1 Introduction
For many complex diseases, there is no “one size fits all” solutions for patients with a particular diagnosis. The proper treatment for a patient depends upon genetic, environmental, and lifestyle choices. The ability to personalize treatment in a scientifically rigorous manner based on these factors is the hallmark of the emerging “precision medicine” paradigm. Nowhere is the potential impact of precision medicine more closely felt than in cancer, where lifesaving treatments for particular patients could prove ineffective or even deadly for other patients based entirely upon the particular genetic mutations in the patient’s tumor(s). Significant effort, therefore, has been devoted to deepening the scientific research surrounding precision medicine. This includes a Precision Medicine Initiative (Collins and Varmus, 2015) launched by former President Barack Obama in 2015, now known as the All of Us Research Program.
A fundamental difficulty with putting the findings of precision medicine into practice is that–by its very nature–precision medicine creates a huge space of treatment options (Frey et al., 2016). These can easily overwhelm clinicians attempting to stay up-to-date with the latest findings, and can easily inhibit a clinician’s attempts to determine the best possible treatment for a particular patient. However, the ability to quickly locate relevant evidence is the hallmark of information retrieval (IR). Further, for three consecutive years the TREC Clinical Decision Support (CDS) track has sought to evaluate IR systems that provide medical evidence to the point-of-care. It was natural, then, to specialize the CDS track to the needs of precision medicine so IR systems can focus on this important issue.
The 2017 Precision Medicine track focused on a single field, oncology, for a specific use case, genetic mutations of cancer. As described above, main idea behind precision medicine is to use detailed patient information (largely genetic information in most current research) to identify the most effective treatments. Improving patient care in precision oncology then requires both (a) a mechanism to locate the latest research relevant to a patient, and (b) a fallback mechanism to locate the most relevant clinical trials when the latest techniques prove ineffective for a patient. In the first part, the track continues the previous Clinical Decision Support track (with a more focused use case), while in the second part expands the task to cover a new type of data (clinical trial descriptions).
The remainder of this overview is organized as follows: Section 2 describes the Clinical Decision Support tracks, including their motivation and data, and how this led to the Precision Medicine track; Section 3 describes the structure of the topics and the process of creating them; Section 4 outlines the retrieval tasks; Section 5 describes the evaluation method; finally, Section 6 details the results of the participant systems.
2 Background
The TREC Clinical Decision Support track (2014–2016) sought to evaluate systems that provided evidence-based information (in the form of full-text literature articles) to clinicians for a specific patient (represented as a case description or admission note). This included information on diagnosing, treating, and testing patients. No attempt was made to limit topics by medical speciality (e.g., cardiology, pediatrics), which in some respects made it difficult to define precise use cases and have a uniform definition of relevance. Despite this, the track was extremely successful in attracting a large and diverse group of participants (ranging from 26 to 36 participating participants in each year). The track was also heavily inspired by the TREC Genomics (Hersh and Voorhees, 2009) and Medical Records (Voorhees and Hersh, 2012) tracks, in addition to the medical case-based retrieval track of ImageCLEF (Seco de Herrera et al., 2013), all of which are no longer active. All of these tracks have demonstrated significant interest in the problem of medical ad hoc retrieval.
To address the needs of a specific, high-profile, and clinically valuable use case, the Clinical Decision Support track was transitioned to the Precision Medicine track. While the Clinical Decision Support track utilized full-text articles from PubMed Central (PMC), the Precision Medicine track utilized shorter MED-LINE abstracts. This is mainly due to PMC being a poor resource for precision medicine: a low proportion of precision medicine-related articles are deposited in PMC. Further, those articles that are deposited are often subject to a 6–24 month embargo evaluation, a significant length of time in a fast-moving field such as precision medicine. Additionally, clinical trials were added as a separate corpus, consistent with the importance of this resource in precision oncology.
3 Topics
The 2017 Precision Medicine track provided 30 topics created by experienced precision oncologists at the University of Texas MD Anderson Cancer Center and the Oregon Health & Science University (OHSU) Knight Cancer Institute. Due to the difficulty in obtaining actual patient data, the topics were synthetically created, though often inspired by actual patients, with modification.1
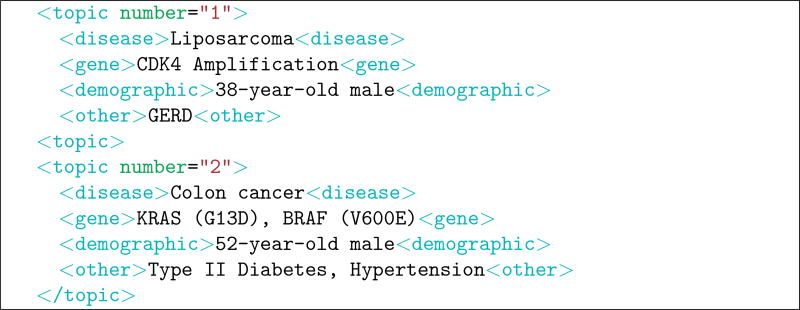
The topics contain four key elements in a semi-structured format to reduce the need to perform natural language processing to identify the key elements. The four key elements are: (1) disease (e.g., type of cancer), (2) genetic variants (primarily the genetic variants in the tumors themselves as opposed to the patient’s DNA), (3) demographic information (e.g., age, sex), and (4) other factors (which could impact certain treatment options). Four topics from the track are shown in Table 1. The first two topics are additionally shown in their corresponding XML format (i.e., what was provided to the participants) in Table 2.
Table 1.
Example topics from the 2017 track.
| Disease: Liposarcoma |
| Variant: CDK4 Amplification |
| Demographic: 38-year-old male |
| Other: GERD |
|
|
| Disease: Colon Cancer |
| Variant: KRAS (G13D), BRAF (V600E) |
| Demographic: 52-year-old male |
| Other: Type II Diabetes, Hypertension |
|
|
| Disease: Cervical Cancer |
| Variant: STK11 |
| Demographic: 26-year-old female |
| Other: None |
|
|
| Disease: Cholangiocarcinoma |
| Variant: IDH1 (R132H) |
| Demographic: 64-year-old male |
| Other: Neuropathy |
Table 2.
