Abstract
Esophageal atresia (EA/TEF) is a common congenital abnormality present in 1 of 4000 births. Here we show that atretic esophagi lack Noggin (NOG) expression, resulting in immature esophagus that contains respiratory glands. Moreover, when using mouse esophageal organoid units (EOUs) or tracheal organoid units (TOUs) as a model of foregut development and differentiation in vitro, NOG determines whether foregut progenitors differentiate toward esophageal or tracheal epithelium. These results indicate that NOG is a critical regulator of cell fate decisions between esophageal and pulmonary morphogenesis, and its lack of expression results in EA/TEF.
Keywords: Development
Keywords: Embryonic development, Human stem cells, Surgery
Introduction
Congenital esophageal atresia (EA) represents a failure of the esophagus to develop as a continuous passage, which causes the esophagus to end in a blind upper pouch rather than connecting normally to the stomach. Tracheal-esophageal fistula (TEF) represents an abnormal connection between the trachea and esophagus. EA/TEF is generally assumed to be a multifactorial condition (1, 2).It is believed that defects in foregut morphogenesis underlie the birth defects that cause EA and TEF. This has been demonstrated in animal models of EA/TEF generated by mutations in both respiratory and gastrointestinal markers (NKX2-1 and SOX2) (3, 4) as well as for factors with more dynamic expression patterns (SHH and NOG) (5, 6). In addition, the Adriamycin animal model that disturbs normal expression of Shh, leading to failed separation of the foregut, provides unique insight into how a nongenetic factor can disrupt the developmental process (7). Genetic and population-based studies have been performed to correlate the animal data to possible causes of EA in humans. These studies have identified single gene mutations that are associated with syndromic EA/TEF, including mutations in SOX2, CHD7, MYCN, GLI3, and MID1 (8, 9). However, most cases of EA are nonsyndromic and occur in isolation. Less is known about the molecular events that lead to isolated EA/TEF.
The BMP and NOG pathway has been shown to be important in the development of esophagus and trachea and several studies have shown that disruption of the pathway in mice results in a foregut phenotype similar to EA/TEF (6, 10, 11). BMPs are secreted proteins that bind to a complex of type I and type II transmembrane receptors and activate a series of downstream cascades that include SMADs. BMP signaling is regulated by extracellular antagonists such as NOG that bind to BMP and inhibit BMP binding and activating its receptor (12).
We used human esophagus at different stages of development as well as tissue from EA/TEF surgical repair to study the expression of genes known to be important in mouse esophageal development. In addition, we used esophageal organoid units (EOUs) and tracheal organoid units (TOUs) as models of human esophageal and tracheal development in vitro to study the interaction of these molecules and their potential role in development of EA/TEF in humans. Here we show that the NOG/BMP signaling cascades influence cell proliferation and plasticity in differentiation of foregut progenitors into either esophagus or trachea/lung cell lineages. In addition, we show that the absence of NOG in the esophageal progenitors switches their fate into respiratory epithelium. Furthermore, we found that the esophageal epithelium of patients with EA/TEF lack NOG expression, and histological evaluation of the tissue indicates the presence of esophageal and respiratory epithelium, making the absence of NOG a likely cause of EA/TEF in humans. Finally, we showed that, in the presence of NOG, EOUs derived from TEF patients grow larger, healthier, and express esophageal markers that they lack if NOG is not replaced in the culture.
Results
NOG is expressed in the developing human esophagus, but not in the respiratory tissue.
To study the normal expression of genes previously shown to be important in foregut development, we obtained paraffin-embedded human tissue at different gestational ages. We examined the expression of NOG, BMP2, BMP4, BMP7, SHH, SOX2, NKX2-1, WNT2, and PAX9 in esophagus and respiratory tissue of neonates at 23 weeks of gestation (preterm neonate) and neonates older than 36 weeks of gestation (term neonates) (Figure 1A). PAX9 was not detected in any of the samples. In both full-term and preterm tissues, BMP2, BMP4, BMP7, SHH, and SOX2 were found in both respiratory and esophageal epithelium. However, NOG was only found in the esophageal tissue, consistent with results in mice. This finding supports the role of NOG as a dorsal morphogen, which specifies esophageal tissue and inhibits respiratory tissue differentiation (6, 13).
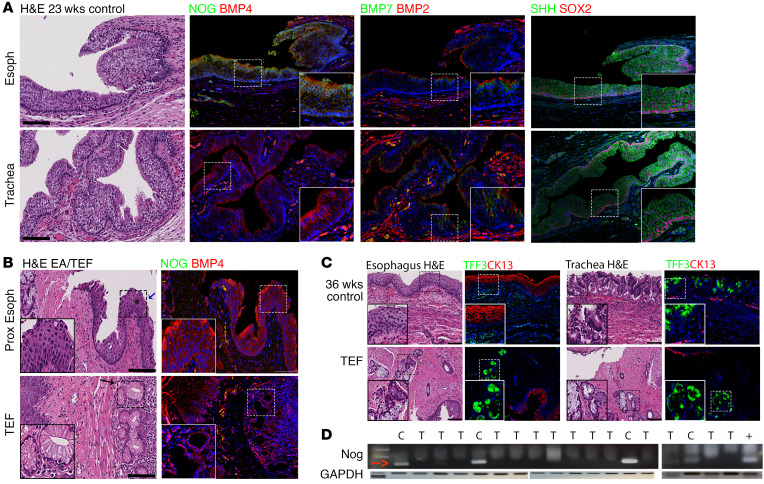
Figure 1. NOG is selectively expressed in developing human esophagus but not in the trachea, and EA/TEF tissue lacks NOG.
(A) Pathology samples from human autopsies stained for genes that have been previously known to be important in foregut development. Paraffin sections were used to perform IHC with NOG, BMP7, SHH (green), BMP4, BMP2, and SOX2 (red). All genes were present in both esophagus and trachea. However, NOG was missing from respiratory tissue. (B) Pathology samples from EA/TEF repair were collected to study NOG and BMP expression using IHC. On H&E staining, a combination of esophageal and respiratory glands were found in atretic esophagus as well as in fistula. NOG is absent from EA/TEF samples when evaluated by IHC as well as RT-PCR (B, D). (C) Human esophageal and respiratory pathology samples stained for CK13 (red) and TFF3 (green). TEF from human neonate expresses both CK13 and TFF3. (D) RNA was isolated from paraffin samples of either esophagus of control neonates, or EA/TEF tissue obtained at the time of surgical repair. Positive control (+) is RNA extracted from Kato III cell line. *Pseudostratified columnar epithelium with cilia. Blue arrow indicates cilia. Black arrow indicates seromucinous glands. Inserts are ×2 magnification. Scale bars: 100 μm. C, control, T, TEF.
Esophagus from EA/TEF patients contains a mixture of squamous cell epithelium and stratified columnar epithelia with cilia, and the tissue lacks NOG expression.
To study the function of the NOG/BMP molecular cascade on development of EA/TEF, we studied the expression of NOG and BMP in pathology samples obtained at the time of EA/TEF repair. When the pathology samples were analyzed using H&E staining, we found that both proximal and distal esophagus contained a combination of immature esophageal epithelium and immature respiratory glands (Figure 1B). Interestingly, respiratory glands were present even when a TEF was not found (patient with long gap EA without TEF) and there was no communication between the esophagus and trachea. Similarly, the TEF, when present, contained a combination of immature esophageal and respiratory glands. Respiratory epithelium was confirmed with TFF3 immunostaining and esophageal epithelium was confirmed with CK13 expression (Figure 1C). In addition, the muscular layer of the atretic esophagus and the TEF contained a combination of disorganized smooth and skeletal muscle.
In contrast, when we studied the expression of NOG and BMP, we found that BMP4 was expressed in the atretic esophagus as well as in the fistula. However, NOG was not present in either atretic esophagus or fistula (Figure 1B). We also observed that BMP7 was minimally expressed in the epithelium and BMP2 was mainly expressed in the muscular layer (data not shown). Therefore, in the rest of the experiments, we focused on BMP4.
In addition, we used paraffin blocks of samples from EA/TEF collected at the time of surgical repair to extract RNA and investigate the amount of NOG mRNA found in the tissue. NOG mRNA was absent from EA/TEF samples when we evaluated it using reverse transcription–polymerase chain reaction (RT-PCR) (Figure 1D). These data suggest that, similar to mice, in humans NOG expression is important for normal esophageal development. Moreover, the absence of NOG during esophageal development may cause abnormal differentiation of the esophagus, resulting in EA/TEF.
Murine EOUs represent a good in vitro model for development of the esophagus.
The development of EOUs was recently reported by Grikscheit and Spence (14–16). The Grikscheit group demonstrated that a mature EOU provides all of the key components of native esophagus including epithelium, subjacent muscularis, differentiated suprabasal and proliferative basal layers of esophageal epithelium, muscle, and nerves. Cultured EOUs grew as an expanding sphere of proliferative basal cells on a neuromuscular network for the first several days, and after approximately 8 days in culture, EOUs demonstrated spontaneous peristalsis consistent with a mature engineered esophagus.
