Abstract
The SARS-CoV-2 causes severe pulmonary infectious disease with an exponential spread-ability. In the present research, we have tried to look into the molecular cause of disease, dealing with the development and spread of the coronavirus disease 2019 (COVID-19). Therefore, different approaches have investigated against disease development and infection in this research; First, We identified hsa-miR-1307-3p out of 1872 pooled microRNAs, as the best miRNA, with the highest affinity to SARS-CoV-2 genome and its related cell signaling pathways. Second, the findings presented that this miRNA had a considerable role in PI3K/Act, endocytosis, and type 2 diabetes, moreover, it may play a critical role in the prevention of GRP78 production and the virus entering, proliferation and development. Third, nearly 1033 medicinal herbal compounds were collected and docked with ACE2, TMPRSS2, GRP78, and AT1R receptors, which were the most noticeable receptors in causing the COVID-19. Among them, there were three common compounds including berbamine, hypericin, and hesperidin, which were more effective and appropriate to prevent the COVID-19 infection. Also, it was revealed some of these chemical compounds which had a greater affinity for AT1R receptor inhibitors can be suitable therapeutic targets for inhibiting AT1R and preventing the adverse side effects of this receptor. According to the result, clinical assessment of these three herbal compounds and hsa-miR-1307-3p may have significant outcomes for the prevention, control, and treatment of COVID-19 infection.
Keywords: COVID-19 receptor blockers, SARS-CoV-2, miRNAs, Medicinal herbal compounds, Bioinformatics, Therapeutic targets
Graphical abstract
Highlights
-
•
Different therapeutic targets were found for inhibiting SARS-CoV-2.
-
•
Molecular docking analysis of medicinal herbal compounds were conducted with SARS-CoV-2 receptors.
-
•
The affinity of different miRNAs to 3′ UTR of SARS-CoV-2 genome were investigated.
-
•
The role of miRNAs in the human cell signaling pathways in which SARS-CoV-2 is involved was investigated.
1. Introduction
The SARS-CoV-2 induces severe respiratory syndrome in humans [1]. The viral major proteins include spike (S) protein, matrix protein (M), small envelope protein (E) protein, and nucleocapsid protein (N) [2]. The location of viral attachment to the host cell resides within the S protein [3]. SARS-CoV-2 is able to bind to angiotensin-converting enzyme 2 (ACE2) [4], transmembrane protease serine 2 (TMPRSS2) [5], glucose regulating protein 78 (GRP78) [6]. There are several strategies to tackle the SARS-CoV-2, including inhibiting virus multiplication, inhibiting virus entry through receptors (blocking receptors), and blocking viral proteins. The first approach is using miRNAs' to attach to the 3′UTR of the SARS-CoV-2 genome, so that viral translation will be inhibited [7,8]. Blocking the aforementioned three receptors is the second approach to prevent SARS-CoV-2. It has suggested that decreasing the levels of ACE2 on the cell surface or inhibiting its function, helps in preventing SARS-CoV-2 infection [9]. By binding the spike protein to the ACE2, the angiotensin 2 is produced in large quantities by the ACE and then binds to the AT1R receptor. This process leads to fibrosis and damage lung tissues. A possible way of combating the infection could be the injection of soluble ACE2 into the bloodstream which will have the two-fold effect; preventing the attachment of the virus to non-infected cells and replenishing ACE2 in infected cells [3]. Secondly, it is assumed that SARS-CoV-2 uses the serine protease TMPRSS2 for S protein priming [10,11]. So, the inhibition of TMPRSS2 protease is required for a robust blockade of viral entry. Thirdly, given the fact that diabetic individuals are more likely to get infected by the SARS-CoV-2 virus as its receptor is highly expressed on the lung and intestine and liver cells of individuals with type 2 diabetes. To our knowledge the role of GRP78 is facilitating the entrance of virus to the host cells [12]. Finally, the AT1R receptor is one of the important receptors in modulating of blood pressure. High blood pressure is directly linked to the SARS-CoV-2 virus, by inhibition of this receptor, presumably we would be able to prohibit lung injury. Researches have shown that some of the AT1R-blockers such as Azilsartan and Olmesartan can overexpress the ACE2 and it could be a risky situation for SARA-CoV-2 entry [13]. The third approach could be to target the respective enzyme to prevent the virus from progressing and surviving in the host cell. In this study, we have surveyed these approaches to address alternative therapeutic methods against viral infection. For this purpose, thirty medicinal herbs that have the most impact on the pathways associated with each receptor were selected to analyze their affinity to bind the ACE2, TMPRSS2, GRP78, and AT1R receptors. Besides, to control the virus genome, a comprehensive investigation was performed on the SARS-CoV-2 genome inhibitors.
2. Material and methods
Information about the virus (structure, reproduction, and genome) was obtained from the Viral Zone database (www.viralzone.expasy.org) [14].
