Abstract
Long non-coding RNA (lncRNA) ZNFX1 antisense RNA 1 (ZFAS1) is upregulated in acute myocardial infarction; however, the role of ZFAS1 in myocardial ischemia/reperfusion (I/R) injury remains unknown. The present study aimed to detect microRNA (miR)-590-3p expression levels in cardiomyocytes subjected to I/R, and to investigate the effects of ZFAS1 on myocardial I/R injury. An in vitro model of I/R injury was established using rat H9c2 cardiomyocytes exposed to hypoxia/reoxygenation (H/R). It was demonstrated that ZFAS1 was upregulated and miR-590-3p was downregulated in the in vitro model of cardiac I/R injury. Western blotting results indicated that the protein expression levels of p50, tumor necrosis factor-α (TNF-α), interleukin (IL)-6, Bax and cleaved caspase-3 were upregulated, and the expression levels of Bcl-2 and pro-caspase-3 were downregulated. Flow cytometry results revealed that downregulation of ZFAS1 reduced H/R-induced apoptosis in H9c2 cells. In addition, downregulation of ZFAS1 significantly increased the expression of miR-590-3p, and p50 was identified as a target gene of miR-590-3p. Furthermore, with 12 h of hypoxia followed by 2 h of reoxygenation in H9c2 cells, ZFAS1 knockdown increased the expression levels of miR-590-3p, Bax and cleaved-caspase-3, and decreased the expression levels of Bcl-2 and pro-caspase-3. It was also found that the miR-590-3p-mimic transfection increased the expression levels of Bax and cleaved-caspase-3, and decreased the protein expression levels of p50, TNF-α, IL-6, Bcl-2 and pro-caspase-3. In addition, TNF-α treatment induced apoptosis of H9c2 cells, and the changes in Bax, Bcl-2, cleaved-caspase-3 and pro-caspase-3 expression levels in a dose-dependent manner. Collectively, the present results suggested that ZFAS1 was upregulated in H9c2 cells subjected to I/R injury, and that ZFAS1 knockdown protected against I/R-induced myocardial cell apoptosis by directly interacting with miR-590-3p, via the NF-κB pathway.
Keywords: long non-coding RNA, ZNFX1 antisense RNA 1, ischemia/reperfusion, microRNA-590-3p
Introduction
Coronary artery disease has one of the highest morbidity and mortality rates of any disease worldwide, and myocardial infarction is the most common coronary artery disease (1,2). Percutaneous coronary intervention (PCI) is the primary effective treatment for myocardial infarction in clinical practice; it can clear narrow lumens and occlude the coronary lumen, but ischemia-reperfusion (I/R) injury is the most important obstacle to PCI treatment (1,2). I/R injury is one of the main mechanisms of arrhythmia, myocardial contractile dysfunction and irreversible damage of cardiomyocytes (3). Increased inflammation induced by myocardial I/R is one of the main causes of myocardial cell apoptosis (4). However, the release of inflammatory mediators also initiates the repair of damaged tissues in the body (5,6). Moreover, inflammatory cytokines can induce cardiomyocyte apoptosis, which further promotes increases in inflammatory cytokine levels (7,8).
Long non-coding RNA (lncRNA) is a type of RNA that is >200 bp in length with no or little open reading frame, which does not encodes a protein (9,10). With the development of gene sequencing, gene chips and genomics, numerous lncRNAs have been revealed to be involved in the regulation of inflammation (9,11). Previous studies had reported that lncRNAs are not only involved in the development of cardiovascular diseases, such as cardiac hypertrophy, myocardial infarction, heart failure and myocardial fibrosis (12,13), but also in inflammation and inflammatory diseases through the regulation of numerous gene expression and signaling pathways (14).
lncRNA ZNFX1 antisense RNA 1 (ZFAS1) is abnormally expressed in patients with acute myocardial infarction (15) and in atherosclerotic model rats (16). Furthermore, lncRNA ZFAS1 was reported to contribute to the impairment of cardiac contractile function in myocardial infarction (17,18). The aim of the present study was to investigate the relationship between lncRNA ZFAS1 and I/R injury using hypoxia/reoxygenation (H/R)-induced H9c2 cardiomyocytes as an in vitro model to examine apoptosis and inflammation. It was found that H/R increased the expression level of ZFAS1 in H9c2 cells, and that ZFAS1 knockdown reduced inflammation and apoptosis by targeting the microRNA (miR)-590-3p/NF-κB pathway.
