Abstract
Citrus peel has been used as a Traditional medicine in Asia to treat coughs, asthma and bronchial disorders. Therefore, the anti-inflammatory effects of 3,5,6,7,3′,4′-hexamethoxyflavone (quercetogetin, QUE) isolated from Citrus unshiu peel were investigated in lipopolysaccharide (LPS)-induced RAW 264.7 macrophage cells. The results showed that QUE repressed the production of prostaglandin E2 and nitric oxide by suppressing LPS-induced expression of cyclooxygenase-2 and inducible nitric oxide synthase. It also suppressed the production of interleukin (IL)-6, IL-1β, and tumor necrosis factor-α cytokines, and decreased the nuclear translocation of NF-κB by interrupting the phosphorylation of NF-κB inhibitor α in macrophage cells. Based on the finding that QUE inhibited the phosphorylation of ERK protein expression in LPS-induced RAW264.7 cells, it was confirmed that inhibition of inflammatory responses by QUE was mediated via the ERK pathway. Therefore, this study suggests that QUE has strong anti-inflammatory effects, making it a promising compound for use as a therapeutic agent in treating inflammatory lung diseases, such as emphysema.
Keywords: quercetogetin, anti-inflammatory, NF-κB, ERK, cyclooxygenase-2, inducible nitric oxide synthase
Introduction
Although inflammation is required for a healthy immune response, it is also responsible for causing a number of diseases, including arthritis, diabetes, cardiovascular disease and cancer. The balance between pro- and anti-inflammatory cytokines is involved in the inflammatory response (1). Acute inflammation is a short-term response characterized by extravasation of plasma, erythrocytes and leukocytes into the injured tissue. On the other hand, chronic inflammation is characterized by tissue infiltration by macrophages and lymphocytes.
During inflammatory processes, macrophages are critical mediators of immune and inflammatory responses that are activated by binding with various stimuli (2). One of the most potent activators of macrophages is lipopolysaccharide (LPS), which binds with Toll-like receptor 4, the activation of which results in the production of proinflammatory cytokines, such as interleukin (IL)-6, tumor necrosis factor-α (TNF-α) and IL-1β, and inflammatory mediators, such as prostaglandin (PGE2) and nitric oxide (NO) (3–5). Uncontrolled production of these mediators and cytokines can lead to chronic inflammatory-derived diseases (6). Accordingly, substances that reduce the production of these mediators have attracted significant attention in the development of anti-inflammatory drugs.
The key targets of anti-inflammatory agents include transcription factors, cytokines and enzymes (7). NF-κB plays an important role in the expression of inflammatory mediators containing NF-κB binding motifs (8). It is involved in the regulation of numerous genes, such as inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2), that are mediators of acute-phase immune and inflammatory responses (9). Additionally, activation of the mitogen-activated protein kinase (MAPK) pathway also plays an essential role in the initiation and development of inflammatory processes that are transmitted by sequential phosphorylation events (10). The three major groups of MAPK cascades are ERK, p38 and c-Jun N-terminal kinase (JNK). Each MAPK is activated by the upstream activation of MAPK kinase (MKK) and MAPK kinase kinase. Once activated by kinases, MAPK can phosphorylate transcription factors or other downstream kinases that promote the expression of proinflammatory mediators of extracellular stimuli (10,11).
Flavonoids are natural compounds with various biological and pharmacological properties, including anticancer, antimicrobial, antiviral, anti-inflammatory and antithrombotic effects (10,11). Specifically, the anti-inflammatory activities of flavonoids have long been known (12). Some flavonoids have been reported to attenuate chronic inflammation in animal models (13). Thus, continued evaluation of the anti-inflammatory activity of flavonoids may be valuable. Notably, Citrus-derived flavonoids and their metabolites have been reported to have important biological activities, including anti-inflammatory, antiviral, anticancer and anti-atherogenic properties (14–16). The highest concentration of polymethoxyflavones (PMFs; 6,7,4′,5′-tetramethoxy-5-monohydroxyflavone, 5,6,8,3′,6′-pentamethoxyflavone, 5,6,7,3′,4′,5′-hexamethoxyflavone) is found in the Citrus peel (17,18). Tangeretin (4′,5,6,7,8-pentamethoxyflavone, TAN) and nobiletin (5,6,7,8,3′,4′-hexamethoxyflavone, NOB) are two PMFs that are relatively common in Citrus peel (19). Previous research demonstrated that PMFs suppress degranulation in antigen-stimulated basophilic leukemia cells (20). Likewise, NOB had inhibitory effects on H2O2-induced apoptosis in SH-SY5Y cells, while a binary mixture of 5-demethylnobiletin and TAN attenuated cell growth (21). Furthermore, the PMF-rich fractions from the peel of Citrus sunki showed stronger antiproliferative activity than single PMFs in human leukemia cells (22). Previously, synergistic anti-inflammatory effects have been reported on RAW264.7 cells treated with a binary mixture of sulforaphane and NOB (23,24). However, pharmacological studies of the biological effects of PMF compounds have been limited.
In the present study, 3,5,6,7,3′,4′-hexamethoxyflavone (quercetogetin, QUE) from extracts of Citrus unshiu peel (CUP) were isolated and identified. As the anti-inflammatory effects of QUE have not been studied, the anti-inflammatory effects and molecular mechanisms regulated by QUE in LPS-induced RAW264.7 cells were examined. These findings suggested that QUE may be beneficial in preventing LPS-induced inflammation and is a promising agent against various inflammatory diseases.
Materials and methods
Cell culture
Murine macrophage, RAW264.7 cells were purchased from the American Type Culture Collection. This cell line was cultured in Dulbecco's modified Eagle's medium (DMEM; WeGene) with 100 µg/ml streptomycin (Thermo Fisher Scientific, Inc.), 100 U/ml penicillin (Gibco; Thermo Fisher Scientific, Inc.) and fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.).
