Abstract
Gene expression alterations in periodontal ligament stem cells (PDLSCs) during bisphosphonate (BP) usage and the transcriptomic mechanism underlying BP-related osteonecrosis of the jaw have not been fully elucidated. In the present study, human PDLSCs were isolated from adults with no history of periodontal disease, and subsequently incubated and treated with zoledronate on days 3 and 5. Subsequently, PDLSCs from all timepoints were screened using an Affymetrix Gene Expression Array. Limma differential expression analysis was performed on a normalized gene expression matrix, followed by cluster analysis, pathway and network analyses. Overall, 906 genes (352 upregulated and 554 downregulated) exhibited differential expression levels between days 0 and 5, and these were termed slow-response genes. These slow-response genes were enriched in cellular stress response signaling pathways (upregulated genes), as well as proliferation- and ossification-associated signaling pathways (downregulated genes). Furthermore, 168 (day 3 vs. 0) and 105 (day 5 vs. 3) genes were differentially expressed between adjacent timepoints. These genes were also enriched in stress response- and proliferation-associated signaling pathways, but not in ossification-associated signaling pathways. Poly(ADP-ribose) polymerase 1 (PARP1) and CYLD lysine 63 deubiquitinase (CYLD) had the most protein-protein interaction partners among the slow-response genes and were connected with both stress- (e.g. caspase-1) and ossification-associated genes [e.g. secreted phosphoprotein 1 and collagen type I α1 chain (COL1A1)]. BP treatment induced stress response-like transcriptional alterations in PDLSCs, followed by inhibition of proliferation and ossification. These alterations may contribute to the onset of jaw osteonecrosis. PARP1 and CYLD may be two key genes involved in this pathological procedure.
Keywords: bisphosphonate, osteonecrosis of the jaw, transcriptome, inflammation, periodontal ligament stem cells, stress response
Introduction
Bisphosphonate (BP) is an effective treatment for osteoporosis, multiple myeloma, Paget's disease and bone-metastatic cancer (1). BP decreases the rate of complications of bone metastasis and minimizes skeletal-associated events in malignancies. However, unwanted side effects, such as osteonecrosis of the jaw and atypical femur fractures, still occur (2). Lo et al (3) used thorough screening to show an increased number of cases of BP-related osteonecrosis of the jaw (BRONJ; prevalence in America in 2006, 0.10%; 95% CI, 0.05–0.20) due to chronic oral BP usage. The highest reported incidence rate of BRONJ is 5–10%, occurring in patients using high doses of BP, which leads to decreased drug prescription and increased occurrence of osteoporosis-associated complications (4,5).
The majority of BRONJ cases occur following dental extraction; incidence rates depend on the severity of dental disease or adjacent active periodontal disease. Marx (6) reported that 85/152 BRONJ cases were associated with dental extraction, and suggested prevention strategies, such as interrupting BP treatment before such procedures. As periodontal disease is present in the majority of patients with osteonecrosis, oral infections and surgical dental treatments are considered to be a triggering event for BRONJ (7). Previous case reports and animal studies have identified periodontal disease as a key risk factor for the development of BRONJ (8,9). Clinically, a number of reports have noted that BRONJ occurs following tooth extraction due to the severity of dental disease or the presence of active periodontal or periapical disease in surrounding tissues (10,11). In 2014, Oteri et al (8) conducted a case-control study and revealed that patients with BRONJ have fewer teeth, greater clinical attachment level and less alveolar bone support than control subjects. Our previous study demonstrated that patients with BRONJ had deep periodontal pockets and severe periodontal bone defects adjacent to the exposed necrotic bone (12). However, at present, the exact molecular signaling pathway of BRONJ pathogenesis, as well as its association with infections or other stress, remains poorly understood.
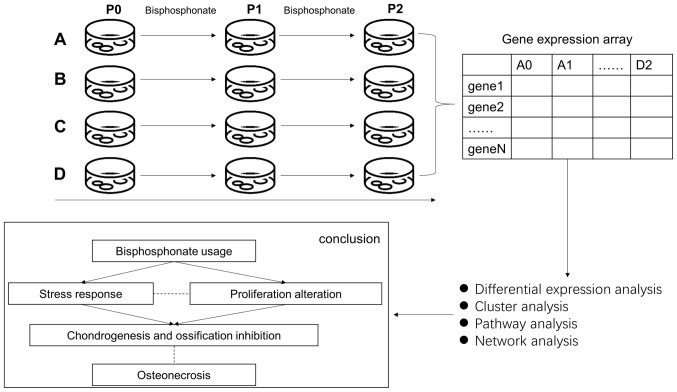
In order to explore the underlying mechanism of BRONJ, the effect of the most widely used BP (zoledronic acid) on periodontal ligament stem cells (PDLSCs) was analyzed along with intracellular molecular changes. Human PDLSCs isolated from human periodontal ligament (PDL) tissue exhibited a capacity for self-renewal and multipotency, thereby serving a key role in regeneration of periodontal tissue. The aim of the present study was to analyze the impact of BP treatment on the transcriptome of PDLSCs and to identify key processes and genes involved in BRONJ that may serve as potential therapeutic targets. A schematic outlining the current experiments is presented in Fig. 1.
Figure 1.
Flowchart of the present study. Top left: Cell culture procedures. Top Right: Expression quantification. Bottom right: Bioinformatic analysis. Bottom left: Conclusion.
