Abstract
The field of human microbiome research has revealed the intimate co-association of humans with diverse communities of microbes in various habitats in the human body, and the necessity of these microbes for the maintenance of human health. Microbial heterogeneity between humans and across spatial and temporal gradients requires multidimensional datasets and a unifying set of theories and statistical tools to analyze the human microbiome and fully realize the potential of this field. Here we consider the utility of community ecology as a framework for the interrogation and interpretation of the human microbiome.
In their visionary perspective from 2016 (ref.1), Byrd and Segre highlighted the need to incorporate microbial communities into a contemporary understanding of human disease. With advances in our understanding of human microbial community dynamics across temporal and spatial gradients, it has become imperative that we embrace a conceptual framework in which findings in the field can be interrogated and interpreted2. Community ecology offers such a framework. Originally developed to understand complex interactions between macro-organism species across space and time, there is increasing interest in the application of existing ecological theory and statistics to the study of human-associated microbial communities. Indeed, the study of the human microbiome, a term coined in 2001 (ref.3), is synonymous with microbial ecology.
Community ecology theory and the statistical tools that support the testing of hypotheses in complex multidimensional datasets offer an ideal framework for interrogation of microbial ecosystem properties. It is impracticable to elucidate the mechanisms underlying microbiome associations with disease without a fundamental comprehension of how microbes behave and interact with each other and their human hosts over spatial and temporal gradients and in response to environmental exposures.
In this Perspective, we introduce a number of ecological terms and discuss key community ecology theories focusing on assembly, succession, disturbance and restoration (Fig. 1) because of their emerging importance to human health. We present a number of recent human microbiome studies indicating that ecological theory offers a feasible framework for interrogation and interpretation of findings in this field.
Fig. 1 |. Microbial ecological concepts.
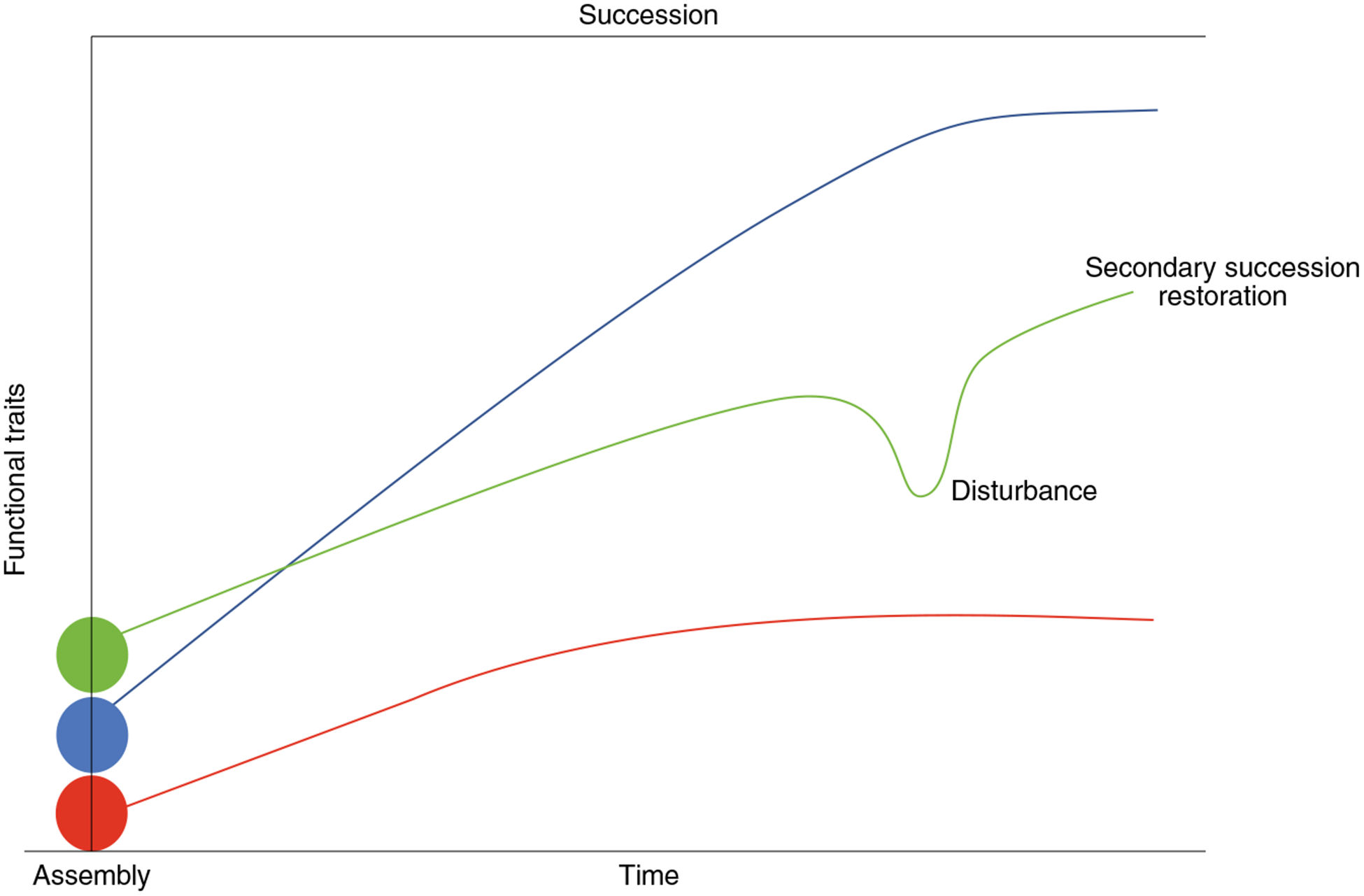
Ecological concepts of initial community assembly (processes that influence community membership; filled dots), succession (accumulation of species and their functional traits into an ecosystem; colored lines indicate distinct successional trajectories), disturbance (perturbation to ecosystem; green line) and restoration (reassemby of the ecosystem following perturbation; green line).
Measuring the microbiome
