Abstract
Brucella ovis is a facultative intracellular bacterium that causes a non-zoonotic ovine brucellosis mainly characterized by male genital lesions and is responsible for important economic losses in sheep farming areas. Studies about the virulence mechanisms of Brucella have been mostly performed with smooth (bearing O-polysaccharide in lipopolysaccharide) zoonotic species, and those performed with B. ovis have revealed similarities but also relevant differences. Except for few strains recently isolated from unconventional hosts, Brucella species are non-motile but contain the genes required to assemble a flagellum, which are organized in three main loci of about 18.5, 6.4, and 7.8 kb. Although these loci contain different pseudogenes depending on the non-motile Brucella species, smooth B. melitensis 16M builds a sheathed flagellum under particular culture conditions and requires flagellar genes for virulence. However, nothing is known in this respect regarding other Brucella strains. In this work, we have constructed a panel of B. ovis PA mutants defective in one, two or the three flagellar loci in order to assess their role in virulence of this rough (lacking O-polysaccharide) Brucella species. No relevant differences in growth, outer membrane-related properties or intracellular behavior in cellular models were observed between flagellar mutants and the parental strain, which is in accordance with previous results with B. melitensis 16M single-gene mutants. However, contrary to these B. melitensis mutants, unable to establish a chronic infection in mice, removal of the three flagellar loci in B. ovis did not affect virulence in the mouse model. These results evidence new relevant differences between B. ovis and B. melitensis, two species highly homologous at the DNA level and that cause ovine brucellosis, but that exhibit differences in the zoonotic potential, pathogenicity and tissue tropism.
Keywords: Brucella ovis, virulence, flagella, deletion mutant, intracellular survival, mouse model
Introduction
The genus Brucella is constituted by six classical species (B. melitensis, B. abortus, B. suis, B. canis, B. ovis, and B. neotomae), that cause brucellosis in terrestrial mammals, and six other species (B. ceti, B. pinnipedialis, B. microti, B. inopinata, B. vulpis, and B. papionis) that have been isolated since the 1990s from other terrestrial mammals or from marine mammals (https://lpsn.dsmz.de/genus/brucella). The Brucella spp. host range has more recently increased to amphibians and fish, with atypical strains isolated from several frog species and a ray (1, 2). Despite the high percentage of DNA-DNA hybridization detected among the classical Brucella species (96 ± 5% when compared to B. melitensis 16M) (3), some differential genetic markers have been found and they differ in several phenotypic characteristics, host preference and pathogenicity. Nevertheless, a common trait is their ability to survive and replicate inside phagocytic cells (4–7). The Brucella species are smooth (S) or rough (R) depending on the presence or absence, respectively, of O-polysaccharide chains in the lipopolysaccharide (LPS). B. ovis and B. canis are the only rough Brucella species but are virulent for their natural hosts (sheep and dogs, respectively), which contrasts with the other Brucella species that are smooth and require S-LPS for full virulence (8–10).
Although studies regarding the virulence of R strains has increased in the last years, most work in this respect has been performed with S Brucella species (mainly with zoonotic B. melitensis, B. abortus, and B. suis). Among the genes involved in the virulence of smooth B. melitensis, flagellar genes are required for the establishment of a chronic infection in mice (11), which constitutes an intriguing trait since B. melitensis is a non-motile species (1). In fact, among the brucellae only B. inopinata and the Brucella atypical strains isolated from frogs and a ray are motile (1, 2, 12–14) and at least frog isolates are able to build a polar flagellum in culture medium (1). Despite the presence of several pseudogenes in the three main flagellar loci (1, 11) and its non-motile character (1), B. melitensis 16M is able to build a sheathed polar flagellum in the early exponential phase of growth (11) and, as mentioned above, flagellar mutants of B. melitensis 16M are attenuated in virulence (11). Although the three flagellar loci are conserved in the genus Brucella, with a different pattern of pseudogenization in most cases, no additional studies have been performed to evaluate the relevance of flagellar genes in the virulence other Brucella species. According to its rough phenotype, its particular outer membrane (OM)-related and virulence characteristics and its shared preference with B. melitensis by the ovine host (15–21), we have selected B. ovis to extend the knowledge about the role of flagellar genes in the virulence of the genus Brucella. With this aim, we have constructed a panel of flagellar mutants in rough virulent B. ovis PA (with one, two or the three flagellar loci deleted) that has been characterized regarding growth characteristics, OM-related properties, intracellular behavior in cellular models of professional and non-professional phagocytes and virulence in the mouse model.
Materials and Methods
Plasmids, Bacterial Strains, and Culture Conditions
Plasmids pGEM-T Easy (Promega, Madison, WI, United States) and pCVDKan-D (18) were used to construct the recombinant plasmids containing the inactivated flagellar loci. They were maintained in Escherichia coli JM109 and CC118 (λpir), respectively. Recombinant E. coli strains were cultured at 37°C in Luria Bertani (LB) medium supplemented with 50 μg/ml ampicillin (pGEM-T Easy derived plasmids) or kanamycin (pCVDKan-D derived plasmids).
Virulent B. ovis PA was used as parental strain to obtain the panel of flagellar mutants and as reference strain for comparisons in the different assays. It was obtained from the bacterial culture collection maintained at the Institut National de la Recherche Agronomique, Nouzilly, France. B. ovis strains were cultured in tryptic soy agar (TSA) or tryptic soy broth (TSB) (Pronadisa-Laboratorios Conda, Torrejón de Ardoz, Spain), supplemented with 0.3% yeast extract (YE) (Pronadisa-Laboratorios Conda, Torrejón de Ardoz, Spain) and 5% horse serum (HS) (Gibco-Life Technologies, Grand Island, NY, United States). When required for the mutagenesis procedure, TSA-YE-HS was supplemented with kanamycin (Kan) at a final concentration of 50 μg/ml or with 5% sucrose (Sigma-Aldrich, St. Louis, MO, United States). B. ovis strains were cultured at 37°C under a 5% CO2 atmosphere.
In silico DNA and Protein Analysis, Primers, and Nucleic Acid Techniques
Genomes of B. melitensis 16M (ATCC 23456) and B. ovis 63/290 (ATCC 25840) were analyzed from GeneBank data (accession numbers AE008917 and AE008918 for B. melitensis 16M chromosome I and II, respectively, and accession numbers NC_009505 and NC_009504 for B. ovis 63/290 chromosomes). Gene data for motile Brucella sp. B13-0095 isolated from a Pac-Man frog were retrieved from the Pathosystems Resource Integration Center (PATRIC; genome ID 1867845.3; https://www.patricbrc.org) (22). Orthologs were analyzed at the Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.kegg.jp) and protein and DNA alignments were performed with LALIGN (https://www.ebi.ac.uk/Tools/psa/lalign/) from the European Bioinformatics Institute (23). PSORTb v3.0.2 (Brinkman Laboratory, Simon Fraser University, British Columbia, Canada; https://www.psort.org/psortb/) was used to predict protein subcellular localization (24). Gene Construction Kit (GCK 4.5; Textco Biosoftware, Raleigh, NC, United States) was used as assistant tool for the analysis of nucleotide sequences and schematic drawing of the flagellar loci.
DNA primers (IDT, Leuven, Belgium) used for gene expression analysis and for the construction and characterization of mutant strains are described in Table 1. PCR amplification was performed with AccuPOL DNA polymerase (VWR, Leuven, Belgium), Red Taq DNA polymerase master mix (VWR, Leuven, Belgium) or ExpandTM Long Template PCR System (Roche, Mannheim, Germany), depending on the experiment. For gene expression studies, RNA was extracted with RNeasy mini kit (Qiagen, Hilden, Germany) from 5 × 109 CFU of B. ovis that had been cultured in liquid medium for 16 or 49 h (t16 or t49; exponential and stationary growth phase, respectively). Residual DNA was removed with RQ1 DNase (Promega, Madison, WI, United States) and cDNA was synthetized with the first strand cDNA synthesis kit for RT-PCR (Roche, Mannheim, Germany) using the random hexamers provided with the kit as primers for reverse transcriptase (RT). Parallel control reactions were performed in the same conditions but omitting RT. Subsequent PCR reactions were performed (using the cDNA as template and a panel of primer pairs targeting genes in the three flagellar loci) either with the ExpandTM Long Template PCR System for end-point RT-PCR or with the KAPA SYBR® Fast Master Mix (Kapa Biosystems, Cape Town, South Africa) for relative quantification by real time RT-PCR (qRT-PCR). Four biological replicates, with three technical replicates each, were used in qRT-PCR assays that were performed in a StepOnePlusTM device (Applied Biosystems, Foster City, CA, United States). Gene expression levels were determined, with the StepOneTM software v2.3, by the 2−ΔΔCt method with the 16S gene as internal reference.
