Abstract
Colorectal cancer (CRC) is one of the most common cancers worldwide, with high mortality. Abnormally expressed microRNAs (miRNAs) are considered novel biomarkers in cancer diagnosis. The aim of this study was to investigate the diagnostic value of miR‐92a‐1 in patients with CRC. Serum samples were collected from 148 patients pathologically diagnosed with CRC and 68 gender‐ and age‐matched healthy volunteers. Quantitative real‐time polymerase chain reaction (qRT‐PCR) was used to measure serum miR‐92a‐1 level. Relationship between miR‐92a‐1 and clinicopathological features of CRC cases was analysed via chi‐square test. Receiver operating characteristic (ROC) curve was plotted to estimate the diagnostic value of miR‐92a‐1 in CRC. Serum miR‐92a‐1 was significantly up‐regulated in CRC patients compared with healthy individuals (P < .001). Moreover, miR‐92a‐1 expression was correlated with TNM stage (P = .02), histological stage (P = .003), lymph node metastasis (P = .003) and distant metastasis (P < .001). ROC analysis showed that the area under the ROC curve (AUC) was 0.914, suggesting high diagnostic accuracy of miR‐92a‐1 in ROC. The optimal cut‐off value was 1.485, with a sensitivity of 81.8% and a specificity of 95.6%. MiR‐92a‐1 is increased in CRC patients and correlated with aggressive clinical characteristics. Serum miR‐92a‐1 may be a potential diagnostic biomarker for CRC.
Keywords: Colorectal cancer, Diagnosis, MiR‐92a‐1
1. INTRODUCTION
Colorectal cancer (CRC) is one of the most common malignancies in the world, with high incidence and mortality. 1 In China, the morbidity and mortality of CRC show rising trends, due to changes in lifestyle and diets. 2 , 3 Although treatment methods for CRC have been improved, the survival rates of CRC patients are still not ideal. 4 , 5 Tumour stage at diagnosis is a major factor influencing therapeutic effects. 6 Reportedly, CRC patients diagnosed at early stage were more likely to be cured, while those in advanced stage frequently faced dismal prognosis. 7 Thus, finding novel and effective biomarkers for early diagnosis of CRC is a valuable way to improve clinical outcome of the patients.
Growing evidences have indicated that the expression patterns of microRNAs (miRNAs) possess close association with tumour development and progression, which might hold the potential to serve as biomarkers for diverse cancers, including CRC. 8 , 9 , 10 MiRNAs are a class of small non‐coding RNAs and play important roles in regulating gene expression. 11 MiRNAs can bind to the 3′ untranslated region (3′ UTR) of their target mRNAs, thus preventing gene translation. 12 Aberrant expression of miRNAs can contribute to various human diseases, like malignancy. 13 In previous tumour investigations, numerous miRNAs were reported to act as oncogenes or tumour suppressors in the progression of CRC. For example, microRNA‐429 (miR‐429), miR‐1260b and miR‐10b played oncogenic roles in CRC development, 14 , 15 , 16 while miR‐526‐3p, miR‐185 and miR‐452 could inhibit aggressive progression of the malignancy. 17 , 18 , 19 Abnormally expressed miRNAs may provide a novel way for CRC diagnosis.
MicroRNA‐92a‐1 (MiR‐92a‐1) is a member of miR‐17‐92 cluster. MiR‐17‐92 cluster includes four families: miR‐17, miR‐18, miR‐19 and miR‐92a. 20 MiR‐92a family includes four members: miR‐25, miR‐92a‐1, miR‐92a‐2 and miR‐363. The members of miR‐92a family play significant roles in the development and progression of CRC, and might serve as biomarkers for the cancer. A study carried out by Li et al demonstrated that the expression of miR‐25 was up‐regulated in CRC tissues and that its overexpression predicted poor prognosis for the patients. 2 In the study of Xu et al, the expression profile of miR‐363 was confirmed to be a novel diagnostic biomarker for CRC. 21 MiR‐92a has ever been reported to be involved in the development and metastasis of CRC and serve as an effective biomarker for the cancer diagnosis and prognosis. 22 , 23 , 24 Therefore, we speculated that the expression of miR‐92a‐1 might also act as an indicator in early detection of CRC.
