Abstract
Polycystic ovary syndrome (PCOS) is a multifactorial disorder characterized by irregular menstrual problems, hyperandrogenism, and presence of polycystic ovaries. Till date, molecular mechanism underlying PCOS remains elusive. Recently mitochondrial displacement loop (D-loop) variants have been identified to be novel players in the pathogenesis of PCOS. At present, rare variants, besides common variants, are also the focus of research as it is believed to make essential contribution to the risk of complex diseases. However, rare and low hetroplasmic variants in mitochondrial D-loop are still not investigated in PCOS women. Furthermore, variants in light-strand origin of DNA replication (OriL) of mitochondrial DNA (mtDNA) have not been explored in PCOS. Hence, in this study, we investigated rare to common mitochondrial D-loop and OriL region variants obtained using mtDNA next-generation sequencing in women with PCOS. Furthermore, we also assessed mtDNA copy number, a biomarker of mitochondrial dysfunction (MD) in women with PCOS, as the variants in mtDNA are known to be associated with low mtDNA copy number in PCOS women. A total of 67 D-loop variants including 6 novel variants were identified in 30 PCOS women. Among 67 variants, 29 variants were reported in PCOS women. A single variant, 5746A was found in OriL region in two PCOS women. Both transition and transversion variants were found but transition variants occur at very high frequency compared with transversions (82.35% vs. 17.64%, respectively). As transition variants in mtDNA are known to arise because of polymerase γ errors, occurrence of high transition rates indicates that most mutation arises because of defect in replication errors that causes mtDNA damage leading to MD. Furthermore, mtDNA copy number was found to be low in women with PCOS compared with healthy control women suggesting that MD may be the contributing factor in the pathogenesis of PCOS.
Keywords: polycystic ovary syndrome, mitochondrial DNA, next-generation sequencing, mitochondrial displacement loop (D-loop), light-strand origin of DNA replication
Introduction
Polycystic ovary syndrome (PCOS) is a complex endocrine disorder affecting 6.3–8.5% women of reproductive age and is characterized by irregular menses, hyperandrogenism (excessive hairs and hyperpigmentation), and polycystic ovaries revealed by ultrasonography (Lauritsen et al., 2014). Furthermore, it poses a substantial risk for developing number of comorbidities including metabolic syndrome, cardiovascular diseases, and type 2 diabetes in women (Cussons et al., 2007). Hormonal investigations revealed that women with PCOS have higher levels of thyroid-stimulating hormone (TSH), total testosterone, luteinizing hormone (LH)/follicle-stimulating hormone (FSH) ratio, and lower levels of FSH, thyroxin (T4), and progesterone compared with control women (Fakhoury et al., 2012; Lauritsen et al., 2014). Dyslipidemia is commonly seen in PCOS women as indicated by low high-density lipoproteins (HDL) and HDL-cholesterol (HDL-C), and increased levels of cholesterol, low-density lipoproteins, very-low-density lipoproteins, and triglycerides (TGs) (Holte et al., 1994; Wild et al., 2011).
At present, the molecular mechanism underlying PCOS remains poorly understood. Recently, oxidative stress has been considered as one of the inducing factor in the pathogenesis of PCOS (Mohammadi, 2019). The role of mitochondria, being a major site of reactive oxygen species (ROS) production and hub of metabolic activities including biosynthesis of steroid hormones (Rigotto and Basso, 2019), in PCOS pathogenesis cannot be excluded.
Mitochondria, also called “powerhouses of the cell,” generate majority of cellular energy in the form of ATP through oxidative phosphorylation (OXPHOS). During ATP generation, oxygen radical that passes through electron transport chain also generates ROS. The mitochondrial genome is a small, circular double-stranded DNA molecule of 16,569 nucleotides that encode 37 genes, 13 of which encode core structural respiratory chain complex subunits, 22 encode transfer RNAs, and 2 encode ribosomal RNAs (Tuppen et al., 2010).
Mitochondrial DNA (mtDNA) that contains one noncoding region (NCR) is a control region of ∼1 kb sequence and contains origin of replication for the H-strand (OriH). At a distant of NCR and downstream of OriH, mtDNA harbors origin of replication for the L-strand (OriL), a ∼30 nt sequence. NCR also contains one transcription promoter for each strand referred as light-strand promoter and heavy-strand promoter (Yasukawa and Kang, 2018). As most of all replication events are terminated early after ∼650 nt at termination-associated sequences located at the end of NCR, the resulting short DNA fragment remains bound to the parental L-strand, whereas the parental H-strand is displaced leading to form a displacement loop (D-loop).
D-loop contains highly conserved sequences and harbors promoters for nuclear encoded transcription factors responsible for mtDNA replication, transcription, and translation. mtDNA replication is accomplished with unique set of enzymes such as polymerase gamma (POLG) encoded by Pol γ gene, Twinkle helicase, and the mitochondrial single-stranded DNA binding protein (mtSSB) (Rusecka et al., 2018). After initiation of mtDNA replication at OriH, replication machinery synthesized two-thirds of leading-strand DNA until it reaches OriL, which initiates the synthesis of L-strand DNA in the opposite direction. After replication initiation at OriL, synthesis of both leading and lagging-strand DNA proceeds continuously, until each strand is replicated into daughter strands (Fuste et al., 2010).
