Abstract
Enzymatic degradation of abundant renewable polysaccharides such as cellulose and starch is a field that has the attention of both the industrial and scientific community. Most of the polysaccharide degrading enzymes are classified into several glycoside hydrolase families. They are often organized in a modular manner which includes a catalytic domain connected to one or more carbohydrate-binding modules. The carbohydrate-binding modules (CBM) have been shown to increase the proximity of the enzyme to its substrate, especially for insoluble substrates. Therefore, these modules are considered to enhance enzymatic hydrolysis. These properties have played an important role in many biotechnological applications with the aim to improve the efficiency of polysaccharide degradation. The domain organization of glycoside hydrolases (GHs) equipped with one or more CBM does vary within organisms. This review comprehensively highlights the presence of CBM as ancillary modules and explores the diversity of GHs carrying one or more of these modules that actively act either on cellulose or starch. Special emphasis is given to the cellulase and amylase distribution within the filamentous microorganisms from the genera of Streptomyces and Aspergillus that are well known to have a great capacity for secreting a wide range of these polysaccharide degrading enzyme. The potential of the CBM and other ancillary domains for the design of improved polysaccharide decomposing enzymes is discussed.
Keywords: carbohydrate-binding module, cellulase, amylase, CAZymes diversity, domain architecture, Aspergillus, Streptomyces
Introduction
Plant biomass contains lignocellulose and starch which represent the most abundant carbohydrate biopolymers in nature. Lignocellulose is a complex polymer mix composed of lignin, an aromatic polymer and two carbohydrate polymers, cellulose and hemicellulose. The main component is cellulose consisting of β-1,4-linked D-glucose. In plant cell walls, cellulose chains interact with each other through hydrogen bonds to form microfibrils (Pérez et al., 2002; Somerville, 2006). Like cellulose, starch is polymeric carbohydrate consisting of solely glucose molecules linked via α-glycosidic bonds, instead of β-glycosidic bonds. Starch is composed of a mixture of two polymers, amylose and amylopectin. Amylose is a linear polysaccharide composed of α-1,4-linked D-glucose. Amylopectin is also a polysaccharide composed of primarily linear α-1,4-linked D-glucose as the backbone with branches that are α-1,6-linked to the backbone (Buleon et al., 1998).
The degradation of the renewable biopolymers cellulose or starch into sugar monomers by industry has a great economic value for its use as a feedstock for the production of various value-added products, such as fuels, chemicals and foods (Zhu et al., 2016). In general, degradation of the polysaccharides into sugar monomers requires the synergistic action of several classes of carbohydrate-active enzymes (CAZymes). Amongst the CAZymes, the glycoside hydrolases (GHs) are the major enzyme family involved in the degradation of polysaccharides such as starch and cellulose (Janeček et al., 2014; Berlemont and Martiny, 2016; Nguyen et al., 2018). However, both cellulose and starch contain a fraction that resists hydrolysis and is indicated as the recalcitrant fraction (Jeoh et al., 2006). Full decomposition of the recalcitrant fraction cannot be obtained by available enzyme cocktails and thus improvement of enzyme performance is necessary (Leggio et al., 2015). One of the options to consider in improving GH characteristics toward the decomposition of the recalcitrant fraction is the engineering of chimeric GH by the addition of one or more carbohydrate-binding modules (CBM).
In the CAZy classification system, the CBMs are a large group of protein domains that are frequently found attached to GH enzymes (Lombard et al., 2014). The CBMs exist as single or multiple (duplicated) domains attached to the C- and/or N-terminus of the catalytic domain. They consist of a relatively small number of amino acids, ranging from approximately 30–200 amino acids. CBMs do not have catalytic activity but function as substrate binding modules (Boraston et al., 2004; Lombard et al., 2014). It has been suggested that the binding characteristic of a CBM improves catalytic function of the CAZymes through targeting the enzyme to the substrate and increasing substrate-enzyme proximity as well as disrupting the crystallinity of the insoluble substrate fraction (Arantes and Saddler, 2010; Reyes-Ortiz et al., 2013; Bernardes et al., 2019; Chalak et al., 2019). As a consequence, the removal of the CBM from the enzyme results in a decreased enzymatic activity (Arai et al., 2003; Cockburn et al., 2017) and a reduced enzyme stability (Bissaro et al., 2017; Courtade et al., 2018; Teo et al., 2019).
Originally, many CBMs were classified as cellulose binding domains (CBDs) because of their affinity for cellulose. The first of these domains was found appended to the cellobiohydrolase CBHI and cellobiohydrolase CBHII in the fungus Trichoderma reesei (Van Tilbeurgh et al., 1986; Tomme et al., 1988). Thereafter, similar CBDs but with different amino acid sequences were also found attached to the cellulases from Cellulomonas fimi (Gilkes et al., 1988). As several modules showed affinity to substrates other than cellulose but still met the criteria as carbohydrate binding domain, the nomenclature CBM was introduced to cover the large variety of carbohydrate binding domains. For example, CBM1 was used for the first discovered fungal CBDs and thereafter also for modules with related amino acid sequences. The CBM2 family contains the CBDs showing high similarity with the bacterial CBDs as found in C. fimi (Gilkes et al., 1988) and other CBM families followed based on their particular ligand specificity, amino acid similarity and structural characteristics (see review Boraston et al., 2004). Like cellulose binding domains, starch binding domains (SBDs) were initially found in fungi as a C-terminal domain of Aspergillus niger glucoamylase (Hayashida et al., 1982; Svensson et al., 1982) and were collected in the CBM20 family (Lombard et al., 2014). As of April 2020, 86 CBM families are listed in the CAZy database (Lombard et al., 2014).
Although it is already very well known that CBM domains are often associated with GHs and are suggested to have a role in enhancing the enzymatic degradation, only limited information is available related to how widely the CBMs are distributed within the GH classes and how these domains are organized in the GHs produced by a given microorganism. Multiple CBMs from different families can also be present in GHs at either the C- or N-terminus. It has been reported that the distribution of the polysaccharide degrading enzymes among the CAZy classes is highly variable within genera, including the GH families (Benoit et al., 2015; Berlemont, 2017). In that regard, mapping the CBM distribution within the GHs may provide novel insight for designing new enzyme architecture with the potential to improve polysaccharide degradation. One of the approaches is to design a chimeric enzyme containing one or more CBMs (Punt et al., 2011; Duan et al., 2017).
This review provides a comprehensive overview on the variation and distribution of CBM domains present in the GH families of cellulases and amylases from filamentous microorganism, particularly Aspergillus and Streptomyces. Filamentous microorganisms, such as Streptomyces (Book et al., 2016; Montella et al., 2017) and Aspergillus (Punt et al., 2002; Yuan et al., 2008; Benoit et al., 2015; Gruben et al., 2017) have a great potential to produce large numbers of CAZymes. Both genera are also well-known for the production of industrially important extracellular enzymes (Fleißner and Dersch, 2010; Sevillano et al., 2016). Many Aspergilli species, mainly the black Aspergillus strains such as Aspergillus niger and Aspergillus oryzae are used for the production of a variety of industrial enzymes such as amylases, pectinases, proteases, β-galactosidase, glucoamylase, lipase, phytase, protease, hemicellulase, and cellulase (Fiedler et al., 2013; Polizeli et al., 2016). Streptomyces strains are also used as production hosts for enzymes. Streptomyces lividans and Streptomyces coelicolor as the examples, have been applied for recombinant extracellular protein production, including hydrolases, proteases/peptidases, chitinases/chitosanases, cellulases/endoglucanases, amylases, and pectate lyases (Anné et al., 2014; Hamed et al., 2018; Ahmed et al., 2020). The striking variety of domain architectures in these organisms calls for a new approach toward enzyme engineering. Examples are discussed where the attention is focused on designing chimeric enzymes with Streptomyces and Aspergillus inspired domain organization for improved cellulose or starch degradation.
Enzyme Families Involved in Starch and Cellulose Degradation
Starch-Degrading Enzymes (α-Amylase, Gluco-Amylase, β-Amylase)
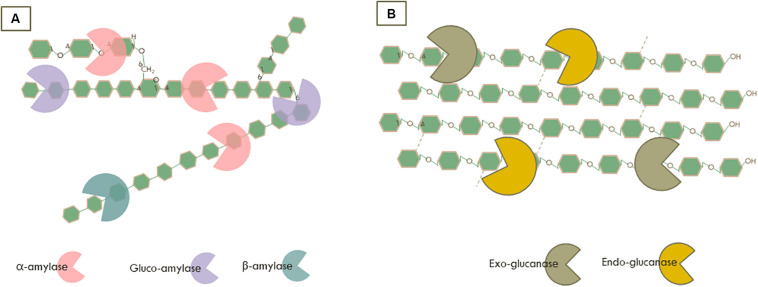
All enzymes capable of degrading starch are collectively indicated as amylolytic enzymes. In this review, we focused on the main endo- and exo-amylolytic enzymes which are involved in saccharification of starch into glucose (Figure 1A). Endo-amylases, mainly α-amylases (1,4-α-D-glucan glucanohydrolase, EC 3.2.1.1) hydrolyse randomly the internal α-1,4-glucan chains to yield small oligosaccharides units (Gupta et al., 2003). Subsequently, gluco-amylases (1,4-α-D-glucan glucohydrolase, EC 3.2.1.3) as the main exo-amylolytic enzymes can hydrolyse linear α-(1,4) as well as the branching sites with an α-(1,6)-linkage at the non-reducing end of starch to release glucose (Sauer et al., 2000). Furthermore, β-amylases are another important class of exo-acting enzymes producing mainly maltose by cleaving starch molecules at the non-reducing end of α-1,4-glucan chains (Ray and Nanda, 1996). Their industrial application is essentially for maltose-rich syrup production (Niu et al., 2018). Overall, amylolytic enzymes have been largely used in a broad range of industrial applications, such as foods, detergents, pharmaceuticals, and the paper as well as textile industries (Ray and Nanda, 1996; de Souza and de Oliveira Magalhães, 2010).
FIGURE 1.
Synergistic action of the enzymatic degradation by (A) Amylases on starch and (B) Cellulases on cellulose.
In general, α-amylases are composed of three domains. Domain A has a TIM-barrel fold containing the active site residues and a chloride ion-binding site. Domain B is a long loop region that contains a calcium-binding site, and domain C is the C-terminal β-sheet domain that shows variability in sequence and length amongst amylases. Several amylases have at least one conserved calcium-binding site, as calcium is essential for enzyme stability, while the chloride binding site present in the active site region is important for the catalytic function. The active center of α-amylases, in general, involves mainly asparagine (Asp) and glutamic acid (Glu) residues (Pujadas and Palau, 2001; Mehta and Satyanarayana, 2016). For glucoamylase, the catalytic domain folds as a twisted (α/α)6-barrel, with the active site in a pocket shape at the N-terminal side of barrels. Two conserved glutamic acid residues are involved in the catalytic mechanism (Sauer et al., 2001; Aleshin et al., 2003). The β-amylase structure exhibits a well conserved (β/α) 8-barrel fold in the core domain with the active site as a deep cleft within the barrel where two glutamic acid (Glu) residues are involved in the hydrolysis (Mikami et al., 1994; Oyama et al., 2003).
