Abstract
There are few studies on the role of ring finger protein (RNF)180 in non-small cell lung cancer (NSCLC). The present study investigated the expression of RNF180 in NSCLC and its associations with the clinical factors and prognosis of NSCLC. The mRNA and protein expression levels of RNF180 were detected via reverse transcription-quantitative PCR and western blotting. Methylation-specific PCR (MSP) analysis was utilized to detect the methylation of RNF180. RNF180 expression levels were analyzed via immunohistochemistry. The protein and mRNA expression levels of RNF180 were lower in NSCLC cell lines compared with in the non-tumor cell line. Immunohistochemistry revealed that 64 patients that were negative for RNF180, while MSP detection analysis demonstrated that 60 patients exhibited RNF180 promoter methylation. The methylation status of RNF180 was significantly associated with RNF180 expression level. Among all factors evaluated, logistic regression analysis indicated that only T stage was significantly associated with RNF180 expression. Cox multivariate analysis demonstrated that RNF180 expression was an independent predictor of overall survival in patients with NSCLC. Methylation in the promoter of RNF180 was shown to reduce its expression levels. In summary, low RNF180 expression levels were associated with poor biological behaviors, thus RNF180 expression level may be used as a clinical marker to predict the prognosis of patients with NSCLC.
Keywords: non-small cell lung cancer, ring finger protein 180, methylation, prognosis
Introduction
Despite the decline in the rate of non-small cell lung cancer (NSCLC) in recent years, it remains the leading cause of cancer mortality worldwide, 1.8 million people are diagnosed with lung cancer, and 1.6 million individuals died as a result of the disease in 2017 worldwide (1). Accurate diagnosis and precise prognostic predictions are critical for targeted treatment and prolonging the overall survival of patients with NSCLC (2). However, commonly used biomarkers, such as carcino-embryonic antigen and Cyfra21-1, offer low sensitivity and specificity and are not appropriate for clinical applications (3,4). Novel molecular or immunohistochemical-based biomarkers may facilitate identification of cancer at an early stage in the future (5). Therefore, there is a need for the development of novel markers.
Ring finger protein (RNF)180 is composed of a RING finger domain, a basic coiled-coil domain, a novel conserved domain and a C-terminal hydrophobic region (6). RNF180 was first reported in the brain, kidney, testis and uterus, and it has important biological roles in the developing lens and brain at the embryonic stage (7). RNF180 is associated with Helicobacter pylori infection and serves as a biomarker for atrophic gastritis (8). A previous report revealed that RNF180 is involved in tumorigenesis in gastrointestinal cancer (9). Deng et al (10) demonstrated that high expression levels of RNF180 inhibit colony formation, proliferation, migration and invasion. As an anti-oncogene, RNF180 serves key roles in suppressing tumor growth and lymphangiogenesis (10). In hepatocellular carcinoma, RNF180 acts as a tumor suppressor during tumorigenesis (11). RNF180 participates in cell growth and apoptosis, and upregulates not only antiproliferative regulators, but also proapoptotic mediators (12). Thus, RNF180 is associated with invasion and metastasis in cancer. However, understanding remains limited regarding the role of RNF180 in the etiology of NSCLC. Therefore, the present study aimed to elucidate the clinical implication of RNF180 expression level and its association with the survival rate of patients with NSCLC.
Materials and methods
Cell culture
NSCLC cell lines (A549 and HCC827) and a non-tumor cell line (MRC-5) were obtained from The Cell Bank of Type Culture Collection of Chinese Academy of Sciences. Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone; Cytiva) and 100 U/ml penicillin-streptomycin (HyClone; Cytiva) at 37°C under a humidified atmosphere of 5% CO2.
Sample collection
Samples were collected from eligible patients from First Teaching Hospital of Tianjin University of Traditional Chinese Medicine (Tianjin, China) with NSCLC between February 2007 and December 2012. The inclusion criteria for the study included i) Pathologically confirmed patients with NSCLC who underwent radical surgery; ii) patients with surgical specimens, which were frozen in −80°C refrigerator; iii) patients diagnosed with Tumor-Node-Metastasis I–IIIA stage NSCLC (13); and iv) patients who did not receive treatment prior to radical surgery, such as chemotherapy or radiation therapy. Following curative surgery, tumor tissue were frozen in liquid nitrogen immediately and stored at −80°C until use, all patients were followed up every 3–6 months for 2 years, then every year until the end of the study or death. The follow-up was completed in September 2018.