Table 1.
Primers used in this work for the construction and verification of B. ovis PA flagellar mutants.
| Primer name | Nucleotide sequence 5'-3'a | Target locus or geneb |
|---|---|---|
| Construction of B. ovis PA flagellar mutants | ||
| Flg1MUT-F | AAATGCCCGGGATCATGT | Locus I |
| Flg1OVL-R | ATTGGCCTTGTTGTCGGA | Locus I |
| Flg1OVL-F | TCCGACAACAAGGCCAATGCCCGATGATCCGCATTA | Locus I |
| Flg1MUT-R | GATTCTGGCTCTTTGACG | Locus I |
| Flg2MUT-F | GCGGCAAGGCCATTTTCT | Locus II |
| Flg2OVL-R | CCTTGCAGCCAGATCGAA | Locus II |
| Flg2OVL-F | TTCGATCTGGCTGCAAGGGGCTGGAACATTCTGGTT | Locus II |
| Flg2MUT-R | TGCAAGCATGAGCGTCAA | Locus II |
| Flg3MUT-F2 | GCTGCCAATGGCAAGACT | Locus III |
| Flg3OVL-R | CGCATCATCAACACACGG | Locus III |
| Flg3OVL-F | CCGTGTGTTGATGATGCGGACAGACAGGCGCAAAAC | Locus III |
| Flg3MUT-R | GGCGCGAGCTTGTATGTC | Locus III |
| Additional primers for the verification of recombinant plasmids and mutants | ||
| Universal-F | GTTTTCCCAGTCACGAC | pGEM-T Easy |
| Universal-R | CAGGAAACAGCTATGAC | pGEM-T Easy |
| Flg1-F | AATGCTTCGTACTGGTCC | Locus I |
| Flg1-R | TCCCTTGAGCTGTTCGAT | Locus I |
| Flg2-F | TGAAGGGGCTCAATCAGA | Locus II |
| Flg2-R | GATCGCTTTGTTCATGCT | Locus II |
| Flg3-F | CCTATCCTTGGTTTCCGC | Locus III |
| Flg3-R | CGATGCAGGATGCAGTTG | Locus III |
| Primers for RT-PCR or qRT-PCR | ||
| FliC RT-F | CAAACTCGTCGGCTCTGA | fliC (locus I) |
| Flg1OVL-R | ATTGGCCTTGTTGTCGGA | fliC (locus I) |
| FliF RT-F | TTGATGGGTGCGATCCTC | fliF (locus I) |
| FliF RT-R | CCTTGCCGATTGGAACGA | fliF (locus I) |
| FtcR RT-F | AGCCTTCCTGATTGGTGA | ftcR (locus I) |
| FtcR RT-R | ATTTCGCGGACATGAACG | ftcR (locus I) |
| FlgE RT-F | CGGAAACGCAATTCTCCT | flgE (locus I) |
| FlgE RT-R | TTGTCCGGCACGAAAGAA | flgE (locus I) |
| FlbT RT-F | CATCAATGGCGCGGTTCT | flbT (locus I) |
| FlbT RT-R | AACATGCCTTTCAGCATC | flbT (locus I) |
| FlgJ RT-F | AGGGCTGACGCAGGATAA | flgJ (locus I) |
| FlgJ RT-R | AAAGTCGCAGTCGTGTCG | flgJ (locus I) |
| FlgG RT-F | TGACGCTTGACGGCAATC | flgG (locus II) |
| FlgG RT-R | GTTCGAGACCGGCTTCAT | flgG (locus II) |
| FlhB RT-F | ATCGAAACCGGCAATGGC | flhb (locus III) |
| FlhB RT-R | CCGCAAGCGTCATCGTCT | flhb (locus III) |
| FlgF RT-F | GCTGATCAAGACCGACAA | flgF (locus III) |
| FlgF RT-R | GACATCGAGGATCGCATT | flgF (locus III) |
| 16S-RT Fw | TCTCACGACACGAGCTGACG | 16S |
| 16S-RT Rv | CGCAGAACCTTACCAGCCCT | 16S |
Underlined sequences in Flg1OVL-F, Flg2OVL-F, and Flg3OVL-F2 correspond to regions overlapping with Flg1OVL-R, Flg2OVL-R, and Flg3OVL-R, respectively.
Target gene is the B. ovis locus to be deleted or PCR-amplified for the verification of mutant strains or for RT-PCR. Primers Universal-F and Universal-R target pGEM-T Easy and its derived recombinant plasmids at both sides of the cloned insert and were used for sequencing of the DNA insert. The remaining primers target the B. ovis genome and were designed according to the published genome sequence of B. ovis 63/290 (ATCC 25840) (accession numbers NC_009505 and NC_009504 for chromosome I and II, respectively). Primers targeting 16S and fliF were those previously described (18, 25).
Mutagenesis Procedure
Mutant strains for the three main flagellar loci (Table 2) were obtained by in-frame deletion with overlapping PCR as described previously (18). Briefly, for removal of the entire locus I, the 5′end and upstream DNA (about 700 bp) was PCR amplified with primers Flg1MUT-F and Flg1OVL-R and AccuPOL DNA polymerase. Similarly, the 3′ end and downstream DNA was amplified with primers Flg1OVL-F and Flg1MUT-R. Both fragments were fused, through the complementary regions of primers Flg1OVL-F and Flg1OVL-R (Table 1), with an overlapping PCR reaction with primers Flg1MUT-F and Flg1MUT-R and the ExpandTM Long Template PCR System. The resulting DNA fragment was ligated in pGEM-T Easy, verified by DNA sequencing, and then cloned in pCVDKan-D, a plasmid that confers resistance to kanamycin and sensibility to sucrose (18). The recombinant plasmid was introduced in parental B. ovis PA by electroporation. B. ovis PA colonies bearing the plasmid integrated in the chromosome, that consequently contains one copy of the wild type locus and one copy of the modified locus, were detected by plating on TSA-YE-HS plates containing kanamycin. Colonies were verified by PCR with appropriate primers to detect both copies of locus I (intermediate strain). Colonies suffering a second recombination event, leading either to the desired mutant strain or to a strain reverting to the wild type genotype, were detected by plating the intermediate strain on TSA-YE-HS plates containing sucrose. The differentiation between the mutant strain lacking flagellar locus I and the intermediate or wild type strains was performed by a series of PCR reactions with Red Taq DNA polymerase master mix and primers located inside and/or outside the deleted region. Mutants lacking the entire locus II or the entire locus III (Table 2) were obtained similarly with their specific primers (Table 1). The single mutants for each flagellar locus served as parental strains for a second round of mutation leading to the deletion of an additional flagellar locus. The double mutants obtained (Table 2) were subsequently used as parental strains to obtain the panel of triple mutants of B. ovis PA lacking the three main flagellar loci (Table 2).
Table 2.
Most relevant B. ovis PA mutants in flagellar loci obtained in this worka.
| B. ovis strainb | Deleted loci and order of deletion | bp deleted |
|---|---|---|
| B. ovis PA single mutants (one entire locus deleted) | ||
| Locus I completely deleted | 18459 | |
| Locus II completely deleted | 6320 | |
| Locus III completely deleted | 7736 | |
| B. ovis PA double mutants (two entire loci deleted) | ||
| B. ovis Δflg1Δflg2 | Loci I and II completely deleted | 24779 |
| B. ovis Δflg1Δflg3 | Loci I and III completely deleted | 26195 |
| B. ovis Δflg2Δflg1 | Loci II and I completely deleted | 24779 |
| B. ovis Δflg2Δflg3 | Loci II and III completely deleted | 14056 |
| B. ovis Δflg3Δflg1 | Loci III and I completely deleted | 26195 |
| B. ovis Δflg3Δflg2 | Loci III and II completely deleted | 14056 |
| B. ovis PA triple mutants (three entire loci deleted) | ||
| Loci I, II, and III completely deleted | 32515 | |
| B. ovis Δflg1Δflg3Δflg2 | Loci I, III, and II completely deleted | 32515 |
| Loci II, I, and III completely deleted | 32515 | |
| B. ovis Δflg2Δflg3Δflg1 | Loci II, III, and I completely deleted | 32515 |
| B. ovis Δflg3Δflg1Δflg2 | Loci III, I, and II completely deleted | 32515 |
| Loci III, II, and I completely deleted | 32515 |
Intermediate strains obtained during mutagenesis are not cited.