The multidimensional datasets generated in microbiome studies are difficult to condense into a thoroughly representative discrete or continuous measurement, as is commonly encountered in univariate analysis. Unfortunately, when researchers are challenged to integrate microbiome data into epidemiological analyses, ecological measures that fail to capture the dimensionality and complexity of microbiome data are commonly employed. For example, alpha diversity (Box 1), while providing a simple metric for assessment of the microbiome, is an incomplete snapshot of the community that is based on a number of discrete, random samples of the microbes present. This snapshot is not comprehensive since current sequencing technology, even with extensive sequencing depth4, only captures a subset of the community. To ameliorate this problem, multiple microbiome datasets must be normalized before comparison to avoid misinterpretation and to reduce the false discovery of compositional units, which are the basic elements of composition used in a database, that are differentially abundant between the components of analysis (a problem that is also associated with both amplicon and metagenomic sequencing data)5.
Box 1 |. definitions and concepts.
Alpha diversity.
A within-sample summary statistic that reduces the complexity of a multi-species community to a single integer that describes how many taxonomic units are present (richness), how they are distributed (evenness) and, in some cases, their phylogenetic relationships (Faith’s phylogenetic diversity).
Community assembly.
The process of developing a mixed-species community of organisms.
Beta diversity.
A between-sample measure of ecosystem dissimilarity.
Composition.
The number of taxonomic or functional units — e.g., taxa, genes, genomes, et cetera — within a community.
Disturbance.
Perturbation of ecosystem composition, functional traits or activities.
Microbiome.
The entire microbial ecosystem within a host, including the microorganisms (bacteria, archaea, single-celled eukaryotes and viruses), their genomes and surrounding environmental conditions.
Microbiota.
The assortment of microbial taxonomic units within a habitat.
Steady-state ecosystem.
The balance between the forces that act to change the composition or function of the ecosystem and those that act to maintain status quo.
Keystone species.
Organisms that when removed from an ecosystem result in a significant change in the ‘state’, often resulting in a breakdown of the steady state.
Pioneer species.
Organisms that initially populate a nascent ecosystem and impact habitat conditions.
Resistance.
Capacity of the ecosystem to withstand change.
Resilience.
Capacity of the ecosystem to maintain or return to a steady state in the presence of or following perturbation.
Restoration.
Process of ecosystem reestablishment.
Succession.
Accumulation of species into an ecosystem. Primary succession describes species accumulation into a nascent ecosystem. Secondary succession describes re-accumulation of species following disturbance of an established ecosystem.