2.1. Calculation of 3′UTR and miRNAs affinity
The nucleotide sequences of all 1872 miRNAs were downloaded from the miRBase database (http://mirbase.org) [15] and their expression levels were determined in normal lung squamous cells and lung tissue through TCGA-Assembler 2 package by R software, version 4.0.0 [16]; non-expressed microRNAs were deleted. Then, the 3′UTR sequence of the SARS-COV-2 RNA was obtained from NCBI database entry MTO49951.1 (www.ncbi.nlm.nih.gov) [17]. The affinity between each miRNA and 3′UTR of the viral genome was computed by the RNAhybrid database (https://bio.tools/rnahybrid) [18].
2.2. Identification of the efficient miRNAs, and transcriptional factors
By using the miRWalk database (http://mirwalk.umm.uni-heidelberg.de/) [19] and the TransmiR database (http://www.cuilab.cn/) [20] the targetome and transcription factors of the best miRNA were achieved. Then, those which had expression in lung tissue were checked and selected by the TCGA-Assembler 2 package of R software. Finally, the signaling pathways of the selected miRNAs, targetome, and transcription factors were separately obtained from the KEGG database (www.kegg.jp) [21]. The schematic figure of the signaling pathways related to both targetome and transcription factors of the best miRNAs were drowned by the Biorender software. The targetome schematic figure was produced by Cytoscape software, version 3.8.0 [22].
2.3. Modeling analysis for each receptor
First, the sequence of ACE2, GRP78, TMPRSS2, and AT1R receptors were acquired from the NCBI database and modeled using SWISS-MODEL modeling tools (https://swissmodel.expasy.org/) [23]. The SARS-CoV-2 structure was downloaded from the RCSB PDB database (https://www.rcsb.org/) [24] and the binding site of each receptor related to SARS-CoV-2 obtained from articles.
2.4. Collecting the herbal phytochemicals
After determining thirty medicinal herbs (Table 1 ), their phytochemical compounds were obtained by reviewing different articles that have used gas chromatography-mass spectrometry (GC-MS) technique. The 3-D structure of 1033 compounds from thirty herbs and that of each receptor blockers were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) [25].
Table 1.
Thirty medicinal herbs investigated in this study.
| Name of medicinal herbs | ||
|---|---|---|
| Salvia officinalis | Apium graveolens | Cuminum cyminum |
| Coriandrum sativum | Hibiscus sabdariffa | Foeniculum vulgare |
| Anethum graveolens dhi | Olea europaea | Juglans regia |
| Trigonella foenumgraecum | Lavandula angustifolia | Punica granatum |
| Thymes | Berberis vulgaris | Curcuma longa |
| Allium sativum | Ziziphora | Crocus sativus |
| Camellia sinensis | Achillea millefolium | Pimpinella anisum |
| Silybum marianum | Rhus | Plantago ovata |
| Taxus baccata | Taraxacum | Artemisia absinthium |
| Matricaria chamomilla | Glycine max | Glycyrrhiza glabra |
2.5. Preparation and molecular docking analysis
The 3-D structure of receptor blockers (Table 2 ) and herbal compounds were prepared for docking by removing all water and H-bond optimization, then, Gasteiger charges were added. After that, molecular docking analysis was performed for each herbal compound and receptor blockers using AutoDock vina. Figures were made by Chimera software version 1.14 [26].
Table 2.
Name of receptor blockers for TMPRSS2, AT1R, and GRP78.
| Name of blocker | Receptor blocker |
|---|---|
| Meclizince | TMPRSS2 |
| Probucol | TMPRSS2 |
| Anthralin | TMPRSS2 |
| 2,3-dihydroxy-6,7- dichloroquinoxaline | TMPRSS2 |
| 50k | TMPRSS2 |
| Azilsartan (Edarbi) | AT1R |
| Candesartan (Atacand) | AT1R |
| Eprosartan | AT1R |
| Irbesartan (Avapro) | AT1R |
| Losartan (Cozaar) | AT1R |
| Olmesartan (Benicar) | AT1R |
| Telmisartan (Micardis) | AT1R |
| Valsartan | AT1R |
| OSU-03012 | GRP78 |
| EGCG | GRP78 |
2.6. Determination of beneficial compounds characteristics
The activity, daily dose carcinogenicity, blood-brain barrier penetration, and antiviral features caught from way 2 drug [27] (www.pharmaexpert.ru/passonline/predict.php), lazar (lazar.in-silico.ch/predict) [28] and AVCpred (http://crdd.osdd.net/servers/avcpred) [29], respectively.
3. Results
3.1. Selecting the most efficient miRNAs
According to affinity examination between 3′UTR of SARS-CoV-2 and all 1872 miRNAs, 42 miRNAs with the highest score were identified (Table 3 ). Among them, hsa-miR-1307 3p had the highest score among miRNAs with −37.6 kCal/mol, and high expression level in lung and squamous tissues (Fig. 1 ).
Table 3.
