Increased dosage of TB1 reduces height and stem internode length in bread wheat without affecting coleoptile length.
Keywords: Architecture, coleoptile, growth, height, TEOSINTE BRANCHED1 (TB1), wheat (Triticum aestivum)
Abstract
Regulation of plant height and stem elongation has contributed significantly to improvement of cereal productivity by reducing lodging and improving distribution of assimilates to the inflorescence and grain. In wheat, genetic control of height has been largely contributed by the Reduced height-1 alleles that confer gibberellin insensitivity; the beneficial effects of these alleles are associated with less favourable effects involving seedling emergence, grain quality, and inflorescence architecture that have driven new research investigating genetic variation of stem growth. Here, we show that TEOSINTE BRANCHED1 (TB1) regulates height of wheat, with TB1 being expressed at low levels in nodes of the main culm prior to elongation, and increased dosage of TB1 restricting elongation of stem internodes. The effect of TB1 on stem growth is not accompanied by poor seedling emergence, as transgenic lines with increased activity of TB1 form longer coleoptiles than null transgenic controls. Analysis of height in a multiparent mapping population also showed that allelic variation for TB1 on the B genome influences height, with plants containing the variant TB-B1b allele being taller than those with the wild-type TB-B1a allele. Our results show that TB1 restricts height and stem elongation in wheat, suggesting that variant alleles that alter the expression or function of TB1 could be used as a new source of genetic diversity for optimizing architecture of wheat in breeding programmes.
Introduction
Height is an important trait in cereals including wheat, rice, and barley, with genetic variation that reduces stem elongation having contributed significantly to the generation of superior yielding cultivars during the ‘Green Revolution’ (Hedden, 2003). Plants with reduced height benefit from improved lodging resistance, and the suppression of growth increases the ability of plants to tolerate higher inputs of nitrogen-based fertilizer; the decrease in stem elongation boosts partitioning of assimilates to the developing inflorescence, facilitating improved floret fertility and grain numbers per ear (Youssefian et al., 1992a; Flintham et al., 1997)
Genetic variation that contributes semi-dwarfing alleles in wheat, barley, and rice mostly involves modification of biosynthesis, metabolism. or signalling of the growth-promoting phytohormone, gibberellic acid (GA) (Hedden, 2003; Pearce et al., 2011). For example, the semi-dwarfing Reduced height-1 (Rht-1; Rht-B1b and Rht-D1b) alleles encode variants of the DELLA protein that no longer confer responsiveness to GA, with the DELLA protein being a negative regulator of GA signalling that represses growth (Peng et al., 1999; de Lucas et al., 2008; Feng et al., 2008; Pearce et al., 2011). In rice and barley, the semi-dwarfing sd1 and sdw1 alleles contain loss-of-function mutations in the key GA biosynthesis enzyme GA 20-oxidase 2 (GA20ox2)—the defective alleles reduce levels of the growth-promoting bioactive GAs (Spielmeyer et al., 2002; Jia et al., 2009; Xu et al., 2017). In addition to the beneficial height reduction effects of the semi-dwarfing Rht-1 alleles, the decreased responsiveness to GA provokes unfavourable pleiotropic phenotypes including reduced seedling emergence, lower photosynthetic rate, and decreased grain size and grain protein content, which has encouraged research of novel Rht-B1 alleles that contain second site mutations and other dwarfing alleles, such as Rht8 and Rht18/Rht25 (Allan, 1980; Youssefian et al., 1992a, b; Schillinger et al., 1998; Rebetzke et al., 1999; Wu et al., 2011; Gasperini et al., 2012; Chandler and Harding, 2013; Casebow et al., 2016; Kowalski et al., 2016; Van De Velde et al., 2017; Ford et al., 2018; Mo et al., 2018; Jobson et al., 2019). Rht18 encodes a GA 2-oxidase, which is responsible for converting bioactive GA into inactive forms of the hormone (Ford et al., 2018). The effect of these Rht alleles on stem elongation and height is accompanied by changes in physical characteristics of the inflorescence; dwarfed plants develop compact spikes with an altered number of spikelets (Mo et al., 2018).
Inflorescence development is closely associated with stem elongation, as elongation of stem internodes 2 and 3 occurs simultaneously with the white anther and terminal spikelet stages, respectively, which are when floret and spikelet numbers are determined (Kirby, 1987; Reynolds et al., 2009; Guo and Schnurbusch, 2015). To identify genes that influence spikelet and floret formation, we and others have studied genes that alter the rate of inflorescence development, spikelet architecture, and/or floret fertility (Boden et al., 2015; Guo and Schnurbusch, 2015; Dixon et al., 2018; Sakuma et al., 2019). We showed that a wheat orthologue of the major maize domestication gene, TEOSINTE BRANCHED1 (TB1), is a key regulator of plant architecture and inflorescence development (Dixon et al., 2018). Increased dosage of TB-D1 (D genome homeologue of TB1) delayed inflorescence development, restricted tiller growth, and promoted formation of paired spikelets, which are supernumerary spikelets characterized by the development of two spikelets at a single rachis node, rather than the typical single spikelet (Boden et al., 2015). These traits are all linked physiologically by the suppression of growth, particularly of axillary meristems including spikelet primordia and tiller buds, with a decrease in the rate of inflorescence development occurring at the terminal spikelet stage associated with elongation of stem internode 3 (Dixon et al., 2018). The effect of TB1 on growth is consistent with it being a class II TCP transcription factor, which regulates expression of genes involved in processes including cell proliferation, the cell cycle, as well as signalling and metabolism of hormones such as GA and jasmonic acid; TB1 and its Arabidopsis homologue, BRANCHED1 (BRC1), inhibit expression of cell cycle genes and those involved in GA signalling (González-Grandío et al., 2013; Dong et al., 2019). Based on the role of TB1/BRC1 controlling cell proliferation, and its key role in regulating the rate of inflorescence development and plant architecture, we hypothesized that TB1 may influence the height of wheat plants.
In this study, we investigated height and stem internode lengths in wheat lines that contain increased dosage of TB-D1, including a pair of near-isogenic lines (NILs) developed from a MAGIC population that are tetrasomic for chromosome 4D and transgenic plants that express TB1 at higher levels (Dixon et al., 2018). We show that increased activity of TB1 restricts height and stem elongation in bread wheat, and that allelic diversity for TB-B1 influences height in an advanced mapping population. These results provide new insights into the genetic regulation of height in wheat, which may help breeders optimize plant architecture.