Variants in the mtDNA D-loop and in the origin of replication may affect DNA replication and transcription of mtDNA proteins, and may influence the mtDNA heteroplasmy. Because of the failure of genome-wide association studies to fully explain the genetic variance for PCOS and as most common mtDNA mutations also failed to show any association with the risk of causing PCOS owing to homoplasmic nature of the variants, it is plausible that there may be some hidden rare variants that can together largely contribute to the disease cause. Hence, identification of rare variants contributing to pathogenesis of PCOS is indeed very important.
Till date, the reports on mitochondrial D-loop variants in PCOS women are very scanty and that too detected by traditional Sanger sequencing methods that greatly limits the detection of low hetroplasmic variants and rare variants owing to its low sensitivity. Therefore, such variants in mitochondrial D-loop remain unidentified. Furthermore, there is no report of variant in OriL region in PCOS women. Next-generation sequencing (NGS) is an excellent approach that can identify both rare, low hetroplasmic and common variants. Variants in the D-loop has been found to be associated with low mtDNA copy number as it is the regulatory region and impacts mitochondrial dynamics (Li et al., 2019; Reddy et al., 2019). Furthermore, these variants may cause mtDNA damage in existing mitochondria by disrupting respiratory chain and inducing ROS leading to dysfunctional mitochondria (Li et al., 2019). These damaged mitochondria get eliminated through a process called “mitophagy” in the cell leading to low mitochondrial copy number (Li et al., 2019). Reduced mtDNA content indicates the involvement of mitochondrial dysfunction (MD) and has been reported in women with PCOS by few groups (Lee et al., 2011; Reddy et al., 2019).
In this study, we analyzed variants in mtDNA D-loop and OriL region in women with PCOS obtained using NGS approach. We also quantitated mtDNA copy number, an indirect measurement of MD in PCOS group and healthy control group women to understand the pathogenesis of PCOS in a better way.
Methodology
Participants
Thirty women with PCOS, 15–40 years of age were recruited from the Infertility Clinic of the National Institute for Research in Reproductive Health (NIRRH), Mumbai, India. In addition, 30 age-matched, regularly menstruating women with no clinical or biochemical signs of hyperandrogenism and having normal ovaries on ultrasound were also recruited. Diagnosis of PCOS was carried out according to Rotterdam 2003 criteria, which includes presence of at least two of the following three features: (i) oligomenorrhea and/or anovulation, (ii) clinical and/or biochemical signs of hyperandrogenemia, and (iii) polycystic ovaries on ultrasound (Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group, 2004). Exclusion criteria were women with other related disorders such as congenital adrenal hyperplasia, Cushing syndrome, thyroid dysfunction, androgen-secreting tumors, and hyperprolactinemia or women taking any medication known to effect reproductive physiology for at least 3 months before entering the study.
Ethical approval was obtained from the institutional ethics committee of the NIRRH (Project No: 334/2018). Anthropometric parameters such as height and weight were measured and body mass index (BMI) was calculated. Detailed personal and family history was taken and blood sample was collected after obtaining written informed consent from all participants.
Hormonal and biochemical estimation
Five milliliters of peripheral blood was collected from all PCOS women and healthy control participants. Plasma and serum were separated from peripheral blood. Serum was stored at −20°C until further biochemical and hormonal analysis. An oral glucose tolerance test was conducted to determine the plasma glucose levels at fasting and 2 h after 75 g glucose load using the enzymatic glucose oxidase method. Fasting serum samples were used to measure the levels of hormones such as FSH, LH, TSH, and total testosterone and total T4 using commercial enzyme-linked immunosorbent assay kits following the manufacturer's protocol (Baillargeon and Carpentier, 2007; Saleh et al., 2015). Lipid profiling including cholesterol, TGs, and HDL-C was performed on an automated biochemistry analyzer using commercial kits. Indices of insulin resistance [homeostasis model assessment of insulin resistance, HOMA-IR was calculated as HOMA-IR = fasting insulin (lU/mL) × plasma glucose (mg/dL)/405] (Yamada et al., 2015).
DNA isolation
Total genomic DNA was isolated from blood using DNA isolation kit (Qiagen). DNA concentration and purity were measured using NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, MA).
NGS of mtDNA
Mitochondrial amplicons were generated using different sets of primers and the resultant amplicons of each sample were pooled based on concentration. Amplicon libraries were prepared with Sure Select QXT whole-genome library prep kit (Agilent, Santa Clara, CA) at Genotypic Technology Pvt. Ltd. (Bangalore, India). In brief, genomic DNA was quantified using Qubit and diluted to 25 ng/μL. DNA was fragmented and adapter-tagged using Sure Select QXT Enzyme mix. Purification of fragmented and adapter-tagged DNA was carried out using HighPrep PCR Beads (#AC-60050; MAGBIO). Purified DNA was amplified and indexed using PCR program (68°C for 2 min, denaturation at 98°C for 2 min), eight cycles of (98°C for 30 s, 56°C for 30 s and 72°C for 1 min). The amplified library was purified with HighPrep Beads and quantity and quality of DNA were assessed using Qubit fluorometer (Thermo Fisher Scientific) and Agilent 2200 Tapestation. Finally, sequencing was performed using the Illumina MiSeq Reagent v2 kit (500 cycles) on the Illumina MiSeq platform as per the manufacturer's instructions (Illumina).