Materials and methods
Cell culture and H/R stress
Rat H9c2 cardiomyocyte cells (cat. no. CRL-1446; American Type Culture Collection) were cultured with DMEM (cat. no. 12491-15; Thermo Fisher Scientific, Inc.) with 10% of FBS (cat. no. 10100-147; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (cat. no. 15640055; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. H/R treatment was used to establish an I/R injury model in H9c2 cells. H9c2 cells were sequentially exposed to hypoxia for 2, 4, 8, 12, 18 and 24 h (95:5 CO2:N2 ratio) at 37°C and re-oxygenated for (95:5 O2:CO2 ratio) 2 h at 37°C (19).
Cell transfection
Small interfering (si)RNA for ZFAS1 knockdown (si-ZFAS1 forward, 5′-UGGAUUUGUACCAUUCUUCUG-3′ and reverse, 5′-GAAGAAUGGUACAAAUCCAAG-3′), negative control knockdown (si-NC forward, 5′-AGUUUCAACCGUCUUAAUCAG-3′ and reverse, 5′-GAUUAAGACGGUUGAAACUAG-3′), hsa-miR-590-3p-inhibitor (5′-AAUUUUCAUAUUCGAUCA-3′), hsa-miR-590-3p-mimic (5′-UUAAAAGUAUAAGCUAGU-3′) and hsa-miR-590-5p-NC (5′-GGAUGGCCAAUCUUCGCGGGCU-3′) were designed and synthesized by Shenggong Bioengineering Co., Ltd. Cells (1×106) were directly transfected with 25 nmol si-RNA, si-NC, miR-inhibitor, miR-mimic or miR-NC using Lipofectamine® 2000 transfection reagent (cat. no. 11668019; Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 72 h. Subsequent experiments were performed at 72 h post-transfection.
Dual-luciferase reporter assay
The StarBase database (starbase.sysu.edu.cn/index.php) was used to identify the binding sites between miR-590-3p and ZFAS1. The wild-type (WT) or mutant (MUT) mRNA 3′-untranslated regions (UTRs) of ZFAS1 and p50 were cloned into the psiCHECK2 vector (Promega Corporation). Cells (5×106) were transfected with psiCHECK2 vectors using Lipofectamine® 2000. The Dual-Lucy Assay kit (cat. no. D00100; Beijing Solarbio Science & Technology Co., Ltd.) was used to detect luciferase activities according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Reverse transcription-quantitative PCR (RT-qPCR)
RT-qPCR was used to detect the mRNA expression levels of U6, miR-590-3p and lncRNA ZFAS1 in cells. TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract the total RNA from H9c2 cells. The extracted RNA was reverse transcribed into cDNA using PrimeScript RT Master mix RT kit (cat. no. RR036B; Takara Bio, Inc.) at 37°C for 15 min and 85°C for 15 sec. qPCR was set up and conducted according to the SYBR Green qPCR Master Mix kit instructions (cat. no. 638320; Takara Bio, Inc.) and amplified using an ABI 7500 fluorescence qPCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for qPCR: Initial denaturation at 94°C for 30 sec; 40 cycles of 93°C for 2 min, 93°C for 1 min and 55°C for 2 min; followed by final extension at 72°C for 1.5 min. PCR primers were as follows: U6, forward 5′-AUAAAUCCCUUUACACCUCTT-3′, reverse 5′-AAUAAAUCCCUUUACACCUCTT-3′; GAPDH, forward 5′-AGGTCGGTGTGAACGGATTTG-3′, reverse, 5′-GGGGTCGTTGATGGCAACA-3′; miR-590-3p, forward 5′-ACACTCCAGCTGGGTGATCGAATATGTAT-3′, reverse 5-TGGTGTCGTGGAGTCG-3; and ZFAS1, forward 5′-ACGTGCAGACATCTACAACCT-3′, reverse 5′-TACTTCCAACACCCGCAT-3′. miRNA and mRNA expression levels were quantified using the 2−∆∆Cq method (20) and normalized to the internal reference genes U6 and GAPDH, respectively.
