Abstract
Growth arrest-specific 5 (GAS5) is a known tumor suppressor which negatively regulates cell survival and malignancy in several cancer cell types. The present study aimed to establish the correlation between GAS5 and unc-51 like autophagy activating kinase (ULK)1/2, two key regulators of autophagy initiation in breast cancer (BC). To address this, expression levels of these genes were quantitively analyzed in BC clinical samples by performing reverse transcription-quantitative PCR. GAS5 was downregulated in BC clinical samples compared with adjacent samples and was positively correlated with ULK1/2. Detection methods including cell cycle analysis, annexin V-FITC/PI double staining and flow cytometry analysis, Transwell cell invasion assay, transfection and western blotting were used for BC cells. In MCF-7 cells, it was also observed that overexpression of GAS5 upregulated ULK1/2 protein levels without disturbing other autophagy initiation-associated proteins and inhibited cell proliferation, invasion and tumor formation. These effects were reversed by blocking autophagy with 3-methyladenine (3-MA). These results demonstrated that the suppressive effects of overexpressed GAS5 were mediated via autophagy induction, at least in part. Overexpression of GAS5 induced chemoresistance to cisplatin, which was not reversed by 3-MA-mediated inhibition of autophagy, indicating that GAS5 promotes chemosensitivity in an autophagy-independent manner. Collectively, these results indicated that GAS5 contributes to the pathogenesis of BC potentially by promoting autophagy. However, the mechanism by which GAS5 functions as a tumor suppressor in an autophagy-independent manner remains unknown.
Keywords: growth arrest-specific 5, autophagy, breast cancer, unc-51-like autophagy activating kinase 1/2, chemosensitivity
Introduction
Long noncoding RNAs (lncRNAs), which are a major class of transcripts >200 nucleotides in length, drive several important physiological processes, including proliferation, metastasis and apoptosis of tumor cells (1,2). Numerous studies have revealed that lncRNAs have critical functions in the regulation of malignant tumor behavior, including in liver (3), pancreatic (4) and breast cancer (5). Growth arrest-specific 5 (GAS5), located at chromosome 1q25.1, is an antitumor lncRNA that is frequently downregulated in various cancer types and associated with clinicopathological characteristics, including tumour size, staging and invasion (6). In bladder cancer cells, GAS5 was revealed to function as a transcriptional factor promoting the transcriptional activity of enhancer of zeste homolog 2 by physically interacting with E2F transcription factor 4, thus inducing apoptosis (7). In gastric cancer cells, GAS5 blocked cell cycle phases by interacting with Y-box binding protein 1 and thus upregulating p21 expression, which has a demonstrable role in inhibiting tumor growth (8).
Autophagy is a regular physiological process of metabolic degradation, and the process occurs in double-membrane autophagosomes (9). Autophagy serves a critical role in numerous biochemical processes, including pro-survival signaling and pro-apoptotic signaling (10). In several cancer cell types, the modulation of autophagy serves a tumor-promoting or tumor-inhibiting role depending on the different modulators and the specific autophagy pathway (11). During cellular stress, including hypoxia and nutrient deprivation, these factors function as tumor-promoting modulators of autophagy by helping cells overcome these stresses (12,13). For example, in RAS-mutated cancer cells, the basal level of autophagy was revealed to be markedly high to compensate for the loss of RAS-modulated signaling pathways, including pathways related to proliferation, survival and metabolism (14–16). In addition, the basal level of autophagy has also been revealed to serve as a regulatory mechanism for tumor suppression by inducing cellular damage and imbalanced cellular homeostasis (17). Studies have revealed that in several cancer types, including cervical squamous-cell carcinoma (18) and hepatocellular carcinoma (19), Beclin1, which is an important protein for autophagosome formation, is upregulated and thus acts as a tumor suppressor.
Unc-51 like autophagy activating kinase (ULK)1 plays a key role in the regulation of autophagy initiation in an autophagy-related (Atg)5- and Atg7-independent macroautophagic manner (20). The human genome contains two ULK1 homologs, ULK1 and ULK2, which are known to initiate autophagy (21,22). Accumulating evidence has demonstrated that ULK1 tightly regulates tumor progression by modulating autophagy in cancer cells. For example, Blessing et al (23) reported that increased autophagy upregulated ULK1 expression, further promoting autophagy in prostate cancer. In glioma, ULK1 and ULK2 were revealed to be methylated and silenced; subsequently, ULK1 and/or ULK2 failed to induce autophagy, which resulted in promotion of proliferation and tumor progression, indicating that downregulation of ULK1/2 is essential for glioma development (24). These results indicated that ULK1/2 may tightly regulate tumor progression by initiating autophagy.
In the present study, the expression levels of GAS5, ULK1 and ULK2 were measured in breast cancer (BC) clinical samples and adjacent samples, also aiming to analyze any correlation between these proteins. Additionally, the present study investigated the possible molecular mechanism of GAS5 involved in the regulation of autophagy in breast cancer cells. The results should increase understanding of the relationship between GAS5 and autophagy in BC cells, and provide new insights into the impact of GAS5 on the chemoresistance to cisplatin, which may facilitate the development of more effective clinical treatments for BC.