Materials and methods
Cell culture
Human PDLSCs were isolated from healthy premolars or third molars extracted from eight young adults (five males and three females, aged 15–23 years) under orthodontic treatment with no history of periodontal disease. Sample collection was conducted at Shanghai Ninth People's Hospital between January, 2018 and April, 2018. The experimental protocols were approved by the Ethics Committee of the Ninth People's Hospital Affiliated with Shanghai Jiao Tong University, School of Medicine (Shanghai, China). All donors provided written informed consent prior to participating in the present study. PDLSCs were isolated and cultured as previously reported, with a slight modification (12). Following extraction, teeth were immediately placed into a solution of PBS containing 100 U/ml penicillin and 100 U/ml streptomycin. After washing thoroughly in PBS (3–5 times) to remove blood components, the PDL tissues were gently scraped from the surface of the middle of the root, minced into 1–3 cubes (2 mm3), and placed into 6-well culture dishes. Subsequently, glass cover slips were placed over the tissues to prevent floating, and PDL tissues were incubated in culture medium (DMEM, HyClone; Cytiva) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin) in a humidified atmosphere of 95% air and 5% CO2 at 37°C. The tissues were subsequently maintained by replacing the medium every 3–4 days until cell density reached 90% confluence. Subsequently, the cells were detached using 0.25% trypsin/EDTA and sub-cultured at a density of 1×105 cells/cm2 in 100-mm dishes. Cells at passage 3–5 were used for subsequent experiments.
Self-renewal and multiple differentiation of PDLSCs in vitro
In order to assess the self-renewal capacity of PDLSCs, cells (200/well) were plated in a 6-well plate. The 14-day cultures were fixed with 4% formalin for 15 min at 37°C and stained with 0.1% crystal violet (Beyotime Institute of Biotechnology) for 15 min at 37°C. Aggregates of ≥50 cells were scored as colonies. In order to investigate the osteogenic ability, PDLSCs at passage 3 were seeded at a density of 5×104 cells/well in 12-well plates and incubated in growth medium (DMEM, HyClone; Cytiva) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin) containing 10 nM dexamethasone (Sigma-Aldrich; Merck KGaA), 5 mM β-glycerophosphate (Sigma-Aldrich; Merck KGaA) and 500 µM ascorbic acid (Sigma-Aldrich; Merck KGaA) for 14 days to induce mineralization. The cells were then stained with 1% Alizarin Red (Sigma-Aldrich; Merck KGaA) for 1 h at 37°C. In order to investigate adipogenic ability, PDLSCs at passage 3 were seeded at a density of 5×104 cells/well in 12-well plates and incubated in basal medium (Gibco; Thermo Fisher Scientific, Inc.) (DMEM with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin) at 37°C supplemented with 0.5 mM methylisobutylxantine, 0.5 mM hydrocortisone, 200 µM indomethacin and 10 µg/ml insulin for 4 weeks and stained with Oil Red O (5 mg/ml; Sigma-Aldrich; Merck KGaA) for 1 h at 37°C. All images of the cells were captured using an inverted contrast-phase light microscope (Nikon Corporation).
Zoledronate treatments
Zoledronate was purchased from Novartis Pharmaceuticals UK Ltd. and diluted in 0.9% NaCl infusion solution. PDLSCs were treated with an oncologic dose of zoledronate (50 µM) at days 3 and 5 at 37°C. Control cells were incubated with the same procedure, except that they were treated with vehicle only (0.9% NaCl). The oncologic dose of zoledronate was selected based on published pharmacokinetic data (13). Calculations of the concentrations of zoledronate exposure equivalents on cells were performed as previously described (14).
RNA extraction, purification and quality control
Total RNA from each cell sample was extracted using TRIzol® (cat. no. 15596-018; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA integrity was evaluated using an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.). Qualified total RNA was further purified using an RNeasy Micro kit (cat. no. 74004; Qiagen GmbH) and RNase-Free DNase Set (cat. no. 79254; Qiagen GmbH). Extracted RNA was analyzed using a NanoDrop ND-2000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.) and Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.). Quality thresholds were set as RNA integrity number >6 and S28/S18 >0.7.
Array hybridization
Prior to hybridization, total RNA was first amplified, labeled and purified using a GeneChip 3′IVT PLUS Reagent kit (cat. no. 902416; Affymetrix; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol to obtain biotin-labeled cRNA. Array hybridization and washing were performed using a GeneChip® Hybridization, Wash and Stain kit (cat. no. 900720; Affymetrix; Thermo Fisher Scientific, Inc.) in a Hybridization Oven 645 (cat. no. 00-0331-220V; Affymetrix; Thermo Fisher Scientific, Inc.) and Fluidics Station 450 (cat. no. 00-0079; Affymetrix; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols.
Data acquisition and pre-processing
Slides were scanned using a GeneChip® Scanner 3000 (Affymetrix; Thermo Fisher Scientific, Inc.) and Command Console software (version 4.0; Affymetrix; Thermo Fisher Scientific, Inc.) using the default settings. Raw data for each sample were filtered using the following thresholds: i) Average background value <100; and ii) for at least one housekeeping gene, 3′/5′signals <3. All samples passed these thresholds. Raw data were quantile-normalized and log2-transformed using the robust multi-array average algorithm and Gene Spring software (Agilent Technologies, Inc., version 12.6.1). A total of 49,282 detected probes were mapped to the GRCh37/hg19 human genome. For each gene, the probe with the maximum average value was selected to represent its expression level. Only probes that were detected in all samples were included in subsequent analysis. Expression level data for 18,384 genes were obtained for downstream analysis.