Alpha diversity, seemingly a simple metric, requires significant interpretation. Measurements of microbiome diversity must take into consideration whether the community being observed is in a steady state (homeostasis) or a disturbed state (dysbiosis), and both these states can be defined on the basis of interactions between microbes and/or between the microbes and the host. For clinical investigation, dysbiosis or perturbation can be defined as a disturbance of the underlying molecular relationships between the host and the microbiome. To measure these dynamics, it is necessary to determine the temporal frequency of sampling required to capture them, and hence the periodicity of events that can influence microbiome dynamics is important.
Community assembly and succession
The integration of large and complementary multidimensional human microbiome datasets that will permit high-resolution phylogenetic and functional assessment of microbial communities remains a challenge for the field. Early evidence suggests that ecological concepts are borne out in human-associated microbiomes. For example, community assembly, the processes that shape the membership of ecological communities, is influenced in part by prevailing ecosystem conditions. For example, the human host influences which microbes are present and their activity in distinct compartments of the healthy gastrointestinal tract via gastric acid, pancreatic and biliary secretions, which modulate transit time of microbes through the gut, and immune activity — for example, through the production of inflammatory cytokines, antimicrobial peptides and IgA, which selectively binds specific microbes6,7. Beyond host factors that govern community assembly, microbial factors, such as the production of antimicrobials8 and bacteriocins9, quorum sensing (intra- and interspecies cell density–dependent biomolecular communication) that regulates phenotypic traits of a microbial population10 and substrate utilization11 also shape microbial communities, though this area of research remains relatively unexplored in human microbiomes.
As the field develops, it is becoming increasingly clear that microbial networks of species and/or metabolic products exist12 and that microbiomes en masse influence host cell gene expression, producing microbiome-to-host feedback loops and selective pressures that dictate microbial and host cellular productivity and thus community assembly. Exploiting next-generation microbiome data to identify factors that influence microbial assembly and specific programs of molecular productivity thus represents a fruitful avenue of investigation and is likely to lead to novel therapeutic strategies to effectively manipulate host microbiomes.
Multiple studies have now demonstrated that distinct early-life microbial assemblages exist in the human gut13–15 and airways16 that relate to subsequent health and disease outcomes in childhood. How early-life microbiome disturbances relate to health outcomes years later in childhood may be explained in part by ecological succession. Succession refers to the temporal change in species or functional trait accumulation that occurs in an ecosystem over time. On a macroecological scale, pioneer species (Box 1) can influence successional trajectories; even slight differences in pioneer colonizer communities may alter successional trajectories, particularly in environments where interspecies competition is likely to be important17. If successional theory holds for development of the human microbiome, one would predict that pioneer microbial species that are present in early life, particularly those that strongly influence ecosystem conditions such as immune tolerance or inflammation and thus community assembly, would impact the types of microbes and their functional traits that subsequently accumulate in the niche over time.
Evidence for microbial succession exists in the human gastrointestinal tract14,18 and at other habitats in the body16. There is also evidence that distinct early-life microbial assemblages are associated with different microbial successional trajectories in humans. For example, the gut microbiome of four-day-old formula-fed neonates delivered via caesarian section (note that diet and delivery method are known to influence gut microbiome assembly) is distinct from that of vaginally delivered breast-fed or vaginally delivered formula-fed neonates and is associated with microbiological differences that persist in longitudinally collected samples from the infants up to 12 months of age18. Similarly, newborns at high risk for asthma exhibit a distinct meconium microbiota and a delayed trajectory of gut microbiota diversification over the first year of life compared with babies at low risk for asthma14. Thus, understanding the early-life microbial species that are associated with health outcomes in later life and the factors that support their assembly may offer novel strategies to promote ecosystem conditions that facilitate accumulation of exogenous symbiotic microbes (i.e., those that form mutually beneficial partnerships with the host) to prevent disease development.
Disturbance and dynamics
Microbiome perturbations are a consistent feature of human diseases. Disturbance is a well-studied ecological phenomenon that typically produces four outcomes: resistance to disturbance (no change in composition or function of the ecosystem); resilience (communities that initially respond to disturbance but subsequently return to the preperturbation state); functional redundancy (communities that change species composition but retain functional traits); or loss of composition or functional traits (ecosystem state change).