Materials and methods
Clinical samples
Paired human breast cancer clinical samples and adjacent normal samples used in the present study were obtained from 39 female patients, aged from 30 to 82 (median age, 52.9) at the Affiliated Hospital of Southwest Medical University (Luzhou, China) between December 2018 and March 2019. The present study was approved by The Medical Ethics Committee of the Institutional Review Board of the Affiliated Hospital of Southwest Medical University (Luzhou, China). Written informed consent was provided by all patients. All types of breast cancer were invasive ductal carcinoma as confirmed by pathologists. Inclusion criteria were histological diagnosis of invasive ductal carcinoma of the breast before operation. None of the patients received chemotherapy or radiotherapy prior to surgical excision Detailed clinicopathological information were obtained, including sex, age, tumor depth, tumor size, differentiation, lymph node invasion and distant metastasis (data not shown).
Cell cultures
Three human cell lines, including breast cancer cell lines MCF-7 and MDA-MB-231, and breast non-tumor cell line MCF-10a, were used in the present study. All cell lines were frozen in liquid nitrogen and cultured in MEM supplemented with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc.), 100 µ/ml penicillin and 100 µ/ml streptomycin (Amresco, LLC) in a 5% CO2 incubator at 37°C. Cells were passaged once every three days. To inhibit autophagy, 2 mM 3-methyladenine (3-MA, Sigma-Aldrich; Merck KGaA) was added into culturing medium for 24 h.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from BC clinical samples and adjacent normal samples using TRIzol® (Thermo Fisher Scientific, Inc). A total of 5 mg of each sample was suspended in 1 ml RNA EasyOut kit (Chengdu Daosheng Biological Technology Co., Ltd.) and sonicated at room temperature for 5 sec using a SoniConvert™ system (Chengdu Daosheng Biological Technology Co., Ltd.), both according to the manufacturer's instructions. For RT-qPCR analysis, 1 µg total RNA was reverse-transcribed into cDNA using an Superscript III RT-qPCR kit (Thermo Fisher Scientific, Inc.). Then the cDNA was used as template under the following conditions: 40 cycles at 95°C for 30 sec and 60°C for 1 min. An ABI 7500 Fast qPCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for qPCR. For amplification, the following procedure was performed: Predenaturation at 98°C for 2 min for each cycle, denaturation at 98°C for 10 sec, annealing and extension at 60°C for 60 sec, 40 cycles; final extension at 60°C for 5 min. The primers used for qPCR were as follows: GAS5 forward, 5′-GAGCAAGCCTAACTCAAGCC-3′ and reverse, 5′-ACACAGTGTAGTCAAGCCGA-3′; ULK1 forward, 5′-GGCAAGTTCGAGTTCTCCCG-3′ and reverse, 5′-CGACCTCCAAATCGTGCTTCT-3′; ULK2 forward, 5′-TGGAGACCTCGCAGATTATTTGC-3′ and reverse, 5′-CTGTGCAGGATTCGCATGG-3′; β-actin forward, 5′-CATGTACGTTGCTATCCAGGC-3′ and reverse, 5′-CTCCTTAATGTCACGCACGAT-3′. The qPCR results were analyzed and expressed relative to the Cq (threshold cycle) values (25) and then converted to fold change values. A fold-change of 2.0 was considered significant.
Western blotting
Cells (1×106) were suspended in 1 ml of RIPA buffer (Sigma-Aldrich; Merck KGaA) and lysed using SoniConvert® Sonicator (Chengdu Daosheng Biological Technology Co., Ltd.). Protein concentration was determined via BCA assay (Sigma-Aldrich; Merck KGaA) according to the manufacturer's instructions. In each lane, 20 µg total protein was applied to a 5–15% SDS-PAGE gel. Fractionated proteins were transferred onto a nitrocellulose membrane and blocked using 5% BSA containing PBS at room temperature for 1 h. Membranes were then incubated with primary antibodies overnight at 4°C. The primary antibodies used were obtained from Abcam and are listed as follows: Rabbit monoclonal anti-ULK1 antibody (1:2,000; cat. no. ab133766), rabbit polyclonal anti-ULK2 antibody (1:3,000; cat. no. ab97695), rabbit monoclonal anti-Atg3 antibody (1:500; cat. no. ab108251), rabbit polyclonal anti-Atg5 antibody (1:2,000; cat. no. ab228668), rabbit monoclonal anti-Atg7 antibody (1:2,000; cat. no. ab52472), rabbit monoclonal anti-Beclin 1 antibody (1:2,000; cat. no. ab207612), rabbit polyclonal anti-LC3B antibody (1:500; cat. no. ab48394) and mouse monoclonal anti-β-actin antibody (1:5,000; cat. no. ab8226). Following three washes with TBST, secondary antibody was incubated with the membrane at room temperature for 1 h. Goat anti-rabbit IgG H&L antibody (HRP-labelled; 1:10,000; cat. no. ab7090) and goat anti-mouse IgG H&L antibody (HRP-labelled; 1:10,000; cat. no. ab97040) were used as the secondary antibodies. Blot bands were quantified via densitometry using ImageJ software (v2.0; National Institutes of Health). β-actin was used as an internal reference.
