Highlights
-
•
Codon analysis reveal natural selection and other undefined factors dominates the overall codon usage bias in SARS-CoV-2 rather than mutational pressure.
-
•
The host adaptation potential of SARS-CoV-2 is more in human as compared to dog.
Keywords: Codon bias, Corona virus, SARS-CoV-2, Selection pressure, Dog
Abstract
Severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) is recognized as one of the life-threatening viruses causing the most destructive pandemic in this century. The genesis of this virus is still unknown. To elucidate its molecular evolution and regulation of gene expression, the knowledge of codon usage is a pre-requisite. In this study, an attempt was made to document the genome-wide codon usage profile and the various factors influencing the codon usage patterns of SARS-CoV-2 in human and dog. The SARS-CoV-2 genome showed relative abundance of A and U nucleotides and relative synonymous codon usage analysis revealed that the preferred synonymous codons mostly end with A/U. The analysis of ENc-GC3s, Neutrality and Parity rule 2 plots indicated that natural selection and other undefined factors dominate the overall codon usage bias in SARS-CoV-2 whereas the impact of mutation pressure is comparatively minor. The codon adaptation index and relative codon deoptimization index of SARS-CoV-2 deciphered that human is more favoured host for adaptation compared to dog. These results enhance our understanding of the factors involved in evolution of the novel human SARS-CoV-2 and its adaptability in dog.
1. Introduction
Coronaviruses belong to the family Coronaviridae and are the largest enveloped single-stranded RNA viruses, ranging from 26 to 31 kilobases in genome size (Lauber et al., 2012). These viruses infect a wide range of avian and mammalian species, and are responsible for enteric or respiratory infections (Woo et al., 2009). Human coronaviruses, viz. severe acute respiratory syndrome-related coronavirus (SARS-CoV) and Middle-East respiratory syndrome coronavirus (MERS-CoV) emerged in the year 2002 and 2012, respectively (Zaki et al., 2012). Both of these viruses have a zoonotic origin and hence emergence of human infections associated with these viruses has emphasized the need of controlling coronaviruses associated with diseases inanimals in close contact with humans (Kin et al., 2016).
A cluster of pneumonia cases of unknown origin were reported from the Wuhan, the capital city of Hubei Province of China in late December 2019. The cases were found to be linked with Huanan Seafood Market and the pathogen was thought to have a zoonotic origin (Andersen et al., 2020). The virus that caused the outbreak was identified as a novel, human-infecting coronavirus, which is closely related to bat coronaviruses, pangolin coronaviruses, and SARS-CoV (Han, 2020; Perlman, 2020). Subsequently, the virus spread globally causing a pathological condition which was termed as coronavirus disease 2019 (COVID-19), and the pathogen was named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The World Health Organization (WHO) declared the outbreak as a Public Health Emergency of International Concern on 30th January 2020 and recognized it as a pandemic in 11th March 2020. Till June 2020, approximately 9.7 million cases of SARS-CoV-2 were reported worldwide with more than 491,960 deaths.
The SARS-CoV-2 genome contains 14 ORFs encoding 27 proteins. The orf1ab and orf1a genes encode proteins, Pp1ab and Pp1a, respectively. The Pp1ab protein contains 15 nsps (nsp1-nsp10 and nsp12-nsp16). The SARS-CoV-2 genome also contains four structural proteins, namely, spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins (Wu et al., 2020). The S protein is the key protein that regulates the attachment of the virus receptor to the host target cell (Cavanagh, 1995), E protein acts as an ion channel and facilitates virion assembly (Ruch and Machamer, 2012), M and E proteins play a role in virus assembly and are involved in biosynthesis of new virus particles (Neuman et al., 2011), while N protein forms the nucleoprotein complex with the virus RNA (Risco et al., 1996). The 9th ORF of SARS-CoV-2 codes for N protein and another unique accessory protein called ORF9b in a different reading frame, whose function is not yet known. The N-protein is a 46 kDa protein composed of 422 amino acids (Rota et al., 2003). It is a multifunctional protein with distinct functions such as enhancing transcription of the viral genome, association with M protein during virion assembly, and disruption of the various activities of the host cell by inducing toxicity (McBride et al., 2014). It is also the most conserved and stable protein among the CoV structural proteins; whereas, the S protein undergoes substantial changes during virus infection. The S glycoprotein harbours a furin cleavage site at the boundary between the S1/S2 subunits, which is processed during biogenesis. Cleavage of S protein activates the protein for membrane fusion via extensive irreversible conformational changes andthus initiates the binding of SARS-CoV-2 with ACE2 receptor and entry to the host system (Walls et al., 2020).
Codon usage bias is an important measure of genome evolution. Factors that could influence the bias in codon usage include mutational pressure including natural selection, G + C content, secondary protein structure and selective transcription replication (Butt et al., 2014). Codon usage is a driving force in the evolution of viruses (Sewatanon et al., 2007). The codon usage bias frequency of RNA viruses is low, such as in the Zaire ebolavirus (Cristina et al., 2015), and the N gene of Rabies virus (He et al., 2017) and Equine influenza virus (Kumar et al., 2016). However, the overall codon usage bias in case of Hepatitis A virus (HAV) is high (Zhang et al., 2011). Investigation of viral gene structure and its composition at the codon or nucleotide level is essential to understand the mechanism of virus-host relationship and evolution of the virus (van Hemert and Berkhout, 2016). Viruses that infect humans, but not those that infect other mammals or aves, show a strong resemblance to most mammalian and avian hosts, in terms of both amino acid and codon preferences. In groups of viruses that infect humans or other mammals, the highest observed level of adaptation of viral proteins to host codon usages is for those proteins that appear abundantly in the virion. In contrast, proteins that are known to participate in host-specific recognition do not necessarily adapt to their respective hosts (Bahir et al., 2009).
The redundancy of the genetic code provides evolution with the opportunity to adjust the efficiency and accuracy of protein production preserving the same amino acid sequence (Stoletzki and Eyre-Walker, 2007). Similarity in codon usage pattern among viruses and their hosts may influence viral fitness, evasion from host’s immune system and evolution (Costafreda et al., 2014). Synonymous triplet codons are generally not used randomly and the main forces that drive this bias from equal usage are natural selection and mutational biases (Musto, 2016). Therefore, the study of codon usage in viruses can reveal important information about virus evolution, regulation of gene expression and protein synthesis (Butt et al., 2014). In addition, codon composition may also influence robustness of translation and, in turn, robustness of folding, which is critical to the capsid stability of hepatitis A viruses (D’Andrea et al., 2019).
The aim of this study was to carry out a comprehensive analysis of codon usage and composition of the severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) genome and to ascertain the possible evolutionary determinants of the biases found.
2. Materials and methods
2.1. Sequence data
Complete genome sequences of SARS-CoV-2 were obtained from the Virus Resource at the National Centre for Biotechnological Information (https://www.ncbi.nlm.nih.gov/labs/virus). The data set comprised of 12 complete coding genome sequences reported from different countries, viz. accession nos. LC529905.1, MT007544.1, MT012098.1, MT066156.1, MT072688.1, MT093571.1, MT126808.1, MT192759.1, MT192765.1, MT192772.1, MT233519 and NC045512.2. The Open reading frames (ORFs) for each genome were concatenated in the following order: ORF1ab + Spike + Envelop + Membrane + Nucleocapsid.