Mutant strains phenotypically characterized in this work are highlighted in blue bold characters. Order of citation of the deleted loci in the strain name corresponds to the order of deletion of each locus (i.e., B. ovis Δflg2Δflg1 was obtained from parental B. ovis PA by deletion of locus II to obtain B. ovis Δflg2 and, then, by deletion of locus I from the Δflg2 single mutant).
Growth, Autoagglutination, and Susceptibility Assays
Growth of mutant strains in solid and liquid medium was analyzed as previously described (26). Briefly, to evaluate growth in solid medium, bacterial suspensions in PBS with values of optical density at 600 nm (OD600) of 0.2 were appropriately diluted and plated on TSA-YE-HS plates to determine the numbers of CFU/ml after 5 days incubation. Growth curves in liquid TSB-YE-HS were also established by measuring the evolution of OD600 scores and log CFU/ml numbers of bacterial suspensions starting at OD600 values of 0.05 and incubated under agitation for 170 h.
To evaluate the autoagglutination ability, bacterial suspensions in TSB-YE-HS of OD600 values of 0.8 (100% OD600) were incubated for 48 h under static conditions to measure the evolution of the OD600 scores (18, 27). Susceptibility to polymyxin B, sodium deoxycholate and H2O2 (all from Sigma-Aldrich) was measured using a disc assay as follows. Bacterial suspensions (100 μl) with OD600 values of 0.2 were plated on TSA-YE-HS. Discs of 0.9-mm diameter (Los Productos de Aldo, Spain) were then placed in the middle of the plate and soaked with 20 μl of polymyxin B (250 000 UI/ml), sodium deoxycholate (10 mg/ml) or 30% H2O2. The diameter of the growth inhibition halo was recorded in quadruplicate for each plate after a 5-day incubation period and the results expressed as mean ± SD of three plates.
Virulence Evaluation in Cellular Models and Mice
Intracellular behavior of mutant strains was studied in J774.A1 macrophages and HeLa cells as previously described (19). Briefly, 2 × 104 J774.A1 macrophages/well or 1.5 × 104 HeLa cells/well were cultured in 96-well plates for 24 h. Bacteria (4 × 106 or 8 × 106 CFU/well for J774.A1 or HeLa cells, respectively) were allowed to internalize for 2 h in the cell lines. Extracellular bacteria were killed with gentamycin and intracellular bacteria were enumerated in three wells per bacterial strain after lysis of the eukaryotic cells and plating on TSA-YE-HS (t0). The remaining wells were incubated in the presence of gentamycin to evaluate intracellular bacterial numbers at 20 and 44 h (t20 and t44) post-infection (p.i.).
Virulence in mice was evaluated in 6-week old female BALB/c mice (Charles River Laboratories, Chatillon-sur-Chalaronne, France) received 1 week before. They were intraperitoneally inoculated with 106 CFU of parental B. ovis PA or the flagellar triple mutants B. ovis Δflg1Δflg2Δflg3, B. ovis Δflg2Δflg1Δflg3 or B. ovis Δflg3Δflg2Δflg1. Bacterial numbers in spleen were determined -as previously described (28)- in 5 mice per group at 3, 7, and 11 weeks p.i. (W3, W7, and W11), which in B. ovis PA corresponds to the peak of infection in the acute phase, to the chronic phase and to the decline phase of infection, respectively (26, 27).
Statistical Analysis
Statistical comparisons were performed with one-way ANOVA and Fisher's Least Significant Differences test on a GraphPad Prims Software (GraphPad Software Inc., San Diego, CA, United States). Statistically significant differences (P < 0.01) were established with a 99% confidence interval.
Results
Genomic Organization and Transcription of the Flagellar Loci in B. ovis
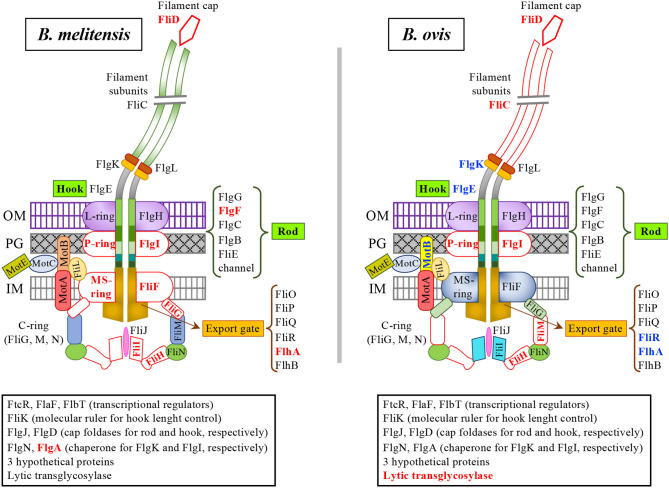
According to the annotated whole genome sequence of the B. ovis reference strain (29), the three flagellar loci of B. ovis (Figure 1A) present a similar organization to that described for B. melitensis 16M (11) and are also located in chromosome II. A search in the PATRIC genome of motile Brucella sp. B13-0095 revealed the presence of additional flagellar genes motA, motB, fliJ, and fliO, that were also detected in chromosome I of B. melitensis 16M and B. ovis 63/290 (Table 3). Hypothetical motA and motB genes were previously identified in locus III and locus I, respectively, of Brucella chromosome II (1, 11, 13) (Table 3) but the four hypothetical flagellar genes detected in chromosome I have not been reported before in studies targeting the Brucella flagellum (1, 11, 13). According to the flagellum structure described for Gram-negative bacteria (30–37), FliO would be part of the export gate (Figure 2) that extends from the membrane-supramembrane (MS ring) of the flagellum to the cytoplasm. FliJ, together with FliI and FliH, would constitute the ATPase complex (Figure 2) of the type III export machinery, although no gene potentially encoding FliH have been detected in the Brucella genomes. Similarly, fliD that encodes the filament cap protein in flagellated bacteria (Figure 2), has not been detected in the genus Brucella.
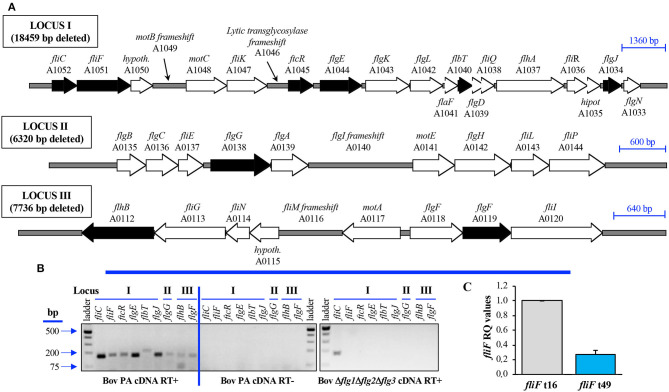
Figure 1.
Genomic organization (A) and transcription (B,C) of the three main flagellar loci detected in the B. ovis genome. Flagellar genes targeted in transcription analysis with B. ovis PA strains (B,C) are represented with a black pattern (A). Mutagenesis procedure deleted 99% of each locus (A) in flagellar mutants listed in Table 2. End-point RT-PCR (B) was performed with cDNA obtained by retrotranscription of RNA extracted at t16 from B. ovis PA or the Δflg1Δflg2Δflg3 triple mutant (RT+ reactions). Reactions of cDNA synthesis lacking RT were used as controls of DNA absence (RT- reactions). Amplification with fliC primers in the Δflg1Δflg2Δflg3 triple mutant (B) was expected, since both primers hybridize adjacent but externally to the deleted fragment (the deletion removes 80% of fliC). qRT-PCR (C) with fliF was performed with RNA extracted at t16 and t49 from B. ovis PA parental strain. Gene expression levels (C) were determined, with the StepOneTM software v2.3, by the 2−ΔΔCt method with the 16S gene as internal reference and t16 results as control condition. Four biological replicates, with three technical replicates each, were analyzed and the results are expressed as means ± SD.
Table 3.