In this study, we sought to investigate the expression level of miR‐92a‐1 in CRC patients, and its association with clinical characteristics. In addition, we explored the diagnostic value of miR‐92a‐1 in CRC.
2. METHODS AND MATERIALS
2.1. Patients and sample collection
The present study was a retrospective investigation. A total of 148 CRC patients were recruited from Cangzhou Central Hospital. Inclusion criteria for CRC patients were as follows: (a) pathophysiologically confirmed; (b) not receiving any anti‐tumour treatments previously; (c) possessing available clinical records. Meeting any one of the following conditions, patients would be excluded: (a) without pathological diagnosis; (b) dying within one month after diagnosis; (c) showing abnormal liver/kidney function and routine blood test results or other associated or co‐existing diseases; (d) with other primary malignancies. In addition, 68 gender‐ and age‐matched healthy volunteers were recruited as healthy controls in this study, who experienced physical examination in the physical examination centre of the hospital. None of the healthy individuals had the history of malignancies or abnormalities in liver/kidney function.
Blood samples were collected from CRC patients and healthy volunteers in the morning after fasting for 8‐10h. The blood samples were centrifuged to isolate serum samples and stored at −80℃ for further studies. Meanwhile, we recorded clinicopathological characteristics of the CRC patients, including age, gender, tumour size, tumour location, TNM stage, histological stage, lymph node metastasis and distant metastasis (Table 1). This study was approved by the ethics committee of the hospital. All participants provided written informed consents for this research.
TABLE 1.
Association of miR‐92a‐1 with the clinicopathological features of CRC patients
| Features | Total N = 148 | MiR‐92a‐1 expression | P‐values | |
|---|---|---|---|---|
| Low (n = 62) | High (n = 86) | |||
| Age (years) | .764 | |||
| ≤60 | 57 | 23 | 34 | |
| >60 | 91 | 39 | 52 | |
| Gender | .721 | |||
| Male | 67 | 27 | 40 | |
| Female | 81 | 35 | 46 | |
| Tumour size (cm) | .471 | |||
| ≤5 | 76 | 34 | 42 | |
| >5 | 72 | 28 | 44 | |
| Location | .785 | |||
| Colon | 64 | 26 | 38 | |
| Rectum | 84 | 36 | 48 | |
| TNM stage | .020 | |||
| I‐II | 74 | 38 | 36 | |
| III‐IV | 74 | 24 | 50 | |
| Histological stage | .003 | |||
| Well; Moderate | 74 | 40 | 34 | |
| Poor | 74 | 22 | 52 | |
| LN metastasis | .003 | |||
| Yes | 72 | 39 | 33 | |
| No | 76 | 23 | 53 | |
| Distant metastasis | <.001 | |||
| Yes | 78 | 22 | 56 | |
| No | 70 | 40 | 30 | |
2.2. RNA extraction and quantitative real‐time polymerase chain reaction (qRT‐PCR)
Total RNAs were extracted from serum samples adopting TRIzol reagent kit (catalogue number: 15596026, Invitrogen) as per the manufacturer's protocols. The ratio of OD260/OD280 (1.9‐2.0) was used to evaluate the purity of isolated RNAs. Total RNA was used for reverse transcription reaction employing TaqMan miRNA reverse transcription kit (catalogue number: 4366596, Applied Biosystems). Obtained cDNA was stored at −20°C until use.
Then, PCR was carried out in ABI 7300 Real‐Time PCR System (Applied Biosystems) with SYBR Green PCR Master Mix (Applied Biosystems). Cycle threshold (CT) was defined as the cycle number of fluorescence signal reaching to threshold. We set threshold level at 0.1‐0.3, and CT values of miRNA samples were automatically calculated based on miRNA abundance. 25 U6 gene served as endogenous control to normalize relative expression level of miR‐92a‐1. All data were calculated with the method of 2−ΔΔCt. Primer sequences were designed based on published data, as follows: miR‐92a‐1, forward‐5'‐ACACAGGTTGGGATCGGTTG‐3', and reverse‐5'‐CAAACTCAACAGGCCGGGA‐3'; U6, forward‐5'‐CTCGCTTCGGCAGCACA‐3', and reverse‐5'‐AACGCTTCACGAATTTGCGT‐3'.