2.2. Analysis of overall nucleotide composition
The nucleotide composition of SARS-CoV-2 was analysed at the third nucleotide position of the codons (A3 %, G3 %, C3 % and U3 %) and the overall composition of nucleotides AU%, AU3 %, GC%, GC12 and GC3 were determined.
2.3. Calculation of relative synonymous codon usage (RSCU)
The RSCU value of a codon is the ratio of its observed frequency to its expected frequency given that all codons for a particular amino acid are used equally (Bera et al., 2017). The RSCU values were calculated using the method described by Kumar et al. (2016) using the following equation:
Where gij is the observed number of the ith codon for the jth amino acid, which has ni kinds of synonymous codons. Codons with RSCU value <1.0, 1.0 and>1.0 represent negative codon usage bias, no bias and positive codon usage bias, respectively.
2.4. Relative dinucleotide abundance analysis
The dinucleotide frequencies of SARS-CoV-2, which is another way of establishing the relation with codon usage bias, were calculated as described by Kumar et al. (2016). The expected dinucleotide values were calculated assuming random association of bases from the observed frequencies of each base for every sequence. The ratio of the observed and the expected dinucleotide frequencies is known as odds ratio. It was used for designation of over-representation (>1.23) or under-representation (<0.78) in terms of relative abundance compared with a random association of mononucleotides.
2.5. Analysis of similarity index
The similarity index analysis was performed to know the result of codon usage by the host and their role in shaping the overall codon usage of the virus. Analysis of codon usage by coding sequences of SARS-COV-2 and its respective hosts (human and dog) was performed using the method of Zhou et al. (2013). The similarity index was calculated using the following formula:
Where ‘ai’ represents RSCU value of a specific codon for the coding sequence of SARS-COV-2 and ‘bi’ indicates RSCU value of host alike codons. The D (A, B) represents overall codon usage of the host on the virus which typically ranges between 0.0 and 1.0. Higher similarity index value indicates substantial influence of the host on codon usage of SARS-COV-2.
2.6. Effective number of codons (ENc) and ENc-plot
Wright (1990) gave the concept of the eff ;ective number of codons (ENc) to recognize the bias in the identical codon usage. The values of ENc range from 20 to 61 and an ENc value of 20 indicates an extreme codon usage bias of a gene, and this means a specific amino acid is denoted by only one codon, despite the availability of synonymous codons. On the contrary, ENc value of 61 indicates no bias in codon usage which means a uniform use of all the synonymous codons. Generally, for a genome or a gene, an ENc value below 35 is known to have a strongly biased codon usage.
An ENc-plot was employed to determine whether the codon usage of SARS-COV-2 (concatenated ORFs) is mainly because of the burden of mutational or selection pressure. The expected ENc plot was generated by plotting the ENc values on x-axis and the GC3 values in y-axis (frequency of either a guanine or cytosine at the third codon position of the synonymous codons) (Wright, 1990). If the predicted ENc value lies on the expected curve, it indicates that codon usage is constrained only by mutation bias, while ENc values below the expected curve indicate that other factors such as selection pressure have affected the codon usage bias.
2.7. Analysis of neutrality and parity plot
A neutrality plot analysis was done to understand the effect of mutational bias and translation selection on codon usage. Neutrality plot was constructed with GC12 on y-axis and GC3 on x-axis, where GC12 stands for the average value of GC contents at the first and the second positions of the codons and GC3 refers to the GC contents at the third position of the codon. A regression line was drawn between contents of GC12 and GC3. The slope of regression line represents the impact of mutational force (Nasrullah et al., 2015). The AT bias [A3/(A3 + T3)] as the ordinate and the GC bias [G3/(G3 + C3)] as the abscissa were used to determine a parity rule 2 (PR2) bias (Wu et al., 2015).
2.8. Calculation of average Hydropathicity (GRAVY) and Aromaticity (AROMO)
The GRAVY value is the total of all amino acids’ hydropathy values in a series separated by the number of residues ranging from −2.0 to +2.0 (Kyte and Doolittle, 1982). Hydrophobicity of a protein is characterized by positive values, whereas negative values are indicative of hydrophilicity. The frequency of the aromatic amino acids, i.e. Phenylalanine, Tyrosine and Tryptophan is known as AROMO value in a given amino acid sequence.
2.9. Analysis of codon adaptation index (CAI)
Codon adaptation index (CAI) analysis is a quantitative value indicating the frequency of a preferred codon utilized by highly expressed genes. This shows the efficiency of translation and is often used to construct nucleotide sequences to get the highest level of protein expression for the purpose of vaccine production (Gustafsson et al., 2012). The value of CAI varies from 0.0 to 1.0; a higher value suggests a greater propensity for gene expression. Alternatively, values close to 1 are shown by the codons with higher RSCU values. In the present study, CAI values were calculated for SARS-COV-2 using an RSCU reference set for human, bat, dog, cat, pig, horse and cattle. The synonymous codon usage data of human, dog, cat, pig, horse and cattle were retrieved from the codon usage database (http://www.kazusa.or.jp/codon/), whereas for bat, the sequence of Pteropus vampyrus (NW_011888782) was retrieved from NCBI (https://www.ncbi.nlm.nih.gov) and by using online program ‘Countcodon’ (available at: http://www.kazusa.or.jp/codon/countcodon.html) the reference codon usage table for bat was prepared.
2.9.1. Relative codon deoptimization index (RCDI)
The Relative Codon Deoptimization Index (RCDI) is used to compare the codon usages of the genes and reference genomes. The viral gene translation rate to a host system is calculated using RCDI value. RCDI value close to one indicates similar codon usages by the host and the pathogen, and a greater adaptation to the host can be predicted (Butt et al., 2016). In the present study, RCDI values of SARS-COV-2 were calculated for human, bat, dog, cat, pig, horse and cattle.
2.10. Different tools and software used
The values for the RSCU and AROMO were estimated using CODONW 1.4 program. Calculation of GRAVY values were done by using the online tool available at http://www.gravy-calculator.de/. CAI and RCDI values were measured using the online tool available at http://genomes.urv.es/CAIcal/ (Puigbo et al., 2008). Another web-based tool was used to obtain the tRNA database (GtRNAdb: Genomic tRNA database).
3. Results
3.1. Nucleotide composition of SARS-CoV-2 genome
The SARS-CoV-2 was found to have comparative abundance of A and U nucleotides in comparison to G and C nucleotides. The nucleotide compositions of SARS-CoV-2 genes were calculated in order to determine the compositional constrains of its genome (Supplementary Table S1). Out of the four nucleotides, the mean percentage of U (32.02 %) was found to be the highest, followed by A (29.94 %) and G (19.78 %), while C (18.25 %) showed the lowest mean value. In the third position of the synonymous codons, U3 (43.87 %) was the highest in frequency, followed by A3 (28.13 %) and C3 (15.36 %), while G3 (12.63 %) was found to be the lowest. The mean AU and GC compositions were 61.96 % and 38.03 %, and the mean AU3 and GC3 compositions were 72 % and 27.99 %, respectively (Supplementary Table S1).
