Abstract
Arrhythmogenic cardiomyopathy is a genetic heart muscle disorder characterized by fibro-fatty replacement of cardiomyocytes leading to life-threatening ventricular arrhythmias, heart failure and sudden cardiac death. Mutations in genes encoding cardiac junctional proteins are known to cause about half of cases, while remaining genetic causes are unknown. Using exome sequencing we identified two missense variants (p.H33N; p.H77Y) that were predicted to be damaging in the integrin-linked kinase (ILK) gene in two unrelated families. The p.H33N variant was found to be de novo.
ILK links integrins and the actin cytoskeleton and is essential for the maintenance of normal cardiac function. Both of the new variants are located in the ILK ankyrin repeat domain, which binds to the first LIM domain of the adaptor proteins PINCH1 and PINCH2. In silico binding studies proposed that the human variants disrupt the ILK-PINCH complex. Recombinant mutant ILK expressed in H9c2 rat myoblast cells shows aberrant prominent cytoplasmic localization compared to the wildtype. Expression of human wild-type and mutant ILK under the control of the cardiac-specific cmlc2 promotor in ipebrafish shows that p.H77Y and p.P70L, a variant previously reported in a dilated cardiomyopathy family, cause cardiac dysfunction and death by about 2–3 weeks of age. Our findings provide genetic and functional evidence that ILK is a cardiomyopathy disease gene and highlight its relevance for diagnosis and genetic counselling of inherited cardiomyopathies.
Background
Arrhythmogenic cardiomyopath (ACM) is a genetic heart muscle disorder, however about half of the genetic causes remain unknown. Finding of novel genetic variants in new disease genes requires evidence for gene-disease association according to the "Clinical Genome Resource" framework. In particular functional data are important.
INTRODUCTION
Arrhythmogenic cardiomyopathy (ACM), also known as arrhythmogenic right ventricular cardiomyopathy (ARVC), is an inherited myocardial disease leading to heart failure and sudden cardiac death, particularly in young and athletic individuals. Fibro-fatty replacement of myocardial tissue is a histological hallmark of the disease [1]. Pathogenic mutations in one or more desmosomal genes (PKP2, DSP, JUP, DSG2, DSC2) occur in about 50% of patients who fulfill criteria for the clinical diagnosis of ACM [2–6]. More rarely, variants in genes encoding structural proteins involved in cell-cell or cell-matrix adhesion have been reported to cause ACM [7, 8]. Incomplete penetrance and variable clinical expression are common in inherited ACM. Even in the same family there can be a wide range in severity and expressivity of phenotypes: some family members develop dilated cardiomyopathy (DCM), while others manifest ACM or even are clinically unaffected [9].
Integrin-linked kinase (ILK) is a serine/threonine protein kinase that plays important roles in cell-matrix interactions and induction of biomechanical signals for cytoskeleton remodeling, angiogenesis, cell growth, proliferation, survival, and differentiation. ILK has recently been identified as a target in various pathways of heart remodeling [10–13]. Indeed, targeted ILK ablation in the murine and zebrafish models causes cardiomyopathy and heart failure, whereas overexpression of ILK improves cardiac remodeling and function [11, 14–16]. Moreover, the ILK-parvin-PINCH (IPP) complex has been shown to play an important role in heart failure via disturbed IPP/protein kinase B-mediated signaling [13, 17, 18].
Little is known about the role of ILK genetic variants in the pathogenesis of inherited forms of cardiomyopathy in humans. Rare ILK variants of uncertain pathogenicity have previously been reported in 5 patients with DCM [14, 19, 20], but none of these variants has been functionally evaluated in detail.
Here, we report novel ILK missense variants (p.H33N; p.H77Y) in two unrelated families with ACM. Both mutations are located in the ILK ankyrin repeat domain (ARD), which binds to the first LIM domain of PINCH1 and PINCH-2, in a region where in silico studies suggest a disruption of the ILK-PINCH complex. Recombinant mutant ILK expressed in H9c2 rat myoblast cells remains stable but shows aberrant cytoplasmic localization compared to the wildtype. Cardiac expression of mutant ILK (p.H77Y or p.P70L, a variant previously reported in a patient with dilated cardiomyopathy [19] in zebrafish causes premature death due to heart failure. Transgenic zebrafish expressing ILK-p.H33N survive until adulthood, but mutant adult cardiomyocytes showed a prolonged action potential. Our data provide evidence that mutations in ILK can cause cardiomyopathy. Our findings have relevance for the genetic diagnosis and counselling of families with inherited cardiomyopathy.
MATERIALS and METHODS
Clinical Evaluation
The study was conformed to the principles outlined in the Declaration of Helsinki and was approved by Institutional Review Boards of the University of Calgary and University of British Columbia (ID-E23515). All involved family members provided informed consent to participate in the research protocol. Whole exome sequencing of three individuals of family A (FORGE 221) was performed under the FORGE research protocol (H10–03215).
Clinical evaluation included 12-lead ECG, 24 h ambulatory Holter monitoring, exercise testing using the Bruce protocol, signal-averaged ECG (SA-ECG) and cardiac magnetic resonance imaging (CMR). The diagnosis of ACM was made in agreement with Task Force Criteria [2]. In some cases, additional investigations and/or records such as electrophysiology studies to induce ventricular tachycardia, two-dimensional echocardiography, rhythm reports from implantable-cardioverter defibrillator (ICD) or pathology reports were obtained. In family A, endomyocardial biopsies from three members (III-1, III-2, III-4) done for clinical purposes were reviewed.
Exome and Sanger Sequencing
Genomic DNA of three affected siblings of family A (III-2, III-3, III-4) underwent whole exome sequencing. Coding exons were captured from isolated genomic DNA using Agilent Sure Select Human All Exon 50Mb Kit (Agilent Technologies) and sequenced using a HiSeq2000 (Illumina). Read alignment, variant calling, and annotation were done with a pipeline based on bcbio (https://github.com/bcbio/bcbio-nextgen), GEMINI [21], methods accumulated in previous FORGE and Care4Rare Canada Projects [22–24] and custom annotation scripts (https://githup.com/naumenko-sa/cre). Minor allele frequencies (MAF) of controls were evaluated using the Genome Aggregation Database v2.1 controls (gnomAD, http://gnomad.broadinstitute.org/) [25]. Rare variants (population frequency <1%) that were shared amongst the three clinically affected individuals were identified as potential causal variants and further validated in the family. Candidate disease-causing variants found in ILK in family A were confirmed by Sanger sequencing. 30 additional index cases with a confirmed diagnosis of ACM were analyzed in all 12 exons and flanking intronic sequences and 5’ and 3’UTRs of ILK (NM_001014795.2) by Sanger sequencing.
Plasmid generation and cell culture
The pEGFP-C2-ILK-WT plasmid was a kind gift from Prof. Dr. Chuanyue Wu (Department of Pathology, University of Pittsburgh, Pittsburgh, PA 15261) and contains full length ILK cDNA.
The ‘QuickChange Lightning Kiť (Agilent Technologies, Santa Clara, USA) was used to insert the variants into the plasmid using appropriate primers. Bases at codon 33 (Cat>Aat), codon 70 (cCc>cTc) and codon 77 (Cat>Tat) of the ILK cDNA were mutated according to the human variants. The pmRuby-N1-PINCH1 and pmRuby-N1-PINCH2 plasmids were cloned by exchange of the DEScDNA in pmRuby-N1-DES against the PINCH1/2 cDNAs [26].
H9c2 and HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, L-Glutamine, 4.5 g/L Glucose, Lonza Group, Basel, Switzerland) supplemented with 10% fetal bovine serum (FBS) and 500 U/mL penicillin and 500 μg/mL streptomycin at 37°C and 5% CO2. Cells were grown on coverslips and transfected using Lipofectamine 2000 (Life Technologies, Carlsbad, USA) or PEI (cellntec) according to the manufacturer’s instructions for transfection.
Confocal microscopy
24 h after transfection cells were washed three times with phosphate buffered saline (PBS, pH 7.4), fixed with methanol (−20°C, 5 min) and embedded with ‘ProLong Gold antifade reagenť (Invitrogen). The ‘LSM 500’ system (Carl Zeiss Microscopy) was used for confocal microscopy as previously described [27].
Co-immunoprecipitation
Co-immunoprecipitation (CoIP) was performed using GFP-NanoTraps (Chromotek) as previously described [28]. Briefly, 1x107 HEK293 cells were transiently co-transfected with pEGFP-ILK and pCMV6-PINCF1 or −2 (Origene) plasmids. Protein lysates were used 24 h after transfection for CoIP in combination with Western blot analysis using rabbit anti-ILK (Millipore, 1:1000) and rabbit anti-PINCH1 (Abcam, 1:1000) or goat anti-PINCH2 antibodies (Abcam, 1:1000).
Generation of zebrafish lines
The University of Calgary Animal Care Committee approved all zebrafish protocols, and these experiments conform to the Guide for the Care and Use of Laboratory Animals in Canada. Additionally, the study was performed in compliance with the recommendations in the Guide and Care for the Use of Laboratory Animals of the University Hospital Wurzburg and was approved by the local government of Lower Franconia (approval no.55.2–2532-2–355).
