SUMMARY:
Deep learning is a form of machine learning using a convolutional neural network architecture that shows tremendous promise for imaging applications. It is increasingly being adapted from its original demonstration in computer vision applications to medical imaging. Because of the high volume and wealth of multimodal imaging information acquired in typical studies, neuroradiology is poised to be an early adopter of deep learning. Compelling deep learning research applications have been demonstrated, and their use is likely to grow rapidly. This review article describes the reasons, outlines the basic methods used to train and test deep learning models, and presents a brief overview of current and potential clinical applications with an emphasis on how they are likely to change future neuroradiology practice. Facility with these methods among neuroimaging researchers and clinicians will be important to channel and harness the vast potential of this new method.
Deep learning is a form of artificial intelligence, roughly modeled on the structure of neurons in the brain, which has shown tremendous promise in solving many problems in computer vision, natural language processing, and robotics.1 It has recently become the dominant form of machine learning, due to a convergence of theoretic advances, openly available computer software, and hardware with sufficient computational power. The current excitement in the field of deep learning stems from new data suggesting its excellent performance in a wide variety of tasks. One benchmark of machine learning performance is the ImageNet Challenge. In this annual competition, teams compete to classify millions of images into discrete categories (tens of different kinds of dogs, fish, cars, and so forth). A watershed year was 2012, when the first neural network–based entry bested the competition and prior years' results by a wide margin.2 Since then, every winning entry has used a deep learning framework, with performance now exceeding that of humans.
Deep learning has the potential to revolutionize entire industries, including medical imaging. Given the centrality of neuroimaging in the diagnosis and treatment of neurologic diseases, deep learning will likely affect neuroradiologists first and most profoundly. This article will introduce deep learning methods, overview their current successes, and speculate on the future evolution of these methods, focusing on their application to neuroradiology.
What is Deep Learning?
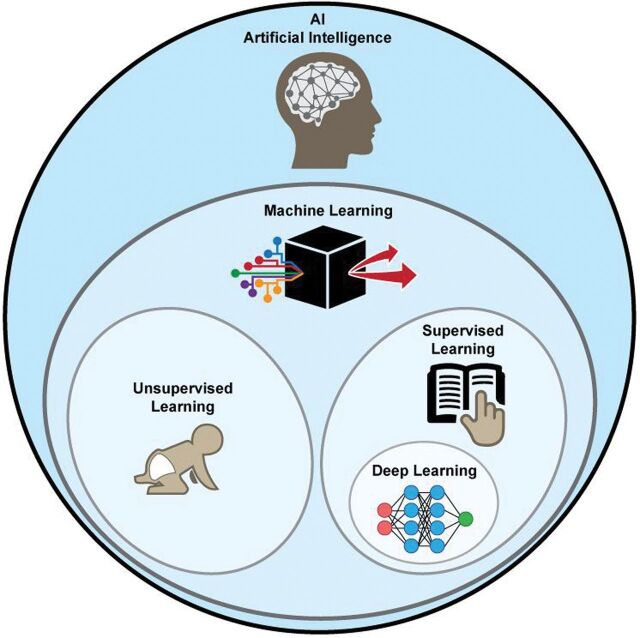
It is useful to consider where deep learning fits into the broader context of artificial intelligence (Fig 1). One definition suggested for artificial intelligence is any computer method that performs tasks normally requiring human intelligence. Machine learning is one type of artificial intelligence that develops algorithms to enable computers to learn from existing data without explicit programming. Examples are classification algorithms such as clustering, logistic regression, and support vector machines.
Fig 1.
Artificial intelligence methods. Within the subset of machine learning methods, deep learning is usually implemented as a form of supervised learning.
Machine learning methods can be further divided into supervised and unsupervised learning. In supervised learning, some “ground truth” exists, which is used to train the algorithms. One example is a collection of brain CT scans that a neuroradiologist has classified into different groups (ie, hemorrhage versus no hemorrhage). In contrast, for unsupervised learning, no criterion standard images or classifications are used—the computer itself must determine the classes. One example is clustering, in which images are placed in multiple groups based on similarity metrics without knowing a priori what is driving the separation.3 While unsupervised learning holds great promise for medical imaging, this review focuses on supervised learning.
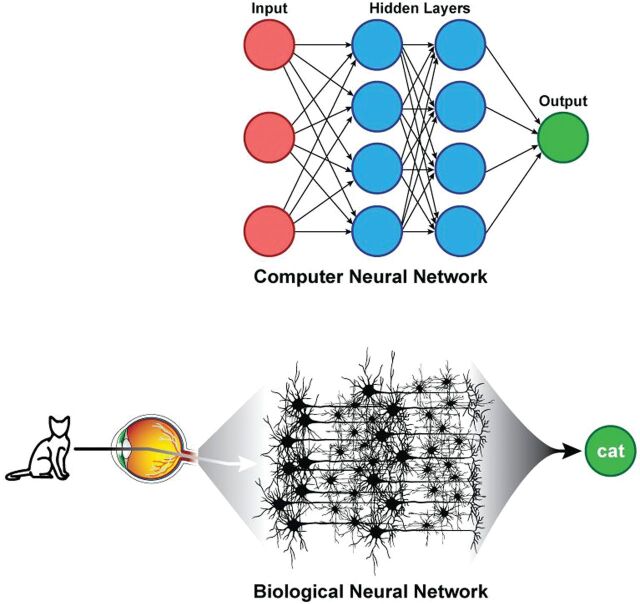
Viewed in this context, deep learning is a supervised machine learning method that uses a specific architecture, namely some form of neural network. The power of these techniques is in their scalability, which is largely based on their ability to automatically extract relevant features. In the past, constructing an image-classification algorithm took years of effort on the part of domain experts and experienced artificial intelligence researchers. Deep learning allows such a classifier to be created automatically from a labeled dataset in days. These neural networks are loosely inspired from how the brain is structured, with hidden layers representing interneurons (Fig 2). While modern neural networks share these similarities to the brain, whether more fidelity to known brain structures would improve performance is an actively debated question.4 For example, in computer vision applications, many of the features to which hidden layers are sensitive (such as edges in different orientations) have correlates in the mammalian visual cortex.5
Fig 2.