In the sequence-based classification of GHs in the CAZy database, α-amylases are represented by four different families, GH13, GH57, GH119, GH126. The main α-amylase family, GH13, is found in a broad range of organisms, including bacteria, archaea, eukaryota, and even viruses. GH57 is found in bacteria, archaea, and very limited in eukaryotes. The other two families, GH119 and GH126 have been found in bacteria only (see review Janeček et al., 2014; Lombard et al., 2014). The glucoamylase family, GH15, does widely occur in prokaryotic and eukaryotic microorganisms including fungi (Aleshin et al., 2003), while the GH97 glucoamylases are predominantly found in bacteria (Lombard et al., 2014). Furthermore, the β-amylase family GH14 is in majority present in plants and bacteria (Lombard et al., 2014).
Cellulose Degrading Enzymes (Exo-Glucanase, Endo-Glucanase)
Almost 50% of lignocellulose is composed of cellulose (Somerville, 2006; Isikgor and Becer, 2015). The degradation of cellulose is accomplished by multiple GHs which are typically acting together as a cocktail with complementary and synergistic modes of action (Payne et al., 2015). For example, endoglucanase (EC 3.2.1.4) cleaves randomly the internal β-glycosidic bonds of cellulose, while exo-glucanases/cellobiohydrolases cleave at both the non-reducing (EC 3.2.1.91) and reducing (EC 3.2.1.176) end of the cellulose chain to release soluble cellobiose (Figure 1B). These enzymes are used on a large scale for different industrial applications and are mainly produced by microorganisms, especially from the fungal and bacterial kingdoms.
Based on the active site topology, the catalytic machinery of cellobiohydrolases is set up in a tunnel-like conformation, enabling the single cellulose chain to be cleaved at the reducing or non-reducing terminus (Parkkinen et al., 2008). In contrast, the endoglucanase catalytic topology is shaped as an open cleft, allowing a linear amorphous cellulose chain to be degraded randomly at any part of the chain (Davies and Henrissat, 1995). Despite the fact that some enzymes share structural similarity, they may exhibit variations in the active site topology as well as the surface loops resulting in different substrate specificities. Based on structural modeling and enzymatic assays of GH6s from Podospora anserina, the exoglucanase PaCel6A has an active-site tunnel topology with the loop corresponding to amino acids 415–429 leading to the formation of the tunnel shape. In contrast the endoglucanase PaCel6B lacks the short 15 amino acid loop at the C-terminus and is predicted to contain a binding cleft rather than a tunnel structure. Those differences in the surface loop influence the mechanism of action toward substrates. Therefore, GH6 PaCel6B showed higher activity on carboxymethylcellulose (CMC) which is a highly specific substrate for endo-acting cellulase. CMC is decrystallized cellulose and thus contains more amorphous sites that are ideal for access to the cellulose chain by endoglucanases that cleave internally. Whereas, the exoglucanase GH6 PaCel6A showed higher activity on the insoluble microcrystalline cellulose Avicel than on CMC, as measured by the liberation of cellobiose from the non-reducing end of the cellulose chain (Poidevin et al., 2013). In addition, deletion of an exo loop in C. fimi cellobiohydrolase allows this enzyme to hydrolyze internal β-1,4-glucosidic bonds, altering its exolytic activity into endolytic activity (Meinke et al., 1995). Moreover, variations in the surface loop conformation were shown to determine the substrate specificity in GH5, such as exo- and endo-mannanase activity (Dias et al., 2004; Kumagai et al., 2015) as well as endoglucanase activity (Tseng et al., 2011). Therefore, in the CAZy classification enzymes could be clustered together as one GH family, although they are different in substrate specificity For example in members of the GH5 family a wide range of cellulase activities have been demonstrated: endo-β-1,4-glucanase, exo-β-1,4-glucanase, as well as non-cellulase activities: endo-β-1,4-xylanase, β-mannosidase, β-glucosylceramidase, glucan β-1,3-glucosidase, glucan endo-1,6-β-glucosidase, mannan endo-β-1,4-mannosidase, cellulose β-1,4-cellobiosidase, chitosanase, xyloglucan-specific endo-β-1,4-glucanase, endo-β-1,6-galactanase, β-1,3-mannanase, endo-β-1,3-glucanase/laminarinase, chitosanase, α-L-arabinofuranosidase (Lombard et al., 2014). In the CAZy database the endoglucanases are classified in 15 different families, GH5, GH6, GH7, GH8, GH9, GH10, GH12, GH26, GH44, GH45, GH48, GH51, GH74, GH124, and GH148. The exoglucanases are divided over 5 GH families, GH5, GH6, GH7, GH9, and GH48. Fungal cellulose degrading enzymes in these families commonly consist of a simple domain arrangement of only a catalytic domain or a catalytic domain with one accessory domain (Berlemont, 2017). Unlike the fungal cellulases, the bacterial cellulases often have a more complex domain structure which can also be involved in assembling multi-enzyme complexes, called cellulosomes (Bayer et al., 2004).
Multi Domain Architecture and Distribution of CBMS in Starch and Cellulose Specific Glycoside Hydrolases From the Genus Streptomyces and Aspergillus
Many proteins within the various GH families show a distinct variation in the domain architecture. As discussed above, in particular the CBM domains are frequently associated with the GH catalytic domain. The abundance of GHs for cellulose degradation varies across bacterial phyla, of which the actinobacteria showed the most extensive variation in GH-cellulase content and are representatives of potential cellulose degraders (Berlemont and Martiny, 2015). For mapping the CBM and the GH diversity, datasets of cellulases and amylases carrying CBMs from Aspergillus and Streptomyces were retrieved from the UniProtKB and the CAZy databases. Subsequently, these datasets were grouped based on the type of CBMs and curated to avoid the inclusion of duplicated sequences by running protein alignment for each GH family in the same species by the Clustal Omega alignment program. Identical sequences from the same species were removed. Eventually, the final datasets were grouped based on the type of CBM associated with selected GHs. In addition, for constructing the graphical domain representation as presented in Figures 2, 3, signal peptides and transmembrane segments were predicted using Phobius (Käll et al., 2004). Subsequently, Pfam domains were determined using the HMMER program (version 3.2.1) against the Pfam database version 31.0 (Eddy, 2011; El-Gebali et al., 2019). Finally, the predictions were programmatically extracted and converted into detailed graphical representations of the various domain organizations, as was previously done for the Auxiliary Activities CAZymes, the Lytic Polysaccharide Mono Oxygenases (LPMO) (Voshol et al., 2017, 2019).
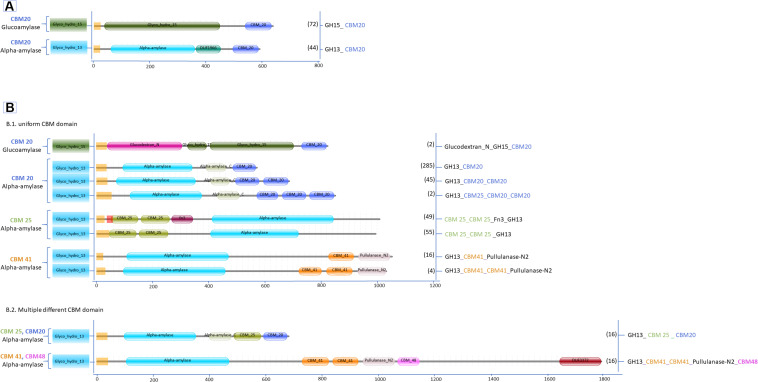
FIGURE 2.
Domain organization of amylases carrying CBM (A) Aspergillus; (B.1) Streptomyces single CBM and (B.2) Streptomyces multiple CBM domains. Between brackets is the number of protein sequences in the various Aspergillus and Streptomyces species retrieved from the UniProtKB and the CAZy databases.
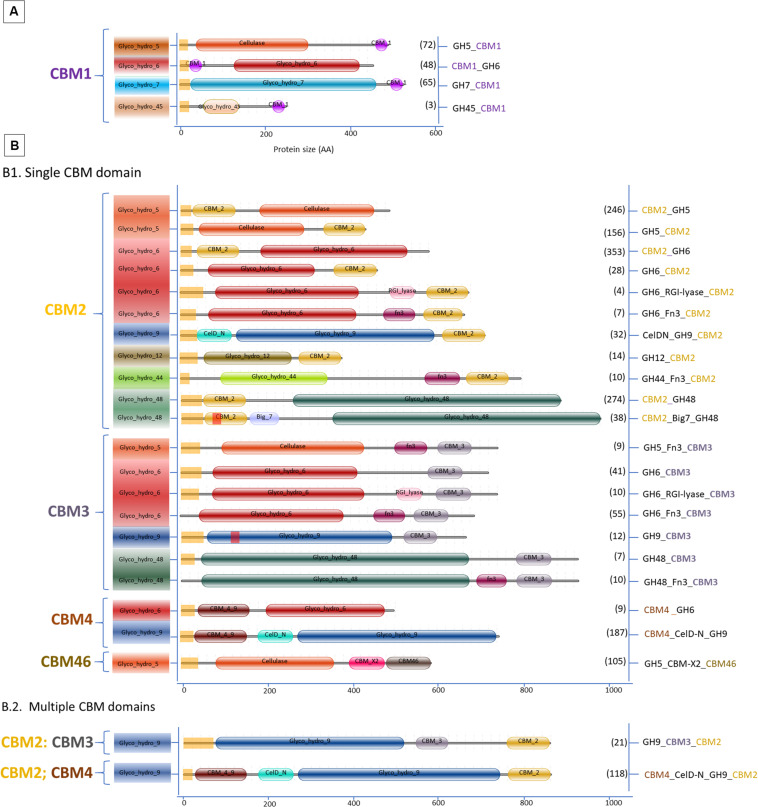
FIGURE 3.
Domain organization of cellulases carrying CBM. (A) Aspergillus; (B.1) Streptomyces single CBM and (B.2) Streptomyces multiple CBM domains. Between brackets is the number of protein sequences in the various Aspergillus and Streptomyces species retrieved from the UniProtKB and the CAZy databases.
