Abstract
Understanding the immunological behavior of COVID-19 cases at molecular level is essential for therapeutic development. In this study, multi-omics and systems pharmacology analyses were performed to unravel the multi-targeted mechanisms of novel bioactives to combat COVID-19. Immuno-transcriptomic dataset of healthy controls and COVID-19 cases was retrieved from ArrayExpress. Phytocompounds from ethnobotanical plants were collected from PubChem. Differentially expressed 98 immune genes associated with COVID-19 were derived through NetworkAnalyst 3.0. Among 259 plant derived compounds, 154 compounds were targeting 13 COVID-19 immune genes involved in diverse signaling pathways. In addition, pharmacological properties of these phytocompounds were compared with COVID-19 drugs prescribed by WHO, and 25 novel phytocompounds were found to be more efficient with higher bioactive scores. The current study unravels the virogenomic signatures which can serve as therapeutic targets and identified phytocompounds with anti-COVID-19 efficacy. However, further experimental validation is essential to bring out these molecules as commercial drug candidates.
Keywords: COVID-19, Cheminformatics, Immuno-transcriptomics, Immune genes, Interactomics, Phytocompounds, Systems pharmacology
Highlights
-
•
This study unraveled COVID-19 human immune targets and mined the novel bioactives against COVID-19 via in silico analyses
-
•
Immuno-transcriptomic analysis revealed 98 immune responsive genes were differentially expressed in COVID-19 cases
-
•
Cheminformatics and interactome study unveiled 154 compounds interacting with 13 COVID-19 associated immune genes
-
•
Identified 13 immune genes could be useful to design competitive antagonists to combat COVID-19
-
•
A 25 potential compounds were found to be more efficacious than the WHO prescribed drugs for the treatment of COVID-19
1. Introduction
Certain viruses belong to Coronaviridae family regularly present in the human population, for example, rhinoviruses which cause mild respiratory infections, a certain type of viruses such as severe acute respiratory syndrome associated coronavirus (SARS-CoV) and the Middle East respiratory syndrome associated coronavirus (MERS-CoV) cause a lethal respiratory syndrom in humans [1,2]. The emergence of novel corona virus (2019-nCoV) has evolved as a serious global threat due to highest transmission from human to human [3,4]. Because of the global spread, the World Health Organization (WHO) officially declared coronavirus disease (COVID-19) as a pandemic disease on March 11, 2020. A new virus 2019-nCoV was identified to be a type of beta coronavirus and as it is closely related to SARS-CoV with 76% genome similarity and it was renamed as SARS-CoV2 by the International Committee on Taxonomy of Viruses [5]. COVID-19 caused by SARS-CoV2 is spreading at an alarming rate across continents and it certainly impacted the health and wealth of the entire world. This viral pandemic is an imminent threat to the human population. The current COVID-19 pandemic condition is that there is no approved drug available to date to treat this disease and the only measure which is being taken is the prevention of disease spread by social distancing [6,7]. Thus, the need of the hour for this world is to identify the potential drug molecules to treat the condition and to understand the in-depth molecular mechanisms and switches involved in virus entry into a host cell to prevent the transmission and infection.
The spike protein of SARS-CoV2 is almost structurally identical to that of SARS-CoV and the entry of SARS-CoV2 into host cells is mediated by the binding of S proteins to the angiotensin-converting enzyme-2 (ACE2) receptor present on the host cell surface followed by membrane fusion [[8], [9], [10]]. The spike protein has two functional subunits namely S1 involved in receptor binding and S2 involved in membrane fusion [11]. In addition, there is a need for spike protein priming which is mediated by the proteolytic activity of transmembrane protease serine 2 (TMPRSS2) to expose the functional subunits [12]. But role of these receptor molecules with their interacting immune responsive genes remain unexplored. The scientific efforts taken so far to overcome the pandemic of COVID-19 are focusing more on ACE2 receptor inhibitors, TMPRSS2 inhibitors and the development of spike protein vaccines. But none of the studies conclusively cataloged the status of phytomolecules and their respective human immune responsive target genes.
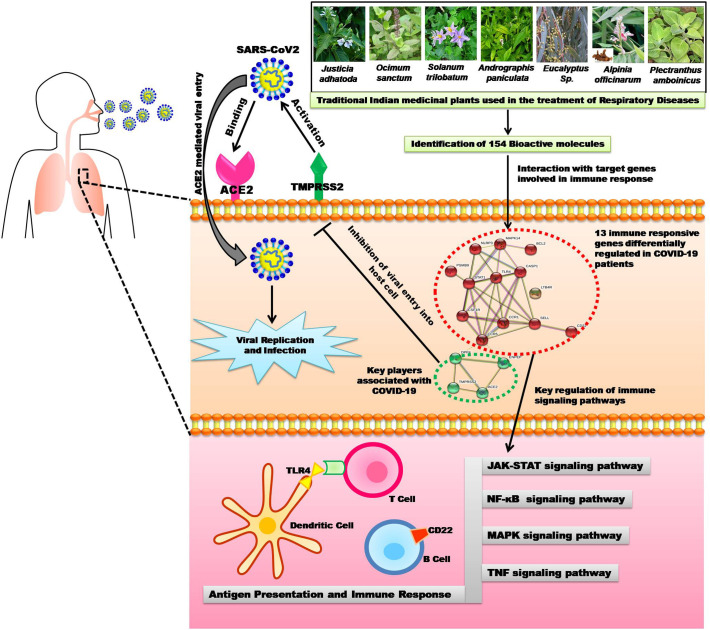
Medicinal plants are the best reservoir of huge numbers of pharmacologically active bioactive compounds and used as a curative medicine for multiple diseases from ancient period. These are all the backbone of traditional medicine; approximately 3.3 billion people in developing countries are still using them on an ordinary basis [13]. These reservoirs contain plenty of pharmaceutical ingredients that can be used in the process of new drug development. Medicinal plants play a crucial role in the development of human cultures around the world. In accordance with the IUCN (International Union for Conservation of Nature) report, nearly 80,000 flowering plant species were worn for medicinal purposes. Justicia adhatoda, Ocimum sanctum, Solanum trilobatum, Andrographis paniculata, Eucalyptus Sp., Alpinia officinarum and Plectranthus amboinicus are some of the essential medicinal plants in traditional Indian medicines. These plants were used in traditional Ayurvedic medicines to treat the various respiratory tract diseases such as pharyngitis, bronchitis, sinusitis, asthma, cough, tuberculosis [[14], [15], [16], [17], [18], [19], [20], [21], [22]]. In addition, these plants are also used to treat skin diseases, wound healing, diabetes, dysentery, digestive and cardiovascular diseases. The selected medicinal plants also possess a wide variety of pharmacological activities including antiviral, adaptogenic, antioxidant, anticancer, analgesic, anti-tussive, anti-inflammatory, antimicrobial, and immune-modulator [[14], [15], [16], [17], [18], [19], [20], [21], [22]].
In spite of the pivotal role of Indian traditional medicine, how the phytomolecules will work and what are their significant immune responsive human targets are still a major bottleneck. The main goal of this present study is to explore the significant immunological mechanism and the pharmaceutical properties and activities of bioactive compounds from J. adhatoda, O. sanctum, S. trilobatum, A. paniculata, Eucalyptus Sp., A. officinarum and P. amboinicus against COVID-19, major issues addressed in the current study are as follows: (i) which human immune responsive genes are differentially regulated in COVID-19? (ii) Which bioactive phytomolecules are involved in the immune regulatory functions for the treatment of this deadly COVID-19 infection? (iii) Which human COVID-19 immune responsive genes are closely associated and modulated by the phytomolecules to accomplish the immunobiological activity and the purpose of curing the respiratory (COVID-19) viral disease? With the advancement of systems pharmacology and pivotal analytical tools such as immuno-transcriptomics, cheminformatics, interactomics analyses allow us to unravel the molecular mechanisms of traditional Indian medicines in treating this deadly infection. Hence, the present study reveals in-depth information into the immunological mechanisms of bioactive molecules and their pharmacological roles. Immuno-transcriptomic profiling identifies the differentially expressed genes associated with COVID-19. Cheminformatics analysis was performed to filter the novel bioactive compounds with essential pharmacological activities and also the dependability of compound – human immune target interactions were predicted. The obtained COVID-19 immunological targets were then imported on to the specialized databases to find out their immunological mechanisms and signaling pathways of active phytocompounds. We hope that the help of human systems pharmacology and the investigation of immunological mechanisms of traditional Indian medicines will significantly promote the development of new drugs and for the treatment of COVID-19 and other respiratory diseases in mere future.
2. Materials and methods
A global multi-omics and systems pharmacology integrated approaches have been applied for the very first time to unravel the significant curative efficacy of potential therapeutic molecules from ethnobotanical plants to combat deadly COVID-19 consisting of: (i) target mining and functional enrichment analysis to identify the phytocompounds – COVID-19 direct immune target network; (ii) systemic network edifice and analysis to demonstrate the molecular machinery of phytocompounds derived from Indian traditional medicinal plants in treating COVID-19; (iii) functional gene ontology and STRING interaction for COVID-19 immune responsive gene targets will pave the way for diverse biological pathway analysis to reveal the functional mode of key players in multiple nodes from an immunological pathway level.
2.1. In silico mining of immune responsive genes in healthy controls and COVID-19 cases from human transcriptome
The human immuno- transcriptomic dataset of healthy controls and COVID-19 cases (E-MTAP-8871) was collected/ retrieved from ArrayExpress database (www.ebi.ac.uk/arrayexpress/). The retrieved dataset was manually curated using MS Excel. Further, this human immuno- transcriptomic data was imported into the Gene Expression Table of NetworkAnalyst 3.0 tool [23]. The dataset was uploaded using (.txt or .zip) input file format and that can be plotted in Excel file with microarray data intensities (gene expression values), corresponding immune responsive genes or healthy controls and COVID-19 cases or samples or time series in columns and rows with gene name. Each column was named as per the type of specific samples and or cases. Filtering and normalization were performed to remove data that are simply erroneous and to ensure that the expression distribution of each sample is similar across the entire experiment, respectively. Followed by differential gene expression analysis was carried out to identify the significant immune responsive genes using Limma statistical model with adjusted P-value 0.05 and Log2 fold change is 1.0. The identified significant immune responsive genes (official gene symbol) were used to study the over-representation analysis (ORA) functional enrichment and tissue specific (whole blood) protein-protein interaction (PPI) analysis through inbuilt KEGG and DifferentialNet databases of NetworkAnalyst 3.0, respectively. It will pave the way to impute the potential human drug targets for further analysis.
2.2. Collection of pharmacologically active phytomolecules
The comprehensive information on 259 pharmacologically active compounds from J. adhatoda, O. sanctum, S. trilobatum, A. paniculata, Eucalyptus Sp., A. officinarum and P. amboinicus were collected from web sources and literature [[14], [15], [16], [17], [18], [19], [20], [21], [22]]. A list of pharmacologically active phytomolecules was given in Table 1 .
Table 1.