Ethics statement
Written informed consent was obtained from all participants prior to participation, according to the Helsinki Declaration. The study protocol [TYLL2018(K) 007] was approved by the Ethics Committee of Ethics Committee of First Teaching Hospital of Tianjin University of Traditional Chinese Medicine (Tianjin, China).
DNA and RNA extraction
RNA and DNA was extracted from cell lines and tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. RNA extracted from the cells lines for RT-qPCR and DNA extracted from the tissues for MSP.
Reverse transcription-quantitative PCR
In total, 1 µg RNA was synthesized to cDNA using PrimeScript™ RT Reagent kit (cat. no. DRR0037A; Takara Biotechnology Co., Ltd.) at 42°C for 30 min and 85°C for 5 min according to the manufacturer's protocol. qPCR was performed on the 7500 Fast Real Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) Primers designed and utilized for RNF180 and GAPDH were as follows: RNF180 forward, 5′-TCTGACTTTCCTGATGGACCTG-3′ and reverse, 5′-CCTGAGTATTTACCCTGCTTCTGT-3′ and GAPDH forward, 5′-TGGGTGTGAACCATGAGAAGT-3′ and reverse, 5′-TGAGTCCTTCCACGATACCAA-3′. The PCR cycling conditions for all sequences were 45 cycles of denaturation at 95°C for 30 sec, annealing for 30 sec, and extension at 53.5°C for 30 sec, followed by a final extension at 57.5°C for 10 min. Annealing was performed at 60.0°C. The relative RNA levels was normalized to the GAPDH value using the 2−ΔΔCq method (14).
Methylation-specific PCR (MSP)
The following RNF180 primers were used to detect the methylated or unmethylated alleles of the RNF180 promoter: Methylated RNF180 forward, 5′-TTTGCGCGGGGTTAAAGTTC and reverse, 5′-CGATACCGATTCGACGAAACG-3′; and unmethylated RNF180 forward, 5′-TGTTTGTTTGTGTGGGGTTAAAGTTT-3′ and reverse, 5′-CAACAACAATACCAATTCAACAAAACA-3′. MSP was performed using Ampli Taq-Gold (Promega Corporation). The PCR reaction volume was 25 µl, and the thermocycling conditions were as follows: 97°C initial denaturation for 5 min, then 25 cycles of denaturation at 95°C for 40 sec, annealing at 60°C for 50 sec and extension for 50 sec at 72°C, then final extension at 72°C for 10 min. PCR amplification was detected using 2% agarose gel (Jingtianmo Technology Development Co., Ltd.) under the external light.
Western blotting
Western blotting was performed using Pierce™ Fast Western Blot kit, ECL Substrate (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Total protein (40 µg, determined by BCA) was loaded onto 12% gels and separated using SDS-PAGE and then transferred to PVDF membranes. Membranes were subsequently blocked with milk or BSA for 60 min at mean room temperature, and incubated with primary antibodies against RNF180 polyclonal antibody (1:1,000; cat. no. ab127548; Abcam) and GAPDH (1:1,000; cat. no. Ab8245; Abcam) at 4°C overnight. Subsequently, the membranes were washed three times with TBS + 0.1% Tween-20 and incubated with secondary anti-rabbit immunoglobulin antibody (1:2,000; cat. no. SP-9001; ZhongShan Biotechnology). Finally, the membranes were washed three times, detected and visualized by an enhanced chemiluminescence detection system and a Gel Imager system (both Asia Xingtai Mechanical and Electrical Equipment Company) was to analyze images and to determine gray values.
Immunohistochemistry analysis
Paraffin blocks (4-µm thick) were fixed in 4% paraformaldehyde at room temperature for 24 h before used, then deparaffinized at 70°C for 15 min. Antigen was retrieval was performed at 95°C for 40 min in 0.01 mol/l sodium citrate buffer (pH 6.0), and endogenous peroxidase was blocked using 3% hydrogen peroxide for 30 min at room temperature. Rabbit anti-RNF180 antibody (1:50; cat. no. ab127548) was purchased from Abcam. The paraffin sections were incubated overnight with primary antibody at 4°C and then treated with peroxidase using a labeled polymer method at 37°C with Zhongshan peroxidase (OriGene Technologies, Inc.) for 30 min. Antibody binding was visualized using the Avidin Biotin Complex (ABC) Elite kit and 3,3′-diaminobenzine according to the manufacturer's instructions (City Key Laboratory of Tianjin Cancer Center). Sections were then counterstained in hematoxylin for 30 sec at mean room temperature. For general negative controls, the primary antibody was replaced with PBS. All sections were scored under a light microscope (Olympus Corporation) and recorded at 100× or 400× magnification.