Parallels between artificial and biologic neural networks. Hidden layers of artificial neural networks can be thought to be analogous to brain interneurons.
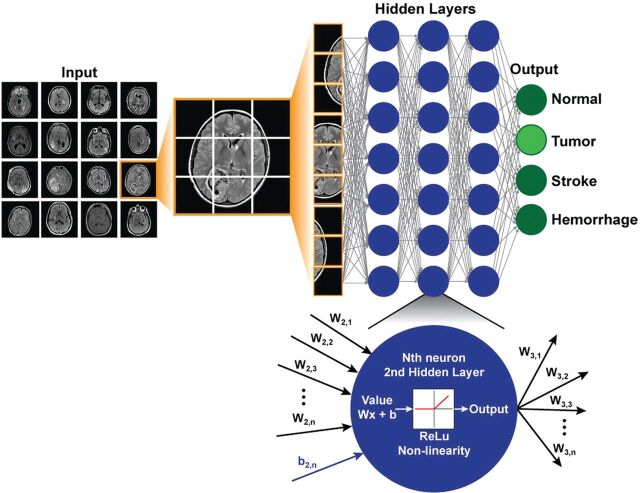
For neuroimaging, a simple deep learning model may accept image data as a vector composed of voxel intensities, with each voxel serving as an input “neuron.” While the examples below assume the use of individual images, more generally, the input can consist of entire imaging series, multiple series, or even multiple modalities. Next, one must determine how many layers (how deep) and how many neurons per layer (how wide) to include; this is known as the network architecture (Fig 3). Each neuron stores a numeric value, and each connection between neurons represents a weight. Weights connect the neurons in different layers and represent the strength of connections between the neurons. A “fully connected” layer in which all neurons in one layer are connected to all neurons in the next can be interpreted and implemented as a matrix multiplication. Finally, it is customary to include a nonlinear “activation function” at the output of the neuron. This introduces nonlinearity into the equations so that complex functions can be represented that would not otherwise be possible. Historically, sigmoids and hyperbolic tangents have been used on the basis of insights from neuroscience; however, researchers have since found that the rectified linear unit is both simpler to implement and more effective. The rectified linear unit function outputs the value of the neuron for positive values and zero for negative values.
Fig 3.
Example of a simple deep network architecture. The goal of this network is to classify MR images into 4 specific diagnoses (normal, tumor, stroke, hemorrhage). Multiple different images form the training set. For each new case, the image is broken down into its constituent voxels, each one of which acts as an input into the network. This example has 3 hidden layers with 7 neurons in each layer, and the final output is the probabilities of the 4 classification states. All layers are fully connected. At the bottom is a zoomed-in view of an individual neuron in the second hidden layer, which receives input from the previous layer, performs a standard matrix multiplication (including a bias term), passes this through a nonlinear function (the rectified linear unit function in this example), and outputs a single value to all the neurons of the next layer.
The choice of network architecture for a specific application is not always obvious, though some typical configurations and assumptions exist. The number of neurons in the hidden layers tends to be larger than that in either the input or the output layers. The final layer encodes the desired outcomes or labeled states. For example, if one wishes to classify an image as “hemorrhage” or “no hemorrhage,” 2 final layer neurons are appropriate. Commonly, the value stored by each final neuron is interpreted as the probability that the training example corresponds to a specific class. The goal of training is to optimize the network weights so that when a new sample image is input, the probabilities measured at the output are heavily skewed to the correct class. For example, if we input an image with hemorrhage, we would like the model to output a high probability for hemorrhage and low probability for other classes. How is this accomplished?
Training Simple Neural Network Deep Learning Models
Neural networks are ideally trained using large numbers of cases that are divided into several groups. Usually the largest fraction is used for model training (50%–60%), with another 10%–20% for validation and 20%–40% for testing. The training cases are used to set the model parameters; large training sets are important because even relatively shallow networks may have 100,000s of free parameters (weights). The training dataset is looped through multiple times (epochs) until the accuracy of the model converges. At first, predictions will be poor. However, the beauty of this setup is that you can compare the output of the model with the ground truth via the use of a “cost function,” a single number that quantifies how far off the model is. Back-propagation, a technique whereby the strength of connections between neurons (weights) can be adjusted on the basis of the value of the cost function, is then used to reinforce correct predictions and penalize incorrect predictions. This procedure is repeated using separate training examples and multiple iterations, thereby optimizing the weights and effectively training the model. Once the model is trained, there are several “hyperparameters” to optimize, including the learning rate and number of epochs. Finally, the testing set is used to assess the model accuracy on data that has not been used for training. This assessment will yield an error rate similar to or higher than that for the training set, and this helps to gauge how well the final model will perform on real world data. While training the model is often time-intensive, the application of the final trained model to new data is usually computationally fast.