Plant active compounds and its abbreviations.
| S. No | Compounds | Abbreviations |
|---|---|---|
| Justicia adhatoda | ||
| 1. | Undecanal | UD |
| 2. | Anisotine | AS |
| 3. | Peganine | PN |
| 4. | Arachidic acid | ARA |
| 5. | Octanal | OCT |
| 6. | Tetradecane | TD |
| 7. | 5-Octadecenal | 5-OD |
| 8. | Scopolamine | SL |
| 9. | Tetradecanol | TD |
| 10. | Megastigmatrienone B | MGM B |
| 11. | Ascorbic acid | AA |
| 12. | Scopoline | SCP |
| 13. | Docosanoic acid | DA |
| 14. | 17-Octadecynoic acid | 17-OCA |
| 15. | Taraxerol | TX |
| 16. | beta-Carotene | β-CT |
| 17. | beta-Sitosterol | β-SS |
| 18. | 9,12-Octadecadienoic acid | 9,12-ODA |
| 19. | Vasicine | VS |
| 20. | Betaine | BT |
| 21. | Vasicinol | VSC |
| 22. | Vasicinolone | VSN |
| 23. | 1-Octene | 1-OC |
| 24. | 1-Pentadecanol | 1-PD |
| 25. | (-)-Verbenone | (-)-VB |
| 26. | Cerotic acid | CA |
| 27. | Eicosane | ES |
| 28. | Phytol | PT |
| 29. | Heptacosane | HEC |
| 30. | Tricosane | TRC |
| 31. | Pentacosane | PEC |
| 32. | 8,11-Octadecadienoic acid | 8,11-OCA |
| 33. | Palmitic acid | PA |
| 34. | Caryophyllene oxide | CAO |
| 35. | Vasicinone | VSC |
| 36. | Isomethyl-alpha-ionone | IAI |
| 37. | Deoxyvasicinone | DOV |
| 38. | Hentriacontane | HC |
| 39. | Tetracontane | TC |
| 40. | Nonacosane | NC |
| 41. | Heneicosane | HC |
| 42. | Vasicol | VC |
| 43. | Fenretinide | FRT |
| 44. | Tetracosanoic acid | TTA |
| 45. | Dodecane | DD |
| 46. | beta-Eudesmol | β-ED |
| 47. | Vasicoline | VSC |
| 48. | Linoleic acid | LA |
| 49. | Vasicolinone | VSL |
| 50. | Lyoniside | LYN |
| Ocimum sanctum | ||
| 1. | Aromadendrene oxide | ADO |
| 2. | Apigenin | AG |
| 3. | Benzaldehyde | BA |
| 4. | Borneol | BN |
| 5. | Bornyl acetate | BA |
| 6. | Campesterol | CP |
| 7. | Caryophyllene oxide | CO |
| 8. | Carvacrol | CC |
| 9. | Cineole | CN |
| 10. | Cirsilineol | CSL |
| 11. | Cirsimaritin | CSM |
| 12. | Cubenol | CB |
| 13. | alpha-terpineol | α-TP |
| 14. | Citral | CT |
| 15. | D-Limonene | D-LM |
| 16. | beta-Elemene | β-EM |
| 17. | Eucalyptol | ECL |
| 18. | Camphor | CP |
| 19. | Eugenol | EG |
| 20. | Eicosane | EC |
| 21. | Germacrene D | GD |
| 22. | Heptanol | HT |
| 23. | Linalol | LL |
| 24. | Linoleic acid | LA |
| 25. | Luteolin | LT |
| 26. | Methyleugenol | MTE |
| 27. | Butyl benzoate | BB |
| 28. | Ocimarin | OM |
| 29. | Oleanolic acid | OLA |
| 30. | Oleic acid | OA |
| 31. | Palmitic acid | PA |
| 32. | Phytol | PT |
| 33. | Rosmarinic acid | ROA |
| 34. | Sabinene | SB |
| 35. | Stearic acid | SA |
| 36. | Selinene | SL |
| 37. | Stigmasterol | SS |
| 38. | Thymol | TM |
| 39. | Ursolic acid | UA |
| 40. | Vanillic acid | VA |
| 41. | Viridiflorol | VDF |
| 42. | Xylose | XL |
| 43. | 1-Octen-3-OL | 1-OCT-3-OL |
| 44. | α-Camphene | α-CP |
| 45. | alpha-Myrcene | α-MRC |
| 46. | alpha-Pinene | α-PN |
| 47. | α-Bisabolene | α-BB |
| 48. | α-humulene | α-HM |
| 49. | Linolenic acid | LLA |
| 50. | α-thujene | α-TJ |
| 51. | β-caryophyllene | β-CP |
| 52. | β-gurjunene | β-GJ |
| 53. | β-Pinene | β-PN |
| 54. | ß-carotene | β-CT |
| 55. | β-guaiene | β-GE |
| 56. | β-bisabolene | β-BB |
| 57. | ß-sitosterol | β-SS |
| 58. | (E)-beta-ocimene | (E)- β-OM |
| 59. | 3-Furaldehyde | 3-FA |
| Solanum trilobatum | ||
| 1. | Sobatum | SB |
| 2. | Solasodine | SSD |
| 3. | Solanine | SN |
| 4. | Tomatidine | TM |
| 5. | Disogenin | DG |
| 6. | β-Solamargine | β-SM |
| 7. | Campesterol | CS |
| 8. | Sitosterol | SS |
| 9. | Soladunalinidine | SDL |
| Andrographis paniculata | ||
| 1. | Andrographolide | ADG |
| 2. | Neoandrographolide | NAG |
| 3. | Isoandrographolide | IAG |
| 4. | Andrographiside | AGP |
| 5. | Andrograpanin | AG |
| 6. | Andrographolactone | AGL |
| 7. | Bisandrographolide A | BAG |
| 8. | Apigenin | AG |
| 9. | 7-O-methylwogonin | 7-O-MW |
| 10. | Onysilin | OS |
| 11. | Andrographidine A | AGA |
| 12. | Andrographidine C | AGC |
| 13. | Luteolin | LT |
| 14. | 14-Deoxyandrographolide | 14- DOA |
| 15. | 14-Deoxy 11, 12-didehydroandrographolide | 14-D-11,12-DAG |
| 16. | 14-Deoxy-11-hydroxyandrographolide | 14-D-11-HAG |
| 17. | 14-Deoxy-12-hydroxyandrographolide | 14-D-12-HAG |
| 18. | 3-O-beta-D-glucopyranosyl 14, 19-dideoxyandrographolide | 3-O β-D-G14,19-DDAG |
| 19. | 8, 17-Epoxy-14-deoxyandrographolide | 8,17-E-14-DAG |
| 20. | 3,4-Dicaffeoylquinic acid | 3,4-DA |
| Alpinia officinarum | ||
| 1. | Chrysin | CS |
| 2. | Pinocembrin | PC |
| 3. | Tectochrysin | TC |
| 4. | Galangin | GAL |
| 5. | Acacetin | AC |
| 6. | Kaempferide | KPF |
| 7. | Isorhamnetin | IR |
| 8. | Apigenin | AG |
| 9. | 3‑O‑methylgalangin | 3-0-MG |
| 10. | Kaempferol | KAF |
| 11. | Quercetin | QC |
| 12. | Rutin | RT |
| 13. | Yakuchinone A | YCA |
| 14. | Hexahydrocurcumin | HHC |
| 15. | Hannokinol | HK |
| 16. | Nootkatone | NK |
| 17. | Luteolin | LT |
| 18. | Izalpinin | IP |
| 19. | Pinobaksin | PB |
| 20. | 5‑hydroxy‑1,7‑diphenyl‑3‑heptanone -3,5‑dihydroxy‑1,7‑diphenylheptane | 5-H-1,7-D-3-H |
| 21. | 1,7‑diphenylhept‑4‑en‑3‑one | 1,7-DP-4-E3O |
| 22. | Zingerone | ZG |
| 23. | 1’‑acetoxychavicol acetate | 1'-ACA |
| 24. | β‑sitosterol | β-SS |
| 25. | p‑Coumaryl alcohol | p-CA |
| 26. | 1,5‑bis‑(4‑hydroxyphenyl)‑2‑(hydroxymethyl)‑4‑penten‑1‑ol | 1,5-B-4H-2HE-4-P-1 |
| 27. | 1,5‑bis‑(4‑hydroxyphenyl)‑1‑methoxy‑2‑(methoxymethyl) ‑4‑pentene | 1,5-B-4H-1M-2-4P |
| 28. | 1,5‑bis‑(4‑hydroxyphenyl)‑1‑ethoxy‑2‑(methoxymethyl)‑4‑pentene | 1,5-B-4H-1E-2-M-4-P |
| 29. | 1,5‑bis‑(4‑hydroxyphenyl)‑1‑[3‑(4‑acetoxyphenyl)‑2‑propenoxy]‑2‑(methoxymethyl)‑4‑pentene | 1,5-B-4H-1-3-4A-2P-2-M-4-P |
| 30. | 1,5‑bis‑(4‑hydroxyphenyl)‑2‑(methoxymethyl)‑4‑penten‑1‑ol | 1,5-B-4H-2-ME-4-P-1 |
| Plectranthus amboinicus | ||
| 1. | Carvacrol | CC |
| 2. | Thymol | TM |
| 3. | Eugenol | EG |
| 4. | Chavicol | CVC |
| 5. | 1,8-Cineole | 1,8-CN |
| 6. | β-Caryophyllene | β-CP |
| 7. | p-Cymene | p-CM |
| 8. | Caryophyllene oxide | CPO |
| 9. | α-Terpinene | α-TP |
| 10. | Spathulenol | STL |
| 11. | Ethyl Salicylate | ES |
| 12. | Terpinen-4-ol | TP-4-ol |
| 13. | α-Terpinolene | α-TPL |
| 14. | Squalene | SQL |
| 15. | Oleic acid | OA |
| 16. | Phytol | PT |
| 17. | β-Cedrene epoxide | β-CE |
| 18. | Tetradecanal | TTD |
| 19. | α-Humulene | α-HM |
| 20. | β-Copaen-4-α-ol | β-CP-4-α-ol |
| 21. | β-Selinene | β-SL |
| 22. | α-Terpineol | α-TP |
| 23. | γ-Terpinene | γ-TP |
| 24. | β-Himachalene oxide | β-HO |
| 25. | Undecanal | UD |
| 26. | α-Calacorene | α-CC |
| 27. | Methyl chavicol | MC |
| 28. | Patchoulane | PC |
| 29. | Terpine-4-ol | TP-4-ol |
| 30. | trans-Caryophyllene | tr-CP |
| 31. | α-Cadinol | α-CD |
| 32. | 1-Octen-3-ol | 1-OCT-3-ol |
| 33. | (Z)-1,3-Hexadiene | (Z)-1,3,-HD |
| 34. | (Z)-3-Hexenol | (Z)-3-HX |
| 35. | α-Muurolene | α-MR |
| 36. | (E,E)-α-Farnesene | (E,E)-α-FS |
| 37. | Camphor | CP |
| 38. | δ-3-Carene | δ-3-CR |
| 39. | Linalool | LL |
| 40. | Nerol acetate | NA |
| 41. | Geranyl acetate | GA |
| 42. | 3-Carene | 3-CR |
| 43. | Caffeic acid | CA |
| 44. | Gallic acid | GA |
| 45. | p-Coumaric acid | p-CA |
| 46. | Rosmarinic acid | ROA |
| 47. | Salvianolic acid A | SAA |
| 48. | Chrysoeriol | CS |
| 49. | Durohydroquinone | DHQ |
| 50. | Dihydro carveol | DHC |
| 51. | Cirsimaritin | CSM |
| 52. | Eriodictyol | ED |
| 53. | Luteolin | LT |
| 54. | Rutin | RT |
| 55. | Salvigenin | SG |
| 56. | Thymoquinone | TQ |
| 57. | Methyl carvacrol | MC |
| 58. | Methyl octanoate | MO |
| 59. | β-Sesquiphellandrene | β-SQP |
| 60. | Quercetin | QC |
| 61. | Methyl eugenol | ME |
| 62. | Ocimene | OM |
| 63. | Geraniol | GN |
| 64. | Germacrene D | GCD |
| Eucalyptus Sp. | ||
| 1. | α- pinene | α- PN |
| 2. | 1,8-cineol | 1,8-CN |
| 3. | α-Terpineol | α-TP |
| 4. | p-Cymene | p-CM |
| 5. | γ-Terpinene | γ-TP |
| 6. | Trans-Pinocarveol | tr-PC |
| 7. | α-Terpinyl acetate | α-TA |
| 8. | Globulol | GB |
| 9. | Limonene | LN |
| 10. | Guaiene | GE |
| 11. | Spathulenol | STL |
| 12. | Terpinene-4-ol | TP-4-Ol |
| 13. | Aromadendrene | AM |
| 14. | Cryptone | CP |
| 15. | Verbenone | VB |
| 16. | Phellandral | PL |
| 17. | p-Cymen-8-ol | P-CM-8-ol |
| 18. | Caryophyllene oxide | CPO |
| 19. | Epiglobulol | EG |
| 20. | Viridiflorol | VIF |
| 21. | Carvacrol | CV |
| 22. | α-Eudesmol | α- ED |
| 23. | β-Eudesmol | β-ED |
2.3. Retrieval of phytochemical information and human target imputations
In total, 259 pharmacologically active plant derived compounds and their canonical SMILES from J. adhatoda, O. sanctum, S. trilobatum, A. paniculata, Eucalyptus Sp., A. officinarum and P. amboinicus were obtained from the PubChem database [24]. The identified plant derived active compounds with their respective canonical SMILES were searched against Homo sapiens in SwissTargetPrediction tool to retrieve the compounds with their corresponding human targets especially on immune responsive genes (www.swisstargetprediction.ch/).