The most important microRNAs which were expressed in lung tissue according to their affinity to the 3′UTR of SARS-COV-2 genome.
| Important miRNAs in COVID-19 with high affinity to SARA-COV-2 genome 3′UTR Affinity (kCal/mol) | |
|---|---|
| hsa-miR-6762-3p | −27.1 |
| hsa-miR -6746-5p | −27.1 |
| hsa-miR −636 | −27.2 |
| hsa-miR -6842-3p | −27.4 |
| hsa-miR −449a | −27.4 |
| hsa-miR -6885-5p | −27.6 |
| hsa-miR -4640-5p | −27.6 |
| hsa-miR -5p | −27.6 |
| hsa-miR -6783-3p | −27.7 |
| hsa-miR -939-5p | −27.8 |
| hsa-miR -4649-5p | −27.8 |
| hsa-miR −181d-5p | −27.8 |
| hsa-miR -615-3p | −27.9 |
| hsa-miR −4674 | −27.9 |
| hsa-miR -6830-3p | −28.0 |
| hsa-miR −5090 | −28.0 |
| hsa-miR -6812-5p | −28.1 |
| hsa-miR −572 | −28.1 |
| hsa-miR -4756-5p | −28.2 |
| hsa-miR −34a-5p | −28.2 |
| hsa-miR -6729-5p | −28.3 |
| hsa-miR -1296-5p | −28.3 |
| hsa-miR -6789-5p | −28.4 |
| hsa-miR −4523 | −28.4 |
| hsa-miR −4469 | −28.6 |
| hsa-miR −484 | −29.0 |
| hsa-miR −449b-5p | −29.0 |
| hsa-miR −1268b | −29.1 |
| hsa-miR -4745-5p | −29.3 |
| hsa-miR −92b-3p | −29.4 |
| hsa-miR −4734 | −29.4 |
| hsa-miR −7974 | −29.6 |
| hsa-miR -6727-5p | −29.6 |
| hsa-miR -2467-5p | −29.9 |
| hsa-miR -1909-3p | −30.2 |
| hsa-miR-7108-5p | −30.3 |
| hsa-miR -381-3p | −30.3 |
| hsa-miR −5087 | −30.6 |
| hsa-miR -6821-5p | −30.7 |
| hsa-miR −4741 | −31.6 |
| hsa-miR −449c-5p | −33.4 |
| hsa-miR -1307-3p | −37.6 kcal/mol |
Fig. 1.
hsa-miR-1307 interaction with 3′UTR of SARS-CoV-2 genome.
3.2. The hsa-miR-1307 3p targetome enrichment study
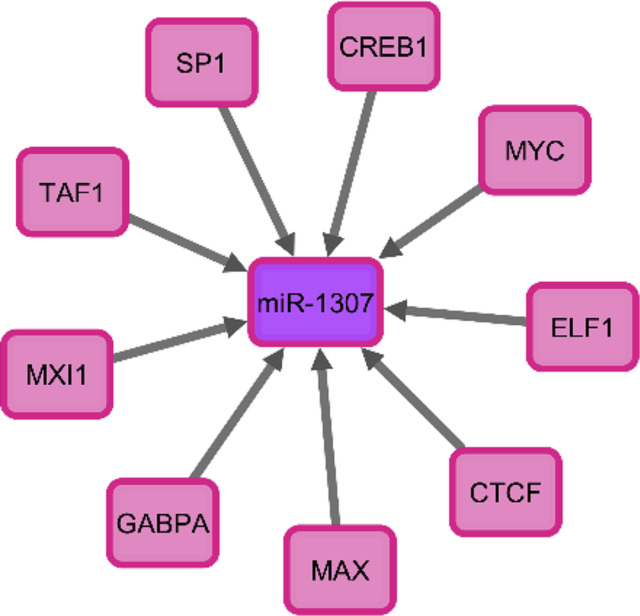
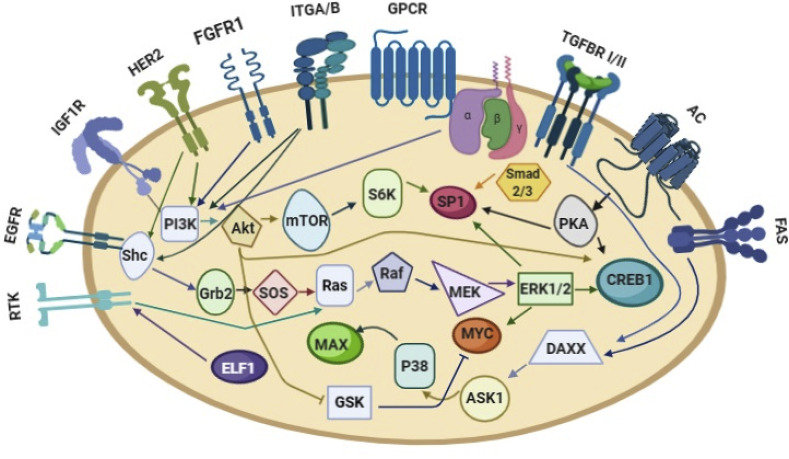
Approximately 377 predicted and valid targets of hsa-miR-1307 which were predicted (Supplementary figure 1). The results revealed hsa-miR-1307-3p had an important role in the “PI3K/Act signaling pathway”, “endocytosis signaling pathway”, and “type 2 diabetes signaling pathway” (Fig. 2 ). SP1, CREB1, MYC, ELF1, CTCF, MAX, GABPA, MAI1, and TAF1, are the most notable of transcription factors (TFs) upstream of hsa-miR-1307 (Fig. 3 A), which the upstream respective signaling pathways responsible for activating these TFs are shown in Fig. 3 B.
