Abstract
New coronavirus SARS-CoV-2 is capable to infect humans and cause a novel disease COVID-19. Aiming to understand a host genetic component of COVID-19, we focused on variants in genes encoding proteases and genes involved in innate immunity that could be important for susceptibility and resistance to SARS-CoV-2 infection.
Analysis of sequence data of coding regions of FURIN, PLG, PRSS1, TMPRSS11a, MBL2 and OAS1 genes in 143 unrelated individuals from Serbian population identified 22 variants with potential functional effect. In silico analyses (PolyPhen-2, SIFT, MutPred2 and Swiss-Pdb Viewer) predicted that 10 variants could impact the structure and/or function of proteins. These protein-altering variants (p.Gly146Ser in FURIN; p.Arg261His and p.Ala494Val in PLG; p.Asn54Lys in PRSS1; p.Arg52Cys, p.Gly54Asp and p.Gly57Glu in MBL2; p.Arg47Gln, p.Ile99Val and p.Arg130His in OAS1) may have predictive value for inter-individual differences in the response to the SARS-CoV-2 infection.
Next, we performed comparative population analysis for the same variants using extracted data from the 1000 Genomes project. Population genetic variability was assessed using delta MAF and Fst statistics. Our study pointed to 7 variants in PLG, TMPRSS11a, MBL2 and OAS1 genes with noticeable divergence in allelic frequencies between populations worldwide. Three of them, all in MBL2 gene, were predicted to be damaging, making them the most promising population-specific markers related to SARS-CoV-2 infection.
Comparing allelic frequencies between Serbian and other populations, we found that the highest level of genetic divergence related to selected loci was observed with African, followed by East Asian, Central and South American and South Asian populations. When compared with European populations, the highest divergence was observed with Italian population.
In conclusion, we identified 4 variants in genes encoding proteases (FURIN, PLG and PRSS1) and 6 in genes involved in the innate immunity (MBL2 and OAS1) that might be relevant for the host response to SARS-CoV-2 infection.
Keywords: COVID-19, SARS-CoV-2, Host genomics, Susceptibility and resistance, Gene variants, Functional prediction, Allele frequencies, Population genomics
Highlights
-
•
Variants in genes for proteases & innate immunity in relation to SARS-CoV-2 infection.
-
•
We analyzed variants in FURIN, PLG, PRSS1, TMPRSS11a, MBL2 and OAS1 genes.
-
•
Functional prediction and comparative population analysis of variants were performed.
-
•
Ten variants are predicted to be relevant for host response to SARS-CoV-2 infection.
1. Introduction
A new coronavirus SARS-CoV-2 capable to infect humans, emerged in mid-December 2019 in Wuhan, China, causing the novel disease COVID-19 (Wu et al., 2020). Clinical manifestations of this viral infection vary from asymptomatic to severe acute respiratory syndrome and death. The number of infected people and affected countries have quickly risen and already in March 2020, World Health Organization declared a pandemic (WHO). Therefore, it became highly important to understand if specific human genes and genetic variants could be associated with susceptibility or resistance to SARS-CoV-2 infection and how frequencies of these variants vary between different populations. Human genes usually associated with susceptibility and resistance to viral infection are those associated with the point of viral entry into the human host cells, such as genes encoding receptors, co-receptors and enzymes that modify receptors (Kenney et al., 2017). Furthermore, various genes involved in the immune response, such as virus sensing, signaling in response to virus, antiviral factors etc. have also been found to be important for the severity and the outcome of viral infections. A recent study which analyzed COVID-19 symptoms in monozygotic and dizygotic twins, reported that 50 (95% confidence intervals 29–70)% of the variance of ‘predicted COVID-19’ phenotype is due to genetic factors (Williams et al., 2020).
Deciphering the RNA sequence of the SARS-CoV-2 genome showed that this new virus belongs to lineage B betacoronaviruses, together with the SARS-CoV virus which emerged in 2002 (Wu et al., 2020). Genetic susceptibility and resistance to SARS-CoV had been extensively studied by genotyping SARS patients with extremely severe and extremely mild clinical manifestations. As a result, variants in OAS1, MX1, MBL2, CCL2, CCL5, ASHG, IFNgamma, CD14 and CD209 genes were associated with genetic susceptibility to SARS-CoV (Chong et al., 2006; He et al., 2006; Ng et al., 2007; Yuan et al., 2007; Chan et al., 2010; Zhu et al., 2011; Tu et al., 2015). Majority of variants that emerged in such studies were located in non-coding parts of the human genome and their effect is observable in the fine tuning of the gene expression. Also, variants residing in coding regions, such as rs1800450 in MBL2 gene (Tu et al., 2015) were particularly interesting as they alter protein structure and function.
SARS-CoV and SARS-CoV-2 share ~76% amino acid sequence identity of the spike (S) protein sequence, a crucial part of the viral envelope which enables specific binding to the receptors at human cells, therefore contributing to viral potential to infect humans (Lu et al., 2015; Chan et al., 2020). It was shown that SARS-CoV-2 binds to the human angiotensin-converting enzyme 2 (ACE2) receptors, as SARS-CoV does, however with the higher affinity (Wrapp et al., 2020). Human transmembrane protease serine 2 (TMPRSS2), an enzyme important for the entry of SARS-CoV (Matsuyama et al., 2010), was recently found to activate the S protein of the SARS-CoV-2 (Hoffmann et al., 2020).
Moreover, it was found that the S protein of the SARS-CoV-2 contains a furin-like cleavage site which is absent in coronaviruses of the same clade (Coutard et al., 2020). Knowing that furin cleavage sites are responsible for the high virulence of human influenza viruses (Chen et al., 1998), it was suggested that furin-like site in the S protein of SARS-CoV-2 represents its advantage in attaching to the human cells expressing ACE2 receptor.
The SARS-CoV-2 relies on the host cell proteases to cut its S protein in two parts thus forming N-terminal part which recognizes ACE2 receptor and C-terminal part involved in the viral entry which must be further cleaved by furin and/or other furin-like enzymes (Coutard et al., 2020). It has been shown that plasmin is also capable to cleave furin sites (Zhao et al., 2020) and that individuals with elevated plasmin demonstrated higher susceptibility to COVID-19 (Ji et al., 2020). In addition to plasmin, S protein of coronaviruses may be cleaved by other airway proteases such as trypsin-1 and TMPRSS11a (Ji et al., 2020).
At the beginning of May 2020, in the full swing of COVID-19 pandemic, the data on genetic susceptibility or resistance to SARS-CoV-2 which would be based on the genotyping of individuals infected by SARS-CoV-2 are still lacking. However, a detailed study using computational prediction methods for protein structure analyses revealed that 17 variants in the coding regions of the ACE2 gene are located at positions important for the binding of ACE2 with the SARS-CoV-2 S protein (Hussain et al., 2020). Based on these predictions, individuals carrying these variants would probably be resistant to SARS-CoV-2 infection. These variants were rare (less than 0.00388 allele frequency) and their association with human diseases or disorders has never been reported (Hussain et al., 2020). Having in mind the rarity of variants that could directly influence the binding of SARS-CoV-2 to ACE2, it is not surprising that comparative genetic analysis of the frequency of ACE2 variants in different populations did not predict the existence of individuals resistant to SARS-CoV-2 infection (Cao et al., 2020).
ACE2 is not the only possible player that could influence the interaction between humans and SARS-CoV-2 virus. The furin could be the second important factor to contribute to high pathogenicity of the novel virus. Furthermore, we can learn from clinical experiences of the SARS-CoV-2 infection which showed that plasmin and other airway proteases could also be important for patients experiencing more severe form of COVID-19 (Ji et al., 2020). In addition, the knowledge based on SARS epidemic shows that MBL2 and OAS1 genes, involved in the innate immune response, could also modulate susceptibility to infection with betacoronaviruses.