Choosing the right cost function is important. For classification, the value of the cost function should be low when the model predicts the correct class and high when its predictions are off. A popular cost function for classification is “cross-entropy loss,” an extension of logistic regression to multiple classes that can be implemented using the softmax function. For image prediction, common cost functions include the root-mean-square error between the predicted and reference images and measures of similarity, such as the structural similarity index metric.6,7 One promising approach is replacing the cost function itself with a network whose goal is to make it optimally difficult to distinguish reference images from predicted images, an approach known as the “generative adversarial network” approach.8 Generative adversarial networks strive to eliminate systematic differences between the predicted and reference images, which is highly desirable in the radiology setting.
From Simple to Convolutional Neural Networks
Fully connected neural networks are computationally expensive because the number of weights is very large, especially with images of typical matrix sizes (256 × 256 = 65,536 voxels). With even just 1 slice, >4 billion weights are required to implement a fully connected layer. Thus, much research in image-based deep learning has moved to using more computationally efficient structures, specifically convolutional neural networks (CNNs).
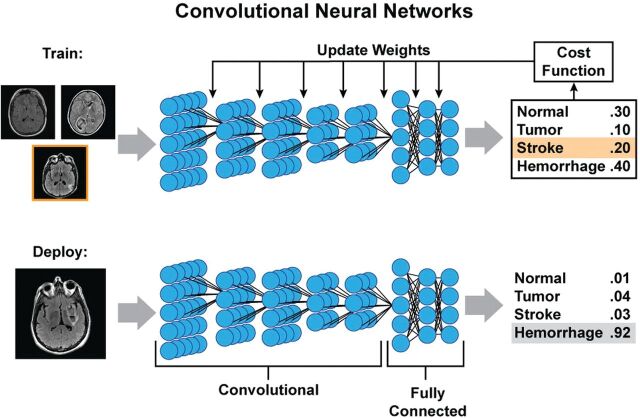
CNNs are well-suited for imaging. Instead of full connections, a small “kernel” of weights is applied at each image position to determine the value of the neuron of the next layer (Fig 4). The approach mimics the mathematic operation of convolution. Between 2 layers, the only weights required are those for the kernel, which is then rastered across the image to obtain the next layer. This method has several advantages. First, it markedly reduces the number of weights. Second, it allows spatial invariance: Image features may occur in different locations, and a CNN allows their identification independent of their precise location. Often, CNNs pool adjacent voxels or slide the kernel across these images at spaced intervals (a hyperparameter known as the “stride length”), so that the dimensions in each subsequent layer are smaller than those in the last one. For each layer, multiple different kernels can be trained, creating multiple “channels” in each layer; such a structure allows the network to learn many location-invariant features, such as edges, textures, and other nonlinear representations of the data. With pooling or increased stride lengths, it is possible to incorporate ever larger features into the hidden layers of the network. Indeed, the ability of CNNs to extract relevant imaging features in a location-invariant way parallels the structure of the visual system of the brain; Hubel and Wiesel5 showed in the 1960s that different regions of the cat brain responded strongly to features such as edges oriented in different directions.
Fig 4.
Example of training and deployment of deep convolutional neural networks. During training, each image is analyzed separately, and at each layer, a small set of weights (convolution kernel) is moved across the image to provide input to the next layer. Each layer can have multiple channels. By pooling adjacent voxels or using larger stride distances between application of the kernel, deeper layers often have smaller spatial dimensions but more channels, each of which can be thought of as representing an abstract feature. In this example, 5 convolutional layers are followed by 3 fully connected layers, which then output a probability of the image belonging to each class. These probabilities are compared with the known class (stroke in the training example) and can be used to measure of how far off the prediction was (cost function), which can then be used to update the weights of the different kernels and fully connected parameters using back-propagation. When the model training is complete and deployed on new images, the process will produce a similar output of probabilities, in which it is hoped that the true diagnosis will have the highest likelihood.
For classification, ≥1 fully connected layer is typically added to reach the final output layer. For image prediction, upsampling layers are used to “re-form” the smaller dimension hidden layers back into the original size of the input image. Such an architecture is called an “encoder-decoder” because it represents the image in terms of increasing abstraction (encoding) in the hidden layers and then uses them to recreate (decode) the image.
Overfitting and Data Augmentation
As described above, typical deep learning models have millions of weights. Analogous to the idea that you need more equations than variables to solve algebraic equations, if a deep network is trained on too few examples, it is possible to perfectly represent the transform between the input and output states. However, this approach will not generalize to new cases, a problem known as “overfitting.” The best solution to overfitting is collecting more training examples, though other solutions such as regularization and drop-out can also be used.7,9 Another potential solution is data augmentation.
Data augmentation is a method of increasing the amount of training data. Because most image data should be recognizable whether offset in the x-y plane, rotated, flipped, or slightly stretched or skewed, it is conventional to perform such image manipulations to augment the training data. While these image alterations do not add more data, they have been shown to improve the robustness of the models, possibly by preventing the model from learning features that occur only in a specific orientation.
Broad Classes of Applications
Deep learning can address many aspects of neuroradiology. The overall flow of work in neuroradiology is a useful framework in which to consider these applications. This starts with referring clinicians ordering studies and then moves to image acquisition. Next, the images are put before radiologists, and tasks surrounding detection and segmentation of lesions and differential diagnosis arise. Each link in this chain can potentially benefit from a deep learning approach.