Here we used P1–P3 murine esophagus to generate EOUs. They were cultured in matrigel for 4–12 days and either embedded in paraffin or processed as whole mounts. Immunostaining was used to detect expression of different esophageal markers at different stages (Figure 2).
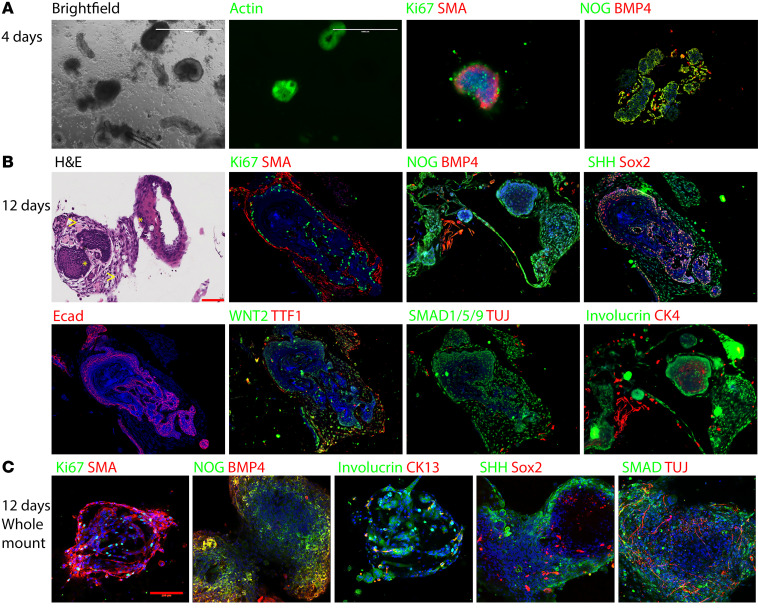
Figure 2. EOU culture is a suitable model for mammalian esophageal differentiation in vitro.
mEOUs cultured for 4–12 days and either embedded in paraffin or taken as whole mount. IHC was used to detect expression of esophageal markers. Red scale bars: 100 μm; white scale bars: 1000 μm. (A) mEOUs 4 days in culture showing that most cells were Ki67+. (B) mEOUs 12 days in culture, larger in size, fewer cells were Ki67+. H&E staining demonstrates squamous epithelium surrounded by stromal components. Squamous epithelium shows maturation from basal layer to superficial layer. Differentiated cells expressed the squamous epithelium markers CK4 and involucrin, smooth muscle marker SMA, neuron marker TUJ, and other molecules important in foregut development: SHH, BMP4, NOG, SMAD1/5/9. (C) mEOUs 12 days in culture examined in whole-mount 3D images showing how different cells types come together to form EOUs.
At 4 days in culture, the EOUs consisted largely of dividing progenitor cells (Ki67-positive cells), with the presence of both NOG and BMP4. In contrast, at 12 days EOUs formed a more mature esophagus-resembling unit containing a layer of smooth muscle (smooth muscle actin [SMA] positive cells), squamous epithelium (CK13 [KRT13], CK4 [KRT4], involucrin expressing cells), nerves (TUJ-1 positive cells), and important developmental proteins (SHH, SMAD, WNT2, SOX2) (Figure 2). In addition, after approximately 10 days in culture the EOUs began contracting spontaneously, consistent with functional innervation.
These data suggest that EOUs are a good model to study development of mammalian esophagus and how the proteins interact to create a normal esophagus versus EA/TEF.
Development of a small molecule that inhibits NOG binding to BMP.
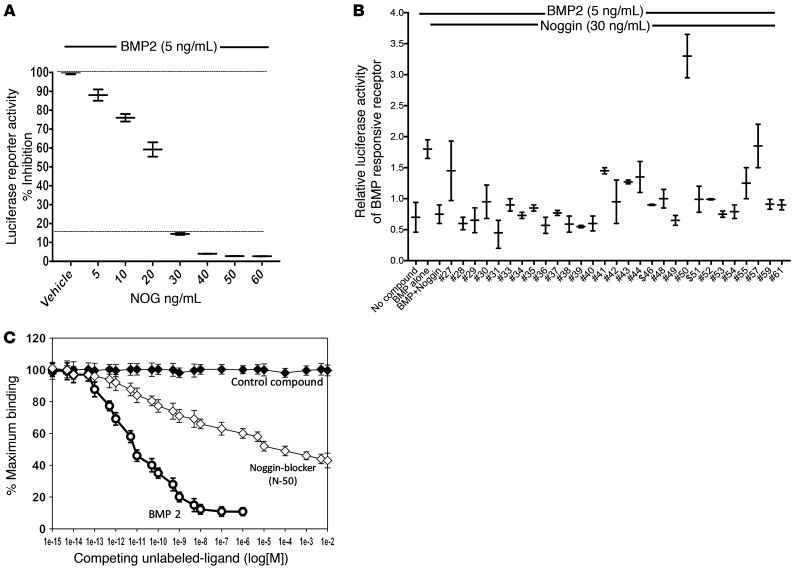
To study the effects of NOG in EOU cultures, we selected a small molecule that would inhibit NOG interaction with BMP. By computational analysis, we identified the binding region of NOG with BMP2 and the binding region of receptors with BMP2 and specifically designed molecules that block BMP-NOG interaction and would not bind to the BMP receptors (17). To screen potential NOG-blocking compounds, we developed a rapid in vitro assay to determine the inhibitory effect of NOG on the BMP2 signaling pathway using mouse myoblastic C2C12 cells, which can be stimulated by BMP toward the osteoblastic phenotype. C2C12 cells were transfected with the 9XGCCG BMP Smad1 reporter construct and control Renilla luciferase vectors for 24 hours. The cells were then treated for 24 hours with BMP2 (5 ng/mL) and various concentrations of NOG and the relative luciferase activity was measured with a luminometer. As expected, NOG inhibited BMP2 induced reporter activity in a dose-dependent manner. The 1:6 ratio of BMP to NOG achieved 85% inhibition of BMP-induced reporter activity (Figure 3A) and was chosen for further screening assays. We then tested the candidate NOG-blocking compounds at a higher dose (1 μg/mL) to see if any were capable of reversing the effect of exogenous NOG beyond the BMP2 baseline response. N-50 was the most potent enhancer above baseline, increasing BMP2 responsiveness 2-fold (Figure 3B). The enhancement above the baseline BMP2 luciferase response suggests that N-50 was the best compound to also inhibit endogenous NOG in addition to reversing the effects of exogenous NOG as in the earlier experiments.
Figure 3. N-50 is a small molecule that inhibits NOG in vitro.
(A) Validation of a BMP2 reporter assay and selection of optimal BMP-NOG ratio for compound screening (1:6 ratio is chosen). The dose-dependent inhibition by NOG of BMP2-induced luciferase activity in C2C12 cells transfected with BMP-specific and Smad1-driven 9 × GCCG reporter plasmid. The reporter responded dose dependently to BMP2. Data points of luciferase activities were determined in triplicate cell culture wells (n = 3). The error bar represents SEM. Statistical significance (P < 0.05) was determined by 1-way ANOVA between the controls and treatments. (B) N-50 inhibition increases BMP efficacy in addition to reversing the effects of exogenously added NOG. N-50 was the most potent enhancer above baseline, increasing BMP2 responsiveness 2-fold (tallest green bar). The enhancement above the baseline BMP2 luciferase response (100%) suggests that N-50 was the best compound to also inhibit endogenous NOG and reverse the effects of exogenously added NOG. Luciferase activities were determined in triplicate. Error bars (±SEM) from triplicate determinations (n = 3) from cell-culture wells. The BMP+NOG group was compared with all other treatment groups using 1-way ANOVA, and statistical significance of P < 0.05 was determined between the control and treatments. Compounds that exhibited undesired effects on cell growth and morphology were dropped from further analyses. (C) In vitro binding assay with purified recombinant BMP2 and NOG proteins. NOG and BMP2 were labeled with biotin and 125iodine, respectively. Constant amounts of biotin-labeled NOG and 125iodine-labeled BMP2 were incubated with or without varying concentrations of unlabeled BMP2 or N-50 as described in Methods. Biotin-labeled NOG was pulled down with neutravidin-agarose beads and the counts associated with pellets were counted. The chemical 2-vinyl-4,6-diamino-1,3,5-triazine was designed as the control based on a different target structure expected not to interfere with BMP-NOG binding.