Bioinformatical analysis
The adapter-clipped, high-quality reads were aligned to the reference mitochondrial genome sequence. The quality of Illumina paired-end raw reads was checked using FastQC. Illumina raw reads were processed using Trim Galore. Processing steps included adapters and low-quality bases trimming Trim Galore version: 0.4.4 with cutoff q30 and minimum read length of 20. The processed reads were aligned to revised Cambridge Reference Sequence (rCRS) of the Human Mitochondrial DNA (NC_012920.1 gi:251831106) with Bwa program.
Variants were analyzed using MToolBox for haplogroup prediction and variant prioritization workflow for recognizing functionally important nonsynonymous variants based on Macro Haplogroup Consensus Sequences (MHCS) pathogenicity of each mutated allele, and the nucleotide variability of each variant site (Calabrese et al., 2014). For each variant allele, additional annotations were performed from HmtDB, MITOMAP resources, dbSNP database, and their frequency among 1000 Genomes Project samples. Variants not reported in Mitochondrial 1000 genome database, MITOMAP, or dbSNP database were considered as novel. Variants with frequency ≥0.01 in 1000 genome dataset were filtered to obtain rare variants.
Quantitation of mtDNA copy number
The mtDNA content was determined by quantitative real-time PCR (qRT-PCR) using the specific primers to amplify the nuclear β-globins and mitochondrial ND1 genes using primers for nuclear β-globin gene; forward, 5′-CTGGGCATGTGGAGACAGAGAAGACT; reverse, 5′-AGGCCATCACTAAAGGCACCGAGC, mitochondrial ND1 gene; forward, 5′-GACGCCATAAAACTCTTCACCAA, reverse, 5′-AGGTTGAGGTTGACCAGGGG as carried out previously (Ding et al., 2017). qRT-PCR reactions were carried out in 12 μL of PCR mix using 1 × Absolute QPCR Mix containing SYBR Green (Thermo Fisher Scientific) in the presence of mtDNA primers and nuclear DNA primers for 45 cycles at annealing temperature of 60°C using thermocycler (Rotor-Gene Q; Qiagen). The relative mtDNA copy number using the Ct value differences to quantify mtDNA copy number relative to the β-globin gene according to the following equation: relative copy number = 2ΔCt, where ΔCt is the Ct β-globin − CtND1.
Statistical analysis
As mtDNA copy number does not show normal distribution, logarithmic transformation of original obtained values was used for relative mtDNA copy number. Values were expressed as mean ± standard deviation. Differences between the two groups, PCOS participants and control participants, were determined using Student's t-test. Value of p < 0.05 was considered significant.
Results
Participant characteristics
PCOS participant's age range was between 20 and 31 years except one participant whose age was 15 years. BMI was classified using the revised consensus guidelines for Asian Indians (Aziz et al., 2014). Based on the revised consensus guidelines for India, participants were categorized as underweight (<18.5 kg/m2), normal or lean BMI (18.5–22.9 kg/m2), overweight (23.0–24.9 kg/m2), and obese (≥25 kg/m2). Among 30 PCOS participants, 16 participants were obese, 2 were overweight, and the remaining were lean (Table 1). Five women (16.6%) had grade 1 nonalcholic fatty liver disease (NAFALD) (Table 1) identified by ultrasound. Eleven of 30 (36.6%) PCOS women had history of irregular menses in sisters of their family. Sixteen of 30 (53.3%) PCOS women had history of diabetes and 8 of 30 (26.6%) had history of cardiovascular disease in their family (Table 1).
Table 1.
Clinical Characteristics, Family History, and Mitochondrial DNA Variants in Polycystic Ovary Syndrome Participants
| S. No. | PCOS ID | Age | Obese/ overweight/ lean | NAFALD | History of irregular menses/diabetes/CVD/of high BP in the family | Haplogroups | mtDNA variants |
|---|---|---|---|---|---|---|---|
| 1 | Ps1 | 21 | Obese | No | No/Yes, diabetes in paternal uncle/No/No | R5a2 | 16330C,16524G,16266T,16356C |
| 2 | Ps2 | 28 | Obese | No | Yes, in sister/No/No/No | R8a1a1a2 | 521G |
| 3 | Ps3 | 27 | Lean | No | Yes/diabetes in paternal grandfather/father/No/No | M5a3 | 16295T,194T,150T |
| 4 | Ps4 | 29 | Obese | No | No/No/No/No | M2a1 | 16524C,16352C,447G,16270T,16172C |
| 5 | Ps5 | 24 | Overweight | No | Yes, insister/Yes, diabetes in sister (gestational) and father/No/No | M18a | 246C,16318T,194T,93G |