Care and breeding of zebrafish were conducted as previously described [29]. Briefly, zebrafish were bred and maintained under standard conditions at 28.5°C. Embryos were kept in embryo medium E3 (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2 and 0.33 mM MgSO4) dissolved in water and staged as described previously in hours post fertilization (hpf) [30].
For cardiac-specific overexpression, human eGFP-C2-ILK constructs with one of the three mutations (p.H77Y; p.H33N; p.P70L) and the human eGFP-C2-ILK wild-type were cloned downstream of the cmlc2/myl7 promoter into the Tol2kit expression system using Gateway technology (Invitrogen) according to Kwan et al., 2007 [31]. The allele numbers studied are pDestTol2PA2 EGFP-ILK-WT, pDestTol2PA2 EGFP-ILK-H77Y, pDestTol2PA2 EGFP-ILK-H33N and pDestTol2PA2 EGFP-ILK-P70L. We co-injected 75 pg of the ILK constructs with 25 pg capped Tol2 transposase mRNA into one-cell-stage zebrafish embroys to establish stable transgenic founder lines. F1 incrosses were used for further analyses.
Survival analysis
Progeny from transgenic F1 in-crossed mating were raised in the same nursery environment. At 3 dpf, larvae were screened for transgene gfp expression and at 5 dpf were grown at a density of 50 larve per 3 L tank. Mortality was determined by recording the number of dead larvae/juvenile fish daily over a time course of 30 days.
Measurement of fractional shortening (FS) and heart rate (HR)
FS and HR were calculated according to Hoage et al. [32]. Briefly, videos of beating hearts from embryos 72 hpf were recorded for 20s using a Leica DM IL LED fluorescent microscope equipped with a Leica DFC camera. Lengths of the long and short axes were measured between the myocardial borders of the ventricle at diastole and systole in three contractile cycles per video. FS was calculated with the formula: [(width at diastole – width at systole) / (width at diastole) ]x 100%. HR was calculated by counting the number of beats in 15 s, from either a video file or in living animals.
Embryonic heart isolation and action potential recording
72 hpf embryos were anaesthetized using cold E3 medium. Embryonic hearts were dissected from the thorax en bloc by using fine forceps and transferred to the recording chamber containing room temperature Tyrode solution (NaCl 145 mM, KCl 5.4 mM, CaCl2 2 mM, 1mM MgCl2, 1 mM Na2HPO4,5 mM N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid (HEPES) and 5.5 mM glucose (pH 7.4, adjusted with NaOH). Only spontaneously beating whole embryonic hearts were studied. Electrodes were fabricated using borosilicate capillary glass tabes (World Precision Instruments; Sarasota, FL). Electrodes had resistances of 1–3 MΩ when filled with filling solution (135 mM KC1, 5 mM ethylene glycol-bis (β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid (EGTA), 10 mM HEPES, pH 7.2, adjusted with KOH). Action potentials from spontaneously beating hearts were recorded by placing an electrode adjacent to the heart and forming a seal by application of suction. Stable spontaneou&action potentials were recorded using an Axopatch 200B clamp amplifier, digitized using Digidata 1322A, and stored using pClamp software (Molecular Devices, Sunnyvale). The duration of multiple action potentials from each heart were measured. Since deteMiination of the exact end of action potentials is prone to error, we opted to measure the interval between onset of action potentials and the time at 90% of repolarization (APD90). The diastolic duration prior to each action potential was determined by measuring the interval between the end of the preceding action potential and the onset of the measured action potential.
Adult cardiac myocyte preparation and APD recording
Adult cardiac myocytes were isolated as previously described [33]. Briefly, adult zebrafish were anaesthetized by thermal shock using ice cold water. The heart was excised and positioned in Tyrode solution. After isolating the heart from other surrounding tissue, a 34-gauge needle (Nodegraf, Tokyo, Japan) was inserted into the ventricle via the bulbous arteriosus and secured in position using a single fiber of surgical silk suture. The cannulated heart was then transferred to a Ca2+ free Tyrode solution and was perfused for 20 min with Ca2+ free Tyrode solution containing 50 μM EGTA and 0.16 mg/ml collagenase (Yakult Pharmaceutical Industries, Tokyo, Japan) and 0.5 mg/mL bovine serum albumin. At the end of the digestion, the atrium was removed and the remaining ventricle was transferred to a Ca2+-free Tyrode’s solution. Ca2+ was gradually increased to 1 mM over 30 min. Isolated cardiac myocytes were immediately used for measurement of action potentials. Membrane potentials were recorded in the whole-cell configuration of the patch clamp technique, using the instruments described above. Action potentials were triggered by 5 ms subthreshold current steps at the frequency of 1 Hz. Representative action potentials for each cell were analyzed for the duration from the overshoot to peak and then at 25%, 50%, 75% and 90% of repolarization (APD10, APD50, APD75 and APD90, respectively).
Histology
Adult zebrafish were euthanized by immersion in MS222 (Sigma-Aldrich) in PBS and decapitated after loss of righting reflexes. Excised hearts were fixed in 10% buffered formalin solution at room temperature overnight, oriented correctly for sagittal 5 μm sectioning through the ventricle, atrium and bulbus arteriosus, and embedded in paraffin wax. Hematoxylin and eosin (H&E) and PSR (1% Sirius red in saturated aqueous picric acid) staining was done according to standard protocols as previously described [34]. For lipid staining, slides or hearts were incubated in filtered 0.3% Oil red O (Sigma-Aldrich) solution for 15 min at room temperature. Embryonic zebrafish were embedded in JB-4 plastic (Polysciences Inc.) according to Sullivan-Brown et al. [35], sectioned and stained with H&E. Sectioning at 5 μm was carried out on a Leica RM 2245 vibratome. All images were captured on a Keyence Biozero BZ-8000K microscope.
Statistical Analyses
Data are presented as means ± SEM. Statistical analyses were done using the one-way ANOVA with p < 0.05 considered as statistically significant; *=p < 0.05, **=p < 0.01, ***=p< 0.001, ****=p<0.0001. GraphPad Prism (Graph Pad Software) was used for statistical analysis.
RESULTS
Patient characteristics
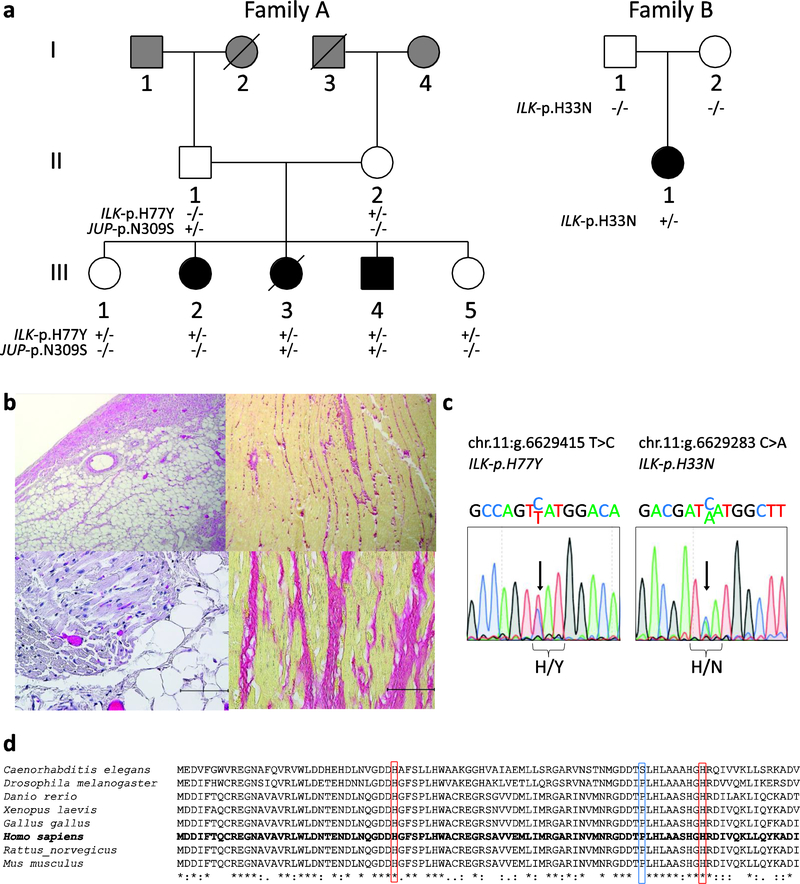
A Caucasian family (family A; Fig 1, A) was evaluated by the Provincial Medical Genetics Program in Vancouver, BC, after a 16-year-old girl (III-3) died due to an unexpected post-operative cardiac arrest after bowel surgery. The autopsy revealed that her right ventricle (RV) was slightly dilated and had a thin free wall (0.3–0.1 cm in thickness). Macroscopic and microscopic findings showed extensive fatty infiltration of the RV, with some fibrosis as well as fatty infiltration of the cardiac conduction system, including the AV-node and His-bundle (Fig 1, B). As these findings are classical hallmarks of ACM, this diagnosis was made post-mortem. Consequently, her siblings and parents were clinically investigated according to the ARVC TFC (Fig 1, A; Table I). Clinically, there were no cardiac symptoms reported nor severe arrhythmias documented in any family member, but two siblings (III-2 and III-4) fulfilled ARVC TFC criteria at the ages of 22 and 14 years, respectively. Another sister (III-1) had minor changes on the cardiac MRI and on 1/6 cardiac biopsy samples that were considered to be suggestive of ARVC. All three of these siblings subsequently underwent implantation of an internal cardioverter defibrillator (ICD) for primary prevention. The fourth living sib (III-5), a ten-year old girl at the time of evaluation, underwent cardiac investigations according to ARVC TFC, and appeared to be normal. Both parents were clinically unaffected, and neither had signs of ACM on clinical investigations (Table I).