Flagellar genes detected in the genomes of Brucella sp. B13-0095, B. melitensis 16M and B. ovis 63/290a.
| Gene identification in genome of Brucella spp. | Protein name (position in flagellum)b | Predicted subcellular localization (PSORTb)c | ||
|---|---|---|---|---|
| B13-0095 | Bme 16M | Bov 63/290 | ||
| Chromosome II Locus I | ||||
| BA060_07860 | BMEII0150 | FliC (filament) | Extracellular | |
| BA060_07855 | BOV_A1051 | FliF (MS-ring) | Cytoplasmic membrane | |
| BA060_07850 | BMEII0153 | BOV_A1050 | Hypothetical (unknown) | Cytoplasm |
| BA060_07845 | BMEII0154 | MotB (stator) | Periplasm/cytoplasmic memb. | |
| BA060_07840 | BMEII0155 | BOV_A1048 | MotC (stator) | Periplasm |
| BA060_07835 | BMEII0156 | BOV_A1047 | FliK (hook molecular ruler) | Unknown |
| BA060_07830 | BMEII0157 | Lytic transglycosylase (unknown) | Not cytoplasm | |
| BA060_07825 | BMEII0158 | BOV_A1045 | FtcR (regulator) | Cytoplasm |
| BA060_07820 | BMEII0159 | FlgE (hook) | Extracellular | |
| BA060_07815 | BMEII0160 | FlgK (hook-filament junction) | Outer membrane | |
| BA060_07810 | BMEII0161 | BOV_A1042 | FlgL (hook-filament junction) | Unknown |
| BA060_07805 | BMEII0162 | BOV_A1041 | FlaF (regulator) | Unknown |
| BA060_07800 | BMEII0163 | BOV_A1040 | FlbT (regulator) | Cytoplasm |
| BA060_07795 | BMEII0164 | BOV_A1039 | FlgD (cap foldase for hook) | Extracellular |
| BA060_07790 | BMEII0165 | BOV_A1038 | FliQ (export gate) | Cytoplasmic membrane |
| BA060_07785 | FlhA (export gate) | Cytoplasmic membrane | ||
| BA060_07780 | BMEII0168 | FliR (export gate) | Cytoplasmic membrane | |
| BA060_07775 | BMEII0169 | BOV_A1035 | Hypothetical (unknown) | Unknown |
| BA060_07770 | BMEII0170 | BOV_A1034 | FlgJ (cap foldase for rod) | Unknown |
| BA060_07765 | BMEII0171 | BOV_A1033 | FlgN (chaperone for FlgK) | Cytoplasm |
| Chromosome II Locus II | ||||
| BA060_08780 | BMEII1089 | BOV_A0135 | FlgB (rod) | Unknown |
| BA060_08785 | BMEII1088 | BOV_A0136 | FlgC (rod) | Periplasm |
| BA060_08790 | BMEII1087 | BOV_A0137 | FliE (rod) | Unknown |
| BA060_08795 | BMEII1086 | BOV_A0138 | FlgG (rod) | Periplasm |
| BA060_08800 | BOV_A0139 | FlgA (chaperone for FlgI) | Cytoplasmic membrane | |
| BA060_08805 | FlgI (P-ring) | Periplasm | ||
| BA060_08810 | BMEII1083 | BOV_A0141 | MotE (chaperone for stator MotC) | Not cytoplasm |
| BA060_08815 | BMEII1082 | BOV_A0142 | FlgH (L-ring) | Outer memb. |
| BA060_08820 | BMEII1081 | BOV_A0143 | FliL (stator) | Cytoplasmic membrane |
| BA060_08825 | BMEII1080 | BOV_A0144 | FliP (export gate) | Cytoplasmic membrane |
| Chromosome II Locus III | ||||
| BA060_08660 | BMEII1114 | BOV_A0112 | FlhB (export gate) | Cytoplasmic membrane |
| BA060_08665 | BOV_A0113 | FliG (C-ring) | Cytoplasm | |
| BA060_08670 | BMEII1112 | BOV_A0114 | FliN (C-ring) | Cytoplasmic membrane |
| BA060_08675 | BMEII1111 | BOV_A0115 | Hypothetical (unknown) | Not cytoplasm |
| BA060_08680 | BMEII1110 | FliM (C-ring) | Cytoplasm | |
| BA060_08685 | BMEII1109 | BOV_A0117 | MotA (stator) | Cytoplasmic membrane |
| BA060_08690 | BMEII1108 | BOV_A0118 | DUF1217 domain protein (unknown) | Unknown |
| BA060_08695 | BOV_A0119 | FlgF (rod) | Periplasm | |
| BA060_08700 | BOV_A0120 | FliI (ATPase complex) | Cytoplasm | |
| Chromosome I genes | ||||
| BA060_12400 | BMEI0948 | BOV_1003 | FliO (export gate) | Unknown |
| BA060_11245 | BMEI0422 | BOV_1543 | FliJ (ATPase complex) | Cytoplasm |
| BA060_01660 | BMEI0325 | MotA (stator) | Cytoplasmic membrane | |
| BA060_01665 | BMEI0324 | MotB (stator) | Cytoplasmic membrane | |
Brucella sp. B13-0095 is a motile strain isolated from a Pac-Man frog (Ceratophrys ornata) (13), B. melitensis 16M is the type strain of the genus and is able to build a sheathed flagellum in particular culture conditions and B. ovis 63/290 is the B. ovis type strain. Red lettering indicates premature stop codons or frameshifts and blue lettering indicates internal in-frame deletions, when compared to the genes of Brucella sp. B13-0095.
Protein identification according to the annotation in the B. ovis 63/290 or Brucella sp. B13-0095 (for chromosome I genes) genomes. Position in flagellum as shown in Figure 2 according to the general structure described for flagella of Gram-negative bacteria (30–37).
PSORTb v3.0.2 predicts subcellular localization of bacterial proteins (https://www.psort.org/psortb/). The analysis was performed with the proteins of B. ovis 63/290 or Brucella sp. B13-0095 (for B. ovis 63/290 frameshifted proteins).
Figure 2.
Schematic representation of the B. melitensis and B. ovis flagellum. The figure was elaborated according to the classic structure of the flagellum of Gram-negative bacteria (30–37) and the flagellar genes detected in the Brucella genomes (39 genes distributed in the three main flagellar loci of chromosome II and 4 genes found in chromosome I). Additional proteins detected in the flagellar loci intervening in flagellum biosynthesis or with unknown function are framed at the bottom of the figure. Flagellar proteins encoded by genes that, when compared to those of motile Brucella sp. B13-0095, contain frameshifts, premature stop codons or mutations affecting the start codon are written in red bold characters and represented by unfilled forms with red borders. This pattern has also been used for FliD and FliH, which have not been found in the Brucella genomes. B. ovis proteins with internal in-frame deletions in the encoding genes are written in blue bold characters. B. ovis motB in chromosome II contains a frameshift; the figure represents a MotB homolog encoded in chromosome I that is shorter than the B. melitensis ortholog due to an internal in-frame deletion in the encoding gene. C-ring (cytoplasmic ring), MS-ring membrane supramembrane ring, P-ring (peptidoglycan ring), L-ring (lipopolysaccharide ring), IM (inner membrane), PG (peptidoglycan), OM (outer membrane).
When compared to the genome of motile flagellated Brucella sp. B13-0095 isolated from a Pac-Man frog (13), B. melitensis 16M and B. ovis exhibited a different pattern of defective genes (Table 3, Figure 2) with the characteristics listed in Table 4. In the B. melitensis 16M genome, six flagellar genes with premature stop codons or frameshifts have been detected (Tables 3, 4). Additionally, the hypothetical start codon and ribosome binding site of the B. melitensis 16M flgA gene -coding for a putative chaperone for the FlgI P-ring protein (whose gene is frameshifted when compared to Brucella sp. B13-0095 flgI)- are lost due to a deletion of 18 nt. In B. ovis 63/290, five flagellar genes contain internal in-frame deletions shortening the encoded protein (Table 3, blue lettering, and Table 4) and six additional genes (Figure 1A, Table 3, red lettering, and Table 4) contain premature stop codons or frameshifts. However, flagellin fliC gene is not annotated as pseudogene in the whole genome sequence of B. ovis. Since in this work we have used virulent B. ovis PA, the possibility of some differences with B. ovis 63/290 cannot be discarded. However, previous genomic studies with B. ovis PA provided the same results as B. ovis 63/290 (18, 19, 38, 39) and all sequences we have determined in this work for the construction of flagellar mutants (including internal sequences not reported here) were identical to those of B. ovis 63/290.
Table 4.