| 6 | Ps6 | 29 | Obese | No | Yes, in cousin sister/No/No/No | M5a2a | 16234T |
| 7 | Ps7 | 21 | Obese | No | No/Yes, diabetes in mother, grandmother, uncle, maternal aunt/Yes, in mother and uncle/No | K2a5b | 324T,280T |
| 8 | Ps8 | 31 | Lean | No | No/No/No/No | M2a1c | 335G,16352C,447G,16176T,16265G,16270T |
| 9 | Ps9 | 22 | Lean | No | Yes, in mother, cousin sister/Diabetes in uncle and maternal grandmother/Yes in uncle/No | M64 | 16081G,205C,302C,16527T |
| 10 | Ps10 | 22 | Lean | No | No/No/No/No | M2a1 | 16524C,16352C,447G,16224C,16270T,16172C |
| 11 | Ps11 | 30 | Obese | No | Yes, in sister/Yes, diabetes in father/Yes, in father/No | M18a | 16081G,205C,246C,16318T,16114T,93G |
| 12 | Ps12 | 34 | Obese | No | No/No/No/No | R5 | 205C,16524G,16526A,16304C |
| 13 | Ps13 | 31 | Obese | No | No/Yes, diabetes in mother/No/No | M52a | 16081G,205C,568G,569G,16355T,16291T |
| 14 | Ps14 | 23 | Obese | No | No/No/No/Yes, high BP in mother | U2b2 | 16352C,234G,16239T,16051G,16209C,16399G,16390T |
| 15 | Ps15 | 22 | Lean | No | No/Yes, diabetes in father/No/No | M18a | 246C,16318T,194T,93G,16192T |
| 16 | Ps16 | 23 | Lean | No | No/Yes, diabetes in maternal grandfather/No/No | M39b1 | 16304C,205C,61G,485C,153G,16304C |
| 17 | Ps17 | 24 | Obese | Yes, grade 1 fatty liver | No/No/Yes, in mother/No | R5a2b | 16524G,16527T,16266T,16309G,16356C,16325C,16304C |
| 18 | Ps18 | 22 | Obese | Yes, hepatomegaly with grade 1 fatty liver | Yes, in grandmother and two aunts/Yes, diabetes in maternal grandmother and grandfather/No/No | N1a1b1 | 16301T,250C,16391A,143A,199C |
| 19 | Ps19 | 27 | Obese | Yes, grade 1 fatty liver | No/Yes, diabetes, in paternal grandmother/Yes, in paternal grandmother/Yes, in paternal grandmother | M2a′b | 447G,337G,16518C,447G,143A,16320T |
| 20 | Ps20 | 24 | Overweight | No | No/No/No/No | W4a | 192C,119C,196C,16286T,194T,143A |
| 21 | Ps21 | 23 | Obese | No | Yes, in sister/No/No/No | U2b2 | 16353T,16352C,234G,16239T,16051G,16209C,16234T |
| 22 | Ps22 | 27 | Overweight | No | Yes, in cousin sister/Yes, diabetes in mother and maternal grandmother/Yes, in mother/No | M2a1 | 447G,16390T,16270T,342G,16308G 16081G,16389C,243G,302C |
| 23 | Ps23 | 21 | Obese | No | No/Yes, diabetes in maternal aunt/No/No | M3c_152 | 16126C,482C |
| 24 | Ps24 | 24 | Lean | No | Yes, cousin sister/Yes, diabetes in maternal grandfather/No/No | R5a2b | 16305G,16266T,16356C,16325C,16304C |
| 25 | Ps25 | 20 | Lean | No | Yes, elder sister/Yes, diabetes in maternal grandparents and paternal grandmother/Yes, in father and maternal grandparents/No | M2a′b | 447G,143A,337G,16518C,447G,16320T |
| 26 | Ps26 | 23 | Obese | Yes, grade 1 fatty liver | No/No/No/No | M5d | None |
| 27 | Ps27 | 30 | Obese | Yes, hepatomegaly with grade 1 fatty liver | No/No/No/No | W6 | 194T,16325C,16390T,16192T,16081G,16389C |
| 28 | Ps28 | 29 | Obese | No | No/No/No/Yes, in mother | R5a2 | 205C,16524G,16526A,16355T,16266T 16399G,16356C,16172C,16304C |
| 29 | Ps29 | 15 | Obese | No | Yes, two cousin sisters/Yes, diabetes in maternal grandmother, paternal grandfather/Yes, elder paternal uncle/Yes, in father, paternal grandmother, paternal uncle | M2a1 | 16524C,16352C,447G,16270T,16172C |
| 30 | Ps30 | 28 | Overweight | No | No/No/No/No | M2a1 | 16172C,16524C,16352C,447G,16270T |
BP, blood pressure; CVD, cardiovascular disease; mtDNA, mitochondrial DNA; NAFALD, nonalcoholic fatty liver disease; PCOS, polycystic ovary syndrome.
Hormonal and biochemical characteristics
All hormonal and cardiometabolic characteristics of the PCOS participants and control women are given in Table 2. Fasting, fasting insulin, testosterone, FSH, and LH/FSH ratio were found to be significantly increased in women with PCOS compared with healthy control women (p < 0.05) (Table 2). Diagnostic indicators of PCOS such as LH/FSH ratio of >2:1 or 3:1 was found in 11 of 30 (46.6%) PCOS participants. Cardiometabolic factors such as HOMA-IR, TG, and cholesterol was found to be significantly increased (p < 0.05) and HDL-C level was found to be decreased in PCOS participants compared with control group as expected (Table 2). Furthermore, HOMA-IR index with cutoff >2.9 shows that 17 of 30 (56.6%) women with PCOS were significantly insulin resistant.
Table 2.