Fig. 2.
The targets of hsa-miR-1307. ITGA, ITGB, RTK, and Cytokine receptors are the most important receptors to activate the PI3K. Plus, CDK as the cell cycle progression regulator and BCL2 as anti-apoptotic protein are the most important proteins in cell survival and division; hsa-miR-1307 plays a critical role in suppressing the tumorgenesis and cell proliferation by inhibiting these two genes. On the other hand, hsa-miR-1307 by repressing Erk, PKC, VDCC, and PDX1 can affect type 2 diabetes signaling pathway. Furthermore, the inhibiting effect of hsa-miR-1307 on EPSIN, AP-2, and PIP5k. Rab10, Rab35, PKC, and Actin has a significant role in endocytosis and maybe exocytosis signaling pathways.
Fig. 3A.
The TFs of hsa-miR-1307-3p in lung tissue.
Fig. 3B.
SP1, Smad2/3, PKA, CREB1, MYC, MAX, ELF1 are the most significant of TFs related to hsa-mir-1307, which present in field shapes. The most important receptors to activate the TFs of hsa-miR-1307 are shown in the cell surface.
3.3. Docking analysis
The best herbal compounds for ACE2, TMPRSS2, GRP78, and AT1R receptors were identified based on their binding energy.
3.4. The common medicinal herbs and blocker drugs
Thirty-five common medicinal herbal compounds and blocker drugs were investigated between ACE2, TMPRSS2, GRP78, and AT1R; with a score of −7 kCal/mol or higher. Among them, Berbamine had the highest binding energy for all four receptors (Supplementary Table 1).
3.5. Selecting top common compounds
Berbamine, Hesperidin, and Hypericin were the best common compounds for interaction with ACE2 (Supplementary Figure2), TMPRSS2 (Supplementary Figure3), GRP78 (Supplementary Figure4), and AT1R (Supplementary Figure5).
The results of molecular docking also showed a high tendency of compounds to receptors. The molecular docking results of the ACE2, TMPRSS2, GRP78, and AT1R receptors with medicinal herbal compounds are presented in Supplementary Table 2.
3.6. Comparison of the binding energy for receptors
The results of molecular docking to compare the binding energy between the available blocking drugs and the herbal compounds investigated from the present study presented that the affinity of herbal compounds to bind to the ACE2, TMPRSS2, GRP78, and ACE2 receptors were higher (Table 4 ).
Table 4.
Comparison between the binding energy of blocker drugs and herbal compounds for AT1R , TMPRSS2, GRP78, and ACE2.
| AT1R blockers | Score (kCal/mol) | TMPRSS2 blockers | Score (kCal/mol) | GRP78 blockers | Score (kCal/mol) | ACE2 blockers | Score (kCal/mol) |
|---|---|---|---|---|---|---|---|
| Losartan (Cozaar) | −13.3 | epigallocatechin-3-gallate | −8.9 | OSU-03012 | −6.8 | Losartan (Cozaar) | −9.3 |
| Telmisartan (Micardis) | −11 | 50K | −8.9 | epigallocatechin-3-gallate | −6.2 | Scutellarin | −8.6 |
| Azilsartan (Edarbi) | −10.5 | meclizine | −7.8 | – | – | baicalin | −8.3 |
| Irbesartan (Avapro) | −9.9 | anthralin | −7.4 | – | – | Hesperetin | −7 |
| Candesartan (Atacand) | −9.8 | 2,3-dihydroxy-6,7-dichloroquinoxaline | −7.1 | – | – | N-[[4-(4-methylpiperazin-1-yl)phenyl]methyl]-1,2-oxazole-5-carboxamide | −6.8 |
| Olmesartan (Benicar) | −9.5 | – | – | – | – | Chloroquine phosphate (ResochinTM) | −5.8 |
| valsartan | −8.9 | – | – | – | – | Nicotianamine | −5.5 |
| Eprosartan |
−8.1 |
– |
– |
– |
– |
N-(2-aminoethyl)-1 aziridine-ethanamine |
−3.7 |
|
Herbal compounds for AT1R |
Score(kCal/mol) |
Herbal compounds for TMPRSS2 |
Score(kCal/mol) |
Herbal compounds for GRP78 |
Score(kCal/mol) |
Herbal compounds for ACE2 |
Score(kCal/mol) |
| Berbamine | −15.7 | Berbamine | −11.8 | Berbamine | −11 | Berbamine | −12.3 |
| Hop-17(21)-en-3β-ol | −11.4 | Hesperidin | −9.9 | Lupeol | −7.7 | Naringin | −8.6 |
| 18β-glycyrrhetinic acid | −11.4 | Berberine | −9.8 | Shinflavanone | −7.7 | Officinatrione | −8.4 |
| Glycyrrhetinic acid | −11.4 | Isosilybin B | −9.8 | Glabrolide | −7.7 | α-Carotene | −8.4 |
| Hispaglabridin B | −11.3 | Plantamajoside | −9.8 | – | – | – | – |
| – | – | Apigenin-7-O-rutinoside | −9.7 | – | – | – | – |
| – | – | Isomartynoside | −9.7 | – | – | – | – |
| – | – | Rutin | −9.6 | – | – | – | – |
| – | – | Silandrin | −9.6 | – | – | – | – |
| – | – | Silyhermin | −9.6 | – | – | – | – |
3.7. Attaining the specifications of top common compounds
The lazar database had no result for Hestridine, but characteristics of other top three common compounds include antitussive, respiratory analeptic, hepatoprotection, antivirus, RNA synthesis inhibitor. Furthermore, features such as blood-brain barrier penetration, carcinogenicity, maximum recommended daily dose, and antiviral ability of each three common herbal compounds were surveyed and presented in Table 5 .