A particularly notable example of microbiome response to disturbance demonstrated the importance of stable pathogenic microbiome states as mechanistic contributors to post-dieting weight gain19. In a mouse model of obesity, an obese-associated gut microbial signature persisted despite caloric restriction, and these persistent microbes contributed to weight gain upon re-exposure of the mice to an obesogenic diet. Ecologically, this could be viewed as resilience of the obesogenic microbiome, and one would therefore predict that a more substantial force must be applied to the stable obesogenic microbiome to shift the microbial community toward a nonobesogenic stable state. The study demonstrated that this could be achieved via administration of flavonoids, which diminish the metabolic abnormalities associated with the obesogenic stable microbial state. The authors posit that the observed species persistence may be an ecological adaptation associated with complex microbe–host interactions that evolved to prevent wild swings in microbial metabolic activity as a result of transient shifts in nutritional availability. It is also feasible that such evolutionary adaptations support microbial stability in multiple human microbial ecosystems as a means to prevent the loss of species that are important for the success of the human superorganism.
To better understand human–microbial metabolic interdependencies, it is essential to appreciate microbial–host codependence. It is, in our opinion, highly likely that coevolution has shaped both the human immune system and the microbiome interdependently and, in the case of the gut microbiome, this coevolution has occurred in the context of historically available nutritional substrates. However, the eco-evolutionary relationship is not as clear-cut as one might expect, and microbiome–host symbiosis should be viewed as reciprocity between microbial competition and host control20. Microbial traits, including metabolic functions, that provide a substantial advantage in terms of host survival would presumably be retained and transmitted vertically and/or be horizontally disseminated across a population. For example, many obligate anaerobes that ferment fiber and synthesize short-chain fatty acids produce an excess of these fatty acids that are sensed and used by human cells, which in turn regulate host metabolism and inflammation. Indeed, fermentative microbes are conserved across healthy human gut microbiomes, and their loss, attributed in part to microbiome disturbances induced by lifestyle changes, is associated with a range of chronic inflammatory diseases21.
Evidence for preserved relationships between the microbiome and host health suggest that conserved ecological states (that is, the microbiome composition in the host) may exist across diverse peoples. Stable, resistant or resilient microbial states, irrespective of whether they are associated with healthy or unhealthy status, typically have keystone community members22, which may be proportionally abundant or rare, but always fulfill an important functional role in ecosystem stability and therefore underpin conservation of ecological states across populations. In a recent study of more than a thousand adults with diverse genetic histories, the authors demonstrated that the inclusion of microbiome data alongside host genetic data significantly improved their capacity to predict host traits, including body mass index and high-density lipoprotein cholesterol and fasting glucose levels, in comparison to using human genome data alone23. However, the intraindividual microbial variability observed over time suggests that to truly understand these states it will be necessary to capture the longitudinal dynamics of the microbiome within individuals across human populations. Shortterm fluctuations in the gut microbiome may have profound implications for disease states by producing temporary changes or loss of function, despite community resilience. For example, patients with inflammatory bowel disease can exhibit substantially greater temporal variability in their microbiome than healthy controls, and these short-term fluctuations are associated with shifts in calprotectin levels (an indicator of inflammation) and medication used to treat disease flares24.
Restoration
Ecosystem restoration in part encompasses strategies to re-establish microbial interactions that support community assembly or reassembly and reinstate microbial functional networks lost through disturbance. From a human health perspective, restoration of a disturbed microbial ecosystem offers a novel approach to the management or prevention of disease. However, there is a fundamental need to understand how human microbial ecosystems (re)assemble over time, and the factors that influence this process, to facilitate rational design of such interventions.
Human birth cohort studies offer the opportunity to identify factors that govern early-life human microbiome assembly and succession. For example, recent studies have identified distinct microbiota assemblages in very early life that relate to increased risk of disease outcomes in childhood and the host and environmental factors related to the presence of these community assemblages13,14. Though nascent, these studies provide insights into modifiable factors that influence the human microbial ecosystem, which may be incorporated into rationally designed microbial interventions to treat or prevent disease.
At present, microbiome manipulation studies in humans are largely confined to FMT and prebiotic (nutrients designed to stimulate the growth of beneficial microbes), probiotic (microbes that confer a health benefit when consumed at sufficient levels) or synbiotic (a combination of a prebiotic and a probiotic) supplementation. Successes include use of Lactobacillus delbrueckii to treat bacterial vaginosis (more effective than antimicrobials25) and FMT for Clostridium difficile infection (CDI)26,27. Leveraging metagenomic and other datasets that capture microbial genomic content and functionally relevant features of such successful interventions across human populations will ultimately permit development of precision therapeutic manipulation of the microbiome.