Evaluation of immunohistochemical staining
In order to avoid potential bias, all sections were assessed blindly by two independent observers; in case of disagreement, a third independent assessment was involved. Protein expression levels were analyzed for each patient based on a randomly selected section. The staining score of each slide was assessed according to staining intensity and the percentage of positive cells. Staining intensity was scored as 0–4, where 0 was a negative value, 1 was weak, 2 was moderate, 3 was strong intensity pattern and 4 was a very strong intensity pattern. The extent of staining was scored as follows: 0, 0–10, 1, 11–30; 2, 31–50; 3, 51–75; and 4, >75%, according to the percentage of positive-staining cells in relation to the total cancer cells. A final staining score >3 was considered to indicate a positive expression level.
Statistical analysis
Data are presented as the mean ± standard deviation and each experiment was repeated three times. Differences between variables were estimated one-way ANOVA followed by Student-Newman-Keuls post hoc test. Categorical variables were analyzed using χ2 test. Survival analysis was performed with the Kaplan-Meier method and log-rank test. Univariate analysis of patient variables was performed by log-rank test, and multivariate Cox regression analysis was performed to identify prognostic indicators. Statistical calculations were performed using SPSS software (version 21; IBM Corp.). P<0.05 was considered to indicate a statistically significant difference.
Results
mRNA and protein expression level analysis of RNF180
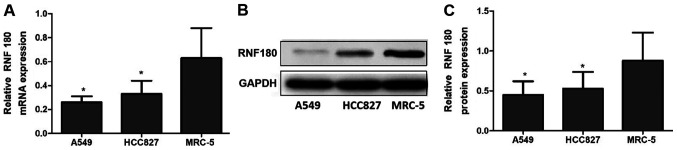
The relative mRNA expression level of RNF180 in the NSCLC cell lines A549 (0.26±0.05; P=0.003) and HCC827 (0.33±0.11; P=0.007) were significantly lower compared with the non-tumor cell line MRC-5 (0.63±0.25). Consistent with mRNA expression levels, the relative protein expression levels of RNF180 in A549 (0.45±0.17; P=0.002) and HCC827 (0.53±0.21; P=0.006) were lower compared with MRC-5 (0.88±0.35; Fig. 1).
Figure 1.
Relative RNF180 mRNA and protein expression levels in cell lines. (A) Relative RNF180 mRNA levels in cell lines. (B) RNF180 protein expression in cell lines analyzed using western blotting. (C) Relative RNF180 protein levels in cell lines *P<0.05. RNF180, ring finger protein 180.
Patient characteristics
Based on the inclusion criteria, 91 patients with NSCLC were eligible for the present study. The 5-year survival rate of the patients with NSCLC was 43.9%. The clinical characteristics of the patients are presented in Table I.
Table I.
Clinicopathological characteristics of patients with non-small cell lung cancer.
| Variable | Cases, n |
|---|---|
| Sex | |
| Male | 55 |
| Female | 36 |
| Age, years | |
| ≤60 | 50 |
| >60 | 41 |
| Smoking status | |
| Yes | 37 |
| No | 54 |
| Histological subtype | |
| Squamous | 28 |
| Adenocarcinoma | 63 |
| Tumor location | |
| Peripheral | 66 |
| Central | 25 |
| T stage | |
| T1 | 33 |
| T2 | 41 |
| T3 | 17 |
| N stage | |
| N0 | 40 |
| N1 | 10 |
| N2 | 41 |
T, tumor; N node.