XML format for the first two topics from Table 1.
4 Tasks
In the Clinical Decision Support track, three types of topics were utilized: diagnosis, treatment, and test. For the Precision Medicine track, only treatment topics were used. However, different types of data may be of interest, namely literature article and clinical trials. In more detail, the two types of results are:
Literature Articles. Because precision medicine is a fast-moving field, keeping up-to-date with the latest literature can be challenging due to both the volume and velocity of scientific advances. Therefore, when treating patients, it would be helpful to present the most relevant scientific articles for an individual patient. The primary literature corpus is therefore a snapshot of MEDLINE abstracts (i.e., what is searchable through the PubMed interface). Relevant literature articles can guide precision oncologists to the best-known treatment options for the patient’s condition. Specifically, this corpus is composed of approximately 26,759,399 MEDLINE abstracts and is supplemented with two additional sets of abstracts: (i) 37,007 abstracts from recent proceedings of the American Society of Clinical Oncology (ASCO), and (ii) 33,018 abstracts from recent proceedings of the American Association for Cancer Research (AACR). These additional datasets were added to increase the set of potentially relevant treatment information. Notably, the latest research is often presented at conferences such as ASCO and AACR prior to submission to journals (thus these proceedings may represent a more up-to-date snapshot of scientific knowledge than MEDLINE).
Clinical Trials. In many oncology patients, no approved treatment is available (or, commonly, none of the available treatments have proven effective). The common recourse in this case is to determine if any potential treatments are undergoing evaluation in a clinical trial. Therefore, in such situations, it would be helpful to automatically identify the most relevant clinical trials for an individual patient. Precision oncology trials typically use a certain treatment (e.g., a form of chemotherapy or radiation) for a certain disease with a specific genetic variant (or set of variants). Such trials can have complex inclusion and/or exclusion criteria that are challenging to match with automated systems (Weng et al., 2011). The corpus is derived from ClinicalTrials.gov, a repository of past, present, and future clinical trials in the U.S. and abroad. A total of 241,006 clinical trial descriptions compose the corpus provided to participants. Note that for the purposes of this track, the state of the trial (e.g., recruiting, active, completed) and geographic location constraints are not considered.
5 Evaluation
The evaluation followed standard TREC evaluation procedures for ad hoc retrieval tasks. Participants submitted (in trec_eval format) a maximum of five automatic or manual runs per task, each consisting of a ranked list of up to 1,000 literature article IDs and 1,000 ClinicalTrials.gov Identifiers per topic. That is, up to 10 total runs: a maximum of 5 literature runs and 5 clinical trial runs per topic.
The highest ranked articles and trials for each topic were pooled and judged by physician graduate students at OHSU and postdoctoral fellows at the National Library of Medicine (NLM), just as in the Medical Records and Clinical Decision Support tracks.
In the previous years of the TREC Clinical Decision Support Track, relevance assessors judged results on a simple scale: “definitely relevant”, “partially relevant”, and “not relevant”. Due to the particular challenges involved in precision medicine, however, this is not necessarily appropriate. Not only is precision medicine a highly specialized field (and thus difficult to get true experts to act as assessors), but the notion of relevance is far more flexible and case-specific. As such, the assessment process was two-tiered: first a manual assessment was made by the human assessors based on several categories for each result (referred to here as Result Assessment), then a relevance score was assigned to the result based on its categorization (referred to here as Relevance Assessment).
5.1 Result Assessment
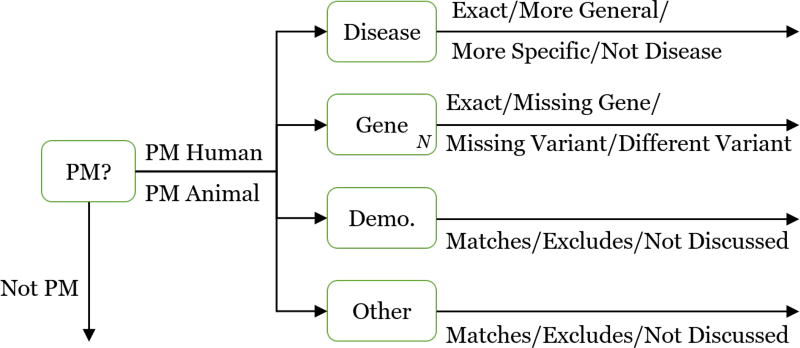
Result assessment can be viewed as a set of multi-class annotations. Judging an individual result, whether an article or trial, proceeds in a cascaded manner with two steps: an initial pass ensures the article/trial is broadly relevant to precision medicine, after which the assessor categorizes the article/trial according to the four fields above.
See Figure 1 for a flow chart style overview of this process. The first step is designed to save assessor time by filtering out unrelated articles/trials, since the second step can be more time-consuming (possibly requiring a more detailed reading of the article/trial). The assessors were free to quickly skim the article/trial in order to make the initial decision. Then, if the article/trial is relevant to precision medicine (by the standard outlined below), a more detailed reading may be necessary in order to accurately assess all fields.
Figure 1.
Two-step result assessment process
Step 1 is to determine whether the article/trial is related to precision medicine. There are three options:
Human PM: The article/trial (1) relates to humans, (2) involves some form of cancer, (3) focuses on treatment, prevention, or prognosis of cancer, and (4) relates in some way to at least one of the genes in the topic.
Animal PM: Identical to Human PM requirements (2)–(4), except for animal research.
Not PM: Everything else. This includes “basic science” that focuses on understanding underlying genomic principles (e.g., pathways), but provides no evidence for treatment.
Step 2 is to determine the appropriate categorization for each of the four fields:
- Disease:
- Exact: The form of cancer in the article/trial is identical to the one in the topic.
- More General: The form of cancer in the article/trial is more general than the one in the topic (e.g., blood cancer vs. leukemia).