Taking all these players into account, we performed genetic analysis of variants in coding regions of FURIN, plasminogen (PLG), trypsin-1 (PRSS1), TMPRSS11a, MBL2 and OAS1 genes in Serbian population aiming to identify possible genetic markers that are capable to impact protein structure/function and thus contribute to the susceptibility or resistance to SARS-CoV-2 infection. We postulate that the variants in genes encoding proteases could be advantageous, while variants in genes encoding proteins involved in the innate immunity add some disadvantage to individuals in combating COVID-19. We also performed comparative genetic analysis in different populations in order to assess the divergence between populations for those variants.
2. Subjects and methods
2.1. Genetic and bioinformatic analysis
In this study, we analyzed the genomic sequence data of unrelated individuals from Serbian population, extracted from the in-house database of Laboratory for Molecular Biomedicine, Institute of Molecular Genetics and Genetic Engineering, University of Belgrade. Written informed consent was obtained from all participants. The study was conducted in accordance with the Helsinki Declaration and approved by the Ethics Committee of Institute of Molecular Genetics and Genetic Engineering, University of Belgrade.
Total of 143 unrelated Serbian individuals (84 males and 59 females) were previously analyzed by NGS approach using the Illumina Clinical Exome Sequencing TruSight One Gene Panel (Illumina, San Diego, CA, USA), as previously described (Skakic et al., 2018). VCF files were further annotated and examined using the Illumina VariantStudio 3.0 Data Analysis Software (Illumina, San Diego, CA, USA). Variants that did not pass variant call quality filters (those with quality score < Q20, read depth < 20, percentage of variant frequency for the minor allele <20% and homopolymer length > 8) in more than 10% of samples were excluded. Variants that were considered for further analysis were either missense, start loss, stop gain, splice region variants and frameshift, while synonymous variants were omitted. For all selected variants, Hardy-Weinberg equilibrium was tested, using the chi square goodness of fit test.
Next, we compared genotype data from Serbian population with European (Italy, Spain, Finland, Great Britain and USA with European ancestry), as well as other 4 super-populations, Eastern Asians, South Asians, African and Ad Mixed American - Central and South American populations (total of 2504 subjects). Genotype data were extracted from the VCF files of Phase 3 variant calls of the 1000 Genomes Project (1kGP) sample collection (https://www.internationalgenome.org/) via Ensembl Data Slicer Tool. Populations and details regarding the 1kGP data were described previously (Genomes Project et al., 2015). Fisher exact test was used to measure significant differences in genotypes distributions between Serbian and 1kGP populations, applying Bonferoni correction for multiple testing.
We examined the level of population genetic variability at each selected locus using: (1) the maximal global differences in minor allele frequencies (delta MAF) calculated by subtracting the maximum and the minimum MAF across analyzed population groups, (2) using Fst statistics (Nei, 1987), which is widely used in population genetics (Holsinger and Weir, 2009).
R software was utilized for genotype data manipulation, statistical calculations as well as graphical presentations. For estimating Fst statistics, R packages adegenet and hierfstat were used.
2.2. In silico prediction analysis
Protein sequences were downloaded from Ensemble, accessed on 2020/04/24 at http://www.ensembl.org. To predict the effect of nonsynonymous amino acid substitutions, we used in silico prediction algorithms: PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2), SIFT/PROVEAN (http://provean.jcvi.org/index.php) and MutPred2 (http://mutpred.mutdb.org/). The Swiss-Pdb Viewer (Swiss Institute of Bioinformatics, at http://www.expasy.org/spdbv) was used to analyze the effects of variants upon the structure of the proteins, using the crystal structures with PDB codes 4ig8.pdb for the structure of human OAS1 in complex with dsRNA and 2′-deoxy ATP, and 4z2a.pdb for the structure of unglycosylated apo human furin (Protein Data Bank-RCSB, accessed on the 2020/04/24, at http://www.rcsb.org/pdb).
3. Results
3.1. Analysis of genetic variants found in Serbian population
Analysis of sequence data of coding regions of FURIN, PLG, PRSS1, TMPRSS11a, MBL2 and OAS1 genes in 143 unrelated individuals from Serbian population identified 22 variants with potential functional effects (Table 1 ). Among identified variants, there were 19 missense variants, 1 start loss, 1 missense/splice region and 1 splice region variant. Majority of the detected variants were rare (8 variants had minor allele frequency of 0.3%, while 4 variants of 0.7%). We detected the highest minor allelic frequency for the OAS1 p.Gly162Ser, being 41%. All detected variants were in Hardy-Weinberg equilibrium.
Table 1.
Genetic variants detected in Serbian population.
| Gene | HGVS nomenclature |
SNV ID | Genotype |
HW p | MAF | Consequence |
In silico prediction |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide change | Amino acid change | ref/ref | ref/alt | alt/alt | SIFT | PolyPhen-2 | MutPred2 | Swiss-Pdb Model | |||||
| FURIN | NM_002569.2:c.97A > G | NP_002560.1:p.Thr33Ala | rs1368782279 | 141 | 2 | 0 | 0.9 | 0.007 | missense | tolerated (0.628) | possibly damaging (0.613) | 0.408 | no |
| NM_002569.2:c.436G > A | NP_002560.1:p.Gly146Ser | rs201551785 | 141 | 2 | 0 | 0.9 | 0.007 | missense | deleterious (0.009) | probably damaging (0.975) | 0.879 | yes | |
| PLG | NM_000301.3:c.163G > C | NP_000292.1:p.Glu55Gln | rs1435341698 | 142 | 1 | 0 | 0.9 | 0.003 | missense | tolerated (0.409) | possibly damaging (0.601) | 0.137 | no |
| NM_000301.3:c.266G > A | NP_000292.1:p.Arg89Lys | rs143079629 | 142 | 1 | 0 | 0.9 | 0.003 | missense | tolerated (0.179) | benign (0.253) | 0.326 | no | |
| NM_000301.3:c.782G > A | NP_000292.1:p.Arg261His | rs4252187 | 142 | 1 | 0 | 0.9 | 0.003 | missense | deleterious (0.012) | probably damaging (0.990) | 0.398 | no | |