Imaging Logistics
After a study is ordered, it needs to be triaged to a specific neuroimaging protocol. This process often involves the precious time of radiologists and relies on their knowledge of imaging protocols and attention to clinicians' specific requests, which are often encapsulated in the order history as free text. Deep learning methods to interpret natural language are already mature, making automation of the protocoling process quite feasible. In theory, this problem is just about classification, with the different protocols being the classes to predict and the input being the order itself and patient metadata. The protocolling application is ideal for deep learning because of the immense amount of training data that already exists; all prior studies that have been protocolled by humans can be used for training.
Another promising application is to triage image review in the order of suspected acuity. For example, if models can be trained to identify critical findings on images, it is possible to prioritize radiologic review of these studies, even if they were not initially ordered as “stat” studies. For large organizations, such triage offers the potential to reduce the time between acquisition and interpretation for critical cases, with presumably positive effects on patient outcome.
Image Acquisition and Improvement
Deep learning methods can be used to perform image reconstruction and improve image quality. Deep learning frameworks are capable of “learning” standard MR imaging reconstruction techniques, such as Cartesian and non-Cartesian acquisition schemes.10 Combining deep learning to k-space undersampling with model-based/compressed sensing reconstruction schemes holds the potential to revolutionize imaging science by optimizing how image data are collected.11–13
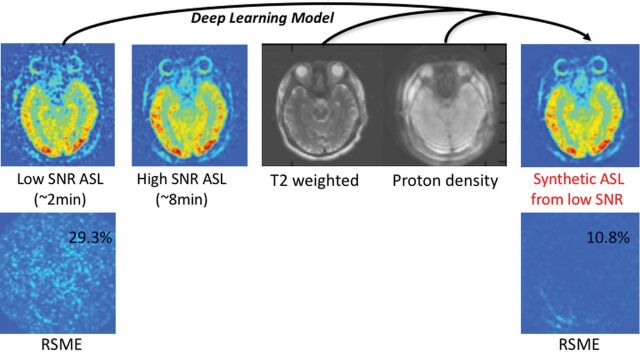
Also, one could apply deep learning methods to improve image quality. If images at low-resolution and high-resolution are available, it is possible to use a deep network for super-resolution.14 If paired image sets of low and high quality are available, learning the optimal nonlinear transformation between them can be considered. This has already been applied to CT imaging and has been demonstrated to be of value on a dataset consisting of normal-dose and simulated low-dose CT.15 Another approach uses paired MR images of the same anatomy, which are acquired at different field strengths. A study using 3T input data and 7T output data showed that a deep network can be trained to create simulated “7T-like” images from 3T data.16 Often obtaining a certain imaging sequence can be very time-consuming; an example is DTI, in which the need for multiple angular directions lengthens the examination beyond what many patients can tolerate. A deep learning approach can reduce imaging duration 12-fold by predicting final parameter maps (fractional anisotropy, mean diffusivity, and so forth) from relatively few angular directions.17 By acquiring paired arterial spin-labeling (ASL) CBF images with 2 and 30 minutes of acquisition time, our group has trained a deep network to boost the SNR of ASL significantly (Fig 5).18
Fig 5.
An example of improving the SNR of arterial spin-labeling MR imaging using deep learning. The model is trained using low-SNR ASL images acquired with only a single repetition, while the reference image is a high-SNR ASL image acquired with multiple repetitions (in this case, 6 repetitions). Proton-density-weighted images (acquired routinely as part of the ASL scans for quantitation) and T2-weighted images are also used as inputs to the model to improve performance. The results of passing the low-SNR ASL image through the model are shown on the right, a synthetic image with improved SNR. In this example, the root-mean-squared error (RSME) between the reference image and the synthetic image compared with the original image is reduced nearly 3-fold, from 29.3% to 10.8%.
Image Transformation
An extension of this is to create images with different contrast or with features of different modalities. For example, using the National Alliance for Medical Imaging Computing data base (http://www.insight-journal.org/midas/community/view/17), Vemulapalli et al19 used a deep network to predict T1 images from T2 images, and vice versa. Another application is to PET/MR imaging; unlike PET/CT, in which CT is used to calculate an attenuation map, MR images do not directly yield attenuation images. However, if there is information about soft tissue, air, and bone in the MR images, these sequences can be used as input to a deep network. Crucially here, the image to predict is no longer another MR image, but rather a coregistered CT scan of the same subject. Proof of principle was recently demonstrated for brain MR imaging attenuation correction, with performance superior to that of competing techniques.20 Another study demonstrated a similar use of MR imaging to create synthetic CT for radiation therapy.21
In clinical trials, situations arise in which patients may not be able to undergo a certain diagnostic technique, such as patients with MR imaging–incompatible implants. Alternatively, they may lack images at specific time points. While statistical techniques can be used to account for such missing data, if enough patients drawn from the same population have completed all imaging examinations, it is possible to train a deep learning network to recreate these data. Li et al22 demonstrated this using the Alzheimer's Disease Neuroimaging Initiative (ADNI; http://www.adni-info.org/). They trained a CNN on patients with both FDG-PET and T1-weighted MR imaging and then used this network on a test set to predict the expected PET images from patients' MR imaging studies,22 showing that the CNN method outperformed more traditional methods.