Tandem mRFP/mCherry-GFP fluorescence microscopy
MCF-7 cells were seeded at 2×105 cells/well in a 12-well plate and attached overnight at 37°C in a 5% CO2 incubator. Cells were transfected using pre-made lentiviral particles expressing a fusion target of GFP-RFP-LC3 (cat. no. GM-1314L204H-S, Genomeditech Co., Ltd.) according to the manufacturer's instructions. After 2 h incubation at 37°C in the dark, the medium was refreshed and cells were incubated in MEM supplemented with 10% FBS at 37°C for 48 h. Cells were visualized under an X71 (U-RFL-T) fluorescence microscope (magnification, ×100; Olympus Corporation) and analyzed with ImageJ software (v2.0; National Institutes of Health).
Double staining of lysotracker green and mitotracker sox red
MCF-7 cells were seeded at 2×105 cells/well in a 12-well plate and incubated with Lysotracker green and mitotracker sox red according to the manufacturer's instructions. After 1 h incubation at 37°C, 1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI) was used to stain nuclei for 5 min at room temperature in the dark. Cellular fluorescence was observed using an X71 (U-RFL-T) fluorescence microscope (magnification, ×100; Olympus Corporation).
Cell Counting Kit (CCK)-8 assay for proliferation
A total of 5×103 cells/well were seeded in 96-well plates and allowed to attach overnight. After addition of 1, 2, 3, 4 and 5 µmol/L of cisplatin (Thermo Fisher Scientific, Inc.), for 1–5 days, the cell proliferation assay was performed by adding 10 µl CCK-8 solution (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's protocol. After 1 h incubation at 37°C, the absorbance of each well was measured at a wavelength of 450–620 nm using a Multiskan spectrum microplate reader (Thermo Fisher Scientific, Inc.).
Cell cycle analysis
A total of 1×106 MCF-7 cells were collected and washed with ice-cold PBS twice, and fixed using 1 ml 70% ethanol and stored at 4°C overnight. After washing with ice-cold PBS three times, the staining solution containing 100 ng/ml RNase A and 5 µg/ml propidium iodide (PI) (Sigma-Aldrich; Merck KGaA) was added to cells and incubated in the dark for 30 min at 4°C. Cells were the screened by a three laser Navios flow cytometer (Beckman Coulter, Inc.) and analyzed by ModFit LT software (v2.0, Verity Software House).
Annexin V-FITC/PI double staining and flow cytometric analysis
MCF-7 Cells were treated with 2 µM cisplatin for 24 h, and untreated cells were considered as control group. Cells were resuspended in trypsin and washed twice with ice-cold PBS. Then, Annexin V-FITC/PI double staining was performed according to the instructions of the Annexin V-FITC/PI apoptosis detection kit (Thermo Fisher Scientific, Inc.). For each sample, ~2×104 cells were collected and scanned by a three laser Navios flow cytometer (Beckman Coulter, Inc.). Data files was analyzed using FlowJo v9.7.6 (FlowJo LLC).
Transwell cell invasion assay
Transwell membranes were precoated with 60 µl Matrigel (8%) in MEM and incubated at room temperature for 2 h. For each well, 1×104 cells were added into upper chamber. Subsequently, 600 µl of MEM supplemented with 10% FBS was added to the lower chamber. After 24 h, medium was removed and 4% paraformaldehyde was used for fixation at room temperature for 10 min. After washing with PBS twice, cells were stained with 0.1% crystal violet (in PBS) at room temperature for 30 min. After staining, the crystal violet solution was removed and the cells were washed with PBS five times. Then the cells in five random views were counted under an X71 fluorescence microscope (magnification, ×100; Olympus Corporation).
Transfection
A total of 0.8 µg GAS5-expressing plasmid or empty vector (Addgene, Inc.) was mixed with 4 µl Lipofectamine™ 2000 transfection reagent (Thermo Fisher Scientific, Inc.) in 0.5 ml OptiMEM medium (Thermo Fisher Scientific, Inc.), and this was added to MCF-7 cells for 4 h followed by replacing the medium. After 24 h, the culture media was refreshed containing 500 µg/ml geneticin sulfate 418 (G418, Sigma-Aldrich; Merck KGaA) for antibiotic selection. Two weeks later, the transfected cells that had survived were collected and maintained in 4,000 µg/ml G418. Individual clones (GAS5 clone-1 and clone-2) were detected, separately.
Tumor formation in soft agar
Low-melting agarose (2 ml 0.6%, Sigma-Aldrich; Merck KGaA) diluted in MEM supplemented with 10% FBS was added to 6-well plates. For each well, a total of 5×103 cells were added in 1 ml of 0.3% (weight/volume) low-melting agarose diluted in MEM supplemented with 10% FBS. After 2 weeks of incubation at 37°C, cells were stained with 10 ng/ml nitro blue tetrazolium at 37°C and colony number and diameter were quantified under an X71 fluorescence microscope (magnification, ×100; Olympus Corporation).
Statistical analysis
Statistical analyses were carried out using SPSS v19.0 for Windows (IMB Corp). Student's paired t-tests and ANOVA were used to compare two or more groups for statistical analysis followed by Tukey's post hoc test, respectively. Pearson's correlation analysis was used for the correlation between GAS5, ULK1 and ULK2 levels. Data are presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference. All experiments were repeated at least three times.