Hormonal and Cardiometabolic Characteristics of Polycystic Ovary Syndrome Participants
| Hormonal/Biochemical parameters | PCOS (N = 30), mean ± SD | Controls (N = 30), mean ± SD | p |
|---|---|---|---|
| Fasting glucose (mg/dL) | 96.66 ± 19.40 | 84.69 ± 12.37 | 0.0092* |
| Glucose (postprandial) (mg/dL) | 111.3 ± 38.6 | 98.42 ± 15.23 | 0.1165 |
| Insulin (fasting) (mU/L) | 13.95967 ± 5.925428 | 11.26 ± 5.030 | 0.0779 |
| HOMA-IR | 3.353667 ± 1.547965 | 2.38 ± 1.308 | 0.0209* |
| Testosterone (ng/dL) | 37.06 ± 12.13 | 19.50 ± 11.587 | 0.0001* |
| Total T4 (μg/dL) | 8.53 ± 1.53 | 9.67 ± 2.53 | 0.0381* |
| TSH (μIU/mL) | 1.92 ± 0.98 | 2.36 ± 1.16 | 0.1268 |
| FSH (mIU/mL) | 5.93 ± 1.73 | 10.32 ± 8.81 | 0.0099* |
| LH (mIU/mL) | 12.12 ± 6.95 | 6.79 ± 4.194 | 0.0024* |
| LH/FSH | 2.06 ± 1.09 | 0.74 ± 0.41 | 0.0001* |
| Cholesterol (mg/dL) | 196.43 ± 45.93 | 148.44 ± 22.21 | 0.0001* |
| Triglycerides (mg/dL) | 120 ± 60.4 | 98.24 ± 47.94205 | 0.1507 |
| HDL-C (mg/dL) | 41.83 ± 8.41 | 43.15 ± 7.23 | 0.625 |
p < 0.05 indicates significance.
FSH, follicle-stimulating hormone; HDL-C, high-density lipoprotein–cholesterol; HOMA-IR, homeostasis model assessment of insulin resistance; LH, luteinizing hormone; SD, standard deviation; Total T4: total thyroxine; TSH, thyroid-stimulating hormone.
Analysis of mtDNA D-loop and OriL region variants
Novel and reported variants
A total of 67 variants in the D-loop region and 1 variant in OriL region were found in 30 PCOS women (Supplementary Table S1). Among 67 variants in D-loop region, 6 variants, 342G, 16308G, 16389C, 205C, 568G, and 569G, were found to be novel (8.95%) (Supplementary Table S1) as not reported in Mitochondrial 1000 genome database, MITOMAP, or dbSNP database. The heteroplasmy levels of the novel mutations ranges from 0.8% to 5.2%. Of 67 variants in D-loop region, 61 variants (91%) were found to be reported in dbSNP or MITOMAP database (Supplementary Table S1). Among 68 variants, 43 variants were found to be rare. One of the already reported variant, 16399G with 89.5% and 91.9% heteroplasmy levels was found to be same in two unrelated PCOS women having obese phenotype and mothers with high blood pressure. A single reported variant, 5746A was found in OriL region with heteroplasmy levels of 0.8% and 1.3% in two PCOS women, both belonging to haplogroup M2a1. This variant has been reported in MITOMAP database with GenBank frequency of 0.1%.
Transition and transversion variants
The mechanism causing mtDNA variants in PCOS is not known yet. It is believed that transition changes in mtDNA generally arise from errors incorporated owing to polymerase γ, the only DNA polymerase in human mitochondria, whereas transversion mutations arises owing to oxidative DNA damage (Ziada et al., 2019). Hence, in our study, we analyzed the frequency of transition versus transversion variants in PCOS women in D-loop and OriL region. We found that transition variants, namely A ↔ G and C↔T, were found to occur at very high frequency compared with transversions (G↔T, T↔A, and C↔G) (82.35% vs. 17.64%) (Supplementary Table S1). Among novel variants in D-loop, all six variants were transversion variants. Among reported variants in D-loop, only six variants were transversions and others were transitions. Variant 5746A, identified in OriL region, was a transition variant. Our results suggests that polymerase γ errors are the major source of mtDNA variants in PCOS and few mtDNA variants do arise from oxidative damage of mtDNA.
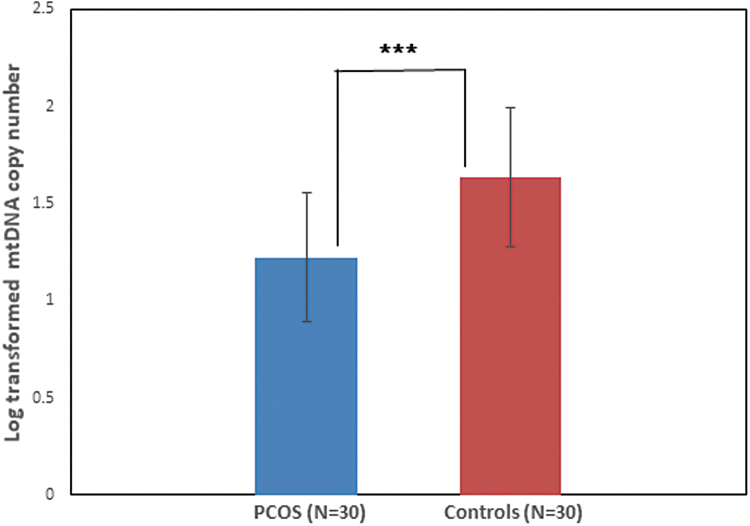
Determination of mtDNA copy number
Determination of mtDNA copy number using qRT-PCR revealed that mtDNA copy is significantly reduced in women with PCOS compared with healthy control women (p < 0.0001) (Fig. 1). The mean mtDNA copy number in PCOS group and control group was 1.22 ± 0.33 and 1.64 ± 0.35, respectively.
FIG. 1.