| NM_000301.3:c.1259G > A | NP_000292.1:p.Gly420Asp | rs139071351 | 142 | 1 | 0 | 0.9 | 0.003 | missense, splice region | tolerated (0.872) | benign (0.035) | 0.146 | no | |
| NM_000301.3:c.1414G > A | NP_000292.1:p.Asp472Asn | rs4252125 | 68 | 64 | 11 | 0.4 | 0.3 | missense | tolerated (0.402) | benign (0) | 0.045 | no | |
| NM_000301.3:c.1481C > T | NP_000292.1:p.Ala494Val | rs4252128 | 141 | 2 | 0 | 0.9 | 0.007 | missense | deleterious (0.014) | probably damaging (0.992) | 0.451 | no | |
| NM_000301.3:c.1567C > T | NP_000292.1:p.Arg523Trp | rs4252129 | 141 | 2 | 0 | 0.9 | 0.007 | missense | tolerated (0.176) | benign (0.037) | 0.279 | no | |
| PRSS1 | NM_002769.4:c.162C > G | NP_002760.1:p.Asn54Lys | rs148440491 | 132* | 3* | 0* | 0.9 | 0.01 | missense | deleterious (0.004) | possibly damaging (0.454) | 0.503 | no |
| NM_002769.4:c.592-8C > T | / | rs200381474 | 140* | 1* | 0* | 0.9 | 0.003 | splice region | / | / | / | no | |
| TMPRSS11a | NM_182606.3:c.3G > A | NP_872412.3:p.Met1Ile | rs977728 | 103 | 33 | 7 | 0.06 | 0.16 | start loss | tolerated (0.074) | benign (0) | 0.395 | no |
| NM_182606.3:c.143A > G | NP_872412.3:p.Lys48Arg | rs139010197 | 128 | 15 | 0 | 0.5 | 0.05 | missense | tolerated (0.583) | benign (0.020) | 0.053 | no | |
| NM_182606.3:c.878A > G | NP_872412.3:p.Gln293Arg | rs353163 | 9 | 55 | 79 | 0.8 | 0.26 | missense | tolerated (1) | benign (0) | 0.057 | no | |
| NM_182606.3:c.983G > A | NP_872412.3:p.Arg328Gln | rs150048717 | 139 | 4 | 0 | 0.8 | 0.01 | missense | tolerated (0.221) | possibly damaging (0.944) | 0.188 | no | |
| MBL2 | NM_000242.2:c.154C > T | NP_000233.1:p.Arg52Cys | rs5030737 | 120 | 22 | 1 | 0.9 | 0.08 | missense | deleterious (0) | probably damaging (1) | 0.259 | no |
| NM_000242.2:c.161G > A | NP_000233.1:p.Gly54Asp | rs1800450 | 106 | 34 | 3 | 0.9 | 0.14 | missense | deleterious (0.003) | probably damaging (1) | 0.849 | no | |
| NM_000242.2:c.170G > A | NP_000233.1:p.Gly57Glu | rs1800451 | 139 | 4 | 0 | 0.8 | 0.01 | missense | deleterious (0) | probably damaging (0.994) | 0.745 | no | |
| OAS1 | NM_001032409.1:c.140G > A | NP_001027581.1:p.Arg47Gln | rs751350524 | 142 | 1 | 0 | 0.9 | 0.003 | missense | tolerated (0.521) | benign (0.064) | 0.034 | yes |
| NM_001032409.1:c.295A > G | NP_001027581.1:p.Ile99Val | rs753837415 | 142 | 1 | 0 | 0.9 | 0.003 | missense | deleterious (0.05) | possibly damaging (0.950) | 0.3 | yes | |
| NM_001032409.1:c.389G > A | NP_001027581.1:p.Arg130His | rs1021340095 | 142 | 1 | 0 | 0.9 | 0.003 | missense | deleterious (0.001) | probably damaging (1) | 0.323 | yes | |
| NM_001032409.1:c.484G > A | NP_001027581.1:p.Gly162Ser | rs1131454 | 24 | 70 | 49 | 0.9 | 0.41 | missense | tolerated (0.343) | benign (0) | 0.057 | no | |
Total of 143 subject from Serbian population were genotyped for selected genes by next generation sequencing approach. Genotype is presented as ref./ref. for the homozygous reference allele, ref./alt for heterozygous and alt/alt for homozygous variant allele. For two variants in PRSS1 gene, variant call quality filters did not pass in all samples, therefore genotyping of variant rs148440491 involved 135, while rs200381474 involved 141 subjects (marked with *). PolyPhen-2, SIFT and MutPred2 prediction of coding nonsynonymous variants are presented with scores, with SIFT score ranging from 1 for benign to 0 for deleterious variants, whereas scores for PolyPhen-2 and MutPred2 range from 0 for benign to 1 for damaging variants (using cutoff value 0.5 for MutPred). Where possible, Swiss-Pdb Model was used for in silico prediction, which was indicated in the table for individual variants. SNV ID - single nucleotide variant identifier, HW p - Hardy-Weinberg equilibrium statistics.
3.2. In silico prediction analysis
Next, we performed in silico prediction analysis of the identified variants' effect, using PolyPhen-2, SIFT and MutPred2 algorithms (Table 1). Nine missense variants were predicted to be deleterious/damaging by both PolyPhen-2 and SIFT (p.Gly146Ser in FURIN; p.Arg261His and p.Ala494Val in PLG; p.Asn54Lys in PRSS1; p.Arg52Cys, p.Gly54Asp and p.Gly57Glu in MBL2; p.Ile99Val and p.Arg130His in OAS1), while MutPred2 provided additional indication of pathogenicity for 4 of them (p.Gly146Ser in FURIN, p.Asn54Lys in PRSS1, p.Gly54Asp and p.Gly57Glu in MBL2). Although variant p.Arg47Gln in OAS1 is predicted to be benign/tolerated by PolyPhen-2, SIFT and MutPred2 algorithms, the analysis in Swiss-Pdb Viewer showed that the side chain of Arg47, unlike the glutamine at this position, was predicted to be important for dsRNA binding.
Among those 10 genetic variants predicted to impact the structure and/or function of proteins, 8 were found to be rare (p.Gly146Ser in FURIN; p.Arg261His and p.Ala494Val in PLG; p.Asn54Lys in PRSS1; p.Gly57Glu in MBL2; p.Arg47Gln, p.Ile99Val and p.Arg130His in OAS1). These variants may have predictive value for inter-individual differences in the response to the SARS-CoV-2 infection.
Other 2 variants with potentially damaging effect, p.Arg52Cys and p.Gly54Glu in MBL2 gene, having allelic frequency of 8% and 14%, respectively, are the most promising population-specific markers to be considered for association study in COVID-19 patients in Serbia.
In total, 4 variants in genes encoding proteases (FURIN, PLG and PRSS1) and 6 in genes involved in the innate immunity (MBL2 and OAS1) might be interesting for further studies in relation to the response to SARS-CoV-2 infection.
3.3. Comparative population analysis
Our next goal was to investigate the allele frequency in populations worldwide for the same 22 genetic variants detected in Serbian population. The minor allele frequencies (MAF) were extracted from the 1 kG project involving European populations (Italy, Spain, Finland, Great Britain and USA with European ancestry) as well as Eastern Asians, South Asians, African and Ad Mixed American (Central and South American populations) (Table 2 ). We denoted MAF as the frequency of the minor allele found in European populations. Majority of variants were with low frequency among all populations (MAF ≤ 0.05). From all detected variants, 17 were shared between at least two populations, while 5 were found only in Serbian population.
Table 2.
Minor allele frequencies (MAF) across Serbian and 1kGP populations.