In general, filamentous fungi represent a richer reservoir of CAZymes compared to bacteria (Berlemont, 2017; Voshol et al., 2019). However, the variability of enzymes associated with CBMs or other accessory domains is much less in fungi than in bacteria (Talamantes et al., 2016; Berlemont, 2017), as exemplified here by the comparison of the genera Aspergillus and Streptomyces. In total 2540 different amylase and cellulase amino acid sequences with CBMs from Aspergillus and Streptomyces were retrieved from the databases, representing 385 different Streptomyces species and 60 different Aspergillus species. Amongst the sequences, approximately 90% of the GHs cellulase and amylase with CBM were identified from Streptomyces. In contrast, GH cellulases without CBM are more widespread in fungi representing about 80% of these cases. Thus, as a first difference between these two genera, many cellulases and amylases from Aspergillus lack any CBM, while Streptomyces enzymes contain many CBM types as well as a very diverse enzyme architecture associated with different CBMs (Figures 2, 3).
Amylases With CBM From Aspergillus and Streptomyces
Among the starch degrading enzymes only two GH families are associated with CBMs in these two genera, the GH13 α-amylases and the GH15 glucoamylases. Neither Aspergillus nor Streptomyces contain genes encoding GH14 β-amylases with a CBM (Figure 2). As reported by the CAZy database and several studies, β-amylases are secreted mainly by plants, followed by several bacteria, and by a very limited number in fungi (Ray, 2004; Derde et al., 2012; Thalmann et al., 2019). Two CBM families were identified in bacterial β-amylases, i.e., CBM20 from species of Bacillus and Clostridium as well as CBM25 from Paenibacillus, and no CBMs were found in the fungal GH14 β-amylases (see review Janeček et al., 2019).
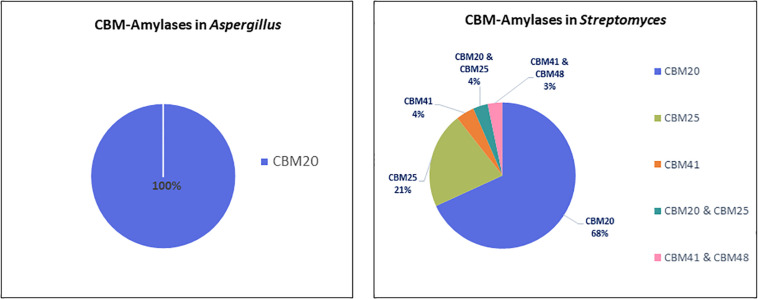
In total, 606 protein sequences with starch binding CBMs were identified in Aspergillus and Streptomyces amylases. Among these sequences, more than half (466 sequences) are associated with CBM20. In Aspergillus amylases, CBM20 was found as the only starch binding domain associated with GH13 α-amylases and GH15 glucoamylases (Figure 2). More CBM families were found in conjunction with amylases from Streptomyces. CBM20 was found as the most frequently domain (68%), followed by CBM25 (21%), leaving the rest (around 4% each) to CBM41 and amylases with a combination of CBMs: CBM20&CBM25 and CBM41&CBM48. Thus, in amylases from these genera, CBM20 is the most dominant CBM (Figure 4).
FIGURE 4.
CBMs associated with amylases from Aspergillus (left) and Streptomyces (right).
In term of diversity of domain architecture, Aspergillus species tend to have a simple domain architecture consisting of only a single CBM20 module located at the C-terminus (Figure 2A). On the other hand, not only higher in the variability of CBM families, but also a more divers domain architecture of amylases with CBMs were identified in Streptomyces. For example, CBM20 and CBM41 are present as a single module, or multiple repeated domains from the same family. Moreover, some of the Streptomyces amylases have combinations of different CBMs attached: CBM20&CBM25 as well as CBM41&CBM48 (Figures 2B1,B2). In general, the binding modules of the CBM20 family are found in a single copy, while only in a few special cases a CBM20 is present together with other CBMs, such as CBM25, CBM34, and CBM48 (Janeček et al., 2019).
Based on the domain position, the CBM20s in α-amylases from Aspergillus and Streptomyces are always located at the C-terminal end (Figure 2). This is also true for other starch active enzymes, such as in GH14 (Oyama et al., 1999), GH15 (Bott et al., 2008; Roth et al., 2018), LPMO AA13 (Vu et al., 2014), and in most of the GH13 enzymes (see review Janeček et al., 2019). Based on the CAZy database, only in a few GH13s, the CBM20 is located at the N-terminal end of the catalytic domain, especially the GH13s from green algae (Lombard et al., 2014). Moreover, in the Streptomycesα-amylases, the CBM25 can be present either N- or C-terminally. Two copies of a CBM25 domain can be present at the N-termini, while co-occurrence of CBM25 with CBM20 at the C-terminus occurs. The other starch binding domains, CBM41 and the combination of CBM41 & CBM48, were found exclusively C-terminally.
Single or two copies of CBM41 can be found in several bifunctional enzymes, such as amylopullulanases (bifunctional with one catalytic center) and bifunctional α-amylase-pullulanases with two catalytic domains, one from α -amylase and one from pullulanase (Figure 2B). According to substrate binding studies, the CBM41 was demonstrated to be able to bind substrate with both 1,4- and 1,6 α-glucosidic linkages (Mikami et al., 2006; Van Bueren and Boraston, 2007). The CBM41s were largely identified in bacterial amylases and in only five members of eukaryotes, including four types of algae and a single species of fungi called Ostreococcus tauri (Lombard et al., 2014). CBM41s are often located at the N-terminus of the protein and present in two copies, while in a few cases, CBM41 is positioned C-terminally (see review Janeček et al., 2019). Interestingly, in bifunctional amylases from Streptomyces, CBM41 is present either as a single module or two repeated modules in between the amylase catalytic domain and the pullulanase domain (Figure 2B1), and even in combination with aCBM48 located C-terminal of the pullanase domain (Figure 2B2).
Other accessory domains present in the amylase architecture were annotated as Fn3, Glucodextran-N, DUF1966 and DUF 3372 (Figures 2A,B). In Streptomyces, the Fn3 domain is found in combination with CBM25. The CBM25-Fn3 domain combination is also present in Microbacterium aurum α-amylase and suggested to improve binding on starch indicated by the alteration of starch granule morphology (Huang et al., 2017). Furthermore, gradual domain truncations were made in the amylopullulanase from Thermoanaerobacterium saccharolyticum NTOU which consists of two Fn3 modules in between the N-terminal catalytic domain and C-terminal CBM20. The amylopullulanase remains active after removal of theCBM20 and the 2nd Fn3 module but further deletion of 1st Fn3 module that is connected with the catalytic module resulted in completely loss of activity (Lin et al., 2011). Generally, very little information is available on the function of the Fn3 modules in amylases. However, there is an indication that Fn3 is involved in binding between the enzyme and the polysaccharide substrates since its removal results in decreased or even loss of enzymatic activity. Another accessory domain, annotated as glucodextran_N in Pfam (El-Gebali et al., 2019) is especially found in bacterial and archaeal glucoamylases and glucodextranases (Mizuno et al., 2004). The domain consists of 17 antiparallel β-strands (Mizuno et al., 2004) which appear to stabilize its structure (Aleshin et al., 2003). This domain is absent in fungal glucoamylases. It was suggested that the eukaryotic glucoamylases may have evolved from prokaryotic glucoamylases together with substitution of the N-terminal glucodextran_N domain with the so-called peripheral subdomain and by the addition of a C-terminal starch-binding domain (Aleshin et al., 2003). The peripheral subdomain is an extra α-helix that is smaller than the other 12 α-helices in the catalytic domain structure and is located between α-helices αH10 and αH11 in the catalytic domain of glucoamylases A. niger (Aleshin et al., 1992, 2003; Lee and Paetzel, 2011). Lastly, several domains of unknown function, so-called DUFs, are present in the GH13 α-amylases of Aspergillus and the bifunctional α-amylase-pullulanase in Streptomyces, as DUF1966 and DUF3372, respectively. This domain folds in a β-barrel structure, however, the exact function has not been determined yet. Moreover, the truncation of this domain in the amylase from Sclerotinia sclerotiorum, ScAmy43, had no effect on the biochemical properties of the enzyme (Abdelmalek et al., 2009).
Cellulases With CBM in Aspergillus and Streptomyces
Based on the datasets retrieved, various cellulase (GH) families do contain a CBM domain in Aspergillus and Streptomyces (Figure 3).
Interestingly, members of two GH families (GH7 and GH45) containing a CBM in Aspergillus are absent in Streptomyces. According to Segato et al. (2012), the genome of almost every Aspergillus encodes two GH7-cellobiohydrolases which can be present with or without a CBM. Meanwhile, three families, GH9, GH44, and GH48, containing a CBM present in Streptomyces are completely lacking in Aspergillus. The GH12 family proteins are present in both genera, but only in Streptomyces the GH12 have a CBM. The GH5 and GH6 cellulases with a CBM are found in both genera.
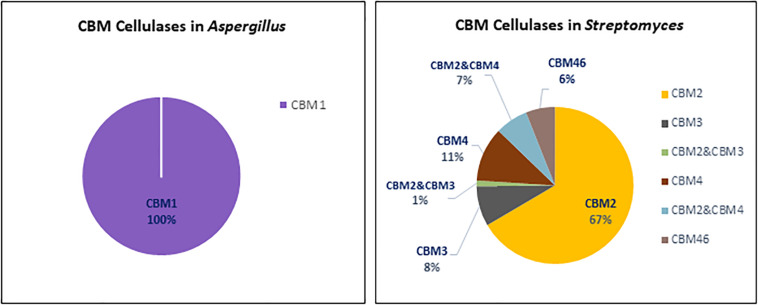
In the Aspergillus and Streptomyces genomes, 1946 sequences of cellulases with CBM domain were identified. Among these sequences 188 were annotated as cellulase with a single CBM in Aspergillus, 1256 as cellulase with a single CBM in Streptomyces and 502 as cellulases with multiple CBMs in Streptomyces. Figure 3 shows that Streptomyces cellulases are not only higher in their diversity of the CBM type, but also more varied in the domain organization compared to Aspergillus. In Streptomyces, four different CBMs associated with the GH cellulase catalytic domain were identified, including CBM2, CBM3, CBM4, and CBM46. Moreover, each CBM is not only present as a single module, but also appears in combination with other accessory domains including another CBM (CBM2&CBM4 and CBM2&CBM3). CBM2 is the most frequently found CBM (67%) among the four different CBMs in Streptomyces cellulases (Figure 5). In contrast to this diversity, in Aspergillus, CBM1 is the only module associated with GH cellulases and always present in a simple arrangement of one catalytic domain plus one CBM. Unlike observed for the amylases, the Aspergillus and Streptomyces cellulases do not share a common CBM.
FIGURE 5.
CBMs associated with cellulases from Aspergillus (left) and Streptomyces (right).