Western blotting
Total protein was extracted from H9c2 cells using RIPA lysis buffer (cat. no. P0013K; Beyotime Institute of Biotechnology) and quantified using the BCA Protein Assay kit (cat. no. P0010S; Beyotime Institute of Biotechnology). Proteins (50 µg) were separated via 12% SDS-PAGE and then transferred to PVDF membranes. The membranes were blocked with 5% skimmed milk powder at room temperature for 2 h. Membranes were incubated overnight at 4°C with the following primary antibodies: Anti-p50 (1:1,000; cat. no. ab32360; Abcam), anti-tumor necrosis factor-α (TNF-α; 1:2,000; cat. no. ab6671; Abcam), anti-interleukin (IL)-6 (1:1,000; cat. no. 12912; Cell Signaling Technology, Inc.), anti-Bax (1:3,000; cat. no. ab32503; Abcam), anti-Bcl-2 (1:500; cat. no. ab692; Abcam), anti-cleaved-caspase 3 (1:5,000; cat. no. ab2302; Abcam), anti-pro-caspase-3 (1:10,000; cat. no. ab32499; Abcam) and anti-GAPDH (1:3,000; cat. no. ab9484; Abcam). The membranes were subsequently incubated with the following horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature: Goat anti-mouse (cat. no. ab6789; 1:3,000; Abcam) or goat anti-rabbit (cat. no. ab6721; 1:3,000; Abcam). Protein bands were visualized using the BeyoECL Plus kit (cat. no. P0018S; Beyotime Institute of Biotechnology). Protein expression levels were quantified using ImageJ software (version 1.8.0; National Institutes of Health) with GAPDH as the loading control.
MTT assay
H9c2 cells (2×104 cells/well) were seeded into a 96-well plate and cultured for 12 h in 5% CO2 at 37°C. Cells were subjected to H/R exposure and then washed twice with PBS. Cell viability was measured using an MTT assay kit (cat. no. C0009; Beyotime Institute of Biotechnology) according to the manufacturer's instructions. The absorbance was measured in a Bio-Rad 680 microplate reader at 490 nm (Bio-Rad Laboratories, Inc.).
Flow cytometry
H9c2 cells (2×106) were treated with 0, 1, 2 and 4 µg/ml TNF-α (cat. no. P6231; Beyotime Institute of Biotechnology) at 37°C for 12 h. H9c2 cells (1×106) were collected and an Annexin V FITC/PI kit (Invitrogen; Thermo Fisher Scientific, Inc.) was used for flow cytometric analysis to detect apoptosis, according to the manufacturer's protocol. A Beckman CytoFLEX flow cytometer (Beckman Coulter, Inc.) and FlowJo software (version 10.0.7; FlowJo LLC) were used to analyze the rate of apoptosis.
Statistical analysis
Data are presented as the mean ± SD, and were analyzed using SPSS 20.0 (IBM Corp.). Student's t-test was used to compare differences between two groups, and one-way ANOVA with Tukey's test was used to compare the difference between multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
H/R induces cell apoptosis and gene expression changes in H9c2 cells
H9c2 cells were cultured under anoxic conditions for 2, 4, 8, 12, 18 and 24 h, and then re-oxygenated for 2 h to establish an in vitro I/R model. MTT assay (Fig. 1A) and flow cytometry (Fig. 1B) results suggested that H/R-induced cell injury decreased cell viability and increased apoptotic rates in a time-dependent manner.
Figure 1.