Chemicals and reagents
LPS, Griess reagent, MTT and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich; Merck KGaA. The antibodies against COX-2 (cat. no. sc-376861), iNOS (cat. no. sc-7271), β-actin (cat. no. sc-8432) and horseradish peroxidase (HRP)-conjugated secondary antibodies [anti-mouse (cat. no. sc-2005), anti-goat (cat. no. sc-2020) and anti-rabbit (cat. no. sc-2004)] were purchased from Santa Cruz Biotechnology, Inc. The antibodies against histone H3 (cat. no. 9715), NF-κB (p65; cat. no. 8242), NF-κB inhibitor α (IκB-α; cat. no. 4812), phosphorylated (p)-IκB-α (cat. no. 2859), p38 (cat. no. 9212), p-p38 (cat. no. 9211), ERK (cat. no. 4695), p-ERK (cat. no. 4377), JNK (cat. no. 9258) and p-JNK (cat. no. 4671) were purchased from Cell Signaling Technology, Inc. The antibody against GAPDH (cat. no. LF-PA0018) was purchased from AbFrontier Co., Ltd.
Isolation of QUE
CUP was harvested along the coast of Jeju Island, Korea in 2010. The dried CUP (3 kg) was extracted three times in 95% (v/v) ethyl alcohol overnight at room temperature. The ethyl alcohol (838 g) extract was evaporated and suspended in distilled water, and then divided into two fractions using chloroform as the non-aqueous phase. The chloroform fraction (155.9 g) was separated on a liquid chromatographic column (15×40 cm) packed with silica gel column (230–400 mesh) using gradient mixtures of CHCl3-MeOH (from CHCl3:MeOH=100:1 to 1:1, v/v) as the mobile phase to obtain nine fractions, F01 to F09, through thin-layer chromatography. F04 (50.2 g) was applied to a liquid chromatographic column (15×40 cm) packed with silica gel column (230–400 mesh) using gradient mixtures of Hexan-EtOAc (from Hexan:EtOAc=80:1 to 1:3, v/v) to obtain 14 sub-fractions (F04-1 to 14). The active fraction F04-11 (271 mg) was fractionated through solid-phase extraction using 100% MeOH as the solvent system to obtain the active compound 3 (3,5,6,7,3′,4′-hexamethoxyhomoflavone, 9.48 mg, QUE). The structure of the isolated QUE was elucidated through spectroscopic analysis [proton nuclear magnetic resonance (1H-NMR) and carbon-13 (13C)-NMR] and comparison with published data (19).
QUE showed the following characteristics: C21H22O8, yellow amorphous powder, electrospray ionization mass spectrometry (ESI-MS) m/z 402.1187 (M-H)+, 1H-NMR (300 MHz, CDCl3) δ (ppm): 7.56 (1H, dd, J=8.5, 2.0 Hz, H-6′), 7.41 (1H, d, J=2.0 Hz, H-2′), 7.00 (1H, d, J=8.5 Hz, H-5′), 6.61 (1H, s, H-3), 4.10, 4.02, 3.98, 3.96, 3.95 X2 (each 3H, s, 6 OMe at C-3, −5, −6, −7, −3′, −4′); 13C NMR (75 MHz, CDCl3) δ (ppm): 177.5 (s, C-4), 161.2 (s, C-2), 152.1 (s, C-4′), 151.6 (s, C-7), 149.5 (s, C-3′), 148.6 (s, C-8), 147.9 (s, C-9), 144.3 (s, C-5), 138.2 (s, C-6), 124.2 (s, C-1′), 119.8 (d, C-6′), 114.6 (s, C-10), 111.4 (d, C-5′), 108.7 (d, C-2′), 107.1 (d, C-3), 62.4, 62.1, 62.0, 61.8, 56.3, 56.2 (6 q, OMe at C-5, −6, −7, −3, −3′, −4′).
Cell viability assay
Cells were seeded in 96-well plates at 5ⅹ103 cells/well and treated with QUE (0, 1, 10 or 100 µM) at 37°C for 24 h. Then, 0.5 mg/ml MTT solution was added to each well and cells were incubated 37°C for 4 h, 100 µl DMSO was added. The absorbance was measured at a wavelength of 570 nm (Tecan Group, Ltd.).
Nitric oxide measurements
Cells were seeded in 48-well plates at 1ⅹ105 cells/well and induced with 1 µg/ml LPS and treated with QUE (0, 1, 10 or 100 µM) at 37°C for 24 h. In brief, 100 µl of culture medium was mixed with the same volume of Griess reagent (Sigma-Aldrich; Merck KGaA) and incubated for 10 min at room temperature. The absorbance was measured at 550 nm and the concentration of nitrite was determined using sodium nitrite as a standard.
Measurement of PGE2, IL-1β, IL-6 and TNF-α
RAW264.7 cells were seeded in 24-well plates at 2ⅹ105 cells/well and then treated with QUE with or without LPS (1 µg/ml) at 37°C for 24 h. The concentrations of IL-6 (cat. no. R600B), PGE2 (cat. no. KGE004B), IL-1β (cat. no. RLB00) and TNF-α (cat. no. RTA00) in the cell culture supernatant were determined using ELISA kits (R&D Systems, Inc.).
Preparation of cytosolic and nuclear extracts
Macrophage cells were plated in 60 mm cell culture plates, pretreated with QUE or DMSO for 1 h, and then stimulated with LPS for 30 min at 37°C. Cells were washed with PBS, scraped from the plates into PBS, and centrifuged at 500 × g for 3 min at room temperature. The pellets were suspended in Cytoplasmic Extraction Reagent (CER) I (with protease inhibitor), and then, the samples were added to cold CER II. The mixture was centrifuged at 16,000 × g for 5 min at 4°C, and the supernatant (cytosolic extract) was collected. The pellets were resuspended in Nuclear Extraction Reagent (with protease inhibitor) for 40 min on ice, and then centrifuged at 16,000 × g for 10 min at 4°C. The supernatant (nuclear extract) was collected.
Western blot analysis
Cells were washed with Dulbecco's PBS (WeGene), and then scraped into 500 µl of 2X RIPA (50 mM Tris-HCl, pH 7.5, 2 mM EDTA, 0.5% deoxycholate, 150 mM NaCl, 1% NP-40, 0.1% SDS and 1X protease inhibitor) buffer. The protein concentration was determined using BSA reagent (Bio-Rad Laboratories, Inc.). Then, proteins (20–40 µg) from each sample were resolved via SDS-PAGE on 10% gel, and the separated proteins were transferred to PVDF membranes (EMD Millipore). The membranes were blocked with 5% (w/v) non-fat milk powder in TBS with Tween-20 (TBST; 50 mM Trish-HCl, 150 mM NaCl and 0.1% Tween-20) for 1 h at room temperature, following which they were incubated with primary antibodies (1:1,000) in blocking solution overnight at 4°C. After three washes in TBST, the membranes were incubated with HRP-conjugated secondary antibodies (1:5,000) for 1 h at room temperature. Each protein was detected using the Western BLoT Hyper substrate kit (Takara Bio, Inc.). Band intensity was semi-quantified by densitometry analysis using ImageJ software (version 1.52a; National Institutes of Health).