Probiotic administration is an attractive approach to restore depleted microbial functions and confer a host health benefit. In a study of 4,556 infants from rural India, treatment with a synbiotic composed of both Lactobacillus plantarum and a fructooligosaccharide was highly effective in reducing the instance of sepsis in this vulnerable population28. While the mechanism of action for its treatment is poorly understood, the synbiotic presumably effectively supports the presence and activities of microbes that protect against pathogenic microbial activity.
However, the pre-existing endogenous microbiome also influences the colonization success of exogenous species — for example, oral administration and engraftment of the probiotic Bifidobacterium longum is dependent on the endogenous microbiome of each human participant. This species exhibited persistence for at least 6 months in only 30% of participants who consumed it29. This suggests that microbial interactions in the gut, and presumably in other body habitats, play a significant role in determining colonization success and thus the efficacy and longevity of such microbial interventions, a critical consideration for microbial therapeutic interventions.
FMT represents an example of ecosystem restoration in which a disturbed microbiome is replaced or replenished with microbial species and functional genes that reduce disease symptoms and risk of relapse. The human microbiome provides colonization and proliferation resistance against pathogens; disturbance of this function through selective loss of protective organisms via perturbing influences, such as diet or antibiotic use, creates an opportunity for CDI30. Secondary succession of the ecosystem, i.e., the process of re-developing the microbial community, may commence following cessation of antimicrobial administration. However, recovery can take several weeks, and, like primary succession, the trajectory of reassembly is governed both by the endogenous microbial species present and the prevailing ecosystem conditions. This was recently demonstrated in a group of healthy human adults who received an antimicrobial treatment followed either by a single auto-FMT (that is, transplant of participants feces collected before antimicrobial administration), 4 weeks of probiotic supplementation or no post-antimicrobial intervention (spontaneous reconstitution). Microbiome recovery was fastest in the auto-FMT group, slower in the spontaneous reconstitution group (taking up to 12 weeks to restore) and impeded by sustained probiotic supplementation31,32.
Administration of a ‘healthy’ microbiome via FMT to patients with CDI restores microbial bile acid metabolism that promotes colonization resistance and protects against C. difficile spore germination33. To model the impact of a common probiotic on bile acid metabolism, in one study, a model microbial ecosystem was created from Clostridium scindens, Collinsella aerofaciens and Blautia obeum and grown under conditions of varying combinations of bile acids and in the presence or absence of the probiotic Lactobacillus acidophilus. Using this approach, novel bile acid metabolism pathways, including those that interacted with the probiotic, were identified. This noncomputational modeling environment provides an example of how to determine the underlying bacterial metabolic pathways that are important for host–microbe interactions, which can then be used to inform computational models to design novel synbiotic interventions for specific diseases.
While FMT has been successful in the treatment of CDI and in a pilot trial of children with autism spectrum disorder34, there are several examples in which FMT had less success, especially in chronic inflammatory diseases (such as inflammatory bowel disease35). This is likely due to a failure to fully understand ecosystem conditions conducive to successful FMT. In particular, an understanding of the rules of microbial engagement and competitive exclusion that influence the ability of exogenous microbial species to form interactive and productive microbial networks that successfully colonize and beneficially modulate immune and physiological features in the gut is lacking in the field.
Because of microbiome-associated ecosystem heterogeneity across patients with a similar disease or disorder, it will likely be necessary to develop distinct strategies for ecosystem (re)engineering to promote microbial restoration efficacy across all patients. For example, patients with chronic rhinosinusitis exhibit four distinct sinus mucosal microbial community structures, each of which induces a distinct inflammatory response36. It is unlikely that all four of these microbial assemblages would respond similarly to the same microbial intervention, but rather each pathogenic microbial assemblage may require a distinct strategy to restore appropriate mucosal microbial colonization and immune function. Microbial restoration within the gut will also require a similar approach and is complicated by the serial influx of nutritional substrates that shape microbial assembly and productivity in this niche37,38. This suggests that, as for other organ transplants, a matrix must be developed for therapeutic microbial restoration strategies, in which the pre-existing microbiome assemblages together with immune or physiological features of dysfunction and nutritional inputs are considered in the rational design of novel therapeutic interventions (microbial and immunological) for chronic diseases. Trials of such interventions would be perfectly suited to adaptive designs in order to accelerate the process of development of tailored microbial therapies for heterogeneous patient populations.