Association between RNF180 expression levels and methylation status of RNF180 promoter
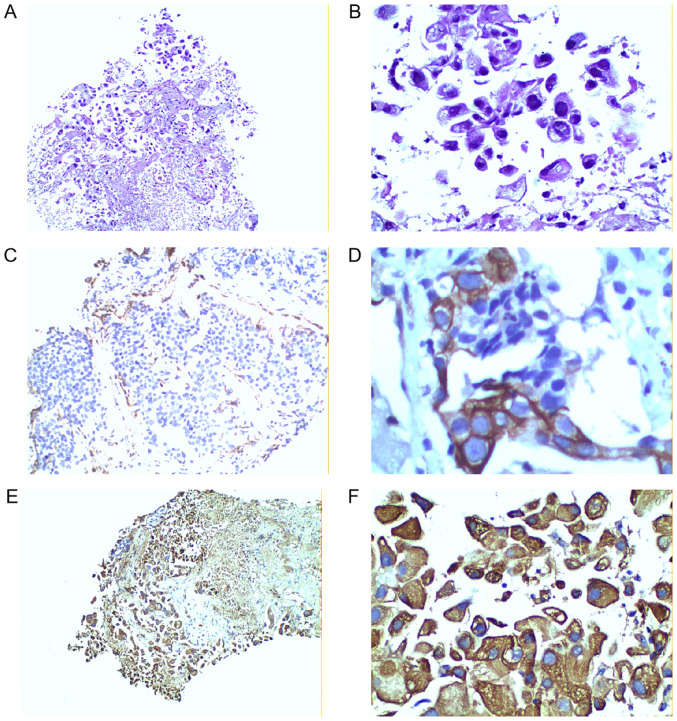
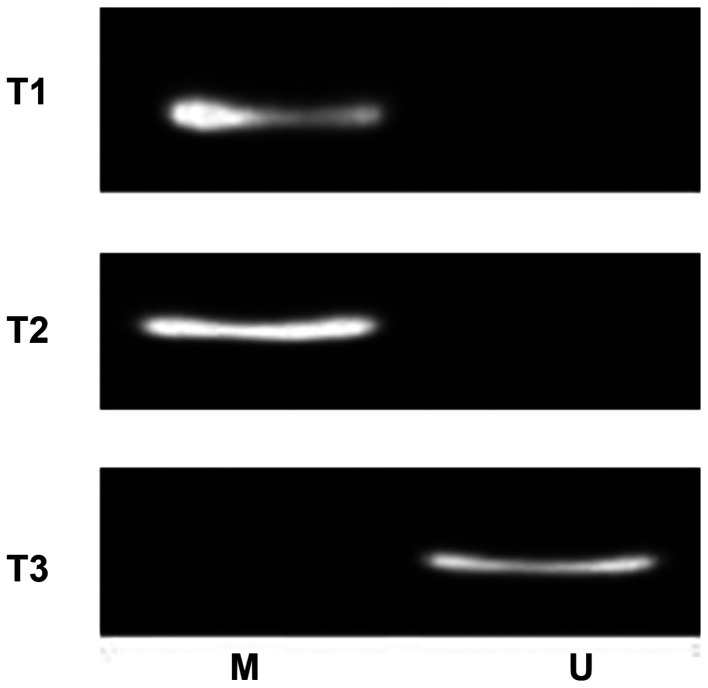
RNF180 expression levels in tissue specimens were observed in the cytoplasm by immunohistochemical staining, as shown in the representative images in Fig. 2. Fig. 2A and B represent negative expression, C and D represent moderate positive expression, and E and F represent strong positive expression. The present results demonstrated that 27 patients were positive for RNF180, whereas 64 patients were RNF180-negative. Furthermore, MSP analysis detected the methylation status of the RNF180 promoter (Fig. 3), showing randomly selected different patients with lung cancer. T1 and T2 were for patients with lung cancer with promoter methylation, while T3 was for lung cancer without promoter methylation). A total of 60 patients exhibited a methylated RNF180 promoter in NSCLC tissues, and the other 31 patients exhibited an unmethylated RNF180 promoter.
Figure 2.
Ring finger protein 180 is expressed in NSCLC tissue samples. Negative RNF180 expression at (A) 100× and (B) 400× magnification. Moderate RNF180 expression at (C) 100× and (D) 400× magnification. Strong positive RNF180 expression at (E) 100× and (F) 400× magnification.
Figure 3.
Methylation-specific PCR detection of ring finger protein 180 promoter methylation in different non-small cell lung cancer tissue samples. M, methylated; U, unmethylated. T1 and T2 were for patients with lung cancer with promoter methylation, while T3 was for lung cancer without promoter methylation.
The correlation between the methylation status of RNF180 promoter and RNF180 expression levels was also investigated. The methylation status of the RNF180 promoter was significantly associated with RNF180 expression levels (χ2=22.528; P<0.001; Table II). However, 8 RNF180-positive patients with methylated DNA and 12 RNF180-negative patients with unmethylated DNA were identified.