The CBM1s are found almost exclusively in fungi, while the CBM2s are mainly found in cellulases from bacterial origin. Based on the domain structure and function, CBM1s are relatively small, consisting of approximately 36 amino acids with 4 conserved cysteine residues involved in the formation of two disulphide bridges required for protein folding (Mattinen et al., 1997). The fold of CBM1 is called a cysteine knot (Boraston et al., 2004). This knotted arrangement is usually associated with a β-sheet structure and highly conducive to protein stability (Cheek et al., 2006). In contrast, the bacterial cellulose binding modules like CBM2 are usually much larger in size, about 110 amino acid residues with a β-sandwich fold providing the ligand recognition site (Boraston et al., 2004). In general, a regular plane surface with three aromatic residues such as tyrosine, as well as polar residues glutamine and asparagine are involved in CBM1-cellulose binding (Beckham et al., 2010). Likewise, the CBM2 β-sandwich fold presents a planar surface containing conserved aromatic residues, usually tryptophan and tyrosine, involved in substrate binding. Although the CBMsin Aspergillus and Streptomyces are different in protein structure, creating a chimeric enzyme by fusing a bacterial CBM with a fungal cellulase has potential to create an active enzyme. As reported by Voutilainen et al. (2014), fusing a bacterial CBM3 to a GH7 cellobiohydrolase from the thermophilic fungus Talaromyces emersonii which in fungi is often associated with a CBM1 domain, resulted in an active chimeric GH7 with higher thermal stability.
CBM1s are mainly located at the C-termini of GH5, GH7, and GH45. Only in the GH6 of Aspergillus, the CBM1 is positioned at the N-terminus (Figure 3A). Similar domain structures are also present in other fungi such as Trichoderma (Rahikainen et al., 2019). In a few special cases multiple CBM1 domains are present in a single protein. For example, five different CBM1 modules are arranged in tandem at the N-terminus of GH45 endoglucanase from the yeast, Pichia pastoris GS115 (Couturier et al., 2011). Furthermore, in Streptomyces the CBM2 is present either at the N- or C-terminus of GH cellulases. Among those, 76% were detected at the N-terminus of the cellulase catalytic domain, such as in CBM2_GH5, CBM2_GH6, and CBM2_GH48. The remaining 16 and 8% of the CBM2 were found at the C-terminus, connecting one catalytic domain with other accessory domains or different CBMs (Figures 3B1,B2, respectively). A CBM2 is widely occurring in cellulases from bacteria but much rarer in eukaryotes with only a few examples from gastropods and nematodes.
In the cellulase domain organization, CBM3 is always located at the C-terminus of the catalytic domain and represents the second most frequently found CBM in the domain architecture of cellulases (Figure 3B). Besides its presence as a single module, CBM3 is also commonly found in a multidomain architecture. In Streptomyces, CBM3s were mostly associated with GH6 in co-occurrence with Fn3 domains and linked with CBM2 in the GH9 multidomain architecture. A CBM3-Fn3-Fn3-CBM2 architecture was also found in the Thermobifida cellulosilytica TB100 GH9-endoglucanase (Tóth et al., 2017). A CBM3 domain consists of 150 amino acids and has a β-sandwich fold with nine strands and one calcium binding site (Tormo et al., 1996; Shimon et al., 2000).
In contrast to CBM3, a single CBM4 domain is always present at the N-terminus in the GH6 and GH9 family from Streptomyces (Figure 3B1). In the GH9 multidomain architecture, CBM4 can also be present together with CBM2 and another accessory domain, CelD-N. In general, the CBM4 modules are found in xylanases and endoglucanases in agreement with their binding affinity toward either xylan or β-glucan (Abou-Hachem et al., 2000; Boraston et al., 2002; Simpson et al., 2002). Several binding studies with CBM4 showed differences in substrate binding preference. The differences in substrate binding affinity of CBM4 is determined by the length and topography of the surface loops. The long shallow groove conformation in CBM4 Cellulomonas fimi provides better binding to 1,4-β-glucan than 1,3-β-glucan (Coutinho et al., 1992).,The prominent U-shape with high-sided walls formed by loops between the β-strands from Thermotoga maritima CBM4 contribute to the higher affinity to 1,3-β-glucan (Zverlov et al., 2001). The conserved aromatic clusters in CBM4 containg tyrosine and/or tryptophan contribute to the surface binding site and are suggested to be involved in binding specificity (Boraston et al., 2002). Furthermore, replacing the CBM1 of a fungal acetyl xylan esterase with the CBM4 from the thermophilic bacterium Rhodothermus marinus Xyn10A enhanced the specific activity and the thermostability of the acetyl xylan esterase (Liu and Ding, 2016). Indicating that creating a chimeric enzyme between a mesophilic carbohydrate esterase and a CBM4 from a thermophile does result in improved thermophilic properties.
Another CBM present in Streptomyces cellulases is CBM46. According to the CAZy database, CBM46 is exclusively found in bacterial cellulases. In GH cellulase family 5, the CBM46 is present in combination with an accessory module called CBM_X2 (PF03442). A similar architecture was also found in GH5s of Bacillus halodurans which are associated with CBM46 and CBM_X2 (Venditto et al., 2015). The CBM_X2 or generally also called the X2 module is the immunoglobulin (Ig)-like domain with capability to bind cellulose and bacterial cell walls (Mosbah et al., 2000; Kosugi et al., 2004; Liberato et al., 2016). Moreover, the catalytic activity of GH5 from Bacillus licheniformis is entirely dependent on its CBM46 and X2 modules (Liberato et al., 2016). Besides in GH5, the X2 module is also present in GH74 endoglucanase from Paenibacillus polymyxa A18 with the arrangement GH74_X2_CBM3. Based on the enzyme activity studies, endoglucanase with X2-CBM3 domains have a several fold increase in activity toward insoluble substrates. In substrate binding studies, the module X2 showed a higher affinity toward phosphoric acid swollen cellulose, whereas CBM3 showed a higher affinity toward crystalline cellulose (Avicel) (Pasari et al., 2017).
Other Accessory Domains: The Fn3 Domain
Other types of accessory modules found in Streptomyces GH cellulases are the Fn3 domain and the variants of the X2 module CelD-N and Big7. According to the Pfam database (El-Gebali et al., 2019), domains annotated as X2, CelD_N and Big7 are a group of bacterial Ig-like domain which are often associated with bacterial GHs cellulases and suggested to be involved in assembling multienzyme complexes called cellulosomes. The Fn3 domain is a domain composed of a seven-stranded β-sandwich, usually occurring in multiple copies in both intracellular and extracellular proteins.
Further investigation of the presence and role of a Fn3(-like) domain in GH cellulases, was performed by querying the genomes of both A. niger and S. leeuwenhoekii, as representatives of the two genera we have focused or research on and mapping the Fn3 domains present in these and other proteins involved in polysaccharide degradation (Table 1). Indeed, besides cellulases there are several proteins with Fn3 domains, including GH3 β-glucosidase, GH3 β-xylosidase, GH5 mannosidase, GH18 chitinase, and chitin biosynthesis proteins with several types of domain organization. This wide occurrence of Fn3 domains, suggests that the Fn3 domain could be of functional significance.
TABLE 1.
Protein domain associated with Fn3 in A. niger and S. leeuwenhoekii.
| Organism | Domain associated with Fn3 | Main activity | Accession number (NCBI) |
| A. niger | GH3_Fn3-like | β-glucosidase | CAK48740.1 |
| GH3_Fn3-like | β-xylosidase | CAK37179.1 | |
| RhgB__Fn3_CBM-like | Rhamnogalacturonat lyase | XP_001400741.2 | |
| Chs5n_Fn3_BRCT-CHS5like | Chitin biosynthesis protein | XP_001395839.2 | |
| S. leeuwenhoekii | GH5_Fn3_CBM2 | Mannosidase | CQR60862.1 |
| CBM2_Fn3_GH18 | Chitinase | CQR61740.1 | |
| CBM4_Fn3_GH18 | Chitinase | WP_029384933.1 |
As can be seen from Table 1, Fn3 is often present between the catalytic domain and a CBM in Streptomyces, in correspondence to the suggestion that it plays a role as stable linker between the enzymatic and substrate binding domain (Jee et al., 2002; Valk et al., 2015). However, the Fn3 domain in Aspergillus GH3 is positioned at the C-terminal end, and not followed by a CBM. Several other fungal β-glucosidases carry Fn3 domains, such as those from Aspergillus aculeatus and Trichoderma reesei (Suzuki et al., 2013), as well as the thermophilic fungus Rasamsonia emersonii (Gudmundsson et al., 2016).
Although Fn3 is suggested as a stable linker (Jee et al., 2002), many studies showed that the Fn3 domain also has relevance to a ligand binding function and is involved in protein-protein interaction (Hansen, 1992; Koide et al., 1998). From scanning electron microscope studies, it is concluded that Fn3 present in a bacterial cellobiohydrolase contributes to cellulose surface disruption, and therefore the presence of Fn3 increases the efficiency of degradation (Kataeva et al., 2002). Similarly, it has also been demonstrated that an isolated CBM2 from Cellulomonas fimi cellulase disrupted the surfaces of cellulose fibers (Din et al., 1991), indicating a potential functional similarity between Fn3 and a genuine CBM. Moreover, removal of the Fn3 domain can result in decreased or loss of enzymatic activity, as we found in a study on the GH3 function (Figure 8). As other examples, removal of the Fn3 domain of a xylanse dramatically decreased xylanolytic activity from Cellulosimicrobium sp. strain HY-13 (Kim et al., 2009), Flavobacterium johnsoniae (Chen et al., 2013), a Marinifilaceae bacterium strain SPP2 (Han et al., 2019) and cellulase activity for the GH6 cellobiohydrolase in another bacterial species (Cerda-Mejía et al., 2017). Deletion of the Fn3 domains also resulted in reduced enzymatic activity of GH55 endoglucanase (Conway et al., 2016) and GH9 endoglucanase (Zhou et al., 2004). Likewise, a CBM-truncated enzyme, in some cases, showed a significant loss of enzymatic activity, such as in GH9 (Burstein et al., 2009) and GH5 (Zheng and Ding, 2012). Surprisingly, our own results suggested that designing a GH3 chimeric enzyme by replacing the Fn3 domain with a Streptomyces CBM2 domain does not result in a functional GH3 enzyme (Figure 8, Sidar et al., in preparation). Thus, a function of the Fn3-like domain may be both to potentiate the activity of the GHs and to increase the substrate proximity to the catalytic domain in a similar fashion as the action of a CBM. Similar as found for CBM1 and CBM2 (see above) the (C-terminus of the) Fn3 domain consists of clusters of aromatic residues, indicating that it could indeed play a role in ligand binding interaction, instead of solely existing as a linker (Lima et al., 2013). Another similarity between Fn3 and CBM is that some CBMs have a binding site for calcium which plays a significant role in the substrate interaction (Tormo et al., 1996; Montanier et al., 2010; Yaniv et al., 2012). Kataeva et al. (2002) also experimentally showed that each Fn3 domain in cellobiohydrolase CbhA binds calcium. The literature evidence for the Fn3 domain performance so far suggest a putative dual role of this domain. We hypothesize that Fn3/Fn3-like domains play a biological role both in potentiation the functionality of the catalytic center and as a functional domain, rather than merely as a simple spacer, and may even represent a novel CBM-like domain.