Statistical and modeling approaches
Computational modeling of multiscale (i.e., from the community to the molecular level) ecological dynamics of the human microbiome should be a research priority for a number of reasons. First, modeling these dynamics will enable us to design better interventions, as appropriate parameterized models should permit prediction of, for example, drug bioavailability or the effects of pharmaceuticals on microbial activities, and hence on host function and outcomes. This is self-evident given recent studies demonstrating that drug bioavailability is related to the presence and activities of microbes with the capacity to catabolize these drugs in the human gut39 and the emerging appreciation of the influence of non-antibiotic medications, for example, psychotropic drugs, on microbial survival and community structure within the human microbiome40. Second, computational modeling of the temporal dynamics of microbial metabolism in humans provides an opportunity to interrogate the underlying mechanisms by which the microbiome interacts with the human host over time to produce an emergent phenotype. This will enhance our understanding of the microbial functional traits that are appropriate for each stage of host development and the consequences of the loss of these traits on health outcomes.
The temporal fluctuation in microbial community structure presents not only considerable analytical challenges, but also new opportunities to develop more sophisticated statistical tools and models41,42. It is immediately necessary to determine how to deal with the replicates and controls that are needed with longitudinal data, and to determine how earlier pioneer microbiomes influence later assemblages and their functions. Many platforms already exist to facilitate longitudinal modeling of microbiome dynamics, including Poisson autoregressive integrated moving average (ARIMA)43, microbial temporal variability linear mixed model (MTV-LMM)43, adaptive generalized principal components analysis (gPCA)44, microbial dynamical systems inference engine for microbiome time-series analyses 2.0 (MDSINE 2.0)45, temporal gaussian process model for compositional data analysis (TGP-CODA)46 and multinomial logistic-normal dynamic linear models (MALLARDs)47. The main feature of these statistical tools is the inclusion of nonlinear effects on microbial components either directly or indirectly as external covariates (for example, nonlinear Bayesian neural networks48). All parameters from experimental design and the method of data generation can influence how data is prepared for analysis (i.e., what mathematical transformation and which tools are applied to identify important statistical associations). However, data treatment and choice of modeling environment are crucial factors in ensuring appropriate interpretation of microbial dynamics, and we refer the readers to a more in-depth review of this topic42. While primarily used for descriptive purposes, modeling the temporal dynamic of microbiomes requires statistical innovation to manage the ever-increasing size and dimensionality of the datasets under consideration.
Cross-sectional and longitudinal studies using DNA-based sequencing data to elucidate and interpret microbial dynamics rely heavily on data preprocessing and statistical modeling. Models that derive mathematical relationships between individual components that can be used to describe mechanism(s) underlying observed trends and predict a systems response to an external influence are still nascent. Innovation in these modeling approaches is necessary if we are to understand and optimize treatment strategies for microbial restoration. One approach that attempts to capture this complexity is flux balance modeling. These models integrate multidimensional datasets to describe and predict the metabolic responses of microbes to an external influence. Constraint-based metabolic modeling can include biochemical activities within the cell and how these facilitate metabolic interactions between constituent members within an ecosystem. These models assume that metabolic interactions are faster than cellular growth and environmental shifts and as such that cellular metabolites are in a steady state that enables prediction of metabolic flux using linear models49. Using these models, metabolic output of each cell in an assemblage as well as its relative growth rate under defined conditions and constraints may be predicted50. These models can also theoretically consider resource competition to permit capture of ecological dynamics at a scale relevant to a microbial community. Single isolates can already be reliably modeled, with growth yields accurately predicted for isolated organisms51; these models can be further augmented by allowing for cross-feeding between cellular models52. By incorporating time-steps, it is possible to model the dynamic metabolic ecology through dynamic flux balance analysis53, which has the potential to create an agent-based model to predict the outcome following a modeled external influence. Overall, through the application of ecological theory and systems-scale models, the mechanistic relationships between microbial species and the human host may be elucidated.
Future directions
Ecological medicine is fast becoming a practical reality, spurred by the integration of microbiome analyses into clinical trials and biomedical research. Currently (as of March 2019), there are approximately 1,400 on-going microbiome-focused clinical trials.