Figure 1. Clinical and genetic characteristics of family A and B with ACM.
(A) Family A carrying an ILK and JUP variant (left) and family B carrying a de novo ILK variant. Squares represent males, circles females, slash across symbol denotes deceased. Black filled symbols indicate a clinical diagnosis of ACM. White filled symbols denote unaffected, whereas gray filled symbols indicate an unknown clinical status. +/− denotes heterozygous carrier status; −/− denotes non-carrier. (B) Myocardial tissue analysis of patient III-3 from family A. H&E staining (left panel) demonstrating fatty replacement of myocardium. Picro Sirius red staining (right panel) demonstrate extensive interstitial fibrosis. Scale bars represent 50 μm. (C) Sanger sequencing results of heterozygous ILK variants from genomic DNA of carriers (III-3, family A left; II-1 family B, right). (D) Conservation status of ILK amino acid sequences across species. Red squares indicate p.H33 and p.H77, the blue square p.P70; all three residues are highly conserved across species.
Table1:
Clinical characteristics of family A and family B.
| ECG/Signal-averaged ECG |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Individual | Age of Investigation | Clinical History | Family History/Genetics | Structural and Functional Alterations by CMR or Echocardiography | Tissue Characterization on RV-Biopsy or Autopsy | Repolarization Abnormalities | Depolarization Abnormalities | Arrhythmias | TF-Score Major/Minor Fullfilled Yes/No |
| Family A | |||||||||
| III-1 | 25 | asympomatic, ICD implanted because of possible diagnosis | (+/−) ILK: p.H77Y; 1st degree relative died suddenly with ACM confirmed on autopsy | Anterior RV wall appears thinned with fat infiltration, no akinesis or dyskineses, normal RVEF | 1/6 biopsy samples showed focal fatty infiltration and fibrosis | Non-specific T-wave abnormalities | None | None | 1M/1m: No |
| 33 | remains asymptomatic. ICD explanted after lead fracture | (+/−) ILK: p.H77Y; 1st degree relative died suddenly with ACM confirmed on autopsy | Preserved left and right ventricular function, estimated RVEF: 55%, RV not dilated | All biopsies non-diagnostic for ARVC | N/A | None | None | 1M: No | |
| III-2 | 22 | clinically diagnosed with ACM, largely asympomatic, ICD implanted | (+/−) ILK: p.H77Y; 1st degree relative died suddenly with ACM confirmed on autopsy | Anterior RV wall is thinned with fat infiltration, no akinesis or dyskineses, mildly reduced RVEF, changes more pronounced than in sibs | Biopsies show focal fatty infiltration and fibrosis | None | 2/3 positive for late potenials on SA-ECG | None | 2M / 1m: Yes |
| III-3 | 16 | cardiac arrest during bowel surgery (ischemic bowel with lymphoid hyperplasia), signs of ACM diagnosed on autopsy | (+/−) ILK: p.H77Y | N/A | RV slightly dilated, RV free wall quite thin (0.3–0.1 cm in thickness). Fatty infiltration into RV free wall. Extensive fatty infiltratiorr in RV, some fibrosis. Focal fatty infiltration of AV node and bundle of his | N/A | N/A | N/A | >2M, confirmed ACM on autopsy: Yes |
| III-4 | 14 | clinically diagnosed with ACM, largely asympomatic, ICD implanted | (+/−) ILK: p.H77Y; 1st degree relative died suddenly with ACM confirmed on autopsy | Anterior RV wall appears thinned with fat infiltration, no akinesis or dyskinesis, normal RVEF | Biopsies with moderate interstitial fibrosis and with fatty infiltration | None | None | Rare PVCs, no arrhythmias | 2M: Yes |
| III-5 | 10 | no clinical signs of ACM | (+/−) ILK: p.H77Y; 1st degree relative died suddenly with ACM confirmed on autopsy | Normal RV size and systolic function. No wall motion abnormalities. Normal LV size, mass and systolic function. | N/A | None | None | None | 1M: No |
| II-1 | 48 | No cardiac concerns | (−/−) ILK: p.H77Y; 1st degree relative died suddenly with ACM confirmed on autopsy | Normal RV and LV Size and systolic function. | N/A | None | 2/3 positive for late potenials on SA-ECG | Rare PVCs, no arrhythmias | 1M/1m: No |
| II-2 | 51 | No cardiac concerns | (+/−) ILK: p.H77Y; 1st degree relative died suddenly with ACM confirmed on autopsy | Normal RV and LV Size and systolic function. | N/A | None | None | None | 1M: No |
| Family B | |||||||||
| II-1 | 14 | clinically diagnosed with ACM, syncope, ICD implanted | (+/−) ILK: p.H33N (de novo) | RV-dyskinesis with reduction of RV ejection fraction (RV-EF: 45%), no aneurysms, no LV impairment | N/A | Negative T-waves in V1-V3 | 2/3 positive for late potentials on SA-ECG | > 500 PVCs and three triplets in 24h Holter | 2M/3m:Yes |
| I-1 | 45 | No cardiac concerns | (−/−)p.H33N; 1st degree relative with ACM | Normal RV and LV size and systolic function. | N/A | None | None | None | 1M: No |
| I-2 | 43 | No cardiac concerns | (−/−)ILK: p.H33N; 1st degree relative with ACM | Normal RV and LV size and systolic function. | N/A | None | None | None | 1M: No |
CMR, cardiac magnetic resonance imaging; RV-EF, right ventricular ejection fraction; LV, left ventricle; RV, right ventricle; PVCs, premature ventricular beats; LBBB, left bundle branch block; RBBB, right bundle branch block; ILK, integrin linked kinase; ICD, implantable cardioverter defibrillator; ACM, arrhythmogenic cardiomyopathy; SA-ECG, signal averaged electrocardiogram; TF-Score, task force score; M, major; m, minor. Diagnostic terminology for revised criteria 2010 (ref. 2): definite diagnosis: 2 major or 1 major and 2 minor criteria or 4 minor from different categories; borderline: 1 major and 1 minor or 3 minor criteria from different categories; possible: 1 major or 2 minor criteria from different categories.
Individual III-1 was clinically reassessed at the age of 33 years, when her ICD generator was depleted. The ICD was explanted because of concern about fracture of the Medtronic Sprint Fidelis lead, and full re-assessment of her cardiac status was undertaken. All investigations were normal at that time; in particular the newly taken biopsies and the cardiac MRI were non-diagnostic according to current TFC for ARVC (2).SibIII-1 is, therefore, now considered to be clinically unaffected, although she remains under clinical surveillance. The second Caucasian family (family B; Fig 1,A) was identified in a cohort of 30 unrelated index patients diagnosed with ACM. The index case(II-1) in this family is a 14-year-old girl who presented with syncope short after physical exercise at school. Bystanders reported that they could not initially feel her pulse, but after a few seconds she recovered. After this event she underwent cardiac investigations as the syncope was suspected to be an arrhythmic event. Her ECG showed negative T-waves in leads V1–3, and the 24 h Holter test showed about 650 single premature ventricular beats(PVCs), including a few couplets, and three triplets. CMR images revealed a reduced RV ejection fraction and RV-dyskinesis but no LV impairment. ARVC was diagnosed on TFC, and an ICD was implanted. Both of her parents (I-1,I-2) had normal cardiac investigations(Table I).
Genetic findings
Targeted next generation sequencing for genes previously associated with ACM (JUP, PKP2, DSP, DSC2, DSG2, TMEM43, RYR2) was performed on individual III-4 of family A. A rare heterozygous missense variant in plakoglobin (JUP) c.926A>G leading to the amino acid change p.N309S (chr.l7:g.39921303 T > C; RefSeq:NM_021991.2; rsl40606359) was identified. This variant was found 15 times with an overall MAF of 0.00005 in gnomAD. In silico prediction tools are consistent with this change being benign. The variant was found to be inherited from the father and was seen in the affected sibs (III-3 and III-4) but not in affecfed sib III-2 or either unaffected sib (III-1, III-5 ).