Defective flagellar genes in B. melitensis 16M and B. ovis 63/290a.
| Gene identification in the genome of Brucella spp. | Protein | Relevant gene defect(s) when compared to motile Brucella sp. B13-0095b | |
|---|---|---|---|
| B. melitensis 16M | |||
| Locus I | BMEII0151-52 | FliF | 1 nt substitution leading to premature stop codon |
| BMEII0166-67 | FlhA | 1 nt substitution leading to premature stop codon | |
| Locus II | BMEII1085 | FlgA | 18 nt deletion involving start codon and probable ribosome binding site |
| BMEII1084 | FlgI | First 6 nt differ affecting start codon 1 nt deletion leading to frameshift | |
| Locus III | BMEII1113 | FliG | Internal 83 nt deletion with frameshift |
| BMEII1107 | FlgF | 1 nt deletion leading to frameshift | |
| BMEII1106-05 | FliI | 1 nt insertion leading to frameshift | |
| B. ovis 63/290 | |||
| Locus I | BOV_A1052 | FliC | Internal 48 nt in-frame deletion C-terminal 203 nt deletion leading to frameshift and affecting the intergenic FliC-FliF region |
| BOV_A1049 | MotB | 35 nt deletion leading to frameshift | |
| BOV_A1046 | Lytic transglyc. | 1 nt deletion leading to premature stop (frameshift) codon 71 nt deletion | |
| BOV_A1044 | FlgE | Internal 57 nt in-frame deletion | |
| BOV_A1043 | FlgK | Internal 42 nt in-frame deletion Internal 18 nt in-frame deletion | |
| BOV_A1037 | FlhA | Internal 36 nt in-frame deletion | |
| BOV_A1036 | FliR | Internal 48 nt in-frame deletion | |
| Locus II | BOV_A0140 | FlgI | First 6 nt differ affecting start codon 1 nt deletion leading to frameshift |
| Locus III | BOV_A0116 | FliM | 1 nt substitution leading to premature stop codon |
| Chrom. I | BOV_1655 | MotA | Internal 31 nt deletion leading to frameshift |
| BOV_1656 | MotB | Internal 87 nt in-frame deletion | |
Compared to the genes of motile Brucella sp. B13-0095 isolated from a Pac-Man frog (Ceratophrys ornata) (13).
Relevant defects included in the table are nucleotide deletions, insertions or substitution leading to in-frame deletions, frameshift or premature stop codons. Nucleotide substituions that do not introduce premature stop codons are not considered as relevant defects.
To evaluate whether flagellar loci are transcribed in B. ovis PA, end-point RT-PCR was performed with RNA extracted at the exponential phase of growth and primers targeting nine genes distributed in the three main loci. Transcription of all evaluated genes was detected in the parental strain B. ovis PA (Figure 1B), while no amplification was observed with cDNA obtained from the Δflg1Δflg2Δflg3 mutant, except for fliC (Figure 1B). This exception was expected because the selected primers for RT-PCR amplify a 164 nt fragment of the 5′-end of fliC that is externally bordering the deleted DNA fragment of locus I (deletion removes 99% of locus I, which includes 80% of fliC). Studies of relative expression in TSB-YE-HS liquid medium of the locus I fliF gene (performed by qRT-PCR using 16S as internal reference gene) showed that fliF is down regulated (with about 3.5-fold reduction) in the stationary growth phase (t49) when compared to the exponential growth phase (t16) (Figure 1C).
Construction, Growth, and OM-Related Properties of B. ovis PA Flagellar Mutants
Three initial B. ovis PA mutants were constructed (B. ovis Δflg1, Δflg2, and Δflg3), each with one of the three main flagellar loci deleted (locus I, II, and III, respectively). Despite the high size of the deleted fragment, mainly in B. ovis Δflg1 (about 18 kb deleted), no difficulties were found to obtain the three mutants. Similarly, double and triple mutants combining the deletion of two or the three flagellar loci, were obtained (32.5 kb deleted in triple mutants). All possible combinations of double and triple mutants were obtained in order to set out a panel of mutants that could be analyzed in case of discovering a differential behavior in one mutant and thus minimize the risk of attributing differences caused by other undesired mutations to the absence of flagellar loci.
No differences in growth in solid medium were observed in single, double or triple mutants that showed similar CFU/ml values for bacterial suspensions of OD600 = 0.2 than the parental strain (data not shown). Equivalent results among strains were also observed in TSB-YE-HS liquid medium with a similar evolution of OD600 values and CFU/ml with time (Supplementary Figure 1). In the autoagglutination assay no differences were found among strains since, as expected for the parental B. ovis PA strain (26), all of them remained in suspension (Supplementary Figure 2). According to these results, only three triple mutants (B. ovis Δflg1Δflg2Δflg3, Δflg2Δflg1Δflg3 and Δflg3Δflg2Δflg1, which have the three flagellar loci deleted in a different order) were initially selected for the remaining studies. The other mutants would only be analyzed if differences were found with the triple mutants.
Properties related to the OM, and that have also been related to survival in the host, were evaluated in the selected triple mutants in comparison with B. ovis PA. Diameters of growth inhibition halos obtained by exposure to H2O2, sodium deoxycholate or polymyxin B did not show relevant differences between the three triple flagellar mutants and the parental strain (Supplementary Figure 3).
Virulence of B. ovis PA Flagellar Mutants
Since B. ovis is an intracellular pathogen (6, 17, 19, 26, 40), the behavior in J774.A1 and HeLa cells of the triple mutants was evaluated in comparison to that of the parental strain. Removal of the three main flagellar loci in B. ovis PA did not affect the internalization of the bacterium or its intracellular evolution (Figures 3A,B). These results were somehow expected since flagellar mutants of B. melitensis 16M did not show an altered intracellular pattern (11), although it must be taken into account that each one of these mutants was only defective in one single gene.
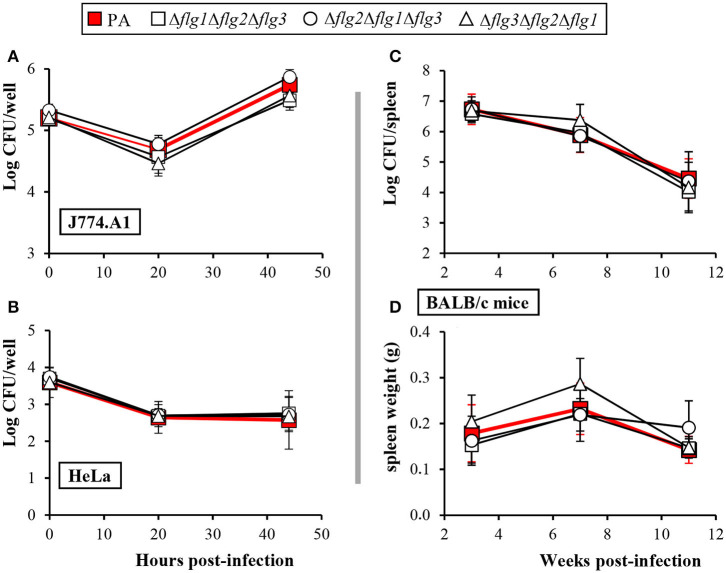
Figure 3.
Virulence of B. ovis PA triple mutants in flagellar loci. Virulence assays were performed in J774.A1 (A) and HeLa (B) cell lines and in BALB/c mice (C,D). Parental B. ovis PA was included in the experiments for comparisons. In cell assays (A,B), the results are expressed as the means ± SD (n = 3) of the log CFU/well at each time point. Virulence in mice is expressed as the means ± SD (n = 5) of the log CFU/spleen at each time point (C). Spleen weight in the same mice is also represented (D).
On the contrary, while single gene mutants of B. melitensis 16M were unable to establish a chronic phase of infection in mice (11), the three triple mutants of B. ovis PA analyzed, lacking the 32.5 kb of the three main flagellar loci, did not show attenuation in BALB/c mice (Figure 3). Thus, both the splenic bacterial counts (Figure 3C) and the spleen weight (Figure 3D) of the flagellar mutants showed the same temporal evolution than those observed with the parental strain.