Table 5.
Different characteristics of common medicinal herbs with high affinity to ACE2, AT1R, TMPRSS2, and GRP78 receptors.
| Name of Compound | activity● | Blood-Brain Barrier Penetration (Human)* | Carcinogenicity (Mouse)* | Maximum Recommended Daily Dose (Human)* | ANTI VIRAL (Predicted percentage)§ |
|---|---|---|---|---|---|
| Berbamin | Antitussive, Apoptosis agonist, Leukopoiesis stimulant, Antinociceptive, Respiratory analeptic, Spasmolytic, Membrane permeability inhibitor. | Penetrating | non-carcinogenic | 0.0064 (mmol/kg-bw/day) 3.9 (mg/kg_bw/day) |
54.176 |
| Hesperidin | Free radical scavenger, Anticarcinogenic, Hepatoprotectant, Membrane permeability inhibitor, Chemopreventive, Vasoprotector, Hemostatic, Membrane integrity agonist, Caspase 3 stimulant, Proliferative diseases treatment, Cardioprotectant, Antihypercholesterolemic, Antiprotozoal (Leishmania), Skin whitener, Antioxidant, Antineoplastic, Laxative, Antifungal, Expectorant, TP53 expression enhancer, Caspase 8 stimulant, Vasodilator, Antihemorrhagic, Antitussive, Antiulcerative, Antiinflammatory, Apoptosis agonist, Antibacterial, Hepatic disorders treatment, Antiinfective, Respiratory analeptic, RNA synthesis inhibitor, Antiviral (Herpes), | non-penetrating | non-carcinogenic | 0.022 (mmol/kg-bw/day) 13.5 (mg/kg_bw/day) |
54.176 |
| Hypericin | Membrane integrity agonist, TP53 expression enhancer, Antineoplastic, Membrane permeability inhibitor, Apoptosis agonist, Caspase 3 stimulant, Antiseborrheic, Antiseptic, Kidney function stimulant, Antimutagenic, Vasoprotector | – | – | – | 58.805 |
● The activity of each active substance gained from the pass online database (http://www.pharmaexpert.ru/passonline/index.php). * The Blood-Brain Barrier, Carcinogenicity, and Maximum Recommended Daily Dose prediction was obtained from the lazar database (https://lazar.in-silico.ch). § The antiviral activity of each active substances predicted by AVCpred database (http://crdd.osdd.net/servers/avcpred).
4. Discussion
The high mortality rate around the world due to COVID-19 has led to the recognition and treatment of the SARS-CoV-2. There has been a lot of research on this disease over the past few months, but there is still a need for a comprehensive study that considers all aspects of the disease [30]. In this bioinformatics study, it was tried to attend different approaches related to the SARS-CoV-2 before and after the infection.
The most important receptors for SARS-Cov-2 are ACE2 [31] TMPRSS2 [32], and GRP78 [6], which by blocking these receptors can protect the body against the virus. In the process of infecting by SARS-CoV-2, binding the virus to ACE2 prevents the attachment of angiotensin 1 and 2 to ACE2. In the lack of ACE2 enzyme, angiotensin I convert to angiotensin II by ACE enzyme. Subsequently, angiotensin II binds to the AT1R, which causes more damage to the lungs. Therefore, targeting the AT1R [33] could be an alternative treatment goal.