Reverse transcription (RT)-semi-quantitative PCR
After isolation of total RNA using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), the reverse transcription of RNA (1 µg) was performed using oligo-dT18 and the ImProm-II™ reverse transcription kit (Promega Corporation). Reactions were incubated at 25°C for 5 min, at 42°C for 60 min, and then for 10 min at 70°C to inactivate the reverse transcriptase. The primers used for PCR were as follows: COX-2 forward, 5′-CACTACATCCTGACCCACTT-3′ and reverse, 5′-ATGCTCCTGCTTGAGTATGT-3′; iNOS forward, 5′-CCCTTCCGAAGTTTCTGGCAGCAGC-3′ and reverse, 5′-GGCTGTCAGAGCCTCGTGGCTTTGG-5′; IL-6 forward, 5′-GTACTCCAGAAGACCAGAGG-3′ and reverse, 5′-TGCTGGTGACAACCACGGCC-3′; IL-1β forward, 5′-CAGGATGAGGACATGAGCACC-3′ and reverse, 5′-CTCTGCAGACTCAAACTCCAC-3′; TNF-α forward, 5′-TTGACCTCAGCGCTGAGTTG-3′ and reverse, 5′-CCTGTAGCCCACGTCGTAGC-3′; GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′. The thermocycling conditions were as follows: Initial denaturation at 95°C for 5 min; 30 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec; and a final extension at 72°C for 10 min. The amplification was performed on a thermal cycler (Takara PCR Thermal Cycler model TP600; Takara Bio, Inc.). The products were visualized using 1.0% agarose gels stained with EtBr. GAPDH was used as a housekeeping gene when indicated. Band intensity was semi-quantified by densitometry analysis using ImageJ software (National Institutes of Health).
Statistical analysis
All data represent the mean ± SD of at least three independent experiments. One-way ANOVA followed by Tukey's test was used for data comparison among groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Chemical structure and cell viability effects of QUE on LPS-induced RAW264.7 cells
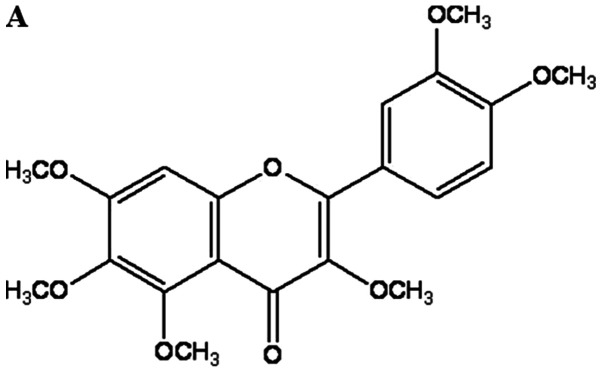
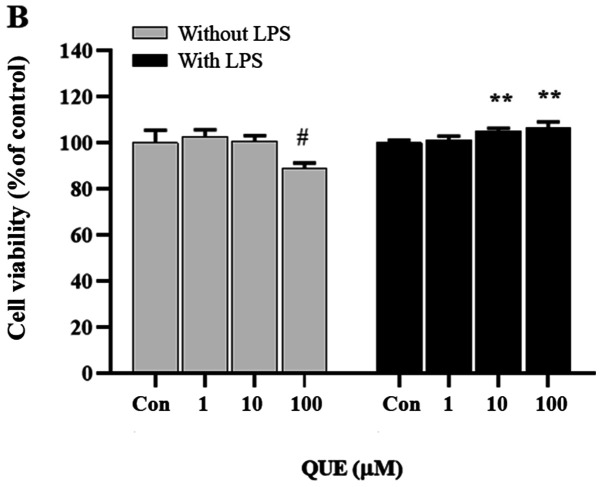
The activity of the extracts and fractions obtained from CUP was assessed using the NO assay in LPS-induced RAW 264.7 cells. Three pure compounds were isolated and identified as the main components driving the inhibition of NO production. The chemical structures of these compounds were determined through spectroscopic methods, including mass and NMR spectroscopic analyses (Figs. S1–S3) (19). To examine the effect of QUE on macrophage cell viability, Raw264.7 cells were treated with various concentrations of QUE (Fig. 1A). The survival curve shown in Fig. 1B indicates that QUE at 1, 10 and 100 µM did not exhibit cytotoxicity on Raw264.7 cells.
Figure 1.
Effect of QUE on the viability of LPS-induced RAW264.7 cells. (A) Chemical structure of QUE. (B) Cells were treated with QUE (1, 10 and 100 µM) and 1 µg/ml LPS for 24 h. The data shown represent the mean ± standard deviation of three experiments. #P<0.05 vs. non-treated group; **P<0.01 vs. LPS-treated group. QUE, quercetogetin; LPS, lipopolysaccharide. (D) The protein expression levels of iNOS and COX-2 were examined using reverse transcription-semi-quantitative PCR and western blotting, respectively. β-actin was used as a loading control. ###P<0.001 vs. non-treated group; *P<0.05, **P<0.01 and ***P<0.001 vs. LPS-treated group. NS, not significant; QUE, quercetogetin; NO, nitric oxide; PGE2, prostaglandin E2; LPS, lipopolysaccharide; iNOS, inducible nitric oxide synthase; COX-2, cyclooxygenase-2.