3.2. Relative synonymous codon usage (RSCU) analysis
RSCU values were analysed to determine the patterns of synonymous codon usage. The average RSCU values were evaluated for all the important five genes of SARS-CoV-2. Twenty four most abundantly used codons in SARS-CoV-2 genes (AGA, GCU, GUU, UUA, GGU, CCU, AGU, AUU, CUU, UCU, ACU, UUU, UGU, CCA, ACA, UCA, CAA, AAA, GAU, GAA, UAU, CGU, CAU and AAU) were A/U-ended (A-ended: 8; U-ended: 16). It was evident from the RSCU analysis that SARS-CoV-2 genomes exhibited higher codon usage bias towards codons ending with A/U compared to that with G/C.
Furthermore, RSCU values were divided into three categories: (A) codons with RSCU values ≤ 0.6 (under-represented), (B) codons with RSCU values between 0.6 and 1.6 (unbiased- represented), and (C) codons with RSCU values ≥ 1.6 (over-represented). Analysis of over-and under- represented codons showed that RCSU values of SARS CoV-2 ranged from 0.6 and 1.6. It was quite interesting to note that over-represented codons were A/U ended and mostly under-represented codons were C/G-ended (Table 1 ).
Table 1.
Relative synonymous codon usage (RSCU) analysis of SARS-CoV-2 and different hosts including human, dog, pig, cattle, pig, cat, horse and Bat. Bold numbers are referred as most preferred codon.
| Amino acids | Codon | SARS CoV-2 | Human | Dog | Cat | Pig | Horse | Cattle | Bat |
|---|---|---|---|---|---|---|---|---|---|
| Phenylalanine | UUU | 1.42 | 0.93 | 1.09 | 0.77 | 0.75 | 0.83 | 0.85 | 0.49 |
| UUC | 0.58 | 1.07 | 0.91 | 1.13 | 1.25 | 1.17 | 1.15 | 1.01 | |
| Leucine | UUA | 1.66 | 0.46 | 1.32 | 0.35 | 0.31 | 0.33 | 0.38 | 0.44 |
| UUG | 1.06 | 0.77 | 0.51 | 0.76 | 0.63 | 0.72 | 0.71 | 0.8 | |
| CUU | 1.75 | 0.79 | 1.22 | 0.67 | 0.69 | 0.73 | 0.7 | 0.85 | |
| CUC | 0.57 | 1.17 | 1.01 | 1.29 | 1.32 | 1.32 | 1.26 | 1.22 | |
| CUA | 0.68 | 0.43 | 1.42 | 0.36 | 0.34 | 0.34 | 0.36 | 0.44 | |
| CUG | 0.28 | 2.37 | 0.51 | 2.57 | 2.72 | 2.56 | 2.59 | 0.37 | |
| Isoleucine | AUU | 1.54 | 1.08 | 1.05 | 0.95 | 0.97 | 0.92 | 0.98 | 1.19 |
| AUC | 0.54 | 1.41 | 0.94 | 1.58 | 1.66 | 1.66 | 1.57 | 1.3 | |
| AUA | 0.92 | 0.51 | 1.01 | 0.47 | 0.37 | 0.42 | 0.45 | 0.49 | |
| Valine | GUU | 1.93 | 0.73 | 1.12 | 0.62 | 0.50 | 0.6 | 0.64 | 0.72 |
| GUC | 0.58 | 0.95 | 0.57 | 1.13 | 1.22 | 1.08 | 1.01 | 0.97 | |
| GUA | 0.89 | 0.47 | 1.67 | 0.38 | 0.26 | 0.35 | 0.4 | 0.51 | |
| GUG | 0.59 | 1.85 | 0.64 | 1.87 | 2.01 | 1.97 | 1.95 | 1.79 | |
| Serine | UCU | 2 | 1.13 | 1.35 | 1.12 | 0.86 | 1.09 | 1.04 | 1.03 |
| UCC | 0.44 | 1.31 | 1.04 | 1.48 | 1.10 | 1.43 | 1.37 | 1.22 | |
| UCA | 1.63 | 0.9 | 1.27 | 0.74 | 1.36 | 0.8 | 0.79 | 0.89 | |
| UCG | 0.11 | 0.33 | 0.39 | 0.38 | 0.42 | 0.34 | 0.39 | 0.29 | |
| AGU | 1.46 | 0.9 | 0.91 | 0.8 | 0.99 | 0.86 | 0.87 | 0.98 | |
| AGC | 0.36 | 1.44 | 1.05 | 1.47 | 1.27 | 1.48 | 1.53 | 1.56 | |
| Proline | CCU | 1.92 | 1.15 | 1.41 | 1.03 | 0.95 | 1.19 | 1.08 | 1.21 |
| CCC | 0.31 | 1.29 | 1.24 | 1.51 | 0.61 | 1.38 | 1.39 | 1.21 | |
| CCA | 1.64 | 1.11 | 0.92 | 0.97 | 0.79 | 0.97 | 1 | 1.23 | |
| CCG | 0.14 | 0.45 | 0.43 | 0.5 | 1.65 | 0.45 | 0.53 | 0.35 | |
| Threonine | ACU | 1.77 | 0.99 | 1.35 | 0.84 | 1.00 | 0.94 | 0.89 | 0.97 |
| ACC | 0.39 | 1.42 | 1.06 | 1.59 | 0.49 | 1.58 | 1.55 | 1.42 | |
| ACA | 1.65 | 1.14 | 1.16 | 0.94 | 1.02 | 0.96 | 1.01 | 1.23 | |
| ACG | 0.19 | 0.46 | 0.43 | 0.63 | 1.49 | 0.52 | 0.56 | 0.37 | |
| Alanine | GCU | 2.19 | 1.06 | 1.07 | 0.96 | 1.15 | 1.05 | 1 | 1.12 |
| GCC | 0.57 | 1.6 | 1.27 | 1.79 | 0.44 | 1.72 | 1.71 | 1.57 | |
| GCA | 1.09 | 0.91 | 1.18 | 0.76 | 0.41 | 0.77 | 0.8 | 0.94 | |
| GCG | 0.15 | 0.42 | 0.48 | 0.5 | 0.70 | 0.45 | 0.48 | 0.35 | |
| Tyrosine | UAU | 1.23 | 0.89 | 1.15 | 0.78 | 0.75 | 0.75 | 0.79 | 0.87 |
| UAC | 0.77 | 1.11 | 0.85 | 1.22 | 1.25 | 1.25 | 1.21 | 1.12 | |