2.4. Computational mining of human targets and encoding features
Identified significant human immune responsive genes/ targets were imported onto the NCBI-Gene database and/or Expression atlas for retrieving the molecular features such as official gene symbol with their full name, exact position of the targets, chromosome number and orthologs of differentially expressed immune responsive genes [25].
2.5. Compound Target Network (C-T-N) construction
C-T-N was constructed to battle the COVID-19 by illuminating the multi-target therapeutic features of the pharmacologically active plant derived compounds. In this molecular interactome, promising immune target proteins and active phytocompounds interacted if the protein is targeted by the phytomolecule. Molecular interactome was visualized by Cytoscape v3.7.2 [26]. In the obtained interactome, node depicts compounds and targets whereas edges denote the molecular interactions between them.
2.6. Identification of features of the phytomolecules
Phytocompounds with their respective canonical SMILES were subjected to Molinspiration (www.molinspiration.com/cgi-bin/properties) [27] tool to obtain the significant calculation on pharmacologically active compounds with their molecular features. In addition to that, the imputation of the bioactive score for the vital targets such as Kinase inhibitor activity (Ki), GPCR ligand activity (GPCR), protease inhibitor activity (Pi), number of violations (nvio), enzymes and nuclear receptors (Ncr) was predicted.
2.7. Drug and phytomolecule comparison
Identified pharmacologically active molecules with their respective molecular and bioactive features were compared with commercially available WHO suggested drugs for the treatment of COVID-19. This comparative analysis was done to unveil the active compounds with the help of nvio, GPCR, Ki, Ei, Pi and Ncr properties.
2.8. Gene Ontology (GO) enrichment analysis
Differentially expressed genes (DEGs) with their encoding gene symbols were uploaded to the GOnet database (https://tools.dice-database.org/GOnet/) [28] to attain ontology against H. sapiens with significant threshold q-value level is <0.05. Immune responsive genes were also pigeonholed as per GO molecular function (MF) and biological process (BP) according to the functional enrichment classification of the GOnet database.
2.9. Immune responsive and human COVID-19 receptor genes network analysis
Forty-two human COVID-19 receptors and essential immunity genes were collected from the literature [29]. The list of these genes and their functions were given in Table S1. Protein- protein interactions (PPI) analysis of these COVID-19 receptors, essential immunity genes and identified significantly active immune responsive genes (official gene symbols) were done by STRING v10.5 with a high confidence score of 0.7. Further, the PPI network enrichment analysis was executed through a significant value of 0.01. This signalome was used to delineate the physical and functional role of the candidate's involved [30].
3. Results
3.1. Immune responsive genes from meta–analysis of human immuno-transcriptome
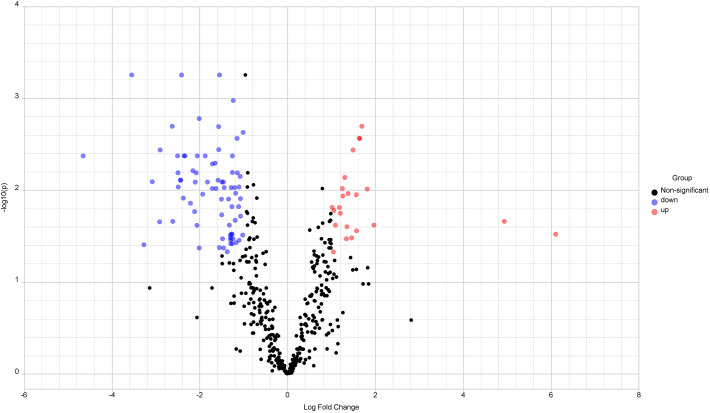
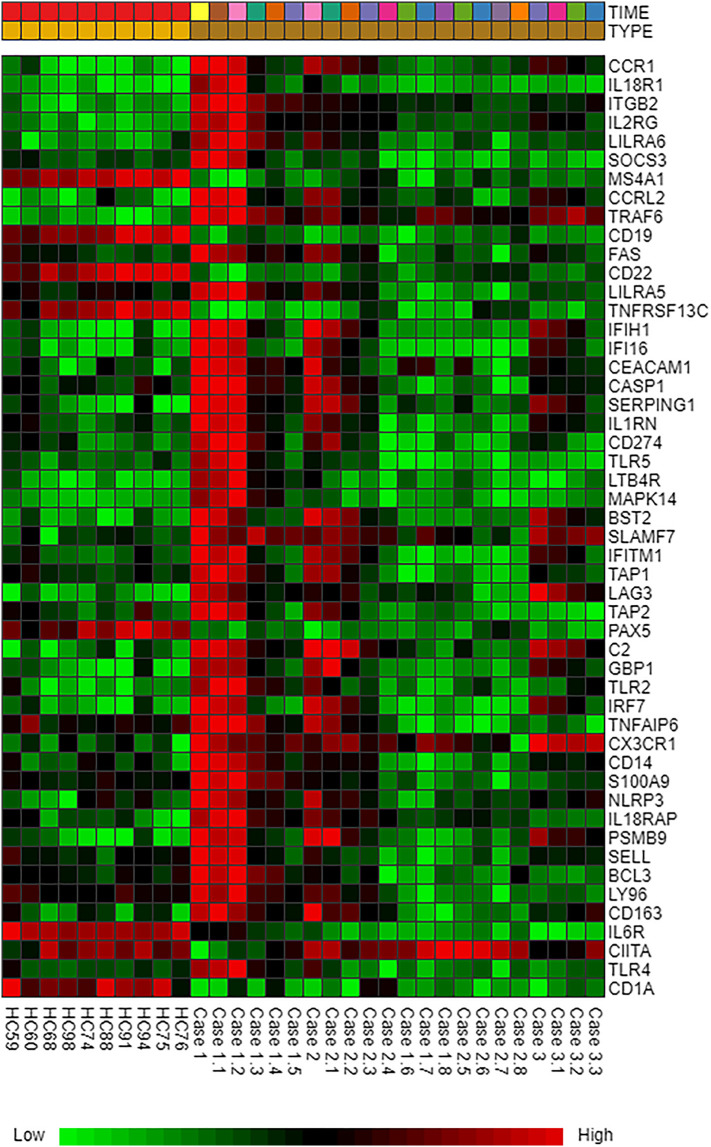
The immuno-transcriptomic dataset contains 579 immune responsive genes of which 549 genes were commonly found in meta–differential (up, down and non-significant) expression (Fig. 1 ) and the remaining 30 genes were unmatched. Heatmap profiling revealed that 98 immune responsive genes were differentially expressed in COVID-19 cases at various time points when compared to healthy controls (Fig. 2 ). Further, these significant genes were involved in tissue specific (whole blood) PPI network. This network had 1228 nodes, 1616 edges and 80 seed proteins (Fig. 3 and Table S2) and the pathway based ORA enrichment network showed the involvement in various biological pathways (Fig. 4 and Table S3).
Fig. 1.
Volcano Plot for human immune differential gene expression. Scattered points encode genes. The x-axis is log2 fold change values of gene expression levels between healthy controls and COVID-19 cases, whereas the y-axis is adjusted P-value based on –log10. Red color dots - upregulation, blue color dots - down regulation represents differentially expressed immune genes based on significant thresholds (i) adjusted P-value <0.05 and (ii) log2 fold change level is 1.0 and black color dots – non significant immune responsive genes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
Heatmap profiling denoting the top 98 genes which are differentially expressed between human healthy controls and COVID-19 cases. Green color, down regulation; Red color, up regulation; Black color, non significant expression. Dataset: E-MTAB-8871- Time: In Days (0, 4,5, 6, 7, 8, 9, 10, 11, 12, 13, 18 and 19) and Type: Healthy Controls and COVID-19 cases. The colored scale bar at bottom represents relative expression, where green and red colors represent down regulation and up regulation, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Tissues Specific protein protein interaction. Red color represents differentially expressed human immune responsive genes/nodes, blue color indicates interacting partners present in human whole blood tissue type. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
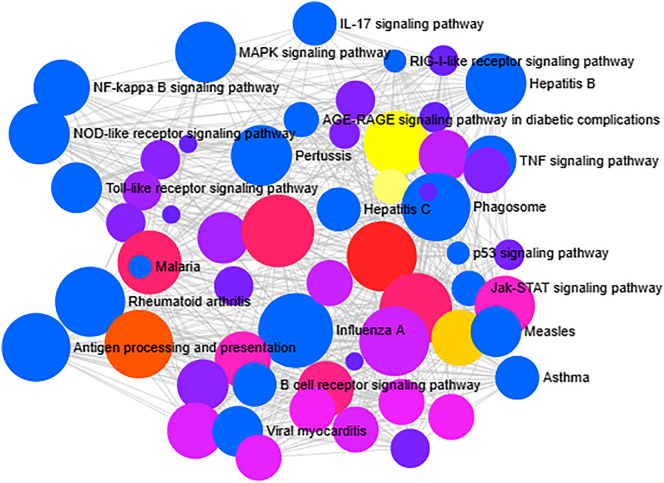
Fig. 4.
Gene set enrichment network analysis. The number of human immune responsive genes falling in each KEGG pathway category is directly proportional to the node size. The nodes are color shaded according to the significance level (adjusted P-value <0.05).
3.2. Phytomolecule information retrieval
A 259 number of phytochemicals were employed as a query in PubChem database to fetch and retrieve the canonical SMILES (Table S4). This obtained SMILES information was used for further biomolecular analyses.
3.3. Identification of pharmacologically active compounds interacting with human targets
Pharmacologically active phytomolecules targeting human immune receptors were imputed through SwissTargetPrediction tool. Among 259 phytomolecules, 154 compounds were significantly targeting 13 out of 98 human immune responsive genes/ receptors which were differentially expressed between healthy controls and COVID-19 cases. A detailed list of phytochemicals and significant human immune responsive gene information are provided in Table S5.
3.4. Properties of human immune responsive genes
One hundred fifty four numbers of potential compounds significantly targets the 13 human immune responsive genes. These immune receptors with their corresponding information such as chromosome number, physical position and full name of the genes and orthologs details were retrieved and given in Table 2 . These immune responsive genes with their attributes will pave the way to delineate their detailed molecular function.
Table 2.