Lesion Detection and Segmentation
Detecting and segmenting lesions is an onerous task for humans but is well-suited to machine learning. While related, they are really 2 different tasks. The former starts with an unlabeled image and marks potential abnormalities. The goal of the latter is to circumscribe the regions encompassing the abnormal structures. Identifying and delineating the margins of a lesion is important because neuroradiologists are often tasked with monitoring the change in size or activity of known lesions across time or in response to treatment. Deep learning also has advantages for segmenting normal brain structures because existing methods are time-consuming and may not generalize to younger or older subjects.23–25 Furthermore, many research projects rely on manual delineation of image lesions. One can train a deep network to take images as input and hand-drawn manually segmented masks as output. Indeed, such an approach has shown great early success. While there are many examples in different neurologic disease conditions, we will reference 3 representative areas: detecting microhemorrhages, identifying infarcts and predicting final infarct volumes in patients with stroke, and segmenting brain tumors.
Dou et al26 described a process to detect brain microhemorrhages by training a CNN on an annotated dataset of susceptibility-weighted images. They proposed a cascaded, 2-step approach, in which candidate lesions are first identified by the CNN and then only these lesions are input to a discriminatory CNN (ie, true microhemorrhage or mimic). With this approach, they achieved a sensitivity of >93%, with an average of about 3 false-positive identifications per subject.
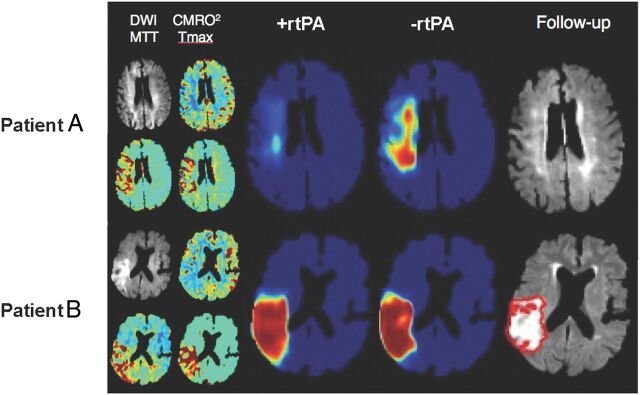
Automatic identification and outlining of infarcted brain tissue would be useful in the acute stroke setting. Chen et al27 used DWI as input to a 2-stage deep learning algorithm and were able to detect 94% of all acute infarcts. Using the Dice coefficient as a marker of accuracy, they showed a mean score of 0.67 in a large cohort of patients 2 days after stroke. Another study using a 3-layer-deep CNN followed by 2 fully connected layers showed similar results and outperformed several other machine learning methods.28 Another intriguing application is to predict final infarct volume from early DWI and/or PWI in patients with acute stroke. Currently, the diffusion-perfusion mismatch approach is the dominant paradigm, which states that DWI lesions represent irreversibly damaged tissue, while PWI identifies tissue at risk of infarction. Rather than using such hand-crafted features, a deep neural network can be trained using initial DWI and PWI maps as input and final infarct size measured several days later as the output. Using such a framework, Nielsen et al29 demonstrated that a deep learning architecture outperforms traditional state-of-the-art lesion prediction methods in acute stroke. They also showed that a 37-layer architecture outperformed a shallower 3-layer architecture, highlighting the importance of the depth feature of the network. One exciting application of this approach is to train separate networks based on different treatments. In stroke, one could train networks in patients who received stroke treatment and those who did not. New predictions using these 2 different models could give insight into whether treatment would lead to a reduced infarct volume (Fig 6).
Fig 6.
An example of the predicted risk of final infarct for 2 patients with acute ischemic stroke using 2 neural networks trained, respectively, on patients with and without rtPA administration. Patient A is a 76-year-old woman with an admission NIHSS score of 10 scanned 1.5 hours after symptom onset. The +rtPA network estimates a negligible permanent lesion, consistent with the acute DWI suggesting little permanent tissue damage at follow-up. The −rtPA network indicates that without treatment, a considerable volume of the acute ischemic region will progress to a permanent lesion. Patient B is a 72-year-old man also with an admission NIHSS score of 10, scanned 2 hours after onset. In this case, the 2 networks indicate little expected impact of treatment, likely due to the progression at the time of imaging of the ischemic event as seen on the DWI. CMRO2 indicates cerebral metabolic rate of oxygen; Tmax, time-to-maximum. Figure courtesy of Kim Mouridsen and Anne Nielsen/Aarhus University, Combat Stroke.
Automated segmentation of brain tumors, not just their enhancing margins, but also other features such as regions of enhancement and necrosis, would be useful for a wide range of indications, including diagnosis, presurgical planning, and follow-up. The Brain Tumor Image Segmentation dataset is a publicly available dataset of brain tumor images with expert manual segmentations that has been a useful proving ground for new segmentation algorithms.30 The highest performance on the Brain Tumor Image Segmentation dataset in 2016 was achieved using a fully convolutional residual neural network, built on the structure that won the 2015 ImageNet Challenge, with Dice coefficients for complete tumor, core tumor, and enhancing tumor between 0.72 and 0.87.31 In a separate study, Korfiatis et al32 trained a deep autoencoder-decoder to segment T2-FLAIR lesions on the Brain Tumor Image Segmentation dataset, which included manually drawn outlines in 186 patients. They then applied their model to a separate group of 135 patients with tumor in which they had 3 expert segmentations and measured a Dice coefficient of 0.88 based on a method that incorporated the individual tracings of the 3 readers. They did note significant variability among their readers' segmentations, pointing out the importance of how the segmentation criterion standard is implemented.