Differential expression analysis
Differential expression analysis was performed using limma version 3.38.3 (15) on R 3.4.1 (16). The limma pipeline for time course experiments was used to compare P2 (5 days after treatment) vs. P0 (day 0), P2 vs. P1 (3 days after treatment) and P1 vs. P0. A linear model was fitted with the linear formula exp-series + timepoint where exp denoted gene expression, series denoted cell line batch and timepoint corresponded to P0, P1 and P2.
The empirical Bayes method (15) was used to estimate the fold change between time points. For P2 vs. P0 and P1 vs. P0, the reference level for timepoint was set as P0, and results were extracted for each comparison with the parameter coef. For P2 vs. P1, another linear model was fitted using only eight samples (discarding four samples of P0) of P2 and P1 and the reference level for timepoint was set as P1. The threshold of differentially expressed genes (DEGs) was set at: i) Benjamini-Hochberg-adjusted P-value <0.05; and ii) |log2 fold change (FC)|>1. Control cells were analyzed in the same way. DEGs that exhibited the same direction in both treatment and control cells were excluded from downstream analysis.
Cluster analysis
Hierarchical cluster analysis based on Euclidean distance was performed on all 12 samples of three timepoints using two sets of features: i) All DEGs from P2 vs. P0; and ii) union of DEGs from P2 vs. P1 and P1 vs. P0. The expression level value of each gene was scaled and centered to 0 before analysis. Cluster analysis and construction of the heatmap were achieved using the heatmap function in R (17).
Gene Ontology (GO) analysis
GO-biological process (GO-BP) pathway analysis (18) was performed using the clusterProfiler R package version 3.16.0 (19). A hypergeometric test was used to investigate whether the DEG lists were enriched in any GO-BP pathways. Gene background was defined as all genes with GO annotation. Only pathways with ≥10 genes were included in the analysis. P-values of hypergeometric tests were adjusted for multiple testing via the Benjamini-Hochberg method. For all pathways with adjusted P-value ≤0.05, non-redundant pathways were selected as results (i.e. no pathway was the parent term of any other pathway).
Protein-protein interaction (PPI) network analysis
Human PPI data was obtained from the Biogrid database (20) (access date, June 1, 2018). A PPI network of all DEGs was constructed using Cytoscape version 3.6.1 (21). Node degree was calculated using the network analyzer in Cytoscape. In order to investigate co-expression patterns in the PPI network, Pearson correlation coefficients (PC) were calculated for all identified PPI pairs using the ‘cor’ function in R.
Results
In vitro self-renewal and multipotent capacities of PDLSCs
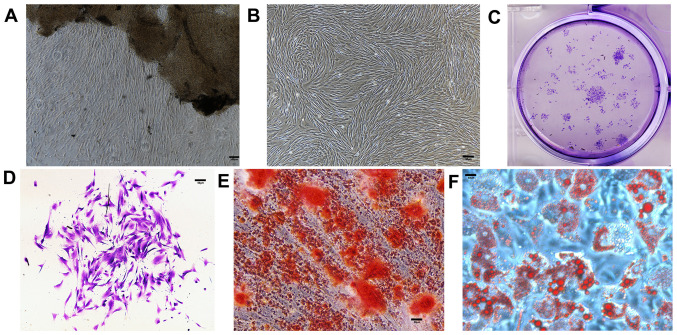
The first step of the present study was to verify whether the in vitro model of PDLSCs could correctly imitate the in vivo physiology of PDL tissues. Cells were found growing around the PDL tissues 5–10 days after initial incubation (Fig. 2A) and proliferated to reach 90% confluence in 7–10 days (Fig. 2B). Similar to other mesenchymal stem cells, PDLSCs formed adherent clonogenic cell clusters of fibroblast-like cells (Fig. 2C and D). The multipotent capacity of PDLSCs was verified by induction in the osteogenic and adipogenic media in vitro. Alizarin Red staining (Fig. 2E) revealed a number of calcified nodules in the cultures after 14 days of induction. After 4 weeks of culture in adipogenic medium, intracellular lipid vacuoles appeared in PDLSCs, the presence of which was confirmed by Oil Red O staining (Fig. 2F).
Figure 2.
In vitro characteristics of cultured PDLSCs. Scale bars=200 µm. (A) PDLSCs grew around periodontal ligament tissues five days after incubation. (B) PDLSCs reached confluence seven days after incubation. (C) Entire plate view of colony-forming PDLSCs, which were plated at a low density and cultured for 7 days before staining with crystal violet. (D) Cell clusters derived from PDLSCs with typical fibroblast-like morphology seven days after incubation. (E) Alizarin Red-S staining 14 days after incubation. (F) Oil Red O staining four weeks after incubation. PDLSC, periodontal ligament stem cell.
Quantification of the PDLSC transcriptome via gene expression level array
In order to evaluate the impact of BP on PDLSCs, genome-wide transcription levels of all samples were measured at three time points (days 0, 3 and 5) using the Affymetrix PrimeView Human Gene Expression Array. A total of 49,282 probes were detected and mapped to 20,608 genes. Following normalization and filtration, the expression levels of 18,384 genes for all samples were obtained. These expression level data enabled further analysis of the biological effects of BP usage.