Discussion
Since the classical Brucella species lack motility (41) and display random patterns of pseudogenization (1), it is tempting to hypothesize that flagellar loci, which are conserved in the genus Brucella, are remnants of an environmental ancestor that no longer required motility after the evolutive adaptation to the animal host and to an intracellular lifestyle. However, the detection of a flagellum in B. melitensis 16M and the involvement of flagellar genes in its virulence in mice (11, 42, 43) and probably in goats (44) raises a new perspective. Moreover, although the relevance for virulence remains unexplored, the recently reported motility of atypical Brucella strains (1, 2, 12–14) and their ability, at least for amphibian isolates, to build a polar flagellum (1) also encourages additional studies to elucidate the function of flagellar genes in the genus Brucella. In this work, we have selected a virulent B. ovis strain to construct and characterize a panel of mutants in flagellar loci with a main focus in virulence.
Although the profile of defective flagellar genes detected in B. ovis (Figure 2, Tables 3, 4) makes the assembly of a flagellum unlikely, B. melitensis 16M also contains defective genes (Figure 2, Tables 3, 4) that would not be compatible with the synthesis of a flagellum. Therefore, it is probable that either the modified proteins are functional or B. melitensis 16M is able to synthetize at least some whole-length molecules by suppression of the stop codons (i.e., fliF and flhA) (11) and compensation of DNA frameshifts (i.e., flgI, fliG, flgF, and fliI) by transcription slippage or ribosomal frameshift (45). Accordingly, the defects observed in some B. ovis flagellar genes do not necessarily imply the impossibility of assembling a complete or partial flagellar structure that could contribute to virulence.
We have detected that B. ovis PA is able to transcribe flagellar genes located in the three main loci and that, as described for B. melitensis 16M (11), the transcription level is higher in the exponential growth phase, at least for fliF (Figures 1B,C). To our knowledge, no other studies have evaluated expression of flagellar genes in B. ovis, either in culture medium or inside phagocytes, and the sole study that analyzed the intracellular transcriptome of B. ovis did not report upregulation or downregulation of flagellar genes (46). However, expression of B. ovis flagellar genes in an intracellular environment, as it has been reported for B. melitensis 16M (11), cannot be discarded. Although this aspect would merit further attention, our results clearly demonstrate that the entire three main flagellar loci of B. ovis PA (accounting for 39 genes) are dispensable for all properties evaluated, including intracellular survival and virulence in the mouse model (Figure 3 and Supplementary Material). Therefore, most likely B. ovis does not build a flagellum or, if it does, it would not be required for the establishment of infection.
Since the mechanism/s responsible for the contribution of flagellum to virulence of B. melitensis 16M has not been elucidated, it is difficult to hypothesize about how the presumed absence of flagellum in B. ovis has influenced the host-pathogen interaction. Flagella may be involved in the four main stages of the infectious process of bacterial pathogens (47): (i) reaching the host or target site (ii) colonization and invasion, (iii) maintenance and replication, and (iv) dispersal to new hosts. The absence of motility (at least in vitro) and of chemotactic systems would exclude the first role in the classical Brucella species. Role in adhesion and invasion of host cells (at least in cell cultures) is also unlikely, since B. melitensis flagellar mutants show no internalization defects in HeLa cells or in bovine peritoneal macrophages (11). The same cellular models also revealed that, even though the fliF promoter is induced intracellularly, the flagellum is not required for intracellular replication of B. melitensis 16M (11). Additionally, attenuation in mice of B. melitensis 16M flagellar mutants was not detected at 1W p.i., but only at later time points (11, 43). However, in vivo mouse imaging technology showed that a luminescent B. melitensis 16M flagellar mutant, lacking four genes coding for rod proteins, had a limited ability to disseminate from the point of intraperitoneal inoculation (48). This impaired dissemination might be related to the impossibility of B. melitensis 16M flagellar mutants to establish a chronic infection in mice (11). But also, if this behavior were reproduced in the natural host, the flagellum could be responsible, at least in part, for the tropism of B. melitensis 16M by the placenta. This statement would be in accordance with the fact that B. melitensis infections frequently induces abortions (21) while B. ovis (that share with B. melitensis the preference for the ovine host) exhibits a marked tropism by the male genital tract and is seldom associated to abortions (20) despite its ability to internalize and replicate in trophoblasts (6). Moreover, a contribution of the flagellum to the zoonotic potential of B. melitensis, which is the highest of the genus, should also be considered. More studies involving flagellar genes in other Brucella species associated with abortions in their preferred hosts or able to infect humans would help to clarify these points.
The exacerbated virulence pattern, accompanied by histological damage in spleen, that was observed in BALB/c mice with a non-polar ΔfliC mutant of B. melitensis 16M constitutes an additional remarkable observation regarding flagellar genes (43). It was proposed that FliC flagellin of B. melitensis 16M triggers the innate immune response and that a tight regulation of flagellar expression in this strain is part of the stealthy strategy that allows to maintain a persistent infection without severely damaging host tissues (43). Some other evidences point to the requirement for a finely tuned regulation of flagellar genes in B. melitensis 16M to establish a persistent infection: (i) the large number of reported direct or indirect regulators of flagellar gene expression: FtcR, FlbT, and FlaF, which are encoded in flagellar locus I (Figure 1A), or VjbR, BlxR, RpoE1, BpdA, and YbeY (48–54), (ii) the results obtained in an in vivo model simulating the onset of B. melitensis 16M infection in cattle (first 4 h) showing that the three main flagellar loci were repressed while transcription of the rpoE1 repressor gene was activated (55), and (iii) the flagellum is sheathed by an extension of the outer membrane ending by a club-like structure that has been suggested to contribute to the assembly of FliC flagellin subunits (42) in the absence of the filament cap protein FliD (Figure 2) that has not been detected in the Brucella genomes; since the Brucella outer membrane is considered as part of its stealthy strategy to establish persistent infections (56), the flagellar sheath could contribute to limit FliC presentation to the immune system.
In the case of B. ovis PA and even if a flagellum is not assembled, flagellin is likely to be synthetized, since this strain is able to transcribe fliC (Figure 1B) and B. melitensis 16M synthesizes FliC even in the absence of deeper flagellar structural proteins such as FliF (basal body protein) or FlgE (hook protein) (53). Therefore, flagellin might be translocated to the cytoplasm of the host cell through the type-IV secretion system (encoded by the virB operon), as it has been suggested for B. melitensis 16M, and induce an innate immune response mediated by cytosolic NCRC4 receptors (43). However, even if B. ovis PA produces flagellin and is able to translocate it into the host cell cytoplasm, it would not be relevant in the induction of a detrimental immune response for the bacterium because flagellar mutants behave as the parental strain (Figure 3). This fact could be related with the differences detected in the C-terminal residues of FliC in B. ovis when compared to the protein of B. melitensis (Tables 3, 4). Both N- and C-terminal domains are involved in the self-polymerization of flagellin subunits (35, 57), but C-end residues have also been proposed as targets for the innate immune response sensed by NLRC4 receptors (43). On the other hand, presentation of assembled flagellin subunits in a surface-exposed flagellar structure could be essential to induce the immune response (and/or to interact with its effectors), and the pattern of defective flagellar genes in B. ovis would not allow this requirement.
Another intriguing observation regarding the B. melitensis 16M flagellum is the contrast between the exacerbated virulence in mice of the ΔfliC mutant (unable to synthetize flagellin and therefore the filament of the flagellum) and the attenuation of the ΔfliF mutant (unable to synthetize the MS-ring proteins) (43). This characteristic suggests that the flagellar export channel of B. melitensis 16M could also be used for the transport of molecules required to maintain a chronic infection in the host. Whether the Type III machinery involved in the specialized export of the flagellar subunits (32, 33) contributes to the export of other molecules in B. melitensis 16M that might participate in virulence has not been elucidated. If this were the case and considering the full virulence of flagellar mutants (Figure 3), this possibility does not seem to occur in B. ovis either by a naturally defective channel or by the absence of the hypothetical virulence determinants.
The demonstration that flagellar genes are dispensable for B. ovis virulence in mice (Figure 3) constitutes a new particular characteristic of this rough species to add to the previously reported differences with other brucellae (16–19). To build a profile of differential characteristics for each Brucella species would contribute to decipher the mechanisms underlaying the differences of pathogenicity and host preference that exist between the classical Brucella species despite their high similarity at the DNA level. However, although the mouse model usually mimics the results obtained in the natural host for attenuated mutants, it has limitations (58, 59), and Brucella mutants exhibiting whole virulence in the mouse model but attenuated in the natural host have been reported (60). Therefore, although unlikely, a role of B. ovis flagellar genes in the natural host cannot be completely discarded. More studies in other Brucella species, including abortifacient and zoonotic Brucella species and the recently isolated motile strains, would help to clarify the relevance of flagellar genes in the genus Brucella.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
Mice experiments were designed according to the Spanish and European legislation for research with animals (RD 53/2013 and directive 86/609/EEC). Microbiological procedures and experimentation with mice were approved by the Biosecurity and Bioethics Committees of the University of Salamanca and certified by the competent authority of Junta de Castilla León, Spain.