To confirm that N-50 actually inhibits BMP and NOG interaction as designed, we optimized an in vitro binding assay with labeled recombinant BMP2 and NOG proteins. NOG and BMP2 were labeled with biotin and 125iodine, respectively, as described in Methods. As expected, unlabeled BMP2 competed off about 90% of labeled BMP2. Our data demonstrate that the BMP2 and NOG interaction is saturable and concentration dependent in this optimized binding assay. The NOG-blocker N-50 competed with BMP2 to prevent more than 50% of the labeled BMP2 from binding to NOG, confirming that our lead compound N-50 disrupts BMP2 binding to NOG (Figure 3C).
Inhibition of NOG during murine EOU culture results in phenotypic changes that resemble human EA/TEF tissue.
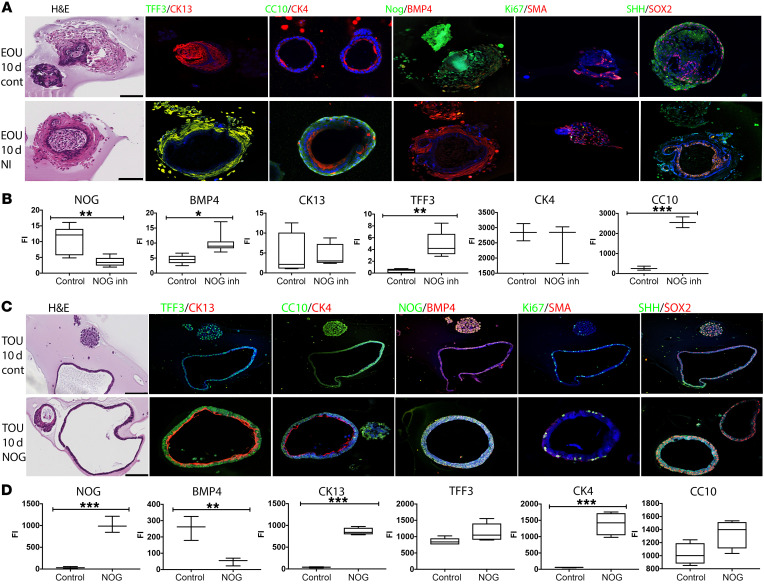
After establishing N-50 as a good in vitro inhibitor of NOG, we tested the effects of inhibition of NOG during esophageal development using the EOU model. Murine P1–P3 esophagus were collected to generate EOUs. NOG inhibitor was added into the culture medium for 24, 48, and 96 hours to investigate the effects of NOG inhibition on EOU. We found that addition of NOG inhibitor for more than 96 hours resulted in large amounts of cell death. We therefore used the inhibitor for 48 hours only, at which point the inhibitor was removed and the culture was allowed to proceed for a total of 10 days. At the end of the culture the EOUs were collected and processed for paraffin sectioning. EOUs treated with NOG inhibitor contained larger epithelial cells with more cuboidal shape (Figure 4A). In addition, immunohistochemical evaluation demonstrated that after 10 days in culture, EOUs treated with NOG inhibitor expressed the respiratory epithelium markers TFF3 and CC10 (SCGB1A1) together with esophageal squamous epithelium markers CK13 (KRT13) and CK4 (KRT4). When the NOG inhibitor was added to the media, expression of BMP4 was higher overall, suggesting that activation of the BMP signaling cascade may trigger expression of respiratory epithelium proteins (Figure 4B).
Figure 4. Cultured murine foregut progenitors change fate depending on presence of NOG.
(A) mEOUs cultured for 10 days. NOG inhibitor was added to the culture for 48 hours, washed, and cultures allowed to grow for 10 days. Tissue was embedded in paraffin and slides analyzed with IHC for expression of esophageal and tracheal markers. (B) Fluorescence intensity of NOG, BMP4, CK13, TFF3, CC10, and CK4 was evaluated using Image J. mEOUs from 6 different litters (n = 4–12). (C) P1–P3 murine trachea were collected to generate TOUs. TOUs were cultured for 10 days with or without NOG, fixed, and paraffin sections were used to evaluate their morphology and protein expression. (D) Fluorescence intensity of NOG, BMP4, CK13, TFF3, CC10, and CK4 was evaluated using Image J. mTOUs from 5 different litters (n = 3–5). Scale bars: 100 μm. There was no change in the expression of SOX2 or SHH. *P < 0.05, **P < 0.005, ***P < 0.0009.
We wanted to validate the expression of respiratory markers in our cultures. Therefore, we tested well-known respiratory markers in murine trachea and esophagus of P1–P3 pups. We found that MUC5AC, CK8 (KRT8), CK18 (KRT18), and TTF1 are expressed in both murine esophageal and tracheal epithelium at earlier postnatal stages (P1–P3) and later become specific in tissues with age. However, CC10 (SCGB1A1), TFF3, and SCGB3A3 were specific to respiratory epithelium (Supplemental Figure 2; supplemental material available online with this article; https://doi.org/10.1172/JCI123597DS1).
To confirm that the effects of NOG inhibitor on expression of respiratory markers in murine EOUs (mEOUs) were indeed due to inhibition of NOG function, we added both compounds together (NOG inhibitor and NOG) for 48 hours, washed the mEOUs, and allowed the culture to continue for an additional 8 days. At the end of the culture, addition of NOG to the NOG inhibitor blocked expression of respiratory makers, confirming that the NOG inhibitor is specific to NOG (Supplemental Figure 3).
Addition of NOG to TOU culture results in organoids containing squamous epithelium.
During embryogenesis, the trachea and esophagus develop from the same progenitor cells in the endoderm (13). We sought to test the effects of alteration of NOG/BMP signaling in progenitor cells of the trachea by creating TOU cultures and adding NOG to the system. The same protocol was used to create organoid units derived from trachea. Murine P1–P3 tracheas were collected to generate TOUs and they were cultured for 12 days. NOG was added to the media for the entire culture time, and TOUs were subsequently fixed and embedded in paraffin. H&E staining of the TOUs treated with NOG showed squamous epithelial cells in the majority of the organoid units, which was confirmed with immunostaining for squamous epithelium markers CK13 (KRT13) and CK4 (KRT4) (Figure 4C). In addition, when NOG was added to the media, expression of BMP4 was significantly lower, suggesting inhibition of BMP signaling may trigger expression of proteins specific to the esophagus (Figure 4D). Minimal cleaved caspase 3 (CASP3) expression was seen in both cultures, suggesting that most cells remained healthy during the culture and there was low cell death (data not shown). These data confirm the BMP signaling cascade as an important regulator of foregut differentiation. When NOG was present and the BMP cascade was inhibited, progenitor cells differentiated toward esophageal epithelium. On the other hand, when NOG was absent and BMP active, progenitors differentiated toward respiratory epithelium.
Inhibition of NOG in EOUs derived from normal pediatric human esophagus results in expression of respiratory markers.
To confirm the function of NOG and BMP in human foregut development, we repeated the experiments using EOUs derived from normal human pediatric esophagus. A total of 5 esophagi from pediatric organ donors were collected and EOUs were cultured for 10 days. NOG inhibitor was added to the media for 48 hours, subsequently washed out and cultures were allowed to grow for 8 more days. As with the mouse EOU cultures, the human EOUs (hEOUs) expressed both NOG and BMP4, shown to be essential in foregut development. In addition, hEOUs expressed markers of mature esophagus such as CK13 (KRT13) and CK4 (KRT4) at the end of a 10-day culture (Figure 5A) demonstrating that hEOUs are a good model for human esophageal differentiation. Furthermore, hEOUs cultured in the presence of the NOG inhibitor coexpressed the respiratory marker TFF3 and CC10 (SCBG1A1) with CK13 (KRT13) and CK4 (KRT4), suggesting the inhibition of NOG was able to change the fate of esophageal progenitor cells to become respiratory epithelium (Figure 5, A and B). To confirm that the effects of NOG inhibitor on expression of respiratory markers were specific to NOG, we added both NOG and NOG inhibitor to the culture and found that NOG was able to rescue the phenotype. The hEOUs did not express respiratory markers at the end of the culture (Supplemental Figure 4).
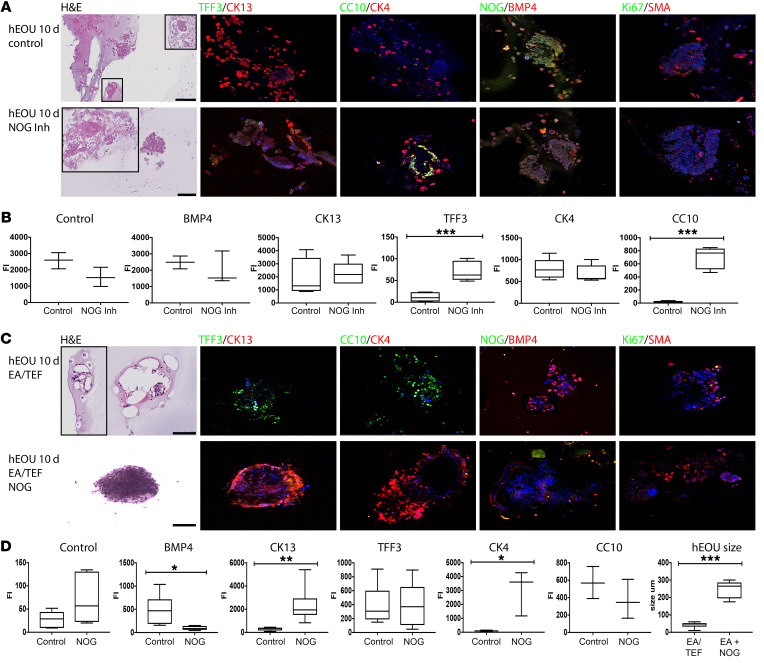
Figure 5. Human EOU culture in the presence of NOG inhibitor resembles EA/TEF.