Genes exhibit different patterns of expression in response to BP
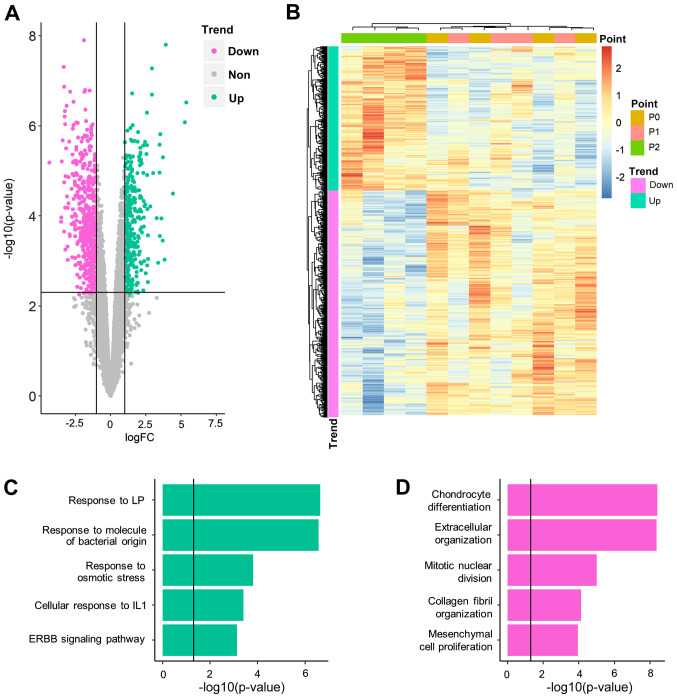
Having quantified the gene expression levels of PDLSCs at different time points (P0, 1 and 2), gene expression levels which were significantly altered following BP exposure were identified. First, gene expression levels were compared at P2 and P0, which represented the longest exposure to BP. A total of 906 genes (352 upregulated and 554 downregulated) were demonstrated to be significantly differentially expressed between P2 and P0 (Fig. 3A; Table SI). Among these genes, two were also differentially expressed in control cell lines (Table SII) and were, therefore, excluded from downstream analysis. The remaining DEGs could distinguish P2 from P1 and P0 samples but could not distinguish between P1 and P0 samples (Fig. 3B-D). It was hypothesized that these genes required a long period of BP exposure to induce significant alteration. The DEGs between P2 and P0 were referred to as slow-response DEGs. One of these DEGs, COL1A1 (logFC, −2.89; P=0.007), was highlighted in a previous genetic association study, which reported that COL1A1 polymorphism increased the risk of BRONJ (22).
Figure 3.
Biological significance of slow-response DEGs. (A) Volcano plot for differential expression analysis at day 5 vs. 0. (B) Hierarchical clustering analysis for all 12 samples, using all DEGs identified. Gene Ontology-Biological Process results for (C) upregulated and (D) downregulated DEGs. DEG, differentially expressed gene; FC, fold change; IL, interleukin; LP, lipopolysaccharide.
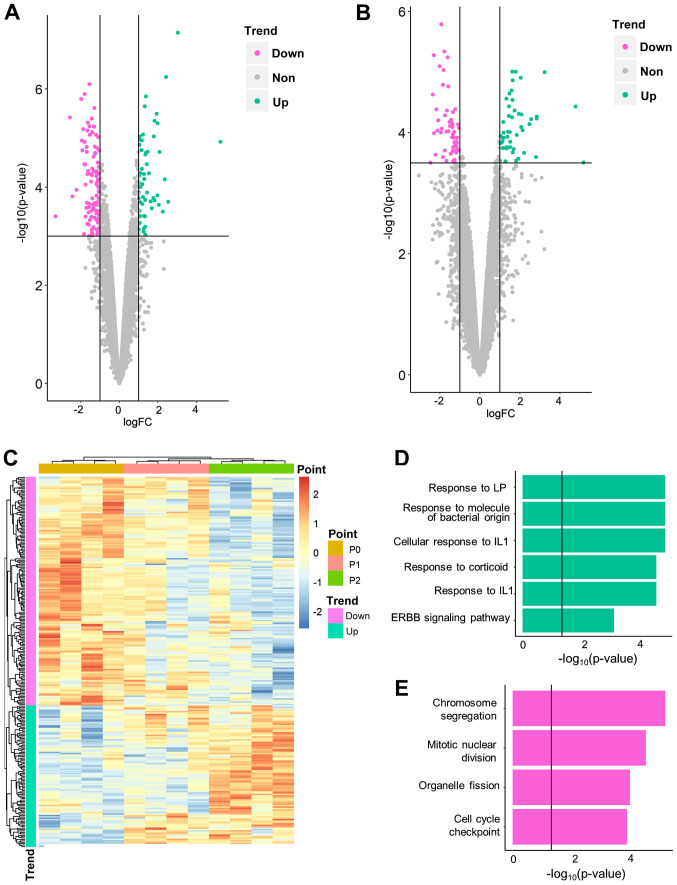
Similarly, expression level data was compared between P2 and P1, and P1 and P0. It was hypothesized that such comparisons would identify genes that were altered rapidly following BP exposure; hence, these DEGs between adjacent time points were referred to as fast-response DEGs. A total of 264 fast-response DEGs, including 168 DEGs for P1 vs. P0 (Fig. 4A; Table SIII) and 105 DEGs for P2 vs. P1 (9 overlapping genes; Fig. 4B; Table SIV), were identified. The majority (229/264) were also slow-response DEGs (i.e. differentially expressed between P0/1 and 2). Among all fast-response DEGs, 163 genes were downregulated and 101 were upregulated over time (Fig. 4C). No DEGs were identified in adjacent comparisons of control cells (Table SII), which indicates that the fast-response DEGs were not driven by experimental procedures. Fast-response DEGs successfully separated three time points in hierarchical clustering analysis (Fig. 4C).