Materials and methods
Plant materials and growth conditions
Hexaploid wheat (Triticum aestivum) germplasm used in this study included the following genotypes: MAGIC line 0053 of the CSIRO four-way multiparent advanced generation intercross (MAGIC) population, from which the NILs termed wild type (WT) and highly-branched (hb) were derived (see Dixon et al., 2018); transgenic lines expressing TB-D1 using the VRN1 promoter generated in the cv. Fielder genetic background (Dixon et al., 2018); and the hexaploid wheat TILLING line, Cadenza1721 (tb-d1) (Krasileva et al., 2017). The hb line is tetrasomic for chromosome 4D and contains an additional haploid copy of TB-D1, relative to its WT NIL that is diploid for 4D (see Dixon et al., 2018). The pVRN1:TB1 transgenic lines expressed the TB-D1 allele from cv. Baxter that is present in hb and WT NILs of the MAGIC line 0053, under the control of the barley VRN1 promoter (Alonso-Peral et al., 2011). The ‘NIAB Elite MAGIC’ bread wheat population has been previously described (Mackay et al., 2014), and height data were collated from published data of plants grown under field conditions at the NIAB experimental farm in Cambridge, UK (52°13'19''N, 0°5'46''E) (Mackay et al., 2014; Scutari et al., 2014).
WT, hb plants, transgenic lines, and the F1 plants of the hb crosses to Cadenza, tb-d1 (Cadenza1721; see Supplementary Fig. S1 at JXB online), that were used for phenotype analysis and gene expression studies were grown in controlled growth chambers under long-day (16 h light/8 h dark) photoperiods at 300 µmol m–2 s–1 [using Plantastar 400-W HQI bulbs (Osram) and Maxim 60 W tungsten bulbs], with a day temperature of 20 °C and a night temperature of 15 °C.
RNA extractions and quantitative real-time PCR analysis
For experiments with WT and hb plants, RNA was extracted from internode 2 stem, prior to internode elongation. For experiments using the pVRN1:TB1 transgenic and null control lines, including cv. Fielder, RNA was extracted from internode 2 stem, prior to initiation of elongation. For transcript analysis of TB-B1 and TB-D1 in cv. Cadenza, RNA was extracted from internodes 1, 2, and 3 and the peduncle prior to initiation of elongation and from tiller buds, which are immature tillers that were transparent or light green in colour for which no leaf blade had begun to emerge. All RNA extractions were performed using the RNeasy Plant Mini Kit (Qiagen). Synthesis of cDNA and quantitative real-time PCR (qRT-PCR) were performed as described previously (Boden et al., 2014). Oligonucleotides for qRT-PCR analysis are provided in Supplementary Table S1. Expression of TB-B1 and TB-D1 was normalized using Traes_6DS_BE8B5E56D.1 (gene ID in IWGSC reference genome is TraesCS6D02G145100) (Borrill et al., 2016). All qRT-PCR data points are the average of at least four biological replicates, with two technical replicates performed in each reaction.
Analysis of coleoptile elongation
Coleoptile length measurements were performed using seeds of equal weight (between 40 mg and 50 mg), with individual seeds sown into pots (dimensions 90×90×180 mm) containing fertile compost-based potting mix at a sowing depth of ~5 cm. The soil was initially watered until saturation (~150 ml), and pots were placed in growth chambers and covered with lids wrapped in aluminium foil. Lights were turned off and the temperature was set to 20 °C/15 °C (day/night). Pots were watered equally every 2 d (15 ml). Coleoptile length was measured using a ruler, 12 d after sowing once the cotyledon had emerged from the coleoptile. Each experiment was performed at least three times, with 12 seeds per genotype per replicate.
Kompetitive allele-specific PCR analysis
Kompetitive allele-specific PCR (KASP-PCR) analysis was performed as per Dixon et al. (2018). Sequences for oligonucleotides used in KASP assays are provided in Supplementary Table S2.
Statistical analysis
Differences between genotypes were tested by two-tailed Student’s t-test. The number of replicates for each experiment are provided elsewhere in the Materials and methods and in the relevant figure legend.
Accession numbers
The gene IDs for TB-A1, TB-B1, and TB-D1 are TraesCS4A02G271300, TraesCS4B02G042700, and TraesCS4D02G040100, respectively.