Exome sequencing of the proband and two affected siblings (family A; III-2, III-3, III-4) resulted in a set of 51 shared rare non-synonymous coding variants. Further bioinformatic analyses and manual examination of the expression and functional annotations of each of these genes eliminated all but 17 of these variants as disease-causing candidates in this family (Supplemental Table 1). Two of the genes (ILK and TUBGCP2) appeared most promising as pathogenic candidates, but only one (ILK) had evidence from animal models that indicated an important role in cardiac structure or function [14, 15, 18, 36]. This heterozygous missense variant found at ILK-c.229C>T (chr.11: g.6629415 T > C;RefSeq: NM_004517.3; rs750788075) encodes an amino acid change from histidine to tyrosine at position 77 of the integrin-linked kinase protein (p.H77Y; Fig 1, C). This position demonstrates high evolutionary conservation (Fig 1, D), and the variant is predicted to be deleterious by several in silico prediction tools. This variant was found only once in the gnomAD v2.1 population database, with an allele frequency of 0.00003. The variant was tested for segregation in the family A, and the mother (II-2) and two clinically unaffected sibs (III-1 and III-5) were also found to be carriers of ILK-p.H77Y.
As pathogenic variants in ILK have been not previously reported for ACM, we searched for novel coding variants in this gene in a cohort of 30 unrelated index patients with confirmed ACM who did not have detectable mutations of known ACM genes. A novel heterozygous ILK variant at genomic position chr.11:6629283 C > A, changing a histidine at position 33 to an asparagine (p.H33N; Fig 1, C) was detected in a 14-year old girl (family B, II-1; Fig 1, A). This variant was absent from gnomAD v2.1, was evolutionarily conserved (Fig 1, D), and was predicted to be damaging by all four in silico tools used. Testing for this variant in both unaffected parents confirmed that the variant is very likely a de novo mutation in the index case.
In vitro analysis of ILK variants
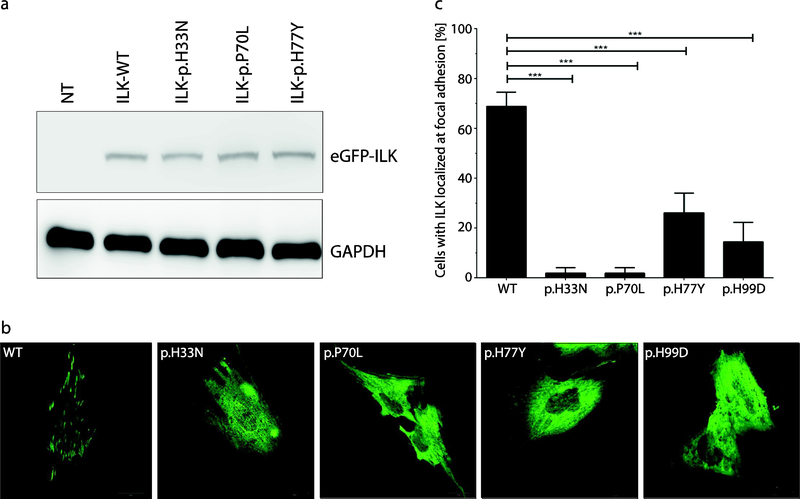
In order to investigate the functional consequence of these novel and potentially pathogenic ILK variants, we mutated the p.H33N and p.H77Y residues in a fusion protein (human ILK with eGFP; eGFP-ILK). We also created constructs for a previously published variant (p.P70L) previously associated with DCM [19] and an artificial variant that disrupts ILK-PINCH binding (p.H99D) [37]. We expressed mutant and wild-type ILK in H9c2 rat myoblasts. Western blot analysis revealed stable expression of mutant and wild-type ILK (Fig 2, A). However, the normal cellular localization of ILK at focal adhesions was disrupted in all three putative pathogenic human variants as well as the artificial control; these mutant ILK proteins were mainly localized in the cytoplasm (Fig 2, B,C).
Figure 2. Expression and cellular localization of mutant recombinant ILK-proteins.
(A) Western blot analysis of eGFP-ILK expression in ILK-wild-type (WT) and three different mutant forms of ILK transfected in H9c2 cells. GAPDH expression was used as a loading control. There was no significant difference between WT and mutant protein expression. (B) Representative fluorescence images of ILK (green) in transfected H9c2 cells. Of note, ILK-WT localizes at the focal adhesions, whereas the four rrhitant forms were mainly localized in the cytoplasm. (C) Quantitative assessment of cells (%) localized at the focal adb|sions fey ILK-WT and the four mutant proteins which show significantly fewer cells with focal adhesion localization compared to the WT (***=p< 0.001). The transfection experiment was repeated three times.
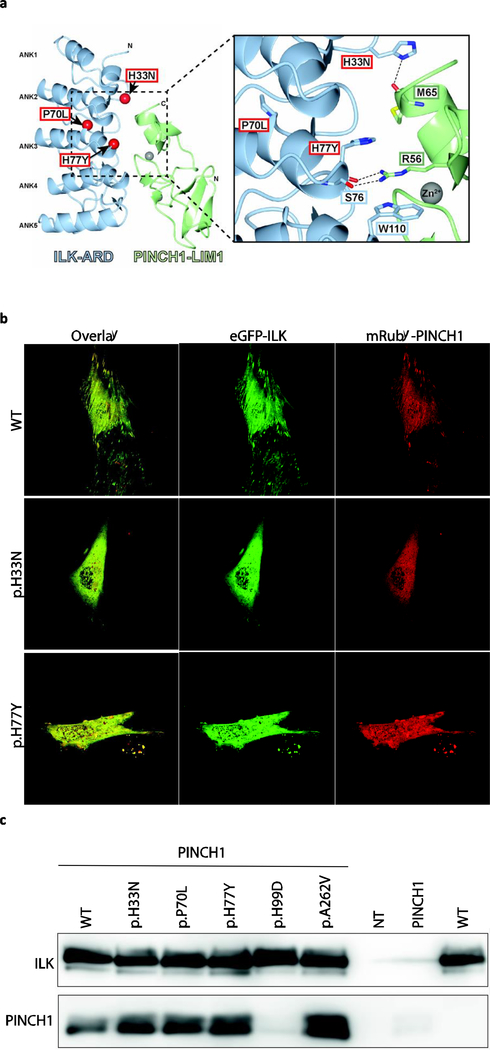
ILK is an important adapter protein localized at the focal adhesions and involved in the connection of the cytoskeleton and the extracellular matrix. All three human variants are located in the ILK ankyrin repeat domain (ARD), which binds to the first LIM domain of PINCH1 and PINCH2 [38]. In order to investigate the binding affinity between mutant ILK-ARD and PINCH we performed an in silico structural analysis. The co-crystal structures of the ILK-PINCH1 and ILK-PINCH2 complexes revealed that the ILK-ARD is comprised of five ANK repeats forming a ’palm’ of alpha helices connected by protruding loops that serve as `fingers’. This ILK-ARD binds to both zinc-fingers of the PINCH LIM1 domain. The ILK variants identified in this study, p.H33N and p.H77Y, are both located at the interface of the complex (Fig 3, A). H33 is located in the ’finger’ portion of ANK2 and forms a hydrogen bond with the carbonyl oxygen of M65 in PINCH. The change of H33 to asparagine could potentially disrupt this hydrogen bond.H77 is located in the ’palm’ region of ANK3 and forms a pi-cation-pi stack with PINCH R56 (which interacts with ILK S76) and W110 in ILK ANK4. The change of H77 to tyrosine, with its larger aromatic sidechain, may destabilize the local structure of ANK3. Both H33 and H77 are largely buried in the interface, with over 70% of the available surface area of H33 and 40% of the H77 surface buried (PISA server [39]). Thus, the mutations identified may disrupt the ILK-PINCH complex (Fig 3, A).
Figure 3. Assessment of mutant ILK and PINCH binding.
(A) Structural analysis of ACM variants in ILK. The molecular structure of the complex between ILK-ARD and PINCH1-LIM1 is shown as a ribbon diagram (left). The localization of p.H33, p.P70 and p.H77 are indicated by red spheres corresponding to the Ca position and are labeled. Grey spheres show the Zn2+-ions. Inset (right): details of the interface involving p.H33 and p.H77. (B) Representative fluorescence images of the localization of eGFP-ILK (green) and mRubyPINCH1 (red) in double transfected H9c2 cells. Of note, wild-type ILK and PINCH1 are co-localized at the focal adhesions (top panel), whereas the expression of mutant ILK leads to an abnormal cytoplasmic co-localization of both proteins and loss of focal adhesion localization (middle, bottom panel). (C) Co-immunoprecipitation analysis. HEK293 cells were co-transfected with plasmids encoding eGFP-ILK and PINCH1 (first six lanes). Non-transfected (NT) and single-transfected cell lysates were used as controls. Nano-Traps recognizing eGFP were used to pull down ILK-protein complexes. Afterwards, the binding of PINCH1 was assessed using Western-blot analysis detecting both proteins.