- More Specific: The form of cancer in the article/trial is more specific than the one in the topic (e.g., squamous cell lung carcinoma vs. lung cancer).
- Not Disease: The article/trial is not about a disease, or is about a different disease (or type of cancer) than the one in the topic.
- Gene [for each particular gene in the topic]
- Exact: The article/trial focuses on the exact gene and variant as the one in the topic. If the topic does not contain a specific variant, then this holds as long as the gene is included. By “focus” this means the gene/variant needs to be part of the scientific experiment of the article/trial, as opposed to discussing related work.
- Missing Gene: The article/trial does not focus the particular gene in the topic. If the gene is referenced but not part of the study, then it is considered missing.
- Missing Variant: The article/trial focuses on the particular gene in the topic, but not the particular variant in the topic. If no variant is provided in the topic, this category should not be assigned.
- Different Variant: The article/trial focuses on the particular gene in the topic, but on a different variant than the one in the topic.
- Demographic
- Matches: The article/trial demographic population matches the one in the topic.
- Excludes: The article/trial demographic population specifically excludes the one in the topic.
- Not Discussed: The article/trial does not discuss a particular demographic population.
- Other
- Matches: The article/trial population matches the one in the topic. If the other field is “None” this category should also be assigned.
- Excludes: The article/trial population specifically excludes the one in the topic.
- Not Discussed: The article/trial does not discuss a population relating to the provided factors.
5.2 Relevance Assessment
Relevance assessment is defined here as the process of mapping the multi-class result assessments described above onto a single numeric relevance scale. This allows for the computation of evaluation metrics (e.g., P@10, infNDCG) as well as the tuning of IR systems to improve their search ranking. As already demonstrated by the need for result assessment above, for the Precision Medicine track the notion of relevance assessment becomes more complex than previous tracks.
One of the factors that makes precision medicine a difficult domain for IR is that different patient cases require different types of flexibility on the above categories. For some patients, the exact type of cancer is not relevant. Other times, the patient’s demographics or other factors might weigh more heavily. Most notably, the very concept of precision medicine acknowledges the uniqueness of the patient, and so it is to be expected that no perfect match is found. Not only do the topics provided to the participants not contain the necessary information to decide what factors are more/less relevant (e.g., the patient’s previous treatments), in many ways it isn’t realistic to assign the IR system this responsibility. Precision medicine requires a significant amount of oversight by clinicians, including the ability to consider multiple treatment options. So it might ultimately make the most sense to allow the relevance assessment to be, at least in part, designed by the clinician to allow the IR system to adjust its rankings to suit. Given the constraints of an IR shared task, however, it is necessary to define a relevance assessment process. As such, a fairly broad notion of relevance based on the above categories was used:
Definitely Relevant: The result should: be either Human PM or Animal PM; have a Disease assignment of Exact or More Specific; have at least one Gene is Exact; have both Demographic and Other assignments are either Exact or Not Discussed.
Partially Relevant: Largely the same as Definitely Relevant, but with the exception that Disease can also be More General and Gene can also be Missing Variant or Different Variant.
Not Relevant: Neither of the above.
The primary evaluation metrics for the literature articles are precision at rank 10 (P@10), inferred normalized discounted cumulative gain (infNDCG), and R-precision (R-prec). For infNDCG, Definitely Relevant has a score of 2, Partially Relevant is 1, and Not Relevant is 0. The primary evaluation metrics for clincial trials is P@5, P@10, and P@15.
6 Results
In total, there were 22,642 judgments for the literature articles and 13,441 judgments for the clinical trials. Table 3 shows basic statistics of the results and relevance assessments. Table 4 shows the number of Definitely Relevant, Partially Relevant, and Not Relevant judgments for each topic. Since each result was judged only once, no inter-rater agreement is available for the judgments. However, the PM assessment (Human, Animal, or Not PM) is independent of the topic, and thus some agreement calculation can be made when the same article/trial is judged for different topics. A Kappa agreement would be difficult to calculate and of limited utility due to the number of assessors (20 assessors, who judged between 1 and 6 topics) and inconsisent rates of duplicate judging (most duplicate judging involved just 2 topics, but two clinical trials were judged in 18 topics). Basic agreement numbers can be calculated, which work out to 84.5% agreement for literature articles and 85.8% agreement for clinical trials. These are by no means desirable agreement numbers (the baseline is effectively 50%), which underscores both the difficulty of assessment as well as the vague description of ‘precision medicine’. More analysis is certainly required as to why such disagreements arise and how to improve similar types of judgments on future tasks.
Table 3.
Descriptive statistics (per-topic) of manual judgments (both results assessment and relevance assessment) for both literature articles and clinical trials.