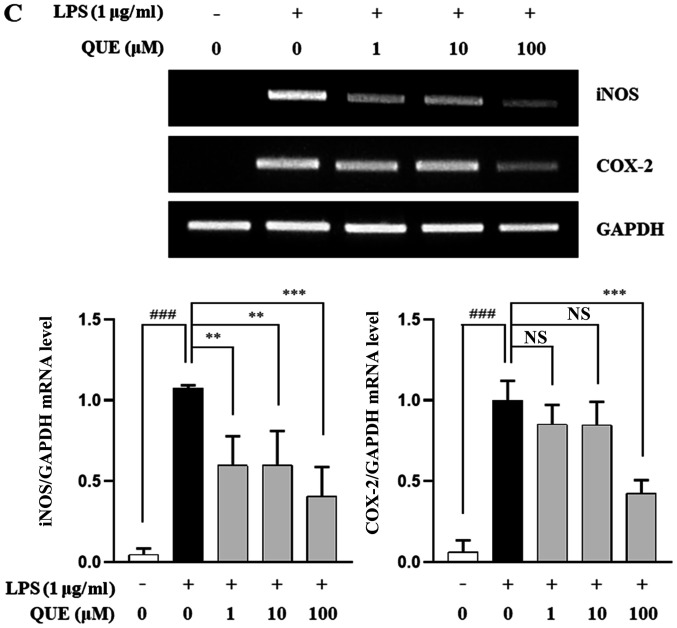
QUE inhibits production of NO and PGE2 by suppressing iNOS and COX-2 expression in LPS-induced RAW264.7 cells
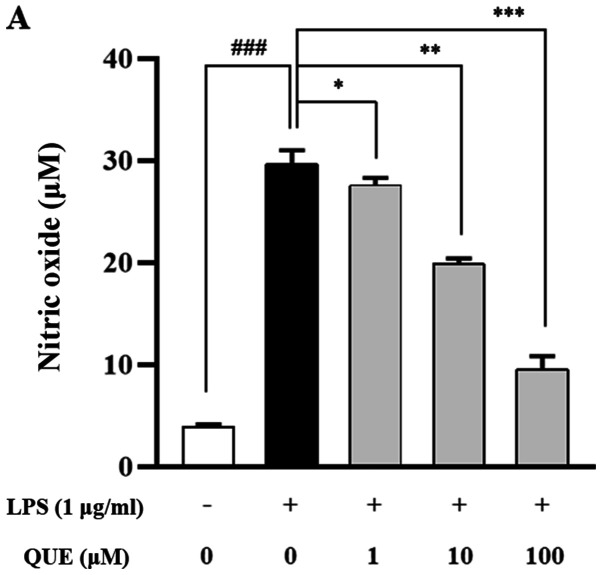
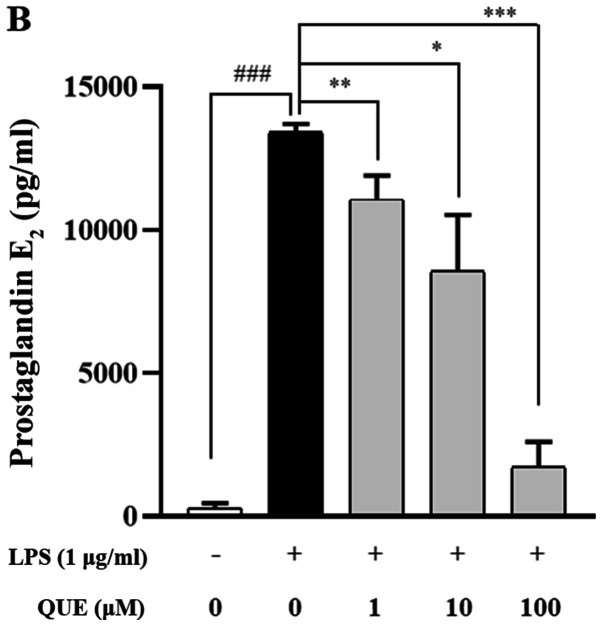
As NO and PGE2 are important inflammatory mediators, the effects of QUE on NO and PGE2 production in LPS-induced RAW264.7 cells were examined. QUE (1, 10 or 100 µM) inhibited NO and PGE2 production in LPS-induced cells in a dose-dependent manner (Fig. 2A and B). To identify the molecular mechanism through which QUE inhibited NO and PGE2 production in response to LPS, expression levels of COX-2 and iNOS were examined using RT-PCR and western blot analyses. The results showed that mRNA and protein expression of iNOS and COX-2 were increased in LPS-induced RAW264.7 cells. Conversely, QUE inhibited LPS-induced mRNA and protein expression in a dose-dependent manner (Fig. 2C and D). These results suggested that decreased NO and PGE2 production is related to inhibition of iNOS and COX-2 expression in LPS-induced RAW264.7 cells.
Figure 2.
Effect of QUE on NO and PGE2 production in LPS-induced RAW264.7 macrophages. Cells were induced with 1 µg/ml LPS and treated with various concentrations (1, 10 and 100 µM) of QUE for 24 h. (A) NO production in the cell culture supernatant was measured using the Griess assay. (B) PGE2 production in the cell cultures supernatant was measured using an ELISA assay. The (C) mRNA expression levels of iNOS and COX-2 were examined using reverse transcription-semi-quantitative PCR and western blotting, respectively.
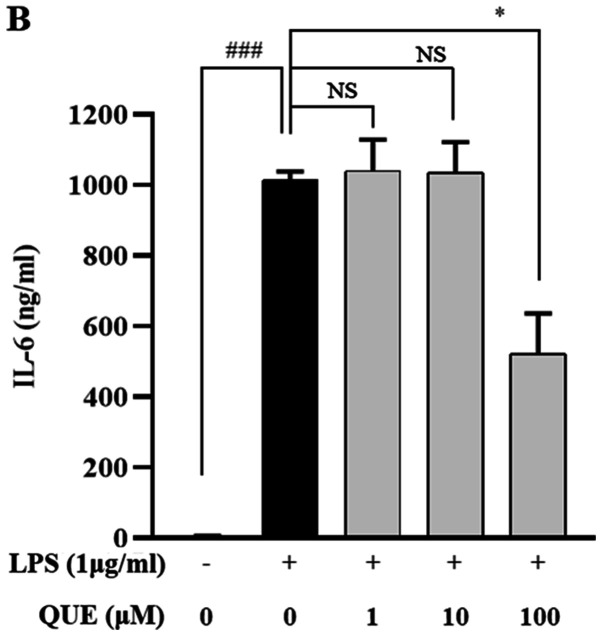
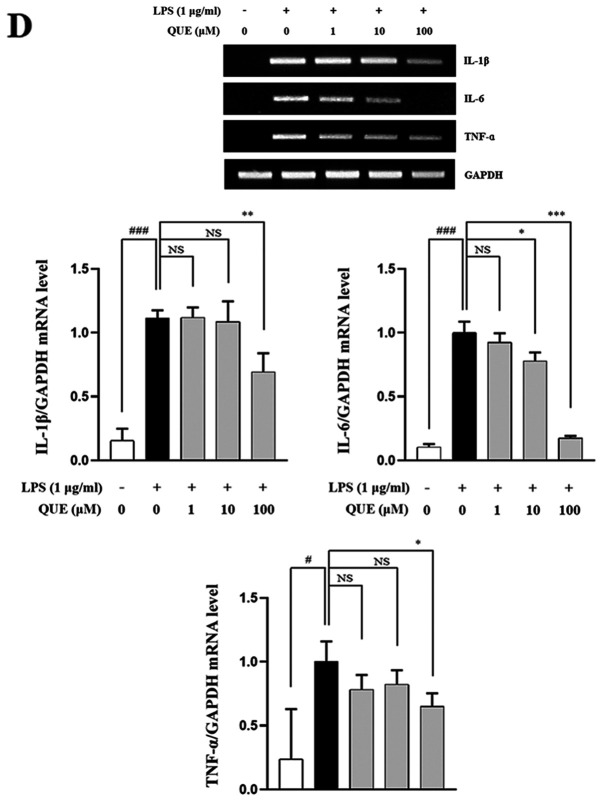
QUE inhibits production and mRNA expression of proinflammatory cytokines in LPS-induced RAW264.7 cells
LPS increases the production of proinflammatory cytokines, such as IL-1β, IL-6, and TNF-α, which play important roles in pathogen-induced inflammatory responses. ELISA and RT-PCR analyses were performed to characterize the effects of QUE on the production of proinflammatory cytokines. As presented in Fig. 3A-C, treatment with 100 µM QUE significantly inhibited LPS-induced IL-1β, IL-6 and TNF-α cytokine levels. To determine whether QUE modulated the production of cytokines at the level of transcription RT-PCR was performed. The mRNA expression levels of cytokines were elevated in LPS-induced cells, but treatment with 100 µM QUE decreased this induction (Fig. 3D). Accordingly, these results suggested that QUE prevents the secretion of proinflammatory cytokines by suppressing gene expression.
