itgb1, Rps3, and Ywhaz can serve as diagnostic and therapeutic targets to predict arthritis resolution and inhibit arthritis progression, respectively. Because secretability was used to select Itgb1, Rps3, and Ywhaz among the hub-like candidates (Supplemental Figure 3A), we postulated that the 3 regulators could be secreted and detected in body fluids, such as serum and urine, reflecting the resolution states of the arthritis. To test this postulation, we examined whether there are differences in the serum levels of Itgb1, Rps3, and Ywhaz between the peak and resolution phases of CIA. Although the serum Itgb1 and Rps3 levels showed no differences (Supplemental Figure 9), the serum Ywhaz level increased at the resolution phase (Figure 6A), suggesting that it could be a surrogate marker for resolution of chronic arthritis. These results, paired with the findings of elevated Ywhaz expression in the synovia at the resolution phase as well as in immunosuppressive Tregs and M2 macrophages, prompted us to investigate whether YWHAZ can reflect resolution states in humans. To this end, we first categorized RA patients (n = 65) into 3 groups (low, moderate, and high activity) based on a disease activity score of 28 for erythrocyte sedimentation rate (DAS28-ESR; Supplemental Table 9). We then compared the abundance of YWHAZ in serum and urine samples among the 3 groups. Pretreatment YWHAZ levels showed no significant difference among the 3 groups (data not shown). However, when serially measured 4 to 6 months after treatment with antirheumatic drugs (n = 60), urinary ΔYWHAZ levels (posttreatment – pretreatment YWHAZ level) were significantly increased in the good-response group (n = 23), unchanged in the moderate-response group (n = 12), and decreased in the no-response group (n = 25) (Figure 6B). However, such changes were not apparent in the serum. No difference was found in YWHAZ levels according to the usage of methotrexate, hydroxychloroquine, leflunomide, tacrolimus, and biologics including TNF-α blocking agents (data not shown). Collectively, these data suggest that YWHAZ represents the resolution of chronic inflammation in humans, and its ΔYWHAZ levels in the urine might be used as a surrogate marker that reflects the resolving state of RA.
Figure 6. YWHAZ as a secretory marker reflecting states of resolution of mouse CIA and human RA.
(A) Protein levels of Ywhaz measured in mouse serum at the peak and resolution phases by ELISAs. Data are the mean ± SEM (peak, n = 16; resolution, n = 29). *P < 0.05 by Student’s t test. (B) Comparison of YWHAZ protein levels before and 4 to 6 months after treatment with antirheumatic drugs in good-response (n = 23), moderate-response (n = 12), and no-response groups (n = 25) categorized based on EULAR response criteria. EULAR response criteria are based on the attained level and change in DAS28-ESR. Data are the median ± SEM. *P < 0.05 as determined by Mann-Whitney U test.
Suppression of CIA by Ywhaz overexpression.
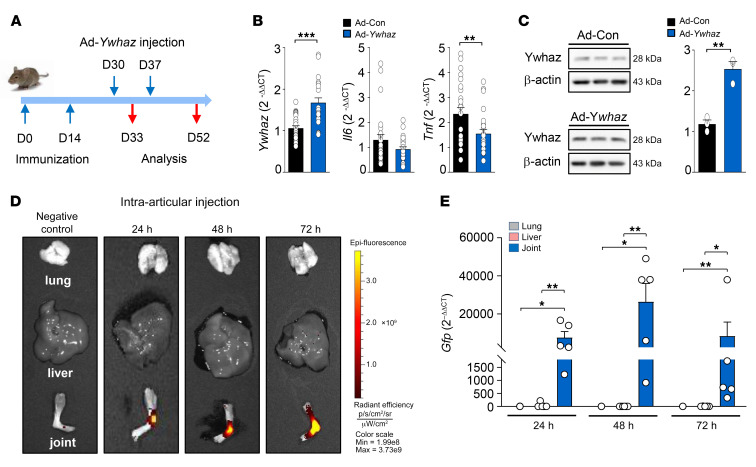
Among the 3 regulators, Ywhaz best reflected the resolution states of chronic inflammation in both mouse and human systems, indicating that Ywhaz may serve as an effective therapeutic target for RA. Therefore, we sought to investigate whether Ywhaz overexpression ameliorates proinflammatory responses and arthritis progression in vivo. To this end, we tested the effects of adenovirus-mediated transfer of the Ywhaz gene (Ad-Ywhaz) tagged with Gfp on the production of proinflammatory cytokines and the progression of arthritis in mice with CIA (Figure 7A). As expected, local transfer of Gfp-tagged Ad-Ywhaz into the joints increased the mRNA and protein levels of Ywhaz in the synovial tissues of mice 3 days (on day 33) after injection compared with the transfer of the empty adenovirus control vector (Ad-Con) (Figure 7, B and C). GFP fluorescence and mRNA expression levels were also much higher in the joints than in the lung and liver as determined by fluorescence imaging and qRT-PCR, respectively, at 1, 2, and 3 days after intra-articular injection of Ad-Ywhaz tagged with Gfp (on day 31, 32, and 33 after the primary immunization) (Figure 7, D and E); in fact, GFP expression in the liver and lung was negligible. By contrast, the liver and/or lung showed significantly higher levels of GFP than the joints after intravenous injection of Ad-Ywhaz, as a control experiment (Supplemental Figure 10). Together, these data indicate that a high amount of Ad-Ywhaz-Gfp specifically accumulated in the injected joints, eliminating concerns that it might overflow or spill out during the joint injection.