Molecular attributes of human immune responsive genes and their orthologs.
| S. No | Human Gene Name | Full name | Chr. No | Start | End | Orthologs (Mus musculus) | Chr. No | Start | End |
|---|---|---|---|---|---|---|---|---|---|
| 1. | TLR4 | toll like receptor 4 | 9 | 117704403 | 117724735 | Tlr4 | 4 | 66827584 | 66930284 |
| 2. | STAT1 | signal transducer and activator of transcription 1 | 2 | 190968989 | 191014250 | Stat1 | 1 | 52119440 | 52161865 |
| 3. | SELL | selectin Lymphocyte | 1 | 169690667 | 169711620 | Sell | 1 | 164061982 | 164084181 |
| 4. | PSMB9 | proteasome 20S subunit beta 9 | 6 | 32854192 | 32859851 | Psmb9 | 17 | 34181987 | 34187764 |
| 5. | CD22 | CD22 molecule | 19 | 35329169 | 35347361 | Cd22 | 7 | 30865402 | 30880342 |
| 6. | CCR1 | C-C motif chemokine receptor 1 | 3 | 46201711 | 46208313 | Ccr1 | 9 | 123962124 | 123968692 |
| 7. | CCR5 | C-C motif chemokine receptor 5 | 3 | 46370142 | 46376206 | Ccr5 | 9 | 124121543 | 124147699 |
| 8. | LTB4R | leukotriene B4 receptor | 14 | 24311502 | 24318036 | – | – | – | – |
| 9. | MAPK14 | mitogen-activated protein kinase 14 | 6 | 36027677 | 36122964 | Mapk14 | 17 | 28691329 | 28748406 |
| 10. | CSF1R | colony stimulating factor 1 receptor | 5 | 150053291 | 150113372 | Csf1r | 18 | 61100598 | 61132149 |
| 11. | BCL2 | BCL2 apoptosis regulator | 18 | 63123346 | 63320280 | Bcl2 | 1 | 106538178 | 106714274 |
| 12. | CASP1 | caspase 1 | 11 | 105025443 | 105035591 | Casp1 | 9 | 5298508 | 5307290 |
| 13. | NLRP3 | NLR family pyrin domain containing 3 | 1 | 247416163 | 247448823 | Nlrp3 | 11 | 59541568 | 59566956 |
3.5. Functional GO enrichment analysis
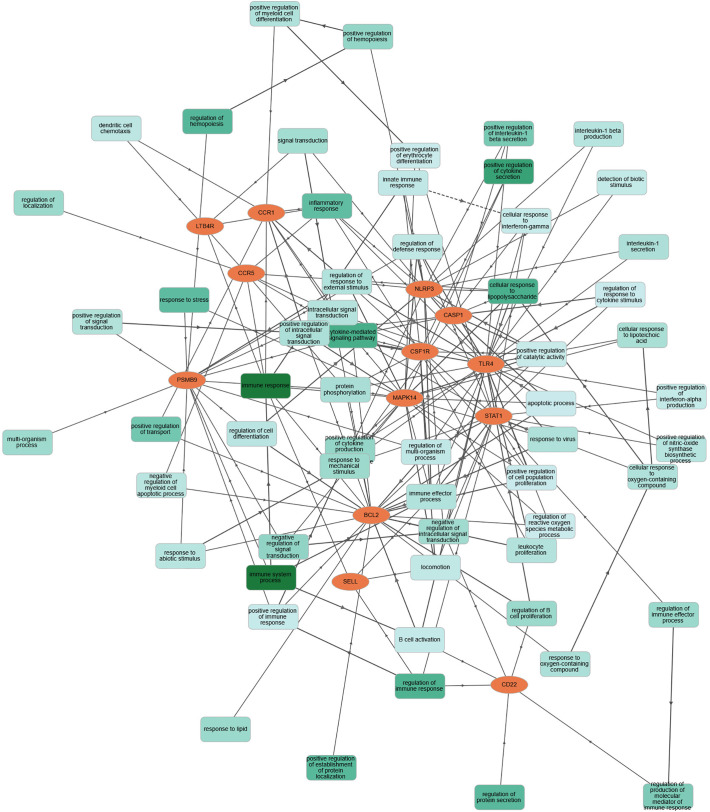
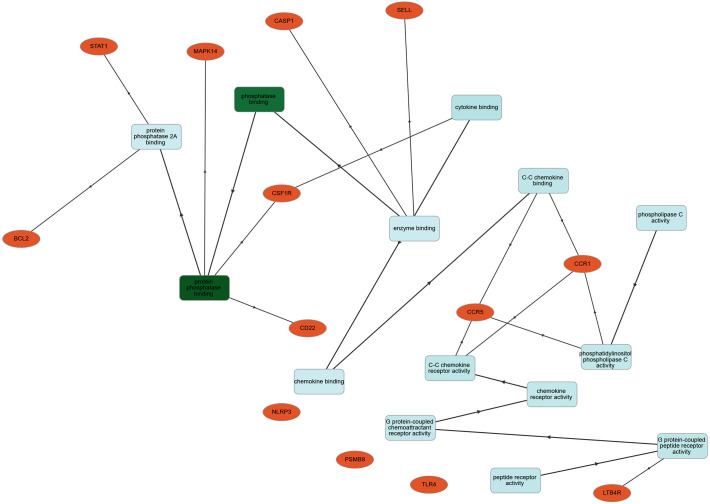
Significant COVID-19 immune responsive gene targets with their corresponding molecular features were analyzed by gene symbols using the GOnet tool which revealed the involvement of these immune proteins in various molecular functions and biological processes. The target immune responsive genes with their corresponding proteins were imputed to be involved in crucial biological regulation of inflammatory response, innate immune response, positive regulation of interferon-α, interlekin-1-β and cytokine secretion, apoptotic process, response to virus, leukocyte and B-Cell proliferation, negative regulation of signal transduction (Fig. 5 ). In molecular functions, human immune targets were involved in phosphatase and chemokine binding, G-protein-coupled chemoattractant receptor and peptide receptor activity (Fig. 6 ).
Fig. 5.
Classification human targets with their biological processes. Orange color encodes human COVID-19 immune targets; green color represents diverse biological processes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Classification human targets with encoding molecular functions. Orange color encodes human COVID-19 immune targets; green color represents significant molecular functions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
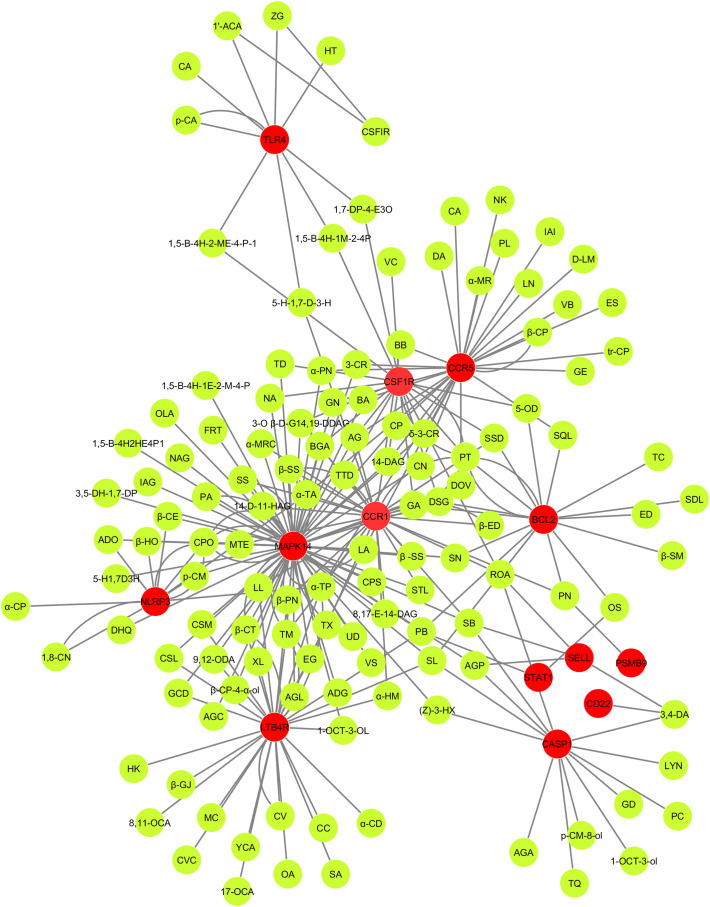
3.6. C-T-N analysis
Fig. 7 showed the C-T-N based cross-talk between 154 active compounds and 13 significant human immune responsive gene targets. Molecular cross-talk between the active compounds and potential immune targets unveiled the multi-target strategy which is the significant feature of herbal medicine. As for the 13 significant human immune targets, 154 phytochemicals (Table S5) had high probability, which revealed the essential therapeutic ability of each natural bioactive molecules present in traditional Indian medicinal plants such as J. adhatoda, O. sanctum, S. trilobatum, A. paniculata, Eucalyptus Sp., A. officinarum and P. amboinicus for combating COVID-19 infection by transducing the transcriptional reprogramming and/or modulating the signals of these possible immune responsive proteins/genes.
Fig. 7.
Compound target network (C-T-N). Green color represents compounds and red color indicates human COVID-19 immune targets. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
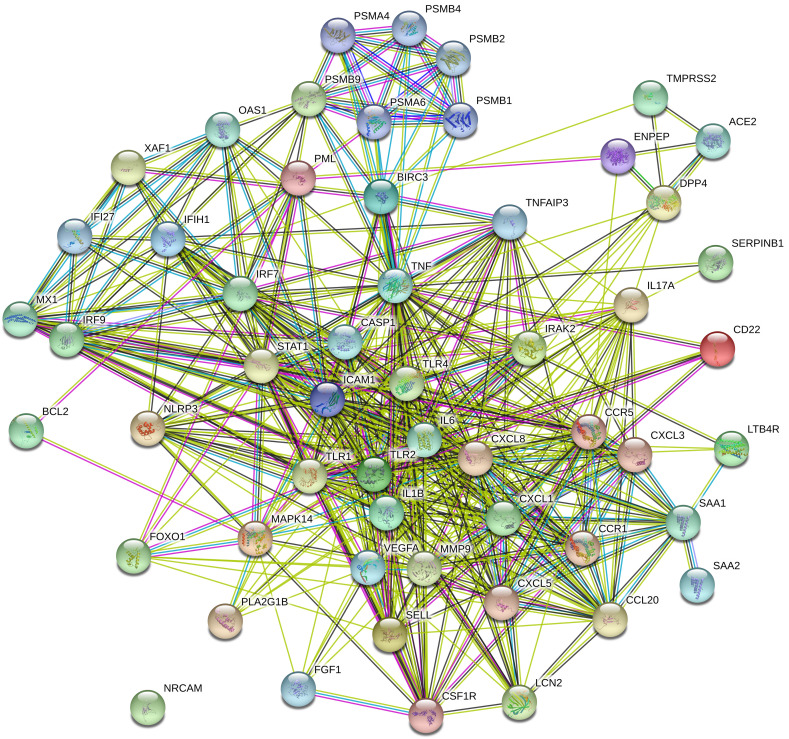
3.7. Molecular interactome analysis
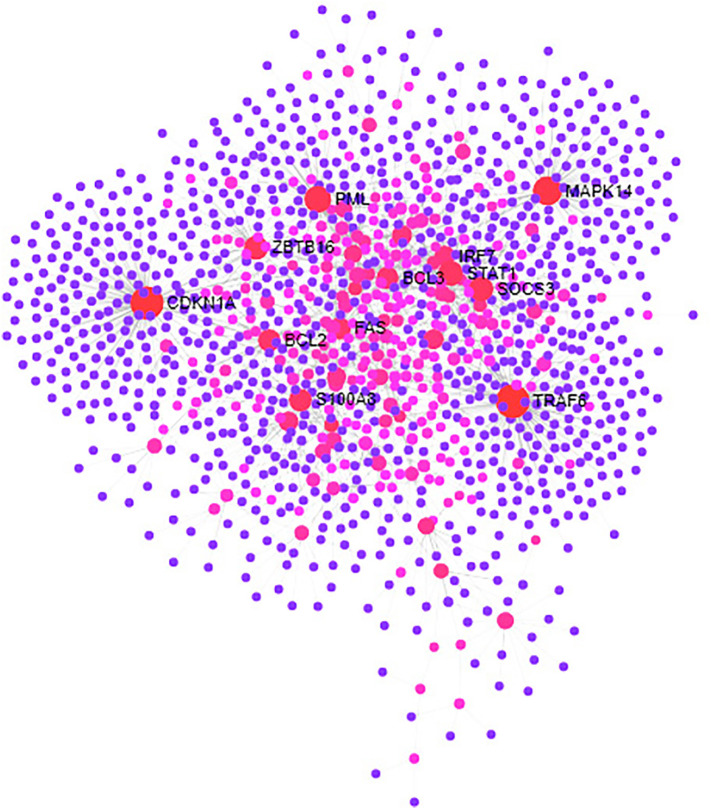
Forty-two numbers of human COVID-19 immune regulators and identified 13 immune responsive genes that are differentially expressed between healthy controls and COVID-19 cases demonstrated molecular cross-talks from H. sapiens. The interactome had 452 edges and 55 nodes (Fig. 8 ). The proteins of the human COVID-19 responsive immune genes with their molecular cross-talks had an average nodal degree of 16.4 in the tightly connected proteins/ immune genes. PPI enrichment of these human COVID-19 responsive immune genes had a P-value score of <0.01. In addition, this interaction showed that the complexity and functionalities of human COVID-19 responsive immune genes, with their cross-talks provide potential targets for therapy against COVID-19 infection.