Figure 3.
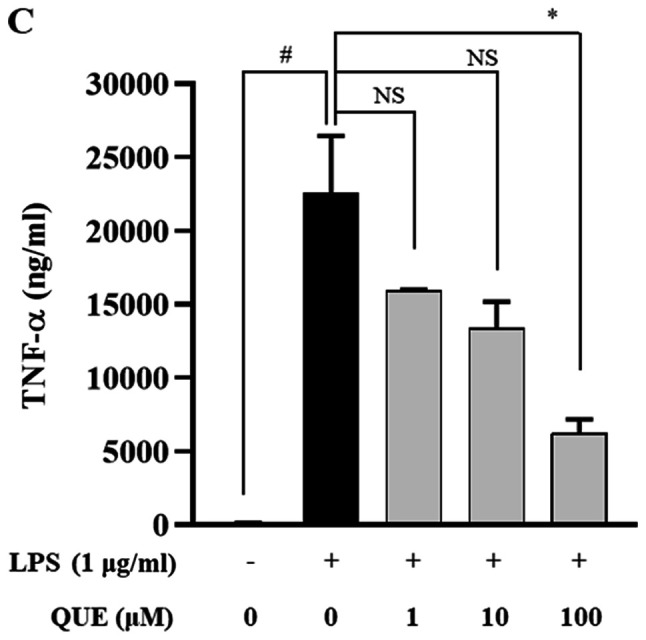
Effect of QUE on IL-1β, IL-6 and TNF-α mRNA expression levels in LPS-induced RAW264.7 macrophages. Cells were induced with 1 µg/ml LPS and treated with various concentrations (1, 10 or 100 µM) of QUE for 24 h. The production of (A) IL-1β, (B) IL-6 and (C) TNF-α in the cell culture supernatants were measured using an ELISA assay. (D) The mRNA expression levels of IL-1β, IL-6 and TNF-α were determined through reverse transcription-semi-quantitative PCR. GAPDH was used as a loading control. The data shown represent the mean ± SD of three experiments. #P<0.05 and ###P<0.001 vs. non-treated group; *P<0.05, **P<0.01 and ***P<0.001 vs. LPS-treated group. NS, not significant; QUE, quercetogetin; LPS, lipopolysaccharide; TNF-α, tumor necrosis factor-α; IL-6, interleukin-6; IL-1β, interleukin-1β.
QUE inhibits phosphorylation of IκB-α and NF-κB binding activity in LPS-induced RAW264.7 cells
NF-κB is an important transcription factor complex that controls the expression of proinflammatory mediators (25). To determine whether QUE regulates the NF-κB pathway, the phosphorylation level of IκB-α was determined using western blot analysis. The results showed that 100 µM QUE inhibited the phosphorylation of IκB-α in LPS-induced RAW264.7 cells (Fig. 4A). As p65 is the major subunit of the NF-κB complex, the nuclear translocation of p65 after its release from IκB-a was investigated. As shown in Fig. 4B, QUE significantly decreased the LPS-induced translocation of p65 to the nucleus. Overall, these results suggested that the NF-κB signaling pathway might be involved in the regulation of inflammatory factors by QUE in RAW264.7 cells.
Figure 4.
Effect of QUE on the phosphorylation of IκB-α and the nuclear translocation of NF-κB in LPS-induced RAW264.7 macrophages. Cells were pre-treated for 2 h with various concentrations (1, 10 or 100 µM) of QUE, and induced for 30 min with 1 µg/ml of LPS. (A) Whole-cell lysates were prepared and subjected to western blot analysis with anti-IκB-α and anti-p-IκB-α antibodies. β-actin was used as a loading control. (B) Cytosolic and nuclear extracts were prepared and subjected to western blot analysis with anti-NF-κB p65 antibody. Histone H3 and GAPDH were used as loading controls. ###P<0.001 vs. non-treated group; **P<0.01 and ***P<0.001 vs. LPS-treated group. NS, not significant; QUE, quercetogetin; LPS, lipopolysaccharide; nuclear, nuclear extracts; cytoplasmic, cytoplasmic extracts; IκB-α, NF-κB inhibitor α; p-, phosphorylated.
QUE inhibits phosphorylation of ERK in LPS-induced RAW264.7 cells
MAPK regulates inflammatory mediators, including proinflammatory cytokines and NO (26). LPS activates the MAPK signaling pathway in RAW264.7 cells (27). To determine whether the inhibition of inflammation by QUE was mediated via MAPK signaling, the effects of QUE on the phosphorylation of ERK, p38, and JNK were observed in LPS-induced cells. Fig. 5 shows that LPS significantly increased the activation of all three MAPK molecules. However, the phosphorylation of ERK was decreased by QUE in a dose-dependent manner, whereas the phosphorylation levels of p38 and JNK were not changed. Collectively, these results indicated that QUE has inhibitory effects on the ERK pathway in LPS-induced macrophage cells.