FIGURE 8.
GH3 β-glucosidase activity determined using an agar plate assay with esculin as substrate (Pointing, 1999).
Designing New Chimeric Enzymes Inspired by the Domain Architecture
In the field of enzyme engineering, the design of chimeric enzymes has been developed as one of the promising approaches to obtain an enzyme with the desired characteristics like improved hydrolysis efficiency. A chimeric enzyme is commonly generated by fusing the GH catalytic domain obtained from one species with another protein domain such as a CBM from another species. In several studies, chimeric enzymes have been shown to improve the catalytic efficiency, thermostability, as well as substrate specificity (Oliveira et al., 2015; Saadat, 2017; Christensen et al., 2020; Gilmore et al., 2020).
As shown above Streptomyces species display a much higher diversity in cellulase domain architecture than Aspergillus. This observation led us to review the Streptomyces domain architecture in more detail as the basis for design of novel chimeric enzymes with cellulolytic activity. As an example, we selected Streptomyces leeuwenhoekii C34 (Busarakam et al., 2014) and Aspergillus niger.
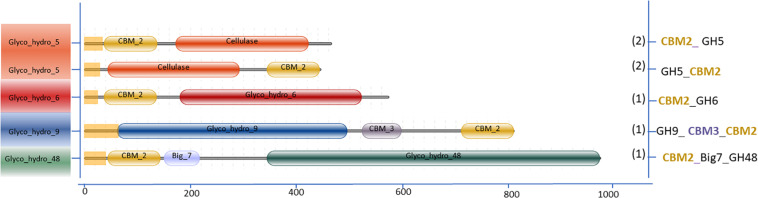
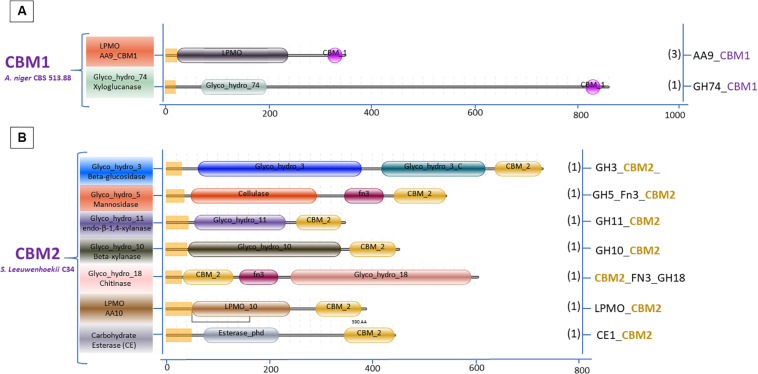
The variation in the domain organization of cellulases tethered to a CBM in S. leeuwenhoekii is presented in Figure 6. The cellulases of the GH5, GH6, GH9, and GH48 families are all associated with CBM2. When expanding our genome mining efforts in S. leeuwenhoekii, to proteins associated with a CBM2, other than cellulases, a considerable group of enzymes such as xylanases, chitinases, LPMOs and an esterase showed up from the genome sequence, in addition to a GH3 family β-glucosidase (Figure 7). The same mapping of enzymes containing a CBM1 in A. niger, resulted in only two other proteins, both related to polysaccharide degradation, being a cellulose active LMPO and a GH74 xyloglucanase (Figure 7).
FIGURE 6.
Cellulases associated with CBM2 in S. leeuwenhoekii C34.
FIGURE 7.
Domain organization of polysaccharide degrading enzymes other than cellulases associated with two dominant CBMs: (A) CBM1 A. niger 513.88 and (B) CBM2 S. leeuwenhoekii C34.
In fungal cellulolytic enzyme cocktails, the β-glucosidases are considered to be the limiting factor in obtaining full conversion of cellulose to glucose. The GH3 family β-glucosidase in filamentous fungi do not have native CBM domains (see Figure 3). Therefore, we suggest that based on the Streptomyces domain architecture, a new concept to design chimeric enzymes could be studied by fusing a CBM2 domain from a Streptomyces GH3 β-glucosidase to a well-known A. niger GH3 β-glucosidase. However, it should be kept in mind that the design of new chimeric enzymes is not as simple as replacing one domain for another as shown by the example of replacing a Fn3 domain by CBM2 (Figure 8 and section “Other Accessory Domains: The Fn3 Domain”).
Future Perspective
As functional domains, the CBMs have attractive characteristics like promoting substrate binding and thus supporting the catalytic function of enzyme. So far, CBMs have been classified in more than 80 families and more CBM families will undoubtedly be discovered. CBMs appear in association with a wide range of proteins in particular the CAZymes and are assembled in various domain organizations in numerous microorganisms. As an example, the diversity in the domain organizations in Aspergillus and Streptomyces is presented in this review. The Streptomyces species harbor cellulases and amylases with a more diverse domain organization, including CBMs and other accessory domains, than found in Aspergillus. This remarkable feature provides inspiration to design chimeric enzymes with a potential for substrate degradation improvement.
Fusions of enzyme domains have been developed in many scientific studies to create improved chimeric enzymes. In one of our studies, the Aspergillus GH3 enzyme that does not have a CBM but does have a C-terminal Fn3-like domain, was redesigned to replace the Fn3 with a CBM2 of Streptomyces. To our surprise these studies revealed that the Fn3-like domain of the fungal GH3 seems to play a pivotal role in the enzymatic activity (Figure 8), This observation might indicate that in addition to the well-known CBMs more domains could play an important role in the activity of CAZymes. Clearly, creating chimeric enzymes by modifying domain arrangement could provide advantages for enhancing enzymatic activity. Overall, the variability in the CBM domain organization of cellulases and amylases, as shown in this review, offers new opportunities to rethink the strategy for designing multiple-domain enzymes. This approach is not limited to chimeric cellulases and amylase to improve degradation of recalcitrant substrate but also holds promise for other CAZymes such as xylanases, pectinases, and oxidative enzymes.
Author Contributions
AS analyzed the data, wrote the review, performed the literature data mining, and designed the figures. EA conducted the cellulases analysis of Streptomyces. GV performed the bioinformatic analysis for the graphical domain annotation. AR presented ideas and edited the review. EV and PP involved in the review outline, ideas, planning, supervision, writing, and editing. All authors reviewed and agreed on the final manuscript.
Conflict of Interest
EA was employed by company Sun Pharmaceutical Industries Europe BV. GV and PP were employed by company Dutch DNA Biotech B.V. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported by a scholarship of the Indonesia Endowment Fund for Education (LPDP) from the Ministry of Finance, Indonesia to AS and by a grant of the Dutch National Organization for Scientific Research NWO, in the framework of an ERA-IB project FilaZyme (053.80.721/EIB.14.021) to EV, GV, and PP.
References
- Abdelmalek I. B., Urdaci M. C., Ali M. B., Denayrolles M., Chaignepain S., Limam F., et al. (2009). Structural investigation and homology modeling studies of native and truncated forms of α-amylases from Sclerotinia sclerotiorum. J. Microbiol. Biotechnol. 19 1306–1318. 10.4014/jmb.0903.03013 [DOI] [PubMed] [Google Scholar]
- Abou-Hachem M., Karlsson E. N., Bartonek-Roxa E., Raghothama S., Simpson P. J., Gilbert H. J., et al. (2000). Carbohydrate-binding modules from a thermostable Rhodothermus marinus xylanase: cloning, expression and binding studies. Biochem. J. 345 53–60. 10.1042/bj3450053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Y., Rebets Y., Estévez M. R., Zapp J., Myronovskyi M., Luzhetskyy A. (2020). Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters. Microb. Cell Fact. 19:5. 10.1186/s12934-020-1277-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleshin A., Golubev A., Firsov L. M., Honzatko R. B. (1992). Crystal structure of glucoamylase from Aspergillus awamori var. X100 to 2.2-Å resolution. J. Biol. Chem. 267 19291–19298. 10.2210/pdb1gly/pdb [DOI] [PubMed] [Google Scholar]
- Aleshin A. E., Feng P.-H., Honzatko R. B., Reilly P. J. (2003). Crystal structure and evolution of a prokaryotic glucoamylase. J. Mol. Biol. 327 61–73. 10.1016/s0022-2836(03)00084-6 [DOI] [PubMed] [Google Scholar]
- Anné J., Vrancken K., Van Mellaert L., Van Impe J., Bernaerts K. (2014). Protein secretion biotechnology in Gram-positive bacteria with special emphasis on Streptomyces lividans. Biochim. Biophys. Acta Mol. Cell Res. 1843 1750–1761. 10.1016/j.bbamcr.2013.12.023 [DOI] [PubMed] [Google Scholar]
- Arai T., Araki R., Tanaka A., Karita S., Kimura T., Sakka K., et al. (2003). Characterization of a cellulase containing a family 30 carbohydrate-binding module (CBM) derived from Clostridium thermocellum CelJ: importance of the CBM to Cellulose Hydrolysis. J. Bacteriol. 185 504–512. 10.1128/jb.185.2.504-512.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes V., Saddler J. N. (2010). Access to cellulose limits the efficiency of enzymatic hydrolysis: the role of amorphogenesis. Biotechnol. Biofuels 3:4. 10.1186/1754-6834-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer E. A., Belaich J. P., Shoham Y., Lamed R. (2004). The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58 521–554. 10.1146/annurev.micro.57.030502.091022 [DOI] [PubMed] [Google Scholar]
- Beckham G. T., Matthews J. F., Bomble Y. J., Bu L., Adney W. S., Himmel M. E., et al. (2010). Identification of amino acids responsible for processivity in a Family 1 carbohydrate-binding module from a fungal cellulase. J. Phys. Chem. B 114 1447–1453. 10.1021/jp908810a [DOI] [PubMed] [Google Scholar]
- Benoit I., Culleton H., Zhou M., DiFalco M., Aguilar-Osorio G., Battaglia E., et al. (2015). Closely related fungi employ diverse enzymatic strategies to degrade plant biomass. Biotechnol. Biofuels 8:107. 10.1186/s13068-015-0285-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlemont R. (2017). Distribution and diversity of enzymes for polysaccharide degradation in fungi. Sci. Rep. 7:222. 10.1038/s41598-017-00258-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlemont R., Martiny A. C. (2015). Genomic potential for polysaccharides deconstruction in bacteria. Appl. Environ. Microbiol. 81 1513–1519. 10.1128/AEM.03718-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlemont R., Martiny A. C. (2016). Glycoside hydrolases across environmental microbial communities. PLoS Comput. Biol. 12:e1005300. 10.1371/journal.pcbi.1005300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes A., Pellegrini V. O. A., Curtolo F., Camilo C. M., Mello B. L., Johns M. A., et al. (2019). Carbohydrate binding modules enhance cellulose enzymatic hydrolysis by increasing access of cellulases to the substrate. Carbohydr. Polym. 211 57–68. 10.1016/j.carbpol.2019.01.108 [DOI] [PubMed] [Google Scholar]
- Bissaro B., Rohr A. K., Müller G., Chylenski P., Skaugen M., Forsberg Z., et al. (2017). Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat. Chem. Biol. 13 1123–1128. 10.1038/nchembio.2470 [DOI] [PubMed] [Google Scholar]