Fig. 8.
Human COVID-19 immune target genes and their molecular interaction. Human immune responsive genes and their cross-talks showing tightly interacting functional components. Colored lines between the seed proteins indicates various types of interacting elements/ signals. Green color- gene neighbourhood; blue color - gene co-occurrence; pink color - experimentally determined/ Post translational modifications; black color - coexpression. Nodes filled with ribbon like structure represents the availability of protein 3D structural information is predicted or known. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.8. Features of natural bioactive molecules and novel compounds
One fifty-four numbers of natural bioactive molecules with their calculated pharmaceutical properties such as GPCR, Pi, Ki, Ncr, Ei, nVio were collected and given in Table 3 . Based on the number of violations and enzyme inhibitor activity, a feature score above 0.5 were considered as a significant level. Further, the WHO prescribed commercial drugs for the treatment of COVID-19 pandemic were compared with the pharmaceutical properties of natural bioactive compounds and 25 potential compounds were found to be more efficacious than the WHO prescribed drugs and are listed in Table 4 .
Table 3.
Phytocompounds and its pharmaceutical features.
| S.No | Compound | GPCR lg | Ki | Ncr | Pi | Ei | nvio |
|---|---|---|---|---|---|---|---|
| Justicia adhatoda (Family:Acanthaceae) | |||||||
| 1. | 17-Octadecynoic acid | 0.22 | −0.11 | 0.42 | 0.23 | 0.47 | 1 |
| 2. | 1-Pentadecanol | −0.24 | −0.35 | −0.22 | −0.31 | 0.03 | 1 |
| 3. | 5-Octadecenal | 0.08 | −0.22 | 0.04 | 0.11 | 0.30 | 1 |
| 4. | 8,11-Octadecadienoic acid | 0.29 | −0.16 | 0.31 | 0.12 | 0.38 | 1 |
| 5. | 9,12-Octadecadienoic acid | 0.29 | −0.16 | 0.31 | 0.12 | 0.38 | 1 |
| 6. | β-Carotene | −0.04 | −0.15 | 0.40 | −0.06 | 0.17 | 2 |
| 7. | β-Eudesmol | −0.02 | −0.62 | 0.60 | −0.10 | 0.48 | 0 |
| 8. | β-Sitosterol | 0.14 | −0.51 | 0.73 | 0.07 | 0.51 | 1 |
| 9. | Caryophyllene oxide | −0.08 | −0.86 | 0.62 | 0.00 | 0.57 | 0 |
| 10. | Cerotic acid | 0.14 | −0.09 | 0.20 | 0.16 | 0.14 | 1 |
| 11. | Deoxyvasicinone | −0.98 | −0.75 | −1.39 | −1.22 | −0.09 | 0 |
| 12. | Docosanoic acid | 0.17 | −0.10 | 0.23 | 0.17 | 0.17 | 1 |
| 13. | Eicosane | −0.04 | −0.14 | −0.05 | −0.11 | 0.03 | 1 |
| 14. | Fenretinide | −0.08 | −0.18 | 0.67 | −0.18 | 0.31 | 1 |
| 15. | Isomethyl-alpha-ionone | −0.50 | −0.91 | −0.01 | −0.96 | 0.13 | 0 |
| 16. | Linoleic acid | 0.29 | −0.16 | 0.30 | 0.12 | 0.38 | 1 |
| 17. | Lyoniside | 0.14 | −0.18 | −0.20 | 0.07 | 0.23 | 3 |
| 18. | Palmitic acid | 0.02 | −0.33 | 0.08 | −0.04 | 0.18 | 1 |
| 19. | Phytol | 0.11 | −0.32 | 0.35 | 0.00 | 0.31 | 1 |
| 20. | Scopolamine | 0.58 | 0.06 | 0.11 | 0.28 | 0.35 | 0 |
| 21. | Scopoline | 0.07 | −0.62 | −1.02 | −0.55 | 0.35 | 0 |
| 22. | Taraxerol | 0.21 | −0.20 | 0.54 | 0.00 | 0.49 | 1 |
| 23. | Tetradecanol | −0.32 | −0.45 | −0.32 | −0.40 | −0.02 | 1 |
| 24. | Undecanal | −0.54 | −0.96 | −0.70 | −0.41 | −0.03 | 1 |
| 25. | Vasicine | 0.03 | −0.43 | −0.59 | −0.36 | 0.24 | 0 |
| 26. | Vasicol | −0.13 | −0.29 | −0.52 | 0.14 | 0.16 | 0 |
| Ocimum sanctum (Family: Lamiaceae) | |||||||
| 1. | 1-Octen-3-OL | −1.78 | −2.59 | −1.68 | −1.86 | −1.09 | 0 |
| 2. | α-Myrcene | −1.11 | −1.65 | −0.70 | −1.29 | −0.22 | 0 |
| 3. | α-terpineol | −0.51 | −1.45 | −0.02 | −0.78 | 0.14 | 0 |
| 4. | Aromadendrene oxide | −0.41 | −1.00 | −0.04 | −0.00 | −0.23 | 0 |
| 5. | Bornyl acetate | −0.32 | −1.33 | −0.59 | −0.44 | −0.12 | 0 |
| 6. | Butyl benzoate | −0.80 | −1.03 | −0.75 | −0.94 | −0.43 | 0 |
| 7. | Campesterol | 0.11 | −0.48 | 0.71 | 0.01 | 0.50 | 1 |
| 8. | Carvacrol | −1.02 | −1.15 | −0.70 | −1.25 | −0.56 | 0 |
| 9. | Caryophyllene oxide | −0.08 | −0.86 | 0.62 | 0.00 | 0.57 | 0 |
| 10. | Cineole | −0.93 | −1.60 | −1.07 | −0.90 | −0.15 | 0 |
| 11. | Cirsilineol | −0.09 | 0.20 | 0.13 | −0.29 | 0.14 | 0 |
| 12. | Cirsimaritin | −0.09 | 0.20 | 0.17 | −0.31 | 0.14 | 0 |
| 13. | D-Limonene | −0.91 | −2.01 | −0.34 | −1.38 | −0.21 | 0 |
| 14. | Eugenol | −0.86 | −1.14 | −0.78 | −1.29 | −0.41 | 0 |
| 15. | Germacrene D | −0.30 | −0.81 | 0.32 | −0.67 | 0.26 | 1 |
| 16. | Heptanol | −2.90 | −3.18 | −2.88 | −2.92 | −2.50 | 0 |
| 17. | Linalool | −0.73 | −1.26 | −0.06 | −0.94 | 0.07 | 0 |
| 18. | Linoleic acid | 0.29 | −0.16 | 0.31 | 0.12 | 0.38 | 1 |
| 19. | Methyleugenol | −0.81 | −1.06 | −0.80 | −1.14 | −0.43 | 0 |
| 20. | Oleanolic acid | 0.28 | −0.40 | 0.77 | 0.15 | 0.65 | 1 |
| 21. | Palmitic acid | 0.02 | −0.33 | 0.08 | −0.04 | 0.18 | 1 |
| 22. | Phytol | 0.11 | −0.32 | 0.35 | 0.00 | 0.31 | 1 |
| 23. | Rosmarinic acid | 0.17 | −0.18 | 0.57 | 0.15 | 0.24 | 0 |
| 24. | Sabinene | −1.15 | −1.79 | −0.69 | −0.78 | −0.60 | 0 |
| 25. | Selinene | −0.26 | −0.94 | 0.35 | −0.48 | 0.29 | 1 |
| 26. | β-carotene | −0.04 | −0.15 | 0.40 | −0.06 | 0.17 | 2 |
| 27. | β-sitosterol | 0.14 | −0.51 | 0.73 | 0.07 | 0.51 | 1 |
| 28. | Stearic acid | 0.11 | −0.20 | 0.17 | 0.06 | 0.20 | 1 |
| 29. | Stigmasterol | 0.12 | −0.48 | 0.74 | −0.02 | 0.53 | 1 |
| 30. | Thymol | −1.05 | −1.29 | −0.78 | −1.34 | −0.57 | 0 |
| 31. | Xylose | −0.77 | −1.34 | −1.61 | −0.83 | 0.25 | 0 |
| 32. | α-Camphene | −1.02 | −1.85 | −1.15 | −1.40 | −0.82 | 0 |
| 33. | β-caryophyllene | −0.34 | −0.78 | 0.13 | −0.60 | 0.19 | 1 |
| 34. | β-guaiene | −0.52 | −1.04 | −0.04 | −0.72 | −0.24 | 0 |
| 35. | β-Pinene | −0.53 | −1.45 | −0.50 | −0.80 | −0.34 | 0 |
| Solanum trilobatum (Family:Solanaceae) | |||||||
| 1. | Sobatum | 0.14 | −0.51 | 0.73 | 0.07 | 0.51 | 1 |
| 2. | Solasodine | 0.24 | −0.66 | 0.36 | 0.01 | 0.60 | 1 |
| 3. | Solanine | −2.38 | −3.44 | −3.13 | −1.82 | −2.61 | 3 |
| 4. | Tomatidine | 0.32 | −0.50 | 0.28 | 0.17 | 0.57 | 1 |
| 5. | Disogenin | 0.05 | −0.57 | 0.58 | −0.06 | 0.61 | 1 |
| 6. | β-Solamargine | −2.45 | −3.52 | −3.22 | −1.92 | −2.59 | 3 |
| 7. | Campesterol | 0.11 | −0.48 | 0.71 | 0.01 | 0.50 | 1 |
| 8. | Sitosterol | 0.14 | −0.51 | 0.73 | 0.07 | 0.51 | 1 |
| 9. | Soladunalinidine | 0.40 | −0.40 | 0.09 | 0.31 | 0.55 | 1 |