| Histidine | CAU | 1.43 | 0.84 | 1.2 | 0.74 | 0.44 | 0.81 | 0.75 | 0.81 |
| CAC | 0.57 | 1.16 | 0.8 | 1.26 | 1.56 | 1.19 | 1.25 | 1.18 | |
| Glutamine | CAA | 1.4 | 0.53 | 1.25 | 0.56 | 0.78 | 0.52 | 0.46 | 0.49 |
| CAG | 0.6 | 1.47 | 0.75 | 1.44 | 1.22 | 1.48 | 1.54 | 1.5 | |
| Asparagine | AAU | 1.36 | 0.94 | 1.18 | 0.82 | 0.77 | 0.84 | 0.81 | 0.92 |
| AAC | 0.64 | 1.06 | 0.82 | 1.18 | 1.23 | 1.16 | 1.19 | 1.07 | |
| Lysine | AAA | 1.29 | 0.87 | 1.37 | 0.86 | 0.74 | 0.79 | 0.78 | 0.84 |
| AAG | 0.71 | 1.13 | 0.63 | 1.14 | 1.26 | 1.21 | 1.22 | 1.15 | |
| Aspartic acid | GAU | 1.29 | 0.93 | 1.13 | 0.84 | 0.83 | 0.83 | 0.84 | 0.98 |
| GAC | 0.71 | 1.07 | 0.87 | 1.16 | 1.17 | 1.17 | 1.16 | 1.01 | |
| Glutamic acid | GAA | 1.45 | 0.84 | 1.17 | 0.86 | 0.77 | 0.76 | 0.78 | 1.88 |
| GAG | 0.55 | 1.16 | 0.83 | 1.14 | 1.23 | 1.24 | 1.22 | 1.05 | |
| Cysteine | UGU | 1.59 | 0.91 | 0.89 | 0.87 | 1.40 | 0.89 | 0.85 | 0.92 |
| UGC | 0.41 | 1.09 | 1.11 | 1.13 | 0.60 | 1.11 | 1.15 | 1.07 | |
| Arginine | CGU | 1.44 | 0.48 | 1.17 | 0.41 | 0.40 | 0.55 | 0.49 | 0.49 |
| CGC | 0.6 | 1.1 | 0.92 | 1.09 | 0.89 | 1.15 | 1.17 | 0.94 | |
| CGA | 0.31 | 0.65 | 0.71 | 0.55 | 0.79 | 0.61 | 0.68 | 0.74 | |
| CGG | 0.2 | 1.21 | 0.48 | 1.19 | 1.94 | 1.08 | 1.32 | 1.18 | |
| AGA | 2.64 | 1.29 | 1.29 | 1.33 | 1.08 | 1.3 | 1.14 | 1.26 | |
| AGG | 0.82 | 1.27 | 1.42 | 1.41 | 0.90 | 1.32 | 1.2 | 1.36 | |
| Glycine | GGU | 2.36 | 0.65 | 1.02 | 0.58 | 0.58 | 0.65 | 0.64 | 0.71 |
| GGC | 0.7 | 1.35 | 1.05 | 1.42 | 1.17 | 1.43 | 1.43 | 1.36 | |
| GGA | 0.81 | 1 | 1.27 | 1.01 | 1.28 | 0.95 | 0.95 | 0.96 | |
| GGG | 0.12 | 1 | 0.66 | 0.99 | 0.97 | 0.97 | 0.99 | 0.95 |
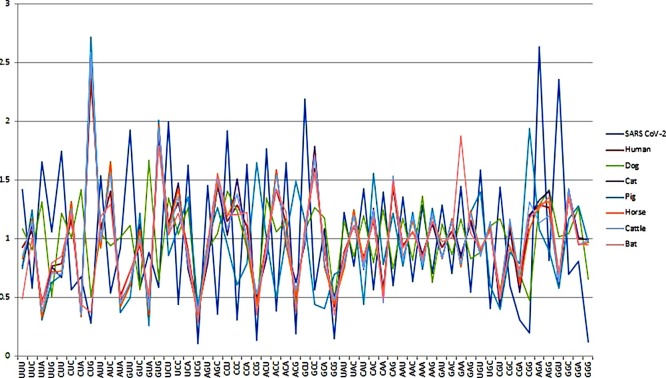
Analysis of RSCU values of SARS-CoV-2 and its different hosts uncovered the codon preferences of SARS-CoV-2, human, dog, cat, pig, horse and cattle (Table 1). The average RSCU of SARS-CoV-2 was compared to that of its normal (human) and accidental (dog) hosts along with other animal species which revealed that the codon preference of SARS-CoV-2 and its hosts (natural, accidental and other) are not similar (Fig. 1 ). Specific preferences in SARS-CoV-2 and its host codon usage suggested that the virus does not compete with the host tRNA array.
Fig. 1.
Comparative analysis of Relative Synonymous Codon Usage (RSCU) patterns of SARS CoV2 with other hosts.
3.3. Significant influence of dinucleotide frequencies in determining the codon usage Bias
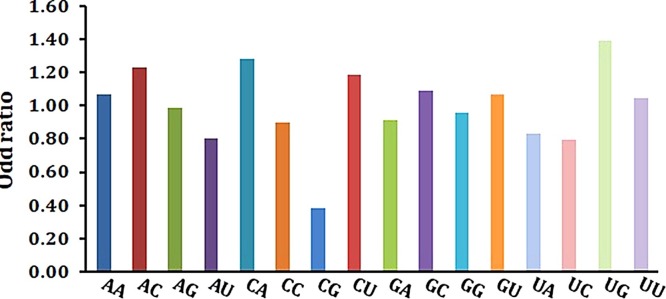
The composition of UpU (8.00 %) and ApA (7.48 %) were obtained as the most abundant dinucleotide in the SARS-CoV-2 genome with odd ratio of 1.04 and 1.07 respectively, while CpC (4.56) and CpG (4.75 %) were the least abundant dinucleotide with the lowest odds ratio (0.38) exhibited by CpG (Supplementary Table S2).
RSCU values of eight codons containing CpG (CCG, UCG, GCG, ACG, CGG, CGC, CGU, and CGA) and six codons containing UpA (UUA, CUA, AUA, GUA, UAU, and UAC) were analysed to determine the possible effects of CpG and UpA representations on codon usage bias. All CpG-containing codons were not over-represented (RSCU ≤ 1.6) and were not preferred for their respective amino acids, while among the UpA containing codons, only UUA (Leucine) was over-represented in case of SARS-CoV-2.The relative abundance of UpG (1.4) and CpA (1.28) dinucleotides also indicated a severe deviation from the normal and these dinucleotides were over-represented compared to others (Fig. 2 ). All the UpG -containing codons were under-represented (RSCU ≤ 1.6) and were not preferred for their respective amino acids. Among the CpA containing codons, ACA (T) and CCA (P) were over-represented (RSCU ≥ 1.6).
Fig. 2.
Relative dinucleotide frequencies in SARS-CoV-2.
