 Meta-substituted benzenesulfonamides specifically inhibit metallo-β-lactamase ImiS with an IC50 value ranging from 0.11 to 0.86 μM.
Meta-substituted benzenesulfonamides specifically inhibit metallo-β-lactamase ImiS with an IC50 value ranging from 0.11 to 0.86 μM.
Abstract
Metallo-β-lactamase (MβL) ImiS contributes to the emergence of carbapenem resistance. A potent scaffold, meta-substituted benzenesulfonamide, was constructed and assayed against MβLs. The twenty-one obtained molecules specifically inhibited ImiS (IC50 = 0.11–9.3 μM); 2g was found to be the best inhibitor (IC50 = 0.11 μM), and 1g and 2g exhibited partially mixed inhibition with Ki of 8.0 and 0.55 μM. The analysis of the structure–activity relationship revealed that the meta-substitutes improved the inhibitory activity of the inhibitors. Isothermal titration calorimetry (ITC) assays showed that 2g reversibly inhibited ImiS. The benzenesulfonamides exhibited synergistic antibacterial effects against E. coli BL21 (DE3) cells with ImiS, resulting in a 2–4-fold reduction in the MIC of imipenem and meropenem. Also, mouse experiments showed that 2g had synergistic efficacy with meropenem and significantly reduced the bacterial load in the spleen and liver after a single intraperitoneal dose. Tracing the ImiS in living E. coli cells by RS at a super-resolution level (3D-SIM) showed that the target was initially associated on the surface of the cells, then there was a high density of uniform localization distributed in the cytosol of cells, and it finally accumulated in the formation of inclusion bodies at the cell poles. Docking studies suggested that the sulfonamide group acted as a zinc-binding group to coordinate with Zn(ii) and the residual amino acid within the CphA active center, tightly anchoring the inhibitor at the active site. This study provides a highly promising scaffold for the development of inhibitors of ImiS, even the B2 subclasses of MβLs.
Introduction
β-Lactam antibiotics remain the most important and frequently used antimicrobial agents, constituting more than 50% of the antibiotics prescribed worldwide.1 However, the effectiveness of β-lactam antibiotics, including penicillins, cephalosporins and carbapenems, has been threatened by the emergence of drug-resistant bacteria that produce β-lactamases.2,3 β-Lactamases are enzymes that inactivate β-lactam antibiotics by breaking the C–N bond of the β-lactam ring and render the drugs ineffective.4 According to the primary sequence homologies, β-lactamases have been categorized into four classes, A–D.5 Class A, C, and D enzymes are called serine β-lactamases (SβLs), which use a common catalytic mechanism where an active site serine nucleophilically attacks the β-lactam carbonyl, leading to a cleaved β-lactam ring.6 Class B enzymes are known as metallo-β-lactamases (MβLs), which use one or two Zn(ii) ions at active sites to mediate the hydrolysis of the β-lactam ring.7 MβLs are further divided into subclasses B1–B3, based on the amino acid sequence homology and Zn(ii) content.8
The B1 and B3 subclasses MβLs hydrolyze almost all known β-lactam antibiotics, leading to multiple-drug resistance in bacteria. In contrast, the B2 subclass enzymes have a narrow substrate profile including carbapenems, which have been called one of the “last resort” antibiotics.9 To combat bacterial drug-resistance, the development of β-lactamase inhibitors to restore the efficacy of the existing β-lactam antibiotics is an essential strategy. The co-administration of β-lactam antibiotics with β-lactamase inhibitors, such as clavulanic acid, tazobactam, and sulbactam, has been successfully used for the treatment of the bacterial infections mediated by SβLs.10 However, there are no MβL inhibitors available for clinical purposes to date.11 Therefore, the development of MβL inhibitors is urgently needed.