| Andrographis paniculata (Family:Acanthaceae) | |||||||
| 1. | 14-Deoxy-11-hydroxyandrographolide | 0.39 | −0.41 | 0.94 | 0.15 | 0.72 | 0 |
| 2. | 3,4-Dicaffeoylquinic acid | 0.19 | −0.01 | 0.50 | 0.16 | 0.40 | 3 |
| 3. | 3-O-beta-D-glucopyranosyl 14, 19-dideoxyandrographolide | 0.39 | −0.28 | 0.48 | 0.15 | 0.68 | 0 |
| 4. | 8, 17-Epoxy-14-deoxyandrographolide | 0.46 | −0.60 | 0.82 | 0.41 | 0.66 | 0 |
| 5. | Andrograpanin | 0.43 | −0.37 | 0.76 | 0.07 | 0.63 | 0 |
| 6. | Andrographidine A | 0.09 | −0.19 | 0.11 | 0.00 | 0.29 | 0 |
| 7. | Andrographidine C | 0.01 | 0.06 | 0.05 | −0.07 | 0.33 | 0 |
| 8. | Andrographiside | 0.36 | 0.08 | 0.55 | 0.28 | 0.80 | 2 |
| 9. | Andrographolactone | 0.39 | −0.46 | 0.19 | −0.20 | 0.20 | 0 |
| 10. | Andrographolide | 0.32 | −0.01 | 0.94 | 0.26 | 0.81 | 0 |
| 11. | Apigenin | −0.07 | 0.18 | 0.34 | −0.25 | 0.26 | 0 |
| 12. | Bisandrographolide A | −0.21 | −0.92 | −0.32 | −0.19 | −0.25 | 1 |
| 13. | Isoandrographolide | 0.32 | −0.01 | 0.94 | 0.26 | 0.81 | 0 |
| 14. | Neoandrographolide | 0.47 | −0.16 | 0.44 | 0.21 | 0.70 | 0 |
| 15. | Onysilin | 0.01 | −0.22 | 0.18 | −0.16 | 0.12 | 0 |
| Eucalyptus Sp., (Family:Myrtaceae) | |||||||
| 1. | α- pinene | −0.48 | −1.50 | −0.62 | −0.85 | −0.34 | 0 |
| 2. | 1,8-cineol | −0.93 | 0.01 | −1.60 | −1.07 | −0.15 | 0 |
| 3. | Terpineol alpha | −0.51 | −1.45 | −0.02 | −0.78 | 0.14 | 0 |
| 4. | α-Terpinyl acetate | −0.35 | −1.14 | 0.00 | −0.50 | 0.28 | 0 |
| 5. | Limonene | −0.91 | −2.01 | −0.34 | −1.38 | −0.21 | 0 |
| 6. | Guaiene | −0.49 | −1.27 | −0.01 | −0.57 | −0.14 | 0 |
| 7. | Spathulenol | −0.42 | −0.68 | 0.28 | −0.36 | 0.06 | 0 |
| 8. | Cryptone | −1.10 | −2.05 | −0.59 | −0.93 | −0.32 | 0 |
| 9. | Verbenone | −0.76 | −1.98 | −0.50 | −0.86 | −0.33 | 0 |
| 10. | Phellandral | −0.78 | −1.27 | −0.17 | −0.59 | −0.13 | 0 |
| 11. | p-Cymen-8-ol | −0.49 | −1.12 | −0.43 | −0.95 | −0.17 | 0 |
| 12. | Caryophyllene oxide | −0.08 | −0.86 | 0.62 | 0.00 | 0.57 | 0 |
| 13. | Carvacrol | −1.02 | −1.15 | −0.70 | −1.25 | −0.56 | 0 |
| 14. | β-Eudesmol | −0.02 | −0.62 | 0.60 | −0.10 | 0.48 | 0 |
| Alpinia officinarum (Family:Zingiberaceae) | |||||||
| 1. | Pinocembrin | −0.00 | −0.32 | 0.37 | −0.17 | 0.21 | 0 |
| 2. | Tectochrysin | −0.14 | 0.12 | 0.23 | −0.31 | 0.18 | 0 |
| 3. | Yakuchinone A | 0.07 | −0.31 | 0.12 | 0.01 | 0.16 | 0 |
| 4. | Hannokinol | 0.34 | 0.13 | 0.54 | 0.34 | 0.45 | 0 |
| 5. | Nootkatone | −0.40 | −1.73 | 0.66 | −0.58 | 0.34 | 0 |
| 6. | Pinobaksin | 0.03 | −0.10 | 0.21 | −0.00 | 0.31 | 0 |
| 7. | 5-hydroxy-1,7-diphenyl-3-heptanone | 0.22 | −0.27 | 0.18 | 0.30 | 0.42 | 0 |
| 8. | 3,5-dihydroxy-1,7-diphenylheptane | 0.30 | 0.06 | 0.40 | 0.32 | 0.45 | 0 |
| 9. | 1,7-diphenylhept-4-en-3-one | 0.10 | −0.40 | 0.15 | 0.09 | 0.30 | 0 |
| 10. | Zingerone | −0.58 | −1.15 | −0.59 | −0.72 | −0.07 | 0 |
| 11. | 1’-acetoxychavicol acetate | −0.37 | −0.68 | −0.12 | −0.47 | −0.04 | 0 |
| 12. | β-sitosterol | 0.14 | −0.51 | 0.73 | 0.07 | 0.51 | 1 |
| 13. | p-Coumaryl alcohol | −0.63 | −0.93 | −0.28 | −1.07 | −0.09 | 0 |
| 14. | 1,5-bis-(4-hydroxyphenyl)-2-(hydroxymethyl)-4-penten-1-ol | 0.40 | 0.15 | 0.51 | 0.05 | 0.48 | 0 |
| 15. | 1,5-bis-(4-hydroxyphenyl)-1-methoxy-2-(methoxymethyl) -4-pentene | 0.24 | −0.02 | 0.31 | 0.04 | 0.32 | 0 |
| 16. | 1,5-bis-(4-hydroxyphenyl)-1-ethoxy-2-(methoxymethyl)-4-pentene | 0.20 | −0.08 | 0.39 | 0.00 | 0.24 | 0 |
| 17. | 1,5-bis-(4-hydroxyphenyl)-2-(methoxymethyl)-4-penten-1-ol | 0.33 | 0.06 | 0.47 | 0.04 | 0.37 | 0 |
| Plectranthus amboinicus (Family:Lamiaceae) | |||||||
| 1. | (Z)-3-Hexenol | −3.04 | −3.44 | −2.82 | −3.13 | −2.79 | 0 |
| 2. | 1,8-Cineole | −0.93 | −1.60 | −1.07 | −0.90 | −0.15 | 0 |
| 3. | 1-Octen-3-ol | −1.78 | −2.59 | −1.68 | −1.86 | −1.09 | 0 |
| 4. | 3-Carene | −1.29 | −1.51 | −1.28 | −1.28 | −0.53 | 0 |
| 5. | Caffeic acid | −0.48 | −0.81 | −0.10 | −0.79 | −0.09 | 0 |
| 6. | Carvacrol | −1.02 | −1.15 | −0.70 | −1.25 | −0.56 | 0 |
| 7. | Caryophyllene oxide | −0.08 | −0.86 | −0.62 | 0.00 | 0.57 | 0 |
| 8. | Chavicol | −0.99 | −1.39 | −0.81 | −1.39 | −0.48 | 0 |
| 9. | Durohydroquinone | −0.81 | −1.00 | −0.69 | −0.94 | −0.33 | 0 |
| 10. | Eriodictyol | 0.07 | −0.22 | 0.46 | −0.09 | 0.21 | 0 |
| 11. | Eugenol | −0.86 | −1.14 | −0.78 | −1.29 | −0.41 | 0 |
| 12. | Gallic acid | −0.77 | −0.88 | −0.52 | −0.94 | −0.17 | 0 |
| 13. | Geraniol | −0.60 | −1.32 | −0.20 | −1.03 | 0.28 | 0 |
| 14. | Geranyl acetate | −0.50 | −1.11 | −0.12 | −0.80 | 0.21 | 0 |
| 15. | Germacrene D | −0.30 | −0.81 | 0.32 | −0.67 | 0.26 | 1 |
| 16. | Methyl chavicol | −0.06 | −0.46 | 0.29 | −0.18 | 0.15 | 1 |
| 17. | Nerol acetate | −0.50 | −1.11 | −0.12 | −0.80 | 0.21 | 0 |
| 18. | Oleic acid | 0.17 | −0.22 | 0.23 | 0.07 | 0.27 | 1 |
| 19. | p-Coumaric acid | −0.56 | −0.91 | −0.12 | −0.87 | −0.15 | 0 |
| 20. | Phytol | 0.11 | −0.32 | 0.35 | 0.00 | 0.31 | 1 |
| 21. | Rosmarinic acid | 0.17 | −0.18 | 0.57 | 0.15 | 0.24 | 0 |
| 22. | Spathulenol | −0.42 | −0.68 | 0.28 | −0.36 | 0.06 | 0 |
| 23. | Squalene | 0.04 | −0.10 | 0.19 | −0.03 | 0.16 | 1 |
| 24. | Tetradecanal | −0.24 | −0.56 | −0.34 | −0.15 | 0.12 | 1 |
| 25. | Thymol | −1.05 | −1.29 | −0.78 | −1.34 | −0.57 | 0 |
| 26. | Thymoquinone | −1.40 | −1.27 | −1.47 | −1.45 | −0.40 | 0 |
| 27. | trans-Caryophyllene | −0.34 | −0.78 | 0.13 | −0.60 | 0.19 | 1 |
| 28. | Undecanal | −0.54 | −0.96 | −0.70 | −0.41 | −0.03 | 1 |
| 29. | α-Cadinol | −0.09 | −0.87 | 0.39 | −0.63 | 0.40 | 0 |
| 30. | α-Humulene | −0.14 | −0.93 | 0.34 | −0.67 | 0.31 | 1 |
| 31. | α-Muurolene | −0.15 | −0.84 | 0.22 | −0.74 | 0.28 | 1 |
| 32. | α-Terpineol | −0.51 | −1.45 | −0.02 | −0.78 | 0.14 | 0 |
| 33. | α-Terpinolene | −0.88 | −1.61 | −0.50 | −1.74 | −0.26 | 0 |
| 34. | β-Caryophyllene | −0.34 | −0.78 | 0.13 | −0.60 | 0.19 | 1 |
| 35. | β-Cedrene epoxide | −0.10 | −1.10 | 0.12 | −0.02 | 0.48 | 0 |
| 36. | β-Copaen-4-α-ol | −0.19 | −0.57 | 0.45 | −0.17 | 0.27 | 0 |
| 37. | β-Himachalene oxide | −0.33 | −0.79 | 0.63 | −0.39 | 0.43 | 0 |
| 38. | δ-3-Carene | −1.29 | −1.51 | −1.28 | −1.28 | −0.53 | 0 |
Table 4.