Figure 7. Expression of Ywhaz and GFP after intra-articular injection of Ad-Ywhaz tagged with Gfp in mice with CIA.
(A) A schematic diagram illustrating the establishment of CIA and treatment of mice with adenoviral vectors. Following primary and secondary immunization on days 0 and 14, respectively, 1 × 108 plaque-forming units (PFU) of the adenoviral vector containing Ywhaz (Ad-Ywhaz) or the control adenoviral vector (Ad-Con) in 10 μL HEPES were injected into the ankle joints of the mice at 30 and 37 days. Mice were then sacrificed at 33 or 52 days for further immunopathologic analyses. (B and C) mRNA expression levels of Ywhaz, Il6, and Tnf (B) and protein expression levels of Ywhaz (C) in the synovial tissues of CIA mice 33 days after primary immunization, which were determined by qRT-PCR and Western blot analysis, respectively. mRNA and protein levels of the target genes were normalized to those of Gapdh and β-actin, respectively. (D and E) GFP imaging (D) and qRT-PCR assays for Gfp (E) performed in the lung, liver, and ankle joint of CIA mice at 24, 48, and 72 hours after intra-articular injection of Ad-Ywhaz tagged with Gfp or those of mice without the injection. The color bar in D represents the gradient of radiant efficiency. Data in D are representative of 2 independent experiments with similar results, and data in B, C, and E are the mean ± SEM of more than 3 mice. For qRT-PCR assays, Gfp levels at each time point were first normalized to Gapdh levels, and then further normalized to the mean mRNA expression levels measured in the lung with intra-articular injection of Ad-Ywhaz tagged with Gfp. *P < 0.05, **P < 0.01, ***P < 0.001 as determined by Student’s t test (B and C) or by Kruskal-Wallis test with a post hoc test (Dunn’s correction; E).
The antiinflammatory effect of Gfp-tagged Ad-Ywhaz injected intra-articularly was revealed by the significant (P < 0.01) downregulation of Tnf mRNA expression level in the synovial tissues of mice at 33 days, in contrast to the upregulation of Ywhaz mRNA expression level (Figure 7B). Moreover, in the same experimental condition, intra-articular injection of Ad-Ywhaz dampened IL-6, TNF, and/or IL-17 protein expression levels in lymph node cells and splenocytes stimulated with LPS or anti-CD3 plus anti-CD28 antibodies (Supplemental Figure 11), confirming that Ywhaz represses proinflammatory cytokine production in vivo.