| Genetic variant | SRB (n = 143) | TSI (n = 107) | IBS (n = 107) | FIN (n = 99) | GBR (n = 91) | CEU (n = 99) | EAS (n = 504) | SAS (n = 489) | AFR (n = 661) | AMR (n = 347) | dMAF | Fst |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FURIN p.Thr33Ala | 0.007 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.007 | 0.0034 |
| FURIN p.Gly146Ser | 0.007 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.0014 | 0.007 | 0.0023 |
| PLG p.Glu55Gln | 0.003 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.003 | 0.0002 |
| PLG p.Arg89Lys | 0.003 | 0.005 | 0.005 | 0.00 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 | 0.001 | 0.02 | 0.0076 |
| PLG p.Arg261His | 0.003 | 0.00 | 0.00 | 0.05 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.01 | 0.05 | 0.0021 |
| PLG p.Gly420Asp | 0.003 | 0.00 | 0.009 | 0.005 | 0.00 | 0.00 | 0.00 | 0.00 | 0.0007 | 0.00 | 0.009 | 0.0109 |
| PLG p.Asp472Asn | 0.30 | 0.31 | 0.34 | 0.3 | 0.26 | 0.27 | 0.00 | 0.08 | 0.15 | 0.18 | 0.34 | 0.0635 |
| PLG p.Ala494Val | 0.007 | 0.009 | 0.00 | 0.00 | 0.00 | 0.00 | 0.003 | 0.003 | 0.02 | 0.02 | 0.02 | 0.0021 |
| PLG p.Arg523Trp | 0.007 | 0.005 | 0.005 | 0.00 | 0.016 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.0052 |
| PRSS1 p.Asn54Lys | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.0007 | 0.00 | 0.01 | 0.0058 |
|
PRSS1 c.592-8C > T |
0.003 | 0.004 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.004 | 0.01 | 0.0042 |
| TMPRSS11a p.Met1Ile | 0.16 | 0.19 | 0.25 | 0.23 | 0.21 | 0.21 | 0.18 | 0.23 | 0.11 | 0.35 | 0.24 | 0.0168 |
| TMPRSS11a p.Lys48Arg | 0.05 | 0.02 | 0.02 | 0.01 | 0.04 | 0.02 | 0.0030 | 0.03 | 0.000 | 0.010 | 0.05 | 0.0109 |
| TMPRSS11a p.Gln293Arg | 0.26 | 0.38 | 0.37 | 0.33 | 0.4 | 0.36 | 0.18 | 0.33 | 0.14 | 0.42 | 0.28 | 0.0353 |
| TMPRSS11a p.Arg328Gln | 0.01 | 0.00 | 0.00 | 0.05 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.001 | 0.05 | 0.0084 |
| MBL2 p.Arg52Cys | 0.08 | 0.04 | 0.05 | 0.11 | 0.04 | 0.06 | 0.001 | 0.051 | 0.002 | 0.033 | 0.109 | 0.0199 |
| MBL2 p.Gly54Asp | 0.14 | 0.15 | 0.16 | 0.13 | 0.11 | 0.15 | 0.148 | 0.153 | 0.014 | 0.219 | 0.205 | 0.0174 |
| MBL2 p.Gly57Glu | 0.01 | 0.00 | 0.01 | 0.01 | 0.03 | 0.01 | 0.00 | 0.036 | 0.259 | 0.024 | 0.259 | 0.1421 |
| OAS1 p.Arg47Gln | 0.003 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.003 | 0.0002 |
| OAS1 Ile99Val | 0.003 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.003 | 0.0002 |
| OAS1 Arg130His | 0.003 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.003 | 0.0002 |
| OAS1 p.Gly162Ser | 0.41 | 0.54 | 0.39 | 0.35 | 0.42 | 0.42 | 0.41 | 0.42 | 0.85 | 0.37 | 0.5 | 0.0746 |
MAF has been denoted as the frequency of the minor allele found in the European populations. Delta MAF (dMAF) was calculated by substracting the maximum and the minimum MAF across analyzed population groups. Fst values represent measures of genetic divergence among selected populations at each analyzed locus. Fst value ranges between 0 and 1. The smaller Fst is, allele frequencies among populations are more similar, while large Fst means that allele frequencies are more divergent. Populations: SRB - Serbian, TSI - Tuscany in Italy, IBS - Iberian populations in Spain, FIN - Finnish in Finland, GBR - British in England and Scotland, CEU - Utah residents with Northern and Western European ancestry, EAS - East Asian, SAS - South Asian, AFR - African, AMR - Central and South American populations.
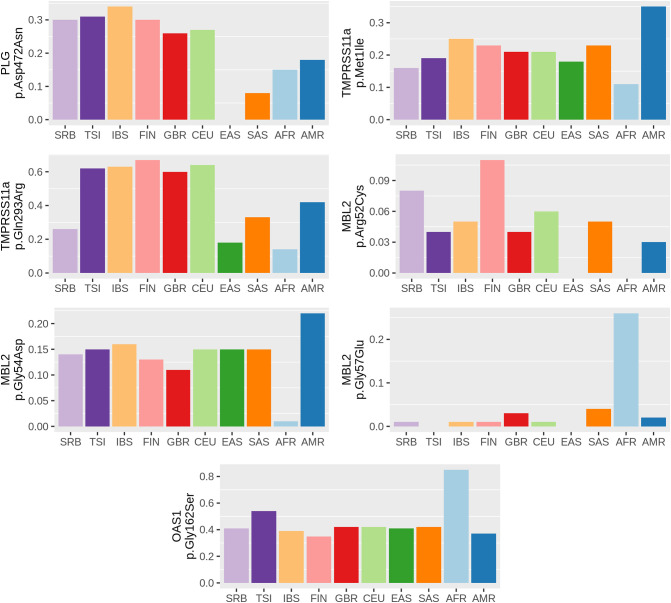
The level of population genetic variability at each selected locus was assessed using two approaches, delta MAF and Fst statistics. Significant correlation between obtained delta MAF and Fst values (r = 0.74, p = 0.000033) was observed. MAF distributions of seven genetic variants (p.Asp472Asn in PLG; p.Met1Ile and p.Gln293Arg in TMPRSS11a; p.Arg52Cys, p.Gly54Asp and p.Gly57Glu in MBL2; p.Gly162Ser in OAS1), that demonstrated delta MAF > 0.1 among analyzed populations, are presented in the Fig. 1 .
Fig. 1.
Distribution of MAF across Serbian and 1kGP populations. Distribution of MAF across Serbian and 1kGP populations for variants that showed highest genetic divergence (delta MAF > 0.1). MAF value (0–1) of each genetic variant is presented on y axis. Populations: SRB - Serbian, TSI - Tuscany in Italy, IBS - Iberian populations in Spain, FIN - Finnish in Finland, GBR - British in England and Scotland, CEU - Utah residents with Northern and Western European ancestry, EAS - East Asian, SAS - South Asian, AFR - African, AMR - Central and South American populations.
Promisingly, three of those variants (p.Arg52Cys, p.Gly54Asp and p.Gly57Glu in MBL2), that showed considerable divergence in frequencies among all analyzed populations, were also predicted to have damaging effect on the protein. Variants p.Gly54Asp and p.Gly57Glu showed extreme MAF values in African compared to other populations (p.Gly54Asp MAF range: 1.4% in African to the 21.9% in Central and South American; p.Gly57Glu MAF range: 0% in Italian and East Asians to the 25.9% in African). MBL2 p.Arg52Cys variant showed variable distribution among analyzed populations, having the highest MAF in Finish (11%), followed by Serbian population (8%), while the lowest MAF was in East Asian and African populations (0%).
Having in mind that variants in proteins involved in the innate immunity could be unfavorable in COVID-19 patients, further studies on variant MBL2 p.Gly54Asp in Central and South American and on variant MBL2 p.Gly57Glu in African populations are indicated.
Furthermore, for these seven most divergent variants, we calculated Fisher exact test statistics of Serbian against 1kGP populations based on their genotypic frequencies (Table 3 ). After Bonferoni correction, significant differences were observed in distribution of PLG p.Asp472Asn and MBL2 p.Arg52Cys compared to Asian populations. In comparison with African populations, all variants, except TMPRSS11a p.Met1Ile, had significantly distinct distributions. Significant differences were observed as well compared to Central and South American populations in distribution of PLG p.Asp472Asn, TMPRSS11a p.Gln293Arg and p.Met1Ile variants.
Table 3.
Comparison of Serbian with 1kGP populations in genotype distribution of selected variants.
| Genetic variant |
p values against 1kGP populations |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| TSI | IBS | FIN | GBR | CEU | EAS | SAS | AFR | AMR | |
| PLG p.Asp472Asn | 0.559 | 0.604 | 0.979 | 0.281 | 0.653 | 2.2*10−6 | 2.2*10−16 | 2.3*10−8 | 1.0*10−4 |
| TMPRSS11a p.Met1Ile | 0.411 | 0.043 | 0.123 | 0.255 | 0.094 | 0.503 | 0.051 | 0.021 | 6.6*10−8 |
| TMPRSS11a p.Gln293Arg | 0.010 | 0.016 | 0.221 | 0.004 | 0.054 | 0.017 | 0.060 | 5.4*10−6 | 9.2*10−6 |
| MBL2 p.Arg52Cys | 0.121 | 0.211 | 0.266 | 0.122 | 0.747 | 3.7*10−15 | 0.070 | 2.6*10−16 | 0.002 |
| MBL2 p.Gly54Asp | 0.726 | 0.645 | 0.914 | 0.461 | 0.957 | 0.956 | 0.903 | 2.2*10−16 | 0.011 |
| MBL2 p.Gly57Glu | 0.137 | 1.000 | 1.000 | 0.315 | 1.000 | 0.002 | 0.175 | 2.2*10−16 | 0.461 |
| OAS1 p.Gly162Ser | 0.010 | 0.534 | 0.352 | 0.528 | 0.937 | 0.793 | 0.860 | 2.2*10−6 | 0.380 |
Fisher exact test was used to measure significant differences in genotypes distributions between Serbian and 1kGP populations. Test was two tailed and p value was considered significant at 0.0007 after Bonferoni correction for multiple testing. Significant differences are indicated in bold. TSI - Tuscany in Italy, IBS - Iberian populations in Spain, FIN - Finnish in Finland, GBR - British in England and Scotland, CEU - Utah residents with Northern and Western European ancestry, EAS - East Asian, SAS - South Asian, AFR - African, AMR - Central and South American populations.