Results
Increased dosage of TB-D1 reduces height and internode length in bread wheat
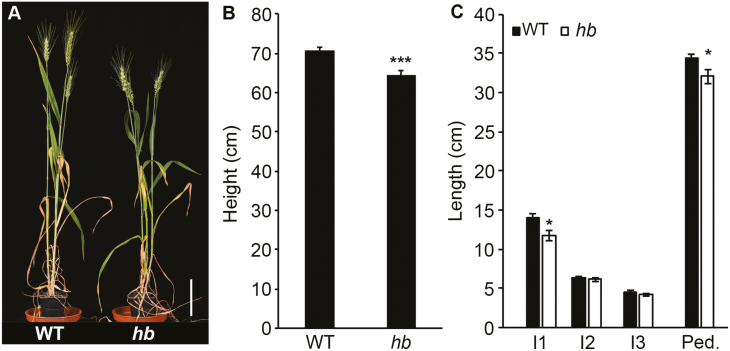
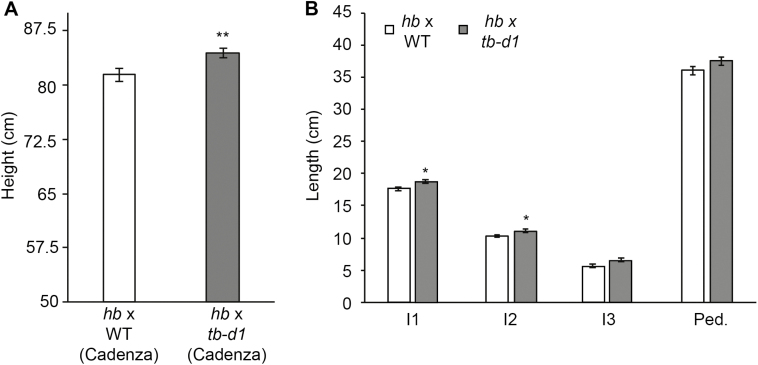
Our previous investigation of inflorescence and plant architecture included analysis of a pair of NILs derived from a single line of a four-way MAGIC population (Dixon et al., 2018). The NIL hb formed multiple paired spikelets on the inflorescence, relative to the WT NIL that formed an inflorescence with single spikelets at each rachis node. The hb line is tetrasomic for chromosome 4D, relative to the euploid WT line. In addition to the inflorescence and tiller phenotypes, the hb line was shorter than the WT. To investigate this trait further and determine if TB1 influences stem growth, we measured height and internode length in hb plants grown under long-day photoperiods, relative to the WT. The hb plants were significantly shorter than the WT (Fig. 1A, B), and lengths of internode 1 and the peduncle were significantly shorter in hb compared with the WT, while there was no significant difference in the length of internodes 2 and 3. This result demonstrates that increased dosage of chromosome 4D restricts plant height in wheat. To test if this effect of tetrasomy 4D is caused by increased dosage of TB1 and not another gene on chromosome 4D, we crossed the hb line to a Cadenza TILLING mutant that contains a premature stop codon in TB-D1 (tb-d1) and analysed height and internode length of the first filial generation (F1) individuals, relative to F1 individuals from a cross between hb and Cadenza (Cadenza contains WT tall alleles of Rht-B1 and -D1). We observed that the F1 individuals of the hb×tb-d1 individuals were taller than those derived from the hb×Cadenza cross (Fig. 2A). The lengths of internodes 1 and 2 were longer in the hb×tb-d1 F1 individuals, relative to individuals of the hb×Cadenza cross, while there was no significant difference in the length of internode 3 (Fig. 2B). Taken together, these results indicate that tetrasomy for chromosome 4D restricts plant height and internode length in wheat, and that increased dosage of TB-D1 contributes to this effect on growth.
Fig. 1.
Tetrasomy for chromosome 4D reduces plant height. (A and B) hb plants that are tetrasomic for chromosome 4D are shorter than wild-type near-isogenic lines. (C) Internode lengths for wild-type and hb plants. I1, internode 1; I2, internode 2; I3, internode 3; Ped., peduncle. Scale bar=10 cm. Values are the mean ±SE of 10 biological replicates. *P<0.05; ***P<0.001.
Fig. 2.
TB-D1 contributes to the reduced height of hb plants. (A) Height and (B) internode length measurements of F1 offspring plants from crosses between hb and Cadenza (WT), and hb and the tb-d1 null mutant line (Cadenza1721). I1, internode 1; I2, internode 2; I3, internode 3; Ped., peduncle. Values are the mean ±SE of 48 biological replicates. *P<0.05; **P<0.01.
Expression analysis of TB-1 in stem tissue
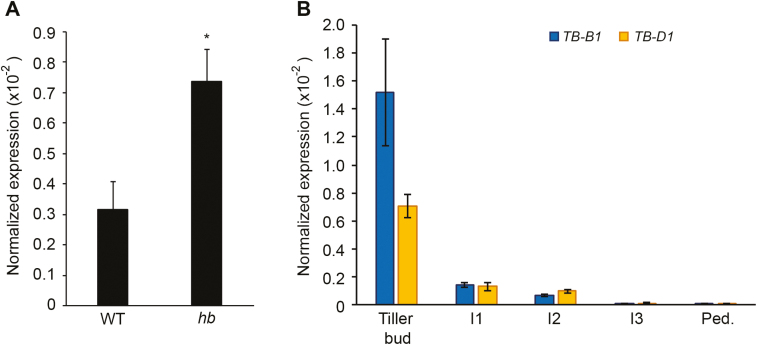
To determine if the effect of TB-D1 on height and internode length in the hb plants could be explained by increased expression of TB-D1, we measured TB-D1 transcript levels in internode 2 stem regions prior to elongation. qRT-PCR analysis detected TB-D1 transcripts in the stem internode 2 in both WT and hb plants, with transcript levels being ~2-fold higher in hb relative to the WT (Fig. 3A). Transcript analysis in Cadenza showed that TB-D1 and TB-B1 are expressed in internodes 1 and 2 prior to elongation of the internodes, and the levels are low relative to those measured in tiller buds which we demonstrated previously to express TB1 (Fig. 3B); transcript levels in Cadenza were slightly lower than those detected in the same internode of the WT NIL. We did not measure TB-A1 expression because our previous analysis has shown that this homoeoallele is not expressed (Dixon et al., 2018). These results show that reduced height and internode lengths of the hb plants are associated with increased expression of TB-D1 in stem internode tissue prior to elongation, and that TB1 is expressed at low levels in stem segments prior to elongation of the internode.
Fig. 3.
TB-B1 and TB-D1 expression in stem tissue. (A) Transcript analysis of TB-D1 in internode 2 stem tissue of WT and hb lines. (B) Quantification of TB-B1 (blue) and TB-D1 (yellow) transcripts in stem tissue and tiller buds. I1, internode 1; I2, internode 2; I3, internode 3; Ped., peduncle. Values are the mean ±SE of four biological replicates. *P<0.05.