Figure 4.
Biological significance of fast-response differentially expressed genes. Volcano plot for differential expression analysis of (A) day 3 vs. 0 and (B) day 5 vs. 3. (C) Hierarchical clustering analysis for all 12 samples, using all fast-response DEG. Gene Ontology-Biological Process results for (D) upregulated and (E) downregulated DEGs. PDLSC, periodontal ligament stem cell; IL, interleukin; LP, lipopolysaccharide.
BP exposure influences stress response and cartilage generation
Having identified genes influenced by BP treatment, the biological consequences of BP usage were investigated. GO-BP analysis was applied separately to upregulated and downregulated DEG lists.
For slow-response DEGs, a notable cellular stress reaction was observed (Fig. 3C; Table SV). Upregulated slow-response DEGs were significantly enriched in response to inflammatory factors, such as lipopolysaccharide (P-adjusted=2.27×10−7), bacterial molecules (P-adjusted=2.69×10−7) and interleukin-1 (P-adjusted=3.93×10−4). Another inflammation-associated signaling pathway, ‘ERBB signaling pathway’ (P-adjusted=7.40×10−4), was also moderately enriched. Downregulated genes (Fig. 3D; Table SVI) inhibited cartilage generation and were notably enriched in ‘chondrocyte differentiation’ (P-adjusted=3.92×10−9) and ‘extracellular organization’ (P-adjusted=4.38×10−9). ‘Ossification’ (P-adjusted=1.36×10−5) was also significantly enriched by downregulated genes, which suggested that it was inhibited during BP treatment. Cell proliferation was also inhibited, demonstrated by enrichment of ‘mitotic nuclear division’ (P-adjusted=9.98×10−6) and ‘mesenchymal cell proliferation’ (P-adjusted=1.12×10−4).
For fast-response DEGs, different enrichment patterns were observed. All enriched pathways were associated with proliferation and cell cycle regulation (e.g. ‘chromosome segregation’; P-adjusted=7.62×10−6; Table VII). For upregulated fast-response DEGs, the induction of cellular stress reaction was similar to that of slow-response DEGs. These results indicated that stress reaction was induced more rapidly than inhibition of cartilage generation and ossification.
CYLD lysine 63 deubiquitinase (CYLD) and poly(ADP-ribose) polymerase 1 (PARP1) function as hub genes in BP pathology
Having identified the key biological processes in BP pathology, it was investigated whether any key genes (hubs) served a central role in these processes. Such hubs may serve as potential interventional targets to prevent osteonecrosis.
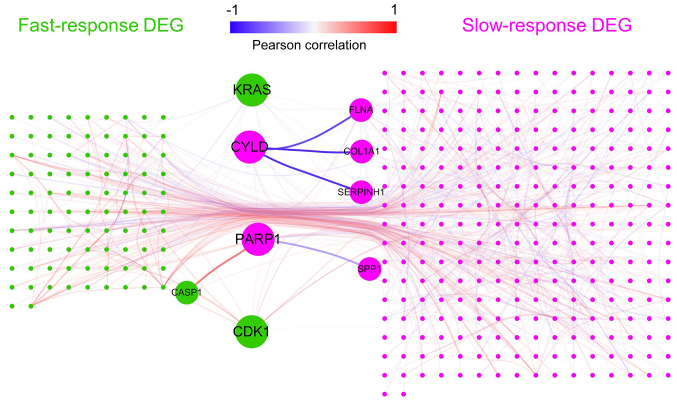
In order to investigate the potential hub genes, a PPI network of all DEGs was constructed (Fig. 5). It was hypothesized that hub genes might act as ‘bridges’ between fast- and slow-response DEGs and connect more slow-response DEGs. If such hub genes exist, they may be potential drug targets. Thus, the number of slow-response DEGs connected by each gene was calculated (Table SVIII). The top gene, PARP1, which connected 14 slow-response DEGs, was an upregulated slow-response DEG. PARP1 was connected and positively correlated with caspase-1 (CASP1), an upregulated fast-response DEG that serves a vital role in response to inflammation and stress (23).
Figure 5.
Network analysis of all DEGs. Green represents fast-response DEGs; purple represents slow-response DEGs. Each line represents a protein-protein interaction connection recorded in Biogrid. Line color represents Pearson correlation coefficients between genes. Key genes (points) and key interactions (lines) are enlarged. DEG, differentially expressed gene; FLNA, filamin A; CYLD, CYLD lysine 63 deubiquitinase; COL1A1, collagen type I α1 chain; SERPINH1, serpin family H member 1; PARP1, poly(ADP-ribose) polymerase 1; SPP1, secreted phosphoprotein 1; CASP1, caspase 1.