H/R induces cell apoptosis and gene expression changes in H9c2 cells. (A) MTT assay for cell viability following H/R exposure. (B) Apoptotic rate of cells was detected by flow cytometry. (C) Reverse transcription-quantitative PCR was used to detect the expression level of ZFAS1, which was normalized by GAPDH, and of miR-590-5p, which was normalized by U6. (D) Representative western blotting images used to determine protein expression levels of (E) p50, TNF-α and IL-6, and (F) Bax, Bcl-2, pro-caspase-3 and cleaved caspase-3. *P<0.05, **P<0.01, ***P<0.001 vs. 0 h. H/R, hypoxia/reoxygenation; IL, interleukin; miR, microRNA; TNF-α, tumor necrosis factor-α; ZFAS1, ZNFX1 antisense RNA 1.
To investigate the underlying molecular mechanisms, the expression levels of genes of interest were assessed. It was demonstrated that, compared with control cells (0 h), H/R increased the expression levels of ZFAS1 (Fig. 1C), as well as the protein expression levels of p50, TNF-α, IL-6, Bax and cleaved-caspase-3 (Fig. 1D-F). Furthermore, H/R decreased the expression levels of miR-590-3p (Fig. 1C), as well as the protein expression levels of Bcl-2 and pro-caspase-3 (Fig. 1D and F). Therefore, the present results suggested that lncRNA ZFAS1 and miR-590-3p may be related to H/R-induced apoptosis of H9c2 cells.
lncRNA ZFAS1 regulates apoptosis in H9c2 cells
The StarBase database was used to identify the binding sites between miR-590-3p and ZFAS1 (Fig. 2A). miR-590-3p-mimic significantly increased the expression of miR-590-3p, whereas miR-590-3p-mimic significantly decreased the expression of miR-590-3p compared with miR-590-3p-NC. Moreover, si-ZFAS1 significantly decreased the expression of ZFAS1 compared with si-NC (Fig. 2B). Results from the luciferase gene reporter assay indicated that infection with miR-590-3p mimic or miR-590-3p inhibitor did not change the expression level of ZFAS1 (Fig. 2C). However, ZFAS1 knockdown increased the expression level of miR-590-3p (Fig. 2D). Furthermore, flow cytometry results suggested that lncRNA ZFAS1 knockdown could affect the protein expression levels of Bax, Bcl-2, cleaved-caspase-3 and pro-caspase-3 (Fig. 2E), as well as decrease H/R-induced apoptosis in H9c2 cells (Fig. 2F). Collectively, the results indicated that ZFAS1 knockdown reduced H/R-induced H9c2 apoptosis, and ZFAS1 knockdown increased miR-590-3p expression.
Figure 2.
ZAFS1 knockdown reduces H/R-induced H9c2 apoptosis. (A) A WT-ZFAS1 3′UTR luciferase reporter vector and a MUT-ZSAF1 3′UTR luciferase reporter vector with mutations on the miR-590-3p binding sites of the ZFAS1 3′UTR were constructed. (B) RT-qPCR was used to detect the expression levels of ZAFS1 or miR-590-3p in H9c2 cells without H/R induction. (C) miR-590-3p-NC, miR-590-3p-mimic and miR-590-3p-inhibitor were transected into H9c2 cells, and luciferase activity was determined. (D) RT-qPCR results of miR-590-3p expression level in H9c2 cells transfected with si-ZFAS1. (E) Western blotting was used to detect the protein expression levels of Bax, Bcl-2, pro-caspase-3 and cleaved caspase-3 in H9c2 cells after si-ZFAS1 transfection and H/R injury. (F) Flow cytometry results of the percentage of apoptotic cells of different transfected groups following H/R induction. ***P<0.001 vs. si-NC group. 3′UTR, 3′untranslated regions; H/R, hypoxia/reoxygenation; miR, microRNA; MUT, mutation; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR; WT, wild-type; ZFAS1, ZNFX1 antisense RNA 1.