Analysis of height in pVRN1:TB-D1 transgenic lines
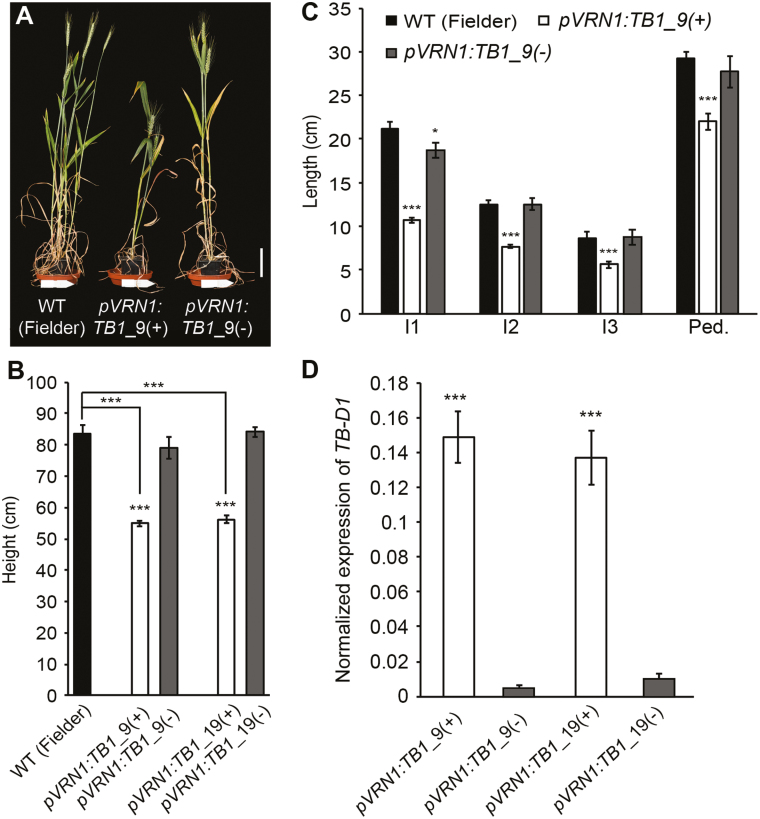
To further test if increased dosage of TB-D1 restricts stem growth and height in wheat, we analysed transgenic lines that contain an additional copy of TB-D1 under control of the VERNALIZATION1 (VRN1) promoter (pVRN1:TB1) (Alonso-Peral et al., 2011). The pVRN1:TB1 transgenic lines were significantly shorter than control null transgenic lines [pVRN1:TB1(–)] and the untransformed plants (cv. Fielder) (Fig. 4A, B). The pVRN1:TB1 transgenic lines had shorter internodes than the null transgenic control lines and cv. Fielder, with length of internodes 1, 2, and 3 and the peduncle being significantly reduced, relative to the control lines (Fig. 4C). The stem growth and height phenotypes are associated with significantly higher transcript levels of TB-D1 in stem segments prior to stem elongation, relative to the null transgenic lines (Fig. 4D). Increased expression of TB-D1 is consistent with VRN1 transcript levels being relatively high in stem tissue compared with leaf tissue where it is expressed robustly (Supplementary Fig. S2). These results show that increased expression of TB-D1 within developing stem tissue restricts internode elongation and plant height in wheat, which support the phenotypes of the hb line relative to the WT NIL.
Fig. 4.
Increased dosage of TB-D1 reduces plant height. (A and B) Height and (C) internode length measurements for two independent pVRN1:TB1 transgenic lines (white bars), relative to control null transgenic lines (grey bars) and the untransformed wild type (cv. Fielder, black bars). (D) TB-D1 expression in internode 2 of the elongating stem of pVRN1:TB1 transgenic plants (white bars), relative to control null transgenic lines (grey bars). I1, internode 1; I2, internode 2; I3, internode 3; Ped., peduncle. Scale bar=10 cm. (A–C) Values are the mean ±SE of 10 biological replicates, and (D) the mean ±SE of five biological replicates. *P<0.05; ***P<0.001.
Increased dosage of TB-D1 does not restrict coleoptile length
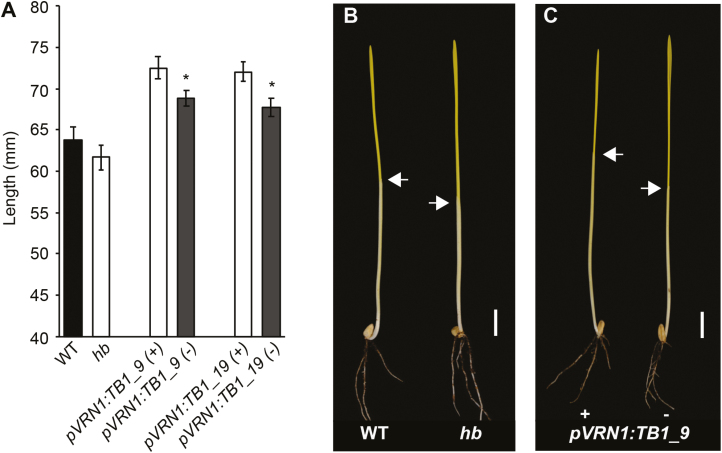
A limitation of using Rht-b1b and Rht-D1b dwarfing alleles in modern wheat is that the benefit of reduced stem elongation to reduce lodging is accompanied by unfavourable pleiotropic traits including poor emergence of seedlings from deep sowing, decreased meristem size, lower grain set, increased susceptibility to diseases, and reduced 1000-grain weight (Allan, 1980; Borner et al., 1996; Schillinger et al., 1998; Rebetzke et al., 1999; Ellis et al., 2004; Saville et al., 2012; Serrano-Mislata et al., 2017). We have shown that increased dosage of TB1 alters spikelet architecture and delays stages of inflorescence development critical for floret formation (Dixon et al., 2018); here, we tested if increased dosage of TB1 reduced coleoptile length to determine if it would restrict seedling emergence from soil. We found that the coleoptile length of hb plants was shorter than that of the WT, but was longer in two independent pVRN:TB1 transgenic lines, relative to null transgenic plants (Fig. 5A–C). These results show that tetrasomy for chromosome 4D reduces coleoptile length, which is consistent with increased dosage of Rht-D1a (also located on 4D); however, increased dosage of TB1 alone does not restrict coleoptile growth or emergence of seedlings. These results indicate that increased activity of TB1 restricts plant height without negatively influencing seedling emergence, suggesting that alleles with increased activity of TB1 could be used as an alternative to Rht-B1b or Rht-D1b semi-dwarfing alleles.
Fig. 5.
Analysis of the effect of TB-D1 on coleoptile length. (A) Coleoptile length measurements for WT (black) and hb (white) lines, and for pVRN1:TB-D1 transgenic lines (grey) relative to the null transgenic control lines (white). Images of representative seedlings for (B) WT and hb and for (C) transgenic pVRN1:TB-D1 transgenic and null control lines. Scale bar=1 cm; white arrows indicate the apical end of the coleoptile. Values are the mean ±SE for 12 biological replicates. * P<0.05.