Given the biomedical importance of MβLs, significant efforts have been made to develop inhibitors of these enzymes,12 such as azolylthioacetamides,13 triazolylthioacetamides,14 bisthiazolidines15 and maleic acid derivatives,16 which exhibit inhibitory activities by binding to the Zn(ii) ions of the target enzymes. Chelating inhibitors, such as aspergillomarasmine A17 and [S,S]-ethylenediamine-N,N′-disuccinic acid (EDDS),18 inactivate clinically-relevant MβLs like VIM-2 and NDM-1 by sequestering the metals of each enzyme. Covalent inhibitors, such as ebselen, inactivate the B1 and B2 subclass MβLs by the formation of a Se–S bond with the cysteine residue at the active site of the enzymes.19 Martin Everett et al. first reported that ANT431, a sulfonamide compound, exhibited inhibition efficacy on MβLs VIM-2, NDM-1 and IMP-1.20 Recently, our studies revealed that azolylthioacetamide was a highly promising scaffold for the development of MβL inhibitors with IC50 values in the submicromolar grade.21
ImiS is a representative of the B2 subclass MβLs; therefore, significant effort has been made in the structural, spectroscopic, mechanistic and inhibition studies on this enzyme.22–24 Recently, our studies showed that the thiazole-substituted azolylthioacetamides specifically inhibited ImiS, with Ki values in the range of 1.2–3.6 μM using imipenem as the substrate,23 and the N-heterocyclic dicarboxylic acid was a competitive inhibitor of ImiS with a Ki value of 3.5 μM.24 The acetazolamide (ACZ),25 pyrrolidinone-substituted benzenesulfonamide (PBS),26 and hydrazido benzenesulfonamide (HBS)27 (Fig. 1) were reported to bind the Zn(ii) ions at the active sites of carbonic anhydrase (CA) with their sulfonamide group, therefore exhibiting strong enzymatic inhibition.27 CA, a monozinc enzyme like ImiS, indicated that the sulfonamides may be used for the development of MβLs, particularly ImiS inhibitors. Our goal is to develop specific inhibitors of MβLs and to use these inhibitors in combination with β-lactam antibiotics to combat bacterial infections mediated by MβLs. Towards this goal, benzenesulfonamides with various aromatic substituents (Fig. 1) were designed, synthesized and characterized.
Fig. 1. Structures of the synthesized benzenesulfonamides and the reported sulfonamides.
The benzenesulfonamides were designed based on two strategies: first, the ortho, meta, and para positions of the aromatic substituents on the benzene ring, relative to the sulfonamide group, were adjusted to define the optimal position for the compound to bind to the active site of the target enzyme, which confers the best inhibitory effect. Second, phenylamide was grafted with different substituents on the molecule to ensure different electronic and lipophilic environments, which could manipulate the activity of the molecules. With these two strategies, twenty-one benzenesulfonamides 1a–j, 2a–h, 3e–f and RS (Fig. 1) were designed and synthesized with previously reported methods.28,29 Briefly, the appropriate benzoic acid was refluxed in SOCl2 for 3 h for conversion into the substituted benzoyl chloride, which reacted with aminobenzensulfonamide in the presence of pyridine to give the desired benzenesulfonamides. All compounds synthesized were characterized by 1H and 13C NMR and confirmed by HRMS (see ESI†). These compounds were tested as inhibitors with the purified MβLs NDM-1, ImiS and L1; their inhibitory modes were investigated by generating Lineweaver–Burk plots and isothermal titration calorimetry (ITC). Also, the antimicrobial activities of these inhibitors in combination with the existing antibiotics against antibiotic-resistant strains were evaluated, and molecular docking was performed to investigate the interactions of inhibitor molecules with the target enzyme.
Results and discussion
Activity evaluation of benzenesulfonamides
To test whether these sulfonamides were MβL inhibitors, the inhibition experiments under steady-state conditions were conducted on an Agilent UV8453 spectrometer using imipenem (40 μM) as the substrate for ImiS, and cefazolin (40 μM) for NDM-1 and L1. The concentrations of inhibitors were varied between 0 and 20 μM. The hydrolysis of imipenem and cefazolin was monitored at 300 and 262 nm, respectively. The initial reaction rates were determined in the absence and presence of inhibitors in triplicate, and the average values were recorded.