3.4. Influence of mutational bias or natural selection pressure on SARS-CoV-2 codon usage patterns
3.4.1. ENc-plot analysis
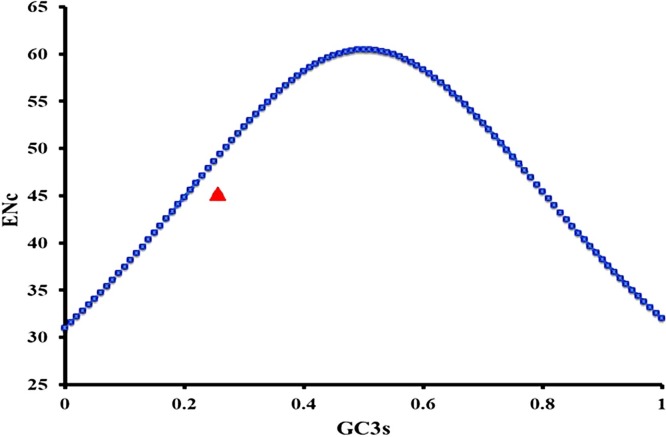
The mutational pressure or selection pressure on a gene or genome due to codon usage is determined by ENc-GC3s plot analysis. The analysis of ENc-plot revealed that all the points of SARS-COV-2 virus lie below the expected curve, indicating the influence of natural selection as the major force in codon usage bias in SARS-COV-2 virus sequences (Fig. 3 ). However, the overall reduction in the percentage of estimated ENc value of all the considered genes of SARS-COV-2 compared to the theoretical value was found to be 8.52 %.
Fig. 3.
ENc–GC3 plot of concatenated CDSs of SARS-CoV-2. The ENc curve is indicating the expected codon usage, if GC compositional constraints only account for the codon usage bias.
3.4.2. Neutrality plot
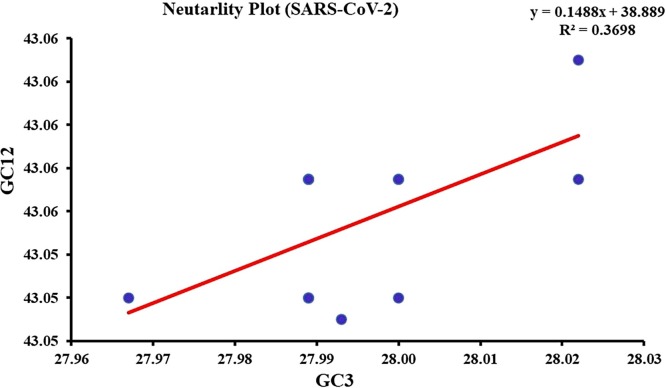
A neutrality plot analysis was done to decipher the degree of influence of natural selection and mutation pressure in shaping the codon usage bias in SARS-COV-2 virus sequences. In case of SARS-COV-2, a weak positive correlation was observed between GC12 and GC3 (r = 0.3). However, the slope of the regression line in respect of SARS-COV-2 was 0.1488 indicating that the relative influence of mutation pressure was 14.88 % and contribution of natural selection was 85.12 % (Fig. 4 ).
Fig. 4.
Neutrality plot: The neutrality plot predicts the influences of mutation bias and translation selection on codon usage. GC12 stands for the average value of GC content at first and second position of codon. GC3 stands for GC content at third position of codon. The slope value indicates the mutational pressure. Blue dots represent concatenated ORFs of SARS-CoV-2.
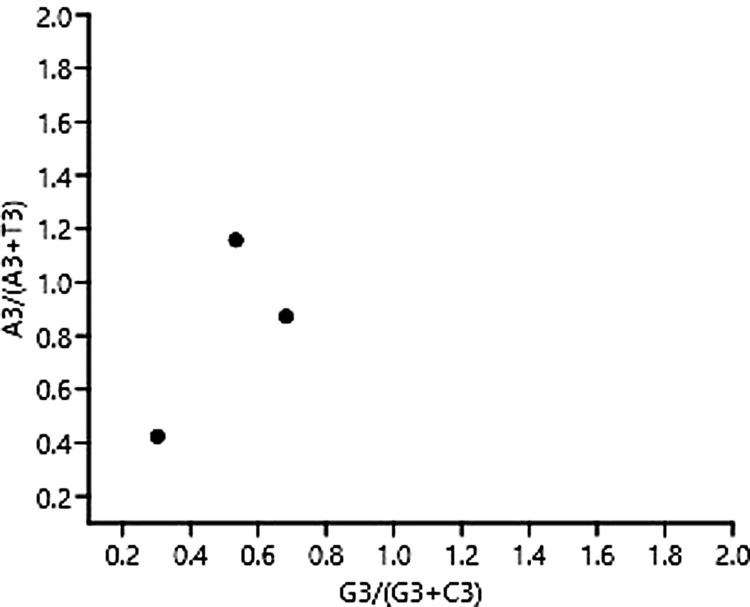
3.4.3. Parity analysis
For parity analysis, we plottedA3/(A3 + T3) and G3/(G3 + C3) as ordinate and abscissa, respectively (Fig. 5 ). The means of AT bias [A3/(A3 + T3)] and GC bias [G3/(G3 + C3)] were found to be 0.39 and 0.451, respectively. A bias value greater than 0.5 suggests a preference for pyrimidine over purine (Zhang et al., 2018). Thus in SARS-CoV-2, T is preferred over A and C is preferred over G.
Fig. 5.
Parity plot showing the presence of AT bias [A3 %/(A3 % + T3 %)] and GC bias [G3 %/(G3 % + C3 %)].
3.4.4. tRNA iso-acceptor
Frequency of tRNA genes in human cells; for a single codon, a variable number of isoacceptor tRNAs are present, which varies across the organisms. Translation selection determines whether most codons preferred by SARS-CoV-2 are recognized by the most abundant isoacceptor tRNAs (Khandia et al., 2019). Out of 18 amino acids (which are encoded by two or more amino acid codons) except for Leucine, Isoleucine, Valine and Proline, non-optimal codon-anticodon base pairs were used (Table 2 ).
Table 2.
Frequency of tRNA genes in human cells for most preferentially used codons in SARS-CoV-2.