- Book A. J., Lewin G. R., McDonald B. R., Takasuka T. E., Wendt-Pienkowski E., Doering D. T., et al. (2016). Evolution of high cellulolytic activity in symbiotic Streptomyces through selection of expanded gene content and coordinated gene expression. PLoS Biol. 14:e1002475. 10.1371/journal.pbio.1002475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004). Carbohydrate binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382 769–781. 10.1042/bj20040892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraston A. B., Nurizzo D., Notenboom V., Ducros V., Rose D. R., Kilburn D. G., et al. (2002). Differential oligosaccharide recognition by evolutionarily-related β-1,4 and β-1,3 glucan-binding modules. J. Mol. Biol. 319 1143–1156. 10.1016/s0022-2836(02)00374-1 [DOI] [PubMed] [Google Scholar]
- Bott R., Saldajeno M., Cuevas W., Ward D., Scheffers M., Aehle W., et al. (2008). Three-dimensional structure of an intact glycoside hydrolase family 15 glucoamylase from Hypocrea jecorina. Biochemistry 47 5746–5754. 10.1021/bi702413k [DOI] [PubMed] [Google Scholar]
- Buleon A., Colonna P., Planchot V., Ball S. (1998). Starch granules: structure and biosynthesis. Int. J. Biol. Macromol. 23 85–112. 10.1016/S0141-8130(98)00040-3 [DOI] [PubMed] [Google Scholar]
- Burstein T., Shulman M., Jindou S., Petkun S., Frolow F., Shoham Y., et al. (2009). Physical association of the catalytic and helper modules of a family-9 glycoside hydrolase is essential for activity. FEBS Lett. 583 879–884. 10.1016/j.febslet.2009.02.013 [DOI] [PubMed] [Google Scholar]
- Busarakam K., Bull A. T., Girard G., Labeda D. P., van Wezel G. P., Goodfellow M. (2014). Streptomyces leeuwenhoekii sp. nov., the producer of chaxalactins and chaxamycins, forms a distinct branch in Streptomyces gene trees. Antonie Van Leeuwenhoek 105 849–861. 10.1007/s10482-014-0139-y [DOI] [PubMed] [Google Scholar]
- Cerda-Mejía L., Valenzuela S. V., Frías C., Diaz P., Pastor F. I. J. (2017). A bacterial GH6 cellobiohydrolase with a novel modular structure. Appl. Microbiol. Biotechnol. 101 2943–2952. 10.1007/s00253-017-8129-4 [DOI] [PubMed] [Google Scholar]
- Chalak A., Villares A., Moreau C., Haon M., Grisel S., d’ Orlando A., et al. (2019). Influence of the carbohydrate-binding module on the activity of a fungal AA9 lytic polysaccharide monooxygenase on cellulosic substrates. Biotechnol. Biofuels 12:206. 10.1186/s13068-019-1548-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheek S., Krishna S. S., Grishin N. V. (2006). Structural classification of small, disulfide-rich protein domains. J. Mol. Biol. 359 215–237. 10.1016/j.jmb.2006.03.017 [DOI] [PubMed] [Google Scholar]
- Chen S., Kaufman M. G., Miazgowicz K. L., Bagdasarian M., Walker E. D. (2013). Molecular characterization of a cold-active recombinant xylanase from Flavobacterium johnsoniae and its applicability in xylan hydrolysis. Bioresour. Technol. 128 145–155. 10.1016/j.biortech.2012.10.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S. J., Badino S. F., Cavaleiro A. M., Borch K., Westh P. (2020). Functional analysis of chimeric TrCel6A enzymes with different carbohydrate binding modules. Protein Eng. Des. Select. 32 401–409. 10.1093/protein/gzaa003 [DOI] [PubMed] [Google Scholar]
- Cockburn D. W., Suh C., Medina K. P., Duvall R. M., Wawrzak Z., Henrissat B., et al. (2017). Novel carbohydrate binding modules in the surface anchored α-amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut. Mol. Microbiol. 107 249–264. 10.1111/mmi.13881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway J. M., Pierce W. S., Le J. H., Harper G. W., Wright J. H., Tucker A. L., et al. (2016). Multidomain, surface layer-associated glycoside hydrolases contribute to plant polysaccharide degradation by Caldicellulosiruptor species. J. Biol. Chem. 291 6732–6747. 10.1074/jbc.m115.707810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtade G., Forsberg Z., Heggset E. B., Eijsink V. G. H., Aachmann F. L. (2018). The carbohydrate-binding module and linker of a modular lytic polysaccharide monooxygenase promote localized cellulose oxidation. J. Biol. Chem. 292 13006–13015. 10.1074/jbc.ra118.004269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho J. B., Gilkes N. R., Warren R. A. J., Kilburn D. G., Miller R. C., Jr. (1992). The binding of Cellulomonas fimi endoglucanase C (CenC) to cellulose and sephadex is mediated by the N-terminal repeats. Mol. Microbiol. 6 1243–1252. 10.1111/j.1365-2958.1992.tb01563.x [DOI] [PubMed] [Google Scholar]
- Couturier M., Feliu J., Haon M., Navarro D., Lesage-Meessen L., Coutinho P. M., et al. (2011). A thermostable GH45 endoglucanase from yeast: impact of its atypical multimodularity on activity. Microb. Cell Fact. 10:103. 10.1186/1475-2859-10-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G., Henrissat B. (1995). Structures and mechanisms of glycosyl hydrolases. Structure 3 853–859. 10.1016/s0969-2126(01)00220-9 [DOI] [PubMed] [Google Scholar]
- de Souza P. M., de Oliveira Magalhães P. (2010). Application of microbial α-amylase in industry - A review. Braz. J. Microbiol. 41 850–861. 10.1590/s1517-83822010000400004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derde L. J., Gomand S. V., Courtin C. M., Delcour J. A. (2012). Characterisation of three starch degrading enzymes: thermostable β-amylase, maltotetraogenic and maltogenic α-amylases. Food Chem. 135 713–721. 10.1016/j.foodchem.2012.05.031 [DOI] [PubMed] [Google Scholar]
- Dias F. M. V., Vincent F., Pell G., Prates J. A. M., Centeno M. S. J., Tailford L. E., et al. (2004). Insights into the molecular determinants of substrate specificity in glycoside hydrolase family 5 revealed by the crystal structure and kinetics of Cellvibrio mixtus mannosidase 5A. J. Biol. Chem. 279, 25517–25526. 10.1074/jbc.m401647200 [DOI] [PubMed] [Google Scholar]
- Din N., Gilkes N., Tekant B., Robert C. M., Anthony J. R. W., Kilburn D. G. (1991). Non–hydrolytic disruption of cellulose fibres by the binding domain of a bacterial cellulase. Nat. Biotechnol. 9 1096–1099. 10.1038/nbt1191-1096 [DOI] [Google Scholar]
- Duan C. J., Huang M. Y., Pang H., Zhao J., Wu C. X., Feng J. X. (2017). Characterization of a novel theme C glycoside hydrolase family 9 cellulase and its CBM-chimeric enzymes. Appl. Microbiol. Biotechnol. 101 5723–5737. 10.1007/s00253-017-8320-7 [DOI] [PubMed] [Google Scholar]
- Eddy S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali J., Mistry A., Bateman S. R., Eddy A., Luciani S. C., Potter M., et al. (2019). Finn. The Pfam protein families database in 2019. Nucleic Acids Res. 47 D427–D432. 10.1093/nar/gky995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler M., Nitsche B., Wanka F., Meyer V. (2013). “Aspergillus: a cell factory with unlimited prospect,” in Applications of Microbial Engineering, eds Gupta V. K., Schmoll M., Mazutti M. A., Maki M., Tuohy M. G. (Boca Raton: CRC Press; ), 1–51. 10.1201/b15250-2 [DOI] [Google Scholar]
- Fleißner A., Dersch P. (2010). Expression and export: recombinant protein production systems for Aspergillus. Appl. Microbiol. Biotechnol. 87 1255–1270. 10.1007/s00253-010-2672-6 [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Warren R. A., Miller R. J., Kilburn D. G. (1988). Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J. Biol. Chem. 263 10401–10407. [PubMed] [Google Scholar]
- Gilmore S. P., Lillington S. P., Haitjema C. H., de Groot R., O’Malley M. A. (2020). Designing chimeric enzymes inspired by fungal cellulosomes. Synth. Syst. Biotechnol. 5 23–32. 10.1016/j.synbio.2020.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruben B. S., Mäkelä M. R., Kowalczyk J. E., Zhou M., Benoit-Gelber I., De Vries R. (2017). Expression-based clustering of CAZyme-encoding genes of Aspergillus niger. BMC Genomics 18:900. 10.1186/s12864-017-4164-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson M., Hansson H., Karkehabadi S., Larsson A., Stals I., Kim S., et al. (2016). Structural and functional studies of the glycoside hydrolase family 3 β-glucosidase Cel3A from the moderately thermophilic fungus Rasamsonia emersonii. Acta Crystallogr. D Struct. Biol. 72(Pt 7), 860–870. 10.1107/S2059798316008482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Gigras P., Mohapatra H., Goswami V. K., Chauhan B. (2003). Microbial α-amylases: a biotechnological perspective. Process Biochem. 38 1599–1616. 10.1016/S0032-9592(03)00053-0 [DOI] [Google Scholar]
- Hamed M. B., Anné J., Karamanou S., Economou A. (2018). Streptomyces protein secretion and its application in biotechnology. FEMS Microbiol. Lett. 365:fny250. 10.1093/femsle/fny250 [DOI] [PubMed] [Google Scholar]
- Han Z., Shang-guan F., Yang J. (2019). Molecular and biochemical characterization of a bimodular xylanase from Marinifilaceae bacterium strain SPP2. Front. Microbiol. 10:1507. 10.3389/fmicb.2019.01507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C. K. (1992). Fibronectin type III-like sequences and a new domain type in prokaryotic depolymerases with insoluble substrates. FEBS Lett. 305 91–96. 10.1016/0014-5793(92)80871-d [DOI] [PubMed] [Google Scholar]
- Hayashida S., Kunisaki S., Nakao M., Flor P. Q. (1982). Evidence for raw starch -affinity site on Aspergillus awamori glucoamylase. I. Agric. Biol. Chem. 46 83–89. 10.1080/00021369.1982.10865024 [DOI] [Google Scholar]
- Huang X. F., Nazarian F., Vincken J. P., Visser R. G. F., Trindade L. M. (2017). A tandem CBM25 domain of α-amylase from Microbacterium aurum as potential tool for targeting proteins to starch granules during starch biosynthesis. BMC Biotechnol. 17:86. 10.1186/s12896-017-0406-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isikgor F. H., Becer C. R. (2015). Lignocellulosic biomass: a sustainable platform for the production of bio-based chemicals and polymers. Polym. Chem. 6:4497 10.1039/C5PY00263J [DOI] [Google Scholar]