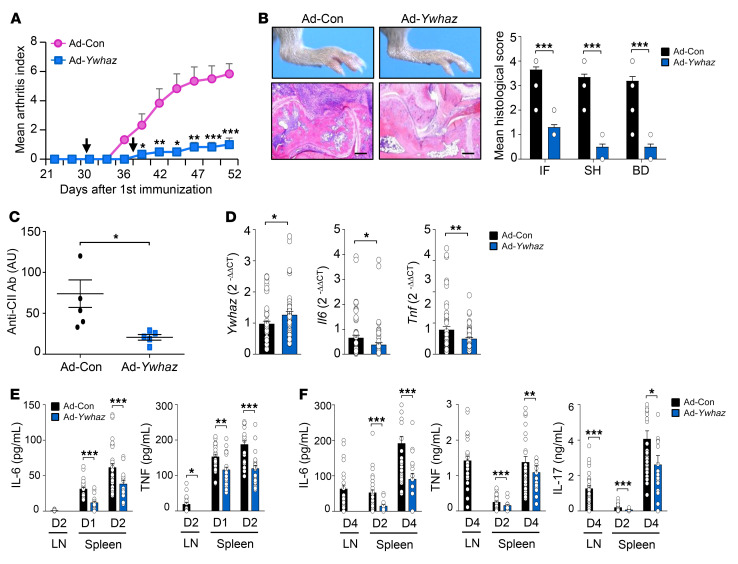
Finally, we investigated the therapeutic effects of Ywhaz overexpression. When intra-articularly injected twice on the 30th and 37th days after the first immunization with CII (Figure 7A), Ad-Ywhaz markedly suppressed the progression of CIA compared with Ad-Con (Figure 8A). In a histologic analysis at 52 days, we also observed a substantial decrease in synovial inflammation and joint destruction in the joints of mice with Ad-Ywhaz (Figure 8B). Moreover, Ywhaz overexpression significantly (P < 0.05) suppressed Il6 and Tnf levels in joint tissues, as well as serum levels of the anti-CII antibody (Figure 8, C and D). Under the same condition, the production of IL-6, TNF, and/or IL-17 in lymph node cells and splenocytes stimulated with LPS or anti-CD3 plus anti-CD28 antibodies was mitigated by the transfer of Ad-Ywhaz (Figure 8, E and F). Because Ywhaz is a secretory protein, Ad-Ywhaz may potentially have systemic effects. Together, these data suggest that Ywhaz overexpression has local and potential systemic effects and ameliorates CIA possibly through the downregulation of proinflammatory cytokine production in vivo.
Figure 8. Ad-Ywhaz inhibition of CIA in mice.
(A) Arthritis severity measured at 2- or 3-day intervals up to 52 days after the primary immunization with type II collagen. Intra-articular injection of 1 × 108 PFU of Ad-Ywhaz or Ad-Con at 30 and 37 days are indicated by arrows (n = 8 per group and per time point). Data are representative of 2 independent experiments with similar results. (B) Ad-Ywhaz suppression of foot swelling (left, top) and chronic inflammation in the ankle joints (left, bottom), which were observed at 52 days. Mean histological severity, as assessed by the extent of inflammatory cell infiltration (IF), synovial hyperplasia (SH), and bone destruction (BD) on H&E staining, is also shown on the right. Scale bars: 200 μm. (C) Reduction of serum levels of the IgG antibody against type II collagen (anti-CII Ab) in mice injected with Ad-Ywhaz. Data are the mean ± SD arbitrary units (AU, n = 5). (D) mRNA expression levels of Ywhaz and proinflammatory cytokines (Il6 and Tnf) in the synovial tissues of CIA mice 52 days after treatment with Ad-Ywhaz or Ad-Con (n = 6 per group). Target gene expression levels were normalized to those of Gapdh (internal control). (E and F) Decrease in proinflammatory cytokine expression by Ad-Ywhaz. On day 52, lymph node (LN) or spleen cells were isolated from mice treated with Ad-Ywhaz (n = 6) or Ad-Con (n = 6), and then stimulated with 10 ng/mL LPS (E) or 1 μg/mL anti-CD3 plus anti-CD28 Abs (F) for indicated times; e.g., D2 indicates day 2. Levels of IL-6, TNF, and IL-17 in the culture supernatants were measured by ELISA. Data are the mean ± SEM for A, B, and D–F. *P < 0.05, **P < 0.01, ***P < 0.001 as determined by Student’s t test.
Together, these results confirm that Ywhaz has an antiinflammatory activity as a hub molecule in the resolution-associated network describing dynamic changes of resolution-related processes, demonstrating that it has therapeutic potential in chronic arthritis.