To reveal disrupted binding in vitro we co-transfected recombinant wild-type and mutant ILK proteins with mRuby-PINCH1 and 2 in C2C12 cells. As expected, wild-type ILK colocalized with PINCH1 at the focal adhesions, whereas both mutant ILK proteins were abnormally localized in the cytoplasm and not at focal adhesions (Fig 3, B). The same results were observed for PINCH2 (data not shown). Next, we used co-immunoprecipitation of both proteins from co-transfected HEK293 cell extracts to test whether binding of PINCH-1 to ILK was disturbed. ILK-p.H99D (previously shown to disrupt the protein-protein interaction with PINCH1) [37] served as a positive control. We observed that only the positive control (eGFP-ILK-p.H99D) was unable to bind PINCH1 after co-immunoprecipitation, whereas all other mutant ILK proteins behaved similarly to wild-type ILK, suggesting that the protein-protein interaction of the ILK-PINCH complex is still intact (Fig 3, C). We obtained the same results with PINCH2 (data not shown). It is possible that even though mutant ILK proteins still bind PINCH1/2, that the KD of this protein-protein interaction is changed for the ILK mutants.
In vivo studies in zebrafish expressing human ILK variants
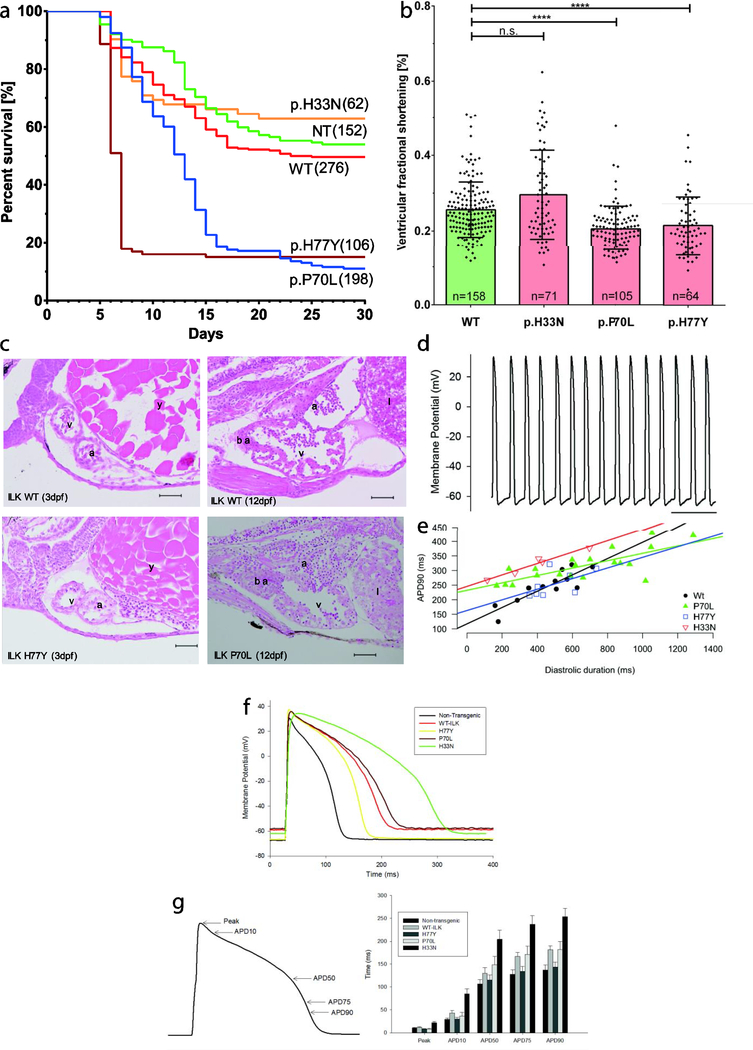
In order to study the effect of our ILK variants in vivo we generated transgenic zebrafish lines overexpressing human ILK as a fusion protein with eGFP (wild-type, p.H33N, p.H77Y and p.P70L) via the Tol2 transposon. Sequencing of DNA from fin clips of each line confirmed germline transmission (Supplementary Figure S2). Multiple alleles were generated for each line showing similar results and transgenic offspring of F1xF1 incrosses were functionally analyzed. No obvious visible embryonic phenotype such as axial defects, looping defects, cardiac edema, or dilated atria or ventricles was observed at 3 dpf. At 120 hpf fish larvae were placed into the larval nursery and survival was observed over the next 30 days. Kaplan-Meier survival analyses indicate that fish expressing ILKp.H77Y extensively died between day 5 and 8, and fish expressing ILKp.P70L died between days 10 and 15 compared to the ILK wild-type, ILKp.H33N and non-transgenic clutchmates. Only 20% of fish of the p.H77Y and p.P70L lines survived beyond day 30. However, fish that survived to 30 days reached adulthood. In comparison, 50–65% of ILK wild-type, mutant line p.H33N and non-transgenic fish survived to adulthood (Fig 4, A).
Figure 4. Survival, structural and functional analyses of transgenic zebrafish.
(A) Kaplan-Meier survival analysis of F1xF1 incrosses of ILK-wild-type (WT) and three different transgenic ILK variants (p.H33N, p.P70L, p.H77Y) or non-transgenic (NT) zebrafish lines. Fewer than 20% of p.P70L and p.H77Y zebrafish survive after day 15. Numbers of investigated fish per line are shown in brackets. (B) Time dependent analysis of fractional shortening (FS) measurements obtained from transgenic fish hearts at 3d pf (left). Data represent mean ventricular FS percentages ± SD. n.s.=not significant; ****=p< 0.0001; n, number of fish hearts investigated per transgenic line. (C) Histological sections of ILK wild-type transgenic and ILK mutant transgenic hearts stained with H&E. Heart morphology appears to be normal in mutant embryos (3dpf) and in fish hearts at 12 dpf compared with wild-type. a, atrium; v, ventricle; y, yolk sack; ba, bulbus arteriosus; 1, liver; scale bars at 50 μm. (D) Typical spontaneous ventricular action potentials from an explanted embryonic zebrafish heart of WT-ILK at 3 dpf. (E) Action potentials were recorded from embryonic ILK wild-type, p.P70L, p.H77Y and p.H33N transgenic zebrafish lines. Multiple APD90 intervals were measured from each heart. These intervals were plotted against diastolic duration, which was determined by measuring the interval between the end of the preceding action potential to the one used for determination of APD90. Lines represented linear regression fitted to each data set. (F) Representative action potentials recorded from isolated adult ventricular myocytes derived from non-transgenic, ILK wild-type, p.H77Y, p.P70L and p.H33N transgenic zebrafish are shown. (G) Time to reach peak membrane potential, APD10, 50,75 and 90 were measured for each action potential. Action potentials measured from cardiac myocytes isolated from ILK p.H33N transgenic zebrafish lines displayed delayed time to peak and prolonged APD when compared to ILK WT. Error bars = SEM.
To determine cardiac function in transgenic animals we measured ventricular fractional shortening (FS) in transgenic fish at 3 dpf by video-microscopy. Interestingly, FS measured in mutant lines p.P70L and p.H77Y was decreased significantly compared to ILK wild-type and the surviving p.H33N lines (Fig 4, B). Reduced FS is an indicator for heart failure. However, histology at 3 dpf or 12 dpf did not show aberrant heart morphology, even in the p.P70L mutant that has been found in a case of DCM (Fig 4, C). Because of pigmentation we were not able to measure FS in the fish at later stages (day 5–15).
Human ACM is often associated with arrhythmias. To assess electrophysiological abnormalities, we performed patch clamp experiments. Spontaneous action potential durations (APDs) were recorded directly from 72 hpf embryonic hearts isolated from all four lines (Fig 4, D). Fig 4, E depicts the measured APD90 plotted as a function of the diastolic period in order to account for heart rate at the time of measurement of each APD. As expected, with decrease in heart rate (i.e. increased diastolic duration), APD measured in wild-type hearts increased. Comparing the mutant and wild-type transgenic hearts revealed no differencmin terms of effect of heart rate on APD. Furthermore, correction of APD for the instantaneous heart rate found no apparent amongst wild-type and mutant transgenic embryos (data not shown).
Furthermore, we also assessed surviving adult fish of each line by patch clamp of isolated cardiomyocytes and histology. We note that since only a few fish survive from p.H77Y and p.P70L mutants, there may be a selection bias of surviving fish compared to their clutchmates. Action potentials recorded from isolated adult cardiac myocytes are shown in Fig 4, F. Time to peak and APDs were not changed in p.H77Y or p.P70L myocytes when compared to cells from wild-type transgenic animals. However, action potentials measured from cardiac myocytes isolated from p.H33N transgenic zebrafish line had delayed time to peak and APD when compared to wild-type (Fig 4, F,G).