Comparison with available drugs and novel bioactive compounds.
| S.No | Drugs | GPCR lg | Ki | Ncr | Pi | Ei | nvio |
|---|---|---|---|---|---|---|---|
| Drugs for COVID-19 treatment prescribed by WHO | |||||||
| 1 | ASC-09 (TMC-310911) | −0.67 | −1.57 | −1.82 | 0.18 | −1.10 | 3 |
| 2 | Camostat | −0.10 | −0.32 | −0.20 | 0.07 | −0.08 | 0 |
| 3 | Chloroquine | 0.32 | 0.38 | −0.19 | 0.05 | 0.11 | 1 |
| 4 | Dapagliflozin | 0.15 | −0.05 | 0.09 | 0.06 | 0.25 | 0 |
| 5 | Famotidine | 0.06 | −0.80 | −1.08 | 0.22 | 0.38 | 1 |
| 6 | Favipiravir | −0.62 | −0.31 | −1.50 | −0.91 | −0.33 | 0 |
| 7 | Fluvoxamine | 0.33 | 0.23 | 0.37 | 0.26 | 0.28 | 0 |
| 8 | Hydroxychloroquine | 0.35 | 0.44 | −0.12 | 0.12 | 0.15 | 0 |
| 9 | Ivermectin | −2.49 | −3.23 | −2.94 | −1.89 | −2.53 | 2 |
| 10 | Lopinavir | 0.04 | −0.55 | −0.66 | 0.42 | −0.37 | 2 |
| 11 | Nafamostat | 0.28 | −0.03 | −0.16 | 0.57 | 0.19 | 1 |
| 12 | Nitazoxanide | −0.55 | −0.19 | −0.73 | −0.66 | −0.31 | 0 |
| 13 | Remdesivir | 0.27 | 0.20 | −0.48 | 0.49 | 0.38 | 2 |
| 14 | Ritonavir | −0.33 | −1.02 | −1.41 | 0.35 | −0.74 | 3 |
| 15 | Umifenovir (arbidol) | −0.19 | −0.39 | −0.34 | −0.46 | −0.07 | 0 |
| Justicia adhatoda (Family:Acanthaceae) | |||||||
| 1 | β-Sitosterol | 0.14 | −0.51 | 0.73 | 0.07 | 0.51 | 1 |
| 2 | Caryophyllene oxide | −0.08 | −0.86 | 0.62 | 0.00 | 0.57 | 0 |
| Ocimum sanctum (Family: Lamiaceae) | |||||||
| 1 | Campesterol | 0.11 | −0.48 | 0.71 | 0.01 | 0.50 | 1 |
| 2 | Caryophyllene oxide | −0.08 | −0.86 | 0.62 | 0.00 | 0.57 | 0 |
| 3 | Oleanolic acid | 0.28 | −0.40 | 0.77 | 0.15 | 0.65 | 1 |
| 4 | β-sitosterol | 0.14 | −0.51 | 0.73 | 0.07 | 0.51 | 1 |
| 5 | Stigmasterol | 0.12 | −0.48 | 0.74 | −0.02 | 0.53 | 1 |
| Solanum trilobatum (Family:Solanaceae) | |||||||
| 1 | Sobatum | 0.14 | −0.51 | 0.73 | 0.07 | 0.51 | 1 |
| 2 | Solasodine | 0.24 | −0.66 | 0.36 | 0.01 | 0.60 | 1 |
| 3 | Tomatidine | 0.32 | −0.50 | 0.28 | 0.17 | 0.57 | 1 |
| 4 | Disogenin | 0.05 | −0.57 | 0.58 | −0.06 | 0.61 | 1 |
| 5 | Campesterol | 0.11 | −0.48 | 0.71 | 0.01 | 0.50 | 1 |
| 6 | Sitosterol | 0.14 | −0.51 | 0.73 | 0.07 | 0.51 | 1 |
| 7 | Soladunalinidine | 0.40 | −0.40 | 0.09 | 0.31 | 0.55 | 1 |
| Andrographis paniculata (Family:Acanthaceae) | |||||||
| 1 | 14-Deoxy-11-hydroxyandrographolide | 0.39 | −0.41 | 0.94 | 0.15 | 0.72 | 0 |
| 2 | 3-O-beta-D-glucopyranosyl 14, 19-dideoxyandrographolide | 0.39 | −0.28 | 0.48 | 0.15 | 0.68 | 0 |
| 3 | 8, 17-Epoxy-14-deoxyandrographolide | 0.46 | −0.60 | 0.82 | 0.41 | 0.66 | 0 |
| 4 | Andrograpanin | 0.43 | −0.37 | 0.76 | 0.07 | 0.63 | 0 |
| 5 | Andrographiside | 0.36 | 0.08 | 0.55 | 0.28 | 0.80 | 2 |
| 6 | Andrographolide | 0.32 | −0.01 | 0.94 | 0.26 | 0.81 | 0 |
| 7 | Isoandrographolide | 0.32 | −0.01 | 0.94 | 0.26 | 0.81 | 0 |
| 8 | Neoandrographolide | 0.47 | −0.16 | 0.44 | 0.21 | 0.70 | 0 |
| Eucalyptus Sp., (Family:Myrtaceae) | |||||||
| 1 | Caryophyllene oxide | −0.08 | −0.86 | 0.62 | 0.00 | 0.57 | 0 |
| Alpinia officinarum (Family:Zingiberaceae) | |||||||
| 1 | β‑sitosterol | 0.14 | −0.51 | 0.73 | 0.07 | 0.51 | 1 |
| Plectranthus amboinicus (Family:Lamiaceae) | |||||||
| 1 | Caryophyllene oxide | −0.08 | −0.86 | −0.62 | 0.00 | 0.57 | 0 |
These novel phytocompounds directly targets the human COVID-19 immune receptors.
4. Discussion
The ongoing COVID-19 pandemic completely disrupted the global homeostasis. The unavailability and ineffectiveness of anti-COVID-19 medicines to deal with this noxious infection make it an urgent need to identify the novel drugs/compounds that are efficacious in the control and treatment of this deadly infection. The effectiveness of traditional Indian medicines was well known for over 1000 years of practice [31]. Despite the fact that the pharmaceutical ingredients of medicinal plants have been extorted and purified for unsullied drug development, this method still ends up in failure due to the higher violation of chemical components and functional drug regulation. Indian traditional medicines were proven to be effective to treat viral diseases such as herpes simplex virus (HSV), measles, influenza, viral carcinogenesis, Hepatitis, coxsackievirus, HIV, etc. [32,33]. It can be also seen as a major complexity and is confronting another complexity, which essentially throw more light in the understanding of the entire human physiological system by controlling the molecular cross-talk between all the immunological key elements within the species. On the other hand, the exact mechanism of the immune responsive targets and biological pathways of herbal medicines remains poorly understood [31]. Hence, in this pilot study, we employed immuno-transcriptomics, cheminformatics, interactomics and integrated pharmacology strategy for the first time to unravel the COVID-19 immune targets and their associated signaling pathways, pharmacological roles of J. adhatoda, O. sanctum, S. trilobatum, A. paniculata, Eucalyptus Sp., A. officinarum and P. amboinicus derived bioactive compounds in the treatment of COVID-19 at molecular system level. In addition, the exploration of plant derived natural bioactive molecules for the management of viral diseases can provide a vital source of therapeutic molecules. Furthermore, the medicinal plant derived active molecules are less toxic when compared to the modern/synthetic drugs which cause numerous side effects. Besides, the findings of the present study will also prescribe the natural and novel bioactive molecules for treating the COVID-19 by altering and or stimulating the immune functional regulations.
This study mainly aimed to identify and understand the host gene expression pattern response to COVID-19/ respiratory viral infections. To the best of our knowledge, this is the first investigation to perform immuno-transcriptomic profiling between healthy controls and COVID-19 cases using publicly available transcriptomic dataset and processed by NetworkAnalyst 3.0 [23]. Heatmap was derived in accordance to the microarray intensities of the COVID-19 immune responsive genes and based on the intensity values of 98 immune responsive genes differentially expressed between the healthy controls and COVID-19 cases at various time points. The identified genes were chosen for subsequent analysis.
On the other hand, with the help of cheminformatics, PubChem and interactomics databases, 259 natural bioactive compounds were identified. Among 259, 154 active compounds strongly interacted with 13 out of 98 differentially expressed COVID-19 immune responsive genes by drug targeting. Notably, these genes were predicted to be involved in various biological/ immunological activities against COVID-19 and have not been reported so far, demonstrating the ability of SwissTargetPrediction and GOnet enrichment evaluation methods. Further, ORA functional and GO enrichment of identified genes that are more specific to COVID-19 were delineated. The analysis of principle compounds that actively targeting the differentially expressed human immune responsive genes through COVID-19 disease highlighted the role of 13 genes (TLR4, STAT1, SELL, PSMB9, CD22, CCR1, CCR5, LTB4R, MAPK14, CSF1R, BCL2, CASP1, and NLRP3). For example, TLR4 (Toll-like receptor 4) protein is being the efficient innate immune receptor that plays a pivotal role in pathogen recognition and activates pro-inflammatory responses. Hence, the cross-talk between the spike protein of SARS-CoV2 and TLR4 might be one of the reasons behind the immuno-pathological expression of COVID-19 [34]. STAT1 immune player is associated with host responses to viral infections and stimulates the type I interferon signaling. But in the case of SARS-CoV2 fled the host interferon dynamism and its ORF6 protein can obstructs the regulation of STAT1-activated genes [35]. NLRP3 (NBS, LRR, and pyrin domain-containing 3) inflammasome complex interacts with apoptosis-associated speck-like protein carrying a caspase activation (ASC) and plays a role in the regulation of immune response, apoptosis and inflammation. Thus, the SARS-CoV2 viroporins (E-protein, ORF3a and ORF8a) are inducing this receptor NLRP3 via lysosomal disruption and ion redistribution mechanisms. It leads to the production of cytokines namely tumor necrosis factor (TNF), IL-1β, IL-6 causing severe tissue inflammation during respiratory problems caused by COVID-19 [36]. Notably, all these 13 COVID-19 genes are linked with other viral infections and have maximum interaction. By selecting closely related genes, an ORA enrichment and STRING network analysis were employed to identify the deeper insights about the function of these genes. It was lucid from the results that the immunological and genetic pathways associated with Influenza A, Hepatitis B and C, Viral myocarditis, Malaria, Rheumatoid arthritis, Measles, tuberculosis, pertussis, viral carcinogenesis and AGE-RAGE signaling mechanism were of significance. The correlation between host immune response to pertussis, Malaria, tuberculosis, Rheumatoid arthritis and COVID-19 has been a wonder to date. Hypothetically, drugs commercially available for the treatment of pertussis, Malaria, tuberculosis and Rheumatoid arthritis may have therapeutic efficacy against COVID-19 [37,38]. Our results revealed that the interaction in the immune response between these infections and the COVID-19 is highly significant at the molecular level.
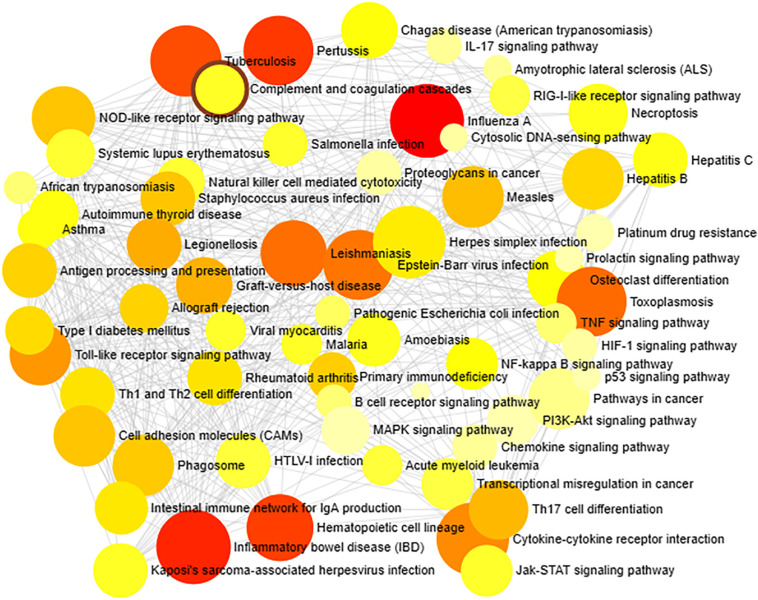
As described earlier, the identified 13 genes were involved in the regulation of various immunological pathways such as Toll-like receptor, IL-17, TNF, NF-kappa B (Nuclear Factor-Kappa B/ NF-kB), MAPK, NOD- like receptor signaling pathway, RIG-I-like receptor signaling pathway, B and T cell receptor signaling pathways, Antigen processing and presentation (Fig. 9 ). In addition, these genes had a strong association with COVID-19 as per ORA functional and GO enrichment and significantly related to TNF and NF-kappa B signaling pathways. The TNF receptors are the key players involved in apoptosis and inflammation. The cross-talk between viral proteins and intracellular components of the TNF receptors demonstrated the critical ability of viral replication machinery to evade the immunological responses [39]. The NF-kB pathway is highly related to pro-oxidant and pro-inflammatory responses and is particularly involved in the inflammatory reactions in acute lung diseases/ injuries. The mechanism of NF-kB activation was projected as a pivotal adjuvant treatment for deadly COVID-19 infection [40].