Discussion
The identification of novel proresolving factors is not only essential for measuring the resolution state of inflammatory arthritis but is also important for understanding the fundamental basis of resolving processes. By analyzing the dynamic gene expression profiles of synovial tissues during the course of CIA progression, we identified the 3 key regulators (Itgb1, Rps3, and Ywhaz) of CIA resolution. Among them, in particular, we demonstrated the diagnostic and therapeutic implications of Ywhaz in arthritis resolution based on the following experimental observations: (a) increased expression of Ywhaz in synovial tissues during the resolution of CIA; (b) its predominant expression in T cells and macrophages, particularly Treg cells and M2 macrophages; (c) activation of anti- and proinflammatory responses by its excess and deficiency, respectively, in cultured splenocytes, macrophages, and synovial fibroblasts; (d) elevated expression of Ywhaz, among the 3 regulators, in the sera during the resolution phase of CIA and in the urine of RA patients with good therapeutic response; and (e) amelioration of CIA by periarticular injection of Ad-Ywhaz. Collectively, through a global transcriptome profiling of synovial tissues, we discovered previously unidentified regulators of the resolution of inflammatory arthritis.
In this study, as key regulator candidates of the resolution of inflammation, we identified 85 hub-like molecules among the 2836 DEGs based on a resolution-associated network model describing the interactions among these DEGs (Supplemental Note 2 and Supplemental Figure 3A). The 85 molecules include 37 that were upregulated at the resolution phase, which can serve as potential regulators of CIA resolution like the 3 key regulators. In order to understand the functional characteristics of the 37 upregulated hub-like genes, we performed GOBP enrichment analysis of these genes and found that they were mainly associated with the processes related to cell proliferation, cell migration, proteostasis, response to oxidative stress, and intracellular signaling (particularly the response to cAMP and TGF-β signaling) (Supplemental Figure 12A). Previously, numerous reports have demonstrated associations of these processes with the resolution of inflammation (3, 16, 22, 23). For instance, TGF-β signaling, T cell proliferation, and neutrophil apoptosis have all been shown to be associated with resolution of inflammation (3). Moreover, the 3 key regulators are involved in cell adhesion and migration (Itgb1) (24), translation (Rps3) (25, 26), and intracellular signaling (Ywhaz) (27, 28). Together with previous reports (3, 16, 22, 23), our data suggest that the upregulated 37 hub-like molecules may serve as a comprehensive resource for potential proresolving factors.
With the end goal of discovering soluble factors that can be used for predicting or inducing resolution of RA activity, we experimentally tested the functional roles of the 3 secretory regulators (Itgb1, Rps3, and Ywhaz). Although the antiinflammatory roles of the 3 secretory regulators are not yet fully understood, several previous studies have suggested potential associations of the 3 regulators with the suppression of chronic inflammation. For example, Itgb1 expressed on the plasma membrane of FLSs from RA patients represses hyperplasia of the cells by inducing Fas-mediated apoptosis (29). Rps3 is secreted as a homodimer by several cancer cell lines (30), and its transduction into skin cells ameliorates ear edema via the inhibition of NF-κB and MAPK activities in mice (26). YWHAZ can be also secreted by monocytes and macrophages (31), and its knockdown increases the production of inflammatory cytokines by squamous cancer cells (32). Moreover, a secretory peptide (PEPITEM) derived from Ywhaz inhibits transendothelial migration of CD3+ T cells in animal models of peritonitis, uveitis, and Sjögren’s syndrome (33). In comparison with the previous findings, our results provide a further comprehensive understanding of the antiinflammatory roles of Itgb1, Rps3, and Ywhaz by demonstrating their cellular sources and immunological targets, in addition to the in vivo regulatory activities of Ywhaz.
One of the emerging issues and challenges in RA treatment is the identification of new surrogate markers in biofluids for predicting disease state. The reduction of proinflammatory markers, including ESR and CRP, has been used previously, but sometimes it cannot accurately reflect RA activity (34, 35), particularly in patients with anti–IL-6 receptor antibodies. Moreover, there has been no trial demonstrating that proresolving or antiinflammatory molecules exhibit prognostic value in clinical settings. Here, our proof-of-concept study showed that the serum Ywhaz level was elevated during resolution of CIA (Figure 6A) and that posttreatment YWHAZ levels in the urine were significantly increased in the good-response group and decreased in the no-response group as compared with pretreatment levels (Figure 6B); although urine is more stable than other biofluids (36), it is unclear why such changes in Ywhaz levels are evident in the urine but not in the sera. Given that Ywhaz was predominantly produced by Tregs and M2 macrophages (Figures 3 and 4), the increase in Ywhaz levels might be a reflection of the numeric expansion of such suppressive cell types in proportion to the improvement of chronic arthritis. We speculate that the change in urinary YWHAZ could be used as a surrogate marker, together with previously known biomarkers, to provide a unique prediction window of the resolving state of RA. Further study on a large scale will be required to clarify this issue.