miR-590-3p regulates inflammation and apoptosis in H9c2 cells
The StarBase was also used to searched for target sites of miR-590-3p in p50 3′UTR (Fig. 3A). To assess whether miR-590-3p can regulate p50 expression level, a luciferase gene reporter assay was performed. It was demonstrated that transfection of miR-590-3p mimic significantly decreased WT type 3′UTR luciferase activity in H9c2 cells (P<0.001; Fig. 3B); however, no effect was observed with the MUT in any group. Furthermore, the miR-590-3p mimic transfection could decrease the protein expression levels of Bax and cleaved-caspase-3, and increase the protein expression levels of Bcl2 and pro-caspase-3 (Fig. 3C). Furthermore, miR-590-3p mimic decreased the expression levels of p50, TNF-α, IL-6 in H9c2 cells following H/R injury (Fig. 3D). In addition, transfection with the miR-590-3p mimic decreased H/R-induced apoptosis (Fig. 3E). Therefore, these results suggested that miR-590-3p may have a protective effect in reducing H/R-induced apoptosis by targeting p50.
Figure 3.
miR-590-3p targets p50 and reduces H/R-induced apoptosis and inflammation. (A) A WT-p50 3′UTR and a MUT-p50 3′UTR luciferase reporter vector, with mutations on miR-590-3p binding sites of the p50 3′UTR were constructed. (B) miR-590-3p-NC, miR-590-3p-mimic or miR-590-3p-inhibitor were transected into H9c2 cells, and luciferase activity was detected. (C) Protein expression levels of Bax, Bcl-2, pro-caspase-3 and cleaved-caspase-3, and (D) p50, TNF-α and IL-6 were detected by western blotting in H/R-induced H9c2 cells. (E) Flow cytometry results of the percentage of apoptotic cells in different transfected groups with H/R. ***P<0.001 vs. miR-590-3p-NC. 3′UTR, 3′untranslated region; H/R, hypoxia/reoxygenation; IL, interleukin; miR, microRNA; MUT, mutation; NC, negative control; TNF-α, tumor necrosis factor-α; WT, wild-type.
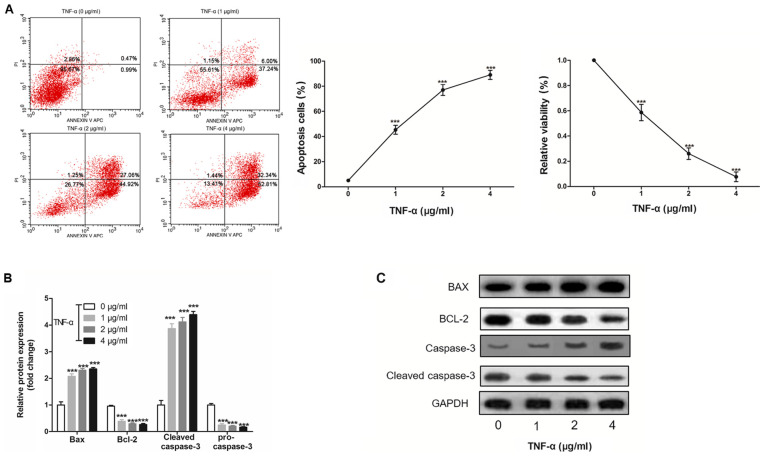
H9c2 cell apoptosis is induced by TNF-α in a dose-dependent manner
To investigate the effect of inflammation on apoptosis, H9c2 cells were treated with TNF-α, and it was revealed that TNF-α induced H9c2 cell apoptosis and cell viability in a dose-dependent manner (Fig. 4A and B). Furthermore, it was demonstrated that TNF-α altered the protein expression levels of Bax, Bcl-2, cleaved-caspase-3 and pro-caspase-3 in a dose-dependent manner (Fig. 4C).
Figure 4.
TNF-α induces H9c2 cell apoptosis in a dose-dependent manner. (A) Apoptotic rate of H9c2 cell was detected by flow cytometry in cells treated with 0, 1, 2 and 4 µg/ml TNF-α for 12 h. (B) MTT was used to detect the viability of H9c2 cells exposed to TNF-α. (C) Western blotting was used to detect the protein expression levels of Bax, Bcl-2, pro-caspase-3 and cleaved-caspase-3. ***P<0.001 vs. 0 µg/ml group, TNF-α, tumor necrosis factor-α.