Allelic variation for TB-B1 influences plant height in bread wheat
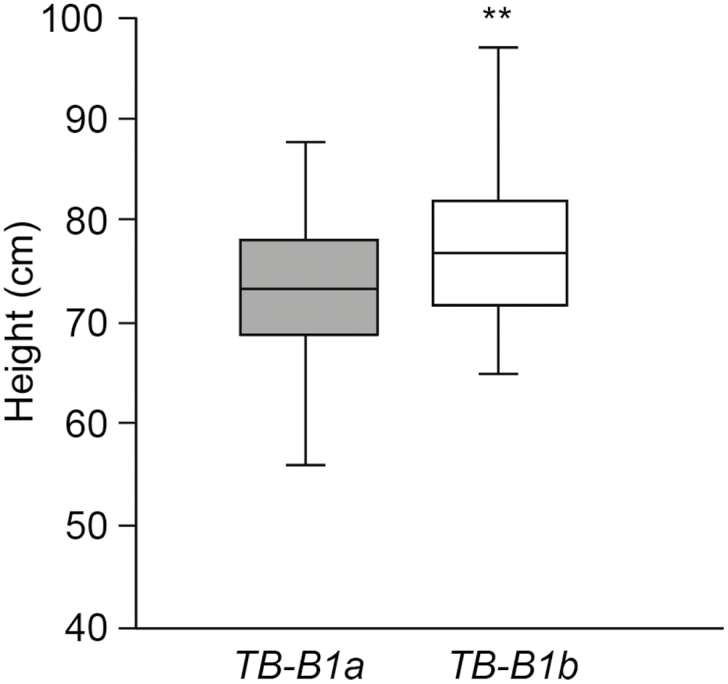
Our previous analysis of TB1-dependent regulation of inflorescence architecture identified a variant allele for TB-B1 (known as TB-B1b), which is predicted to be a weak loss-of-function allele that contains three amino acid substitutions and two synonymous mutations, relative to the reference allele (Dixon et al., 2018). Based on our results showing that increased activity of TB-D1 reduces height (Figs. 1, 4), and TB-B1 being expressed equally to TB-D1 (Fig. 3), we hypothesized that the TB-B1b allele would increase height of bread wheat, relative to the TB-B1a allele. To test this hypothesis, we analysed the height of lines from the eight-way UK MAGIC population that were fixed for the semi-dwarfing allele of Rht-D1 (Rht-D1b; aka Rht-2) but differed with respect to the TB-B1 allele (i.e. Rht-D1b:TB-B1a, or Rht-D1b:TB-B1b) (Scutari et al., 2014; Mackay et al., 2014). We did not analyse any lines with two dwarfing alleles (Rht-B1b/Rht-D1b) or any of the Rht-B1b semi-dwarf lines because they all contained the TB-B1a allele, which is consistent with Rht-B1 and TB-B1 being in close genetic linkage with each other (Dixon et al., 2018). We found that lines containing the TB-B1b allele were significantly taller than lines with the TB-B1a allele (Fig. 6), with TB-B1b genotypes being 4.6% taller (3.4±0.91 cm; P<0.01) than TB-B1a genotypes. These results support the hypothesis that TB-B1b alleles are weak loss-of-function alleles that can partially restore the height of lines that contain dwarfing alleles of Rht-1.
Fig. 6.
Analysis of allelic variation for TB-B1 on height phenotypes. Boxplots showing the height of semi-dwarfed eight-way MAGIC lines (genotype Rht-D1b) that contain either the TB-B1a (grey) or the TB-B1b (white) allele. Boxes extend from the lower to the upper quartile, with a line marking the median, and whiskers extend to 1.5 times the interquartile range. **P<0.01.
Discussion
The genetic regulation of height has contributed significantly to improvement of crop productivity, especially in wheat, rice, and barley, by reducing stem elongation to improve lodging resistance and enhance partitioning of resources to the inflorescence (Hedden, 2003). In wheat, these benefits have largely been provided by the semi-dwarfing Rht-B1b and Rht-D1b alleles (Peng et al., 1999; Pearce et al., 2011); however, due to their pleiotropic effects on seedling emergence, spikelet number, grain weight, and quality (for example), research has continued to investigate novel sources of genetic variation for control of height (Allan, 1980; Youssefian et al., 1992a, b; Schillinger et al., 1998; Rebetzke et al., 1999; Wu et al., 2011; Gasperini et al., 2012; Kowalski et al., 2016; Ford et al., 2018; Mo et al., 2018). Here, we have shown that increased expression of TB1 reduces stem elongation in bread wheat, and that allelic variation for TB-B1 influences plant height.
TB1 and its homologue in eudicots—BRANCHED1 (BRC1)—have been studied extensively due to their role in regulating shoot branching and apical dominance (Doebley et al., 1997; Aguilar-Martínez et al., 2007). TB1/BRC1 of maize, rice, wheat, barley, tomato, pea, Arabidopsis, switchgrass, and cucumber have conserved roles in regulating outgrowth of lateral branches, with loss-of-function mutations increasing lateral branch number and length, while gain-of-function alleles suppress their growth (Doebley et al., 1997; Takeda et al., 2003; Aguilar-Martínez et al., 2007; Weber et al., 2007; Minakuchi et al., 2010; Martín-Trillo et al., 2011; Ramsay et al., 2011; Braun et al., 2012; Seale et al., 2017; Bennett et al., 2016; Dixon et al., 2018; Liu et al., 2018; Zwirek et al., 2019; Shen et al., 2019). A conserved role for TB1/BRC1 in controlling height and stem elongation is, however, less clear; for example, brc1 and brc1/brc2 Arabidopsis mutants are shorter than the WT, as are tomato plants with reduced expression of BRC1, potentially due to reduced apical dominance (Finlayson et al., 2010; Martín-Trillo et al., 2011; Bennett et al., 2016; Seale et al., 2017). The effect of tb1/brc1 loss-of-function alleles in eudicots is maintained in rice, with loss-of-function mutants of TB1, known as FINE CULM1 (FC1), shorter than WT plants (Minakuchi et al., 2010). Analysis in maize did not detect a role for allelic diversity of Tb1 in controlling height, and there is no statistical difference in height for barley and switchgrass lines that contain alleles of reduced TB1/VRS5 expression (Liu et al., 2018; Zwirek et al., 2019). These reports seemingly show that wheat differs from other plants, as our results show that increased expression of TB1 reduces height, and that alleles with reduced function are likely to partially restore height in semi-dwarfed plants (Figs. 1, 4, 6). However, research in model and crop species has mostly involved analysis of reduced function alleles, relative to the gain-of-function alleles tested here. This conclusion is supported by reports showing that overexpression of the maize Tb1 gene in wheat reduces plant height, and induced expression of BRC1 leads to severe arrested growth of Arabidopsis seedlings (Lewis et al., 2008; González-Grandío et al., 2013). The results presented here suggest that further investigation of TB1 variant alleles, particularly those that influence expression through gene editing of cis-regulatory regions, may prove to be useful in regulating stem elongation and height.