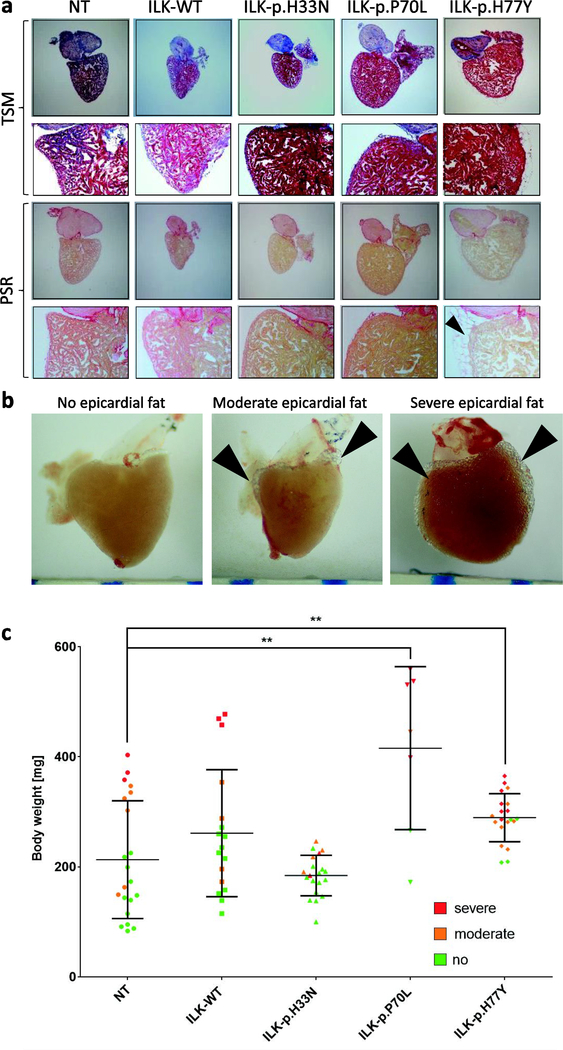
Finally, we assessed adult (> 3 months) hearts macroscopically and performed histology to investigate morphological changes and fibrotic remodeling. Heart size was normalized to body weight. Overall, we did not detect significant differences in morphology, ventricular size or fibrotic pattern between the three mutants and control lines (Fig 5, A; Supplementary Figure S3). Some adult hearts of all genotypes developed severe epicardial fat tissue, but when this was assessed in relation to the size of the fish, it correlated rather with the body weight of the fish than with the expression of mutant ILK (Fig 5, B,C).
Figure 5. Adult histology and cardiac morphology.
(A) Histology of transverse sections of transgenic and non-transgenic hearts (3–4 months). TCM = Trichrome staining after Masson. PCR = Piero Sirius Red staining. Epicardial fat tissue in p.H77Y fish is indicated by black arrows. (B) Examples of native hearts with different degrees of epicardial fat tissue. (C) Quantification of epicardial fat tissue per genotype and of non-transgenic hearts according to no (green), moderate (orange) or severe (red) degree of epicardial fat tissue respectively. Of note, the degree of epicardial fat correlates with body weight and the p.P70L and p.H77Y lines show a higher percentage of hearts with severe epicardial fat tissue (**=p< 0.01). Scale bars represent 1 mm.
DISCUSSION
Our combined clinical, genetic and functional studies provide evidence that two novel missense variants (p.H33N: p.H77Y) of ILK found in our patients can cause ACM.
ACM is a disease of the myocardium associated with fibro-fatty replacement of myocardium and that is clinieally diagnosed by defined TFC [2]. A particular hallmark of ACM, usually not found in any other form of cardiomyopathy [1], is fat tissue replacement of cardiac muscle, which was seen in the myocardium of the deceased patient III-3 (Fig 1B) and in two of her clinically affected sibs, confirming ACM as the phenotype in our families. After excluding known disease genes for ACM, we performed exome analysis in affected members of family A and identified the p.H77Y variant in the ILK gene. This histidine residue is highly conserved across species, and its alteration is predicted to be deleterious and is extremely rare in the gnomAD reference database [25] and predicted to be deleterious. When we performed segregation studies in the parents who are both clinically unaffected, we found that the 51-year old mother is a carrier of the ILK p.H77Y variant, as are two of the proband’s clinically unaffected sisters of the proband, III-1 and III-5. III-5 was only 10 years old at the time of her cardiac evaluation, and an ACM phenotype might not be expected at this young age. Individual III-1 had some findings suggestive of ACM on CMR and cardiac biopsy but did not meet the ARVC TFC for diagnosis, but her mothers’ cardiac evaluation was normal. Therefore, if ILK p.H77Y does cause ACM in this family, the variant must be incompletely penetrant. ACM caused by pathogenic variants of other genetic loci is characterized by remarkably incomplete penetrance and variable expressivity, although the reasons are complex and still not fully understood [3, 40, 41]. Possible contributors to disease expression are environmental factors such as inflammatory processes, epigenetics and genetic modifiers [42, 43].
The second variant in ILK (p.H33N) was found in an affected 14-year old adolescent after screening 30 additional ACM cases. Interestingly, neither parent showed any clinical signs of the disease and neither carried the mutation, indicating a likely de novo event. De novo mutations have a higher prior probability of being disease causing, particularly if a similar phenotype has already been reported as a causal association with other pathogenic variants in the same gene.
Our genetic data suggest a causal effect for ACM in our families, but to validate those variants on a functional level and provide more insights into the mechanisms of ILK as a possible disease gene we performed functional studies. Establishing ILK as a disease gene has consequences on clinical genetic testing because only established or likely disease-associated variants can be used for clinical care. Presently, ILK is not an established disease gene for any form of human cardiomyopathy. A few human missense variants in ILK have been previously reported, and some of them are in association with a phenotype of dilated cardiomyopathy [14, 19, 20]. However, most published DCM-associated variants do not have supporting evidence from functional studies. Only the variant p.A262V, which is located in the kinase domain of ILK, has been further investigated. This variant showed in vitro reduced kinase activity and was unable to rescue an ilk loss of function cardiac phenotype in zebrafish embryos [14]. One putatively pathogenic variant, ILK-p.P70L, was identified in a 44 year-old man and his 21 year-old daughter who both had clinical signs of DCM [19]. No functional studies were reported on this variant, but because it is located in close proximity to our mutations within the ARD of ILK, we included this variant in the functional assessment.
ILK is essential for the maintenance of normal cardiac function; loss of ILK in murine and zebrafish hearts leads to cardiomyopathy with heart failure and arrhythmia [14, 15, 18, 36], whereas its upregulation appears to be protective in diseased or injured myocardium [11, 12, 44]. ILK links β-integrins and the actin cytoskeleton and mediates signaling to downstream effectors [45]. It contains two main domains, the ankyrin repeat domain (ARD), which interacts with the first LIM domain of PINCH1/−2, and the (pseudo)kinase domain that interacts with parvins and integrins. Disturbed ILK-parvin-PINCH MANUSC(IPP)/PKBsignalingdisrupts PKBphosphorylation and signaling through loss of PINCH, parvin or ILK, resulting in progressive heart failure [18, 36, 46, 47]. However, in vivo studies using D. melanogaster, C. elegans and mice showed that the kinase activity may not be essential [45, 48–50]. These studies are controversial [49, 50], but they may indicate that the critical function of ILK is as a scaffold protein that brings different downstream proteins into close proximity.
We were intrigued, therefore, that expression of all ILK variants in this study disrupted ILK localization to focal adhesions in cell lines. These data suggest that the variants may perturb ILK function. Furthermore, our three variants are located in the ARD, which mediates the binding to PINCH1/2 [51]; therefore, we probed binding affinity of the ILK variants to PINCH. All three variants expressed in H9c2 cells demonstrated in vitro loss of focal adhesion location determined by binding to PINCH. Despite structural modelling suggesting there would be disruption of the ILK-PINCH complex for the mutants, the protein-protein interactions of ILK and PINCH1/−2 in vitro were unaltered, which may suggest a more subtle defect. As Co-IP assays are not quantitative, we could not exclude that the KD of PINCH-ILK binding is affected by ILK mutants.
Therefore, we explored the effect of these dominant variants in vivo in the zebrafish model to understand consequences of the variants on heart structure and function. The zebrafish has been previously used to investigate the IPP-complex with focus on kinase function but not to study mutants affecting the ARD that may cause a cardiac phenotype. Overall, we showed that cardiac expression of p.H77Y or p.P70L leads to remarkably decreased survival of the F2-generation and decreased cardiac function (Fig 4, A,B). The cardiac expression of the p.H33N mutant did not demonstrate an obvious cardiac phenotype in zebrafish embryos but showed a delayed time to peak duration and a prolonged action potential duration in adult cardiomyocytes. Both decreased survival and altered action potential may be indicators for subclinical electrical abnormalities. A potential arrhythmic contribution in the p.P70L and p.H77Y lines cannot be excluded because the action potential was investigated before they died (at 3 dpf). We note that a few variant-expressing fish survive to adulthood without further obvious cardiac defects. These surviving fish may indicate a potential limitation of the dominant inheritance model as transgene expression might be silenced with age. Similarly, the genetic variability in the background of fish may play a role in allowing dominant mutants to survive, as the genetic background of zebrafish is very heterogeneous. Variable expression of the transgene, variations of the environmental conditions such as housing, feeding, etc. may also contribute to survival of variant-expressing zebrafish. However, the fact that the dominant transgene is only expressed in the myocardium, coupled with the fact that the majority of variant-expressing fish died during larvals tages, suggests that the variants strongly influence cardiac function. While it is possible that gene dosage in our overexpression model of ILK mutants does not mimic a physiological gene dosage and that different animals may have different expression of the transgene, variable gene dosage (or expression) may be a factor explaining incomplete penetrance in humans, and thus the fish model is a reasonable model of the human situation.