- Janeček Š, Mareček F., MacGregor E. A., Svensson B. (2019). Starch-binding domains as CBM families–history, occurrence, structure, function and evolution. Biotechnol. Adv. 37:107451. 10.1016/j.biotechadv.2019.107451 [DOI] [PubMed] [Google Scholar]
- Janeček Š, Svensson B., MacGregor E. A. (2014). α -Amylase – an enzyme specificity found in various families of glycoside hydrolases. Cell. Mol. Life Sci. 71 1149–1170. 10.1007/s00018-013-1388-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee J. G., Ikegami T., Hashimoto M., Kawabata T., Ikeguchi M., Watanabe T., et al. (2002). Solution structure of the fibronectin type III domain from Bacillus circulans WL-12 chitinase A1. J. Biol. Chem. 277 1388–1397. 10.1074/jbc.M109726200 [DOI] [PubMed] [Google Scholar]
- Jeoh T., Wilson D. B., Walker L. P. (2006). Effect of cellulase mole fraction and cellulose recalcitrance on synergism in cellulose hydrolysis and binding. Biotechnol. Prog. 22, 270–277. 10.1021/bp050266f [DOI] [PubMed] [Google Scholar]
- Käll L., Krogh A., Sonnhammer E. L. L. (2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338 1027–1036. 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Kataeva I. A., Seidel R. D., Shah A., West L. T., Li X. L., Ljungdahl L. G. (2002). The fibronectin type 3-Like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl. Environ. Microbiol. 68 4292–4300. 10.1128/aem.68.9.4292-4300.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y., Han M. K., Park D. S., Lee J. S., Oh H. W., Shin D. H., et al. (2009). Novel GH10 xylanase, with a fibronectin type 3 domain, from Cellulosimicrobium sp. Strain HY-13, a bacterium in the gut of Eisenia fetida. Appl. Environ. Microbiol. 75 7275–7279. 10.1128/aem.01075-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide A., Bailey C. W., Huang X., Koide S. (1998). The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 284 1141–1151. 10.1006/jmbi.1998.2238 [DOI] [PubMed] [Google Scholar]
- Kosugi A., Amano Y., Murashima K., Doi R. H. (2004). Hydrophilic domains of scaffolding protein CbpA promote glycosyl hydrolase activity and localization of cellulosomes to the cell surface of Clostridium cellulovorans. J. Bacteriol. 186 6351–6359. 10.1128/JB.186.19.6351-6359.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y., Yamashita K., Tagami T., Uraji M., Wan K., Okuyama M., Yao M., Kimura A., Hatanaka T. (2015). The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition. FEBS J. 282, 4001–4014. 10.1111/febs.13401 [DOI] [PubMed] [Google Scholar]
- Lee J., Paetzel M. (2011). Structure of the catalytic domain of glucoamylase from Aspergillus niger. Acta Crystallogr. 67 188–192. 10.1107/s1744309110049390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggio L. L., Simmons T. J., Poulsen J. C. N., Frandsen K. E. H., Hemsworth G. R., Stringer M. A., et al. (2015). Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat. Commun. 6. 10.1038/ncomms6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberato M. V., Silveira R. L., Prates E. T., De Araujo E. A., Pellegrini V. O. A., Camilo C. M., et al. (2016). Molecular characterization of a family 5 glycoside hydrolase suggests an induced-fit enzymatic mechanism. Sci. Rep. 6 1–16. 10.1038/srep23473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima M. A., Oliveira-Neto M., Kadowaki M. A., Rosseto F. R., Prates E. T., Squina F. M., et al. (2013). Aspergillus niger β-glucosidase has a cellulase-like tadpole molecular shape: insights into glycoside hydrolase family 3 (GH3) β-glucosidase structure and function. J. Biol. Chem. 288 32991–33005. 10.1074/jbc.M113.479279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. P., Ma H. Y., Lin H. J., Liu S. M., Tzou W. S. (2011). Biochemical characterization of two truncated forms of amylopullulanase from Thermoanaerobacterium saccharolyticum NTOU1 to identify its enzymatically active region. Appl. Biochem. Biotechnol. 165 1047–1056. 10.1007/s12010-011-9319-7 [DOI] [PubMed] [Google Scholar]
- Liu S., Ding S. (2016). Replacement of carbohydrate binding modules improves acetyl xylan esterase activity and its synergistic hydrolysis of different substrates with xylanase. BMC Biotechnol. 16:73. 10.1186/s12896-016-0305-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42 D490–D495. 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattinen M. L., Linder M., Teleman A., Annila A. (1997). Interaction between cellohexaose and cellulose binding domains from Trichoderma reesei cellulases. FEBS Lett. 407 291–296. 10.1016/s0014-5793(97)00356-6 [DOI] [PubMed] [Google Scholar]
- Mehta D., Satyanarayana T. (2016). Bacterial and archaeal α-amylases: diversity and amelioration of the desirable characteristics for industrial applications. Front. Microbiol. 7:1129. 10.3389/fmicb.2016.01129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke A., Damude H. G., Tomme P., Kwan E., Kilburn D. G., Miller R. C., et al. (1995). Enhancement of the endo-β-1, 4-glucanase activity of an exocellobiohydrolase by deletion of a surface loop. J. Biol. Chem. 270 4383–4386. 10.1074/jbc.270.9.4383 [DOI] [PubMed] [Google Scholar]
- Mikami B., Degano M., Hehre E. J., Sacchettini J. C. (1994). Crystal structures of soybean. beta.-amylase reacted with. beta.-maltose and maltal: active site components and their apparent roles in catalysis. Biochemistry 33 7779–7787. 10.1021/bi00191a005 [DOI] [PubMed] [Google Scholar]
- Mikami B., Iwamoto H., Malle D., Yoon H.-J., Demirkan-Sarikaya E., Mezaki Y., et al. (2006). Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site. J. Mol. Biol. 359 690–707. 10.1016/j.jmb.2006.03.058 [DOI] [PubMed] [Google Scholar]
- Mizuno M., Tonozuka T., Suzuki S., Uotsu-Tomita R., Kamitori S., Nishikawa A., et al. (2004). Structural insights into substrate specificity and function of glucodextranase. J. Biol. Chem. 279 10575–10583. 10.1074/jbc.M310771200 [DOI] [PubMed] [Google Scholar]
- Montanier C., Flint J. E., Bolam D. N., Xie H., Liu Z., Rogowski A., et al. (2010). Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules. J. Biol. Chem. 285 31742–31754. 10.1074/jbc.m110.142133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montella S., Ventorino V., Lombard V., Henrissat B., Pepe O., Faraco V. (2017). Discovery of genes coding for carbohydrate-active enzyme by metagenomic analysis of lignocellulosic biomasses. Sci. Rep. 7:42623. 10.1038/srep42623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbah A., Belaich A., Bornet O., Belaich J. P., Henrissat B., Darbon H. (2000). Solution structure of the module X2 1 of unknown function of the cellulosomal scaffolding protein CipC of Clostridium cellulolyticum. J. Mol. Biol. 304 201–217. 10.1006/jmbi.2000.4192 [DOI] [PubMed] [Google Scholar]
- Nguyen S. T. C., Freund H. L., Kasanjian J., Berlemont R. (2018). Function, distribution, and annotation of characterized cellulases, xylanases, and chitinases from CAZy. Appl. Microbiol. Biotechnol. 102 1629–1637. 10.1007/s00253-018-8778-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu C., Zheng F., Li Y., Liu C., Li Q. (2018). Process optimization of the extraction condition of β-amylase from brewer’s malt and its application in the maltose syrup production. Biotechnol. Appl. Biochem. 65 639–647. 10.1002/bab.1650 [DOI] [PubMed] [Google Scholar]
- Oliveira C., Carvalho V., Domingues L., Gama F. M. (2015). Recombinant CBM-fusion technology. Applications overview. Biotechnol. Adv. 33 358–369. 10.1016/j.biotechadv.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Oyama T., Kusunoki M., Kishimoto Y., Takasaki Y., Nitta Y. (1999). Crystal structure of β-amylase from Bacillus cereus var. mycoides at 2.2 Å resolution. J. Biochem. 125, 1120–1130. 10.1093/oxfordjournals.jbchem.a022394 [DOI] [PubMed] [Google Scholar]
- Oyama T., Miyake H., Kusunoki M., Nitta Y. (2003). Crystal structures of beta-amylase from Bacillus cereus var mycoides in complexes with substrate analogs and affinity-labeling reagents. J. Biochem. 133 467–474. 10.1093/jb/mvg061 [DOI] [PubMed] [Google Scholar]
- Parkkinen T., Koivula A., Vehmaanperä J., Rouvinen J. (2008). Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding. Protein Sci. 17 1383–1394. 10.1110/ps.034488.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasari N., Adlakha N., Gupta M., Bashir Z., Rajacharya G. H., Verma G., et al. (2017). Impact of module-X2 and carbohydrate binding module-3 on the catalytic activity of associated glycoside hydrolases towards plant biomass. Sci. Rep. 7:3700. 10.1038/s41598-017-03927-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C. M., Knott B. C., Mayes H. B., Hansson H., Himmel M. E., Sandgren M., et al. (2015). Fungal cellulases. Chem. Rev. 115 1308–1448. 10.1021/cr500351c [DOI] [PubMed] [Google Scholar]
- Pérez J., Muñoz-Dorado J., de la Rubia T., Martínez J. (2002). Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview. Int. Microbiol. 5 53–63. 10.1007/s10123-002-0062-3 [DOI] [PubMed] [Google Scholar]
- Poidevin L., Feliu J., Doan A., Berrin J. G., Bey M., Coutinho P. M., et al. (2013). Insights into exo- and endoglucanase activities of family 6 glycoside hydrolases from Podospora anserina. Appl. Environ. Microbiol. 79 4220–4229. 10.1128/aem.00327-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointing S. B. (1999). Qualitative methods for the determination of lignocellulolytic enzyme production by tropical fungi. Fungal Divers. 2 17–33. [Google Scholar]
- Polizeli M. L. T. M., Vici A. C., Scarcella A. S. A., Cereia M., Pereira M. G. (2016). “Enzyme system from Aspergillus in current industrial uses and future applications in the production of second-generation ethanol,” in New and Future Developments in Microbial Biotechnology and Bioengineering, ed. Gupta V. K. (Amsterdam: Elsevier; ), 127–140. 10.1016/b978-0-444-63505-1.00009-9 [DOI] [Google Scholar]