2.3. Statistical analysis
Continuous data were present as mean ± SD and compared adopting Student's t test. Chi‐square test was used to examine the correlation of miR‐92a‐1 with clinicopathological features of CRC patients. Receiver operating characteristic (ROC) analysis was used to evaluate the diagnostic value of miR‐92a‐1 in CRC. All data analyses were performed in SPSS 21.0 statistical software. P‐value less than .05 was considered as significant level.
3. RESULTS
3.1. Expression level of miR‐92a‐1
According to qRT‐PCR, the expression of miR‐92a‐1 was significantly increased in CRC patients, compared with healthy controls (Figure 1).
FIGURE 1.
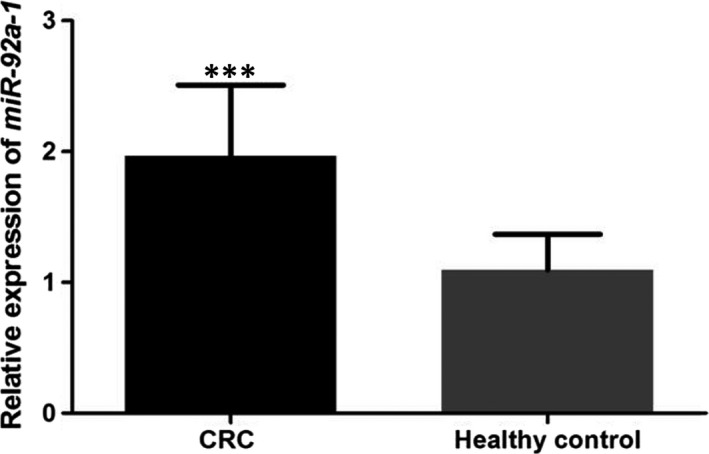
Serum level of miR‐92a‐1 in CRC patients and healthy individuals. The expression of miR‐92a‐1 was significantly up‐regulated in patients with CRC compared with the healthy individuals. ***: suggested P < .001
3.2. Association of miR‐92a‐1 with clinicopathological characteristics of CRC
In the current study, CRC patients were divided into high (n = 86) and low (n = 62) expression groups, according to their median expression of miR‐92a‐1 in CRC tissues. Chi‐square test was used to analyse the relationship between miR‐92a‐1 expression and clinicopathological profiles of CRC. Analysis results indicated that the overexpression of miR‐92a‐1 was correlated with advanced TNM stage (P = .02), poor histological stage (P = .003), positive lymph node metastasis (P = .003) and distant metastasis (P < .001). There was no significant relationship between miR‐92a‐1 and age, gender, or tumour size or location (all P > .05, Table 1).
3.3. Diagnostic value of miR‐92a‐1 in CRC
To explore diagnostic value of miR‐92a‐1 in CRC, ROC analysis was performed and showed an area under the ROC curve (AUC) of 0.914, suggesting its high diagnostic accuracy in CRC. The cut‐off value of miR‐92a‐1 for CRC diagnosis was 1.485, with a sensitivity of 81.8% and a specificity of 95.6% (Figure 2).
FIGURE 2.