Analysis of mtDNA copy number using qRT-PCR revealed reduced mtDNA copy number in women with PCOS as compared with healthy control women (***p < 0.0001). mtDNA, mitochondrial DNA; PCOS, polycystic ovary syndrome; qRT-PCR, quantitative real-time PCR.
Discussion
The contribution of mtDNA variants in PCOS pathogenesis remains unclear and requires further exploration. Unlike nuclear DNA, mtDNA is more prone to damage by environmental carcinogens, as it has no protective histones and is exposed continuously to endogenous ROS arising in its vicinity (Navarro and Boveris, 2007). In this study, we analyzed variants in D-loop and OriL region of mtDNA obtained by NGS in women with PCOS. We focus on D-loop because, first, it is the mutational hotspot in the mtDNA. Second, it is the region where mtDNA replication starts. Third, because promoters for various transcriptional factors needed for the transcription of mitochondrial genes are present in D-loop. Fourth, various nuclear hormonal receptors such as glucocorticoid receptors, estrogen receptors, and thyroid (T3) hormone receptor binds to the D-loop and exert their function (Lee et al., 2008; Hunter et al., 2016). It has been demonstrated that mitochondrial protein(s) binds to putative estrogen responsive elements present in the D-loop of mtDNA (Chen and Yager, 2004). Therefore, mutations in the D-loop are likely to significantly influence the mtDNA regulation, copy number, and the heteroplasmy of mtDNA (Nicholls and Minczuk, 2014).
Recently, variants in mtDNA D-loop has been identified in PCOS women by few groups using Sanger sequencing method that has its limitations of detecting variants only >10% hetroplasmic level. However, rare variants in mitochondrial D-loop have not yet reported to the best of our knowledge. In addition, variants in OriL region, an important region having role in mtDNA replication, has not been studied in PCOS women. Investigation of rare variants is important because, although rare, these variants can be penetrating and can produce larger effect to cause complex diseases. Standard evolutionary and quantitative genetic theory suggests contribution of both rare and common variants in complex diseases. Although common variants are important to study, these variants may be responsible to establish the background liability to many complex diseases and may provide extra impetus that pushes an individual over the disease threshold (Gibson, 2012).
NGS is a high-throughput and sensitive method and has the ability to detect any DNA variants even if they are present at low level and hence can detect rare variants with low heteroplasmy levels (Duan et al., 2018). Hence, in this study we have performed NGS of mtDNA using MiSeq in 30 PCOS women and focus on D-loop variants and variants in OriL region to understand its contribution in PCOS pathogenesis. The variants obtained were investigated for the allele frequency of same variant available from mitochondrial 1000 genome database of healthy controls. We also quantitate mtDNA copy number that is a relative measure of the cellular number of mitochondria and is a biomarker of MD (Ashar et al., 2017). Reduced mitochondrial copy number has been reported by few groups in women with PCOS (Lee et al., 2011; Zhuo et al., 2012; Reddy et al., 2019).
The present study identifies 6 novel variants, 342G, 16308G, 16389C, 205C, 568G, and 569G in PCOS women and 61 reported mtDNA variants in the D-loop region. The reported variants were also found to be associated with other pathologies and disorders as revealed by MITOMAP database such as Cyclic Vomiting Syndrome, Cyclic Vomiting Syndrome with Migraine, Melanoma, Longevity/Cervical Carcinoma/human papilloma virus (HPV) infection risk, elderly Down's syndrome, mitochondrial neurogastrointestinal encephalomyopathy (MNGIE), POLG/progressive external ophthalmoplegia (PEO), pancreatic cancer, gastric carcinoma, glioblastoma, ovarian carcinoma, head/neck tumor, esophageal, breast and prostate tumors, elderly fibroblasts/leukocytes, and lung and thyroid tumors (Supplementary Table S1). Some variants occurred in more than one PCOS women, whereas some were present in single PCOS women.
Among these 61 reported variants, 29 variants were reported earlier in PCOS women (Zhuo et al., 2012; Reddy et al., 2019). Zhuo et al. (2012) carried out mtDNA mutation analysis in 57 women with PCOS and identified 16 D-loop variants. Sequencing of mitochondrial D-loop of 118 women with PCOS and 114 healthy controls of south Indian origin revealed 158 distinct sequence variants including 3 novel mutations (Reddy et al., 2019). Among 158 variants, nucleotide substitutions were most frequent followed by deletions and insertions. Among these variants, two SNPs, A189G (p = 0.018) and D310 (p = 0.042), showed statistically significant difference between PCOS participants and healthy control women. Furthermore, investigation of mtDNA copy number revealed that mtDNA copy number was significantly reduced in PCOS women carrying D310 and 189G alleles compared with noncarriers (p = 0.001 and 0.006, respectively). In addition, the D310 carriers showed significantly increased LH/FSH ratio (p = 0.026).
This suggests that mtDNA D-loop variants and copy number alterations may be inheritable risk factors for PCOS in these women. Mutation analysis of whole mitochondrial genome in a family with three matrilineal relatives exhibiting metabolic syndrome and manifesting PCOS in third generation identified six D-loop variants (Ding et al., 2018).