- Pujadas G., Palau J. (2001). Evolution of α-amylases: architectural features and key residues in the stabilization of the (β/α)8 scaffold. Mol. Biol. Evol. 18 38–54. 10.1093/oxfordjournals.molbev.a003718 [DOI] [PubMed] [Google Scholar]
- Punt P. J., Levasseur A., Visser H., Wery J., Record E. (2011). Fungal protein production: design and production of chimeric proteins. Annu. Rev. Microbiol. 65 57–69. 10.1146/annurev.micro.112408.134009 [DOI] [PubMed] [Google Scholar]
- Punt P. J., van Biezen N., Conesa A., Albers A., Mangnus J., van den Hondel C. (2002). Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 20 200–206. 10.1016/s0167-7799(02)01933-9 [DOI] [PubMed] [Google Scholar]
- Rahikainen J., Ceccherini S., Molinier M., Holopainen-Mantila U., Reza M., Väisänen S., et al. (2019). Effect of cellulase family and structure on modification of wood fibres at high consistency. Cellulose 26 5085–5103. 10.1007/s10570-019-02424-x [DOI] [Google Scholar]
- Ray R. R. (2004). β-amylases from various fungal strains. Acta Microbiol. Immunol. Hung. 51 85–95. 10.1556/amicr.51.2004.1-2.6 [DOI] [PubMed] [Google Scholar]
- Ray R. R., Nanda G. (1996). Microbial β-amylases: biosynthesis, characteristics, and industrial applications. Crit. Rev. Microbiol 22 181–199. 10.3109/10408419609106459 [DOI] [PubMed] [Google Scholar]
- Reyes-Ortiz V., Heins R. A., Cheng G., Kim E. Y., Vernon B. C., Elandt R. B., et al. (2013). Addition of a carbohydrate-binding module enhances cellulase penetration into cellulose substrates. Biotechnol. Biofuels 6:93. 10.1186/1754-6834-6-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C., Moroz O. V., Ariza A., Skov L. K., Ayabe K., Davies G. J., et al. (2018). Structural insight into industrially relevant glucoamylases: flexible positions of starch -binding domains. Acta Crystallogr. D Struct. Biol. 74 463–470. 10.1107/S2059798318004989 [DOI] [PubMed] [Google Scholar]
- Saadat F. (2017). A review on chimeric xylanases: methods and conditions. 3 Biotech 7:67. 10.1007/s13205-017-0660-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer J., Christensen T., Frandsen T. P., Mirgorodskaya E., McGuire K. A., Driguez H., et al. (2001). Stability and Function of Interdomain Linker Variants of Glucoamylase 1 from Aspergillus niger. Biochemistry 40 9336–9346. 10.1021/bi010515i [DOI] [PubMed] [Google Scholar]
- Sauer J., Sigurskjold B. W., Christensen U., Frandsen T. P., Mirgorodskaya E., Harrison M., et al. (2000). Glucoamylase: structure/function relationships, and protein engineering. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1543 275–293. 10.1016/s0167-4838(00)00232-6 [DOI] [PubMed] [Google Scholar]
- Segato F., Damasio A. R., Gonçalves T. A., Murakami M. T., Squina F. M., Polizeli M., et al. (2012). Two structurally discrete GH7-cellobiohydrolases compete for the same cellulosic substrate fiber. Biotechnol. Biofuels 5:21. 10.1186/1754-6834-5-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevillano L., Vijgenboom E., van Wezel G. P., Díaz M., Santamaría R. I. (2016). New approaches to achieve high level enzyme production in Streptomyces lividans. Microb. Cell Fact. 15:28. 10.1186/s12934-016-0425-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimon L. J. W., Pagès S., Belaich A., Belaich J.-P., Bayer E. A., Lamed R., et al. (2000). Structure of a family IIIa scaffoldin CBD from the cellulosome of Clostridium cellulolyticum at 2.2 Å resolution. Acta Crystallogr. Sec. D Biol. Crystallogr. 56 1560–1568. 10.1107/s0907444900012889 [DOI] [PubMed] [Google Scholar]
- Simpson P. J., Jamieson S. J., Abou-Hachem M., Karlsson E. N., Gilbert H. J., Holst O., et al. (2002). The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase. Biochemistry 41 5712–5719. 10.1021/bi012093i [DOI] [PubMed] [Google Scholar]
- Somerville C. R. (2006). Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22 53–78. 10.1146/annurev.cellbio.22.022206.160206 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Sumitani J.-I., Nam Y.-W., Nishimaki T., Tani S., Wakagi T., et al. (2013). Crystal structures of glycoside hydrolase family 3 β-glucosidase 1 from Aspergillus aculeatus. Biochem. J. 452 211–221. 10.1042/bj20130054 [DOI] [PubMed] [Google Scholar]
- Svensson B., Pedersen T. G., Svendsen I., Sakai T., Ottesen M. (1982). Characterization of two forms of glucoamylase from Aspergillus niger. Carlsberg Res. Commun. 47 55–69. 10.1007/BF02907797 [DOI] [Google Scholar]
- Talamantes D., Biabini N., Dang H., Abdoun K., Berlemont R. (2016). Natural diversity of cellulases, xylanases, and chitinases in bacteria. Biotechnol. Biofuels 9:133. 10.1186/s13068-016-0538-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo S. C., Liew K. J., Shamsir M. S., Chong C. S., Bruce N. C., Chan K.-G., et al. (2019). Characterizing a halo-tolerant GH10 Xylanase from Roseithermus sacchariphilus strain RA and its CBM-truncated variant. Int. J. Mol. Sci. 20:2284. 10.3390/ijms20092284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann M., Coiro M., Meier T., Wicker T., Zeeman S. C., Santelia D. (2019). The evolution of functional complexity within the β-amylase gene family in land plants. BMC Evol. Biol. 19:66. 10.1186/s12862-019-1395-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., et al. (1988). Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur. J. Biochem. 170 575–581. 10.1111/j.1432-1033.1988.tb13736.x [DOI] [PubMed] [Google Scholar]
- Tormo J., Lamed R., Chirino A. J., Morag E., Bayer E. A., Shoham Y., et al. (1996). Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 15 5739–5751. 10.1002/j.1460-2075.1996.tb00960.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth Á, Baka E., Luzics S., Bata-Vidács I., Nagy I., Bálint B., et al. (2017). Plant polysaccharide degrading enzyme system of Thermobifida cellulosilytica TB100T revealed by de novo genome project data. Acta Alimentaria 46 323–335. 10.1556/066.2016.0014 [DOI] [Google Scholar]
- Tseng C. W., Tzu P. K., Rey T. G., Jian W. H., Hao C. W., Chun H. H., et al. (2011). Substrate binding of a GH5 endoglucanase from the ruminal fungus Piromyces rhizinflata. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 67 1189–1194. 10.1107/S1744309111032428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk V., Eeuwema W., Sarian F. D., van der Kaaij R. M., Dijkhuizen L. (2015). Degradation of granular starch by the bacterium Microbacterium aurum strain B8. A involves a modular α-amylase enzyme system with FNIII and CBM25 domains. Appl. Environ. Microbiol. 81 6610–6620. 10.1128/AEM.01029-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bueren A. L., Boraston A. B. (2007). The structural basis of α-glucan recognition by a family 41 carbohydrate-binding module from Thermotoga maritima. J. Mol. Biol. 365 555–560. 10.1016/j.jmb.2006.10.018 [DOI] [PubMed] [Google Scholar]
- Van Tilbeurgh H., Tomme P., Claeyssens M., Bhikhabhai R., Pettersson G. (1986). Limited proteolysis of the cellobiohydrolase I from Trichoderma reesei. FEBS Lett. 204 223–227. 10.1016/0014-5793(86)80816-X [DOI] [Google Scholar]
- Venditto I., Najmudin S., Luís A. S., Ferreira L. M. A., Sakka K., Knox J. P., et al. (2015). Family 46 carbohydrate-binding modules contribute to the enzymatic hydrolysis of xyloglucan and β-1,3–1,4-glucans through distinct mechanisms. J. Biol. Chem. 290 10572–10586. 10.1074/jbc.m115.637827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voshol G. P., Punt P. J., Vijgenboom E. (2019). Profile Comparer Extended: phylogeny of lytic polysaccharide monooxygenase families using profile hidden Markov model alignments. F1000Research 8:1834. 10.12688/f1000research.21104.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voshol G. P., Vijgenboom E., Punt P. J. (2017). The discovery of novel LPMO families with a new hidden markov model. BMC Res. Notes 10:105. 10.1186/s13104-017-2429-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutilainen S. P., Nurmi-Rantala S., Penttilä M., Koivula A. (2014). Engineering chimeric thermostable GH7 cellobiohydrolases in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 98 2991–3001. 10.1007/s00253-013-5177-2 [DOI] [PubMed] [Google Scholar]
- Vu V. V., Beeson W. T., Span E. A., Farquhar E. R., Marletta M. A. (2014). A family of starch -active polysaccharide monooxygenases. Proc. Natl. Acad. Sci. U.S.A. 111 13822–13827. 10.1073/pnas.1408090111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv O., Petkun S., Shimon L. J. W., Bayer E. A., Lamed R., Frolow F. (2012). A single mutation reforms the binding activity of an adhesion-deficient family 3 carbohydrate-binding module. Acta Crystallogr. Sect. D Biol. Crystallogr. 68 819–828. 10.1107/s0907444912013133 [DOI] [PubMed] [Google Scholar]
- Yuan X. L., van der Kaaij R. M., van den Hondel C. A. M. J. J., Punt P. J., van der Maarel M. J. E. C., Dijkhuizen L., et al. (2008). Aspergillus niger, genome-wide analysis reveals a large number of novel alpha-glucan acting enzymes with unexpected expression profiles. Mol. Genet. Genomics 279 545–561. 10.1007/s00438-008-0332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F., Ding S. (2012). Processivity and enzymatic mode of a glycoside hydrolase family 5 endoglucanase from Volvariella volvacea. Appl. Environ. Microbiol. 79 989–996. 10.1128/aem.02725-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Irwin D. C., Escovar-Kousen J., Wilson D. B. (2004). Kinetic studies of thermobifida fuscaCel9A active site mutant enzymes. Biochemistry 43 9655–9663. 10.1021/bi049394n [DOI] [PubMed] [Google Scholar]
- Zhu Y., Romain C., Williams C. K. (2016). Sustainable polymers from renewable resources. Nature 540 354–362. 10.1038/nature21001 [DOI] [PubMed] [Google Scholar]
- Zverlov V. V., Volkov I. Y., Velikodvorskaya G. A., Schwarz W. H. (2001). The binding pattern of two carbohydrate-binding modules of laminarinase Lam16A from Thermotoga neapolitana: differences in beta-glucan binding within family CBM4. Microbiology 147 621–629. 10.1099/00221287-147-3-621 [DOI] [PubMed] [Google Scholar]