Analyses of TB1 in wheat have demonstrated roles for this gene in regulating inflorescence development and plant architecture, including height and tillering (Dixon et al., 2018). A link between inflorescence architecture genes and height has also been shown in other crops. For example, in barley, a major inflorescence trait is row-type architecture, with cultivated barley forming spikes with either two or six row arrangements of spikelets; this trait is regulated by at least five genes, VRS1– VRS5 (VRS5 is a TB1 homologue) (reviewed in Gauley and Boden, 2019). Analysis of plant architecture in the vrs mutants has shown that some of the VRS row-type alleles influence plant height—vrs4 and vrs4/5 mutants are significantly taller than WT (cv. Bowman) plants, while vrs1/5 and vrs3/5 double mutants are significantly shorter (Zwirek et al., 2019). Moreover, barley deficiens mutants that maintain protein stability of VRS1 display reduced plant height, relative to NILs with WT VRS1 alleles (Sakuma et al., 2017). In domesticated wheat, a mutation in the miRNA172- (miR172) binding site of an AP2-like transcription factor that underpins the Q locus have contributed to development of a compact spike and a free-threshing phenotype, relative to the elongated rachis of speltoid wheat (Debernardi et al., 2017; Greenwood et al., 2017). Further mutations that disrupt binding of miR172 modify spike compactness further, and are associated with significant reduction in plant height (Debernardi et al., 2017; Greenwood et al., 2017). Inflorescence architecture and height are also associated in rice; for example, the SQUAMOSA PROMOTER BINDING-LIKE PROTEIN-14 (SPL14) gene that underpins the IDEAL PLANT ARCHITECTURE (IPA)/WEALTHY FARMERS PANICLE (WFP) locus influences panicle branching and plant height (Jiao et al., 2010; Miura et al., 2010). Mutations in a miR156-binding site increase expression of SPL14 to promote formation of a more highly branched panicle with improved lodging resistance and reduced tillering. Taken together, these results demonstrate an important link between the genetic regulation of inflorescence and plant architecture, and highlight the importance of considering pleiotropic effects of genes when determining genetic variation that may be used to improve crop performance.
The influence of TB1 on height in wheat, including our analysis suggesting that the variant TB-B1b allele partially restores stem length in semi-dwarfed germplasm, provides an opportunity to investigate genetic variation that will help improve plant architecture in breeding programmes (Fig. 6). Analysis of Rht-B1b and Rht-D1b alleles has shown that semi-dwarfism is associated with unfavourable pleiotropic traits, including reduced seedling emergence from deep sowing, decreased grain size, and lower grain protein content, which has stimulated investigation of alternative Rht alleles, such as Rht8, Rht12, and Rht18/25 (Youssefian et al., 1992a, b; Rebetzke et al., 1999; Gasperini et al., 2012; Kowalski et al., 2016; Mo et al., 2018). Rht8 is particularly interesting as it is likely to encode a protein that is independent of the GA metabolism and signalling pathways, and does not restrict coleoptile growth (Rebetzke et al., 1999; Gasperini et al., 2012). Our results suggest that increased expression of TB1 has a similar effect—elevated activity of TB1 reduced height and stem internode length without negatively affecting coleoptile growth (Figs 4, 5). Alleles that increase expression of TB1 may therefore be useful in restricting stem elongation and tillering without decreasing seedling emergence, providing an alternative to other Rht alleles. The Rht-B1b and Rht-D1b alleles reduce height to ~86% and 83% of tall controls, respectively, and Rht8 dwarfing alleles decrease height by 11%, relative to tall NILs (Flintham et al., 1997; Kowalski et al., 2016). Based on the results shown here, alleles that increase expression of TB1 have potential to reduce height similarly to Rht8 (91% of the height of WT controls) in tetrasomic plants, while the more highly expressed pVRN1:TB1 transgenes decreased height by ~30% (Figs 1, 4). Alleles that increase expression of TB-B1 or -D1 may therefore benefit breeding programmes by providing a comparable reduction in height to existing Rht alleles. Alternatively, TB1 alleles that reduce functionality may also benefit wheat breeding. For example, the effect of TB1 on tillering involves hormones such as abscisic acid, jasmonic acid, and GA, indicating that TB1-dependent regulation of growth is at least partially separable from the molecular role of Rht1 (Peng et al., 1999; de Lucas et al., 2008; Feng et al., 2008; Dong et al., 2019). The TB-B1b allele, or variant TB-D1b allele, could be introduced into Rht-B1b and Rht-D1b semi-dwarfed backgrounds to help fine-tune plant architecture traits including height, tillering, and spikelet number for improved crop performance (Dixon et al., 2018). Due to the genetic linkage of TB1 and Rht1 on chromosome group 4, the most efficient breeding strategy would most probably involve combining reciprocal alleles on the B and D genomes, to form genotypes such as Rht-D1b:TB-B1b or Rht-B1b:TB-D1b. Further investigation of variant TB1 alleles in different semi-dwarfed backgrounds will be necessary to determine the optimal allelic combinations for improved plant architecture.
In summary, our results demonstrate a new role for TB1 in wheat, where increased activity reduces internode elongation and height (Figs. 1, 2, 4). Together with our previous report showing that TB1 controls inflorescence development and tillering, these results show that TB1 is an important regulator of plant architecture—further investigation of TB1 will be critical for identifying pathways that act downstream to regulate tissue-specific phenotypes, which may provide opportunities to optimize key yield-related traits including tillering, spikelet number, and height.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Schematic of the hb×tb-d1 cross and the control hb×Cadenza cross.