Deep Learning for Image-Based Diagnosis
The “Holy Grail” of machine learning in radiology is the so-called “end-to-end” solution, in which images are used as input and the output is a draft radiology report encompassing all the salient features of the image that an expert radiologist would include. As improbable as this might seem, progress is being made on this front with deep learning. Such approaches require tremendous amounts of annotated training data, which exist currently in many heterogeneous forms. Structured reporting, use of standard lexicons (such as RadLex; http://www.rsna.org/RadLex.aspx), and the standardization of electronic medical record platforms are all steps that are enabling the formation of the required large imaging/diagnosis datasets. This section will deal with some early applications of deep learning to neuroradiologic diagnosis.
Gao et al33 classified 285 noncontrast brain CT examinations with a deep network into 1 of 3 categories (normal aging, lesion [such as tumor], or Alzheimer disease [AD]). With an average classification accuracy of 88%, the approach performed marginally better than other approaches that relied on hand-crafted features. Another study34 showed that a multimodal stacked deep polynomial network using the ADNI dataset could classify patients into different binary groups (ie, AD versus healthy control [NC], or mild cognitive impairment converters [MCI-C] versus nonconverters [MCI-nonconverters]). Instead of using images as input, they used the volumes of 93 structures segmented from T1-weighted images along with the [18F] FDG-PET signal intensity in these same regions. For distinguishing patients with AD from NCs, they showed an impressive area under the curve of 0.97. For the more challenging task of predicting MCI converters from nonconverters, they still showed areas under the curve of >0.80. Suk et al35 showed that similar features could be combined with CSF data in the ADNI dataset using a deep-weighted sparse multitask learning framework to improve classification, showing 95% accuracy to distinguish patients with AD from NCs. However, when trying to predict among 3 classes (AD, NC, and MCI), the accuracy dropped to 63%, which further dropped to 54% when trying to classify among 4 groups (AD, NC, MCI-C, and MCI-nonconverter). This latter point speaks to the challenges of moving beyond simple binary classifications into the kinds of tasks in which many diagnostic groups are possible, a situation familiar to neuroradiologists.
Another application is to use deep networks to identify the presence of hemorrhage on noncontrast brain CT. While general machine learning techniques have been applied to these cases with great success,36,37 only recently have deep learning methods been evaluated. Phong et al38 demonstrated a particularly interesting approach, using several “pretrained” deep networks as starting points for their training. Specifically, they used the optimal weights from the GoogLeNet (http://deeplearning.net/tag/googlenet/) or Inception-ResNet (https://keras.rstudio.com/reference/application_inception_resnet_v2.html) that were trained on nonmedical images and then used their data to train a final fully connected layer. This approach (called “transfer learning”) is attractive, given that it allows networks to be trained with less data than if they were trained from scratch.39 Trained on 80 cases and tested on 20 cases, they showed classification accuracies of >98%.
Plis et al40 examined both structural and functional MR imaging as input to deep networks for predicting various neurologic diseases. For structural imaging, they showed that they could distinguish patients with schizophrenia and Huntington disease from healthy subjects. For fMRI, they showed that deep networks performed similar to independent component analysis for identifying functional networks but tended to preserve edge details better. Similar results using a temporal-autoencoding neural network to predict the next time point in a resting-state fMRI time-series were applied to the Human Connectome Project data and showed a similar ability to identify task-specific networks.41
Impact of Deep Learning on Neuroradiology Practice
One concern is that if these approaches are successful, some work that radiologists have traditionally performed may become obsolete. Recently, a framework for inputting medical images (in this case, pathology slides) and outputting diagnostic text-based reports has been reported.42 While this technology is still very rudimentary, it is not difficult to imagine training a similar network with CT scans and their reports. While deep learning does hold much promise to automate tasks that radiologists find unpleasant, ways of checking and verifying results will be needed. Another concern with deep learning is that we have little insight into the inner workings of the models; they work well for prediction, but precisely how they accomplish this is unclear. This contrasts with much prior radiology research, which relied heavily on domain knowledge and the building of realistic models. Understanding how and why deep networks perform so well is an active area of research in the artificial intelligence community.
Also, the application of deep learning is still limited by requirements for large amounts of annotated training datasets and the challenge of keeping models current as source data and practice patterns change. Among applications that are amenable to disruption, it is still unclear which applications are supported by sufficient clinical need to drive widespread adoption. Thus, it is important that radiologists remain engaged with artificial intelligence scientists to both understand the capabilities of existing methods and direct future research in an intelligent way. Although this technology is still developing, state-of-the art deep learning models show little evidence that they could replace all functions of radiologists, though this issue is contentious. For the foreseeable future, they will likely serve as powerful image-processing and decision-support tools that will augment the accuracy and efficiency of radiologists.
Outlook
The wealth of applications of deep learning will likely lead to increased use of this technology. How fast this will happen depends on a few key factors: Deep learning can learn more from larger datasets, usually by adding additional hidden layers. As larger labeled datasets become available, the power of deep learning approaches will increase. The importance of data-sharing initiatives such as ADNI and the Cancer Imaging Archive (https://public.cancerimagingarchive.net/ncia/legalRules.jsf) cannot be overstated. Second, the computer hardware required to run these methods continues to improve and become less expensive. The availability of open-source software frameworks, such as Caffe, Tensorflow, PyTorch, and Keras, is greatly facilitating progress. However, before these methods become a routine part of clinical practice, vendors will need to provide “turn-key” systems that integrate well into current workflow patterns. As more neuroradiology researchers and practitioners become comfortable with these methods, through exposure at medical imaging conferences and conversations with colleagues, applications will branch out from the current “low-hanging fruit” to address more complex and specialized questions. Therefore, we can expect further advances in this field with accompanying benefits in many areas of radiology.