| Amino acids | Most preferred codons in SARS CoV-2 | tRNA isotypes in human cells | Total count | |||||
|---|---|---|---|---|---|---|---|---|
| Ala(A) | GCU(A) | AGC(22) | GGC | CGC(4) | UGC(8) | 34 | ||
| Gly(G) | GGU(G) | ACC | GCC(14) | CCC(5) | UCC(9) | 28 | ||
| Pro(P) | CCU(P) | AGG(9) | GGG | CGG(4) | UGG(7) | 20 | ||
| Thr(T) | ACU(T) | AGU(9) | GGU | CGU(5) | UGU(6) | 20 | ||
| Val(V) | GUU(V) | AAC(9) | GAC | CAC(11) | UAC(5) | 25 | ||
| Ser(S) | UCU(S) | AGA(9) | GGA | CGA(4) | UGA(4) | ACU | GCU(8) | 25 |
| Arg(R) | AGA(R) | ACG(7) | GCG | CCG(4) | UCG(6) | CCU(5) | UCU(6) | 28 |
| Leu(L) | CUU(L) | CAA(9) | GAG | CAG(9) | UAG(3) | CAA(6) | UAA(4) | 31 |
| Phe(F) | UUU(F) | AAA | GAA(10) | 10 | ||||
| Asn(N) | AAU(N) | AUU | GUU(20) | 20 | ||||
| Lys(K) | AAA(K) | CUU(15) | UUU(12) | 27 | ||||
| Asp(D) | GAU(D) | AUC | GUC(13) | 13 | ||||
| Glu(E) | GAA(E) | CUC(8) | UUC(7) | 15 | ||||
| His(H) | CAU(H) | AUG | GUG(10) | 10 | ||||
| Gln(Q) | CAA(Q) | CUG(13) | UUG(6) | 19 | ||||
| Ilu(I) | AUU(I) | AAU(14) | GAU(3) | CAU | UAU(5) | 22 | ||
| Tyr(Y) | UAU(Y) | AUA | GUA(13) | 13 | ||||
| Cys(C) | UGU(C) | ACA | GCA(29) | 29 | ||||
| Trp(W) | UGG | CCA(7) | 7 | |||||
| Met(M) | AUG | CAU(9/10) | 19 | |||||
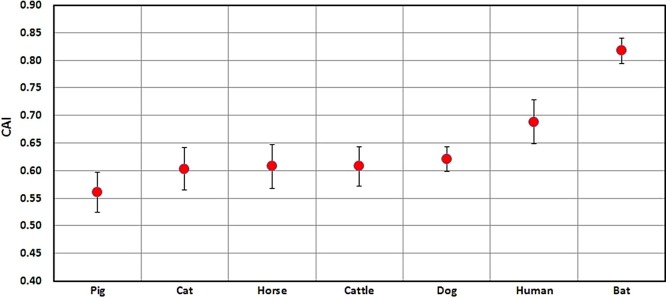
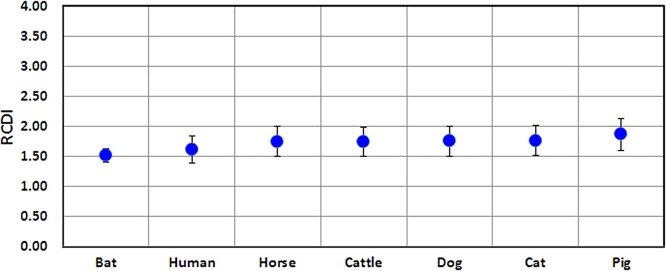
3.5. Adaptation of SARS-CoV-2 (CAI and RCDI)
The average codon adaptation index (CAI) values for all the five genes was found to be highest in bat (0.817) and human (0.698) followed by dog, cattle, horse, cat and pig, respectively (Fig. 6 ). The cumulative effect of codon biases on gene expression was determined by relative codon deoptimization index (RCDI) values. The average RCDI values of all genes indicated that SARS-COV-2 was more adapted to bat (1.518) and human (1.61) as compared to dog or other animals (Fig. 7 ). Higher adaptation increases the infectivity and vice versa (Supplementary table S3).
Fig. 6.
Codon Adaptation Index (CAI) for five coding sequences of SARS-CoV-2 with reference to different hosts. CAI values range between 0.0 and 1.0; with higher values indicating a higher gene expression potential.
Fig. 7.
Relative Codon Deoptimization Index (RCDI) for five coding sequences of SARS-CoV-2 with reference to different hosts. RCDI values provide an estimate of the rate of viral gene translation in a host genome.
Correlation among various parameters such as ENc, GC3, CAI, Laa, AROMO and GRAVY was also studied (Table 3 ). A positive correlation of GC3 was observed with CAI, GRAVY and AROMO, while a negative correlation was observed with Laa and ENc. The correlation analysis among CAI, GRAVY and AROMO was done to determine the effect of GRAVY and AROMO (indicators of natural selection) on expressivity of gene (indicated by CAI). However, no correlation was observed.
Table 3.
Correlation analysis between GC3 %, ENc, CAI, L_aa, GRAVY and AROMO.
| GC3 % | ENc | CAI | L_aa | Gravy | |
|---|---|---|---|---|---|
| Nc | 0.445 | ||||
| CAI | 0.34 | −0.192 | |||
| L_aa | −0.238 | 0.435 | 0.089 | ||
| Gravy | 0.219 | 0.178 | 0.257 | −0.237 | |
| Aromo | 0.033 | −0.183 | −0.05 | 0.088 | −0.208 |
3.6. Similarity index
A similarity (SiD) analysis was performed to ascertain the function of different hosts in framing the codon usage pattern of SARS-CoV-2.The investigation of similarity indices revealed that human has more (0.117) impact than dog (0.05) on SARS-CoV-2 codon usage bias.
4. Discussion
In the present investigation, we studied codon bias and codon usage of SARS-COV-2 by characterizing them with different parameters. The SARS-COV-2 genomes were found have relative abundance of A and U nucleotides and a preference of A/U ending codons over G/C ending codons. Similar results were reported in a previous study on SARS-COV-2 (Dilucca et al., 2020; Tort et al., 2020). It was reported that the N gene of Coronavirus has higher AT% than GC% with an effective number of codons ranging from 40.43 to 53.85 indicating a slight codon bias (Sheikha et al., 2019). The codon usage in RNA viruses is affected by the relative abundance of dinucleotide (Belalov and Lukashev, 2013). CpG depletion is considered to be a selective force that influences the frequency of codons that contain CpG. Low relative abundance of CpG may be attributed to unmethylated CpG-containing sequences, which are recognized as pathogenic signatures and methylation of cytosine residues by innate hosts’ defence systems (Li and Zhang, 2014). It was found that the RSCU value of six codons containing CpG (CCG, GCG, CGG, UCG, ACG and CGA) were under-represented (RSCU < 0.6). This indicates that the selection pressure influences significantly on the codon usage in SARS-COV-2. Our observations were in agreement with those of previous studies on equine influenza virus (Kumar et al., 2016) and Nipah virus (Khandia et al., 2019).
It was reported that TpA and UpA containing dinucleotides were also under-represented in the genome of DNA and RNA viruses (Kumar et al., 2016). Higher cytoplasmic RNase susceptibility to UpA helps to maintain mRNA turnover within the cell (Beutler et al., 1989). However, in case of SARS-COV-2, UpA and TpA were not under-represented, which may be due to the nucleotide A rich genome. UpA-COntaining codons (UUA, GUA, AUA, UAU, CUA and UAC) had RSCU values were not below 0.6 with an odds ratio of 0.83, which indicates utilization of unbiased UpA-COntaining codons, except for UUA which was found to be over-represented with RSCU values >1.6. Among the CpA-COntaining codons (UCA, CCA, ACA, GCA, CAA, CAG, CAU and CAC), three codons (CCA, ACA and UCA) were over-represented (RSCU > 1.6). Among five codons containing UpG (UUG, CUG, GUG, UGU and UGC), three were found to be under-represented (RSCU < 1.6) and only two of them were the preferential codons (UUG and UGU) for their respective amino acids. Relative abundance of UpG and CpA in different organisms is a result of the under-represented CpG dinucleotides (Kumar et al., 2016). Our results suggested that dinucleotide compositions play a significant role in determining the codon usage patterns in SARS-COV-2 genome. This also suggests that selection pressure leading to low UpA frequencies is not directly involved in SARS-COV-2 codon usage patterns; rather these patterns are primarily regulated by compositional constraints, since SARS-COV-2 genome is rich in A and U nucleotides. This result is consistent with the earlier findings of Khandia ; et al.; (2019), who reported that codon bias is primarily due to the direct effect of dinucleotide bias.