Addition of NOG restores viability and stimulates expression of esophageal markers. (A) hEOUs grown for 10 days from 5 different patients. NOG inhibitor was added to the culture for 48 hours, washed, and cultures were allowed to grow for 10 days. IHC analysis for expression of esophageal and tracheal markers. (B) Fluorescence intensity of CK13, TFF3, CC10, CK4, NOG, and BMP was evaluated using Image J (n = 3–5), ***P < 0.0009. (C) hEOUs isolated from EA/TEF patients at the time of surgical repair and cultured for 10 days with or without NOG. hEOUs were fixed and paraffin sections were used to evaluate their morphology and protein expression. (D) Fluorescence intensity of CK13, TFF3, CC10, CK4, NOG, and BMP was evaluated using Image J (n = 3–8), *P < 0.05, **P < 0.005. Scale bars: 100 μm.
EOUs derived from patients with EA/TEF lack NOG expression, grow slowly, and express mainly respiratory markers, but this phenotype is reversed when NOG is added to the culture.
Small full-thickness sections from atretic esophagus and or TEF were collected at the time of EA/TEF surgical repair. The tissues used were pieces that were trimmed at the time of surgery in order for the anastomosis to be attached properly, and that otherwise would have been discarded. EOUs were derived from the tissue and placed in culture for 10 days. H&E staining demonstrated small organoid units with a large number of pyknotic cells and stroma. Most epithelial cells expressed TFF3 and CC10 (SCBG1A1) with minimal squamous cell marker expression (CK13, CK4). In addition, NOG was not present, and BMP4 was highly expressed (Figure 5C).
In contrast, when NOG was added to the culture for the entire 10 days, the organoid units were on average 5 times larger as compared with controls (5.859 ± 0.352, n = 10, P < 0.001). Furthermore, the majority of the epithelia expressed squamous cell markers (CK13, CK4, and involucrin), even though some cells did express CC10 (SCBG1A1) and TFF3. In addition, BMP expression was markedly reduced as compared with controls (Figure 5, C and D). These data suggest that lack of NOG in the epithelium of the esophagus results in aberrant elevation of BMP signaling, which in turn triggers expression of respiratory epithelium proteins seen in EA/TEF. Addition of NOG restored the esophageal phenotype.
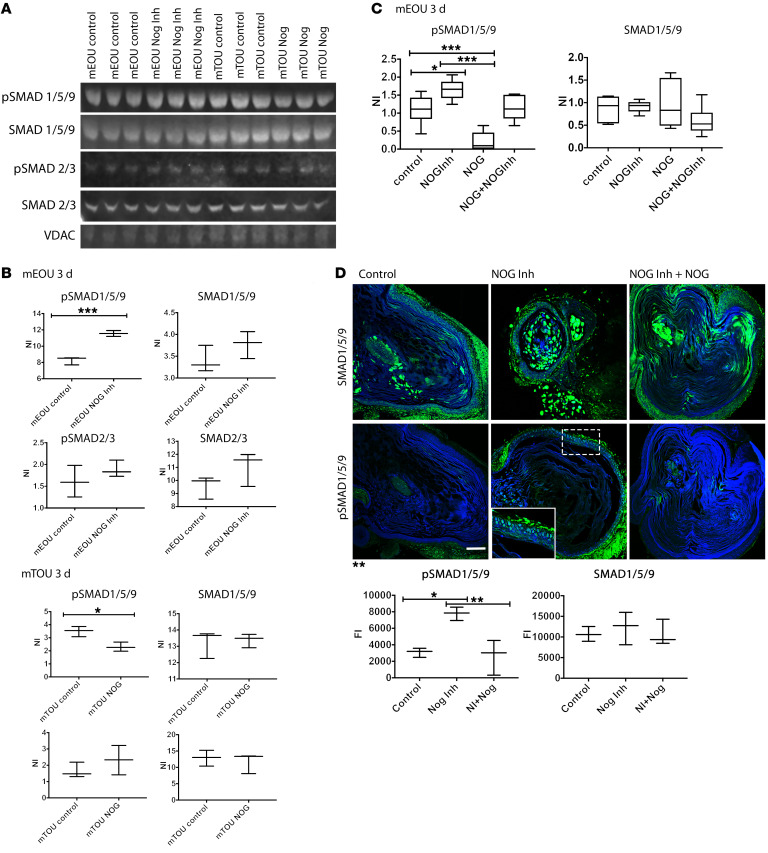
Increased BMP signaling in foregut epithelium correlates with phosphorylation of SMAD1/5/9.
To investigate whether the inhibition of NOG resulted in activation of the BMP cascade, we used Western blots to examine the amount of activated SMADs found in culture. Murine P1–P3 esophagi were collected to generate EOUs and cultured with or without NOG inhibitor for 48 hours, at which time protein was collected. A Western blot was run to determine the amount of phosphorylated/activated SMAD1/5(Ser 463/465)/9(Ser 465/467) found in the cells. Phosphorylated SMAD2(Ser465/467)/SMAD3(Ser423/425) was used as a control.We found that the amount of phosphorylated SMAD2/3 did not change after addition of NOG inhibitor. However, the phosphorylation of SMAD1/5/9 was increased by about 30% (Figure 6, A and B). To confirm that inhibition of BMP during differentiation of TOUs was also involved in the aberrant expression of esophageal markers, we investigated the difference in phosphorylated SMADs between TOUs cultured in the presence or absence of NOG. We collected protein from mouse TOUs that were grown with and without NOG for 10 days and amounts of phosphorylated SMAD 1/5/9 and phosphorylated SMAD 2/3 were studied. Once again, addition of NOG did not change phosphorylation state of SMAD 2/3. However, NOG decreased the amount of phosphorylated SMAD 1/5/9 by about 30% at the end of the culture, consistent with the hypothesis that NOG inhibits BMP activation and phosphorylation of SMAD 1/5/9 (Figure 6, A and B). These data confirmed the changes in the amount of BMP seen with immunostaining of the EOUs and TOUs grown in the different conditions. The presence of NOG inhibitor resulted in activation of the cascade downstream of BMP in mEOUs, and the presence of NOG decreased activation of the BMP cascade in mTOUs.
Figure 6. Activation of BMP signaling cascade results in increased phosphorylation of SMAD1/5/9.
Mouse EOUs were cultured for 2 days with or without NOG inhibitor. Mouse TOUs were cultured for 10 days with or without NOG. (A) Protein was collected at the end of culture and Western blots were run for pSMADs 1/5/9 and pSMADs 2/3. (B) Quantification of Western blot intensity using Image J. (C) To confirm that the activity of NOG inhibitor was indeed specific to NOG, we added NOG and NOG inhibitor together to the culture to examine whether the effects of NOG inhibitor on phosphorylated SMAD 1/5/9 were blocked. Experiments were done with mEOUs (6 different litters). Phosphorylated SMAD protein was normalized to total SMAD protein and VDAC was used as loading controls. (D) mEOUs were cultured for 10 days and IHC was used to test for total or phosphorylated SMAD 1/5/9 expression. All images were obtained at ×20. Scale bar: 100 μm. Fluorescence intensity of total SMAD1/5/9 and pSMAD1/5/9 was evaluated using Image J (n = 3–6), *P < 0.05, **P < 0.005, ***P < 0.001.
To confirm that the effects of NOG inhibitor were specific to blocking NOG function, we repeated the experiments with mEOUs in the presence of both NOG and NOG inhibitor. As shown in Figure 6C, when NOG was added to the culture, the amount of pSMAD1/5/9 fell to almost undetectable. However, when NOG was added together with NOG inhibitor, the amount of phosphorylated SMAD 1/5/9 returned to control levels, demonstrating the specificity of N-50 to NOG.
Furthermore, using IHC we investigated the changes in phosphorylated SMAD 1/5/9 in mEOUs after 10 days in culture in the presence of NOG inhibitor alone or NOG inhibitor plus NOG. The mEOUs cultured in the presence of NOG inhibitor contained 3 times more phosphorylated SMAD1/5/9 compared with control. Moreover, addition of NOG to the culture blocked the increase in phosphorylation (Figure 6D).These data demonstrate that activation of downstream proteins such as SMAD 1/5/9 may be responsible for tissue fate specification in the foregut epithelium.