We also identified one variant, 5746A in OriL region in two PCOS women. OriL region is an important region for mitochondrial function as it has been observed that mutations in OriL were selected against in a mutational saturation study of mtDNA in mutator mice. Since the region is critical for mtDNA replication and maintenance, the OriL region is strongly conserved throughout vertebrates and appears to be well protected from the incidence of mutations (Wanrooij et al., 2012). Experimental evidence indicates that mutation at 5172 nucleotide position in the OriL region correlate with impaired glucose metabolism in mice and higher level of heteroplasmy correlated with reduced mtDNA copy number and altered expression of mtDNA-encoded genes (Hirose et al., 2018).
Our results that showed greater frequency of transversions compared with transition changes suggest that polymerase γ errors and not the mtDNA damage because of ROS are the major source of mtDNA variants in D-loop in PCOS women. However, we cannot rule out the potential contribution of ROS-induced oxidative damage to mtDNA fully. As Pol γ is the only replicative DNA polymerase known to exist in mammalian mitochondria, it is likely to produce these spontaneous errors. The human catalytic subunit of Pol γ has high base substitution fidelity and the intrinsic 3′→5′ exonuclease activity (Longley et al., 2001). Homozygous knock-in mtDNA mutator mouse expressing a proofreading-deficient version of Pol γ appears normal through adolescence but develops alopecia and graying, reduced subcutaneous fat, reduced fertility, anemia, cardiac hypertrophy, and other aging-related features.
Furthermore, impaired OXPHOS enzyme activities and decreased ATP production was also observed in these mice (Trifunovic et al., 2004). Incorporation of nucleotide analogs followed by proofreading failure leads to mtDNA variants and hence causing MD. A study by Ding et al. (2017) revealed that plasma 8-OHdG and malondialdehyde levels, markers of oxidative stress were higher in PCOS women with insulin resistance compared with healthy control women.
Taken together, in this study, we reported for the first time both rare and common variants in D-loop and OriL region using high-throughput sequencing of mtDNA. Reduced mtDNA copy number in PCOS women is an indicator that MD has been associated with the pathogenesis of PCOS. Despite the given association of D-loop and OriL region variants with MD, it is presently unclear whether these variants are actually the cause or effect of MD. There is a need to further elucidate the functional relevance of these variants in larger samples to understand their contribution in the pathogenesis of PCOS.
Supplementary Material
Authors' Contributions
P.S. had made substantial contributions to conceptualize and design the study, performed the experiments, analyses and interpretation of data. S.M. was involved in biochemical and hormone analysis and edited the article critically. A.P. was involved in revising and ensuring the accuracy of clinical data interpretation.
Ethical Approval
This study was approved by NIRRH ethics committee (Project No: 334/2018).
Disclosure Statement
No competing financial interests exist.
Funding Information
The authors acknowledge Indian Council of Medical Research-National Institute for Research in Reproductive Health (ICMR-NIRRH) for providing funding through Intramural NIRRH grant (RA/837/11-2019).
Supplementary Material
References
- Ashar F.N., Zhang Y., Longchamps R.J., Lane J., Moes A., Grove M.L., et al. (2017). Association of mitochondrial DNA copy number with cardiovascular disease. JAMA Cardiol 11, 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz N., Kallur S.D., and Nirmalan P.K. (2014). Implications of the revised consensus body mass indices for Asian Indians on clinical obstetric practice. J Clin Diagn Res 8, Oc01–Oc03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillargeon J.P., and Carpentier A. (2007). Role of insulin in the hyperandrogenemia of lean women with polycystic ovary syndrome and normal insulin sensitivity. Fertil Steril 884, 886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese C., Simone D., Diroma M.A., Santorsola M., Guttà C., Gasparre G., et al. (2014). MToolBox: a highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing. Bioinformatics 30, 3115–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.Q., and Yager J.D. (2004). Estrogen's effects on mitochondrial gene expression: mechanisms and potential contributions to estrogen carcinogenesis. Ann N Y Acad Sci 1028, 258–272 [DOI] [PubMed] [Google Scholar]
- Cussons A.J., Stuckey B.G., and Watts G.F. (2007). Metabolic syndrome and cardiometabolic risk in PCOS. Curr Diab Rep 7, 66–73 [DOI] [PubMed] [Google Scholar]
- Ding Y., Xia B.H., Zhang C.J., and Zhuo G.C. (2018). Mitochondrial tRNA (Leu(UUR)) C3275T, tRNA(Gln) T4363C and tRNA(Lys) A8343G mutations may be associated with PCOS and metabolic syndrome. Gene 642, 299–306 [DOI] [PubMed] [Google Scholar]
- Ding Y., Xia B.H., Zhang C.J., and Zhuo G.C. (2017). Mutations in mitochondrial tRNA genes may be related to insulin resistance in women with polycystic ovary syndrome. Am J Transl Res 9, 2984–2996; eCollection 2017. [PMC free article] [PubMed] [Google Scholar]
- Duan M., Tu J., and Lu Z. (2018). Recent advances in detecting mitochondrial DNA heteroplasmic variations. Molecules 23, pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhoury H., Tamim H., Ferwana M., Siddiqui I.A., Adham M., and Tamimi W. (2012). Age and BMI adjusted comparison of reproductive hormones in PCOS. J Family Med Prim Care 1, 132–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuste J.M., Wanrooij S., Jemt E., Granycome C.E., Cluett T.J., Shi Y., et al. (2010). Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol Cell 37, 67–78 [DOI] [PubMed] [Google Scholar]