Some studies have shown that AT1R blockers result in more ACE2 production. ACE2, in turn, is not harmful to normal lung cells, but it can play a leading role in SARS-Cov-2 infection, as the spike virus uses this receptor to enter the cell. Therefore, in this study, the effect of blocking the active compounds of medicinal plants on AT1R was investigated, and compounds with the high binding affinity to AT1R were identified. Some compounds have a much higher affinity than AT1R common blockers such as Losartan, Olmesartan, Telmisartan. Berbamine, which is Berberis vulgaris phytochemical compound, has the highest affinity to AT1R, with an affinity higher than of losartan. Moreover, recent studies stated that some blockers of AT1R lead to enhance ACE2 expression. The 3-D structure of Berbamin differs from the common blockers, which may not lead to an increase in the expression of ACE2; hence, fewer ACE2 could be available to the SARS-CoV-2 virus. As mentioned earlier, blocking ACE2, GRP78, and TMPRSS2 could be a suggestive therapeutic option. In the present study, phytochemical compounds of each medicinal herbs were surveyed in terms of their blocking ability for each receptor. The binding energy of Berbamine is about −12.3 kCal/mol, −11 kCal/mol, and −11.8 kCal/mol for ACE2, GRP78, and TMPRSS2 respectively. This indicates the efficiency of this compound in a competitive circumstance with the virus. Thirty-five effective compounds with proper scores for ACE2, GRP78, AT1R, and TMPRSS2 receptors were obtained through screening the 1033 medicinal herbal compounds and it was found that the Berbamine, Losartan, Hypericin, and Hesperidin with the highest rate of affinity are common to all four receptors. Moreover, recent studies have shown that Hypericin [34] and Hesperidin [35,36] are important in the ACE2 receptor and Berbamine [37] is related to increasing secretion of ACE2 and DPP receptors and decreasing the amount of these receptors on the plasma membrane. Therefore, blocking receptors can prevent the progression of infection at the lung or it can decrease even the risk of infection for people exposing the virus. As previously stated, Losartan may increase the expression of ACE2, thus, it was not examined in the subsequent investigation in this study. But, Berbamine, Hypericin, and Hesperidin compounds were included in this study to investigate their pharmacological properties. It has been found that Berbamine and Hypericin have a completely different 3-D structure from those of conventional AT1R blockers, which it seems that probably not lead to enhancing ACE2. Patients with higher blood pressure are more likely at risk of COVID-19, so there is a doubt about prescribing anti-blood pressure drugs. Accordingly, the mentioned herbal compounds may downregulate the blood pressure and lung injury by blocking the AT1R receptor without activating ACE2. Among the aforementioned components, Berbamine, Hypericin, and Hesperidin are not carcinogenic. Berbamine has other properties such as antitussive, respiratory protection, non-toxic, antibacterial, which makes the use of this substance as a very safe respiratory tract. By examining the essential oils and extracts of various medicinal plants, it was found that the extracts of Berberis vulgaris (Berbamine), Ziziphora (Hesperidin), and Hibiscus sabdariffa (Hypericin) can play the best role in the prevention and treatment of COVID-19. Finally, Silybum marianum, Glycyrrhiza glabra, and Achillea millefolium had the highest effects on all four receptors.
If the lung cell is infected with the SARS-CoV-2, preventing the virus from replicating is another treatment option [38]. Many studies have shown that miRNAs play an important role in inhibiting viruses' replication by preventing the translation of the viral genome [7]. In this study, among 1872 miRNAs, has-miR-1307-3p was selected as the best miRNA because of its high affinity to SARS-CoV-2 genome 3′UTR and high expression in lung tissue [39]. Increased expression of hsa-miR-1307-3p may lead to a reduction in SARS-CoV-2 replication through binding to the 3′UTR site of the virus genome. miRNAs can be synthesized for therapeutic purposes and inserted into the target area, but their delivery is a complicated and meticulous process. Therefore, the use of TFs to induce the expression of the desired miRNA by lung cells may be a better choice. After obtaining all the signaling pathways related to the hsa-miR-1307-3p transcription factors, the receptors responsible for activating the TFs were also examined. As a result, using the ligand of each of these receptors can lead to the expression of the TFs genes; eventually triggers the induction of the hsa-miR-1307-3p production.
Of important, hsa-miR-1307-3p can affect receptors that are responsible for survival and proliferation. The hsa-miR-1307-3p also affects anti-apoptotic proteins like BCL2 and inhibits them to induce apoptosis, leading to the prohibition of the proliferation. Investigations have proven that the PI3K signaling pathway is so critical for virus survival and spread [40]. The hsa-miR-1307-3p inhibits the PI3K pathway by suppressing the activators of PI3K, and eventually, the cell cycle proliferation is prevented. On the other hand, hsa-miR-1307-3p can ban clathrin-dependent endocytosis by inhibiting AP-2, PIP5K, and might suppress exocytosis by inhibiting of actin. It has been confirmed that endocytosis and exocytosis are associated with virus entry and spread, therefore, controlling these pathways by hsa-miR-1307-3p could be an effective strategy for SARS-CoV-2 infection.