Factors affecting codon usage: The average GC and GC3 contents of SARS-COV-2 genome were 38.03 and 27.99, respectively. In the case of codon usage that is influenced only by the genome’s GC3 content, the ENc values lie just above the predicted ENc curve indicating mutational pressure (He et al., 2016). The ENc values were below the predicted ENc curve in the SARS-COV2 genome indicating the dominant role of selection pressure. A neutrality plot analysis was performed to determine the role of selection pressure. The weak positive correlation between GC12 and GC3 and slope of the regression line closing to zero (regression line slope, y = 0.1488x + 38.889, R 2 = 0.3698) observed in the present study indicated that selection pressure was the dominant factor in shaping the codon usage pattern of SARS-COV-2. It was also observed that the concatenated CDS of SARS-COV-2 were away from the slope of the regression line which further suggested that selection pressure was the major force and mutational pressure was the minor force influencing SARS-COV-2 codon usages. No association between ENc and GRAVY or ENc and AROMO was found, suggesting that hydrophobicity or aromaticity does not affect codon usage bias. In addition, no association between CAI, GRAVY and AROMO was observed to suggest an impact of GRAVY and AROMO on gene expression. Negative correlation between Laa and ENc indicated that the number of amino acids does not have any influence on codon usage bias, which might be due to the effect of natural selection in synonymous codon usage pattern (Wei et al., 2014).
4.1. Hosts effect on SARS-COV-2 codon usage
Analysis of similarity index showed that the human genome has more effect on SARS-COV-2 codon usage than that of dog. Previously, the similarity index analysis was reported for chikungunya virus and Zika virus (Butt et al., 2014, 2016). However, higher similarity indices were observed in dog and African green monkey than human host for Nipah virus (Khandia et al., 2019). Evolutionary analysis suggested that SARS-COV-2 has the highest similarity to bat coronovirus and has the most similar codon usage bias with snake (Ji et al., 2020). Relatively low average values of D (A, B) [where D (A, B), indicate the potential role of the overall use of codon by the host over that of SARS-COV-2] suggested that SARS-COV-2 can replicate efficiently in the host without having much effect on the host codon usage. Rousettus aegyptiacus, an Egyptian fruit bat was reported to display greater similarity index for Marburg virus as compared to human host (Nasrullah et al., 2015). Codon usage can be shaped by many different selection forces including certain host factors. It was hypothesized that the codon usage in SARS-CoV-2 maybe directly correlated to the codon usage of its host (Ji et al., 2019).
4.2. Dog as a host for SARS-COV2
Deoptimization analysis is conducted by contrasting the use of codon in a virus to that of its host. The RCDI values provide an insight into potential virus and host genome co-evolution. Lower RCDI value indicates a virus being more adaptable to its host. Here in our study, human showed lesser mean RCDI value (1.61) than dog (1.753) indicating better adaptation of the virus in human compared to dog. Lower the RCDI value higher is the CAI value. Higher RCDI value may indicate gene expression during latency period or low translation rate maintenance to achieve error-proof translation (Puigbo et al., 2010). Higher average CAI values of human compared to dog observed in the present study indicated that dog is less susceptible to COVID 19 than human. However, till now cross-transmission of SARS-CoV-2 between human and dog hasnot been well-understood. The present study was conducted to compare SARS-CoV-2 adaptation in human and dog hosts. The findings of this study may be useful to evaluate and determine the role of other animal species serving as a host to the virus for their potential. It also highlighted the emerging health hazards to human as a result of living in close contact with animals, which may serve as carriers of a pandemic virus like SARS-CoV-2 and a potential source of infection.
5. Conclusion
SARS-CoV-2 is the recently identified emerging virus causing a serious public health emergency across the globe. There is an urgent need to develop an effective vaccine and to identify possible measures for its control. In this study, we compared humans and dogs as the hosts for SARS-CoV-2 on the basis of codon usage patterns. Based on the CAI and RCDI values, SARS-CoV-2 sequences were found to be highly human-adapted. Knowledge of the pattern of codon usage of a virus is helpful to optimize the expression of its protein. Information on enhanced protein expression would be useful in developing a suitable SARS-CoV-2 vaccine candidate by expressing it in various prokaryotic/eukaryotic systems. Detailed information of codon usage may also be used to evolve effective methods to reduce the synthesis of SARS-CoV-2 protein during pathogen replication. Moreover, it may be useful to obtain analogous information for other viruses.
Authors’ contributions
Rupam Dutta and Lukumoni Buragohain substantially contributed to the conception, design, analysis, interpretation of data, checking and approving final version of the manuscript. Probodh Borah helped in writing and finalization of the manuscript.
Ethical approval
No human or animal samples were handled in the present study. All the sequences were downloaded from the viral database. Therefore, ethical committee approval is not required.
Data sharing
No new data has been generated in our study. All the data were obtained from the Virus Resource at the National Center for Biotechnological Information (https://www.ncbi.nlm.nih.gov/labs/virus). Accession no. of the SARS-CoV-2 genome utilized were (LC529905.1, MT007544.1, MT012098.1, MT066156.1, MT072688.1, MT093571.1, MT126808.1, MT192759.1, MT192765.1, MT192772.1, MT233519 and NC045512.2).