Author Contributions
RS-M and NV conceived the study and wrote the manuscript. RS-M, CT, and NV participated in the experimental work, the discussion of the results, and the revision of the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff of the DNA sequencing and animal experimentation facilities of the University of Salamanca for their efficient collaboration.
Footnotes
Funding. This work was financed by Grant SA151G18 that was awarded by Consejería de Educación, Junta de Castilla y León, Spain.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00441/full#supplementary-material
References
- 1.Al Dahouk S, Köhler S, Occhialini A, Jiménez de Bagüés MP, Hammerl JA, Eisenberg T, et al. Brucella spp. of amphibians comprise genomically diverse motile strains competent for replication in macrophages and survival in mammalian hosts. Sci Rep. (2017) 7:44420. 10.1038/srep44420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisenberg T, Risse K, Schauerte N, Geiger C, Blom J, Scholz HC. Isolation of a novel 'atypical' Brucella strain from a bluespotted ribbontail ray (Taeniura lymma). Antonie Van Leeuwenhoek. (2017) 110:221–34. 10.1007/s10482-016-0792-4 [DOI] [PubMed] [Google Scholar]
- 3.Verger JM, Grimont F, Grimont PA, Grayon M. Brucella, a monospecific genus as shown by deoxyribonucleic-acid hybridization. Int J Syst Bacteriol. (1985) 35:292–5. 10.1099/00207713-35-3-292 [DOI] [Google Scholar]
- 4.von Bargen K, Gorvel JP, Salcedo SP. Internal affairs: investigating the Brucella intracellular lifestyle. FEMS Microbiol Rev. (2012) 36:533–62. 10.1111/j.1574-6976.2012.00334.x [DOI] [PubMed] [Google Scholar]
- 5.Chacón-Díaz C, Altamirano-Silva P, González-Espinoza G, Medina MC, Alfaro-Alarcón A, Bouza-Mora L, et al. Brucella canis is an intracellular pathogen that induces a lower proinflammatory response than smooth zoonotic counterparts. Infect Immun. (2015) 83:4861–70. 10.1128/IAI.00995-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidhu-Muñoz RS, Sancho P, Vizcaíno N. Evaluation of human trophoblasts and ovine testis cell lines for the study of the intracellular pathogen Brucella ovis. FEMS Microbiol Lett. (2018) 365:1–9. 10.1093/femsle/fny278 [DOI] [PubMed] [Google Scholar]
- 7.Waldrop SG, Sriranganathan N. Intracellular invasion and survival of Brucella neotomae, another possible zoonotic Brucella species. PLoS ONE. (2019) 14:e0213601. 10.1371/journal.pone.0213601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monreal D, Grilló MJ, González D, Marín CM, De Miguel MJ, López-Goñi I, et al. Characterization of Brucella abortus O-polysaccharide and core lipopolysaccharide mutants and demonstration that a complete core is required for rough vaccines to be efficient against Brucella abortus and Brucella ovis in the mouse model. Infect Immun. (2003) 71:3261–71. 10.1128/IAI.71.6.3261-3271.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González D, Grilló MJ, De Miguel MJ, Ali T, Arce-Gorvel V, Delrue RM, et al. Brucellosis vaccines: assessment of Brucella melitensis lipopolysaccharide rough mutants defective in core and O-polysaccharide synthesis and export. PLoS ONE. (2008) 3:e2760. 10.1371/journal.pone.0002760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouahrani-Bettache S, Jiménez De Bagüés MP, De La Garza J, Freddi L, Bueso JP, Lyonnais S, et al. Lethality of Brucella microti in a murine model of infection depends on the wbkE gene involved in O-polysaccharide synthesis. Virulence. (2019) 10:868–78. 10.1080/21505594.2019.1682762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fretin D, Fauconnier A, Köhler S, Halling S, Léonard S, Nijskens C, et al. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol. (2005) 7:687–98. 10.1111/j.1462-5822.2005.00502.x [DOI] [PubMed] [Google Scholar]
- 12.Eisenberg T, Hamann HP, Kaim U, Schlez K, Seeger H, Schauerte N, et al. Isolation of potentially novel Brucella spp. from frogs. Appl Environ Microbiol. (2012) 78:3753–5. 10.1128/AEM.07509-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soler-Lloréns PF, Quance CR, Lawhon SD, Stuber TP, Edwards JF, Ficht TA, et al. A Brucella spp. isolate from a pac-man frog (Ceratophrys ornata) reveals characteristics departing from classical Brucellae. Front Cell Infect Microbiol. (2016) 6:116. 10.3389/fcimb.2016.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mühldorfer K, Wibbelt G, Szentiks CA, Fischer D, Scholz HC, Zschöck M, et al. The role of ‘atypical' Brucella in amphibians: are we facing novel emerging pathogens? J Appl Microbiol. (2017) 122:40–53. 10.1111/jam.13326 [DOI] [PubMed] [Google Scholar]
- 15.Burgess GW. Ovine contagious epididymitis: a review. Vet Microbiol. (1982) 7:551–75. 10.1016/0378-1135(82)90049-9 [DOI] [PubMed] [Google Scholar]
- 16.Martín-Martín AI, Sancho P, Tejedor C, Fernández-Lago L, Vizcaíno N. Differences in the outer membrane-related properties of the six classical Brucella species. Vet J. (2011) 189:103–5. 10.1016/j.tvjl.2010.05.021 [DOI] [PubMed] [Google Scholar]
- 17.Silva TM, Paixão TA, Costa EA, Xavier MN, Sá JC, Moustacas VS, et al. Putative ATP-binding cassette transporter is essential for Brucella ovis pathogenesis in mice. Infect Immun. (2011) 79:1706–17. 10.1128/IAI.01109-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martín-Martín AI, Sancho P, de Miguel MJ, Fernández-Lago L, Vizcaíno N. Quorum-sensing and BvrR/BvrS regulation, the type IV secretion system, cyclic glucans, and BacA in the virulence of Brucella ovis: similarities to and differences from smooth brucellae. Infect Immun. (2012) 80:1783–93. 10.1128/IAI.06257-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sidhu-Muñoz RS, Sancho P, Vizcaíno N. Brucella ovis PA mutants for outer membrane proteins Omp10, Omp19, SP41, and BepC are not altered in their virulence and outer membrane properties. Vet Microbiol. (2016) 186:59–66. 10.1016/j.vetmic.2016.02.010 [DOI] [PubMed] [Google Scholar]
- 20.OIE Chapter 3.7.7: Ovine epididymitis (Brucella ovis). In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. (2019). Available online at: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.07.07_OVINE_EPID.pdf (accessed July 15, 2020).
- 21.OIE Chapter 3.1.4: Brucellosis (Brucella abortus, B. melitensis, and B. suis) (infection with B. abortus, B. melitensis, and B. suis). In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. (2019). Available online at: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.01.04_BRUCELLOSIS.pdf (accessed July 15, 2020).