The WHO has stated that obesity and diabetes mellitus are other risk factors for developing COVID-19. In obesity that leads to type 2 diabetes, hyperglycemia leads to inhibition of the immune system [41] as well as the activation of PKC, as a result of its activity, inhibits IRS2, and leads to indirect inhibition of ACT, which in turn leads to glycogenesis and glucose uptake. On the other hand, hyperglycemia increases the expression of GRP78 on the cell surface, which can activate the ACT pathway to reduce blood sugar [42]. Activating the tyrosine kinase receptor activates the ERK protein, and this protein inhibits IRS1 that also indirectly prevents the ACT and its downstream pathway. Studies have implicated that GRP78, which is increased in response to hyperglycemia in diabetes, is a receptor for COVID-19. The hsa-miR-1307-3p can induce glycogenesis and glucose uptaking by inhibiting PKC and ERK, which are inhibitors of RS1 and RS2. Thus, this miRNA indirectly reduces GRP78 production by lowering blood sugar. On the other hand, hsa-miR-1307-3p inhibits insulin expression by inhibiting PDX1 and VDCC, which activates insulin, increasing insulin-dependent proliferative pathways in type 2 diabetes. MicroRNA 1307 also inhibits insulin expression by inhibiting insulin-activated PDX1 and VDCC, resulting in insulin-dependent proliferative pathways in type 2 diabetes, thus controlling insulin-dependent proliferative pathways in type 2 diabetes. Studies have shown [40] that in COVID-19, with increased cytokine storms, lung tissue damage increases. For example, one of the treatment strategies for receptor inhibition is cytokines such as interleukin 6 receptor. Therefore, hsa-miR-1307-3p can be a therapeutic solution to inhibit tissue damage by inhibiting cytokine receptors.
5. Conclusion
The results of this study have indicated the inhibiting effect of Berbamine, Hypericin, and Hesperidin on ACE2, TMPRSS2, GRP78, and AT1R receptors. Also, enrichment analysis of hsa-mir-1307-3p revealed the suppressing role of this miRNA on proliferative, endocytosis, exocytosis, and diabetes signaling pathways. Additionally, hsa-miR-1307-3p targets genes in type 2 diabetes indirectly by repressing the GRP78 receptor expression. Overall, further clinical assessment of these herbal compounds and hsa-miR-1307-3p is required to validate the treating ability of these compounds over COVID-19 infection.
Funding sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
Negar Balmeh, Samira Mahmoudi, Niloufar Mohammadi, Anasik Karabedianhajiabadi have a similar contribution to the first authors. All authors equally contributed to this work, outlined the manuscript, critically updated, and approved the last version before submission. All authors have studied and acknowledged the published version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The study was conducted with the personal interest of all authors and without the support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2020.100407.
Abbreviation
- ACE2
angiotensin-converting enzyme 2
- AT1R
Angiotensin 1 receptor
- AP-2
complex subunit alpha
- BCL2
B-cell lymphoma 2
- CDK
Cyclin-dependent kinase
- CREB1
CAMP responsive element binding protein 1
- CTCF
CCCTC-binding factor
- ELF1
E74-like factor 1
- GRP78
Binding immunoglobulin protein
- GABPA
GA binding protein transcription factor
- GC-MS
Gas chromatography-mass spectrometry
- ITGA
Integrin Subunit Alpha
- ITGB
Integrin Subunit Beta
- MERS-CoV
Middle East respiratory syndrome coronavirus
- MYC
proto-oncogene protein
- MAX
myc-associated factor X
- MXI1
MAX Interactor 1
- PDX1
pancreatic and duodenal homeobox 1
- PIP5k
Phosphatidylinositol-4-phosphate 5-kinase
- PKC
Protein kinase C
- PKA
Protein kinase A
- PI3K
phosphoinositide 3-kinase
- RTK
Receptor tyrosine kinase
- Rab10
Ras-related protein Rab-10
- RAB35
Ras-related protein Rab-35, SARS-CoV, severe acute respiratory syndrome coronavirus
- SP1
specificity protein 1
- SMAD2
Mothers against decapentaplegic homolog 2, SMAD3, Mothers against decapentaplegic homolog 3
- TMPRSS2
Transmembrane protease serine 2
- TAF1
TATA-Box Binding Protein Associated Factor 1
- VDCC
Voltage-dependent calcium channels
- WHO
World Health Organization
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Shen M., Zhou Y., Ye J., AL-maskri A.A.A., Kang Y., Zeng S. Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal. 2020:97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fields B.N., Knipe D.M., Howley P.M., Griffin D.E. Lippincott Williams & Wilkins; Philadelphia: 2001. Fields' virology. [Google Scholar]
- 3.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. BioRxiv. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [Google Scholar]