Finally, we analyzed genetic variation related to selected loci between Serbian and 1kGP populations using pairwise Fst calculation (Nei, 1987) (Table 4 ). Fst value ranges between 0 and 1. The smaller Fst indicates similar allele frequencies among populations, and vice versa, large Fst means that allele frequencies are more divergent. Comparing Serbian to other populations, the highest level of genetic differentiation related to selected loci was observed with African (Fst = 0.147), followed by East Asian (Fst = 0.054), Central and South American populations (0.036) and South Asian population (0.026). When compared with European populations, the highest divergence was observed with Italian population (Fst = 0.012).
Table 4.
Pairwise Fst values across Serbian and 1kGP populations.
| SRB | TSI | IBS | CEU | GBR | FIN | EAS | SAS | AFR | AMR | |
|---|---|---|---|---|---|---|---|---|---|---|
| SRB | NA | |||||||||
| TSI | 0.012 | NA | ||||||||
| IBS | 0.007 | 0.007 | NA | |||||||
| CEU | 0.003 | 0.003 | −0.001 | NA | ||||||
| GBR | 0.009 | 0.005 | 0.001 | −0.003 | NA | |||||
| FIN | 0.004 | 0.016 | 0.000 | 0.000 | 0.004 | NA | ||||
| EAS | 0.054 | 0.081 | 0.078 | 0.056 | 0.062 | 0.066 | NA | |||
| SAS | 0.026 | 0.033 | 0.030 | 0.016 | 0.016 | 0.025 | 0.020 | NA | ||
| AFR | 0.147 | 0.135 | 0.179 | 0.159 | 0.160 | 0.185 | 0.184 | 0.157 | NA | |
| AMR | 0.036 | 0.031 | 0.015 | 0.015 | 0.015 | 0.019 | 0.064 | 0.018 | 0.202 | NA |
Fst values matrix showing genetic divergence between each population using genotype data for all analyzed loci (22 selected variants). Fst value ranges between 0 and 1 (negative values are considered as zeros). The smaller Fst is, allele frequencies among populations are more similar, while large Fst means that allele frequencies are more divergent. Populations: SRB - Serbian, TSI - Tuscany in Italy, IBS - Iberian populations in Spain, FIN - Finnish in Finland, GBR - British in England and Scotland, CEU - Utah residents with Northern and Western European ancestry, EAS - East Asian, SAS - South Asian, AFR - African, AMR - Central and South America.
4. Discussion
While mutations in the RNA of betacoronavirus opened the possibility to jump to a new specie, variants in various human genes may contribute to increased susceptibility to a new pathogen, or could have a protective role. One of the most illustrious examples are variants in CD4 receptor which contribute to susceptibility to some strains of HIV infection, while variants in CCR5 gene provide resistance to its carriers (Marmor et al., 2006; Oyugi et al., 2009).
Recent studies addressed rare variants in ACE2 gene which could directly influence the binding to S protein of the SARS-CoV-2 and the population-specific differences of more frequent ACE2 gene variants (Cao et al., 2020; Hussain et al., 2020). However, having in mind complexity of the interaction between the virus and the host, which besides the S protein – ACE2 receptor interaction, includes the role of different host proteases and at least some elements of innate immunity with antiviral activity, in this study we selected additional genes that could be important for the susceptibility or resistance to the SARS-CoV-2 viral infection. We focused on genetic variants in genes FURIN, PLG, PRSS1, TMPRSS11a, that encode proteases, as well as variants in genes MBL2 and OAS1, that encode proteins involved in innate immunity, attempting to find promising candidate alleles that might be included in the future genetic test related to new waves of the SARS-CoV-2 pandemics.
To systematically examine the coding variants in selected genes and differences in the allele frequency between populations, we performed comparative analysis of 22 variants found in FURIN, PLG, PRSS1, TMPRSS11a, MBL2 and OAS1 genes from Serbian population database with different European populations and super-populations extracted from 1kGP database (Genomes Project et al., 2015). Also, we used bioinformatic tools to predict the effect of these genetic variants on the structure and/or function of proteins encoded by FURIN, PLG, PRSS1, TMPRSS11a, MBL2 and OAS1 genes.
It could be possible that the existence of variants in genes encoding proteases provide advantage while variants in genes for proteins involved in the innate immunity add some disadvantage to individuals in combating COVID-19.
4.1. Variants in genes for host proteases
Host proteases, such as furin, plasmin, trypsin-1 and TMPRSS11a, are crucial in the process of cutting the S protein of the SARS-CoV and SARS-CoV-2 envelope at the S1/S2 cleavage site, which is a necessary event needed to release the S1 fragment (N-terminal part) and the S2 fragment (C-terminal part) (Coutard et al., 2020). The S1 fragment recognizes ACE2 receptor at the surface of the human cells, and the S2 fragment is involved in the viral entry into the cells (Wan et al., 2020). Furthermore, the spike protein of the SARS-CoV-2 must be cleaved at both, S1/S2 cleavage site and at the furin-like cleavage site inside the S2 fragment in order to enable viral entry (Coutard et al., 2020). It was previously shown that the cleavage by host proteases plasmin, trypsin-1 and TMPRSS11a at both sites of the S protein is mandatory for the entry of SARS-CoV into human bronchial epithelial cells in vitro (Kam et al., 2009). Interestingly, the furin like S2’ cleavage site is identical between SARS-CoV and SARS-CoV-2 (Coutard et al., 2020) thus implying the capability of furin, plasmin, trypsin-1 and TMPRSS11a to perform activation steps and enable viral entry into the human cells. The efficiency of all these four proteases contribute to regulation of cellular tropism and determination of viral pathogenesis. Thus, genetic variants that would impact the structure and/or function of furin, plasmin, trypsin-1 and TMPRSS11 may lead to inter-individual differences in the response to the SARS-CoV-2 infection.
FURIN encodes a type 1 membrane bound protease, one of the seven basic amino acid-specific members which cleave their substrates at single or paired basic residues. First, furin is autocatalytically processed in the endoplasmatic reticulum and then transported to the trans-Golgi network where a second autocatalytic event takes place and the catalytic activity is acquired. In addition to having affinity for various substrates in human tissues, it is probably one of the proteases responsible for the activation of SARS-CoV-2 envelope S glycoproteins (Coutard et al., 2020). Two rare variants, p.Thr33Ala and p.Gly146Ser, were detected in FURIN. Thr33 residue is located in the pro-domain of a peptidase and therefore cleaved off in the process of activation of the furin enzyme, whereas Gly146 residue is located within the peptidase domain of the enzyme (Dahms et al., 2016). A substitution of Gly146 to Ser is predicted to be probably damaging/deleterious by PolyPhen-2 and SIFT algorithms, while MutPred2 software has several hypotheses, including altered metal binding and altered catalytic site. Also, when we analyzed the effect of this variant upon the structure of furin protein in Swiss-Pdb Viewer, a change to Ser at position 146 is predicted to cause steric clashes with His145, which may lead to an unstable protein. Taken altogether, p.Gly146Ser variant may amend the activity of furin in its proprotein convertase function, and may change its ability to cleave furin-like sites in the S protein of the SARS-CoV-2.