| Literature Articles | Clinical Trials | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | Class | Total | Mean | Median | Min | Max | Total | Mean | Median | Min | Max |
| PM | Human PM | 8,738 | 291 | 277 | 65 | 627 | 3,959 | 132 | 119 | 18 | 428 |
| Animal PM | 536 | 18 | 17 | 2 | 71 | 2 | 0 | 0 | 0 | 1 | |
| Not PM | 13,368 | 446 | 435 | 92 | 881 | 9,480 | 316 | 314 | 80 | 565 | |
|
| |||||||||||
| Disease | Exact | 4,149 | 138 | 120 | 9 | 506 | 1,093 | 36 | 26 | 0 | 139 |
| More Specific | 1,273 | 42 | 20 | 0 | 358 | 723 | 24 | 6 | 0 | 249 | |
| More General | 938 | 31 | 18 | 0 | 139 | 679 | 23 | 17 | 0 | 92 | |
| Not Disease | 2,914 | 97 | 85 | 0 | 275 | 1,466 | 49 | 41 | 0 | 179 | |
|
| |||||||||||
| 1st Gene | Exact | 4,421 | 147 | 154 | 10 | 331 | 1,486 | 50 | 37 | 0 | 230 |
| Missing Variant | 1,419 | 47 | 3 | 0 | 464 | 452 | 15 | 5 | 0 | 108 | |
| Different Variant | 560 | 19 | 10 | 0 | 110 | 243 | 8 | 2 | 0 | 91 | |
| Missing Gene | 2,874 | 96 | 60 | 0 | 378 | 1,780 | 59 | 35 | 0 | 391 | |
|
| |||||||||||
| 2nd Gene | Exact | 540 | 18 | 0 | 0 | 287 | 119 | 4 | 0 | 0 | 55 |
| Missing Variant | 230 | 8 | 0 | 0 | 218 | 127 | 4 | 0 | 0 | 121 | |
| Different Variant | 91 | 3 | 0 | 0 | 83 | 17 | 1 | 0 | 0 | 14 | |
| Missing Gene | 964 | 32 | 0 | 0 | 264 | 579 | 19 | 0 | 0 | 170 | |
|
| |||||||||||
| 3rd Gene | Exact | 104 | 3 | 0 | 0 | 104 | 47 | 2 | 0 | 0 | 47 |
| Missing Variant | 6 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | |
| Different Variant | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | |
| Missing Gene | 136 | 5 | 0 | 0 | 136 | 124 | 4 | 0 | 0 | 124 | |
|
| |||||||||||
| Demographics | Matches | 658 | 22 | 10 | 0 | 91 | 3,221 | 107 | 105 | 0 | 402 |
| Not Discussed | 7,736 | 258 | 226 | 37 | 578 | 376 | 13 | 1 | 0 | 197 | |
| Excludes | 880 | 29 | 17 | 0 | 141 | 364 | 12 | 3 | 0 | 57 | |
|
| |||||||||||
| Other | Matches | 789 | 26 | 2 | 0 | 220 | 1,114 | 37 | 3 | 0 | 208 |
| Not Discussed | 8,320 | 277 | 283 | 0 | 646 | 2,736 | 91 | 81 | 0 | 425 | |
| Excludes | 165 | 6 | 1 | 0 | 45 | 111 | 4 | 0 | 0 | 63 | |
|
| |||||||||||
| Relevance | Definitely Relevant | 2,022 | 67 | 44 | 1 | 221 | 436 | 15 | 5 | 0 | 98 |
| Partially Relevant | 1,853 | 62 | 27 | 1 | 476 | 735 | 25 | 15 | 0 | 120 | |
| Not Relevant | 18,767 | 626 | 624 | 259 | 1,209 | 12,270 | 409 | 418 | 165 | 606 | |
Note: only 6 topics had a 2nd Gene and only 1 had a 3rd Gene, but means are still provided across 30 topics.
Table 4.
Counts of Definitely Relevant (DR), Partially Relevant (PR), and Not Relevant (NR) results for each topic.
| literature articles | clinical trials | |||||
|---|---|---|---|---|---|---|
| Topic | DR | PR | NR | DR | PR | NR |
| 1 | 48 | 14 | 377 | 5 | 12 | 313 |
| 2 | 156 | 205 | 330 | 17 | 120 | 323 |
| 3 | 45 | 6 | 535 | 5 | 19 | 221 |
| 4 | 221 | 46 | 625 | 31 | 26 | 519 |
| 5 | 69 | 21 | 677 | 35 | 1 | 375 |
|
| ||||||
| 6 | 24 | 104 | 486 | 0 | 27 | 345 |
| 7 | 159 | 187 | 623 | 98 | 107 | 333 |
| 8 | 119 | 1 | 563 | 54 | 7 | 418 |
| 9 | 14 | 476 | 259 | 2 | 60 | 165 |
| 10 | 3 | 94 | 614 | 0 | 0 | 422 |
|
| ||||||
| 11 | 39 | 3 | 656 | 4 | 15 | 489 |
| 12 | 165 | 55 | 448 | 25 | 14 | 382 |
| 13 | 1 | 24 | 935 | 2 | 32 | 369 |
| 14 | 1 | 30 | 568 | 0 | 7 | 310 |