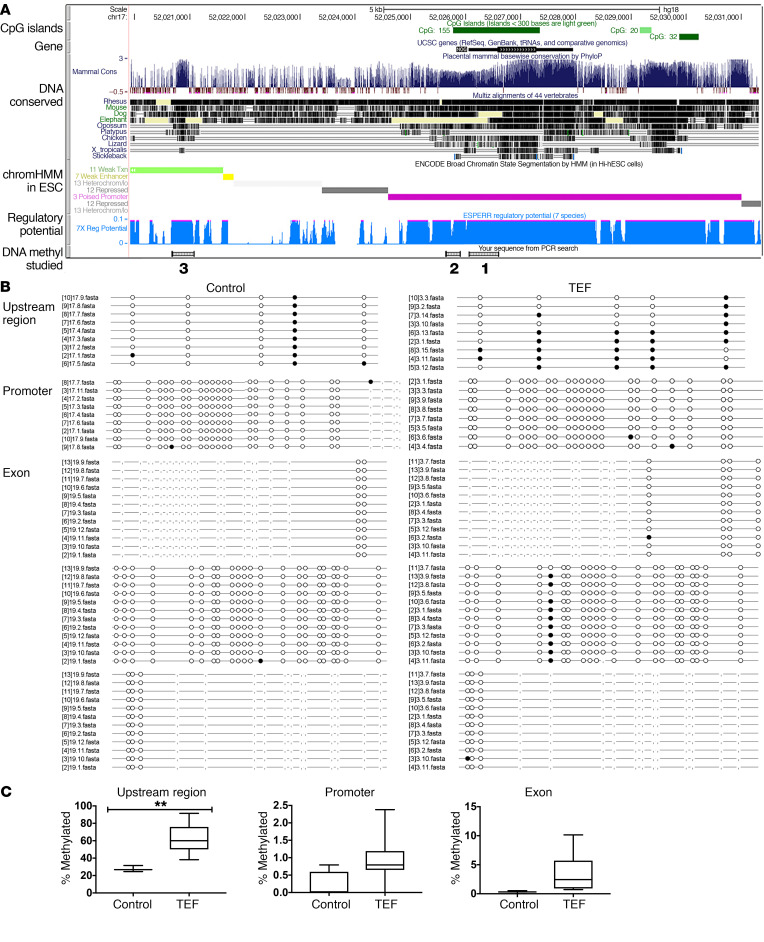
DNA methylation of the NOG locus is increased in patients with EA/TEF.
Tissue (and temporal) specific gene regulation is essential for proper gene expression that drives human development. This regulation is controlled by binding of transcriptional factors and changes in epigenetic modifications in gene regulatory regions such as promoters and enhancers. To investigate a mechanism that might cause abnormal silencing of the NOG gene in EA/TEF patients, we searched the NOG gene locus for underlying epigenetic mechanisms that may be involved in regulating the expression of the gene. The NOG locus contained a large number of CpG islands and thus we investigated the DNA methylation status of the gene locus in EA/TEF patients and compared it with healthy controls. DNA from paraffin sections of tissue collected at the time of EA/TEF surgical repair was extracted to analyze methylation patterns of the NOG exon, promoter, and an upstream regulatory area. We evaluated a 230-bp region 4.5 kb upstream of the promoter that had high variability in DNA methylation among 15 different tissues and cell lines, which suggested methylation in this area as a way to regulate NOG expression in different tissues. The DNA sequence in this region is highly conserved among vertebrates and mammals, suggesting high regulatory potential for this area (see corresponding tracks in Figure 7A). Additionally, Encyclopedia of DNA Elements (ENCODE) chromatin segmentation in human embryonic stem cells (ESCs) (ChromHMM), an automated computational system for learning chromatin states, characterizing their biological functions and visualizing the resulting genome-wide maps of chromatin-state annotations (18, 19), demonstrated that this area is transcribed in ESCs and it adjusts by potential poised enhancer (Figure 7A). We were able to collect samples from 16 TEF patients and 3 control subjects. Esophagus from autopsies of neonates who have died of reasons other than foregut anomalies were used as controls. The DNA was subsequently treated with bisulfite and samples were submitted for Sanger sequencing. We looked at 226 bp in the exon area, 162 bp in the promoter, and 125 bp in the upstream area. When compared with controls, DNA from TEF patients overall was more methylated. The promoter of both groups had the least methylation, with control samples having 0.2% of CpG islands analyzed methylated versus 1% of CpG islands in the TEF group. The CpG islands in the exon region had more methylation in the TEF group when compared with controls; however, the increase in methylation did not reach statistical significance (0.1% in control vs. 3.5% in TEF, P = 0.24). Finally, the upstream area had the most methylation of all the regions studied. In the control group, 27% of the CpG islands were methylated versus 63% in the TEF group (P < 0.006) (Figure 7, B and C). Overall, the DNA methylation pattern in control samples was consistent among different individuals, as opposed to TEF samples, where the methylation pattern was different among individuals with high variability, with some samples having 91% of CpG islands methylated in the upstream area to some samples having only 40% methylated. An increase in DNA methylation of the presumable NOG regulatory region suggests deactivation of this regulatory region, which could lead to premature downregulation of NOG gene expression and abnormal foregut development in EA/TEF patients.
Figure 7. DNA methylation of NOG locus is increased in patients with EA/TEF.
(A) Combined genome-wide tracks for NOG gene locus from UCSC human genome browser hg18. From top to bottom: CpG islands are in dark green; NOG transcript is in black; DNA conserved in mammals is dark blue; invertebrates is in black. ENCODE BROAD chromatin segmentation by HMM in H1-hESC: weak transcription is in light green, weak/poised enhancer is in yellow, repressed chromatin is in gray, and poised promoter is in purple. Regulatory potential of DNA is light blue. The 3 DNA areas of NOG locus that were assessed for DNA methylation are marked as numbered small boxes at the bottom of the panel. 1, exon of NOG gene; 2, promoter of NOG; 3, upstream regulatory region. (B) Lollipop plots of DNA methylation of NOG locus CpG islands from 16 EA/TEF patients and 3 controls. Promoter, exon, and the region 10 kb upstream of promoter were analyzed. (C) Quantification of percentage of CpG islands methylated (n = 6–12), **P < 0.005.
Discussion
EA/TEF is a common congenital abnormality that affects 1 in 4000 live births worldwide. However, the mechanism that results in such anatomy remains unknown. Animal models have been very important to elucidate normal esophageal development in the mammalian system; however, human esophageal development is not well understood.
We have demonstrated that EOUs are a good system to study differentiation of the midgut progenitors into mature esophagus in vitro, and changes in protein function can be successfully studied. Our model shows a group of dividing progenitor cells in the first 5 days of culture that express important developmental proteins such as NOG, BMP, WNT, and further differentiates into mature esophageal epithelium expressing involucrin (IVL), CK13 (KRT13), and CK4 (KRT4). The system also contains an innervated muscular layer that begins contracting at day 10 in culture. Further studies can be carried out to study the function of other proteins in the development of human esophagus using this model.
We have characterized expression of developmental genes in normal human esophageal development, and we have correlated their expression with the formation of EA/TEF. We discovered that NOG expression is absent in esophageal tissue collected from patients with EA/TEF. These data provide evidence for the importance of NOG in human esophageal development and how its abnormal expression may be responsible for the formation of EA/TEF. In our in vitro culture of esophageal and tracheal organoid units, NOG serves as an important cell fate signal that determines whether a foregut progenitor will become trachea or esophagus. The absence of NOG in esophageal progenitors can reprogram them to express respiratory genes. Similarly, the presence of NOG in tracheal progenitors can reprogram them to express esophageal genes. We are unable to completely block the NOG or BMP signaling cascade in our organoid culture and therefore, our EOUs coexpress respiratory genes. Moreover, addition of NOG to TOUs does not completely eliminate BMP signaling and again we have coexpression of respiratory and esophageal proteins. This may be due to intrinsic programming in the progenitor cells that cannot be changed after birth, in addition to involvement of other signaling that we have not studied and may also be important in the development of EA/TEF. For example, well-known inhibitors of BMP such as chordin and follistatin have been shown to modulate BMP signaling (20). Our immunohistochemistry data not shown here detected very low levels of chordin in the esophagus and further investigation will be required to elucidate their function in foregut development. Furthermore, trachea-esophageal morphogenesis is a very complex process that requires other molecular cascades that will not only optimize cell division and differentiation, but will also allow for anatomical division and construction of 3D functional structures that we have not addressed in this study. For instance, Nasr et al. (21) recently demonstrated the importance of endosome-mediated epithelial remodeling that is required for proper tracheoesophageal separation. Further studies will be necessary to reveal other molecular cascades that may be clinically relevant as well.