Blockade of TNF or IL-6 has been effectively applied as a therapeutic for RA (5). However, these drugs can increase the risk of susceptibility to infectious diseases and are less likely to restore immune tolerance after the withdrawal of therapies (37). In addition, antiinflammatory cytokines, such as IL-4, -10, and -13, have been suggested as potential therapeutic candidates, but they have failed to treat RA in clinical settings (38). Thus, there has been an unmet need for the identification of new candidates that enhance antiinflammatory and proresolving responses beyond the currently used drugs that suppress proinflammatory responses. Our findings provide evidence that Itgb1, Rps3, and Ywhaz repress the effector functions of macrophages and synovial fibroblasts by blocking the production of proinflammatory cytokines and chemokines, including Il6, Tnf, and Il8. Ywhaz regulation of proinflammatory cytokines seems not to be dependent on the PEPITEM peptide sequence because PEPITEM, unlike the whole Ywhaz protein, failed to inhibit IL-6 production in both splenocytes and FLSs (Supplemental Figure 13). Moreover, intra-articular overexpression of the Ywhaz gene substantially reduced the development of CIA, whereas i.p. or intra-articular injection of PEPITEM (100 μg, twice a week for 3 weeks) had no effect (data not shown). Taken together, our data indicate that the 3 regulators (particularly Ywhaz), singly or in combination with antiinflammatory agents, could be used for the treatment of RA in the hope of inducing homeostatic repairs as well as of promoting antiinflammation.
Proresolving lipid mediators, including lipoxins, resolvins, and protectins, have been shown to inhibit the functions of proinflammatory cells (23). They are synthesized and released from M2 macrophages (39, 40) and then exert their protective activity at multiple levels in a variety of cell types during resolution of inflammation. These proresolving lipid mediators also promote the functions of suppressive immune cells, increasing generation of Tregs and/or polarizing macrophages toward a proresolving phenotype (41, 42). Similar to the proresolving lipid mediators, Itgb1, Rps3, and Ywhaz were highly expressed in Tregs and M2 macrophages and shown to directly act on multiple effector cells of RA, such as splenocytes, macrophages, and synovial fibroblasts (Figure 5), leading to the inhibition of proinflammatory responses and/or promotion of antiinflammatory responses. These data suggest that Itgb1, Rps3, and Ywhaz, possibly in combination with proresolving lipid mediators, may serve as therapeutic or diagnostic molecular signatures of RA resolution (see Supplemental Figure 12B). Further clinical studies on a large scale are required to clarify this issue.
In summary, our systems approach to resolution dynamics reveals previously unidentified proresolving hub regulators, Itgb1, Rps3, and Ywhaz, which are secreted from suppressive immune cells and target a broad spectrum of RA effector cells, thereby serving as therapeutic or diagnostic molecules for resolution of arthritis. Moreover, our data also provide a comprehensive resource of potential regulators whose roles in the resolution of inflammation can be experimentally tested. This resource can be useful for those who investigate molecular signatures and mechanisms for the resolution of chronic inflammatory diseases, including RA. We believe that this approach can be applied to other autoimmune disorders in order to find new resolution-associated molecules and signaling pathways.
Methods
Additional details can be found in the supplemental material.
Animals.
Male DBA/1 and C57BL/6 mice (Jackson Laboratory) and male Lewis rats (Charles River Laboratories) at 6 to 8 weeks of age were used for experiments. The mice were maintained under specific pathogen–free conditions and used according to the guidelines of the Institutional Animal Care and Use Committee.
Induction of CIA in mice and rats.
Male DBA/1 mice were immunized with bovine CII (Chondrex), as previously described (43). Briefly, CII (1 mg/mL in 0.01 N acetic acid) was emulsified with CFA (ratio 1:1). The mice were injected intradermally at the base of the tail with 0.1 mL of the emulsion (containing 100 μg of CII) as a primary immunization. Two weeks after the primary immunization, booster injections with 50 μg of CII in incomplete Freund’s adjuvant were given through the footpad. Arthritis severity was determined by visual inspection, as previously described (43). In some experiments, a rat model of CIA also was established, as previously described (44).