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgment
The authors acknowledge DBT, Govt of India for providing financial support to the host department.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.198113.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahir I., Fromer M., Prat Y., Linial M. Viral adaptation to host: a proteome‐based analysis of codon usage and amino acid preferences. Mol. Syst. Biol. 2009;5:311. doi: 10.1038/msb.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belalov I.S., Lukashev A.N. Causes and implications of codon usage bias in RNA viruses. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera B.C., Virmani N., Kumar N., Anand T., Pavulraj S., Rash A., Elton D., Rash N., Bhatia S., Sood R., Singh R.K., Tripathi B.N. Genetic and codon usage bias analyses of polymerase genes of equine influenza virus and its relation to evolution. BMC Genomics. 2017;18:652. doi: 10.1186/s12864-017-4063-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E., Gelbartt T., Hans J., Koziolt J.A., Beutler B. Evolution of the genome and the genetic code: selection at the dinucleotide level by methylation and polyribonucleotide cleavage (nucleas/stop codon) Proc. Natl. Acad. Sci. USA. 1989;86:192. doi: 10.1073/pnas.86.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt A.M., Nasrullah I., Tong Y. Genome-wide analysis of codon usage and influencing factors in chikungunya viruses. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt A.M., Nasrullah I., Qamar R., Tong Y. Evolution of codon usage in Zika virus genomes is host and vector specific. Emerg. Microbes Infect. 2016;5:e107. doi: 10.1038/emi.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. The coronavirus surface glycoprotein. In: Siddell S.G., editor. The Coronaviridae, The Viruses. Springer; Boston, MA: 1995. pp. 73–113. [DOI] [Google Scholar]
- Costafreda M.I., Pérez-Rodriguez F.J., D’Andrea L., Guix S., Ribes E., Bosch A., Pintó R.M. Hepatitis A virus adaptation to cellular shutoff is driven by dynamic adjustments of codon usage and results in the selection of populations with altered capsids. J. Virol. 2014;88:5029–5041. doi: 10.1128/JVI.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristina J., Moreno P., Moratorio G., Musto H. Genome-wide analysis of codon usage bias in Ebolavirus. Virus Res. 2015;196:87–93. doi: 10.1016/j.virusres.2014.11.005. [DOI] [PubMed] [Google Scholar]
- D’Andrea L., Pérez-Rodríguez F.J., Castellarnau M., Guix S., Ribes E., Quer J., Gregori J., Bosch A., Pintó R.M. The critical role of codon composition on the translation efficiency robustness of the hepatitis a virus capsid. Genome Biol. Evol. 2019;11:2439–2456. doi: 10.1093/gbe/evz146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilucca M., Forcelloni S., Georgakilas A.G., Giansanti A., Pavlopoulou A. Codon usage and phenotypic divergences of SARS-CoV-2 genes. Viruses. 2020;12(5):498. doi: 10.3390/v12050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C., Minshull J., Govindarajan S., Ness J., Villalobos A., Welch M. Engineering genes for predictable protein expression. Protein Expr. Purif. 2012;83:37–46. doi: 10.1016/j.pep.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.Z. Pangolins harbor SARS-CoV-2-Related coronaviruses. Trends Microbiol. 2020;28:515–517. doi: 10.1016/j.tim.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Dong H., Jiang C., Cao F., Tao S., Xu L.A. Analysis of codon usage patterns in Ginkgo biloba reveals codon usage tendency from A/U-ending to G/C-ending. Sci. Rep. 2016;6:35927. doi: 10.1038/srep35927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Zhang H., Zhang Y., Wang R., Lu S., Ji Y., Chang L., Yuan P., Su S. Codon usage bias in the N gene of rabies virus. Infect. Genet. Evol. 2017;54:458–465. doi: 10.1016/j.meegid.2017.08.012. [DOI] [PubMed] [Google Scholar]
- Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2019;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandia R., Singhal S., Kumar U., Ansari A., Tiwari R., Dhama K. Analysis of Nipah virus codon usage and adaptation to hosts. Front. Microbiol. 2019;10:886. doi: 10.3389/fmicb.2019.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kin N., Miszczak F., Diancourt L., Caro V., Moutou F., Vabret A. Comparative molecular epidemiology of two closely related coronaviruses, bovine coronavirus (BCoV) and human coronavirus OC43 (HCoV-OC43), reveals a different evolutionary pattern. Infect. Genet. Evol. 2016;40:186–191. doi: 10.1016/j.meegid.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Bera B.C., Greenbaum B.D., Bhatia S., Sood R., Selvara J.P. Revelation of influencing factors in overall codon usage bias of equine influenza viruses. PLoS One. 2016;11:e0154376. doi: 10.1371/journal.pone.0154376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lauber C., Ziebuhr J., Junglen S., Drosten C., Zirkel F., Nga P.T. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch. Virol. 2012;157:1623–1628. doi: 10.1007/s00705-012-1295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Zhang Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014;6 doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musto H. What we know and what we should know about codon usage. J. Mol. Evol. 2016;82:245–246. doi: 10.1007/s00239-016-9742-z. [DOI] [PubMed] [Google Scholar]
- Nasrullah I., Butt A.M., Tahir S., Idrees M., Tong Y. Genomic analysis of codon usage shows influence of mutation pressure, natural selection, and host features on Marburg virus evolution. BMC Evol. Biol. 2015;15:174. doi: 10.1186/s12862-015-0456-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S. Another decade, another coronavirus. N. Engl. J. Med. 2020;382:760–762. doi: 10.1056/NEJMe2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P., Bravo I.G., Garcia-Vallve S. CAIcal: a combined set of tools to assess codon usage adaptation. Biol. Direct. 2008;3:38. doi: 10.1186/1745-6150-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P., Aragones L., Garcia-Vallve S. RCDI/eRCDI: a web-server to estimate codon usage deoptimization. BMC Res. Notes. 2010;3:87. doi: 10.1186/1756-0500-3-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C., Antón I.M., Enjuanes L., Carrascosa J.L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol. 1996;70:4773–4777. doi: 10.1128/JVI.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Ruch T., Machamer C. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewatanon J., Srichatrapimuk S., Auewarakul P. Compositional bias and size of genomes of human DNA viruses. Intervirology. 2007;50:123–132. doi: 10.1159/000098238. [DOI] [PubMed] [Google Scholar]
- Sheikha A., Al-Taherb A., Al-Nazawib M., Al-Mubarakc A., Kandeel M. Analysis of preferred codon usage in the coronavirus N genes and their implications for genome evolution and vaccine design. J. Virol. Methods. 2019;277 doi: 10.1016/j.jviromet.2019.113806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N., Eyre-Walker A. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol. Biol. Evol. 2007;24:374–381. doi: 10.1093/molbev/msl166. [DOI] [PubMed] [Google Scholar]
- Tort F.L., Castells M., Cristina J. A comprehensive analysis of genome composition and codon usage patterns of emerging coronaviruses. Virus Res. 2020;283 doi: 10.1016/j.virusres.2020.197976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert F., Berkhout B. Nucleotide composition of the Zika virus RNA genome and its codon usage. Virol. J. 2016;13:95. doi: 10.1186/s12985-016-0551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., He J., Jia X., Qi Q., Liang Z., Zheng H., Ping Y., Liu S., Sun J. Analysis of codon usage bias of mitochondrial genome in Bombyx mori and its relation to evolution. BMC Evol. Biol. 2014;14:262. doi: 10.1186/s12862-014-0262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhao D., Tao J. Analysis of codon usage patterns in herbaceous paeony (Paeoni alactiflora Pall.) based on transcriptome data. Genes. 2015;6:1125–1139. doi: 10.3390/genes6041125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu Y., Liu W., Zhou J., Chen H., Wang Y., Ma L., Ding Y., Zhang J. Analysis of synonymous codon usage in Hepatitis A virus. Virol. J. 2011;8:174. doi: 10.1186/1743-422X-8-174. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.P., Cai Y.Y., Yu D.N., Storey K.B., Zhang J.Y. Gene characteristics of the complete mitochondrial genomes of Paratoxodera polyacantha and Toxodera hauseri (Mantodea: toxoderidae. Peer J. 2018;6:e4595. doi: 10.7717/peerj.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.H., Zhang J., Sun D., Ma Q., Chen H., Ma L. The distribution of synonymous codon choice in the translation initiation region of dengue virus. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.