Fig. S2. Tissue-specific analysis of VRN1 expression in wild-type wheat (cv. Cadenza)
Table S1. Oligonucleotide sequences used for qRT-PCR assays.
Table S2. Oligonucleotide sequences used for KASP-PCR assays
Table S3. TB-B1 genotype information of MAGIC elite lines used to investigate height.
Acknowledgements
We acknowledge the Biotechnology and Biological Sciences Research Council (BB/P016855/1) and the Royal Society (UF150081) for funding this research. The authors thank Dr James Cockram (NIAB) for providing Rht-B1 and Rht-D1 genotype information for the eight-way MAGIC lines.
References
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. 2007. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. The Plant Cell 19, 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan RE. 1980. Influence of semidwarfism and genetic background on stand establishment of wheat. Crop Science 20, 634–638. [Google Scholar]
- Alonso-Peral MM, Oliver SN, Casao MC, Greenup AA, Trevaskis B. 2011. The promoter of the cereal VERNALIZATION1 gene is sufficient for transcriptional induction by prolonged cold. PLoS One 6, e29456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T, Liang Y, Seale M, Ward S, Müller D, Leyser O. 2016. Strigolactone regulates shoot development through a core signalling pathway. Biology Open 5, 1806–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden SA, Cavanagh C, Cullis BR, Ramm K, Greenwood J, Jean Finnegan E, Trevaskis B, Swain SM. 2015. Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nature Plants 1, 14016. [DOI] [PubMed] [Google Scholar]
- Boden SA, Weiss D, Ross JJ, Davies NW, Trevaskis B, Chandler PM, Swain SM. 2014. EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. The Plant Cell 26, 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner A, Plaschke J, Korzun V, Worland AJ. 1996. The relationships between the dwarfing genes of wheat and rye. Euphytica 89, 69–75. [Google Scholar]
- Borrill P, Ramirez-Gonzalez R, Uauy C. 2016. expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiology 170, 2172–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot JP, et al. 2012. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiology 158, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casebow R, Hadley C, Uppal R, Addisu M, Loddo S, Kowalski A, Griffiths S, Gooding M. 2016. Reduced height (Rht) alleles affect wheat grain quality. PLoS One 11, 1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Harding CA. 2013. ‘Overgrowth’ mutants in barley and wheat: new alleles and phenotypes of the ‘Green Revolution’ DELLA gene. Journal of Experimental Botany 64, 1603–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi JM, Lin H, Chuck G, Faris JD, Dubcovsky J. 2017. microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 144, 1966–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. 2008. A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. [DOI] [PubMed] [Google Scholar]
- Dixon LE, Greenwood JR, Bencivenga S, Zhang P, Cockram J, Mellers G, Ramm K, Cavanagh C, Swain SM, Boden SA. 2018. TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum). The Plant Cell 30, 563–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. 1997. The evolution of apical dominance in maize. Nature 386, 485–488. [DOI] [PubMed] [Google Scholar]
- Dong Z, Xiao Y, Govindarajulu R, Feil R, Siddoway ML, Nielsen T, Lunn JE, Hawkins J, Whipple C, Chuck G. 2019. The regulatory landscape of a core maize domestication module controlling bud dormancy and growth repression. Nature Communications 10, 3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MH, Rebetzke GJ, Chandler P, Bonnett D, Spielmeyer W, Richards RA. 2004. The effect of different height reducing genes on the early growth of wheat. Functional Plant Biology 31, 583–7. [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, et al. 2008. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson SA, Krishnareddy SR, Kebrom TH, Casal JJ. 2010. Phytochrome regulation of branching in Arabidopsis. Plant Physiology 152, 1914–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flintham JE, Borner A, Worland AJ, Gale MD. 1997. Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. Journal of Agricultural Science 128, 11–25. [Google Scholar]
- Ford BA, Foo E, Sharwood R, et al. 2018. Rht18 semidwarfism in wheat is due to increased GA 2-oxidaseA9 expression and reduced GA content. Plant Physiology 177, 168–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini D, Greenland A, Hedden P, Dreos R, Harwood W, Griffiths S. 2012. Genetic and physiological analysis of Rht8 in bread wheat: an alternative source of semi-dwarfism with a reduced sensitivity to brassinosteroids. Journal of Experimental Botany 63, 4419–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauley A, Boden SA. 2019. Genetic pathways controlling inflorescence architecture and development in wheat and barley. Journal of Integrative Plant Biology 61, 296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Grandío E, Poza-Carrión C, Sorzano CO, Cubas P. 2013. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. The Plant Cell 25, 834–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood JR, Finnegan EJ, Watanabe N, Trevaskis B, Swain SM. 2017. New alleles of the wheat domestication gene Q reveal multiple roles in growth and reproductive development. Development 144, 1959–1965. [DOI] [PubMed] [Google Scholar]
- Guo Z, Schnurbusch T. 2015. Variation of floret fertility in hexaploid wheat revealed by tiller removal. Journal of Experimental Botany 66, 5945–5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. 2003. The genes of the Green Revolution. Trends in Genetics 19, 5–9. [DOI] [PubMed] [Google Scholar]
- Jia Q, Zhang J, Westcott S, Zhang XQ, Bellgard M, Lance R, Li C. 2009. GA-20 oxidase as a candidate for the semidwarf gene sdw1/denso in barley. Functional & Integrative Genomics 9, 255–262. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, et al. 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics 42, 541–544. [DOI] [PubMed] [Google Scholar]
- Jobson EM, Johnston RE, Oiestad AJ, Martin JM, Giroux MJ. 2019. The impact of the wheat Rht-B1b semi-dwarfing allele on photosynthesis and seed development under field conditions. Frontiers in Plant Science 10, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby EJM, Appelyard M. 1987. Cereal development guide. Kenilworth, UK: Arable Unit, National Agricultural Centre. [Google Scholar]