- 5.Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 Spike-host cell receptor GRP78 binding site prediction. J Infect. 2020:554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haasnoot J., Berkhout B. Antiviral RNAi. Springer; 2011. RNAi and cellular miRNAs in infections by mammalian viruses; pp. 23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peele K.A., Chandrasai P., Srihansa T., Krupanidhi S., Sai A.V., Babu D.J. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: a computational study. Inf Med Unlocked. 2020:100345. doi: 10.1016/j.imu.2020.100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;80– doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84(24):12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85(2):873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu H., Chan C.-M., Zhang X., Wang Y., Yuan S., Zhou J. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J Biol Chem. 2018;293(30):11709–11726. doi: 10.1074/jbc.RA118.001897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurwitz D. Angiotensin receptor blockers as tentative SARS‐CoV‐2 therapeutics. Drug Dev Res. 2020:537–540. doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulo C., De Castro E., Masson P., Bougueleret L., Bairoch A., Xenarios I. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res. 2011;39(suppl_1):D576–D582. doi: 10.1093/nar/gkq901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(D1):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y., Qiu P., Ji Y. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods. 2014;11(6):599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coordinators N.R. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2017;45(Database issue):D12. doi: 10.1093/nar/gkw1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ison J., Rapacki K., Ménager H., Kalaš M., Rydza E., Chmura P. Tools and data services registry. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sticht C., De La Torre C., Parveen A., Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PloS One. 2018;13(10) doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong Z., Cui Q., Wang J., Zhou Y. TransmiR v2. 0: an updated transcription factor-microRNA regulation database. Nucleic Acids Res. 2019;47(D1):D253–D258. doi: 10.1093/nar/gky1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hm berman Bank PD., westbrook j., feng z., gilliland g. Tn bhat, h. weissig, in shindyalov, pe bourne. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 27.Filimonov D.A., Druzhilovskiy D.S., Lagunin A.A., Gloriozova T.A., Rudik A.V., Dmitriev A.V. Computer-aided prediction of biological activity spectra for chemical compounds: opportunities and limitations. Biomed Chem Res Methods. 2018;1(1) e00004–e00004. [Google Scholar]
- 28.Maunz A., Gütlein M., Rautenberg M., Vorgrimmler D., Gebele D., Helma C. Lazar: a modular predictive toxicology framework. Front Pharmacol. 2013;4:38. doi: 10.3389/fphar.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qureshi A., Kaur G., Kumar M. AVC pred: an integrated web server for prediction and design of antiviral compounds. Chem Biol Drug Des. 2017;89(1):74–83. doi: 10.1111/cbdd.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azim K.F., Ahmed S.R., Banik A., Khan M.M.R., Deb A., Somana S.R. Screening and druggability analysis of some plant metabolites against SARS-CoV-2: an integrative computational approach. Inf Med Unlocked. 2020:100367. doi: 10.1016/j.imu.2020.100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun M.L., Yang J.M., Sun Y.P., Su G.H. Inhibitors of RAS might be a good choice for the therapy of COVID-19 pneumonia. Zhonghua jie he he hu xi za zhi= Zhonghua jiehe he huxi zazhi= Chinese. J Tuberc Respir Dis. 2020;43 doi: 10.3760/cma.j.issn.1001-0939.2020.0014. E014–E014. [DOI] [PubMed] [Google Scholar]
- 34.Smith M., Smith J.C. 2020. Repurposing therapeutics for COVID-19: supercomputer-based docking to the SARS-CoV-2 viral spike protein and viral spike protein-human ACE2 interface. [Google Scholar]
- 35.Meneguzzo F., Ciriminna R., Zabini F., Pagliaro M. Review of evidence available on hesperidin-rich products as potential tools against COVID-19 and hydrodynamic cavitation-based extraction as a method of increasing their production. Processes. 2020;8(5):549. [Google Scholar]
- 36.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang L, Li H, Yuen TT-T, Ye Z, Fu Q, Sun W, et al. Berbamine inhibits the infection of SARS-CoV-2 and aviviruses by compromising TPRMLs-mediated endolysosomal tra cking of viral receptors.
- 38.Krishnan D.A., Sangeetha G., Vajravijayan S., Nandhagopal N., Gunasekaran K. Structure-based drug designing towards the identification of potential anti-viral for COVID-19 by targeting endoribonuclease NSP15. Inf Med Unlocked. 2020:100392. doi: 10.1016/j.imu.2020.100392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soremekun O.S., Omolabi K.F., Soliman M.E.S. Identification and classification of differentially expressed genes reveals potential molecular signatures associated with SARS-CoV-2 infection in lung adenocarcinoma cells. Inf Med Unlocked. 2020:100384. doi: 10.1016/j.imu.2020.100384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kindrachuk J., Ork B., Hart B.J., Mazur S., Holbrook M.R., Frieman M.B. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob Agents Chemother. 2015;59(2):1088–1099. doi: 10.1128/AAC.03659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh A.K., Gupta R., Ghosh A., Misra A. Diabetes in COVID-19: prevalence, pathophysiology, prognosis and practical considerations. Diabetes Metab Syndr Clin Res Rev. 2020:303–310. doi: 10.1016/j.dsx.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Krieken R., Mehta N., Wang T., Zheng M., Li R., Gao B. Cell surface expression of 78-kDa glucose-regulated protein (GRP78) mediates diabetic nephropathy. J Biol Chem. 2019;294(19):7755–7768. doi: 10.1074/jbc.RA118.006939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.