The mechanism by which NOG expression is inhibited during human development remains to be discovered. Epidemiological studies have linked several conditions to EA, such as advanced maternal age, diabetes, exposure to tobacco products, and use of illicit drugs. All of these conditions have in common high oxidative states, which have been linked to DNA methylation and epigenetic errors. Our DNA methylation studies show higher NOG DNA methylation overall in the esophagus of patients with EA, suggesting that a premature deregulation of NOG expression may be responsible for decreasing its expression. We acknowledge that our studies are preliminary specifically because we lack a sample large enough to represent the human population. Collection of human samples with tests and identification of proper controls are very difficult and complex tasks. More studies should be undertaken to fully understand the mechanism by which NOG is silenced in these patients.
Clinically, EA/TEF remains a difficult condition to treat. Even though more than 95% of patients survive after surgery, morbidity due to esophageal strictures and recurrent TEF remains high. A recent study showed postoperative complications in about 62% of patients undergoing surgical repair with anastomotic stricture requiring dilation being the most common complication in about 40% of patients. Anastomotic leak is also a frequent complication found in about 18% of patients and recurrent fistula was seen in about 5% of patients (22). We hypothesize that the remaining progenitors at the anastomosis site continue to lack NOG expression, making them prone to become respiratory epithelium rather than esophagus epithelium. Our organoid unit culture from patients with EA/TEF in the presence of NOG grew 4–10 times larger and healthier, suggesting that the epithelium at the anastomosis site might also grow healthier and stronger if we could supply NOG at the time of surgical repair in the form of a slow release depot that would limit NOG exposure locally to esophagus mucosa. A healthier esophageal epithelium at the site of surgical anastomosis may prevent strictures and leaks and reduce morbidity after repair.
Methods
Table 1 contains a list of reagents, commercial assays, cell lines, experimental models with the appropriate source and identifier.
Table 1. List of reagents.
Human esophageal tissue.
Human control esophagus specimens were obtained from pediatric organ donors approved and supervised by Tennessee Donor Services. Esophageal tissue from EA/TEF patients was collected at the time of surgical repair. Control subjects included 4 females (ages 0 days [38 weeks gestation] and 5, 6, and 8 years) and 2 males (ages 14 months and 12 years). TEF subjects included 4 females (ages 2 and 3 days) and 3 males (ages 3 months and 2 and 3 days).
Animals.
Animals were maintained in a temperature-regulated environment on a 12-hour light dark cycle and with ad libitum chow. Wild-type C57BL/6 mice obtained from The Jackson Laboratory and pups from P1–P3 were used to obtain trachea or esophagus for cultures.
RNA studies.
We acquired surgical pathology samples from 14 TEF patients and 4 controls. The control subjects were day of birth 23 weeks gestation, day of birth 36 weeks gestation, 8 months old, and 2 years old. The tissue from TEF patients was obtained at the time of EA/TEF surgical repair (proximal esophagus, distal esophagus, and fistulae when present) to study the expression of NOG and other molecules previously known to be important in esophageal development. RNA was isolated from paraffin-embedded surgical pathology samples obtained at the time of TEF repair, using the Qiagen RNeasy FFPE kit (catalog 73504). cDNA library was made using the Invitrogen Superscript II kit (catalog 18064-014). NOG, BMP4, BMP7, BMP2, SHH, GLI2, GLI3, TTF1, CHRD, SMAD1, SMAD5, BMPR1A, CER1, SOX2, WNT2, and BARX1 primers were obtained from MilliporeSigma (KiCqStart Primers).
C2C12 cell culture.
Mouse C2C12 immortalized myoblast cells and Dulbecco’s modified Eagle’s medium (DMEM) were purchased from ATCC. Non–heat inactivated fetal bovine serum (FBS) was purchased from HyClone Laboratories, Inc. The C2C12 cells at passages 5–7 were subcultured in T-75 cm2 flasks in DMEM supplemented with 10% FBS at 37°C in 5% CO2 with humidification. When the flasks reached 70%–80% confluence, the cells were trypsinized and seeded in triplicate at 50,000 cells/well in a 12-well plate for the dual-luciferase reporter assays.
NOG inhibitor.
Compound was custom synthesized (AA ChemBio) and was at least about 99% pure based on mass spectrometric analyses. N-50 is of the chemical formula 3-(3-chlorophenyl)-2-(pyridin-2-yl)acrylonitrile; N50-062 is 2-(4-chlorophenyl)-3-(2,5-dimethoxyphenyl)acrylonitrile. A brief compound synthesis is found in Supplemental Figure 1.
Protein iodination and biotinylation.
Recombinant BMP2 protein was iodinated by the chloramine-T method. Briefly, Na125I (0.5 mCi) and chloramine-T (0.1 mg) were added to 1–3 pg BMP2. After the reaction mixture was shaken gently for 1 minute, the reaction was quenched with 0.125 mg Na metabisulfite. The iodinated protein was then separated from free iodine by using a 5-mL Sephadex G50 (Pharmacia) column. The BMP2 protein was eluted using 50 mM Tris-CI-500 mM NaCl, pH 8.5, and the radioactivity was measured on a Beckman Gamma 4000 counter. Typically, the specific activities were 43.35 mCi/mg input protein. Biotinylation of NOG was performed following the manufacturer’s instructions (Thermo Fisher Scientific). Recombinant human BMP2 (rhBMP2) was obtained from Medtronic.
Alkaline phosphatase (ALP) assay.
The C2C12 cells were plated at 200,000 cells/well in 6-well plates and grown overnight in DMEM containing 10% FBS. On day 2, the culture medium was replaced with DMEM containing 2% FBS and the cells were treated with 0.5 μM or the indicated concentration of compound for 24 hours in 2 mL culture medium. On day 3, the cells were treated with a suboptimal final concentration of 50 ng/mL BMP2 with or without compound in DMEM medium containing 2% FBS for 72 hours. The cells were washed with phosphate-buffered saline (PBS) and lysed by the addition of lysis buffer (10 mM Tris-HCl pH 8.0, 1 mM MgCl2, and 0.5 % Triton X-100). The cell lysates were centrifuged for 5 minutes at 13,000g. The supernatant was removed, and the aliquots were assayed for ALP activity and protein amount. The ALP activity was measured in triplicate using an ALP assay kit (Sigma-Aldrich) in microtiter plates. The protein amount was determined with Bio-Rad protein assay reagent (Bio-Rad) using bovine serum albumin (BSA) as a standard. The ALP activity (nmol of p-nitrophenol per mL) was normalized to the protein amount (nmol of p-nitrophenol per μg).
In vitro osteoblast differentiation.
Mouse preosteoblastic MC3T3-E1 cells were purchased from ATCC and differentiated to osteoblasts in α-MEM (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals) 1% l-glutamine (Invitrogen) and 1% penicillin/streptomycin (Thermo Fisher Scientific) and 50μg l-ascorbate and 10 mM β-glycerophosphate. Medium was changed every 2–3 days and continued for 14 days for bone nodule mineralization.
Bone mineralization assay.
To quantify the calcium deposition (mineralization) of osteogenic samples, MC3T3 cells were harvested on day 14. After washing the samples twice with PBS and disrupting the monolayer with a cell scraper, calcium ions were dissolved from the extracellular matrix by shaking in 500 μL/well 0.5 N HCl at 4°C for 4 hours. Calcium content was determined in technical triplicates with the QuantiChrom calcium assay kit (BioAssay Systems) according to the manufacturer’s protocol. Protein content was measured in technical triplicates using Roti-Quant (Carl Roth) according to the manufacturer’s instructions. Calcium concentrations were normalized to the protein content.
Assay for NOG binding using BMP2.
Radioiodinated BMP2 (20,000 cpm) was incubated at room temperature for 30 minutes with or without various concentrations of unlabeled BMP2, with or without various concentrations of compound, in 0.1 mL PBS containing 0.1% BSA. Biotinylated NOG was added in all conditions for pull-down assays. The biotinylated NOG was pulled down with 50 μL of 50% slurry of neutravidin-agarose. The samples were then centrifuged for 5 minutes at 13,000g, and the unbound BMP2 was removed by aspiration. The pellet containing neutravidin-agarose and the bound BMP-NOG complex was washed twice with 0.5 mL PBS containing 0.1% BSA. Bound radioactivity was then counted in a Beckman Gamma 4000 detector. Homologous competition experiments were performed as described above with various amounts of unlabeled BMP2 (0.01–1000 nM) added.
rhBMP2 was obtained from Medtronic. Since we had access to milligram quantities of purified protein, we used the protein as a representative of BMP family for all in vitro and in vivo screening for binding and activity evaluations. Noggin inhibitors are available upon request in a small quantity (few milligrams).
Dual-luciferase reporter assay.