Plasminogen is a protease which belongs to peptidase family S1 and it is encoded by the PLG gene. Proteolysis of plasminogen results in multiple forms of the active plasmin. Plasminogen is present in the human lung tissues, more precisely in the airway, alveolar type I and II epithelial cells as well as in endothelial cells (Ji et al., 2020). Six very rare variants were identified in the PLG gene: p.Glu55Gln, p.Arg89Lys, p.Arg261His, p.Gly420Asp, detected only on one chromosome (MAF 0.003), and p.Ala494Val, p.Arg523Trp on two chromosomes (MAF 0.007). Variants p.Arg261His and p.Ala494Val are predicted to be probably damaging/deleterious, while p.Glu55Gln, p.Arg89Lys, p.Gly420Asp, and p.Arg523Trp are mostly predicted to be benign/tolerated by PolyPhen-2, SIFT and MutPred2 algorithms. Residues Glu55 and Arg89 are located in the Pan-Apple domain, while Arg261, Gly420A, Ala494 and Arg523 reside in different Kringle domains of the proenzyme precursor plasminogen (Law et al., 2012). Given that the Pan-Apple domains mediate protein-protein or protein-carbohydrate interactions, whereas Kringle domains play a role in binding mediators, these variants may be involved in fine tuning of plasmin activity. Having in mind that individuals with elevated plasmin had greater susceptibility to COVID-19 and more severe clinical manifestations (Ji et al., 2020), rare variants in PLG, such as p.Arg261His and p.Ala494Val may be recognized as potential markers of inter-individual differences in susceptibility to coronavirus.
Trypsin-1 is encoded by the PRSS1 gene which is a member of the trypsin-1 family of serine proteases. It is active on peptide linkages involving the carboxyl group of lysine or arginine. This enzyme is secreted by the pancreas and cleaved to its active form in the small intestine. It is also present in airway and alveolar type I and II epithelial cells (Ji et al., 2020). Two rare variants in the PRSS1 gene, c.592-8C > T and p.Asn54Lys, were detected. Variant c.592-8C > T was previously detected in patients with cystic fibrosis presenting with chronic pancreatitis (Sofia et al., 2018). Given that this is a splice region variant, it may affect splicing and therefore the level of trypsin-1 protein. Trypsin-1 variant p.Asn54Lys leads to the substitution of polar asparagine residue to a positively charged lysine. This variant is predicted to be possibly damaging/deleterious by PolyPhen-2 and SIFT algorithms, while MutPred2 software suggested that this change may alter the catalytic site. Given that the catalytic triad of trypsin-1 enzyme is consisted of His57, Asn102 and Ser195 (Polgar, 2005), the proximity of Asn54 to His57 might modify the enzymatic activity. The same codon is affected by an alternative variant, p.Asn54Ser, which was classified pathogenic by UniProt, and associated with chronic pancreatitis (Teich et al., 2005; Richards et al., 2015). Nevertheless, having in mind that the variant p.Asn54Lys was observed in healthy adults, this variant is not regarded as a disease causing in the context of Mendelian diseases. Its mechanism might be through the fine modification of trypsin-1 enzymatic activity, which could contribute to the resistance of some individuals to SARS-CoV-2 infection.
Transmembrane serine protease 11A, encoded by the TMPRSS11a gene is one of the members of type II transmembrane serine proteases and it is present in upper respiratory tract (pharynx and trachea) and digestive tract (Kam et al., 2009). Two rare variants were detected in the TMPRSS11A gene: p.Lys48Arg and p.Arg328Gln. For both variants, PolyPhen-2, SIFT and MutPred2 algorithms predict benign/tolerated effect. Given the scarce data on the protein structure of TMPRSS11A, the effect of this variants is yet to be ascertained.
4.2. Variants in genes for proteins involved in the host innate immunity
Innate immunity is important in defending organism from viral infections. After SARS-CoV outbreak it was found that type I interferons could inhibit the replication of this virus and that these interferons further induce different proteins with antiviral activity (Cinatl et al., 2003). One of such proteins is encoded by the OAS1 gene. OAS1 synthesizes 2′,5′-oligoadenylates, and as a consequence activates latent RNase L which cuts single-stranded RNAs thus leading to the viral RNA degradation and inhibition of viral replication (Rebouillat and Hovanessian, 1999). Three rare variants were detected in the OAS1 gene: p.Arg47Gln, p.Ile99Val and p.Arg130His. Interestingly, human OAS1 protein recognizes two adjacent minor grooves of dsRNA with C-terminal lobe and N-terminal lobe, where the Arg47 residue is one of the residues involved in the recognition (Donovan et al., 2013). When analyzed in Swiss-Pdb Viewer, the side chain of Arg47 residue was predicted to form hydrogen bond with dsRNA, while a change to glutamine at this position was predicted to abolish this hydrogen bond. Furthermore, Gln at this position is predicted to form hydrogen bonds with Cys45 and Phe46, and to cause steric clashes with Glu43 and Arg44. Nevertheless, variant p.Arg47Gln is predicted to be benign/tolerated by PolyPhen-2, SIFT and MutPred2 algorithms, so its effect on the OAS1 protein seems not to affect the structure itself, but it may be apparent upon the binding of the dsRNA. Variant p.Ile99Val is predicted to be possibly damaging/deleterious by prediction algorithms, but the analysis in Swiss-Pdb Viewer revealed that the backbone of the Ile99 residue is predicted to form hydrogen bonds with Arg95, Gly96 and Arg103, so the change to Val at this position did not change the existing bonds, nor did it form novel ones. Variant p.Arg130His is predicted to be probably damaging/deleterious by prediction algorithms, and the analysis in Swiss-Pdb Viewer showed that in case of Arg130, a change to His at this position was predicted to result in the formation of novel hydrogen bonds with Thr188 and also a steric clash with Glu185. These rare variants may have an effect on the OAS1 structure and/or function through the formation of novel hydrogen bonds and steric clashes, leading to a less stable protein. In case of variants located at the protein/RNA interface formed upon the binding, its effects reflect on the formation of (to some extent) weaker bond with the RNA thus lowering its 2′-5′-oligoadenylate synthetase activity, as it was shown that variants at the protein/RNA interface impair OAS1 activity by 60- to 2500-fold (Donovan et al., 2013). Furthermore, having in mind that the active form of human OAS1 is tetrameric, any variant potentially impairing the oligomerization process would also effect on the activity of the enzyme, the synthesis of 2′,5′-oligoadenylates, and consequently the activation of the latent RNase and the degradation of viral RNA of SARS-CoV-2. Knowing that variants in OAS1, such as p.Gly397Arg were previously associated with susceptibility to SARS-CoV infection in some populations (He et al., 2006), the effects of p.Arg47Gln, p.Ile99Val and p.Arg130His might be interesting for further investigation.
4.3. Comparative population analysis
The comparative population analysis pointed to the 7 coding variants in PLG, TMPRSS11a, MBL2 and OAS1 with noticeable divergence in allelic frequencies between analyzed populations.