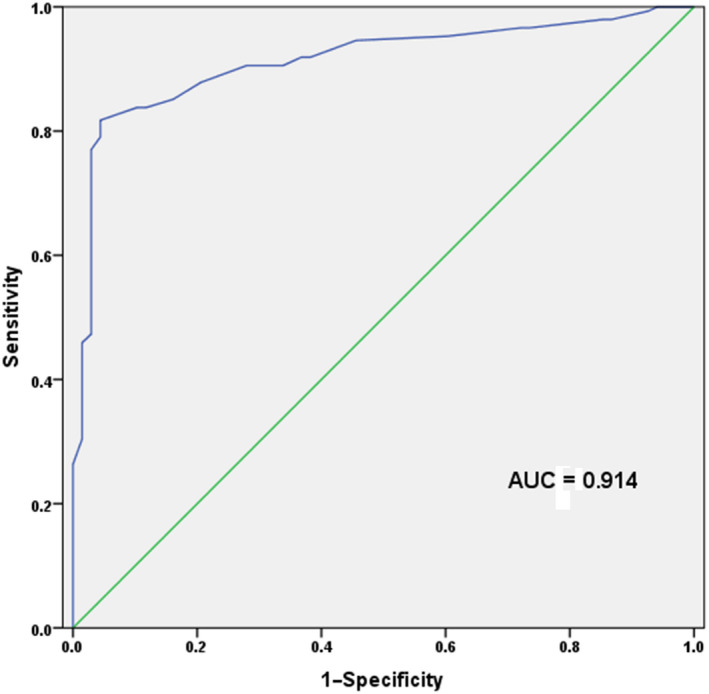
ROC curve constructed based on the expression levels of miR‐92a‐1 in CRC patients and healthy individuals. Analysis results demonstrated that miR‐92a‐1 could discriminate between CRC patients and healthy individuals, with the AUC of 0.914, combing with the sensitivity of 81.8% and the specificity of 95.6%. The cut‐off value of miR‐92a‐1 for CRC diagnosis was 1.485
4. DISCUSSION
CRC is a prevalent malignant tumour in the world. 26 Despite significant improvements in surgical, neoadjuvant chemotherapy and radiotherapy, the survival rates of patients with CRC are still dismal. 27 , 28 Early diagnosis is essential for survival in CRC patients. 29 Until now, the gold standard for CRC diagnosis is colonoscopy, but its invasive nature limits its wide application in clinical practices. 30 Commonly used serum biomarkers for CRC diagnosis include CEA and CA19‐9. However, both of them only show low accuracy in CRC diagnosis. 31 Consequently, novel biomarkers with high sensitivity and specificity are urgently required for CRC diagnosis.
MiRNAs, short non‐coding RNAs, play important roles in physiological and pathological conditions. Their expression profiles show close association with tumour development, progression and treatment response, suggesting their indicative functions in human malignancy. 32 In addition, the expressions of miRNAs are stable and can be easily detected in archived tissue specimens and body fluids. 33 According to existing documents, miRNAs may provide promising approaches for cancer diagnosis. In previous studies, several miRNAs have been confirmed to play predictive roles in the processes of CRC. Wang et al reported that circulating miR‐210 level was significantly different between CRC patients and healthy individuals, which showed diagnostic and prognostic capability for the cancer. 34 MiR‐223, as another example, was reportedly up‐regulated in CRC tissues, and its elevated expression was correlated with positive metastasis and poor prognosis in the patients. 35 In a word, abnormally expressed miRNAs are involved in the aetiology of CRC and might serve as indicators for the cancer.
As a member of miR‐17‐92 cluster, miR‐92a was correlated with the progression of CRC. 22 , 23 , 24 Therefore, we speculated that miR‐92a‐1, the precursor of miR‐92a, might play a crucial role in the progression of CRC. In the current study, we investigated serum level of miR‐92a‐1 in CRC patients and healthy individuals via qRT‐PCR. The results suggested that serum miR‐92a‐1 was up‐regulated in CRC patients compared to healthy individuals. Moreover, increased expression of miR‐92a‐1 was closely linked to advanced TNM stage, poor histological stage and positive lymph node metastasis and distant metastasis, which suggested that miR‐92a‐1 might be involved in CRC development and progression. Alterations in miR‐92a‐1 expression among CRC patients were normal, in comparison with healthy controls, and in further study, we will check the role of miR‐92a‐1 in the progression of CRC. Experimental data revealed that miR‐92a‐1, an oncogene, could promote aggressive progression of CRC. However, specific mechanisms for miR‐92a‐1 affecting CRC were not investigated in the present study. Further analyses are still required.
The members of miR‐92a family play significant roles in the development and progression of CRC and might serve as biomarkers for the cancer. 36 However, diagnostic performance of miR‐92a‐1 in CRC remained unclear. To examine diagnostic value of miR‐92a‐1, ROC curve was constructed in the current study. The results demonstrated that miR‐92a‐1 could distinguish CRC patients from healthy individuals with high sensitivity and specificity. MiR‐92a‐1 might be a potential diagnostic biomarker for CRC. It was worth noting that the sample size in the current study was relatively small, and the application value of miR‐92a‐1 in CRC diagnosis requires further identifications. Future studies should be performed to verify our findings and investigate relevant mechanism both in vivo and in vitro.