- Gibson G. (2012). Rare and common variants: twenty arguments. Nat Rev Genet 13, 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose M., Schilf P., Gupta Y., Zarse K., Kunstner A., Fahnrich A., et al. (2018). Low-level mitochondrial heteroplasmy modulates DNA replication, glucose metabolism and lifespan in mice. Sci Rep 8, 5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holte J., Bergh T., Berne C., and Lithell H. (1994). Serum lipoprotein lipid profile in women with the polycystic ovary syndrome: relation to anthropometric, endocrine and metabolic variables. Clin Endocrinol 41, 463–471 [DOI] [PubMed] [Google Scholar]
- Hunter R.G., Seligsohn M., Rubin T.G., Griffiths B.B., Ozdemir Y., Pfaff D.W., et al. (2016). Stress and corticosteroids regulate rat hippocampal mitochondrial DNA gene expression via the glucocorticoid receptor. Proc Natl Acad Sci U S A 113, 9099–9104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritsen M.P., Bentzen J.G., Pinborg A., Loft A., Forman J.L., Thuesen L.L., et al. (2014). The prevalence of polycystic ovary syndrome in a normal population according to the Rotterdam criteria versus revised criteria including anti-Mullerian hormone. Hum Reprod 29, 791–801 [DOI] [PubMed] [Google Scholar]
- Lee J., Sharma S., Kim J., Ferrante R. J., and Ryu H. (2008). Mitochondrial nuclear receptors and transcription factors: who's minding the cell? J Neurosci Res 86, 961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H, Chung D.J., Lee H.S., Kim T.J., Kim M.H., Jeong H.J., et al. (2011). Mitochondrial DNA copy number in peripheral blood in polycystic ovary syndrome. Metab Clin Exp 60, 1677–1682 [DOI] [PubMed] [Google Scholar]
- Li H., Slone J., Fei L., and Huang T. (2019). Mitochondrial DNA variants and common diseases: a mathematical model for the diversity of age-related mtDNA mutations. Cells 8, pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley M.J., Nguyen D., Kunkel T.A., and Copeland W.C. (2001). The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. J Biol Chem 276, 38555–38562 [DOI] [PubMed] [Google Scholar]
- Mohammadi M. (2019). Oxidative stress and polycystic ovary syndrome: a brief review. Int J Prev Med 10, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., and Boveris A. (2007). The mitochondrial energy transduction system and the aging process. Am J Physiol Cell Physiol 292, C670–C686 [DOI] [PubMed] [Google Scholar]
- Nicholls T.J., and Minczuk M. (2014). In D-loop: 40 years of mitochondrial 7S DNA. Exp Gerontol 56, 175–181 [DOI] [PubMed] [Google Scholar]
- Reddy T.V., Govatati S., Deenadayal M., Sisinthy S., and Bhanoori M. (2019). Impact of mitochondrial DNA copy number and displacement loop alterations on polycystic ovary syndrome risk in south Indian women. Mitochondrion 44, 35–40 [DOI] [PubMed] [Google Scholar]
- Rigotto G., and Basso E. (2019). Mitochondrial dysfunctions: a thread sewing together Alzheimer's disease, diabetes, and obesity. Oxid Med Cell Longev 2019, 7210892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. (2004). Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril 81, 19–25 [DOI] [PubMed] [Google Scholar]
- Rusecka J., Kaliszewska M., Bartnik E., and Tonska K. (2018). Nuclear genes involved in mitochondrial diseases caused by instability of mitochondrial DNA. J Appl Genet 59, 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh B.O., Ibraheem W.F., and Ameen N.S. (2015). The role of anti-Mullerian hormone and inhibin B in the assessment of metformin therapy in women with polycystic ovarian syndrome. Saudi Med J 36, 562–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., et al. (2004). Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 [DOI] [PubMed] [Google Scholar]
- Tuppen H.A., Blakely E.L., Turnbull D.M., and Taylor R.W. (2010). Mitochondrial DNA mutations and human disease. Biochim Biophys Acta 1797, 113–128 [DOI] [PubMed] [Google Scholar]
- Wanrooij S., Miralles Fuste J., Stewart J.B., Wanrooij P.H., Samuelsson T., Larsson N.G., et al. (2012). In vivo mutagenesis reveals that OriL is essential for mitochondrial DNA replication. EMBO Rep 13, 1130–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild R.A., Rizzo M., Clifton S., and Carmina E. (2011). Lipid levels in polycystic ovary syndrome: systematic review and meta-analysis. Fertil Steril 95, 1073–1079.e1–e11. [DOI] [PubMed] [Google Scholar]
- Yamada C., Kondo M., Kishimoto N., Shibata T., Nagai Y., Imanishi T., et al. (2015). Association between insulin resistance and plasma amino acid profile in non-diabetic Japanese subjects. J Diabetes Investig 6, 408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T., and Kang D. (2018). An overview of mammalian mitochondrial DNA replication mechanisms. J Biochem 164, 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo G., Ding Y., Feng G., Yu L., and Jiang Y. (2012). Analysis of mitochondrial DNA sequence variants in patients with polycystic ovary syndrome. Arch Gynecol Obstet 286, 653–659 [DOI] [PubMed] [Google Scholar]
- Ziada A.S., Lu M.Y., Ignas-Menzies J., Paintsil E., Li M., Ogbuagu O., et al. (2019). Mitochondrial DNA somatic mutation burden and heteroplasmy are associated with chronological age, smoking, and HIV infection. Aging Cell 18, e13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.