PARP1 was connected and negatively correlated with secreted phosphoprotein 1 (SPP1), a gene involved in the attachment of osteoclasts to the mineralized bone matrix (24). The CYLD gene, which connected 13 slow-response DEGs, had three PPI partners that are involved in collagen biosynthesis [serpin family H member 1 (SERPINH1) (24), filamin A (FLNA) (25) and COL1A1]. A negative correlation was observed between CYLD and these collagen-associated genes, which indicated that CYLD is upregulated by fast-response genes and may inhibit downstream extracellular matrix generation of bone.
PARP1 and CYLD were slow-response DEGs. It was further determined whether any fast-response DEGs could serve as hub genes. KRAS and CDK1, which connect 11 slow-response DEGs, were the top genes among fast-response DEGs. They both connected a number of genes involved in ‘cell proliferation’ and ‘cell cycle’ (Table SVI). KRAS was negatively corelated with PPI partners WW and C2 domain containing 1 (PC, −0.70503), kinetochore scaffold 1 (PC, −0.56551) and cell division cycle associated 8 (PC, −0.7047), whereas CDK1 exhibited a notable positive correlation with a number of cell cycle genes, such as cyclin B1 (PC, 0.922838) and non-SMC condensin I complex subunit G (PC, 0.975156; Table SIX).
Discussion
Osteonecrosis of the jaw is one of the most severe complications in patients using BP (26). The present study reports the results of a genome-wide landscape of transcriptomic alteration in PDLSCs during BP usage, which may increase the understanding of the mechanism of BRONJ and putative drug targets for BRONJ.
With the advantage of time series designation, the present analysis demonstrated that genes respond to BP exposure at different rates. The majority of BP-associated genes (676/941) exhibited significant differential expression levels only at day 5 vs. 0, indicating that they responded to BP in a slow and gradual manner. The distinct expression level patterns of fast- and slow-response genes built a ‘two-wave’ alteration, which is in accordance with the clinical observation that osteonecrosis of the jaw follows a chronic course following BP exposure (27).
The two-wave expression level alteration affected different biological functions. As indicated by the GO-BP analysis, fast-response DEGs were notably enriched in inflammatory and stress responses, as well as in cell proliferation signaling pathways. However, slow-response DEGs exhibited enrichment in ossification and cartilage-associated signaling pathways. As reported by Aghaloo et al (27) and Lorenzo-Pouso et al (28), inflammation and inhibition of bone remodeling have been suggested as putative mechanisms of BRONJ. While isolated evidence of resorption-generation imbalance (29), periodontal disease (7) and infection (30) has been discovered in pathological research of BRONJ, these hypotheses are treated as being exclusive and require validation (31). The present results indicated that these signaling pathways may be different phases of a single uniform pathological procedure and therefore should not be considered separately.
Another debate regarding BRONJ pathology is the mutual association between inflammatory damage and infection. Infection can directly damage the bone structure and exposed bones are further susceptible to bacterial colonization (32). To date, the causative factors from this mutual association have not been fully elucidated. Based on the present sterile in vitro experiments, it was observed that inflammation and stress responses can arise in the absence of infection. Despite being a sterile environment, PDLSCs exhibited signs of fighting against infection (e.g., enrichment in ‘response to molecules of bacterial origin’). It can be inferred that BP can induce inflammatory reactions in PDLSCs in the absence of other causative factors. Therefore, it was hypothesized that inflammation and stress response are causative factors in the pathology of BRONJ, and prevention of BP-induced inflammation is a potential target for the management of BRONJ.
As the stress response occurs prior to inhibition of ossification, it was hypothesized that the former process gives rise to the latter via key intermediate molecules, namely hubs. PPI network analysis identified two potential hub genes: PARP1 and CYLD. PARP1 encodes a chromatin-associated enzyme, poly(ADP-ribosyl)transferase (33), and acts as a bridge between fast-response inflammation and slow-response ossification inhibition by interacting with CASP1 and SPP1, respectively (23,24). This bridge-like structure suggests that PARP1-based intervention may have a decoupling effect on the inflammation-ossification inhibition cascade. Another potential hub, CYLD, encodes a cytoplasmic protein with three cytoskeletal-associated protein-glycine-conserved domains that functions as a deubiquitinating enzyme (34). Previous studies have linked CYLD with extracellular matrix generation (35) and bone formation/resorption balance (36). In PDLSCs, CYLD was observed to interact with slow-response DEGs (SERPINH1, FLNA and COL1A1) that are involved in collagen biogenesis, a key process in extracellular generation (37) and bone formation (38). Although no evidence was found to support the association between CYLD and inflammation in PDLSCs, the strong connection between CYLD and collagen function make it a putative hub gene in the network of BP pathology. As for the fast-response gene, the upregulation of KRAS and downregulation of CDK1 supported the hypothesis that cell proliferation was inhibited by BP usage. KRAS and CDK1 may be key drivers of such inhibition.
The present study has certain limitations. The duration of BP treatment was relatively short, and all PDLSC samples were collected from patients aged <30 years. The present results cannot be extrapolated to older patients or those receiving long-term BP treatment. Furthermore, the present study only analyzed BRONJ in terms of transcriptomes, so does not represent the overall picture of BRONJ-associated pathology. Further functional validation experiments are required to verify the key processes and hub genes identified by the present analysis.