In summary, miR‐92a‐1 is up‐regulated in CRC patients and correlated with malignant tumour development and progression. Serum miR‐92a‐1 may be a novel diagnostic biomarker for CRC.
CONFLICT OF INTEREST
None.
AUTHOR CONTRIBUTIONS
YS conceived and designed the experiments; ZL conceived and performed the experiments; and YS prepared figures. YS and ZL wrote the main manuscript text. All authors reviewed the manuscript. With the approval of Cangzhou Central Hospital Ethics Committee, written informed consent was obtained from every patient.
ACKNOWLEDGEMENTS
None.
Shi Y, Liu Z. Serum miR‐92a‐1 is a novel diagnostic biomarker for colorectal cancer. J Cell Mol Med. 2020;24:8363–8367. 10.1111/jcmm.15282
DATA AVAILABILITY STATEMENT
All data generated or analysed during this study are included in this article.
REFERENCES
- 1. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277‐300. [DOI] [PubMed] [Google Scholar]
- 2. Li X, Yang C, Wang X, Zhang J, Zhang R, Liu R. The expression of miR‐25 is increased in colorectal cancer and is associated with patient prognosis. Med Oncol. 2014;31:781. [DOI] [PubMed] [Google Scholar]
- 3. Leslie A, Carey FA, Pratt NR, Steele RJ. The colorectal adenoma‐carcinoma sequence. Brit J Surg. 2002;89:845‐860. [DOI] [PubMed] [Google Scholar]
- 4. Colucci G, Gebbia V, Paoletti G, et al. Phase III randomized trial of FOLFIRI versus FOLFOX4 in the treatment of advanced colorectal cancer: a multicenter study of the Gruppo Oncologico Dell'Italia Meridionale. J Clin Oncol. 2005;23:4866‐4875. [DOI] [PubMed] [Google Scholar]
- 5. Power DG, Kemeny NE. Role of adjuvant therapy after resection of colorectal cancer liver metastases. J Clin Oncol. 2010;28:2300‐2309. [DOI] [PubMed] [Google Scholar]
- 6. Narayanan V, Peppelenbosch MP, Konstantinov SR. Human fecal microbiome‐based biomarkers for colorectal cancer. Cancer Prev Res (Phila). 2014;7:1108‐1111. [DOI] [PubMed] [Google Scholar]
- 7. Sumbul AT, Gogebakan B, Bayram S, Batmaci CY, Oztuzcu S. MicroRNA 211 expression is upregulated and associated with poor prognosis in colorectal cancer: a case‐control study. Tumour Biol. 2015;36:9703‐9709. [DOI] [PubMed] [Google Scholar]
- 8. Wang S, Jiao B, Geng S, Ma S, Liang Z, Lu S. Combined aberrant expression of microRNA‐214 and UBC9 is an independent unfavorable prognostic factor for patients with gliomas. Med Oncol. 2014;31:767. [DOI] [PubMed] [Google Scholar]
- 9. Dong G, Liang X, Wang D, et al. High expression of miR‐21 in triple‐negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol. 2014;31:57. [DOI] [PubMed] [Google Scholar]
- 10. Fateh A, Feizi MA, Safaralizadeh R, Azarbarzin S, Ravanbakhsh R. Diagnostic and Prognostic Value of miR‐1287 in Colorectal Cancer. J Gastrointest Cance. 2016;47(4):399–403. [DOI] [PubMed] [Google Scholar]
- 11. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522‐531. [DOI] [PubMed] [Google Scholar]
- 12. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discovery. 2010;9:775‐789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu H, Huang C, Wu L, Wen B. Effect of evodiamine and berberine on miR‐429 as an oncogene in human colorectal cancer. OncoTargets Ther. 2016;9:4121‐4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu DR, Guan QL, Gao MT, Jiang L, Kang HX. miR‐1260b is a Potential Prognostic Biomarker in Colorectal Cancer. Med Sci Monit. 2016;22:2417‐2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y, Li Z, Zhao X, Zuo X, Peng Z. miR‐10b promotes invasion by targeting HOXD10 in colorectal cancer. Oncol Lett. 2016;12:488‐494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang R, Zhao J, Xu J, Wang J, Jia J. miR‐526b‐3p functions as a tumor suppressor in colon cancer by regulating HIF‐1alpha. Am J Transl Res. 2016;8:2783‐2789. [PMC free article] [PubMed] [Google Scholar]