Results
GAS5 is downregulated in BC clinical samples and BC cell lines
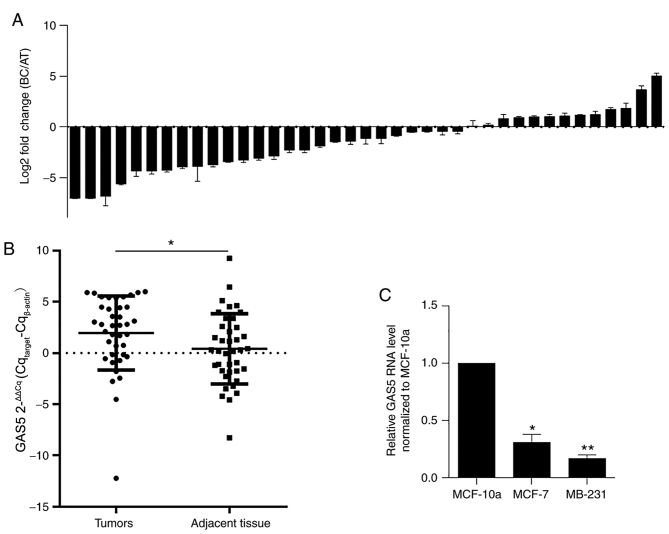
The present study detected the expression levels of GAS5 in paired BC clinical samples and adjacent normal tissues by performing RT-qPCR. As revealed in Fig. 1A, among the 39 cases of BC, ~66.7% exhibited lower levels of GAS5 compared with matched controls, and GAS5 was significantly downregulated in BC tissue samples compared with those in adjacent normal tissues (1.95±0.36 vs. 0.40±0.22, P=0.002, Fig. 1B). To confirm whether similar expression patterns of GAS5 were presented in BC cell lines, GAS5 expression levels were analyzed in MCF-7 and MDA-MB-231 cells compared with the non-tumor cell line MCF-10a. As anticipated, the expression levels of GAS5 in both MCF-7 and MDA-MB-231 was significantly lower compared with those in MCF-10a cells (*P=0.037; **P 0.006, Fig. 1C), indicating that GAS5 may exert antitumor effects in BC.
Figure 1.
Expression levels of GAS5 in 39 cases of BC clinical samples. (A) Using reverse transcription quantitative PCR analysis, the expression levels of GAS5 in BC clinical samples and adjacent normal samples were assessed. (B) Reduced expression of GAS5 in BC clinical samples was compared with adjacent samples (the expression ratio of breast cancer to adjacent tissue, BC/adjacent tissues). *P<0.05 vs. adjacent tissues. (C) Relative GAS5 levels in MCF-7 and MDA-MB-231 cells were measured and normalized to the non-tumor cell line, MCF-10a. *P<0.05, **P<0.01, vs. MCF-10a. GAS5, growth arrest-specific 5; BC, breast cancer.
Expression levels of GAS5, ULK1 and ULK2 are significantly correlated in BC clinical samples
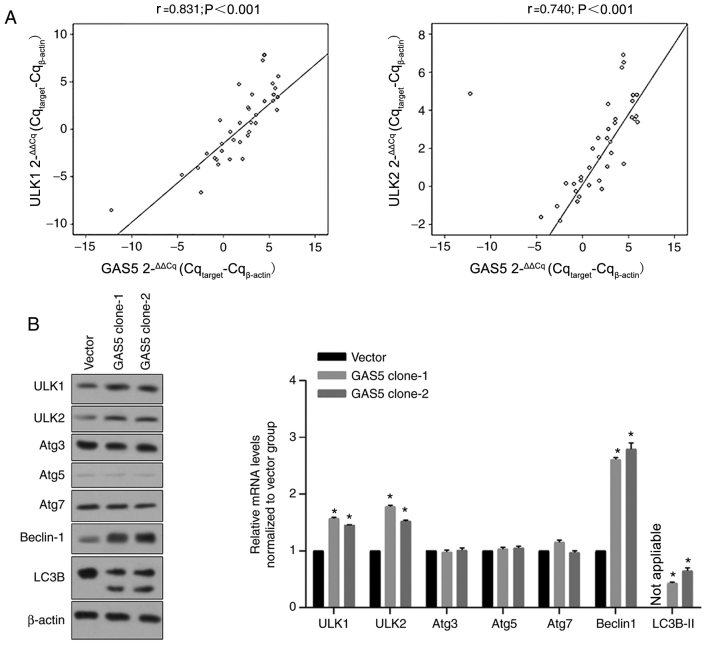
The present study also assessed the expression levels of ULK1 and ULK2 in BC clinical samples and adjacent samples. Results revealed that GAS5 expression in BC clinical samples was positively and significantly correlated with ULK1 and ULK2 levels (Fig. 2A). To confirm whether the expression levels of GAS5 positively regulated ULK1, ULK2 and other autophagy-associated proteins, including AuTophaGy-related (Atg)3, Atg5, Atg7, Beclin-1 and microtubule-associated proteins 1A/1B light chain (LC)3B, cells stably overexpressing GAS5 were selected and analyzed for the expression levels of the aforementioned proteins. As revealed in Fig. 2B, ULK1 and ULK2 protein levels were both significantly upregulated in GAS5 clone-1 and clone-2 (*P<0.05), which was consistent with the tendency observed in clinical samples. In addition, GAS5 overexpression induced an increase in Beclin-1 and LC3B levels with no effect on Atg3, Atg5 and Atg7, indicating that GAS5 potentially initiates autophagy by upregulating ULK1 and ULK2, but not Atgs (data not shown).