Table II.
Association between methylation status of RNF180 promoter and RNF180 expression level.
| Methylation status of RNF180 promoter | ||||
|---|---|---|---|---|
| RNF180 expression level | Methylated | Unmethylated | χ2 value | P-value |
| Negative | 52 | 12 | 22.528 | <0.001 |
| Positive | 8 | 19 | ||
RNF180, ring finger protein 180.
Association between RNF180 expression levels and clinical parameters
The associations between RNF180 expression level and various parameters, such as sex, age, smoking status, histological subtype, tumor location, T stage and N stage, were analyzed. According to univariate analysis, T stage (P<0.001) and N stage (P=0.043) were significantly associated with RNF180 expression (demonstrated by χ2 test). Following multivariate analysis, only T stage was identified to be independently associated with RNF180 expression level (P=0.003; Table III).
Table III.
Association between RNF180 expression level and variables of patients with non-small cell lung cancer.
| RNF180 expression level | P-value | |||
|---|---|---|---|---|
| Variable | Negative, n | Positive, n | Univariate analysis | Multivariate analysis |
| Sex | 0.277 | |||
| Male | 41 | 14 | ||
| Female | 23 | 13 | ||
| Age, years | 0.318 | |||
| ≤60 | 33 | 17 | ||
| >60 | 31 | 10 | ||
| Smoking status | 0.063 | |||
| Yes | 30 | 7 | ||
| No | 34 | 20 | ||
| Histological subtype | 0.400 | |||
| Squamous | 18 | 10 | ||
| Adenocarcinoma | 46 | 17 | 0.214 | |
| Tumor location | ||||
| Peripheral | 44 | 22 | ||
| Central | 20 | 5 | ||
| T stage | <0.001 | 0.003 | ||
| T1 | 15 | 18 | ||
| T2 | 35 | 6 | ||
| T3 | 14 | 3 | 0.043 | 0.075 |
| N stage | ||||
| N0 | 25 | 15 | ||
| N1 | 5 | 5 | ||
| N2 | 34 | 7 | ||
RNF180, ring finger protein 180; T, tumor; N, node.
Survival analysis of patients with NSCLC
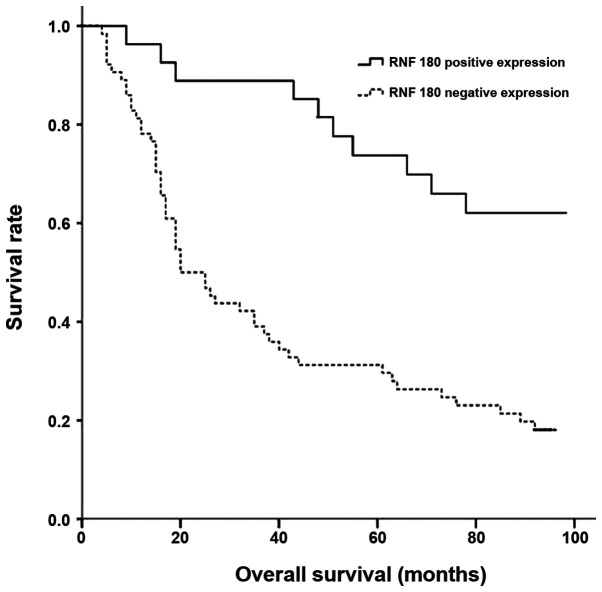
Univariate analysis revealed significant associations between overall survival and the methylation status of the RNF180 promoter, RNF180 expression level, T stage and N stage (P<0.05; Table IV). However, no association with sex, age, smoking status, histological subtype or tumor location was detected (P>0.05). Patients with negative RNF180 expression exhibited a significantly shorter overall survival compared with patients with positive RNF180 expression (Fig. 4).
Table IV.