- 18. Dong‐Xu W, Jia L, Su‐Juan Z. MicroRNA‐185 is a novel tumor suppressor by negatively modulating the Wnt/beta‐catenin pathway in human colorectal cancer. Indian J Cancer. 2015;52(Suppl 3):E182‐E185. [DOI] [PubMed] [Google Scholar]
- 19. He Z, Xia Y, Pan C, Ma T, Liu B, Wang J, Chen L,Chen Y. Up‐Regulation of MiR‐452 Inhibits Metastasis of Non‐Small Cell Lung Cancer by Regulating BMI1. Cell Physiol Biochem. 2015;37:387‐398. [DOI] [PubMed] [Google Scholar]
- 20. He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828‐833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu X, Wu X, Wu S, et al. [Study on miR‐490‐5p and miR‐363 as novel biomarkers for the diagnosis of colorectal cancer]. Zhonghua wei chang wai ke za zhi = Chinese J Gastrointest Surg. 2014;17:45‐50. [PubMed] [Google Scholar]
- 22. Zhou T, Zhang G, Liu Z, Xia S, Tian H. Overexpression of miR‐92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Int J Colorectal Dis. 2013;28:19‐24. [DOI] [PubMed] [Google Scholar]
- 23. Liu GH, Zhou ZG, Chen R, et al. Serum miR‐21 and miR‐92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 2013;34:2175‐2181. [DOI] [PubMed] [Google Scholar]
- 24. Yang X, Zeng Z, Hou Y, et al. MicroRNA‐92a as a potential biomarker in diagnosis of colorectal cancer: a systematic review and meta‐analysis. PLoS ONE. 2014;9:e88745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmittgen TD, Livak KJ. Analyzing real‐time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101‐1108. [DOI] [PubMed] [Google Scholar]
- 26. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10‐29. [DOI] [PubMed] [Google Scholar]
- 27. Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62(4):220‐241. [DOI] [PubMed] [Google Scholar]
- 28. Wolpin BM, Meyerhardt JA, Mamon HJ, Mayer RJ. Adjuvant treatment of colorectal cancer. CA Cancer J Clin. 2007;57:168‐185. [DOI] [PubMed] [Google Scholar]
- 29. Zhan JF, Chen LH, Chen ZX, et al. A functional variant in microRNA‐196a2 is associated with susceptibility of colorectal cancer in a Chinese population. Arch Med Res. 2011;42:144‐148. [DOI] [PubMed] [Google Scholar]
- 30. Du M, Liu S, Gu D, et al. Clinical potential role of circulating microRNAs in early diagnosis of colorectal cancer patients. Carcinogenesis. 2014;35:2723‐2730. [DOI] [PubMed] [Google Scholar]
- 31. Fang Z, Tang J, Bai Y, et al. Plasma levels of microRNA‐24, microRNA‐320a, and microRNA‐423‐5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res. 2015;34:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143‐159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol. 2016;231:25‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang W, Qu A, Liu W, et al. Circulating miR‐210 as a diagnostic and prognostic biomarker for colorectal cancer. Eur J Cancer Care. 2016;26(4):e12448. [DOI] [PubMed] [Google Scholar]
- 35. Li ZW, Yang YM, Du LT, et al. Overexpression of miR‐223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31:256. [DOI] [PubMed] [Google Scholar]
- 36. Elshafei A, Shaker O, Abd El‐Motaal O, Salman T. The expression profiling of serum miR‐92a, miR‐375, and miR‐760 in colorectal cancer: An Egyptian study. Tumour Biol. 2017;39:1010428317705765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this article.