Figure 5.
Effect of QUE on the phosphorylation of MAPK in LPS-induced RAW264.7 macrophages. Cells were pre-treated for 2 h with various concentrations (1, 10 or 100 µM) of QUE and then induced for 30 min with 1 µg/ml of LPS. Total proteins from the cells were subjected to detection of the phosphorylated and total forms of three MAPK molecules, ERK, p38 and JNK. #P<0.05, ##P<0.01 and ###P<0.001 vs. non-treated group; *P<0.05 vs. LPS-treated group. NS, not significant; QUE, quercetogetin; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; JNK, Jun N-terminalkinase; p-, phosphorylated.
Discussion
CUP is used as a traditional medicine in Asia to treat coughs, asthma and bronchial disorders. It is a rich source of a number of flavanones and PMFs, which are also found in smaller quantities in other plants (28). PMFs are usually flavone aglycones with ≥4 methoxy substituents. NOB and TAN belong to this class of flavonoids and are commonly found in CUP (19). Also, these compounds are of commercial interest due to their diverse applications in the pharmaceutical and food industries. Previously, several studies have reported that some PMFs possess anti-inflammatory and anticancer properties (29–31). However, the human health-related activities of the QUE, including its anti-inflammatory effects, are relatively unknown. Therefore, the anti-inflammatory mechanism of this compound was investigated.
In the present study, compounds from extracts of CUP that inhibited NO production and had anti-inflammatory effects were isolated and characterized, and the molecular mechanisms underlying these effects in LPS-stimulated RAW 264.7 cells were identified. Based on the screening of NO production from CUP extracts, PMF compounds were isolated from the chloroform phase, including NOB, TAN and QUE. These compounds inhibited NO production in LPS-induced cells in a dose-dependent manner. Recently, we reported that QUE suppresses cigarette smoke extract-induced mitochondrial dysfunction and mitophagy in two human bronchial epithelial cell lines, Beas-2B and NHBE (32). Inflammation is regulated by numerous inflammatory mediators, such as NO, PGE2 and cytokines. Excessive production of iNOS-derived NO and COX-2-derived PGE2 can cause chronic diseases involving inflammatory and autoimmune disorders (33). These mediators regulate a variety of pathological and physiological processes related to immune responses and inflammation (34). The regulation of inflammatory mediators is therefore important for the development of novel anti-inflammatory drugs. In the present study, it was observed that QUE (10–100 µM) inhibited iNOS and COX-2 expression, thereby suppressing iNOS-induced NO production and COX-2-induced PGE2 production. Moreover, QUE reduced TNF-α, IL-6 and IL-1β expression in LPS-induced macrophage cells. These results suggested that QUE has anti-inflammatory effects that are mediated via the suppression of proinflammatory cytokines and inflammatory mediators.
In the NF-κB signaling pathway, the p50/p65 heterodimer is the most common dimer found (35). In general, NF-κB is inactivated by an interaction with IκB, which sequesters it in the cytoplasm. However, when IκB is phosphorylated, NF-κB is released and translocated to the nucleus (36). The present results indicated that the nuclear translocation of p65 and LPS-induced phosphorylation of IκB-α are significantly reduced after pretreatment with QUE. These results suggested that QUE regulates inflammatory events via suppression of the NF-κB pathway.
MAPKs play a pivotal role in the regulation of cellular stress responses and activation of NF-κB (37). They regulate cellular response to extracellular stimulation and various cellular activities, such as apoptosis, differentiation, inflammation and gene expression (38,39). The MAPK signaling pathway is known to be involved in the regulation of iNOS and COX-2, as well as the production of proinflammatory cytokines by LPS (40). The inhibition of the ERK, p38 and JNK pathways is able to suppress the induction of proinflammatory mediators by LPS (41). The ERK pathway plays a critical role in mediating pain and inflammation in joints (42). Goodrige et al (43) suggested that ERK might be associated with the production of proinflammatory cytokines by macrophages. p38 is a potential therapeutic target among MAPKs for inflammatory diseases because it is involved in the regulation of a number of inflammatory processes (44). JNK is activated by proinflammatory cytokines and mediators, and plays a crucial role in immune system signaling (45). To examine whether the inhibition of inflammation by QUE was regulated via MAPK signaling, the effect of QUE on the phosphorylation of ERK, p38 and JNK in LPS-stimulated RAW264.7 cells was investigated. It significantly suppressed the phosphorylation of ERK in a dose-dependent manner, but did not affect the phosphorylation of p38 and JNK.