Three variants were found in genes encoding proteases. Variant p.Asp472Asn in PLG gene showed notable MAF discrepancy between European and Asian populations, being the highest in Spain (0.34) and the lowest in East Asians (0.0). This substitution of neutral to acidic amino acid arises in a loop region that connects two Kringle domains of plasminogen and it was presumed to have a functional effect through altering the alignment of Kringle domains (Zaas et al., 2008). This variant was shown previously to influence susceptibility to invasive aspergillosis (Zaas et al., 2008). Having in mind the MAF difference between Spanish and East Asian populations, it would be interesting to further investigate whether it also reflects the remarkable differences in susceptibility and morbidity to COVID-19 between these populations. Two variants in TMPRSS11A showed interesting data for Serbian population, with both variants showing lower MAF in Serbian population (0.16 for p.Met1Ile and 0.26 for p.Gln293Arg) compared to other European populations - average MAF 0.22 for p.Met1Ile and 0.37 for p.Gln293Arg. As a start loss variant, p.Met1Ile leads to start of the translation from the neighboring Met2 residue. Start loss variants can range from disease causing to benign start codon variants, so with the high frequency of p.Met1Ile in all analyzed populations, its effect is presumed to be benign. While variant p.Gln293Arg was described as a cumulative risk factor for esophageal squamous cell carcinoma (Suo et al., 2019), its effect on the protease activity and COVID-19 susceptibility remains to be elucidated.
Four variants were found in genes involved in the innate immunity. Variant p.Gly162Ser in OAS1 was shown to have highest MAF in African populations - 0.85, more than double the average MAF of European populations (0.42). This variant was previously weakly associated with type I diabetes (Qu et al., 2009) and multiple sclerosis (Fedetz et al., 2006), but a more recent study showed that this variant was found not to interfere with OAS1 enzyme activity (Kjaer et al., 2014).
Another important element of the innate immune system is mannose-binding protein (soluble mannose-binding lectin) which is encoded by the MBL2 gene. The protein recognizes mannose and N-acetylglucosamine expressed on the surface of many microorganisms, and is capable of activating the classical complement pathway. Deficiencies of this gene have been associated with increased susceptibility to SARS-CoV and other autoimmune and infectious diseases (Tu et al., 2015). Interestingly, out of 7 genetic variants that demonstrated delta MAF > 0.1 in our study, only p.Arg52Cys, p.Gly54Asp and p.Gly57Glu in MBL2 were predicted to be probably damaging/deleterious by prediction algorithms. Moreover, these three variants were already extensively studied and functionally characterized - they showed compromised oligomerization and thus the activity of the final protein (Larsen et al., 2004). In addition to promoter variants, these coding variants have been shown to influence the stability and serum concentration of the protein, where, consequently, low levels of MBL2 have been associated with increased susceptibility to infections (Garred et al., 2006). Therefore, the influence of these three variants was studied in various immunological sceneries, from autoimmune to infections, including SARS-CoV (Ip et al., 2005; Tu et al., 2015). Nevertheless, all three variants are found in all analyzed populations, with relatively high MAF values. Interestingly, p.Gly54Asp and p.Gly57Glu showed extreme MAF values in African compared to other populations: p.Gly54Asp with MAF ranging from 0.014 in African to 0.219 in Central and South American and p.Gly57Glu with MAF ranging from 0.0 in Italian and East Asians to 0.259 in African. Variant p.Arg52Cys showed variable distribution among analyzed populations, having the highest MAF in Finish (0.11), followed by Serbian population (0.08), while the lowest MAF was in East Asian and African populations (0.0). Further studies in the populations in which those variants are frequent could contribute to design of prediction model of the SARS-CoV-2 susceptibility in the carriers of the variants.
Taken altogether, comparing Serbian to other populations, it was found that the highest level of genetic differentiation related to selected loci was observed with African, followed by East Asian and South Asian populations. When compared with European populations, the highest divergence was observed with Italian population.
5. Conclusions
In conclusion, results of our analysis showed that variants predicted to have altering effect to the proteins are very rare in each of the selected European populations as well as super-populations. Thus, although it is not likely to perform massive genetic testing aiming to detect these variants in order to predict prognosis to COVID-19, they may provide answers for the inter-individual differences in the clinical course of disease among patients of the same age and the same genetic background which received identical medical treatment.
On the other hand, variants which have divergent allele frequencies between populations were mostly predicted to lack effect on the structure and/or the function of the proteins. However, few variants, such as those in MBL2 gene were predicted to have some functional effect and their contribution to population differences regarding COVID-19 should be further evaluated.
In general, findings of this study may lead us to conclude that 4 coding variants in genes encoding proteases (FURIN, PLG and PRSS1) and 6 in genes involved in the innate immunity (MBL2 and OAS1) could be considered as candidates in forthcoming studies aiming to explain differences in clinical manifestations, recovery rate and mortality rate of COVID-19 which vary between different populations. Further genetic analysis of variants in the non-coding regions (contributing to the fine tuning of the gene expression) of these genes, as well as variants in other genes, are needed to complement our study and give the complete insight about inter-individual and population-specific genetic susceptibility and resistance to the SARS-CoV-2 infection. A global genetic initiative holds promise that the full spectrum of human genetic factors determining susceptibility, severity and outcomes of COVID-19 will be determined in future.
Declarations of Competing Interest
None.
Acknowledgements
This work was supported by Ministry of Education, Science and Technological Development Republic of Serbia, EB: 451-03-68/2020-14/200042.
References
- Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., Wen F., Huang X., Ning G., Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discover. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.Y., Xu M.S., Ching J.C., Chan V.S., Ip Y.C., Yam L., Chu C.M., Lai S.T., So K.M., Wong T.Y., Chung P.H., Tam P., Yip S.P., Sham P., Lin C.L., Leung G.M., Peiris J.S., Khoo U.S. Association of a single nucleotide polymorphism in the CD209 (DC-SIGN) promoter with SARS severity. Hong Kong Med. J. 2010;16:37–42. http://www.ncbi.nlm.nih.gov/pubmed/20864747 [PubMed] [Google Scholar]
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Lee K.H., Steinhauer D.A., Stevens D.J., Skehel J.J., Wiley D.C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- Chong W.P., Ip W.K., Tso G.H., Ng M.W., Wong W.H., Law H.K., Yung R.W., Chow E.Y., Au K.L., Chan E.Y., Lim W., Peiris J.S., Lau Y.L. The interferon gamma gene polymorphism +874 A/T is associated with severe acute respiratory syndrome. BMC Infect. Dis. 2006;6:82. doi: 10.1186/1471-2334-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/s0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahms S.O., Arciniega M., Steinmetzer T., Huber R., Than M.E. Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism. Proc. Nat. Acad. Sci. U S A. 2016;113:11196–11201. doi: 10.1073/pnas.1613630113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J., Dufner M., Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc. Nat. Acad. Sci. U S A. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedetz M., Matesanz F., Caro-Maldonado A., Fernandez O., Tamayo J.A., Guerrero M., Delgado C., Lopez-Guerrero J.A., Alcina A. OAS1 gene haplotype confers susceptibility to multiple sclerosis. Tissue Antigen. 2006;68:446–449. doi: 10.1111/j.1399-0039.2006.00694.x. [DOI] [PubMed] [Google Scholar]