- 22.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. (2017) 45:D535–42. 10.1093/nar/gkw1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. (2019) 47:W636–41. 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. (2010) 26:1608–15. 10.1093/bioinformatics/btq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elhassanny AE, Anderson ES, Menscher EA, Roop RM II. The ferrous iron transporter FtrABCD is required for the virulence of Brucella abortus 2308 in mice. Mol Microbiol. (2013) 88:1070–82. 10.1111/mmi.12242 [DOI] [PubMed] [Google Scholar]
- 26.Sidhu-Muñoz RS, Sancho P, Cloeckaert A, Zygmunt MS, de Miguel MJ, Tejedor C, et al. Characterization of cell envelope multiple mutants of Brucella ovis and assessment in mice of their vaccine potential. Front Microbiol. (2018) 9:2230. 10.3389/fmicb.2018.02230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caro-Hernández P, Fernández-Lago L, de Miguel MJ, Martín-Martín AI, Cloeckaert A, Grill, ó MJ, et al. Role of the Omp25/Omp31 family in outer membrane properties and virulence of Brucella ovis. Infect Immun. (2007) 75:4050–61. 10.1128/IAI.00486-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sancho P, Tejedor C, Sidhu-Muñoz RS, Fernández-Lago L, Vizcaíno N. Evaluation in mice of Brucella ovis attenuated mutants for use as live vaccines against B. ovis infection. Vet Res. (2014) 45:61. 10.1186/1297-9716-45-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsolis RM, Seshadri R, Santos RL, Sangari FJ, García Lobo JM, de Jong MF, et al. Genome degradation in Brucella ovis corresponds with narrowing of its host range and tissue tropism. PLoS ONE. (2009) 4:e5519. 10.1371/journal.pone.0005519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eggenhofer E, Haslbeck M, Scharf B. MotE serves as a new chaperone specific for the periplasmic motility protein, MotC, in Sinorhizobium meliloti. Mol Microbiol. (2004) 52:701–12. 10.1111/j.1365-2958.2004.04022.x [DOI] [PubMed] [Google Scholar]
- 31.Evans LD, Hughes C, Fraser GM. Building a flagellum outside the bacterial cell. Trends Microbiol. (2014) 22:566–72. 10.1016/j.tim.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans LD, Hughes C, Fraser GM. Building a flagellum in biological outer space. Microb Cell. (2014) 1:64–6. 10.15698/mic2014.01.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altegoer F, Bange G. Undiscovered regions on the molecular landscape of flagellar assembly. Curr Opin Microbiol. (2015) 28:98–105. 10.1016/j.mib.2015.08.011 [DOI] [PubMed] [Google Scholar]
- 34.Zhu S, Kumar A, Kojima S, Homma M. FliL associates with the stator to support torque generation of the sodium-driven polar flagellar motor of Vibrio. Mol Microbiol. (2015) 98:101–10. 10.1111/mmi.13103 [DOI] [PubMed] [Google Scholar]
- 35.Imada K. Bacterial flagellar axial structure and its construction. Biophys Rev. (2018) 10:559–70. 10.1007/s12551-017-0378-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin TS, Zhu S, Kojima S, Homma M, Lo CJ. FliL association with flagellar stator in the sodium-driven Vibrio motor characterized by the fluorescent microscopy. Sci Rep. (2018) 8:11172. 10.1038/s41598-018-29447-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura S, Minamino T. Flagella-driven motility of bacteria. Biomolecules. (2019) 9:279. 10.3390/biom9070279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vizcaíno N, Kittelberger R, Cloeckaert A, Marín CM, Fernández-Lago L. Minor nucleotide substitutions in the omp31 gene of Brucella ovis result in antigenic differences in the major outer membrane protein that it encodes compared to those of the other Brucella species. Infect Immun. (2001) 69:7020–8. 10.1128/IAI.69.11.7020-7028.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vizcaíno N, Caro-Hernández P, Cloeckaert A, Fernández-Lago L. DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. (2004) 6:821–34. 10.1016/j.micinf.2004.04.009 [DOI] [PubMed] [Google Scholar]
- 40.Macedo AA, Silva AP, Mol JP, Costa LF, Garcia LN, Araújo MS, et al. The abcEDCBA-encoded ABC transporter and the virB operon-encoded type IV secretion system of Brucella ovis are critical for intracellular trafficking and survival in ovine monocyte-derived macrophages. PLoS ONE. (2015) 10:e0138131. 10.1371/journal.pone.0138131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alton GG, Jones LM, Pietz DE. Laboratory Techniques in Brucellosis. World Health Organization; (1975). p. 1–163. [PubMed] [Google Scholar]
- 42.Ferooz J, Letesson JJ. Morphological analysis of the sheathed flagellum of Brucella melitensis. BMC Res Notes. (2010) 3:333. 10.1186/1756-0500-3-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terwagne M, Ferooz J, Rolán HG, Sun YH, Atluri V, Xavier MN, et al. Innate immune recognition of flagellin limits systemic persistence of Brucella. Cell Microbiol. (2013) 15:942–60. 10.1111/cmi.12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zygmunt MS, Hagius SD, Walker JV, Elzer PH. Identification of Brucella melitensis 16M genes required for bacterial survival in the caprine host. Microbes Infect. (2006) 8:2849–54. 10.1016/j.micinf.2006.09.002 [DOI] [PubMed] [Google Scholar]
- 45.Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. Ribosomal frameshifting and transcriptional slippage: from genetic steganography and cryptography to adventitious use. Nucleic Acids Res. (2016) 44:7007–78. 10.1093/nar/gkw530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiao H, Li B, Zheng Z, Zhou Z, Li W, Gu G, et al. Transcriptome landscape of intracellular Brucella ovis surviving in RAW264.7 macrophage immune system. Inflammation. (2020). 10.1007/s10753-020-01239-4. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaban B, Hughes HV, Beeby M. The flagellum in bacterial pathogens: for motility and a whole lot more. Semin Cell Dev Biol. (2015) 46:91–103. 10.1016/j.semcdb.2015.10.032 [DOI] [PubMed] [Google Scholar]
- 48.Petersen E, Chaudhuri P, Gourley C, Harms J, Splitter G. Brucella melitensis cyclic di-GMP phosphodiesterase BpdA controls expression of flagellar genes. J Bacteriol. (2011) 193:5683–91. 10.1128/JB.00428-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delrue RM, Deschamps C, Leonard S, Nijskens C, Danese I, Schaus JM, et al. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell Microbiol. (2005) 7:1151–61. 10.1111/j.1462-5822.2005.00543.x [DOI] [PubMed] [Google Scholar]
- 50.Léonard S, Ferooz J, Haine V, Danese I, Fretin D, Tibor A, et al. FtcR is a new master regulator of the flagellar system of Brucella melitensis. 16M with homologs in Rhizobiaceae. J Bacteriol. (2007) 189:131–41. 10.1128/JB.00712-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rambow-Larsen AA, Rajashekara G, Petersen E, Splitter G. Putative quorum-sensing regulator BlxR of Brucella melitensis regulates virulence factors including the type IV secretion system and flagella. J Bacteriol. (2008) 190:3274–82. 10.1128/JB.01915-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferooz J, Lemaire J, Delory M, De Bolle X, Letesson JJ. RpoE1, an extracytoplasmic function sigma factor, is a repressor of the flagellar system in Brucella melitensis. Microbiology. (2011) 157:1263–8. 10.1099/mic.0.044875-0 [DOI] [PubMed] [Google Scholar]
- 53.Ferooz J, Lemaire J, Letesson JJ. Role of FlbT in flagellin production in Brucella melitensis. Microbiology. (2011) 157:1253–62. 10.1099/mic.0.044867-0 [DOI] [PubMed] [Google Scholar]
- 54.Budnick JA, Sheehan LM, Colquhoun JM, Dunman PM, Walker GC, Roop RM, II, et al. Endoribonuclease YbeY is linked to proper cellular morphology and virulence in Brucella abortus. J Bacteriol. (2018) 200:e00105–18. 10.1128/JB.00105-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rossetti CA, Drake KL, Lawhon SD, Nunes JS, Gull T, Khare S, et al. Systems biology analysis of temporal in vivo Brucella melitensis and bovine transcriptomes predicts host:pathogen protein-protein interactions. Front Microbiol. (2017) 8:1275. 10.3389/fmicb.2017.01275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barquero-Calvo E, Chaves-Olarte E, Weiss DS, Guzmán-Verri C, Chacón-Díaz C, Rucavado A, et al. Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. PLoS ONE. (2007) 2:e631. 10.1371/journal.pone.0000631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. (2003) 424:643–50. 10.1038/nature01830 [DOI] [PubMed] [Google Scholar]
- 58.Kahl-McDonagh MM, Ficht TA. Evaluation of protection afforded by Brucella abortus and Brucella melitensis unmarked deletion mutants exhibiting different rates of clearance in BALB/c mice. Infect Immun. (2006) 74:4048–57. 10.1128/IAI.01787-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grill ó MJ, Blasco JM, Gorvel JP, Moriyón I, Moreno E. What have we learned from brucellosis in the mouse model? Vet Res. (2012) 43:29. 10.1186/1297-9716-43-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bellaire BH, Elzer PH, Hagius S, Walker J, Baldwin CL, Roop RMII. Genetic organization and iron-responsive regulation of the Brucella abortus 2,3-dihydroxybenzoic acid biosynthesis operon, a cluster of genes required for wild-type virulence in pregnant cattle. Infect Immun. (2003) 71:1794–803. 10.1128/IAI.71.4.1794-1803.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.