In the present study, QUE was isolated from CUP and its anti-inflammatory effects via the inhibition of NF-κB and ERK signaling pathways were demonstrated in RAW264.7 macrophage cells. These findings suggested that QUE may be a potential anti-inflammatory agent for the treatment of inflammatory diseases.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This study was supported by Gachon University Gil Medical Center (grant no. FRD2017-10-02) and the Korea Research Foundation (grant no. NRF-2019R1A2C2089867).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
ESS made substantial contributions to conception and design, analysis and interpretation of data, and drafting of the manuscript. JWP contributed to data acquisition and interpretation, drafted the manuscript and revised it critically for important intellectual input. WH, OCK and JYN made substantial contributions to data acquisition and interpretation, and the extraction and isolation of quercetogetin. HRP and SHK made substantial contributions to design, and data analysis and interpretation, and revised the manuscript critically for important intellectual input. SHJ interpreted the data, drafted the manuscript and revised it critically for important intellectual input. CSL supervised the manuscript, and made substantial contributions to conception and design, and data analysis and interpretation. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gori AM, Cesari F, Marcucci R, Giusti B, Paniccia R, Antonucci E, Gensini GF, Abbate R. The balance between pro- and anti-inflammatory cytokines is associated with platelet aggregability in acute coronary syndrome patients. Atherosclerosis. 2009;202:255–262. doi: 10.1016/j.atherosclerosis.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Fujiwara N, Kobayashi K. Macrophages in inflammation. Curr Drug Targets Inflamm Allergy. 2005;4:281–286. doi: 10.2174/1568010054022024. [DOI] [PubMed] [Google Scholar]
- 3.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 4.Fujihara M, Muroi M, Tanamoto K, Suzuki T, Azuma H, Ikeda H. Molecular mechanisms of macrophage activation and deactivation by lipopolysaccharide: Roles of the receptor complex. Pharmacol Ther. 2003;100:171–194. doi: 10.1016/j.pharmthera.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Kim YH, Lee SH, Lee JY, Choi SW, Park JW, Kwon TK. Triptolide inhibits murine-inducible nitric oxide synthase expression by down-regulating lipopolysaccharide-induced activity of nuclear factor-kappa B and c-Jun NH2-terminal kinase. Eur J Pharmacol. 2004;494:1–9. doi: 10.1016/j.ejphar.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 6.Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354:610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- 7.Kiemer AK, Hartung T, Huber C, Vollmar AM. Phyllanthus amarus has anti-inflammatory potential by inhibition of iNOS, COX-2, and cytokines via the NF-kappaB pathway. J Hepatol. 2003;38:289–297. doi: 10.1016/S0168-8278(02)00417-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhai XT, Zhang ZY, Jiang CH, Chen JQ, Ye JQ, Jia XB, Yang Y, Ni Q, Wang SX, Song J, et al. Nauclea officinalis inhibits inflammation in LPS-mediated RAW 264.7 macrophages by suppressing the NF-κB signaling pathway. J Ethnopharmacol. 2016;183:159–165. doi: 10.1016/j.jep.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 9.He J, Lu X, Wei T, Dong Y, Cai Z, Tang L, Liu M. Asperuloside and asperulosidic acid exert an anti-inflammatory effect via suppression of the NF-κB and MAPK signaling pathways in LPS-induced RAW 264.7 Macrophages Int J Mol Sci. 2018;19 doi: 10.3390/ijms19072027. 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Havsteen B. Flavonoids, a class of natural products of high pharmacological potency. Biochem Pharmacol. 1983;32:1141–1148. doi: 10.1016/0006-2952(83)90262-9. [DOI] [PubMed] [Google Scholar]
- 11.Middleton E, Jr, Kandaswami C, Theoharides TC. The effects of plant flavonoids on mammalian cells: Implications for inflammation, heart disease, and cancer. Pharmacol Rev. 2000;52:673–751. [PubMed] [Google Scholar]
- 12.Kim HP, Son KH, Chang HW, Kang SS. Anti-inflammatory plant flavonoids and cellular action mechanisms. J Pharmacol Sci. 2004;96:229–245. doi: 10.1254/jphs.CRJ04003X. [DOI] [PubMed] [Google Scholar]
- 13.Gil B, Sanz MJ, Terencio MC, Ferrándiz ML, Bustos G, Payá M, Gunasegaran R, Alcaraz MJ. Effects of flavonoids on Naja naja and human recombinant synovial phospholipases A2 and inflammatory responses in mice. Life Sci. 1994;54:PL333–PL338. doi: 10.1016/0024-3205(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 14.Manthey DE, Teichman J. Nephrolithiasis. Emerg Med Clin North Am. 2001;19(viii):633–654. doi: 10.1016/S0733-8627(05)70207-8. [DOI] [PubMed] [Google Scholar]
- 15.Wu YQ, Zhou CH, Tao J, Li SN. Antagonistic effects of nobiletin, a polymethoxyflavonoid, on eosinophilic airway inflammation of asthmatic rats and relevant mechanisms. Life Sci. 2006;78:2689–2696. doi: 10.1016/j.lfs.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 16.Xu JJ, Liu Z, Tang W, Wang GC, Chung HY, Liu QY, Zhuang L, Li MM, Li YL. Tangeretin from citrus reticulate inhibits respiratory syncytial virus replication and associated inflammation in vivo. J Agric Food Chem. 2015;63:9520–9527. doi: 10.1021/acs.jafc.5b03482. [DOI] [PubMed] [Google Scholar]
- 17.Kanes K, Tisserat B, Berhow M, Vandercook C. Phenolic composition of various tissues of Rutaceae species. Phytochemistry. 1993;32:967–974. doi: 10.1016/0031-9422(93)85237-L. [DOI] [Google Scholar]
- 18.Rouseff RL, Ting SV. Quantitation of polymethoxylated flavones in orange juice by high-performance liquid chromatography. J Chromatogr A. 1979;176:75–87. doi: 10.1016/S0021-9673(00)92088-0. [DOI] [PubMed] [Google Scholar]
- 19.Li S, Lo CY, Ho CT. Hydroxylated polymethoxyflavones and methylated flavonoids in sweet orange (Citrus sinensis) peel. J Agric Food Chem. 2006;54:4176–4185. doi: 10.1021/jf060234n. [DOI] [PubMed] [Google Scholar]
- 20.Itoh T, Ohguchi K, Iinuma M, Nozawa Y, Akao Y. Inhibitory effects of polymethoxy flavones isolated from Citrus reticulate on degranulation in rat basophilic leukemia RBL-2H3: Enhanced inhibition by their combination. Bioorg Med Chem. 2008;16:7592–7598. doi: 10.1016/j.bmc.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 21.Akao Y, Itoh T, Ohguchi K, Iinuma M, Nozawa Y. Interactive effects of polymethoxy flavones from Citrus on cell growth inhibition in human neuroblastoma SH-SY5Y cells. Bioorg Med Chem. 2008;16:2803–2810. doi: 10.1016/j.bmc.2008.01.058. [DOI] [PubMed] [Google Scholar]