| 15 | 4 | 6 | 738 | 3 | 1 | 506 |
|
| ||||||
| 16 | 116 | 26 | 622 | 2 | 3 | 508 |
| 17 | 111 | 5 | 620 | 10 | 23 | 472 |
| 18 | 43 | 151 | 639 | 3 | 35 | 543 |
| 19 | 13 | 22 | 802 | 7 | 16 | 506 |
| 20 | 21 | 28 | 398 | 0 | 5 | 263 |
|
| ||||||
| 21 | 84 | 120 | 409 | 11 | 56 | 289 |
| 22 | 131 | 11 | 818 | 48 | 54 | 508 |
| 23 | 178 | 17 | 609 | 21 | 9 | 519 |
| 24 | 22 | 42 | 761 | 4 | 14 | 525 |
| 25 | 15 | 39 | 741 | 1 | 39 | 409 |
|
| ||||||
| 26 | 14 | 6 | 1029 | 1 | 4 | 606 |
| 27 | 72 | 4 | 771 | 11 | 17 | 433 |
| 28 | 9 | 46 | 735 | 0 | 2 | 332 |
| 29 | 19 | 23 | 665 | 2 | 6 | 418 |
| 30 | 106 | 41 | 714 | 34 | 4 | 449 |
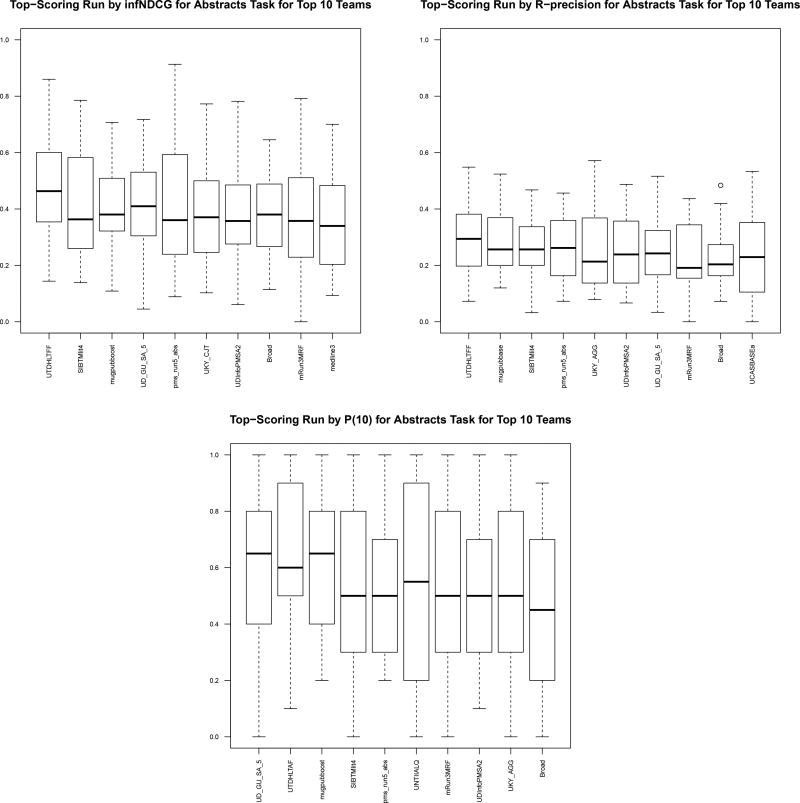
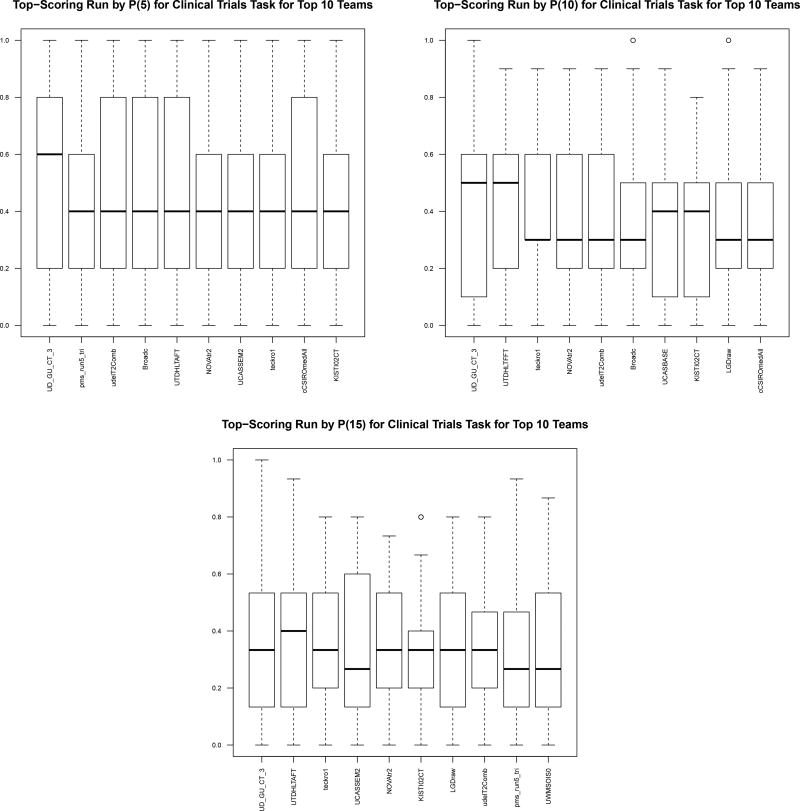
There were a total of 32 participants in the track. For the literature articles, 29 participants submitted 125 runs (122 automatic, 3 manual). For the clinical trials, 31 participants submitted 133 runs (131 automatic, 2 manual). See Table 5 for a list of the participants and numbers of runs. Table 6 shows the top 10 runs (top run per participant) for each metric on each corpus. Figures 2 and 3 show box-and-whisker plots for the top 10 runs. Finally, Tables 7 and 8 show the per-topic aggregate results.
Table 5.
Participating teams and submitted runs. Numbers in parentheses indicate manual runs.
| # Runs | |||
|---|---|---|---|
| Team ID | Affiliation | Articles | Trials |
| BiTeM | BiTeM Group | 5 | 5 |
| cbnu | Chonbuk National University | 3 | 3 |
| CSIROmed | Commonwealth Science and Industry Research Org. | 5 | 5 |
| DA_IICT | Dhirubhai Ambani Inst. of Info. and Comm. Tech. | - | 4 |
|
| |||
| DUTIRL | Information Retrieval Laboratory of Dalian Univ. of Tech. | 1 | 1 |
| ECNUica | East China Normal University | 5 | 5 |
| ETH | ETH Zurich | 5 | 5 |
| FDUDMIIP | School of Computer Science, Fudan University | 5 | 4 |
|
| |||
| GravityWave | GravityWave Technologies | 1 (1) | 1 (1) |
| HokieGo | Virginia Tech | 2 | 2 |
| ielab-CSIRO-QUT | CSIRO and Queensland University of Technology | 5 | - |
| imi_mug | Medical University of Graz | 5 | 5 |
|
| |||
| iris | University of Pittsburgh | 5 | 5 |
| kaist-kse | KAIST Knowledge Service Engineering | 3 | 3 |
| KISTI | Korea Institute of Science and Technology Information | 5 | 5 |
| MayoNLPTeam | Mayo Clinic | 5 | 5 |
|
| |||
| NaCTeM | University of Manchester | 5 (1) | 5 (1) |
| NOVASearch | Universidade NOVA Lisboa | 3 | 5 |
| POZNAN_SEMMED | Poznan University of Technology | 3 | 5 |
| prna-mit-suny | Philips Research North America / MIT / SUNY | 5 | 5 |
|
| |||
| SDSFU | School of Data Science, Fudan University | 5 | 5 |
| teckro | teckro | - | 5 |
| TREC_UB | University at Buffalo | - | 2 |
| UCAS | University of Chinese Academy of Sciences | 5 | 5 |
|
| |||
| udel | University of Delaware | 5 | 5 |
| udel_fang | Infolab at University of Delaware | 5 | 5 |
| UD_GU_BioTM | University of Delaware / Georgetown University | 5 | 5 |
| UKNLP | University of Kentucky | 5 (1) | 4 |
|
| |||
| UMich_MedIER | University of Michigan | 4 | 4 |
| UNTIIA | University of North Texas | 5 | 5 |
| UTDHLTRI | University of Texas at Dallas | 5 | 5 |
| UWMSOIS | University of Wisconsin-Milwaukee | 5 | 5 |
|
| |||
| Total | 125 (3) | 133 (2) | |
Table 6.