Figure 2.
Correlation between GAS5, ULK1 and ULK2 levels. (A) Interactions between GAS5, ULK1 and ULK2 levels were assessed using Pearson's correlation analysis. (B) After stable expression of GAS5 in MCF-7 cells, the autophagic proteins, including ULK1, ULK2, Atg3, Atg5, Atg7, Beclin-1 and LC3B, were analyzed using western blotting (left panel). The bands were analyzed and normalized to the Vector group. *P<0.05, vs. the Vector group. GAS5, growth arrest-specific 5; ULK, unc-51-like autophagy activating kinase; Atg, autophagy-related; LC3B, microtubule-associated proteins 1A/1B light chain 3B.
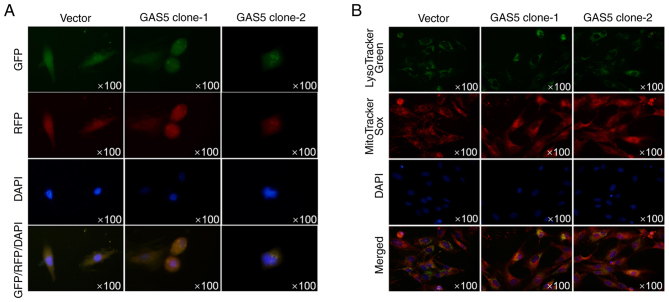
To further confirm whether autophagosome formation was promoted by overexpressed GAS5, images of MCF-7 cells stably overexpressing GAS5 and transiently expressed LC3-RFP-GFP were captured using confocal microscopy to view the autophagosomes. As revealed in Fig. 3A, without autophagy promoter treatment, overexpression of GAS5 promoted the formation of autophagosomes. Double staining of lysotracker green and mitotracker sox red also demonstrated that overexpression of GAS5 promoted colocalization of mitochondria and lysosomes, indicating the promoting effect of GAS5 on autophagy/mitophagy (Fig. 3B).
Figure 3.
Observation of autophagosomes in GAS5-overexpressing MCF-7 cells. (A) After introduction of LC3B-GFP-RFP, GAS5 localization was analyzed. (B) GAS5-overexpressing cells were double stained with lysotracker green and mitotracker sox red. GAS5, growth arrest-specific 5; LC3B, microtubule-associated proteins 1A/1B light chain 3B.
GAS5 exerts antitumor effects partially by inducing autophagy in MCF-7 cells
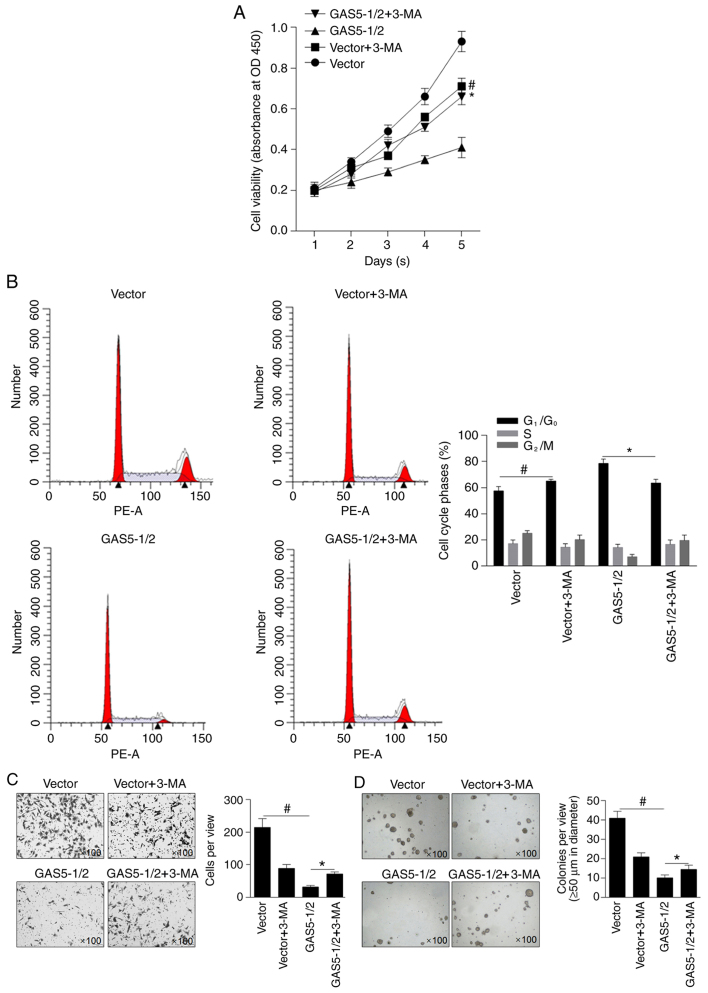
To evaluate the effects of overexpressed GAS5 on the malignant behaviors of MCF-7 cells, cell viability was analyzed. As revealed in Fig. 4A, mixed clone-1 and clone-2 (GAS5-1/2) inhibited cell viability compared with the vector-transfected clone (P<0.05). Pre-treatment of 10 µM 3-MA was performed to demonstrate whether autophagy induced by GAS5 affected cell viability. 3-MA treatment notably decreased cell viability in the vector group, indicating the inhibitory effect of 3-MA on proliferation alone, and this inhibitory effect on cell viability was reversed by GAS5 overexpression (Fig. 4A). By performing cell cycle analysis, it was observed that GAS5-induced block of the cells at G1/G0 was reversed by 3-MA treatment. The effects of GAS5 on invasion and colony formation in MCF-7 cells was then investigated, and results demonstrated that invasion (Fig. 4C) and colony formation (Fig. 4D) of MCF-7 cells were significantly inhibited by GAS5 overexpression compared with vector group and partially reversed by inhibiting autophagy compared with GAS5-1/2 group, indicating that GAS5 may exert antitumor effects by partially inducing autophagy in MCF-7 cells.