Clearly, there are many open mechanistic questions regarding the observed phenotypes in humans and mutant fish that are beyond the scope of this study. One of our findings was the development of epicardial fat tissue in hearts (Fig 5) – an effect that has not been reported in zebrafish but replicates a common human feature of ACM. However, our data suggest that epicardial fat positively correlates with the body weight of the fish but not with the genotype or phenotype. In any case, the observation of extensive fat on the adult zebrafish heart should be kept in mind when investigating adult fish hearts in future studies.
Overall, we provide further evidence for the important role of ILK in inherited cardiomyopathies. We propose that the combined genetic and functional dataset for the two novel variants identified in ACM, in combination with existing variants previously found for DCM and functionally validated for the p.P70L variant in our study, provides further evidence supporting ILK as a disease gene for cardiomyopathy. The evidence for gene-disease association according to the Clinical Genome Resource framework [52] is “moderate” based on the combination of our current study and previously published data on the p.A262V and p.P70L variants [14, 19]. If ILK is established as a disease gene, some cardiomyopathy-associated ILK variants that currently are reported as variants of uncertain significance, such as those listed in ClinVar [https://www.ncbi.nlm.nih.gov/clinvar/?term=ILK%5Bgene%5D], may be re-classified as ’likely pathogenic’ or even ’pathogenic’ [53]. In conclusion, we present genetic and functional evidence that ILK mutations may have relevance for diagnosis and genetic counseling of inherited cardiomyopathies.
Supplementary Material
Translational Significance.
Here, we report potentially disease-causing variants in the integrin-linked kinase (ILK) in patients with ACM. We provide functional data obtained by in vitro and in vivo experiments suggesting that ILK is a cardiomyopathy disease gene and highlighting its relevance for diagnosis and genetic counselling of inherited cardiomyopathies.
ACKNOWLEDGEMENTS
Wae want to thank all patients for their help and participation in our study. This work was selected for study by the FORGE Canada Steering Committee consisting of K. Boycott (University of Ottawa), J. Friedman (University of British Columbia), J. Michaud (Université de Montreal), F. Bernier (University of Calgary ), M. Brudno (University of Toronto), B. Fernandez (Memorial University), B. Knoppers (McGill University), M. Samuels (Universite de Montréal), and S. Scherer (University of Toronto). We would like to thank Janet Marcadier (Clinical Coordinator) and Chandree Beaulieu (Project Manager) for their contribution to the infrastructure of the FORGE Canada Consortium. We are also thankful to Laura Arbour who helped us with updating clinical records. We thank in addition Dr. Yong Xiang Chen (University of Calgary) for his help with the histology, Kristina Martens (University of Calgary), Susanna Schraut and Miriam Zink (University of Wurzburg) for their technical help in the lab and the fish facility. Additionally, we would like to thank Katelin Townsend (BCCHR DNA Biobank) for sample collection and Sanger sequencing validation. The authors would like to thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about. We are grateful for the support of FORGE (Finding of Rare Disease Genes) Canada which was provided by the government of Canada through Genome Canada, the Canadian Institutesrof Health Research and (grant no. FRN: 123351) and of Alberta Innovates Health Solutions (grant no. 201200822). Support was also provided by the Rare Diseases Models and Mechanisms Network from the Institute of Genetics at the Canadian Institutes of Health Research (to SJC), and the Libin Cardiovascular Institute of Alberta. Additionally, support was also provided by the Interdisciplinary Center for Clinical Research (IZKF) Wurzburg, Germany (E-338). Additionally, support was also provided by the German Ministry of Education and Research (BMBF), Berlin, Germany: grant no: 01E01504.
List of abbreviations
- ILK
integrin linked kinase
- PINCH1/2
particularly interesting new Cys-His protein 1/2
- myl7
myosin light chain 7
- ACM
arrhythmogenic cardiomyopathy
- ARVC
arrhythmogenic right ventricular cardiomyopathy
- PKP2
plakophilin-2
- DSP
desmoplakin
- JUP
junctional protein plakoglobin
- DSG2
desmoglein-2
- DSC2
desmocollin-2
- DCM
dilated cardiomyopathy
- IPP
ILK-parvin-PINCH complex
- ARD
ankyrin repeat domain
- FORGE
finding of rare disease genes
- CMR
cardiac magnetic resonance imaging
- TFC
task force criteria
- SA-ECG
signal averaged electrocardiogram
- ECG
electrocardiogram
- EF
ejection fraction
- MAF
minor allele frequency
- gnomAD
genome aggregation database
- SNV
single nucleotide variant
- WT
wild-type
- DMEM
dulbecco’s modified eagle’s medium
- FBS
fetal bovine serum
- DES
desmin
- CoIP
co-immunoprecipitation
- APD
action potential duration
- FS
fractional shortening
- HR
heart rate
- AP
action potential
- PBS
phosphate buffered saline
- HE
hematoxylin and eosin stain
- PSR
picro-sirius red stain
- TCM
trichrome masson stain
- ICD
internal cardioverter defibrillator
- RV
right ventricle
- AV-node
atrioventricular node
- PVCs
premature ventricular complexes
- LV
left ventricle
- RYR2
ryanodine receptor 2
- TMEM43
transmembrane protein 43
- hpf
hours post fertilization
- dpf
days post fertilization
- PKB
pseudokinase domain
- ACMG
American College of Medical Genetics and Genomics
- PM
plasma membrane
- ECM
extracellular matrix
Footnotes
Conflicts of Interest: All authors have read the journal’s policy on disclosure of potential conflicts of interest and have none to declare. All authors have read the journal’s authorship agreement. The manuscript has been reviewed by and approved by all named authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Thiene G, Corrado D, Basso C. Arrhythmogenic right ventricular cardiomyopathy/dysplasia. Orphanet journal of rare diseases. 2007;2:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Marcus FI, McKenna WJ, Sherrill D, Basso C, Bauce B, Bluemke DA, et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the task force criteria. Circulation. 2010;121:1533–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gerull B, Heuser A, Wichter T, Paul M, Basson CT, McDermott DA, et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nature genetics. 2004;36:1162–4. [DOI] [PubMed] [Google Scholar]
- [4].McKoy G, Protonotarios N, Crosby A, Tsatsopoulou A, Anastasakis A, Coonar A, et al. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease). Lancet (London, England) 2000;355:2119–24. [DOI] [PubMed] [Google Scholar]
- [5].Bauce B, Basso C, Rampazzo A, Beffagna G, Daliento L, Frigo G, et al. Clinical profile of four families with arrhythmogenic right ventricular cardiomyopathy caused by dominant desmoplakin mutations. European heart journal. 2005;26:1666–75. [DOI] [PubMed] [Google Scholar]
- [6].Gehmlich K, Syrris P, Reimann M, Asimaki A, Ehler E, Evans A,et al. Molecularchangesintheheart ofaseverecase of arrhythmogenic right ventricular cardiomyopathy caused by a desmoglein-2 null allele. Cardiovasc Pathol. 2012;21:275–82. [DOI] [PubMed] [Google Scholar]
- [7].Mayosi BM, Fish M, Shaboodien G, Mastantuono E, Kraus S, Wieland T, et al. Identification of Cadherin 2 (CDH2) Mutations in Arrhythmogenic Right Ventricular Cardiomyopathy. Circulation Cardiovascular genetics. 2017;10. [DOI] [PubMed] [Google Scholar]
- [8].van Hengel J, Calore M, Bauce B, Dazzo E, Mazzotti E, De Bortoli M, et al. Mutations in the area composita protein alphaT-catenin are associated with arrhythmogenic right ventricular cardiomyopathy. European heart journal. 2013;34:201–10. [DOI] [PubMed] [Google Scholar]
- [9].van der Zwaag PA, van Rijsingen IA, Asimaki A, Jongbloed JD, van Veldhuisen DJ, Wiesfeld AC, et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur J Heart Fail. 2012;14:1199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hannigan GE, Coles JG, Dedhar S. Integrin-linked kinase at the heart of cardiac contractility, repair, and disease. Circ Res. 2007;100:1408–14. [DOI] [PubMed] [Google Scholar]
- [11].Ding L, Dong L, Chen X, Zhang L, Xu X, Ferro A, et al. Increased expression of integrin-linked kinase attenuates left ventricular remodeling and improves cardiac function after myocardial infarction. Circulation. 2009;120:764–73. [DOI] [PubMed] [Google Scholar]
- [12].Traister A, Aafaqi S, Masse S, Dai X, Li M, Hinek A, et al. ILK induces cardiomyogenesis in the human heart. PLoS One. 2012;7:e37802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bendig G, Grimmler M, Huttner IG, Wessels G, Dahme T, Just S, et al. Integrin-linked kinase, a novel component of the cardiac mechanical stretch sensor, controls contractility in the zebrafish heart. Genes & development. 2006;20:2361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Knoll R, Postel R, Wang J, Kratzner R, Hennecke G, Vacaru AM, et al. Laminin-alpha4 and integrin-linked kinase mutations cause human cardiomyopathy via simultaneous defects in cardiomyocytes and endothelial cells. Circulation. 2007;116:515–25. [DOI] [PubMed] [Google Scholar]