Top overall systems (best run per participant).
| Literature Articles | Clinical Trials | ||||
|---|---|---|---|---|---|
| infNDCG | P @ 5 | ||||
| Team | Run | Score | Team | Run | Score |
|
|
|
||||
| UTDHLTRI | UTDHLTFF | 0.4647 | UD_GU_BioTM | UD_GU_CT_3 | 0.5448 |
| BiTeM | SIBTMlit4 | 0.4175 | prna-mit-suny | pms_run5_tri | 0.4552 |
| imi_mug | mugpubboost | 0.4158 | udel | udelT2GeMeSH | 0.4552 |
| UD_GU_BioTM | UD_GU_SA_5 | 0.4135 | UTDHLTRI | UTDHLTAFT | 0.4483 |
| prna-mit-suny | pms_run5_abs | 0.4070 | NaCTeM | Broadc | 0.4483 |
| UKNLP | UKY_CJT | 0.3897 | NOVASearch | NOVAtr2 | 0.4414 |
| udel_fang | UDInfoPMSA2 | 0.3897 | UCAS | UCASSEM2 | 0.4345 |
| NaCTeM | Broad | 0.3800 | teckro | teckro1 | 0.4276 |
| iris | mRun3MRF | 0.3758 | CSIROmed | cCSIROmedAll | 0.4183 |
| FDUDMIIP | medline3 | 0.3555 | KISTI | KISTI02CT | 0.4000 |
|
|
|
||||
| R-prec | P @ 10 | ||||
| Team | Run | Score | Team | Run | Score |
|
|
|
||||
| UTDHLTRI | UTDHLTFF | 0.2993 | UD_GU_BioTM | UD_GU_CT_3 | 0.4448 |
| imi_mug | mugpubbase | 0.2772 | UTDHLTRI | UTDHLTFFT | 0.4172 |
| BiTeM | SIBTMlit4 | 0.2687 | teckro | teckro1 | 0.4000 |
| prna-mit-suny | pms_run5_abs | 0.2622 | NOVASearch | NOVAtr2 | 0.3966 |
| UKNLP | UKY_AGG | 0.2518 | udel | udelT2Comb | 0.3793 |
| udel_fang | UDInfoPMSA2 | 0.2503 | UCAS | UCASSEM2 | 0.3724 |
| UD_GU_BioTM | UD_GU_SA_5 | 0.2477 | NaCTeM | Broadc | 0.3724 |
| iris | mRun3MRF | 0.2374 | KISTI | KISTI02CT | 0.3690 |
| NaCTeM | Broad | 0.2287 | POZNAN_SEMMED | LGDraw | 0.3690 |
| UCAS | UCASBASEa | 0.2282 | BiTeM | SIBTct3 | 0.3586 |
|
|
|
||||
| P @ 10 | P @ 15 | ||||
| Team | Run | Score | Team | Run | Score |
|
|
|
||||
| UD_GU_BioTM | UD_GU_SA_5 | 0.6400 | UD_GU_BioTM | UD_GU_CT_3 | 0.3885 |
| UTDHLTRI | UTDHLTAF | 0.6300 | UTDHLTRI | UTDHLTAFT | 0.3816 |
| imi_mug | mugpubboost | 0.6267 | teckro | teckro1 | 0.3632 |
| BiTeM | SIBTMlit4 | 0.5500 | UCAS | UCASSEM2 | 0.3471 |
| prna-mit-suny | pms_run5_abs | 0.5300 | NOVASearch | NOVAtr2 | 0.3448 |
| UNTIIA | UNTIIALQ | 0.5233 | POZNAN_SEMMED | LGDraw | 0.3356 |
| iris | mRun3MRF | 0.5133 | KISTI | KISTI02CT | 0.3356 |
| udel_fang | UDInfoPMSA2 | 0.5067 | udel | udelT2Comb | 0.3333 |
| UKNLP | UKY_AGG | 0.4933 | UWMSOIS | UWMSOIS0 | 0.3172 |
| NaCTeM | Broad | 0.4667 | prna-mit-suny | pms run5 tri | 0.3172 |
|
|
|
||||
Figure 2.
Top-performing runs (showing only best run per participant) on literature articles.
Figure 3.
Top-performing runs (showing only best run per participant) on clinical trials.
Table 7.
Per-topic statistics (over 125 runs) for 29 topics on literature articles.