Figure 4.
Effects of GAS5 on malignant behaviors in MCF-7 cells. (A) Cell viability of MCF-7 was analyzed using a CCK-8 assay. (B) After PI staining, cells were analyzed using flow cytometry. (C) A Transwell assay was performed to detect invasive ability. (D) Tumor formation in soft agar was assessed. *P<0.05 vs. the GAS5-1/2+3-MA group, #P<0.05 vs. Vector group. GAS5, growth arrest-specific 5; 3-MA, 3-methyladenine.
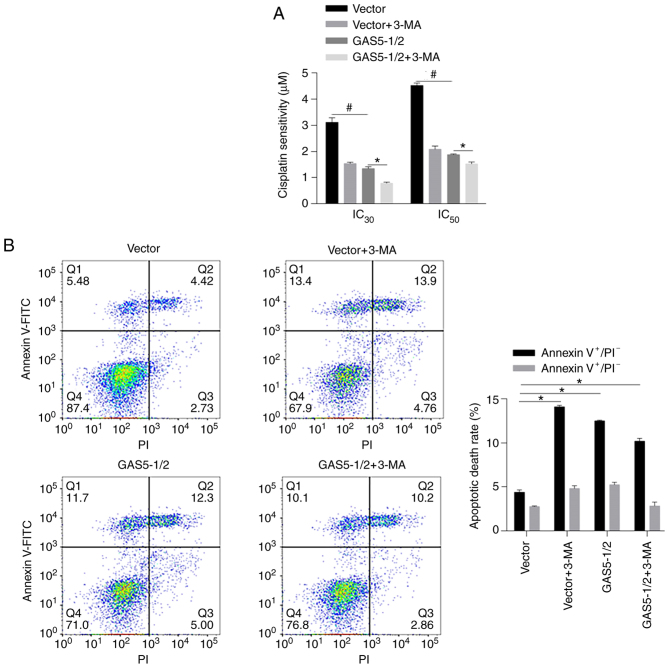
GAS5 sensitizes MCF-7 cells to chemotreatment in an autophagy-independent manner
Since autophagy is a critical physiological process for chemosensitivity in cancer cells, it was then evaluated whether GAS5 affects chemosensitivity by regulating autophagy in MCF-7 cells. By performing chemosensitivity on cisplatin, as anticipated, both 3-MA treatment and overexpression of GAS5 desensitized MCF-7 cells to cisplatin (Fig. 5A, *P<0.05, vs. GAS5-1/2 group; #P<0.05, vs. Vector group). However, 3-MA treatment decreased chemosensitivity in MCF-7 cells overexpressing GAS5 instead of reversing the promoting effect of GAS5 on chemosensitivity. To analyze the apoptotic rate induced by cisplatin, Annexin V-FITC/PI double staining was performed after 24-h treatment with 2 µM cisplatin. Autophagy inhibition significantly increased apoptotic cell death compared with mock group, and GAS5-promoted apoptosis was not notably affected by autophagy inhibition, indicating that the effect of GAS5 on chemosensitivity may be autophagy-independent (Fig. 5B).
Figure 5.
Effect of GAS5 on chemosensitivity in MCF-7 cells. (A) CCK-8 assay was performed to detect chemosensitivity in MCF-7 after GAS5 overexpression. (B) After 24-h treatment of 2 µM Cisplatin, the apoptotic rate of cells was analyzed using flow cytometry after Annexin V-FITC and PI double staining. *P<0.05 vs. the GAS5-1/2+3-MA group, #P<0.05 vs. Vector group. GAS5, growth arrest-specific 5; 3-MA, 3-methyladenine; IC, inhibitory concentration; Annexin V+/PI-, early apoptotic cells; Annexin V+/PI+, late apoptotic cells.
Discussion
The present study is the first to demonstrate the positive correlation between GAS5 and ULK1/2 levels in BC pathogenesis, to the best of the authors' knowledge. GAS5 is not only expressed in multiple cancers including breast cancer, but also widely expressed in a variety of normal tissues, such as lung, esophagus, colon, ovary and kidney (26). GAS5 is involved in the regulation of physiological processes; for example, GAS5 transcriptionally regulates mTOR and thus activates downstream signaling, which promotes cell proliferation by promoting entry into the G2/M phase of the cell cycle (27). GAS5 is also well known as a tumor suppressor in various cancer types, including gastric (8), lung (2), pancreatic (14) and BC (7). In these cancer types, GAS5 was revealed to be transcriptionally downregulated, and GAS5 upregulation inhibited malignant behaviors, including proliferation, migration, invasion and tumor formation. Given these reports, several studies investigating GAS5 have aimed to increase cellular GAS5 levels via specific methods to control the proliferation of cancer cells (26,7,14). In the present study, it was reported that the expression levels of GAS5 were significantly downregulated in BC clinical samples compared to those in adjacent normal samples, and the expression of GAS5 was positively correlated with the expression levels of ULK1 and ULK2, which are key initiators of autophagy (20–22). Further experiments also revealed that GAS5 exerted suppressive effects on the malignant behaviors of BC cells, including proliferation, invasion and tumor formation, partially by inducing autophagy. However, one limitation of the present study is that no triple-negative breast cancer tissues were collected; thus, the functions of GAS5 in these tissues is still unknown and further studies should focus on the effect of GAS5 in triple-negative breast cancer.