BP induced stress response-like transcriptional alterations in PDLSCs, followed by inhibition of proliferation and ossification. Such alterations may contribute to the onset of osteonecrosis of the jaw. PARP1 and CYLD may be two key genes in this pathological procedure. The present findings support an anti-inflammatory strategy in management of BP-induced osteonecrosis and indicate that PARP1 and CYLD may be potential targets in drug therapy of BRONJ.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 81271114 and 31700848) and Science and Technology Commission of Shanghai Municipality (grant nos. 19411962000 and 18441903000). The funding organizations had no role in the design or conduct of this research.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Github repository, github.com/WeiCSong/PDLSC.
Authors' contributions
SW designed and supervised the study. ML and YY collected the samples. HH and ML conducted cellular experiments. YS collected and analyzed the data and wrote the manuscript. YZ and WZ interpreted the data and revised the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
The experimental protocols were approved by the ethics committee of Ninth Peoples Hospital affiliated to Shanghai Jiao Tong University, School of Medicine (approval no. SH9H-2020-T37-2). Written informed consent was obtained from the participants prior to participation in the study. Informed consent was obtained from the parents of the one patient who was a minor.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Woo SB, Hellstein JW, Kalmar JR. Systematic review: Bisphosphonates and osteonecrosis of the jaws. Ann Intern Med. 2006;144:753. doi: 10.7326/0003-4819-144-10-200605160-00009. [DOI] [PubMed] [Google Scholar]
- 2.Holen I, Coleman RE. Bisphosphonates as treatment of bone metastases. Curr Pharm Des. 2010;16:1262–1271. doi: 10.2174/138161210791034003. [DOI] [PubMed] [Google Scholar]
- 3.Lo JC, O'Ryan FS, Gordon NP, Yang J, Hui RL, Martin D, Hutchinson M, Lathon PV, Sanchez G, Silver P, et al. Prevalence of osteonecrosis of the jaw in patients with oral bisphosphonate exposure. J Oral Maxillofac Surg. 2010;68:243–253. doi: 10.1016/j.joms.2009.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elsayed R, Abraham P, Awad ME, Kurago Z, Baladhandayutham B, Whitford GM, Pashley DH, McKenna CE, Elsalanty ME. Removal of matrix-bound zoledronate prevents post-extraction osteonecrosis of the jaw by rescuing osteoclast function. Bone. 2018;110:141–149. doi: 10.1016/j.bone.2018.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz HC. American association of oral and maxillofacial surgeons position paper on medication-related osteonecrosis of the jaw-2014 update and CTX. J Oral Maxillofac Surg. 2015;73:377. doi: 10.1016/j.joms.2014.10.035. [DOI] [PubMed] [Google Scholar]
- 6.Marx RE. Oral and intravenous bisphosphonate-induced osteonecrosis of the jaws: History etiology, prevention, and treatment. Royal College of Surgeons of England. 2009 [Google Scholar]
- 7.Aghaloo TL, Kang B, Sung EC, Shoff M, Ronconi M, Gotcher JE, Bezouglaia O, Dry SM, Tetradis S. Periodontal disease and bisphosphonates induce osteonecrosis of the jaws in the rat. J Bone Miner Res. 2011;26:1871–1882. doi: 10.1002/jbmr.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oteri G, Bramanti E, Nigrone V, Cicciù M. Periodontal health in osteoporotic patients affected by BRONJ: An observational study. J Osteoporos. 2013;2013:231289. doi: 10.1155/2013/231289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li CL, Lu WW, Seneviratne CJ, Leung WK, Zwahlen RA, Zheng LW. Role of periodontal disease in bisphosphonate-related osteonecrosis of the jaws in ovariectomized rats. Clin Oral Implants Res. 2016;27:1–6. doi: 10.1111/clr.12502. [DOI] [PubMed] [Google Scholar]
- 10.Thumbigere-Math V, Michalowicz BS, Hodges JS, Tsai ML, Swenson KK, Rockwell L, Gopalakrishnan R. Periodontal disease as a risk factor for bisphosphonate-related osteonecrosis of the jaw. J Periodontol. 2014;85:226–233. doi: 10.1902/jop.2013.130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krimmel M, Ripperger J, Hairass M, Hoefert S, Kluba S, Reinert S. Does dental and oral health influence the development and course of bisphosphonate-related osteonecrosis of the jaws (BRONJ)? Oral Maxillofac Surg. 2014;18:213–218. doi: 10.1007/s10006-013-0408-3. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Yu Y, Shi Y, Yuqiong Zhou Y, Zhang W, Hua H, Ge J, Zhang Z, Ye D, Yang C, Wang S. Decreased osteogenic ability of periodontal ligament stem cells leading to impaired periodontal tissue repair in BRONJ patients. Stem Cells Dev. 2020;29:156–168. doi: 10.1089/scd.2019.0151. [DOI] [PubMed] [Google Scholar]
- 13.Russell RGG, Watts NB, Ebetino FH, Rogers MJ. Mechanisms of action of bisphosphonates: Similarities and differences and their potential influence on clinical efficacy. Osteoporos Int. 2008;19:733–759. doi: 10.1007/s00198-007-0540-8. [DOI] [PubMed] [Google Scholar]