Statistics.
Statistical analysis was performed for qRT-PCR, Western blotting, ELISA, immunohistochemistry, and flow cytometry experiments using Prism v8 (GraphPad Software). Two-tailed, unpaired Student’s t test or Mann-Whitney U test was applied for comparisons between the 2 groups, and 1-way ANOVA with a post hoc test (Tukey’s or Dunnett’s correction) or Kruskal-Wallis test with a post hoc test (Dunn’s correction) was applied for comparisons of more than 2 groups, as indicated in the figure legends. Across all analyses, a P value of less than 0.05 was considered statistically significant. The statistical methods used for microarray analysis, including deconvolution, functional enrichment, and network analysis, are detailed in the relevant sections of the supplemental methods.
The raw and normalized microarray data have been deposited in the NCBI’s Gene Expression Omnibus with the accession number GSE99087. More detailed information regarding the experimental and computational procedures used in this study is provided in the supplemental methods.
Study approval.
This study was performed with the approval of the institutional review board (KC14TASI0898) at Seoul St. Mary’s Hospital of The Catholic University of Korea.
Author contributions
WUK, CSC, and DH designed the research. JSK, SAY, KMK, and YJB performed in vivo and in vitro experiments. YJP performed disease activity analysis of RA patients. JHP and DH performed microarray experiments and data analyses. DH and WUK interpreted the results from data analyses and experiments. JSK, JHP, CSC, DH, and WUK wrote the manuscript.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2015R1A3A2032927) and Institute for Basic Science (IBS-R013-A1) from the Ministry of Science, ICT and Technology of Korea. We thank the members of the Center for Integrative Rheumatoid Transcriptomics and Dynamics at the Catholic University of Korea for their assistance.
Version 1. 05/14/2020
In-Press Preview
Version 2. 06/22/2020
Electronic publication
Version 3. 08/03/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(8):3974–3986.https://doi.org/10.1172/JCI126866.
Contributor Information
Jin-Sun Kong, Email: matilde00@naver.com.
Ji-Hwan Park, Email: kp706@postech.ac.kr.
Seung-Ah Yoo, Email: youcap78@hanmail.net.
Ki-Myo Kim, Email: jokisa@naver.com.
Yeung-Jin Bae, Email: baeyj0507@hanmail.net.
Yune-Jung Park, Email: jwas@catholic.ac.kr.
Chul-Soo Cho, Email: chocs@catholic.ac.kr.
Daehee Hwang, Email: dhwang@dgist.ac.kr.
Wan-Uk Kim, Email: wan725@catholic.ac.kr.
References
- 1.Prince FH, et al. Sustained rheumatoid arthritis remission is uncommon in clinical practice. Arthritis Res Ther. 2012;14(2):R68. doi: 10.1186/ar3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miles EA, Calder PC. Influence of marine n-3 polyunsaturated fatty acids on immune function and a systematic review of their effects on clinical outcomes in rheumatoid arthritis. Br J Nutr. 2012;107(Suppl 2):S171–S184. doi: 10.1017/S0007114512001560. [DOI] [PubMed] [Google Scholar]
- 3.Ortega-Gómez A, Perretti M, Soehnlein O. Resolution of inflammation: an integrated view. EMBO Mol Med. 2013;5(5):661–674. doi: 10.1002/emmm.201202382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shields AM, Panayi GS, Corrigall VM. Resolution-associated molecular patterns (RAMP): RAMParts defending immunological homeostasis? Clin Exp Immunol. 2011;165(3):292–300. doi: 10.1111/j.1365-2249.2011.04433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isaacs JD. The changing face of rheumatoid arthritis: sustained remission for all? Nat Rev Immunol. 2010;10(8):605–611. doi: 10.1038/nri2804. [DOI] [PubMed] [Google Scholar]
- 6.Prevoo ML, van ‘t Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38(1):44–48. doi: 10.1002/art.1780380107. [DOI] [PubMed] [Google Scholar]