Existing studies provide considerable evidence that application of ecological principles offers a useful framework for improved understanding of the relationship of the human microbiome with human health. Importantly, innovation and application of ecological statistical models to determine how microbial dynamics interact with host physiology may elucidate the underlying principles behind a number of previously uninterpretable clinical outcomes. In a number of cases, this has proven effective, such as for the recurrent obesogenic phenotype.
The ability to predict a disease state appears to be positively correlated with the number of longitudinal samples collected from an individual, as the day-to-day variation in microbial structure can hide statistically relevant trends. Reducing noise and increasing statistical power through increased sampling frequency should be a priority for future microbiome studies aimed at identifying relevant microbial taxonomic, genetic or metabolic features of disease or health or those associated with clinical outcomes. Importantly, increased temporal observation density should be accompanied by advances in the statistical methods used to analyze and interpret longitudinal trends in multidimensional datasets, which therefore also represents a mathematical research priority for the field. By using ecology to better describe and interpret the host–microbiome interactions that underpin health and disease, it is becoming possible to integrate the principles of ecosystem restoration into modern clinical practice, thereby advancing therapeutic avenues that were previously unavailable. While there is a fundamental need to more fully integrate ecological theoretical and statistical frameworks into human microbiome research, early data indicate that the interpretation of findings through these lenses will continue to lead to improved understanding of microbe–host interactions, novel therapies aimed at disease prevention and tailored therapy for existing disease.
Acknowledgements
The authors would like to thank a number of colleagues for suggestions on articles to include in this review. Specifically, L. P. Coelho; B. Prithiviraj; E. Jasarevic; J. Pickard; M. Yassour; and E. Weinstein. S.V.L. is supported by awards from the National Institutes of Health (AI089473, AI113916, AI133765, PO515267, DH082147, DA040532) and the Crohn’s and Colitis Foundation of America.
Footnotes
Competing interests
J.A.G. reports roles as Chief Scientific Advisor, 4inno; Scientific Advisory Board, Biome Makers, Inc.; Scientific Advisory Board, DayTwo Inc.; Scientific Advisory Board, Growcentia Inc.; Scientific Advisory Board, Holobiome Inc.; and Scientific Advisory Board, Valent BioSciences. S.V.L. reports research grants from the NIH/NIAID, NIH/NIDA, NIH/NICHD, NIH/Office of the Director and the Crohn’s and Colitis Foundation of America and the following patents and royalties: ‘Reductive prodrug cancer chemotherapy (Stan449-PRV)’; ‘Combination antibiotic and antibody therapy for the treatment of Pseudomonas aeruginosa infection (WO2010091189A1)’ licensed with royalties by KaloBios Inc.; ‘Therapeutic microbial consortium for induction of immune tolerance’ licensed with royalties by Siolta Therapeutics; ‘Systems and methods for detecting antibiotic resistance (WO2012027302A3)’ issued; ‘Nitroreductase enzymes (US7687474B2)’ issued; ‘Sinusitis diagnostics and treatments (WO2013155370A1)’ licensed by Reflourish, LLC; and ‘Methods and systems for phylogenetic analysis (US20120264637A1)’ issued. S.V.L. is a board member of and consultant for Siolta Therapeutics and reports personal fees for this consultancy outside the submitted work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. © Springer Nature America, Inc. 2019
References
- 1.Byrd AL & Segre JA Infectious disease. Adapting Koch’s postulates. Science 351, 224–226 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Gilbert JA Ecological medicine. Environ. Microbiol 20, 1917–1919 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Lederberg JMA ‘Ome Sweet ‘Omics — a genealogical treasury of words. The Scientist (2001). [Google Scholar]
- 4.Gibbons SM et al. Evidence for a persistent microbial seed bank throughout the global ocean. Proc. Natl Acad. Sci. USA 110, 4651–4655 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandal S et al. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis 26, 27663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belkaid Y & Hand TW Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donaldson GP et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donia MS et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Gutierrez E, Mayer MJ, Cotter PD & Narbad A Gut microbiota as a source of novel antimicrobials. Gut Microbes 10, 1–21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abisado RG, Benomar S, Klaus JR, Dandekar AA & Chandler JR Bacterial quorum sensing and microbial community interactions. MBio 9, e02331–e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yatsunenko T et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannigan GD, Duhaime MB, Koutra D & Schloss PD Biogeography and environmental conditions shape bacteriophage-bacteria networks across the human microbiome. PLOS Comput. Biol 14, e1006099 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimura KE et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med 22, 1187–1191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durack J et al. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat. Commun 9, 707 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arrieta MC et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med 7, 307ra152 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Teo SM et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17, 704–715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukami T & Morin PJ Productivity–biodiversity relationships depend on the history of community assembly. Nature 424, 423–426 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Bäckhed F et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Thaiss CA et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 540, 544–551 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Foster KR, Schluter J, Coyte KZ & Rakoff-Nahoum S The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch SV & Pedersen O the human intestinal microbiome in health and disease. N. Engl. J. Med 375, 2369–2379 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK & Knight R Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothschild D et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Halfvarson J et al. Dynamics of the human gut microbiome in inflammatory bowel disease. New Microbiol 2, 17004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsson PG et al. Extended antimicrobial treatment of bacterial vaginosis combined with human lactobacilli to find the best treatment and minimize the risk of relapses. BMC Infect. Dis 11, 223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Nood E, Dijkgraaf MG & Keller JJ Duodenal infusion of feces for recurrent Clostridium difficile. N. Engl. J. Med 368, 401–415 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Kassam Z, Lee CH, Yuan Y & Hunt RH Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am. J. Gastroenterol 108, 500–508 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Panigrahi P et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 548, 407–412 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Maldonado-Gómez MX et al. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe 20, 515–526 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Jernberg C, Löfmark S, Edlund C & Jansson JK Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 1, 56–66 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Suez J et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 174, 1406–1423.e16 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Zmora N et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174, 1388–1405.e21 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Winston JA & Theriot CM Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 41, 44–50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang DW et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome 5, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halkjær SI et al. Faecal microbiota transplantation alters gut microbiota in patients with irritable bowel syndrome: results from a randomised, double-blind placebo-controlled study. Gut 67, 2107–2115 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Cope EK, Goldberg AN, Pletcher SD & Lynch SV Compositionally and functionally distinct sinus microbiota in chronic rhinosinusitis patients have immunological and clinically divergent consequences. Microbiome 5, 53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David LA et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turnbaugh PJ et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med 1, 6ra14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haiser HJ, Seim KL, Balskus EP & Turnbaugh PJ Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes 5, 233–238 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cussotto S et al. Differential effects of psychotropic drugs on microbiome composition and gastrointestinal function. Psychopharmacology (Berl) 1–15 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Gerber GK The dynamic microbiome. FEBS Lett 588, 4131–4139 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Silverman J, Shenhav L, Halperin E, Muklherjee S & David L Statistical considerations in the design and analysis of longitudinal microbiome studies Preprint at https://www.biorxiv.org/content/10.1101/448332v1 2018).
- 43.Shenhav L, Furman O, Mizrahi I & Halperin E Modeling the temporal dynamics of the gut microbial community in adults and infants Preprint at 10.1101/362822 (2017). [DOI] [PMC free article] [PubMed]
- 44.Fukuyama J et al. Multidomain analyses of a longitudinal human microbiome intestinal cleanout perturbation experiment. PLOS Comput. Biol 13, e1005706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bucci V et al. MDSINE: Microbial Dynamical Systems INference Engine for microbiome time-series analyses. Genome Biol 17, 121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Äijö T, Müller CL & Bonneau R Temporal probabilistic modeling of bacterial compositions derived from 16S rRNA sequencing. Bioinformatics 34, 372–380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silverman JD, Durand HK, Bloom RJ, Mukherjee S & David LA Dynamic linear models guide design and analysis of microbiota studies within artificial human guts. Microbiome 6, 202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larsen PE, Field D & Gilbert JA Predicting bacterial community assemblages using an artificial neural network approach. Nat. Methods 9, 621–625 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Varma AP & Palssom BO Metabolic flux balancing: basic concepts, scientific and practical use. Bio/Technol 12, 994 (1994). [Google Scholar]
- 50.Yang L, Yurkovich JT, King ZA & Palsson BO Modeling the multi-scale mechanisms of macromolecular resource allocation. Curr. Opin. Microbiol 45, 8–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ibarra RU, Edwards JS & Palsson BO Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420, 186–189 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Zomorrodi AR & Maranas CD OptCom: a multi-level optimization framework for the metabolic modeling and analysis of microbial communities. PLOS Comput. Biol 8, e1002363 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahadevan R, Edwards JS & Doyle FJ III Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys. J 83, 1331–1340 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]