- Garred P., Larsen F., Seyfarth J., Fujita R., Madsen H.O. Mannose-binding lectin and its genetic variants. Gene. Immun. 2006;7:85–94. doi: 10.1038/sj.gene.6364283. [DOI] [PubMed] [Google Scholar]
- Genomes Project, C, Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Feng D., de Vlas S.J., Wang H., Fontanet A., Zhang P., Plancoulaine S., Tang F., Zhan L., Yang H., Wang T., Richardus J.H., Habbema J.D., Cao W. Association of SARS susceptibility with single nucleic acid polymorphisms of OAS1 and MxA genes: a case-control study. BMC Infect. Dis. 2006;6:106. doi: 10.1186/1471-2334-6-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger K.E., Weir B.S. Genetics in geographically structured populations: defining, estimating and interpreting F(ST). Nature reviews. Genetics. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., Aziz B. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. 2020 doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip W.K., Chan K.H., Law H.K., Tso G.H., Kong E.K., Wong W.H., To Y.F., Yung R.W., Chow E.Y., Au K.L., Chan E.Y., Lim W., Jensenius J.C., Turner M.W., Peiris J.S., Lau Y.L. Mannose-binding lectin in severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 2005;191:1697–1704. doi: 10.1086/429631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H.L., Zhao R., Matalon S., Matthay M.A. Elevated plasmin(ogen) as a common risk factor for COVID-19 susceptibility. Physiol. Rev. 2020;100:1065–1075. doi: 10.1152/physrev.00013.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y.W., Okumura Y., Kido H., Ng L.F., Bruzzone R., Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS One. 2009;4:e7870. doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney A.D., Dowdle J.A., Bozzacco L., McMichael T.M., St Gelais C., Panfil A.R., Sun Y., Schlesinger L.S., Anderson M.Z., Green P.L., Lopez C.B., Rosenberg B.R., Wu L., Yount J.S. Human genetic determinants of viral diseases. Ann. Rev. Genet. 2017;51:241–263. doi: 10.1146/annurev-genet-120116-023425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaer K.H., Pahus J., Hansen M.F., Poulsen J.B., Christensen E.I., Justesen J., Martensen P.M. Mitochondrial localization of the OAS1 p46 isoform associated with a common single nucleotide polymorphism. BMC Cell Biol. 2014;15:33. doi: 10.1186/1471-2121-15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen F., Madsen H.O., Sim R.B., Koch C., Garred P. Disease-associated mutations in human mannose-binding lectin compromise oligomerization and activity of the final protein. J. Biol. Chem. 2004;279:21302–21311. doi: 10.1074/jbc.M400520200. [DOI] [PubMed] [Google Scholar]
- Law R.H., Caradoc-Davies T., Cowieson N., Horvath A.J., Quek A.J., Encarnacao J.A., Steer D., Cowan A., Zhang Q., Lu B.G., Pike R.N., Smith A.I., Coughlin P.B., Whisstock J.C. The X-ray crystal structure of full-length human plasminogen. Cell Rep. 2012;1:185–190. doi: 10.1016/j.celrep.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor M., Hertzmark K., Thomas S.M., Halkitis P.N., Vogler M. Resistance to HIV infection. J. Urban health. 2006;83:5–17. doi: 10.1007/s11524-005-9003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Columbia University Press; 1987. Molecular Evolutionary Genetics. [Google Scholar]
- Ng M.W., Zhou G., Chong W.P., Lee L.W., Law H.K., Zhang H., Wong W.H., Fok S.F., Zhai Y., Yung R.W., Chow E.Y., Au K.L., Chan E.Y., Lim W., Peiris J.S., He F., Lau Y.L. The association of RANTES polymorphism with severe acute respiratory syndrome in Hong Kong and Beijing Chinese. BMC Infect. Dis. 2007;7:50. doi: 10.1186/1471-2334-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyugi J.O., Vouriot F.C., Alimonti J., Wayne S., Luo M., Land A.M., Ao Z., Yao X., Sekaly R.P., Elliott L.J., Simonsen J.N., Ball T.B., Jaoko W., Kimani J., Plummer F.A., Fowke K.R. A common CD4 gene variant is associated with an increased risk of HIV-1 infection in Kenyan female commercial sex workers. J. Infect. Dis. 2009;199:1327–1334. doi: 10.1086/597616. [DOI] [PubMed] [Google Scholar]
- Polgar L. The catalytic triad of serine peptidases. Cell. Mol. Life Sci. 2005;62:2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu H.Q., Polychronakos C., Type I.D.G.C. Reassessment of the type I diabetes association of the OAS1 locus. Gene. Immun. 2009;10(Suppl. 1):S69–S73. doi: 10.1038/gene.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebouillat D., Hovanessian A.G. The human 2′,5′-oligoadenylate synthetase family: interferon-induced proteins with unique enzymatic properties. J. Interfer. Cytokine Res. 1999;19:295–308. doi: 10.1089/107999099313992. [DOI] [PubMed] [Google Scholar]
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H.L., Committee A.L.Q.A. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skakic A., Djordjevic M., Sarajlija A., Klaassen K., Tosic N., Kecman B., Ugrin M., Spasovski V., Pavlovic S., Stojiljkovic M. Genetic characterization of GSD I in Serbian population revealed unexpectedly high incidence of GSD Ib and 3 novel SLC37A4 variants. Clin. Genet. 2018;93:350–355. doi: 10.1111/cge.13093. [DOI] [PubMed] [Google Scholar]
- Sofia V.M., Surace C., Terlizzi V., Da Sacco L., Alghisi F., Angiolillo A., Braggion C., Cirilli N., Colombo C., Di Lullo A., Padoan R., Quattrucci S., Raia V., Tuccio G., Zarrilli F., Tomaiuolo A.C., Novelli A., Lucidi V., Lucarelli M., Castaldo G., Angioni A. Trans-heterozygosity for mutations enhances the risk of recurrent/chronic pancreatitis in patients with cystic fibrosis. Mol. Med. 2018;24:38. doi: 10.1186/s10020-018-0041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo C., Qing T., Liu Z., Yang X., Yuan Z., Yang Y.J., Fan M., Zhang T., Lu M., Jin L., Chen X., Ye W. Differential cumulative risk of genetic polymorphisms in familial and nonfamilial esophageal squamous cell carcinoma. Cancer Epidemiol. Biomarkers Prevent. 2019;28:2014–2021. doi: 10.1158/1055-9965.EPI-19-0484. [DOI] [PubMed] [Google Scholar]
- Teich N., Nemoda Z., Kohler H., Heinritz W., Mossner J., Keim V., Sahin-Toth M. Gene conversion between functional trypsinogen genes PRSS1 and PRSS2 associated with chronic pancreatitis in a six-year-old girl. Human Mutat. 2005;25:343–347. doi: 10.1002/humu.20148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X., Chong W.P., Zhai Y., Zhang H., Zhang F., Wang S., Liu W., Wei M., Siu N.H., Yang H., Yang W., Cao W., Lau Y.L., He F., Zhou G. Functional polymorphisms of the CCL2 and MBL genes cumulatively increase susceptibility to severe acute respiratory syndrome coronavirus infection. J. Infect. 2015;71:101–109. doi: 10.1016/j.jinf.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO.
- Williams F.M., Freydin M., Mangino M., Couvreur S., Visconti A., Bowyer R.C., Le Roy C.I., Falchi M., Sudre C., Davies R., Hammond C., Menni C., Steves C., Spector T. Self-reported symptoms of covid-19 including symptoms most predictive of SARS-CoV-2 infection, are heritable. medRxiv. 2020 doi: 10.1101/2020.04.22.20072124. 2020.2004.2022.20072124. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F.F., Boehm I., Chan P.K., Marks K., Tang J.W., Hui D.S., Sung J.J., Dyer W.B., Geczy A.F., Sullivan J.S. High prevalence of the CD14-159CC genotype in patients infected with severe acute respiratory syndrome-associated coronavirus. Clin. Vacc. Immunol. 2007;14:1644–1645. doi: 10.1128/CVI.00100-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaas A.K., Liao G., Chien J.W., Weinberg C., Shore D., Giles S.S., Marr K.A., Usuka J., Burch L.H., Perera L., Perfect J.R., Peltz G., Schwartz D.A. Plasminogen alleles influence susceptibility to invasive aspergillosis. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Ali G., Nie H.G., Chang Y., Bhattarai D., Su X., Zhao X., Matthay M.A., Ji H.L. Plasmin improves oedematous blood-gas barrier by cleaving epithelial sodium channels. Br. J. Pharmacol. 2020 doi: 10.1111/bph.15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang Y., Zhang H., Liu X., Chen T., Yang R., Shi Y., Cao W., Li P., Ma Q., Zhai Y., He F., Zhou G., Cao C. Genetic variation of the human alpha-2-Heremans-Schmid glycoprotein (AHSG) gene associated with the risk of SARS-CoV infection. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023730. [DOI] [PMC free article] [PubMed] [Google Scholar]