- 7.van der Helm-van Mil AH, Knevel R, Cavet G, Huizinga TW, Haney DJ. An evaluation of molecular and clinical remission in rheumatoid arthritis by assessing radiographic progression. Rheumatology (Oxford) 2013;52(5):839–846. doi: 10.1093/rheumatology/kes378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nistala K, Wedderburn LR. Th17 and regulatory T cells: rebalancing pro- and anti-inflammatory forces in autoimmune arthritis. Rheumatology (Oxford) 2009;48(6):602–606. doi: 10.1093/rheumatology/kep028. [DOI] [PubMed] [Google Scholar]
- 9.Marston B, Palanichamy A, Anolik JH. B cells in the pathogenesis and treatment of rheumatoid arthritis. Curr Opin Rheumatol. 2010;22(3):307–315. doi: 10.1097/BOR.0b013e3283369cb8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laria A, Lurati A, Marrazza M, Mazzocchi D, Re KA, Scarpellini M. The macrophages in rheumatic diseases. J Inflamm Res. 2016;9:1–11. doi: 10.2147/JIR.S82320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asquith DL, Miller AM, McInnes IB, Liew FY. Animal models of rheumatoid arthritis. Eur J Immunol. 2009;39(8):2040–2044. doi: 10.1002/eji.200939578. [DOI] [PubMed] [Google Scholar]
- 12.van den Broek MF, van den Berg WB, van de Putte LB, Severijnen AJ. Streptococcal cell wall-induced arthritis and flare-up reaction in mice induced by homologous or heterologous cell walls. Am J Pathol. 1988;133(1):139–149. [PMC free article] [PubMed] [Google Scholar]
- 13.Brand DD, Latham KA, Rosloniec EF. Collagen-induced arthritis. Nat Protoc. 2007;2(5):1269–1275. doi: 10.1038/nprot.2007.173. [DOI] [PubMed] [Google Scholar]
- 14.Malfait AM, Williams RO, Malik AS, Maini RN, Feldmann M. Chronic relapsing homologous collagen-induced arthritis in DBA/1 mice as a model for testing disease-modifying and remission-inducing therapies. Arthritis Rheum. 2001;44(5):1215–1224. doi: 10.1002/1529-0131(200105)44:5<1215::AID-ANR206>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 15.Marinova-Mutafchieva L, Gabay C, Funa K, Williams RO. Remission of collagen-induced arthritis is associated with high levels of transforming growth factor-beta expression in the joint. Clin Exp Immunol. 2006;146(2):287–293. doi: 10.1111/j.1365-2249.2006.03204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perretti M, Cooper D, Dalli J, Norling LV. Immune resolution mechanisms in inflammatory arthritis. Nat Rev Rheumatol. 2017;13(2):87–99. doi: 10.1038/nrrheum.2016.193. [DOI] [PubMed] [Google Scholar]
- 17.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178(1):280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 19.Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9(21):4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, et al. 14-3-3ζ delivered by hepatocellular carcinoma-derived exosomes impaired anti-tumor function of tumor-infiltrating T lymphocytes. Cell Death Dis. 2018;9(2):159. doi: 10.1038/s41419-017-0180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Han CC, Cui D, Li Y, Ma Y, Wei W. Is macrophage polarization important in rheumatoid arthritis? Int Immunopharmacol. 2017;50:345–352. doi: 10.1016/j.intimp.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 22.Serhan CN, et al. Resolution of inflammation: state of the art, definitions and terms. FASEB J. 2007;21(2):325–332. doi: 10.1096/fj.06-7227rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol. 2008;8(5):349–361. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwata S, Ohashi Y, Kamiguchi K, Morimoto C. Beta 1-integrin-mediated cell signaling in T lymphocytes. J Dermatol Sci. 2000;23(2):75–86. doi: 10.1016/S0923-1811(99)00096-1. [DOI] [PubMed] [Google Scholar]
- 25.Wan F, et al. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131(5):927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Ahn EH, et al. Transduced PEP-1-ribosomal protein S3 (rpS3) ameliorates 12-O-tetradecanoylphorbol-13-acetate-induced inflammation in mice. Toxicology. 2010;276(3):192–197. doi: 10.1016/j.tox.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Liu YC, Elly C, Yoshida H, Bonnefoy-Berard N, Altman A. Activation-modulated association of 14-3-3 proteins with Cbl in T cells. J Biol Chem. 1996;271(24):14591–14595. doi: 10.1074/jbc.271.24.14591. [DOI] [PubMed] [Google Scholar]