Discussion
The present study established an I/R injury in vitro model by H/R exposure in H9c2 rat cardiomyocytes, and found that H/R increased the expression levels of ZFAS1, p50, TNF-α and IL-6, decreased miR-590-3p expression, and induced apoptosis of H9c2 cells in a time-dependent manner. There have been an increasing number of studies investigating the potential role of lncRNA in heart disease, which have reported that lncRNAs serve a key role in the regulation of heart disease, such as cardiac hypertrophy, cardiac graft rejection and ischemic heart failure (21,22). Moreover, lncRNA ZFAS1 is abnormally expressed in patients with acute myocardial infarction (15) and atherosclerotic model rats (16), and contributes to the impairment of cardiac contractile function in myocardial infarction (17,18).
lncRNAs are non-coding RNAs that exert biological functions by regulating the expression levels of other genes (9,10). Previous studies have shown that there are several mechanisms by which lncRNAs can regulate gene expression (23,24). The interaction mechanism between lncRNAs and miRNAs is an important way that lncRNAs regulate gene expression (25). However, lncRNA not only acts as a target for miRNAs to inhibit their binding to target genes, but also an endogenous miRNA sponge that can inhibit the expression of miRNAs and indirectly inhibits the negative control of miRNAs to target genes (9,10). Moreover, RNA competes with miRNAs to bind to the 3′UTR of target gene mRNA (9,10). In addition, miRNAs can target a large number of protein-coding genes and lncRNAs. The present results suggested that ZFAS1 could directly bind to miR-590-3p and inhibit its expression. There had been a number of previous studies investigating the relationship between miR-590-3p and the NF-κB signaling pathway. For example, Zhao et al (26) found that miR-590-3p is a novel miRNA in myocarditis by targeting NF-κB in vivo. In addition, Bao et al (27) showed that miR-590 protects against oxidized low-density lipoprotein-induced endothelial cell apoptosis via the p53/NF-κB pathway. The present study results indicated that p50 was a target gene of miR-590-3p, and that the miR-590-3p-mimic could downregulate the protein expression levels of p50, TNF-α and IL-6. Moreover, it was found that ZFAS1 knockdown or miR-590-3p overexpression attenuated H/R-induced apoptosis in H9c2 cardiomyocytes.
The important role of lncRNA regulation of the NF-κB signaling pathway has been a focus of research into inflammatory diseases. The lncRNA Lethe binds directly to the NF-κB heterodimeric subunit RelA and inhibits the DNA-binding activity of NF-κB (28). Thus, Lethe acts as a negative feedback regulator of the TNF-α pathway and regulates the inflammatory response (29). lncRNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1) inhibits DNA binding activity of NF-κB, reduces inflammatory cytokine production and downregulates the autoimmune inflammatory response (30). Furthermore, knockdown of MALAT1 upregulates lipopolysaccharide-induced TNF-α and IL-6 expression (30). In I/R injury, the inflammatory response plays a key role in myocardial I/R injury and occurs during the whole process of myocardial cell injury (31). Moreover, adhesion molecules and cytokines involved in the inflammatory reaction have the same NF-κB gene initiation site (32), and NF-κB is activated to mediate overexpression of these factors, thus aggravating the inflammatory response after myocardial I/R (33).
p50 protein is an important part of the NF-κB signaling pathway, and the dimeric complex consisting of p50-p65 is called the standard NF-κB protein complex (34). A previous study found that deletion of the NF-κB subunit p50 reduces I/R injury in vivo (35). The present results suggested that the miR-590-3p-mimic decreased the protein expression level of p50, thus miR-590-3p may inhibit the activation of the NF-κB signaling pathway. In addition, it was found that H9c2 cardiomyocyte apoptosis was induced by TNF-α in a dose-dependent manner. Therefore, downregulation of lncRNA ZFAS1 may protect H9c2 cardiomyocytes from I/R-induced apoptosis via the miR-590-3p/NF-κB pathway. However, there are some limitations to the present study, including the lack of in vivo experiments and the absence of clinical data.