Fig. 9.
ORA enrichment analysis of significant immune responsive genes. The number of human immune responsive genes falling in each KEGG pathway category is directly proportional to the node size. The nodes are color shaded according to the significance level (adjusted P-value <0.05).
Cytoscape analysis results revealed the molecular interaction of bioactive compounds and immune responsive genes network. The C-T-N results exhibited that there are 154 bioactive compounds interacting with or targeting the 13 immune responsive genes. It also unveils the diverse mechanism and plausible mode of action used by active phytochemicals to exert their curative effects and makes the traditional Indian medicines more fruitful and efficacious to the society and to come out of this pandemic situation.
Identified natural bioactive compounds that are targeting/ altering the COVID-19 immune response signaling pathways especially on TNF, JAK-STAT, NF-kB, and MAPK pathways by inhibiting or suppressing the 13 key receptors with their functions and could induce the production of anti-COVID-19 antibodies via antigen presenting cells (Fig. 10 ). Since synthetic drugs are incapable of producing a breakthrough to date in the treatment of this deadly virus disease, it is the need the hour to shift our focus to traditional Indian medicinal plants derived bioactive compounds in confronting this deadly COVID-19 infection.
Fig. 10.
Schematic representation of the current study.
5. Conclusion
The current findings revealed that the Indian traditional medicinal plants exhibited diverse immunological stimulants to treat/ altering the replication machinery of human COVID-19 infection. Biomedical research in traditional Indian medicinal plants notably on J. adhatoda, O. sanctum, S. trilobatum, A. paniculata, Eucalyptus Sp., A. officinarum and P. amboinicus is still bottleneck, significantly, our results on novel bioactive compounds with their pharmacological properties, human COVID-19 immune receptors/targets, diverse biological processes and their molecular interactions will pave the way to open the advanced molecular biological and COVID-19 research floodgates with the combination of Ayurveda to modern medicine era. This is the first and foremost holistic study identified several pivotal aspects of the host immune response to COVID-19 infection. Identified immune responsive genes and their associated diverse signaling pathways could be used for unraveling the pathogenesis of COVID-19. These significant genetic factors/immune responsive genes and pathways have been identified which is used to characterize the immune-pathology of human COVID-19 infection. This study hypothesizes that natural bioactive compounds and in combination with other substances, as is recommended by the traditional (according to Ayurveda) and modern medicine system (prescribed by WHO), may result in synergistic effects and need to be investigated further to develop the new drug to treat the COVID-19. In addition, this study suggested that obtained molecular interaction results can be useful to design competitive 13 immune targets antagonists which will pave the unparalleled way to combat COVID-19. Overall, this holistic study will stand as the platform to improve our understanding of the immunobiology of SARS-CoV2 and the acquired essential knowledge could be helpful in implementing an accurate intervention strategy in mere future. The ethical implications of drugs that enhancing immunity and inhibiting the human COVID-19 pathogenic processes are significant but it should be appropriately alleviated with ethical, legal and social considerations as field researchers enter the advanced world of drug development and provide valid targets for pivotal therapy against COVID-19.
Ethics
No animal or human subjects were used in this study.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgements
The authors thank RUSA 2.0 [F. 24-51/2014-U, Policy (TN Multi-Gen), Dept of Edn, GoI]. The authors also thankfully acknowledge DST-FIST (Grant No. SR/FST/LSI-639/2015(C)), UGC-SAP (Grant No. F.5-1/2018/DRS-II (SAP-II)) and DST-PURSE (Grant No. SR/PURSE Phase 2/38 (G)) for providing instrumentation facilities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ygeno.2020.08.003.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary material 5
References
- 1.Corman V.M., Lienau J., Witzenrath M. Coronaviruses as the cause of respiratory infections. Internist (Berl). 2019;60:1136–1145. doi: 10.1007/s00108-019-00671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections-more than just the common cold. JAMA. 2020 doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 3.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England). 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D., Sidorov I.A., Sola I., Ziebuhr J. Severe acute respiratory syndrome-related coronavirus–the species and its viruses, a statement of the coronavirus study group. BioRxiv. 2020 doi: 10.1101/2020.02.07.937862. 2020.02.07.937862. [DOI] [Google Scholar]
- 6.Sohrabi C., Alsafi Z., O’Neill N., Khan M., Kerwan A., Al-Jabir A., Iosifidis C., Agha R. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKibbin W.J., Fernando R. The global macroeconomic impacts of COVID-19: seven scenarios. SSRN Electron. J. 2020 doi: 10.2139/ssrn.3547729. [DOI] [Google Scholar]
- 8.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., Aziz B. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. 2020 doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann M., Hofmann-Winkler H., Pöhlmann S. 2018. Priming Time: How Cellular Proteases Arm Coronavirus Spike Proteins, Act. Viruses by Host Proteases; pp. 71–98. [DOI] [Google Scholar]
- 13.Davidson-Hunt I. Ecological Ethnobotany: stumbling toward new practices and paradigms. MASA J. 2000;16:1–13. [Google Scholar]
- 14.Elaissi A., Rouis Z., Ben Salem N.A., Mabrouk S., ben Salem Y., Salah K.B.H., Aouni M., Farhat F., Chemli R., Harzallah-Skhiri F., Khouja M.L. Chemical composition of 8 eucalyptus species’ essential oils and the evaluation of their antibacterial, antifungal and antiviral activities. BMC Complement. Altern. Med. 2012;12 doi: 10.1186/1472-6882-12-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasanth V., Palakeerti S. In Silico studies of Justicia adhatoda, Ocimum sanctum plant compounds as Mycobacterium tuberculosis Ftsz inhibitors. Int. J. Bio. 2012;01:22–25. http://www.ebioscholar.com/ijb/index.php/ijb/article/view/9 [Google Scholar]
- 16.Naquvi K.J., Dohare S.L., Shuaib M., Ahmad M.I. CHEMICAL composition of volatile oil of Ocimum sanctum Linn. Int. J. Biomed. Adv. Res. 2012;3:129–131. doi: 10.7439/ijbar.v3i2.290. [DOI] [Google Scholar]
- 17.Hossain M.S., Urbi Z., Sule A., Rahman K.M. Hafizur. Andrographis paniculata (Burm. F.) wall. Ex Nees: a review of ethnobotany, phytochemistry, and pharmacology. Sci. Wor. J. 2014;2014 doi: 10.1155/2014/274905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebei K., Sakouhi F., Herchi W., Khouja M.L., Boukhchina S. Chemical composition and antibacterial activities of seven Eucalyptus species essential oils leaves. Biol. Res. 2015;48:7. doi: 10.1186/0717-6287-48-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balakrishnan P., Ansari T., Gani M., Subrahmanyam S. Review article a perspective on bioactive compounds from Solanum trilobatum. Res. J. Chem. Pharm. 2015;7:507–512. [Google Scholar]
- 20.Arumugam G., Swamy M.K., Sinniah U.R. Plectranthus amboinicus (lour.) Spreng: botanical, phytochemical, pharmacological and nutritional significance. Molecules. 2016;21:369. doi: 10.3390/molecules21040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shukla S., Hegde S., Kumar A., Chaudhary G., Tewari S.K., Upreti D.K., Pal M. Chemical composition and antibacterial activity of essential oil from leaves of Justicia adhatoda against methicillian resistant and sensitive strain along with their clinical isolates. J. Essent. Oil Bear. Plants. 2017;20:116–122. doi: 10.1080/0972060X.2016.1260061. [DOI] [Google Scholar]
- 22.Basri A.M., Taha H., Ahmad N. A review on the pharmacological activities and phytochemicals of Alpinia officinarum (galangal) extracts derived from bioassay-guided fractionation and isolation. Pharmacogn. Rev. 2017;11:43–56. doi: 10.4103/phrev.phrev_55_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou G., Soufan O., Ewald J., Hancock R.E.W., Basu N., Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47:W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolton E.E., Wang Y., Thiessen P.A., Bryant S.H. In: Chapter 12 PubChem: Integrated Platform of Small Molecules and Biological Activities. B.V, editor. Elsevier; 2008. [DOI] [Google Scholar]
- 25.Petryszak R., Keays M., Tang Y.A., Fonseca N.A., Barrera E., Burdett T., Füllgrabe A., Fuentes A.M.-P., Jupp S., Koskinen S., Mannion O., Huerta L., Megy K., Snow C., Williams E., Barzine M., Hastings E., Weisser H., Wright J., Jaiswal P., Huber W., Choudhary J., Parkinson H.E., Brazma A. Expression atlas update--an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44:D746–D752. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smoot M.E., Ono K., Ruscheinski J., Wang P.-L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma C.S., Mishra S.S., Singh H.P., Kumar N. In silico adme and toxicity study of some selected antineoplastic drugs. Int. J. Pharm. Sci. Drug Res. 2016;8:65–67. doi: 10.25004/IJPSDR.2016.080110. [DOI] [Google Scholar]
- 28.Pomaznoy M., Ha B., Peters B. GOnet: a tool for interactive gene ontology analysis. BMC Bioinformatics. 2018;19:470. doi: 10.1186/s12859-018-2533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alsamman A.M., Zayed H. The transcriptomic profiling of COVID-19 compared to SARS, MERS, Ebola, and H1N1. BioRxiv. 2020 doi: 10.1101/2020.05.06.080960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeyasri R., Muthuramalingam P., Suba V., Ramesh M., Chen J.-T. Bacopa monnieri and their bioactive compounds inferred multi-target treatment strategy for neurological diseases: a cheminformatics and system pharmacology approach. Biomolecules. 2020;10 doi: 10.3390/biom10040536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin L.-T., Hsu W.-C., Lin C.-C. Antiviral natural products and herbal medicines. J. Tradit. Complement. Med. 2014;4:24–35. doi: 10.4103/2225-4110.124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akram M., Tahir I.M., Shah S.M.A., Mahmood Z., Altaf A., Ahmad K., Munir N., Daniyal M., Nasir S., Mehboob H. Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: a systematic review. Phytother. Res. 2018;32:811–822. doi: 10.1002/ptr.6024. [DOI] [PubMed] [Google Scholar]
- 34.Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. n/a. 2020 doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prasad K., Khatoon F., Rashid S., Ali N., AlAsmari A.F., Ahmed M.Z., Alqahtani A.S., Alqahtani M.S., Kumar V. Targeting hub genes and pathways of innate immune response in COVID-19: a network biology perspective. Int. J. Biol. Macromol. 2020;163:1–8. doi: 10.1016/j.ijbiomac.2020.06.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah A. Novel coronavirus-induced NLRP3 Inflammasomeactivation: a potential drug target in the treatment of COVID-19. Front. Immunol. 2020;11:1021. doi: 10.3389/fimmu.2020.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Favalli E.G., Ingegnoli F., De Lucia O., Cincinelli G., Cimaz R., Caporali R. COVID-19 infection and rheumatoid arthritis: faraway, so close! Autoimmun. Rev. 2020;19 doi: 10.1016/j.autrev.2020.102523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao J., Tian Z., Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 39.Herbein G., O’Brien W.A. Tumor necrosis factor (TNF)-alpha and TNF receptors in viral pathogenesis. Proc. Soc. Exp. Biol. Med. Soc. Exp. Biol. Med. (New York, N.Y.). 2000;223:241–257. doi: 10.1046/j.1525-1373.2000.22335.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang R., Wang X., Ni L., Di X., Ma B., Niu S., Liu C., Reiter R.J. COVID-19: melatonin as a potential adjuvant treatment. Life Sci. 2020;250 doi: 10.1016/j.lfs.2020.117583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary material 5