Survival analysis of patients with non-small cell lung cancer.
| Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|
| Variable | 5-year survival rate, % | χ2 value | P-value | Hazard ratio | P-value |
| Sex | 0.048 | 0.826 | |||
| Male | 45.5 | ||||
| Female | 41.7 | ||||
| Age, years | 0.131 | 0.717 | |||
| ≤60 | 39.8 | ||||
| >60 | 48.8 | ||||
| Smoking status | 1.766 | 0.184 | |||
| Yes | 35.1 | ||||
| No | 49.9 | ||||
| Histological subtype | 1.013 | 0.314 | |||
| Squamous | 50.0 | ||||
| Adenocarcinoma | 41.2 | ||||
| Tumor location | 1.935 | 0.164 | |||
| Peripheral | 48.4 | ||||
| Central | 32.0 | ||||
| T stage | 6.255 | 0.044 | 1.269 | 0.226 | |
| T1 | 54.5 | ||||
| T2 | 41.5 | ||||
| T3 | 29.4 | ||||
| N stage | 23.003 | <0.001 | 1.670 | <0.001 | |
| N0 | 70.0 | ||||
| N1 | 62.5 | ||||
| N2 | 19.5 | ||||
| RNF180 expression level | 17.027 | <0.001 | 2.254 | 0.033 | |
| Negative | 31.3 | ||||
| Positive | 73.7 | ||||
| Methylation status of RNF180 promoter | 13.791 | <0.001 | 1.988 | 0.035 | |
| Methylated | 30.0 | ||||
| Unmethylated | 70.8 | ||||
T, tumor; N, node; RNF180, ring finger protein 180.
Figure 4.
Survival curve of patients with non-small cell lung cancer according to RNF180 expression level (negative or positive). RNF180, ring finger protein 180.
The variables that were significant in univariate analysis were included in the multivariate Cox proportional hazard analysis to adjust for the effects of covariates. According to this analysis, the methylation status of the RNF180 promoter [hazard ratio (HR), 1.988; P=0.035], RNF180 promoter expression levels (HR, 2.254; P=0.033) and N stage (HR, 1.670; P<0.001) were identified as independent factors of overall survival (Table IV).
Discussion
Protein ubiquitylation is initiated by enzymatic cascades that involve E3 ubiquitin ligases (15). RNF180 is an E3 ubiquitin ligase that serves a key role in the function of the ubiquitin-proteasome system by determining the specificity and timing of ubiquitination and subsequent degradation of its substrates (16). RNF180 is either downregulated or absent in different types of cancer, and is regarded as a suppressor gene. Cheung et al (9) revealed that RNF180 is silenced in the majority of gastric cancer cell lines and is significantly downregulated in gastric cancer compared with normal tissue. Results from a previous study suggest mRNA expression levels of RNF180 in non-tumor tissue are ~3.60-fold higher compared with in cancer tissue (17). However, little is understood regarding the expression levels of RNF180 in NSCLC. In the present study, significantly decreased levels of RNF180 mRNA and protein were observed in lung cancer cell lines compared with normal control cell lines. The results of the present study suggest that the low expression level of RNF180 is modulated by methylation of the RNF180 promoter region in the etiology of NSCLC. The promoter region of the RNF180 gene is composed of abundant CpG islands, which are primarily located in −202/+372 promoter region of RNF180 (17). DNA methylation regulates transcription and acts on CpG islands (18–20). However, unmethylated DNA results in either loss or low levels of gene expression. The present study identified 8 patients with methylated RNF180 that exhibited a high expression level and 12 patients with unmethylated RNF180 that exhibited a lower level of expression. It was hypothesized that methylation of only core CpG islands may change the expression levels of the RNF180 gene. The present results were consistent with a previous report (10). Immunohistochemical staining detected negative RNF180 expression level in 70.3% of cases, and 18.75% of cases contained unmethylated CpG sites. It was hypothesized that the abnormal expression levels of RNF180 in NSCLC may result from DNA promoter methylation.
A previous study suggested that the methylation of RNF180 promoter is associated with lymph node metastasis by downregulating RNF180-mediated expression levels of hepatocyte growth factor, matrix metalloproteinase (MMP)-2, MMP-14 and vascular endothelial growth factor C/D (10). Similarly, a previous study had also demonstrated that negative RNF180 expression levels are observed more frequently in patients with advanced TNM stages (7). Therefore, low RNF180 expression level is associated with poor biological behavior of cancer. Similarly, the present study identified that a negative RNF180 expression level was associated with advanced T stage and increased N stage. These results suggested that RNF180 expression levels were associated with the invasion and metastasis of NSCLC.