- 28.Acuto O, Di Bartolo V, Michel F. Tailoring T-cell receptor signals by proximal negative feedback mechanisms. Nat Rev Immunol. 2008;8(9):699–712. doi: 10.1038/nri2397. [DOI] [PubMed] [Google Scholar]
- 29.Nakayamada S, Saito K, Fujii K, Yasuda M, Tamura M, Tanaka Y. beta1 integrin-mediated signaling induces intercellular adhesion molecule 1 and Fas on rheumatoid synovial cells and Fas-mediated apoptosis. Arthritis Rheum. 2003;48(5):1239–1248. doi: 10.1002/art.10941. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y, Kim HD, Youn B, Park YG, Kim J. Ribosomal protein S3 is secreted as a homodimer in cancer cells. Biochem Biophys Res Commun. 2013;441(4):805–808. doi: 10.1016/j.bbrc.2013.10.132. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi R, et al. 14-3-3 zeta protein secreted by tumor associated monocytes/macrophages from ascites of epithelial ovarian cancer patients. Cancer Immunol Immunother. 2009;58(2):247–258. doi: 10.1007/s00262-008-0549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han X, Han Y, Jiao H, Jie Y. 14-3-3ζ regulates immune response through Stat3 signaling in oral squamous cell carcinoma. Mol Cells. 2015;38(2):112–121. doi: 10.14348/molcells.2015.02101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chimen M, et al. Homeostatic regulation of T cell trafficking by a B cell-derived peptide is impaired in autoimmune and chronic inflammatory disease. Nat Med. 2015;21(5):467–475. doi: 10.1038/nm.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aizu M, et al. Changes in serum interleukin-6 levels as possible predictor of efficacy of tocilizumab treatment in rheumatoid arthritis. Mod Rheumatol. 2017;28(4):592–598. doi: 10.1080/14397595.2017.1370766. [DOI] [PubMed] [Google Scholar]
- 35.Kay J, et al. Clinical disease activity and acute phase reactant levels are discordant among patients with active rheumatoid arthritis: acute phase reactant levels contribute separately to predicting outcome at one year. Arthritis Res Ther. 2014;16(1):R40. doi: 10.1186/ar4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Decramer S, et al. Urine in clinical proteomics. Mol Cell Proteomics. 2008;7(10):1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Smilek DE, Ehlers MR, Nepom GT. Restoring the balance: immunotherapeutic combinations for autoimmune disease. Dis Model Mech. 2014;7(5):503–513. doi: 10.1242/dmm.015099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smolen JS, Steiner G. Therapeutic strategies for rheumatoid arthritis. Nat Rev Drug Discov. 2003;2(6):473–488. doi: 10.1038/nrd1109. [DOI] [PubMed] [Google Scholar]
- 39.Ramon S, et al. The protectin PCTR1 is produced by human M2 macrophages and enhances resolution of infectious inflammation. Am J Pathol. 2016;186(4):962–973. doi: 10.1016/j.ajpath.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werz O, et al. Human macrophages differentially produce specific resolvin or leukotriene signals that depend on bacterial pathogenicity. Nat Commun. 2018;9(1):59. doi: 10.1038/s41467-017-02538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiurchiù V, et al. Proresolving lipid mediators resolvin D1, resolvin D2, and maresin 1 are critical in modulating T cell responses. Sci Transl Med. 2016;8(353):353ra111. doi: 10.1126/scitranslmed.aaf7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid M, Gemperle C, Rimann N, Hersberger M. Resolvin D1 polarizes primary human macrophages toward a proresolution phenotype through GPR32. J Immunol. 2016;196(8):3429–3437. doi: 10.4049/jimmunol.1501701. [DOI] [PubMed] [Google Scholar]
- 43.Yoo SA, et al. Placental growth factor regulates the generation of TH17 cells to link angiogenesis with autoimmunity. Nat Immunol. 2019;20(10):1348–1359. doi: 10.1038/s41590-019-0456-4. [DOI] [PubMed] [Google Scholar]
- 44.Bakharevski O, Stein-Oakley AN, Thomson NM, Ryan PF. Collagen induced arthritis in rats. Contrasting effect of subcutaneous versus intradermal inoculation of type II collagen. J Rheumatol. 1998;25(10):1945–1952. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.