- [15].Quang KL, Maguy A, Qi XY, Naud P, Xiong F, Tadevosyan A, et al. Loss of cardiomyocyte integrin-linked kinase produces an arrhythmogenic cardiomyopathy in mice. Circulation Arrhythmia and electrophysiology. 2015;8:921–32. [DOI] [PubMed] [Google Scholar]
- [16].Lu W, Xie J, Gu R, Xu B. Expression of integrin-linked kinase improves cardiac function in a swine model of myocardial infarction. Experimental and therapeutic medicine. 2017;13:1868–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Troussard AA, Mawji NM, Ong C, Mui A, St -Arnaud R, Dedhar S. Conditional knock-out of integrin-linked kinase demonstrates an essential role in protein kinase B/Akt activation. J Biol Chem. 2003;278:22374–8. [DOI] [PubMed] [Google Scholar]
- [18].Meder B, Huttner IG, Sedaghat-Hamedani F, Just S, Dahme T, Frese KS, et al. PINCH proteins regulate cardiac contractility by modulating integrin-linked kinase-protein kinase B signaling. Molecular and cellular biology. 2011;31:3424–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Meder B, Haas J, Keller A, Heid C, Just S, Borries A, et al. Targeted next-generation sequencing for the molecular genetic diagnostics of cardiomyopathies. Circulation Cardiovascular genetics. 2011;4:110–22. [DOI] [PubMed] [Google Scholar]
- [20].Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. European heart journal. 2015;36:1123–35. [DOI] [PubMed] [Google Scholar]
- [21].Paila U, Chapman BA, Kirchner R, Quinlan AR. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS computational biology 2013;9:e1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kernohan KD, Hartley T, Naumenko S, Armour CM, Graham GE, Nikkel SM, et al. Diagnostic clarity of exome sequencing following negative comprehensive panel testing in the neonatal intensive care unit. American journal of medical genetics Part A. 2018;176:1688–91. [DOI] [PubMed] [Google Scholar]
- [23].Beaulieu CL, Majewski J, Schwartzentruber J, Samuels ME, Fernandez BA, Bernier FP, et al. FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am J Hum Genet. 2014;94:809–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Srour M, Hamdan FF, Schwartzentruber JA, Patry L, Ospina LH, Shevell MI, et al. Mutations in TMEM231 cause Joubert syndrome in French Canadians. J Med Genet. 2012;49:636–41. [DOI] [PubMed] [Google Scholar]
- [25].Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brodehl A, Dieding M, Biere N, Unger A, Klauke B, Walhorn V, et al. Functional characterization of the novel DES mutation p.L136P associated with dilated cardiomyopathy reveals a dominant filament assembly defect. J Mol Cell Cardiol. 2016;91:207–14. [DOI] [PubMed] [Google Scholar]
- [27].Brodehl A, Ferrier RA, Hamilton SJ, Greenway SC, Brundler MA, Yu W, et al. Mutations in FLNC are Associated with Familial Restrictive Cardiomyopathy. Hum Mutat. 2016;37:269–79. [DOI] [PubMed] [Google Scholar]
- [28].Pichler G, Leonhardt H, Rothbauer U. Fluorescent protein specific Nanotraps to study protein-protein interactions and histone-tail peptide binding. Methods in molecular biology (Clifton, NJ). 2012;911:475–83. [DOI] [PubMed] [Google Scholar]
- [29].Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). Eugene, University of Oregon Press; 2000. [Google Scholar]
- [30].Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental dynamics: an official publication of the American Association of Anatomists. 1995;203:253–310. [DOI] [PubMed] [Google Scholar]
- [31].Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Developmental dynamics: an official publication of the American Association of Anatomists. 2007;236:3088–99. [DOI] [PubMed] [Google Scholar]
- [32].Hoage T, Ding Y, Xu X. Quantifying cardiac functions in embryonic and adult zebrafish. Methods in molecular biology (Clifton, NJ). 2012;843:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang PC, Llach A, Sheng XY, Hove-Madsen L, Tibbits GF. Calcium handling in zebrafish ventricular myocytes. American journal of physiology Regulatory, integrative and comparative MANUSCRIPTphysiology.2011;300:R56–66. [DOI] [PubMed] [Google Scholar]
- [34].Brodehl A, Belke DD, Garnett L, Martens K, Abdelfatah N, Rodriguez M, et al. Transgenic mice overexpressing desmocollin-2 (DSC2) develop cardiomyopathy associated with myocardial inflammation and fibrotic remodeling. PLoS One. 2017;12:e0174019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sullivan-Brown J, Bisher ME, Burdine RD. Embedding, serial sectioning and staining of zebrafish embryos using JB-4 resin. Nature protocols. 2011;6:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].White DE, Coutu P, Shi YF, Tardif JC, Nattel S, St Arnaud R, et al. Targeted ablation of ILK from the murine heart results in dilated cardiomyopathy and spontaneous heart failure. Genes & development. 2006;20:2355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chiswell BP, Stiegler AL, Razinia Z, Nalibotski E, Boggon TJ, Calderwood DA. Structural basis of competition between PINCH1 and PINCH2 for binding to the ankyrin repeat domain of integrin-linked kinase. J Struct Biol. 2010;170:157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chiswell BP, Zhang R, Murphy JW, Boggon TJ, Calderwood DA. The structural basis of integrin-linked kinase-PINCH interactions. Proc Natl Acad Sci U S A. 2008;105:20677–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372:774–97. [DOI] [PubMed] [Google Scholar]
- [40].Syrris P, Ward D, Asimaki A, Evans A, Sen-Chowdhry S, Hughes SE, et al. Desmoglein-2 mutations in arrhythmogenic right ventricular cardiomyopathy: a genotype-phenotype characterization of familial disease. European heart journal. 2007;28:581–8. [DOI] [PubMed] [Google Scholar]
- [41].van der Zwaag PA, Cox MG, van der Werf C, Wiesfeld AC, Jongbloed JD, Dooijes D, et al. Recurrent and founder mutations in the Netherlands: Plakophilin-2 p.Arg79X mutation causing arrhythmogenic right ventricular cardiomyopathy/dysplasia. Netherlands heart journal: monthly journal of the Netherlands Society of Cardiology and the Netherlands Heart Foundation. 2010;18:583–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Xu T, Yang Z, Vatta M, Rampazzo A, Beffagna G, Pilichou K, et al. Compound and digenic heterozygosity contributes to arrhythmogenic right ventricular cardiomyopathy. Journal of the American College of Cardiology. 2010;55:587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bhonsale A, Te Riele A S J M, Sawant AC, Groeneweg JA, James CA, Murray B, et al. Cardiac phenotype and long-term prognosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia patients with late presentation. Heart rhythm. 2017;14:883–91. [DOI] [PubMed] [Google Scholar]
- [44].Mu D, Zhang XL, Xie J, Yuan HH, Wang K, Huang W, et al. Intracoronary Transplantation of Mesenchymal Stem Cells with Overexpressed Integrin-Linked Kinase Improves Cardiac Function in Porcine Myocardial Infarction. Scientific reports. 2016;6:19155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wu C, Dedhar S. Integrin-linked kinase (ILK) and its interactors: a new paradigm for the coupling of extracellular matrix to actin cytoskeleton and signaling complexes. J Cell Biol. 2001;155:505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liang X, Sun Y, Ye M, Scimia MC, Cheng H, Martin J, et al. Targeted ablation of PINCH1 and PINCH2 from murine myocardium results in dilated cardiomyopathy and early postnatal lethality. Circulation. 2009;120:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hirth S, Buhler A, Buhrdel JB, Rudeck S, Dahme T, Rottbauer W, et al. Paxillin and Focal Adhesion Kinase (FAK) Regulate Cardiac Contractility in the Zebrafish Heart. PLoS One 2016;11:e0150323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lange A, Wickstrom SA, Jakobson M, Zent R, Sainio K, Fassler R. Integrin-linked kinase is an adaptor with essential functions during mouse development. Nature. 2009;461:1002–6. [DOI] [PubMed] [Google Scholar]
- [49].Wickstrom SA, Lange A, Montanez E, Fassler R. The ILK/PINCH/parvin complex: the kinase is dead, long live the pseudokinase! The EMBO journal. 2010;29:281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ghatak S, Morgner J, Wickstrom SA. ILK: a pseudokinase with a unique function in the integrin-actin linkage. Biochemical Society transactions. 2013;41:995–1001. [DOI] [PubMed] [Google Scholar]
- [51].Fukuda T, Chen K, Shi X, Wu C. PINCH-1 is an obligate partner of integrin-linked kinase (ILK) functioning in cell shape modulation, motility, and survival. J Biol Chem. 2003;278:51324–33. [DOI] [PubMed] [Google Scholar]
- [52].Strande NT, Riggs ER, Buchanan AH, Ceyhan-Birsoy O, DiStefano M, Dwight SS, et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am J Hum Genet. 2017;100:895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine: official journal of the American College of Medical Genetics. 2015;17:405–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.