There have been limited studies investigating the prognostic potential of RNF180 expression levels in patients with NSCLC. In a previous study, Xie et al (21) reported patients who possess ≤7 hypermethylated CpG sites of the RNF180 DNA promoter exhibit improved overall survival. A previous study identified methylated gene Dishevelled-associated antagonist of β-catenin 1 expression levels as an independent predictor of patient survival (22). A previous study had demonstrated that the number of methylated CpG sites can provide distinct survival discrimination of patients (23). However, a cheaper and simpler method to predict the prognosis of patients with NSCLC is required. Assessing methylated CpG site counts and concrete CpG islands by bisulfite genomic sequencing method is expensive and laborious and is not widely used in clinical applications (24). In the present study, the methylation status of the RNF180 promoter and RNF180 expression levels were detected by MSP and immunohistochemistry; and their potential prognostic value for patients with NSCLC was assessed using Cox regression analysis. The present study demonstrated that the methylation status of the RNF180 promoter was an independent factor of the overall survival. However, RNF180 expression level had a higher HR value compared with the methylation status of RNF180 promoter, and hence it was speculated that RNF180 expression levels are more accurate in predicting prognosis. This result indicated that RNF180 expression level detected by immunohistochemistry was a simpler optimal predictor of prognosis for patients with NSCLC compared with methylation status.
In conclusion, the present study demonstrated that a low level of methylated RNF180 promoter is associated with low RNF180 expression level, which is associated with poor biological behavior. Furthermore, the level of RNF180 can be used as a marker for the clinical prediction of the prognosis of NSCLC.
Acknowledgements
Not applicable.
Funding
The present work was supported by the Scientific Research Project of Tianjin Municipal Education Commission (grant no. 2018KJ015).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
All authors were responsible for the conception and design of the present study. HGL and PYY were responsible for the provision of the study materials and the collection and assembly of the data. HGL, PYY, XJL and YJJ performed the data analysis and interpretation and contributed to writing of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all participants prior to participation, according to the Helsinki Declaration. The study protocol [TYLL2018(K) 007] was approved by the Ethics Committee of Ethics Committee of First Teaching Hospital of Tianjin University of Traditional Chinese Medicine (Tianjin, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Miranda-Filho A, Piñeros M, Bray F. The descriptive epidemiology of lung cancer and tobacco control: A global overview 2018. Salud Publica Mex. 2019;61:219–229. doi: 10.21149/10140. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Zhu H, Sun L, Xu W, Wang X. Prognostic value of site-specific metastases in lung cancer: A population based study. J Cancer. 2019;10:3079–3086. doi: 10.7150/jca.30463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao W, Yu H, Han Z, Gao N, Xue J, Wang Y. Clinical significance of joint detection of serum CEA, SCCA, and bFGF in the diagnosis of lung cancer. Int J Clin Exp Pathol. 2015;8:9506–9511. [PMC free article] [PubMed] [Google Scholar]
- 4.Fu L, Wang R, Yin L, Shang X, Zhang R, Zhang P. CYFRA21-1 tests in the diagnosis of non-small cell lung cancer: A meta-analysis. Int J Biol Markers. 2019;34:251–261. doi: 10.1177/1724600819868234. [DOI] [PubMed] [Google Scholar]
- 5.Qin K, Chen Y, Long H, Chen J, Wang D, Chen L, Liang Z. The biomarkers and potential pathogenesis of lung cancer related cerebral hemorrhage. Medicine (Baltimore) 2019;98:e15693. doi: 10.1097/MD.0000000000015693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogawa M, Mizugishi K, Ishiguro A, Koyabu Y, Imai Y, Takahashi R, Mikoshiba K, Aruga J. Rines/RNF180, a novel RING finger gene-encoded product, is a membrane-bound ubiquitin ligase. Genes Cells. 2008;13:397–409. doi: 10.1111/j.1365-2443.2008.01169.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Zhang X, Sun B, Lu H, Wang D, Yuan X, Huang Z. Detection of aberrant promoter methylation of RNF180, DAPK1 and SFRP2 in plasma DNA of patients with gastric cancer. Oncol Lett. 2014;8:1745–1750. doi: 10.3892/ol.2014.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han F, Sun LP, Liu S, Xu Q, Liang QY, Zhang Z, Cao HC, Yu J, Fan DM, Nie YZ, et al. Promoter methylation of RNF180 is associated with H. pylori infection and serves as a marker for gastric cancer and atrophic gastritis. Oncotarget. 2016;7:24800–24809. doi: 10.18632/oncotarget.8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung KF, Lam CN, Wu K, Ng EK, Chong WW, Cheng AS, To KF, Fan D, Sung JJ, Yu J. Characterization of the gene structure, functional significance, and clinical application of RNF180, a novel gene in gastric cancer. Cancer. 2012;118:947–959. doi: 10.1002/cncr.26189. [DOI] [PubMed] [Google Scholar]