In conclusion, it was demonstrated that ZFAS1 was upregulated and miR-590-3p was downregulated in H9c2 cells subjected to I/R injury. Furthermore, the present results suggested that downregulation of ZFAS1 protected against I/R-induced myocardial cell apoptosis by increasing miR-590-3p expression via the NF-κB signaling pathway.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LY and YS conceived and designed the present study. PH and DY performed the experiments and analyzed the data. LY substantially contributed to drafting the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Levine GN, Bates ER, Blankenship JC, Bailey SR, Bittl JA, Cercek B, Chambers CE, Ellis SG, Guyton RA, Hollenberg SM, et al. 2011 ACCF/AHA/SCAI Guideline for Percutaneous Coronary Intervention: Executive summary: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines and the Society for Cardiovascular Angiography and Interventions. Circulation. 2011;124:2574–2609. doi: 10.1161/CIR.0b013e31823a5596. [DOI] [PubMed] [Google Scholar]
- 2.Lee CH, Sethi R, Li R, Ho HH, Hein T, Jim MH, Loo G, Koo CY, Gao XF, Chandra S, et al. Obstructive sleep apnea and cardiovascular events after percutaneous coronary intervention. Circulation. 2016;133:2008–2017. doi: 10.1161/CIRCULATIONAHA.115.019392. [DOI] [PubMed] [Google Scholar]
- 3.Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: A neglected therapeutic target. J Clin Invest. 2013;123:92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bang C, Batkai S, Dangwal S, Gupta SK, Foinquinos A, Holzmann A, Just A, Remke J, Zimmer K, Zeug A, et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. 2014;124:2136–2146. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michaels AD, Gibson CM, Barron HV. Microvascular dysfunction in acute myocardial infarction: Focus on the roles of platelet and inflammatory mediators in the no-reflow phenomenon. Am J Cardiol. 2000;85:50B–60B. doi: 10.1016/S0002-9149(00)00811-0. [DOI] [PubMed] [Google Scholar]
- 6.Maier W, Altwegg LA, Corti R, Gay S, Hersberger M, Maly FE, Sütsch G, Roffi M, Neidhart M, Eberli FR, et al. Inflammatory markers at the site of ruptured plaque in acute myocardial infarction: Locally increased interleukin-6 and serum amyloid A but decreased C-reactive protein. Circulation. 2005;111:1355–1361. doi: 10.1161/01.CIR.0000158479.58589.0A. [DOI] [PubMed] [Google Scholar]
- 7.Pop M, Qi X, Barry J, Strauss BH, Wright GA, Ghugre NR. Hemorrhage promotes inflammation and myocardial damage following acute myocardial infarction. J Cardiovascular Magnetic Resonance. 2014;16:1–2. doi: 10.1186/1532-429X-16-S1-O72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westman PC, Lipinski MJ, Luger D, Waksman R, Bonow RO, Wu E, Epstein SE. Inflammation as a driver of adverse left ventricular remodeling after acute myocardial infarction. J Am Coll Cardiol. 2016;67:2050–2060. doi: 10.1016/j.jacc.2016.01.073. [DOI] [PubMed] [Google Scholar]
- 9.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, et al. An atlas of human long non-coding RNAs with accurate 5′ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta SC, Tripathi YN. Potential of long non-coding RNAs in cancer patients: From Bio-markers to therapeutic targets. Int J Cancer. 2017;140:1955–1967. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 12.Archer K, Broskova Z, Bayoumi AS, Teoh JP, Davila A, Tang Y, Su H, Kim IM. Long non-coding RNAs as master regulators in cardiovascular diseases. Int J Mol Sci. 2015;16:23651–23667. doi: 10.3390/ijms161023651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 14.Mathy NW, Chen XM. Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J Biol Chem. 2017;292:12375–12382. doi: 10.1074/jbc.R116.760884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Sun L, Xuan L, Pan Z, Li K, Liu S, Huang Y, Zhao X, Huang L, Wang Z, et al. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep. 2016;6:22384. doi: 10.1038/srep22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Yao H, Hui JY, Ding SH, Fan YL, Pan YH, Chen KH, Wan JQ, Jiang JY. Global transcriptomic study of atherosclerosis development in rats. Gene. 2016;592:43–48. doi: 10.1016/j.gene.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 17.Sun G, Wang Y, Zhang J, Lin N, You Y. MiR-15b/HOTAIR/p53 form a regulatory loop that affects the growth of glioma cells. J Cell Biochem. 2018;119:4540–4547. doi: 10.1002/jcb.26591. [DOI] [PubMed] [Google Scholar]
- 18.Vervliet T, Robinson EL, Roderick HL. Lnc'ing Ca2+, SERCA and cardiac disease. Cell Calcium. 2018;72:132–134. doi: 10.1016/j.ceca.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, Xing Y, Xu Y, Huang C, Bao H, Hong K, Cheng X. Pim-2 protects H9c2 cardiomyocytes from hypoxia/reoxygenation-induced apoptosis via downregulation of Bim expression. Environ Toxicol Pharmacol. 2016;48:94–102. doi: 10.1016/j.etap.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Gu G, Huang Y, Wu C, Guo Z, Ma Y, Xia Q, Awasthi A, He X. Differential expression of long noncoding RNAs during cardiac allograft rejection. Transplantation. 2017;101:83–91. doi: 10.1097/TP.0000000000001463. [DOI] [PubMed] [Google Scholar]
- 22.Kühl C, Frey N. Springer International Publishing; 2016. Long noncoding RNAs in heart disease. [DOI] [Google Scholar]
- 23.Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al. LncRNA-dependent mechanisms of androgen receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 25.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 26.Zhao S, Yang G, Liu PN, Deng YY, Zhao Z, Sun T, Zhuo XZ, Liu JH, Tian Y, Zhou J, et al. miR-590-3p is a novel MicroRNA in myocarditis by targeting nuclear factor Kappa-B in vivo. Cardiology. 2015;132:182–188. doi: 10.1159/000433596. [DOI] [PubMed] [Google Scholar]
- 27.Bao MH, Li JM, Zhou QL, Li GY, Zeng J, Zhao J, Zhang YW. Effects of miR-590 on oxLDL-induced endothelial cell apoptosis: Roles of p53 and NF-κB. Mol Med Rep. 2016;13:867–873. doi: 10.3892/mmr.2015.4606. [DOI] [PubMed] [Google Scholar]
- 28.Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallach D. The cybernetics of TNF: Old views and newer ones. Semin Cell Dev Biol. 2016;50:105–114. doi: 10.1016/j.semcdb.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 30.Zhao G, Su Z, Song D, Mao Y, Mao X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-κB. FEBS Lett. 2016;590:2884–2895. doi: 10.1002/1873-3468.12315. [DOI] [PubMed] [Google Scholar]
- 31.Liang R, Zhao Q, Jian G, Cheng D, Wang N, Zhang G, Wang F. Tanshinone IIA attenuates contrast-induced nephropathy via Nrf2 activation in rats. Cell Physiol Biochem. 2018;46:2616–2623. doi: 10.1159/000489688. [DOI] [PubMed] [Google Scholar]
- 32.Wang F, Zhang G, Lu Z, Geurts AM, Usa K, Jacob HJ, Cowley AW, Wang N, Liang M. Antithrombin III/SerpinC1 insufficiency exacerbates renal ischemia/reperfusion injury. Kidney Int. 2015;88:796–803. doi: 10.1038/ki.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Z, Lian A, Zhang F. Nuclear factor-κB activation inhibitor attenuates ischemia reperfusion injury and inhibits Hmgb1 expression. Inflamm Res. 2014;63:919–925. doi: 10.1007/s00011-014-0765-x. [DOI] [PubMed] [Google Scholar]
- 34.Ghosh G, Van Duyne G, Ghosh S, Sigler PB. Structure of NF-kappaB p50 homodimer bound to a kappa B site. Nature. 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 35.Frantz S, Tillmanns J, Kuhlencordt PJ, Schmidt I, Adamek A, Dienesch C, Thum T, Gerondakis S, Ertl G, Bauersachs J. Tissue-specific effects of the nuclear Factor kappaB subunit p50 on myocardial ischemia-reperfusion injury. Am J Pathol. 2007;171:507–512. doi: 10.2353/ajpath.2007.061042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.