- Kowalski AM, Gooding M, Ferrante A, Slafer GA, Orford S, Gasperini D, Griffiths S. 2016. Agronomic assessment of the wheat semi-dwarfing gene Rht8 in contrasting nitrogen treatments and water regimes. Field Crops Research 191, 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva KV, Vasquez-Gross HA, Howell T, et al. 2017. Uncovering hidden variation in polyploid wheat. Proceedings of the National Academy of Sciences, USA 114, E913–E921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JM, Mackintosh CA, Shin S, Gilding E, Kravchenko S, Baldridge G, Zeyen R, Muehlbauer GJ. 2008. Overexpression of the maize Teosinte Branched1 gene in wheat suppresses tiller development. Plant Cell Reports 27, 1217–1225. [DOI] [PubMed] [Google Scholar]
- Liu Y, Merrick P, Zhang Z, Ji C, Yang B, Fei SZ. 2018. Targeted mutagenesis in tetraploid switchgrass (Panicum virgatum L.) using CRISPR/Cas9. Plant Biotechnology Journal 16, 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IJ, Bansept-Basler P, Barber T, et al. 2014. An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: creation, properties, and validation. G3 4, 1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M, Grandío EG, Serra F, Marcel F, Rodríguez-Buey ML, Schmitz G, Theres K, Bendahmane A, Dopazo H, Cubas P. 2011. Role of tomato BRANCHED1-like genes in the control of shoot branching. The Plant journal 67, 701–714. [DOI] [PubMed] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N, et al. 2010. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant & Cell Physiology 51, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M. 2010. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nature Genetics 42, 545–549. [DOI] [PubMed] [Google Scholar]
- Mo Y, Vanzetti LS, Hale I, Spagnolo EJ, Guidobaldi F, Al-Oboudi J, Odle N, Pearce S, Helguera M, Dubcovsky J. 2018. Identification and characterization of Rht25, a locus on chromosome arm 6AS affecting wheat plant height, heading time, and spike development. Theoretical and Applied Genetics 131, 2021–2035. [DOI] [PubMed] [Google Scholar]
- Pearce S, Saville R, Vaughan SP, et al. 2011. Molecular characterization of Rht-1 dwarfing genes in hexaploid wheat. Plant Physiology 157, 1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, et al. 1999. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Ramsay L, Comadran J, Druka A, et al. 2011. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nature Genetics 43, 169–172. [DOI] [PubMed] [Google Scholar]
- Rebetzke GJ, Richards RA, Fischer VM, Mickelson BJ. 1999. Breeding long coleoptile, reduced height wheats. Euphytica 106, 159–168. [Google Scholar]
- Reynolds M, Foulkes MJ, Slafer GA, Berry P, Parry MA, Snape JW, Angus WJ. 2009. Raising yield potential in wheat. Journal of Experimental Botany 60, 1899–1918. [DOI] [PubMed] [Google Scholar]
- Sakuma S, Golan G, Guo Z, et al. 2019. Unleashing floret fertility in wheat through the mutation of a homeobox gene. Proceedings of the National Academy of Sciences, USA 116, 5182–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma S, Lundqvist U, Kakei Y, et al. 2017. Extreme suppression of lateral floret development by a single amino acid change in the VRS1 transcription factor. Plant Physiology 175, 1720–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville RJ, Gosman N, Burt CJ, Makepeace J, Steed A, Corbitt M, Chandler E, Brown JK, Boulton MI, Nicholson P. 2012. The ‘Green Revolution’ dwarfing genes play a role in disease resistance in Triticum aestivum and Hordeum vulgare. Journal of Experimental Botany 63, 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillinger WF, Donaldson E, Allan RE, Jones SS. 1998. Winter wheat seedling emergence from deep sowing depths. Agronomy Journal 90, 582–586. [Google Scholar]
- Scutari M, Howell P, Balding DJ, Mackay I. 2014. Multiple quantitative trait analysis using Bayesian networks. Genetics 198, 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale M, Bennett T, Leyser O. 2017. BRC1 expression regulates bud activation potential but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development 144, 1661–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Mislata A, Bencivenga S, Bush M, Schiessl K, Boden S, Sablowski R. 2017. DELLA genes restrict inflorescence meristem function independently of plant height. Nature Plants 3, 749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zhang Y, Ge D, Wang Z, Song W, Gu R, Che G, Cheng Z, Liu R, Zhang X. 2019. CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. Proceedings of the National Academy of Sciences, USA 116, 17105–17114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM. 2002. Semidwarf (sd-1), ‘green revolution’ rice, contains a defective gibberellin 20-oxidase gene. Proceedings of the National Academy of Sciences, USA 99, 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C. 2003. The OsTB1 gene negatively regulates lateral branching in rice. The Plant Journal 33, 513–520. [DOI] [PubMed] [Google Scholar]
- Van De Velde K, Chandler PM, Van Der Straeten D, Rohde A. 2017. Differential coupling of gibberellin responses by Rht-B1c suppressor alleles and Rht-B1b in wheat highlights a unique role for the DELLA N-terminus in dormancy. Journal of Experimental Botany 68, 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Clark RM, Vaughn L, Sánchez-Gonzalez Jde J, Yu J, Yandell BS, Bradbury P, Doebley J. 2007. Major regulatory genes in maize contribute to standing variation in teosinte (Zea mays ssp. parviglumis). Genetics 177, 2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Kong X, Wan J, et al. 2011. Dominant and pleiotropic effects of a GAI gene in wheat results from a lack of interaction between DELLA and GID1. Plant Physiology 157, 2120–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jia Q, Zhou G, Zhang XQ, Angessa T, Broughton S, Yan G, Zhang W, Li C. 2017. Characterization of the sdw1 semi-dwarf gene in barley. BMC Plant Biology 17, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssefian S, Kirby EJM, Gale MD. 1992a Pleiotropic effects of the GA-insensitive Rht dwarfing genes in wheat. 1. Effects on development of the ear, steam and leaves. Field Crops Research 28, 179–190. [Google Scholar]
- Youssefian S, Kirby EJM, Gale MD. 1992b Pleiotropic effects of the GA-insensitive Rht dwarfing genes in wheat. 2. Effects on leaf, stem, ear and floret growth. Field Crops Research 28, 191–210. [Google Scholar]
- Zwirek M, Waugh R, McKim SM. 2019. Interaction between row-type genes in barley controls meristem determinacy and reveals novel routes to improved grain. New phytologist 221, 1950–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.