- 10.Deng J, Liang H, Zhang R, Hou Y, Liu Y, Ying G, Pan Y, Hao X. Clinical and experimental role of ring finger protein 180 on lymph node metastasis and survival in gastric cancer. Br J Surg. 2016;103:407–416. doi: 10.1002/bjs.10066. [DOI] [PubMed] [Google Scholar]
- 11.Udali S, Guarini P, Ruzzenente A, Ferrarini A, Guglielmi A, Lotto V, Tononi P, Pattini P, Moruzzi S, Campagnaro T, et al. DNA methylation and gene expression profiles show novel regulatory pathways in hepatocellular carcinoma. Clin Epigenetics. 2015;7:43. doi: 10.1186/s13148-015-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han F, Liu S, Jing J, Li H, Yuan Y, Sun LP. Identification of high-frequency methylation sites in RNF180 promoter region affecting expression and their relationship with prognosis of gastric cancer. Cancer Manag Res. 2020;12:3389–3399. doi: 10.2147/CMAR.S246995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akhurst T. Staging of non-small-cell lung cancer. PET Clin. 2018;13:1–10. doi: 10.1016/j.cpet.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Ottis P, Toure M, Cromm PM, Ko E, Gustafson JL, Crews CM. Assessing different E3 ligases for small molecule induced protein ubiquitination and degradation. ACS Chem Biol. 2017;12:2570–2578. doi: 10.1021/acschembio.7b00485. [DOI] [PubMed] [Google Scholar]
- 16.Ning Y, Hui N, Qing B, Zhuo Y, Sun W, Du Y, Liu S, Liu K, Zhou J. ZCCHC10 suppresses lung cancer progression and cisplatin resistance by attenuating MDM2-mediated p53 ubiquitination and degradation. Cell Death Dis. 2019;10:414. doi: 10.1038/s41419-019-1635-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng J, Guo J, Guo X, Hou Y, Xie X, Sun C, Zhang R, Yu X, Liang H. Mediation of the malignant biological characteristics of gastric cancer cells by the methylated CpG islands in RNF180 DNA promoter. Oncotarget. 2016;7:43461–43474. doi: 10.18632/oncotarget.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Potabattula R, Dittrich M, Schorsch M, Hahn T, Haaf T, El Hajj N. Male obesity effects on sperm and next-generation cord blood DNA methylation. PLoS One. 2019;14:e0218615. doi: 10.1371/journal.pone.0218615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang ZH, Dang YQ, Ji G. Role of epigenetics in transformation of inflammation into colorectal cancer. World J Gastroenterol. 2019;25:2863–2877. doi: 10.3748/wjg.v25.i23.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutton LP, Jeffreys SA, Phillips JL, Taberlay PC, Holloway AF, Ambrose M, Joo JE, Young A, Berry R, Skala M, et al. DNA methylation changes following DNA damage in prostate cancer cells. Epigenetics. 2019;14:989–1002. doi: 10.1080/15592294.2019.1629231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie XM, Deng JY, Hou YC, Cui JL, Wu WP, Ying GG, Dong QP, Hao XS, Liang H. Evaluating the clinical feasibility: The direct bisulfite genomic sequencing for examination of methylated status of E3 ubiquitin ligase RNF180 DNA promoter to predict the survival of gastric cancer. Cancer Biomark. 2015;15:259–265. doi: 10.3233/CBM-150466. [DOI] [PubMed] [Google Scholar]
- 22.Deng J, Liang H, Zhang R, Ying G, Xie X, Yu J, Fan D, Hao X. Methylated CpG site count of dapper homolog 1 (DACT1) promoter prediction the poor survival of gastric cancer. Am J Cancer Res. 2014;4:518–527. [PMC free article] [PubMed] [Google Scholar]
- 23.Deng J, Liang H, Ying G, Li H, Xie X, Yu J, Fan D, Hao X. Methylation of ras association domain protein 10 (RASSF10) promoter negative association with the survival of gastric cancer. Am J Cancer Res. 2014;4:916–923. [PMC free article] [PubMed] [Google Scholar]
- 24.Paun O, Verhoeven KJF, Richards CL. Opportunities and limitations of reduced representation bisulfite sequencing in plant ecological epigenomics. New Phytol. 2019;221:738–742. doi: 10.1111/nph.15388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.