The BMP-specific Smad1-driven 9 × GCCG (a consensus binding sequence for Smad1) reporter plasmid was provided by Kohei Miyazono (Tokyo University, Tokyo, Japan). The C2C12 cells were trypsinized and seeded in triplicate wells at 50,000 cells/well in 12-well plates on day 1. On day 2, the cells were cotransfected with the 9 × GCCG reporter construct and the Renilla luciferase control vector using SuperFect (Qiagen) for 24 hours. A total of 1 μg plasmids was used for cotransfection in each well and the concentration of Renilla luciferase vector was one-fifteenth of the 9 × GCCG-reporter plasmid. On day 3, medium was replaced with DMEM containing 2% FBS and the cells were treated with various concentrations of compound. On day 4, the cells were treated with BMP2. On day 5, the luciferase activities were measured in 20 μL cell lysate using the dual luciferase assay system (Promega) with a luminometer (LumiCount, Packard Bioscience) following the manufacturer’s instructions. The luciferase activity was expressed as relative units of luciferase (RUL, a ratio of firefly luciferase to Renilla luciferase activity).
Organoid unit culture.
Fresh esophagus and trachea from mice P1–P3 were used to create mEOUs, which simulate esophageal development in vitro, using a protocol previously published by the Grikscheit lab (14, 15, 23).
Recombinant murine NOG and human NOG were purchased from Peprotech (catalog 250-38 and 120-10C) and used at 1 ng/mL. P1–P3 neonatal mice (female and male) were euthanized by exposure to carbon dioxide (CO2) in an inhalation chamber, then decapitated. The mouse esophagi were harvested. Human surgical samples were obtained as discarded tissue collected at the time of TEF surgical correction.
All esophageal tissue was washed 3–4 times with 4°C Hank’s balanced salt solution (Invitrogen) on ice, sedimenting between washes, to remove debris and mucous. Each lavaged esophagus was minced into pieces smaller than 1 mm3 with sterile scissors. Tissue fragments were digested with 0.12 mg/mL dispase (Invitrogen) and 800 U/mL collagenase type 1 (Worthington) at 37°C on an orbital shaker for 20 minutes. Digestion was stopped with 4°C 4% sorbitol (Sigma-Aldrich), 10% FBS (Invitrogen) in high glucose DMEM (Invitrogen). After centrifugation at 39g for 10 minutes, the supernatant was discarded, the pellet was resuspended in 4°C 10% FBS in DMEM, and then centrifuged at 100g for 5 minutes. The resulting pellet contained the isolated OU, heterogeneous multicellular clusters of epithelium and mesenchyme.
The pellet was resuspended in growth factor–reduced Matrigel (BD Biosciences). A quantity of 100 μL of the mixture was put into one well of a 24-well plate and incubated at 37°C for 30 minutes until the Matrigel solidified. A quantity of 2 mL DMEM, 10% FBS with Mycozap (Lonza) was added in each well and incubated at 37°C throughout the culture. Media was changed every 3 days. No additional growth factors were added to the media, except in the experimental samples where NOG or NOG inhibitor was added as described. NOG inhibitor was used at a concentration of 10 μM and NOG at 100 ng/mL in all experiments.
The protocol for isolation of hEOUs from TEF and controls was slightly modified since some of the samples took more than 24 hrs to transport to our lab and place in culture. The tissue was placed in media and minced to pieces smaller than 1 mm3 at the time of isolation. No further digestion was performed if the tissue was out of the body for more than 12 hours.
Protein immunofluorescence.
Paraffin-embedded tissue was collected from autopsies of neonates that died from causes other than defects in the foregut to examine the expression of proteins important in esophageal development in the normal human esophagus and trachea. The samples were collected from neonates at gestational ages ranging from 23–39 weeks and days 0–3 of life. Paraffin-embedded tissue from surgical pathology collected at the time of EA/TEF repair was used in a similar manner to investigate the same proteins as previously described.
Organoid unit sections were fixed in 4% PFA for 1–2 hours, washed in PBS for 5 minutes, and mounted in HistoGel (Richard-Allan Scientific), and paraffin blocks and sections were made.
Sections were deparaffinized in Histoclear (Electron Microscopy Science) and rehydrated in a series of ethanol washes. Then, antigen retrieval was performed using target retrieval solution, pH 6 (Dako), using a pressure cooker. After incubating paraffin sections in serum-free protein block solution (Dako) at room temperature for 1.5 hours, primary antibodies diluted in antibody diluent with background reducing components (Dako) were incubated at 4°C overnight.
Cy2-conjugated AffiniPure donkey anti-rabbit and Cy3-conjugated AffiniPure donkey anti-mouse (catalog 711-225-152 and 715-166-151 respectively; Jackson Immuno Research Laboratories) were used as secondary antibodies. DNA was visualized by staining with DAPI.
Primary antibodies used for detection of proteins investigated using immunocytochemistry were as follows: neuron-specific B-III tubulin (1:500; R&D Systems, catalog MAB1195), SCGB1A1 (Clara cells 10KD CC10) (1:2000; MilliporeSigma, catalog HPA031828), Involucrin (1:200; Abcam 53112), cytokeratin 13 (1:50; Abcam 90096), PAX9 (1:500; Abcam 28538), cytokeratin 8 (1:200; MilliporeSigma, catalog HPA049866), chordin (1:500; MilliporeSigma, catalog HPA035827), TFF3 (1:1000; MilliporeSigma, catalog HPA035464), NOG (1:200; MilliporeSigma, catalog N4538), BMP4 (1:200; MilliporeSigma, catalog SAB5300528), BMP7 (1:200; MilliporeSigma, catalog WH0000655M1), BMP2 (1:200; MilliporeSigma, catalog SAB2108413), SHH (1:200; Millipore, catalog MABD175), TTF1 (NKX2-1; 1:200; Millipore, catalog MAB5460), WNT2B (1:500; Thermo Fisher Scientific, catalog 710888), Ki67 (1:500; Cell Signaling, catalog 12202), SMAD 1/5/9 (1:1000; Abcam 66737), pSMAD 1(Ser463/465)/5((Ser463/465)/9(Ser 465/467) (1:1000; Cell Signaling, catalog 13820).
Western blots.
Organoid cultures were disrupted in lysis buffer (1% CHAPS, 0.5 mM EDTA, 20 mM magnesium acetate, 30 mM Tris, pH 7.5, 150 mM NaCl) supplemented with protease (P8340) and phosphatase (P0044, P5726) inhibitors for 10 minutes on ice. For all samples, protein concentrations were measured by DirectDetect (EMD Millipore), and 20 μg protein was loaded onto a 10% Laemmli polyacrylamide gel. The proteins were transferred onto Odyssey nitrocellulose membranes (LI-COR). Blots were blocked for 1 hour in Odyssey Tris-buffered saline (TBS) Blocking Buffer (LI-COR), and probed with primary antibodies for 18 hour in 0.2% Tween-20/Odyssey TBS Blocking Buffer at 4°C. Blots were washed in TBS/0.05% Tween (TBS-T), followed by a 1-hour incubation with secondary antibodies labeled with 680- or 800-nm fluorescent dyes (LI-COR) and diluted as the primary antibody. Blots were washed 3 times in TBS-T and fluorescence was detected using the Odyssey Fc (LI-COR). The resulting images were opened in ImageJ and the area under the peak was calculated. The densities were then adjusted to the control protein (VDAC) and statistical significance was determined by an unpaired 2-way ANOVA using Prism Graphpad Software. A P value less than 0.05 was considered significant.
Primary antibodies used were anti-SMAD 1/5/9 (1:1000; Abcam 66737), anti-pSMAD 1(Ser463/465)/5((Ser463/465)/9(Ser 465/467) (1:1000; Cell Signaling, catalog 13820), anti-pSMAD 2(S465/467)/3(S423/425) (1:1000; Cell Signaling, catalog 8828S), and anti-SMAD2 (1:1000; Cell Signaling, catalog L16D3).
DNA methylation studies.
NOG DNA sequence was retrieved from the UCSC genome browser database. DNA was extracted from paraffin blocks using the QIAamp DNA FFPE tissue kit (catalog 56404). Bisulfite modification of DNA was performed using EpiTect bisulfite kit from Qiagen (catalog 59104) and amplified by PCR using the following bisulfite sequencing–specific primers:NOG exon forward GYGGAGGGAAGTGTTTTTAGAATTAG,reverse TCCTCTCCCRAATCTACTAAAAAAAAC; NOG promoter forward TGGTGGGGGAGAGAGAAAATTG,reverse ACAACRCCCAAAACTATACCC;NOG upstream sequence forward TTAGTGYGGAGTTAGATGGGG,reverse CRAAATTTAAAACCAAAAAAAAAAAAAAACTTCAAC. Primer sequences were provided by Zymo Research.
The amplified DNA fragment was subcloned into Invitrogen Topo TA 2.1 vector (Invitrogen, catalog K450002). Clones were screened and plasmid DNA was sequenced by Genewiz. Bisulfite sequencing data were analyzed using BiQ Analyzer (Max Planck Institute for Informatics and Saarlan University).
Statistics.