lncRNAs are also involved in autophagy regulation, including autophagy initiation (28), phagophore nucleation (29), autophagosome elongation/closure (30) and autolysosome fusion (31). Several types of lncRNAs are involved in these processes, including GAS5 (32). Song et al (32), reported that in osteoarthritis, GAS5 was overexpressed and thus stimulated apoptosis and suppressed autophagic responses, which was determined by the detection of downregulated Beclin 1, Atg7, Atg12 and LC3B. In the present study, GAS5 was positively correlated with the expression levels of ULK1/2 in BC clinical samples, but no correlation was observed between GAS5 and with Atgs (data not shown). It was also demonstrated that overexpression of GAS5 in MCF-7 cells caused upregulation of ULK1 and ULK2, but it was not determined whether ULK1/2 levels were regulated directly or indirectly. Moreover, GAS5 overexpression did not affect Atg3, Atg5 or Atg7 levels. This may be due to the different expression patterns of microRNA-21, which is suppressed by GAS5 and regulates Atg expression (32). As a result of overexpressed GAS5, autophagy was promoted via increased levels of LC3B and Beclin-1 and the formation of autophagosomes. It is still unclear how GAS5 affects the expression of ULK1 and ULK2; however, the present study demonstrated the promoting effect of GAS5 on autophagy, and GAS5 exerted antitumor effects by promoting autophagy, at least in part.
lncRNA-induced autophagy is involved in modulating chemosensitivity. Hu et al (33), reported that metastasis-associated lung adenocarcinoma transcript 1, which is widely involved in the regulation of biological and cellular processes, including autophagy, induced chemoresistance by modulating autophagy. In bladder cancer cells, the lncRNA gallbladder cancer drug resistance-associated lncRNA1, which is upregulated in bladder cancer cells, promoted autophagic flux and thus induced chemoresistance (34). In the present study, it was reported that both overexpression of GAS5 and autophagy inhibition by 3-MA pretreatment desensitized MCF-7 cells to cisplatin, and that 3-MA pretreatment failed to disturb GAS5-induced chemoresistance, indicating that GAS5 may induce chemoresistance in an autophagy-independent manner. The results demonstrated that GAS5 may not be a promising indicator for predicting chemosensitivity in BC.
Collectively, the present study may provide a possible mechanism by which GAS5 regulates breast cancer by promoting autophagy and affecting proliferation, invasion and tumor formation.
Acknowledgements
Not applicable.
Funding
The present study was funded by Initiation Fund for Doctors of the Affiliated Hospital of Southwest Medical University (grant no. 18110) and The Undergraduate Training Program for Innovation and Entrepreneurship of the Southwest Medical University (grant no. 2019272)
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
GL and LQ performed cellular and molecular experiments. XT and YC contributed to molecular experiments and were responsible for data collection. ZZ performed cellular experiments, analyzed and interpreted the data. CZ conceived and designed the study. ZZ and CZ performed molecular experiments and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by The Medical Ethics Committee of The Institutional Review Board of the Affiliated Hospital of Southwest Medical University (Luzhou, China; approval no. KY2019040). Written informed consent was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.D'Angelo E, Agostini M. Long non-coding RNA and extracellular matrix: The hidden players in cancer-stroma cross-talk. Noncoding RNA Res. 2018;3:174–177. doi: 10.1016/j.ncrna.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu T, Wang Y, Chen D, Liu J, Jiao W. Potential clinical application of lncRNAs in non-small cell lung cancer. OncoTargets Ther. 2018;11:8045–8052. doi: 10.2147/OTT.S178431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Chen L, Gu J, Zhang H, Yuan J, Lian Q, Lv G, Wang S, Wu Y, Yang YT, et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun. 2017;8:14421. doi: 10.1038/ncomms14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang YT, Xu XH, Yang XD, Hao J, Cao H, Zhu W, Zhang SY, Cao JP. Role of non-coding RNAs in pancreatic cancer: The bane of the microworld. World J Gastroenterol. 2014;20:9405–9417. doi: 10.3748/wjg.v20.i28.9405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Z, Chen C, Liu Y, Wu C. 17β-Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem Biophys Res Commun. 2014;445:388–393. doi: 10.1016/j.bbrc.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Pickard MR, Williams GT. Molecular and Cellular Mechanisms of action of tumour suppressor GAS5 LncRNA. genes (Basel) 2015;6:484–499. doi: 10.3390/genes6030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M, Guo C, Wang L, Luo G, Huang C, Li Y, Liu D, Zeng F, Jiang G, Xiao X. Long noncoding RNA GAS5 promotes bladder cancer cells apoptosis through inhibiting EZH2 transcription. Cell Death Dis. 2018;9:238. doi: 10.1038/s41419-018-0264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang G, Zhang Y. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep. 2015;5:10159. doi: 10.1038/srep10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Wang P, Wan L, Xu S, Pang D. The emergence of noncoding RNAs as Heracles in autophagy. Autophagy. 2017;13:1004–1024. doi: 10.1080/15548627.2017.1312041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky DJ, Kotb A, Akihisa A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Guo YF, Liu YZ, Liu YJ, Xiong DH, Liu XG, Wang L, Yang TL, Lei SF, Guo Y, et al. Pathway-based genome-wide association analysis identified the importance of regulation-of-autophagy pathway for ultradistal radius BMD. J Bone Miner Res. 2010;25:1572–1580. doi: 10.1002/jbmr.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo T, Fu J, Xu A, Su B, Ren Y, Li N, Zhu J, Zhao X, Dai R, Cao J, et al. PSMD10/gankyrin induces autophagy to promote tumor progression through cytoplasmic interaction with ATG7 and nuclear transactivation of ATG7 expression. Autophagy. 2016;12:1355–1371. doi: 10.1080/15548627.2015.1034405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu M, Jiang L, Fu X, Wang W, Ma J, Tian T, Nan K, Liang X. Cytoplasmic liver kinase B1 promotes the growth of human lung adenocarcinoma by enhancing autophagy. Cancer Sci. 2018;109:3055–3067. doi: 10.1111/cas.13746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su C-C. Tanshinone IIA can inhibit MiaPaCa-2 human pancreatic cancer cells by dual blockade of the Ras/Raf/MEK/ERK and PI3K/AKT/mTOR pathways. Oncol Rep. 2018;40:3102–3111. doi: 10.3892/or.2018.6670. [DOI] [PubMed] [Google Scholar]
- 15.Zhu D, Zhou J, Zhao J, Jiang G, Zhang X, Zhang Y, Dong M. ZC3H13 suppresses colorectal cancer proliferation and invasion via inactivating Ras-ERK signaling. J Cell Physiol. 2019;234:8899–8907. doi: 10.1002/jcp.27551. [DOI] [PubMed] [Google Scholar]
- 16.Natan S, Tsarfaty G, Horev J, Haklai R, Kloog Y, Tsarfaty I. Interplay between HGF/SF-Met-Ras signaling, tumor metabolism and blood flow as a potential target for breast cancer therapy. Oncoscience. 2013;1:30–38. doi: 10.18632/oncoscience.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang D, Kang R, Livesey KM, Cheh C-W, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, III, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai M, Hu Z, Liu J, Gao J, Liu C, Liu D, Tan M, Zhang D, Lin B. Beclin 1 expression in ovarian tissues and its effects on ovarian cancer prognosis. Int J Mol Sci. 2014;15:5292–5303. doi: 10.3390/ijms15045292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu DM, Wang GL, Chen L, Xu YY, He S, Cao XL, Qin J, Zhou JM, Zhang YX, Qun E. The expression of beclin-1, an autophagic gene, in hepatocellular carcinoma associated with clinical pathological and prognostic significance. BMC Cancer. 2014;14:327. doi: 10.1186/1471-2407-14-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Corrigendum: Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2016;533:130. doi: 10.1038/nature16538. [DOI] [PubMed] [Google Scholar]
- 21.Ganley IG, Lam H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 23.Blessing AM, Rajapakshe K, Reddy Bollu L, Shi Y, White MA, Pham AH, Lin C, Jonsson P, Cortes CJ, Cheung E, et al. Transcriptional regulation of core autophagy and lysosomal genes by the androgen receptor promotes prostate cancer progression. Autophagy. 2017;13:506–521. doi: 10.1080/15548627.2016.1268300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shukla S, Patric IRP, Patil V, Shwetha SD, Hegde AS, Chandramouli BA, Arivazhagan A, Santosh V, Somasundaram K. Methylation silencing of ULK2, an autophagy gene, is essential for astrocyte transformation and tumor growth. J Biol Chem. 2014;289:22306–22318. doi: 10.1074/jbc.M114.567032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Yu Y, Hann SS. Novel tumor suppressor lncRNA growth arrest-specific 5 (GAS5) in human cancer. OncoTargets Ther. 2019;12:8421–8436. doi: 10.2147/OTT.S221305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirna MM, Hasan AM, Farzin F, Williams Gwyn T. Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5) Pharmacol. 2010;78:19–28. doi: 10.1124/mol.110.064055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunlop EA, Tee AR. mTOR and autophagy: A dynamic relationship governed by nutrients and energy. Semin Cell Dev Biol. 2014;36:121–129. doi: 10.1016/j.semcdb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 29.He C, Beth L. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 31.McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell. 2015;57:39–54. doi: 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Song J, Ahn C, Chun C-H, Jin E-J. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J Orthop Res. 2014;32:1628–1635. doi: 10.1002/jor.22718. [DOI] [PubMed] [Google Scholar]
- 33.Hu YR, Yu YC, You SW, Li KQ, Tong XC, Chen SR, Chen ED, Lin XZ, Chen YY. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174. doi: 10.1186/s12943-017-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai Q, Wang S, Jin L, Weng M, Zhou D, Wang J, Tang Z, Quan Z. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol Cancer. 2019;18:82. doi: 10.1186/s12943-019-1016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.