- 22.Ko HC, Jang MG, Kang CH, Lee NH, Kang SI, Lee SR, Park DB, Kim SJ. Preparation of a polymethoxyflavone rich fraction (PRF) of Citrus sunki Hort. ex Tanaka and its antiproliferative effects. Food Chem. 2010;123:484–488. doi: 10.1016/j.foodchem.2010.04.028. [DOI] [Google Scholar]
- 23.Guo S, Qiu P, Xu G, Wu X, Dong P, Yang G, Zheng J, McClements DJ, Xiao H. Synergistic anti-inflammatory effects of nobiletin and sulforaphane in lipopolysaccharide-stimulated RAW 264.7 cells. J Agric Food Chem. 2012;60:2157–2164. doi: 10.1021/jf300129t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harasstani OA, Moin S, Tham CL, Liew CY, Ismail N, Rajajendram R, Harith HH, Zakaria ZA, Mohamad AS, Sulaiman MR, et al. Flavonoid combinations cause synergistic inhibition of proinflammatory mediator secretion from lipopolysaccharide-induced RAW 264.7 cells. Inflamm Res. 2010;59:711–721. doi: 10.1007/s00011-010-0182-8. [DOI] [PubMed] [Google Scholar]
- 25.Fan GW, Gao XM, Wang H, Zhu Y, Zhang J, Hu LM, Su YF, Kang LY, Zhang BL. The anti-inflammatory activities of Tanshinone IIA, an active component of TCM, are mediated by estrogen receptor activation and inhibition of iNOS. J Steroid Biochem Mol Biol. 2009;113:275–280. doi: 10.1016/j.jsbmb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Feng CG, Kullberg MC, Jankovic D, Cheever AW, Caspar P, Coffman RL, Sher A. Transgenic mice expressing human interleukin-10 in the antigen-presenting cell compartment show increased susceptibility to infection with Mycobacterium avium associated with decreased macrophage effector function and apoptosis. Infect Immun. 2002;70:6672–6679. doi: 10.1128/IAI.70.12.6672-6679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao KM, Meighan T, Bowman L. Role of mitogen-activated protein kinase activation in the production of inflammatory mediators: Differences between primary rat alveolar macrophages and macrophage cell lines. J Toxicol Environ Health A. 2002;65:757–768. doi: 10.1080/00984100290071027. [DOI] [PubMed] [Google Scholar]
- 28.Nogata Y, Sakamoto K, Shiratsuchi H, Ishii T, Yano M, Ohta H. Flavonoid composition of fruit tissues of citrus species. Biosci Biotechnol Biochem. 2006;70:178–192. doi: 10.1271/bbb.70.178. [DOI] [PubMed] [Google Scholar]
- 29.Lai CS, Li S, Chai CY, Lo CY, Ho CT, Wang YJ, Pan MH. Inhibitory effect of citrus 5-hydroxy-3,6,7,8,3′,4′-hexamethoxyflavone on 12-O-tetradecanoylphorbol 13-acetate-induced skin inflammation and tumor promotion in mice. Carcinogenesis. 2007;28:2581–2588. doi: 10.1093/carcin/bgm231. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Sang S, Pan MH, Lai CS, Lo CY, Yang CS, Ho CT. Anti-inflammatory property of the urinary metabolites of nobiletin in mouse. Bioorg Med Chem Lett. 2007;17:5177–5181. doi: 10.1016/j.bmcl.2007.06.096. [DOI] [PubMed] [Google Scholar]
- 31.Pan MH, Lai YS, Lai CS, Wang YJ, Li S, Lo CY, Dushenkov S, Ho CT. 5-Hydroxy-3,6,7,8,3′,4′-hexamethoxyflavone induces apoptosis through reactive oxygen species production, growth arrest and DNA damage-inducible gene 153 expression, and caspase activation in human leukemia cells. J Agric Food Chem. 2007;55:5081–5091. doi: 10.1021/jf070068z. [DOI] [PubMed] [Google Scholar]
- 32.Son ES, Kim SH, Ryter SW, Yeo EJ, Kyung SY, Kim YJ, Jeong SH, Lee CS, Park JW. Quercetogetin protects against cigarette smoke extract-induced apoptosis in epithelial cells by inhibiting mitophagy. Toxicol In Vitro. 2018;48:170–178. doi: 10.1016/j.tiv.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Palmer RM, Ashton DS, Moncada S. Vascular endothelial cells synthesize nitric oxide from L-arginine. Nature. 1988;333:664–666. doi: 10.1038/333664a0. [DOI] [PubMed] [Google Scholar]
- 34.Griswold DE, Adams JL. Constitutive cyclooxygenase (COX-1) and inducible cyclooxygenase (COX-2): Rationale for selective inhibition and progress to date. Med Res Rev. 1996;16:181–206. doi: 10.1002/(SICI)1098-1128(199603)16:2<181::AID-MED3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 35.Karin M, Greten FR. NF-κB: Linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 36.Viatour P, Merville MP, Bours V, Chariot A. Phosphorylation of NF-κB and IκB proteins: Implications in cancer and inflammation. Trends Biochem Sci. 2005;30:43–52. doi: 10.1016/j.tibs.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Gong Q, Guo W, Kan X, Xu D, Ma H, Fu S, Liu J. Farrerol relieve lipopolysaccharide (LPS)-induced mastitis by inhibiting AKT/NF-κB p65, ERK1/2 and P38 signaling pathway. Int J Mol Sci. 2018;19:1770. doi: 10.3390/ijms19061770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arthur JSC, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 39.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 40.Uto T, Fujii M, Hou DX. 6-(Methylsulfinyl)hexyl isothiocyanate suppresses inducible nitric oxide synthase expression through the inhibition of Janus kinase 2-mediated JNK pathway in lipopolysaccharide-activated murine macrophages. Biochem Pharmacol. 2005;70:1211–1221. doi: 10.1016/j.bcp.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 41.Yoon WJ, Moon JY, Song G, Lee YK, Han MS, Lee JS, Ihm BS, Lee WJ, Lee NH, Hyun CG. Artemisia fukudo essential oil attenuates LPS-induced inflammation by suppressing NF-κB and MAPK activation in RAW 264.7 macrophages. Food Chem Toxicol. 2010;48:1222–1229. doi: 10.1016/j.fct.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 42.Cruz CD, Neto FL, Castro-Lopes J, McMahon SB, Cruz F. Inhibition of ERK phosphorylation decreases nociceptive behaviour in monoarthritic rats. Pain. 2005;116:411–419. doi: 10.1016/j.pain.2005.05.031. [DOI] [PubMed] [Google Scholar]
- 43.Goodridge HS, Harnett W, Liew FY, Harnett MM. Differential regulation of interleukin-12 p40 and p35 induction via Erk mitogen-activated protein kinase-dependent and -independent mechanisms and the implications for bioactive IL-12 and IL-23 responses. Immunology. 2003;109:415–425. doi: 10.1046/j.1365-2567.2003.01689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korb A, Tohidast-Akrad M, Cetin E, Axmann R, Smolen J, Schett G. Differential tissue expression and activation of p38 MAPK alpha, beta, gamma, and delta isoforms in rheumatoid arthritis. Arthritis Rheum. 2006;54:2745–2756. doi: 10.1002/art.22080. [DOI] [PubMed] [Google Scholar]
- 45.Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.