ABBREVIATIONS:
- AD
Alzheimer disease
- ADNI
Alzheimer's Disease Neuroimaging Initiative
- ASL
arterial spin-labeling
- CNN
convolutional neural network
- MCI
mild cognitive impairment
- NC
normal control
Footnotes
Disclosures: Greg Zaharchuk—UNRELATED: Grants/Grants Pending: GE Healthcare, National Institutes of Health*; OTHER RELATIONSHIPS: Subtle Medical Inc, cofounder and equity relationship. Enhao Gong—UNRELATED: Board Membership: GE Healthcare, Comments: research grant*; Patents (Planned, Pending or Issued): Subtle Medical Inc; Stock/Stock Options: Subtle Medical Inc. Max Wintermark—UNRELATED: Board Membership: GE-NFL Advisory Board. Daniel Rubin—UNRELATED: Grants/Grants Pending: National Institutes of Health*. Curtis P. Langlotz—OTHER RELATIONSHIPS: Montage Healthcare Solutions, Comments: founder, shareholder, board member; received consulting fees and travel reimbursement. *Money paid to the institution.
References
- 1. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–44 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 2. Krizhevsky A, Sutskever I, Hinton G. ImageNet classification with deep convolutional neural networks. http://www.cs.toronto.edu/~fritz/absps/imagenet.pdf. Accessed January 23, 2018.
- 3. Mwangi B, Soares JC, Hasan KM. Visualization and unsupervised predictive clustering of high-dimensional multimodal neuroimaging data. J Neurosci Methods 2014;236:19–25 10.1016/j.jneumeth.2014.08.001 [DOI] [PubMed] [Google Scholar]
- 4. Hassabis D, Kumaran D, Summerfield C, et al. . Neuroscience-inspired artificial intelligence. Neuron 2017;95:245–58 10.1016/j.neuron.2017.06.011 [DOI] [PubMed] [Google Scholar]
- 5. Hubel D, Wiesel T. Receptive fields, binocular interaction and functional architecture of the cat's visual cortex. J Physiol (London) 1962;160:106–54 10.1113/jphysiol.1962.sp006837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Z, Bovik A, Sheikh H, et al. . Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 2004;13:600–12 10.1109/TIP.2003.819861 [DOI] [PubMed] [Google Scholar]
- 7. Zhou H, Gallo O, Frosio I, et al. . Loss functions for image restoration with neural networks. IEEE Transactions on Computational Imaging 2017;3:47–57 10.1109/TCI.2016.2644865 [DOI] [Google Scholar]
- 8. Goodfellow I, Pouget-Abadie J, Mirza M, et al. . Generative adversarial nets. https://papers.nips.cc/paper/5423-generative-adversarial-nets.pdf. Accessed January 23, 2018.
- 9. Srivastava N, Hinton G, Krizhevsky A, et al. . Dropout: a simple way to prevent neural networks from overfitting. J Machine Learning Research 2014;15:1929–58 [Google Scholar]
- 10. Zhu B, Liu JZ, Rosen BR, et al. . Image reconstruction by domain transform manifold learning. https://arxiv.org/pdf/1704.08841.pdf. Accessed January 23, 2018. [DOI] [PubMed]
- 11. Schlemper J, Caballero J, Hajnal JV, et al. . A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE Trans Med Imaging 2018;37:491–503 10.1109/TMI.2017.2760978 [DOI] [PubMed] [Google Scholar]
- 12. Yang Y, Sun J, Li H, et al. . Deep admm-net for compressive sensing MRI. In: Proceedings of the 30th Conference on Neural Information Processing Systems, Barcelona, Spain December 6–10, 2016 [Google Scholar]
- 13. Wang S, Su Z, Ying L, et al. . Accelerating magnetic resonance imaging via deep learning. In: Proceedings of the IEEE 13th International Symposium on Biomedical Imaging, Prague, Czech Republic April 9–16, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dong C, Loy C, He K, et al. . Learning a deep convolutional network for image super-resolution. In: Proceedings of the European Conference on Computer Vision, Zurich, Switzerland September 6–12, 2014 [Google Scholar]
- 15. Chen H, Zhang Y, Kalra MK, et al. . Low-dose CT with a residual encoder-decoder convolutional neural network (RED-CNN). https://arxiv.org/pdf/1702.00288.pdf. Accessed January 23, 2018. [DOI] [PMC free article] [PubMed]
- 16. Bahrami K, Shi F, Zong X, et al. . Reconstruction of 7T-like images from 3T MRI. IEEE Trans Med Imaging 2016;35:2085–97 10.1109/TMI.2016.2549918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Golkov V, Dosovitskiy A, Sperl JI, et al. . Q-space deep learning: twelve-fold shorter and model-free diffusion MRI scans. IEEE Trans Med Imaging 2016;35:1344–51 10.1109/TMI.2016.2551324 [DOI] [PubMed] [Google Scholar]
- 18. Gong E, Pauly J, Zaharchuk G. Boosting SNR and/or resolution of arterial spin label (ASL) imaging using multi-contrast approaches with multi-lateral guided filter and deep networks. In: Proceedings of the Annual Meeting of the International Society for Magnetic Resonance in Medicine, Honolulu, Hawaii April 22–27, 2017 [Google Scholar]
- 19. Vemulapalli R, Nguyen H, Zhou S. Deep networks and mutual information maximization for cross-modal medical image synthesis. In: Zhou S, Greenspan H, Shen D, eds. Deep Learning for Medical Image Analysis. London: Elsevier; 2017 [Google Scholar]