- 14.Otto S, Pautke C, Opelz C, Westphal I, Drosse I, Schwager J, Bauss F, Ehrenfeld M, Schieker M. Osteonecrosis of the jaw: Effect of bisphosphonate type, local concentration, and acidic milieu on the pathomechanism. J Oral Maxillofac Surg. 2010;68:2837–2845. doi: 10.1016/j.joms.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 15.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R Core Team. R Foundation for Statistical Computing; Vienna, Austria: 2019. R: A language and environment for statistical computing. [Google Scholar]
- 17.Raivo Kolde. Pheatmap: Pretty Heatmaps. https://CRAN.R-project.org/package=pheatmap. R package version 1.0.8. 2018 [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatr-aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, O'Donnell L, Oster S, Theesfeld C, Sellam A, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45:D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz J, Gong Y, Salmasinia D, Hou W, Burkley B, Ferreira P, Casanova O, Langaee TY, Moreb JS. Genetic polymorphisms and other risk factors associated with bisphosphonate induced osteonecrosis of the jaw. Int J Oral Maxillofac Surg. 2011;40:605–611. doi: 10.1016/j.ijom.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Franchi L, Eigenbrod T, Muñoz-Planillo R, Nuñez G. The inflammasome: A caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chellaiah MA, Kizer N, Biswas R, Alvarez U, Strauss-Schoenberger J, Rifas L, Rittling SR, Denhardt DT, Hruska KA. Osteopontin deficiency produces osteoclast dysfunction due to reduced CD44 surface expression. Mol Biol Cell. 2003;14:173–189. doi: 10.1091/mbc.e02-06-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falet H, Pollitt AY, Begonja AJ, Weber SE, Duerschmied D, Wagner DD, Watson SP, Hartwig JH. A novel interaction between FlnA and Syk regulates platelet ITAM-mediated receptor signaling and function. J Exp Med. 2010;207:1967–1979. doi: 10.1084/jem.20100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khosla S, Burr D, Cauley J, Dempster DW, Ebeling PR, Felsenberg D, Gagel RF, Gilsanz V, Guise T, Koka S, et al. Bisphosphonate-associated osteonecrosis of the jaw: Report of a task force of the American Society for bone and mineral research. J Bone Miner Res. 2007;22:1479–1491. doi: 10.1359/jbmr.0707onj. [DOI] [PubMed] [Google Scholar]
- 27.Aghaloo T, Hazboun R, Tetradis S. Pathophysiology of osteonecrosis of the jaws. Oral Maxillofac Surg Clin North Am. 2015;27:489–496. doi: 10.1016/j.coms.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorenzo-Pouso AI, Pérez-Sayáns M, González-Palanca S, Chamorro-Petronacci C, Bagán J, García-García A. Biomarkers to predict the onset of biphosphonate-related osteonecrosis of the jaw: A systematic review. Med Oral Patol Oral Cir Bucal. 2019;24:e26–e36. doi: 10.4317/medoral.22763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coleman RE, Major P, Lipton A, Brown JE, Lee KA, Smith M, Saad F, Zheng M, Hei YJ, Seaman J, Cook R. Predictive value of bone resorption and formation markers in cancer patients with bone metastases receiving the bisphosphonate zoledronic acid. J Clin Oncol. 2005;23:4925–4935. doi: 10.1200/JCO.2005.06.091. [DOI] [PubMed] [Google Scholar]
- 30.Ruggiero SL, Dodson TB, Fantasia J, Goodday R, Aghaloo T, Mehrotra B, O'Ryan F, American Association of Oral and Maxillofacial Surgeons American association of oral and maxillofacial surgeons position paper on medication-related osteonecrosis of the jaw-2014 update. J Oral Maxillofac Surg. 2014;72:1938–1956. doi: 10.1016/j.joms.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 31.Allen MR, Burr DB. The pathogenesis of bisphosphonate-related osteonecrosis of the jaw: So many hypotheses, so few data. J Oral Maxillofac Surg. 2009;67(Suppl 5):S61–S70. doi: 10.1016/j.joms.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Reid IR, Cornish J. Epidemiology and pathogenesis of osteonecrosis of the jaw. Nat Rev Rheumatol. 2012;8:90–96. doi: 10.1038/nrrheum.2011.181. [DOI] [PubMed] [Google Scholar]
- 33.Zhu X, Ma X, Hu Y. PARP1: A promising target for the development of PARP1-based candidates for anticancer intervention. Curr Med Chem. 2016;23:1756–1774. doi: 10.2174/0929867321666140915143516. [DOI] [PubMed] [Google Scholar]
- 34.Mathis BJ, Lai Y, Qu C, Janicki JS, Cui T. CYLD-mediated signaling and diseases. Curr Drug Targets. 2015;16:284–294. doi: 10.2174/1389450115666141024152421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Lai Y, Mathis BJ, Wang W, Li S, Qu C, Li B, Shao L, Song H, Janicki JS, et al. Deubiquitinating enzyme CYLD mediates pressure overload-induced cardiac maladaptive remodeling and dysfunction via downregulating Nrf2. J Mol Cell Cardiol. 2015;84:143–153. doi: 10.1016/j.yjmcc.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 36.Vriend J, Reiter RJ. Melatonin, bone regulation and the ubiquitin-proteasome connection: A review. Life Sci. 2016;145:152–160. doi: 10.1016/j.lfs.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Pascual F, Slatter DA. Collagen cross-linking: Insights on the evolution of metazoan extracellular matrix. Sci Rep. 2016;6:37374. doi: 10.1038/srep37374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saito M, Marumo K. Effects of collagen crosslinking on bone material properties in health and disease. Calcif Tissue Int. 2015;97:242–261. doi: 10.1007/s00223-015-9985-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Github repository, github.com/WeiCSong/PDLSC.