Fluorescently immunostained tissue slides were imaged on a Nikon A1R confocal microscope (Vanderbilt Epithelial Biology Center). Tissue sections were imaged at ×20 magnification to a resolution of 0.323 μm/pixel. Images were analyzed using the Image J analysis program. To quantify fluorescence intensity, we measured total positive pixels on the entire slide and divided by total area of tissue scanned. Mouse experiments were done using 2–3 litters per experiment (control and treated) for a total of 3 different experiments. Human organoid experiments contained 6 pediatric control esophagi and 7 EA/TEF samples. Experimental groups contained 4–7 organoid units per condition. All graphs and statistics were completed in Prism GraphPad, using a Mann-Whitney nonparametric test if comparing 2 groups or with nonparametric analysis with Kruskal-Wallis test followed by Dunn’s test for post hoc analysis of group significance. Statistical analysis in methylation studies was done by counting the number of methylated CpG islands divided by the total number of CpG islands present in the region studied. Six to 12 clones were sequenced per patient. A total of 4 control and 16 patients were studied. All graphs and statistics were completed in GraphPad Prism. A P value less than 0.05 was considered significant.
Study approval.
IRB approval was obtained for use of human tissues (Vanderbilt University IRB 161525, University of Iowa Health Care IRB 201712783), which were collected with written informed consent. All experimental animal protocols were in accordance with and approved by the Institutional Animal Care and Use Committee (M1600124-01).
Author contributions
CPG, EYP, and JRG conceptualized the study. CPG and SS devised the methodology. CPG, WJH, KMR, and EM performed study investigation. CPG and EYP wrote the original draft of the manuscript. JRG and JSS reviewed and edited the manuscript. CPG, EYP, JRG, and SS acquired funding for the study. CPG, SS, WJH, JSS, and MSA contributed resources. JRG supervised the study.
Supplementary Material
Acknowledgments
These studies were supported by a Department of Veterans Affairs Merit Review Award (I01 BX000930), DOD grant CA160479, and NIH grant R01 DK071590 (to JRG). CPG was supported by The Helen and Nicholas Abumrad Research Fund, NIH grant T32 CA106183, and a grant from the Vanderbilt Institute for Clinical and Translational Research (VR15139.1). This work was supported by core resources of the Vanderbilt Digestive Disease Center (P30 DK058404) and the Vanderbilt-Ingram Cancer Center (P30 CA68485), and imaging in the Vanderbilt Digital Histology Shared Resource was supported by a VA Shared Instrumentation grant (1IS1 BX003097). The design and optimization of NOG inhibitor compounds were performed at the Atlanta Veterans Affairs Medical Center and partly supported by NIH grant R21TR001751 and an Emory Orthopaedics Seed Grant (to SS). We thank Emily Hodges for guidance in performing studies on DNA methylation. We also thank Scott D. Boden for his valuable suggestions in biooptimization of NOG inhibitors.
Version 1. 05/19/2020
In-Press Preview
Version 2. 07/20/2020
Electronic publication
Version 3. 08/03/2020
Print issue publication
Funding Statement
R21TR001751
Footnotes
Conflict of interest: SDB and SS are inventors on a patent held by Emory University on N-50. SDB is a consultant for Seaspine Inc.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(8):4396–4410.https://doi.org/10.1172/JCI123597.
Contributor Information
Carolina Pinzon-Guzman, Email: carolina.pinzon-guzman@vumc.org.
Sreedhara Sangadala, Email: ssangad@emory.edu.
Katherine M. Riera, Email: katherine.m.riera@vumc.org.
Evgenya Y. Popova, Email: eyp1@psu.edu.
Elizabeth Manning, Email: elizabeth.manning@vumc.org.
Won Jae Huh, Email: won.jae.huh@vanderbilt.edu.
Matthew S. Alexander, Email: matthew-alexander@uiowa.edu.
Julia S. Shelton, Email: julia-shelton@uiowa.edu.
Scott D. Boden, Email: sboden@emory.edu.
James R. Goldenring, Email: jim.goldenring@vanderbilt.edu.
References
- 1.Nassar N, et al. Prevalence of esophageal atresia among 18 international birth defects surveillance programs. Birth Defects Res Part A Clin Mol Teratol. 2012;94(11):893–899. doi: 10.1002/bdra.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen RN, Calzolari E, Husby S, Garne E, EUROCAT Working group Oesophageal atresia: prevalence, prenatal diagnosis and associated anomalies in 23 European regions. Arch Dis Child. 2012;97(3):227–232. doi: 10.1136/archdischild-2011-300597. [DOI] [PubMed] [Google Scholar]
- 3.Que J, et al. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134(13):2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(-/-) mouse embryos. Dev Biol. 1999;209(1):60–71. doi: 10.1006/dbio.1999.9234. [DOI] [PubMed] [Google Scholar]
- 5.Litingtung Y, Lei L, Westphal H, Chiang C. Sonic hedgehog is essential to foregut development. Nat Genet. 1998;20(1):58–61. doi: 10.1038/1717. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Litingtung Y, Ten Dijke P, Chiang C. Aberrant Bmp signaling and notochord delamination in the pathogenesis of esophageal atresia. Dev Dyn. 2007;236(3):746–754. doi: 10.1002/dvdy.21075. [DOI] [PubMed] [Google Scholar]
- 7.Arsić D, Keenan J, Quan QB, Beasley S. Differences in the levels of Sonic hedgehog protein during early foregut development caused by exposure to Adriamycin give clues to the role of the Shh gene in oesophageal atresia. Pediatr Surg Int. 2003;19(6):463–466. doi: 10.1007/s00383-003-0959-8. [DOI] [PubMed] [Google Scholar]
- 8.Murphy AJ, Li Y, Pietsch JB, Chiang C, Lovvorn HN. Mutational analysis of NOG in esophageal atresia and tracheoesophageal fistula patients. Pediatr Surg Int. 2012;28(4):335–340. doi: 10.1007/s00383-011-3022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reutter H, Ludwig M. VATER/VACTERL association: evidence for the role of genetic factors. Mol Syndromol. 2013;4(1-2):16–19. doi: 10.1159/000345300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez P, Da Silva S, Oxburgh L, Wang F, Hogan BL, Que J. BMP signaling in the development of the mouse esophagus and forestomach. Development. 2010;137(24):4171–4176. doi: 10.1242/dev.056077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Que J, Choi M, Ziel JW, Klingensmith J, Hogan BL. Morphogenesis of the trachea and esophagus: current players and new roles for noggin and Bmps. Differentiation. 2006;74(7):422–437. doi: 10.1111/j.1432-0436.2006.00096.x. [DOI] [PubMed] [Google Scholar]
- 12.Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19(23):2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 13.Rosekrans SL, Baan B, Muncan V, van den Brink GR. Esophageal development and epithelial homeostasis. Am J Physiol Gastrointest Liver Physiol. 2015;309(4):G216–G228. doi: 10.1152/ajpgi.00088.2015. [DOI] [PubMed] [Google Scholar]
- 14.Trecartin A, et al. Establishing proximal and distal regional identities in murine and human tissue-engineered lung and trachea. Tissue Eng Part C Methods. 2016;22(11):1049–1057. doi: 10.1089/ten.tec.2016.0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finkbeiner SR, et al. Generation of tissue-engineered small intestine using embryonic stem cell-derived human intestinal organoids. Biol Open. 2015;4(11):1462–1472. doi: 10.1242/bio.013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spurrier RG, Speer AL, Hou X, El-Nachef WN, Grikscheit TC. Murine and human tissue-engineered esophagus form from sufficient stem/progenitor cells and do not require microdesigned biomaterials. Tissue Eng Part A. 2015;21(5-6):906–915. doi: 10.1089/ten.tea.2014.0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmed S, Metpally RP, Sangadala S, Reddy BV. Virtual screening and selection of drug-like compounds to block noggin interaction with bone morphogenetic proteins. J Mol Graph Model. 2010;28(7):670–682. doi: 10.1016/j.jmgm.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28(8):817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balemans W, Van Hul W. Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev Biol. 2002;250(2):231–250. doi: 10.1006/dbio.2002.0779. [DOI] [PubMed] [Google Scholar]
- 21.Nasr T, et al. Endosome-mediated epithelial remodeling downstream of Hedgehog-Gli is required for tracheoesophageal separation. Dev Cell. 2019;51(6):665–674.e6. doi: 10.1016/j.devcel.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. doi: 10.1016/j.jpedsurg.2017.05.024. Lal DR, et al. Challenging surgical dogma in the management of proximal esophageal atresia with distal tracheoesophageal fistula: Outcomes from the Midwest Pediatric Surgery Consortium. J Ped Surgery . [DOI] [PubMed]
- 23.Gregory S, et al. Murine and human tissue-engineered esophagus form from sufficient stem/progenitor cells and do not require microdesigned biomaterials. Tissue Eng Part A. 2015;21(5-6):906–915. doi: 10.1089/ten.tea.2014.0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.