- 20. Liu F, Jang H, Kijowski R, et al. . Deep learning MR-based attenuation correction for PET/MRI. Radiology 2018;286:676–84 10.1148/radiol.2017170700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys 2017;44:1408–19 10.1002/mp.12155 [DOI] [PubMed] [Google Scholar]
- 22. Li R, Zhang W, Suk HI, et al. . Deep learning based imaging data completion for improved brain disease diagnosis. In: Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Boston, Massachusetts September 14–18, 2014;17:305–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wachinger C, Reuter M, Klein T. DeepNAT: deep convolutional neural network for segmenting neuroanatomy. Neuroimage 2018;170:434–45 10.1016/j.neuroimage.2017.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mehta R, Majumdar A, Sivaswamy J. BrainSegNet: a convolutional neural network architecture for automated segmentation of human brain structures. J Med Imaging (Bellingham) 2017;4:024003 10.1117/1.JMI.4.2.024003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen H, Dou Q, Yu L, et al. . VoxResNet: deep voxelwise residual networks for brain segmentation from 3D MR images. Neuroimage 2018;170:446–55 10.1016/j.neuroimage.2017.04.041 [DOI] [PubMed] [Google Scholar]
- 26. Dou Q, Chen H, Yu L, et al. . Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging 2016;35:1182–95 10.1109/TMI.2016.2528129 [DOI] [PubMed] [Google Scholar]
- 27. Chen L, Bentley P, Rueckert D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. Neuroimage Clin 2017;15:633–43 10.1016/j.nicl.2017.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maier O, Schröder C, Forkert ND, et al. . Classifiers for ischemic stroke lesion segmentation: a comparison study. PloS One 2015;10:e0145118 10.1371/journal.pone.0145118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nielsen A, Mouridsen K, Hansen M, et al. . Deep learning: utilizing the potential in data bases to predict individual outcome in acute stroke. In: Proceedings of the Annual Meeting of the International Society for Magnetic Resonance in Medicine, Honolulu, Hawaii April 22–27, 2017;5665 [Google Scholar]
- 30. Menze BH, Jakab A, Bauer S, et al. . The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 2015;34:1993–2024 10.1109/TMI.2014.2377694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chang P. Fully convolutional deep residual neural networks for brain tumor segmentation. In: Proceedings of the International Workshop on Brain Lesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Athens, Greece October 17, 2016;108–18 [Google Scholar]
- 32. Korfiatis P, Kline TL, Erickson BJ. Automated segmentation of hyperintense regions in FLAIR MRI using deep learning. Tomography 2016;2:334–40 10.18383/j.tom.2016.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao XW, Hui R, Tian Z. Classification of CT brain images based on deep learning networks. Comput Methods Programs Biomed 2017;138:49–56 10.1016/j.cmpb.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 34. Shi J, Zheng X, Li Y, et al. . Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer's disease. IEEE J Biomed Health Inform 2018;22:173–83 10.1109/JBHI.2017.2655720 [DOI] [PubMed] [Google Scholar]
- 35. Suk HI, Lee SW, Shen D; Alzheimer's Disease Neuroimaging Initiative. Deep sparse multi-task learning for feature selection in Alzheimer's disease diagnosis. Brain Struct Funct 2016;221:2569–87 10.1007/s00429-015-1059-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scherer M, Cordes J, Younsi A, et al. . Development and validation of an automatic segmentation algorithm for quantification of intracerebral hemorrhage. Stroke 2016;47:2776–82 10.1161/STROKEAHA.116.013779 [DOI] [PubMed] [Google Scholar]
- 37. Al-Ayyoub M, Alawad D, Al-Darabsah K, et al. . Automatic detection and classification of brain hemorrhages. WSEAS Transactions on Computers 2013;10:395–405 [Google Scholar]
- 38. Phong T, Duong H, Nguyen H, et al. . Brain hemorrhage diagnosis by using deep learning. In: Proceedings of the International Conference on Machine Learning and Soft Computing, Ho Chi Minh City, Vietnam January 13–16, 2017 [Google Scholar]
- 39. Esteva A, Kuprel B, Novoa RA, et al. . Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115–18 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Plis SM, Hjelm DR, Salakhutdinov R, et al. . Deep learning for neuroimaging: a validation study. Front Neurosci 2014;8:229 10.3389/fnins.2014.00229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee JH, Wong E, Bandettini P. Temporal-autoencoding neural network revealed the underlying functional dynamics of fMRI data: evaluation using the Human Connectome Project data. In: Proceedings of the Annual Meeting of the International Society for Magnetic Resonance in Medicine, Honolulu, Hawaii April 22–27, 2017 [Google Scholar]
- 42. Zhang Z, Xie Y, Xing F, et al. . MDNet: a semantically and visually interpretable medical image diagnosis network. http://arxiv.org/pdf/1707.02485.pdf. Accessed January 23, 2018.