| infNDCG | P @ 10 | R-prec | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Topic | Best | Median | Worst | Best | Median | Worst | Best | Median | Worst |
| 1 | 0.6068 | 0.4604 | 0.0000 | 1.0000 | 0.6000 | 0.0000 | 0.5000 | 0.3387 | 0.0000 |
| 2 | 0.8794 | 0.5978 | 0.0000 | 1.0000 | 0.9000 | 0.0000 | 0.4404 | 0.2742 | 0.0000 |
| 3 | 0.5130 | 0.2789 | 0.0000 | 0.9000 | 0.3000 | 0.0000 | 0.4118 | 0.1961 | 0.0000 |
| 4 | 0.8268 | 0.4085 | 0.0000 | 1.0000 | 0.7000 | 0.0000 | 0.4157 | 0.2097 | 0.0000 |
| 5 | 0.3011 | 0.1607 | 0.0000 | 0.5000 | 0.2000 | 0.0000 | 0.2111 | 0.1222 | 0.0000 |
| 6 | 0.5652 | 0.4208 | 0.0000 | 0.9000 | 0.6000 | 0.0000 | 0.4219 | 0.3047 | 0.0000 |
| 7 | 0.7042 | 0.3364 | 0.0000 | 1.0000 | 0.5000 | 0.0000 | 0.3786 | 0.1705 | 0.0000 |
| 8 | 0.5613 | 0.2662 | 0.0000 | 0.9000 | 0.3000 | 0.0000 | 0.3417 | 0.2333 | 0.0000 |
| 9 | 0.8506 | 0.6355 | 0.0360 | 1.0000 | 0.8000 | 0.1000 | 0.4612 | 0.3490 | 0.0020 |
| 10 | 0.3937 | 0.1660 | 0.0000 | 0.6000 | 0.2000 | 0.0000 | 0.2680 | 0.1340 | 0.0000 |
| 11 | 0.7937 | 0.2129 | 0.0000 | 0.9000 | 0.2000 | 0.0000 | 0.5952 | 0.1429 | 0.0000 |
| 12 | 0.7985 | 0.5116 | 0.0000 | 1.0000 | 0.8000 | 0.0000 | 0.4591 | 0.2409 | 0.0000 |
| 13 | 0.3238 | 0.0588 | 0.0000 | 0.5000 | 0.1000 | 0.0000 | 0.2400 | 0.0400 | 0.0000 |
| 14 | 0.6704 | 0.0300 | 0.0000 | 0.8000 | 0.0000 | 0.0000 | 0.5484 | 0.0000 | 0.0000 |
| 15 | 0.5073 | 0.1314 | 0.0000 | 0.3000 | 0.1000 | 0.0000 | 0.3000 | 0.1000 | 0.0000 |
| 16 | 0.6899 | 0.4070 | 0.0000 | 1.0000 | 0.6000 | 0.0000 | 0.4648 | 0.2606 | 0.0000 |
| 17 | 0.5334 | 0.2586 | 0.0000 | 1.0000 | 0.3000 | 0.0000 | 0.3707 | 0.2328 | 0.0000 |
| 18 | 0.5024 | 0.3072 | 0.0000 | 1.0000 | 0.5000 | 0.0000 | 0.2990 | 0.1546 | 0.0000 |
| 19 | 0.4655 | 0.1916 | 0.0000 | 0.9000 | 0.2000 | 0.0000 | 0.3429 | 0.0857 | 0.0000 |
| 20 | 0.4810 | 0.1715 | 0.0000 | 0.7000 | 0.2000 | 0.0000 | 0.3469 | 0.1224 | 0.0000 |
| 21 | 0.5998 | 0.3809 | 0.0000 | 0.9000 | 0.5000 | 0.0000 | 0.3922 | 0.2990 | 0.0049 |
| 22 | 0.6420 | 0.1840 | 0.0000 | 0.9000 | 0.4000 | 0.0000 | 0.4155 | 0.1127 | 0.0000 |
| 23 | 0.9132 | 0.5070 | 0.0000 | 1.0000 | 0.6000 | 0.0000 | 0.5744 | 0.3128 | 0.0000 |
| 24 | 0.5551 | 0.2497 | 0.0000 | 1.0000 | 0.4000 | 0.0000 | 0.4375 | 0.1719 | 0.0000 |
| 25 | 0.5310 | 0.2583 | 0.0000 | 0.7000 | 0.3000 | 0.0000 | 0.3704 | 0.1667 | 0.0000 |
| 26 | 0.6342 | 0.0900 | 0.0000 | 0.8000 | 0.1000 | 0.0000 | 0.5000 | 0.0500 | 0.0000 |
| 27 | 0.3064 | 0.0966 | 0.0000 | 0.8000 | 0.1000 | 0.0000 | 0.2105 | 0.0921 | 0.0000 |
| 28 | 0.4787 | 0.1223 | 0.0000 | 0.9000 | 0.2000 | 0.0000 | 0.4000 | 0.0727 | 0.0000 |
| 29 | 0.4628 | 0.2118 | 0.0000 | 1.0000 | 0.3000 | 0.0000 | 0.4048 | 0.1429 | 0.0000 |
| 30 | 0.4755 | 0.1857 | 0.0000 | 0.9000 | 0.2000 | 0.0000 | 0.3265 | 0.1497 | 0.0000 |
Table 8.
Per-topic statistics (over 133 runs) for 28 topics on clinical trials. (Topic 10 had no relevant trials.)
| P @ 5 | P @ 10 | Median | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Topic | Best | Median | Worst | Best | Median | Worst | Best | Median | Worst |
| 1 | 1.0000 | 0.8000 | 0.0000 | 0.9000 | 0.4000 | 0.0000 | 0.6667 | 0.2667 | 0.0000 |
| 2 | 1.0000 | 0.6000 | 0.0000 | 0.9000 | 0.6000 | 0.0000 | 0.9333 | 0.6000 | 0.0000 |
| 3 | 1.0000 | 0.6000 | 0.0000 | 1.0000 | 0.6000 | 0.0000 | 0.8667 | 0.4667 | 0.0000 |
| 4 | 1.0000 | 0.4000 | 0.0000 | 0.9000 | 0.4000 | 0.0000 | 0.8000 | 0.3333 | 0.0000 |
| 5 | 0.8000 | 0.2000 | 0.0000 | 0.5000 | 0.2000 | 0.0000 | 0.5333 | 0.2000 | 0.0000 |
| 6 | 0.8000 | 0.6000 | 0.0000 | 0.8000 | 0.5000 | 0.0000 | 0.6667 | 0.4000 | 0.0000 |
| 7 | 1.0000 | 0.6000 | 0.0000 | 1.0000 | 0.6000 | 0.0000 | 1.0000 | 0.6667 | 0.0000 |
| 8 | 0.8000 | 0.2000 | 0.0000 | 0.7000 | 0.2000 | 0.0000 | 0.6667 | 0.2667 | 0.0000 |
| 9 | 1.0000 | 0.6000 | 0.0000 | 1.0000 | 0.7000 | 0.0000 | 1.0000 | 0.6667 | 0.0000 |
| 11 | 0.8000 | 0.4000 | 0.0000 | 0.7000 | 0.2000 | 0.0000 | 0.6667 | 0.2000 | 0.0000 |
| 12 | 0.8000 | 0.2000 | 0.0000 | 0.7000 | 0.2000 | 0.0000 | 0.6000 | 0.2000 | 0.0000 |
| 13 | 0.6000 | 0.0000 | 0.0000 | 0.6000 | 0.0000 | 0.0000 | 0.6667 | 0.0000 | 0.0000 |
| 14 | 1.0000 | 0.4000 | 0.0000 | 0.6000 | 0.3000 | 0.0000 | 0.4000 | 0.2000 | 0.0000 |
| 15 | 0.4000 | 0.0000 | 0.0000 | 0.2000 | 0.0000 | 0.0000 | 0.1333 | 0.0000 | 0.0000 |
| 16 | 0.4000 | 0.2000 | 0.0000 | 0.3000 | 0.1000 | 0.0000 | 0.2667 | 0.0667 | 0.0000 |
| 17 | 0.6000 | 0.2000 | 0.0000 | 0.6000 | 0.2000 | 0.0000 | 0.5333 | 0.2000 | 0.0000 |
| 18 | 0.6000 | 0.0000 | 0.0000 | 0.4000 | 0.1000 | 0.0000 | 0.3333 | 0.0667 | 0.0000 |
| 19 | 0.8000 | 0.2000 | 0.0000 | 0.5000 | 0.1000 | 0.0000 | 0.4000 | 0.1333 | 0.0000 |
| 20 | 0.4000 | 0.0000 | 0.0000 | 0.3000 | 0.0000 | 0.0000 | 0.2667 | 0.0667 | 0.0000 |
| 21 | 1.0000 | 0.2000 | 0.0000 | 1.0000 | 0.3000 | 0.0000 | 0.9333 | 0.2667 | 0.0000 |
| 22 | 1.0000 | 0.4000 | 0.0000 | 1.0000 | 0.3000 | 0.0000 | 0.9333 | 0.2000 | 0.0000 |
| 23 | 0.8000 | 0.2000 | 0.0000 | 0.7000 | 0.1000 | 0.0000 | 0.5333 | 0.1333 | 0.0000 |
| 24 | 1.0000 | 0.6000 | 0.0000 | 1.0000 | 0.5000 | 0.0000 | 0.8667 | 0.3333 | 0.0000 |
| 25 | 1.0000 | 0.4000 | 0.0000 | 1.0000 | 0.3000 | 0.0000 | 0.8000 | 0.2667 | 0.0000 |
| 26 | 0.2000 | 0.0000 | 0.0000 | 0.3000 | 0.0000 | 0.0000 | 0.2000 | 0.0000 | 0.0000 |
| 27 | 0.8000 | 0.0000 | 0.0000 | 0.6000 | 0.1000 | 0.0000 | 0.4667 | 0.1333 | 0.0000 |
| 28 | 0.0000 | 0.0000 | 0.0000 | 0.1000 | 0.0000 | 0.0000 | 0.0667 | 0.0000 | 0.0000 |
| 29 | 0.8000 | 0.2000 | 0.0000 | 0.6000 | 0.1000 | 0.0000 | 0.4000 | 0.0667 | 0.0000 |
| 30 | 1.0000 | 0.2000 | 0.0000 | 0.7000 | 0.2000 | 0.0000 | 0.5333 | 0.1333 | 0.0000 |
7 Conclusion
This was the first year of the Precision Medicine track. The goal of the track is to inform the creation of information retrieval systems to support clinicians working in precision medicine (specifically oncologists in this track) in making better treatment decisions for individual patients. Participants were provided with synthetic patient data consisting of a type of cancer, one or more genetic variants, patient demographics, and other potentially relevant patient factors. Given this, participants were challenged with retrieving
Acknowledgments
The organizers would like to thank Kate Fultz Hollis for managing the assessment process. KR is supported by the National Institutes of Health (NIH) grant 2R00LM012104-02. DDF is supported by the Intramural Research Program of the U.S. National Library of Medicine, NIH. Finally, the organizers are grateful to the National Institute of Standards and Technology (NIST) for funding the assessment process.
Footnotes
Note that while clinical data is frequently de-identified for research purposes without the need for patient permission, genomic data is fundamentally difficult to de-identify. So to be safe, synthetic data was used.
Contributor Information
Kirk Roberts, School of Biomedical Informatics, The University of Texas Health Science Center, Houston, TX.
Dina Demner-Fushman, Lister Hill National Center for Biomedical Communications, U.S. National Library of Medicine, Bethesda, MD.
Ellen M. Voorhees, Information Technology Laboratory, National Institute of Standards and Technology, Gaithersburg, MD
William R. Hersh, Department of Medical Informatics & Clinical Epidemiology, Oregon Health and Science University, Portland, OR
Steven Bedrick, Department of Medical Informatics & Clinical Epidemiology, Oregon Health and Science University, Portland, OR.
Alexander J. Lazar, Departments of Pathology & Genomic Medicine, The University of Texas MD Anderson Cancer Center, Houston, TX
Shubham Pant, Department of Investigational Cancer Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, TX.
References
- Collins FS, Varmus H. A New Initiative on Precision Medicine. New England Journal of Medicine. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey LJ, Bernstam EV, Denny JC. Precision medicine informatics. Journal of the American Medical Informatics Association. 2016;23:668–670. doi: 10.1093/jamia/ocw053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh W, Voorhees E. TREC genomics special issue overview. Information Retrieval. 2009;12:1–15. [Google Scholar]
- Seco de Herrera AG, Kalpathy-Cramer J, Demner-Fushman D, Antani S, Müller H. Overview of the ImageCLEF 2013 medical tasks. CLEF 2013 Working Notes 2013 [Google Scholar]
- Voorhees EM, Hersh W. Overview of the TREC 2012 Medical Records Track; Proceedings of the Twenty-First Text REtrieval Conference.2012. [Google Scholar]
- Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. Journal of the American Medical Informatics Association. 2011;18(Suppl 1):i116–i124. doi: 10.1136/amiajnl-2011-000321. [DOI] [PMC free article] [PubMed] [Google Scholar]