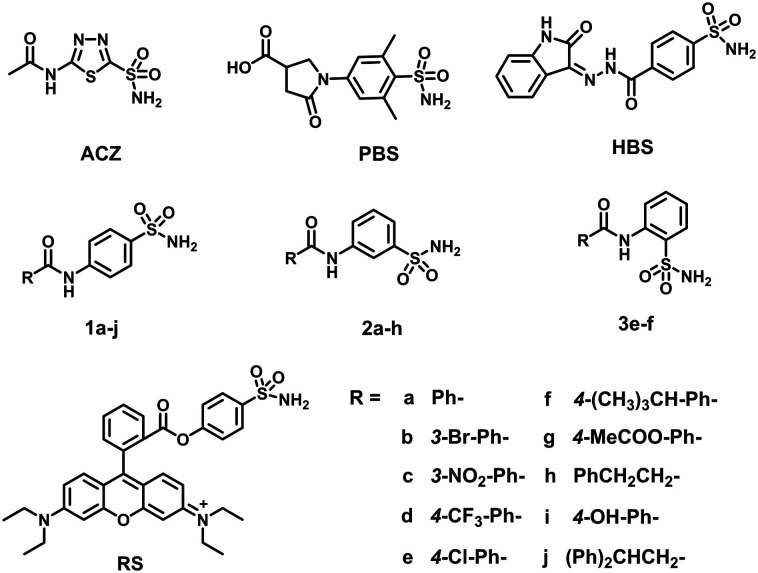
The percent inhibition, defined as enzyme activity without inhibitor (100%) minus residual activity with inhibitor, of benzenesulfonamide derivatives on ImiS, NDM-1 and L1 is shown in Fig. 2. It can be clearly observed that the sulfonamides exhibited more than 70% inhibitory activities against ImiS at a concentration of 20 μM; 2g in particular showed 95% inhibition. However, the compounds had less than 20% inhibition against NDM-1 and L1 at a concentration of up to 20 μM. Therefore, for the remainder of this work, we focused on inhibition studies of ImiS.
Fig. 2. Percent inhibition of benzenesulfonamide derivatives (20 μM) against MβLs. Imipenem was used as substrate for ImiS, and cefazolin was used as substrate for NDM-1 and L1.
The inhibitor concentrations causing 50% decrease in the enzyme activity (IC50) of all sulfonamides against ImiS were determined in 30 mM Tris (pH 7.0) using imipenem as the substrate. The collected IC50 data (Table 1) indicated that all of these compounds had inhibitory efficacy against ImiS, exhibiting an IC50 value range of 0.11–9.3 μM, and 2g was found to be the most potent inhibitor (IC50 = 0.11 μM). It should be noted that 2a–h had an inhibitory potency on ImiS (IC50 < 1 μM) that was an order of magnitude higher as compared to 1a–1j (IC50 = 2.7–9.3 μM) and 3e–3f (IC50 = 4.6–5.4 μM). These IC50 values of sulfonamides are consistent with their percent inhibitions, as shown in Fig. 2. The analysis of these IC50 data revealed a structure–activity relationship (SAR), where the meta-substitutes of the phenyl ring more significantly improved the inhibitory activity of benzenesulfonamides against ImiS as compared to the same substitutes at the ortho- and para-positions.
Table 1. The inhibitory activities (IC50, μM) of benzenesulfonamide derivatives against MβL ImiS a .
| P-Substituted compd. | IC50 | M-Substituted compd. | IC50 | O-Substituted compd. | IC50 |
| 1a | 7.5 ± 0.3 | 2a | 0.15 ± 0.04 | 3e | 5.4 ± 0.2 |
| 1b | 2.7 ± 0.4 | 2b | 0.14 ± 0.07 | 3f | 4.6 ± 0.2 |
| 1c | 4.8 ± 0.1 | 2c | 0.86 ± 0.04 | RS | 6.8 ± 0.5 |
| 1d | 9.3 ± 0.3 | 2d | 0.31 ± 0.05 | ||
| 1e | 5.1 ± 0.5 | 2e | 0.44 ± 0.01 | ||
| 1f | 3.3 ± 0.1 | 2f | 0.23 ± 0.08 | ||
| 1g | 7.3 ± 0.2 | 2g | 0.11 ± 0.02 | ||
| 1h | 3.5 ± 0.2 | 2h | 0.16 ± 0.08 | ||
| 1i | 5.6 ± 0.7 | ||||
| 1j | 4.1 ± 0.6 |
aThe substrate used was imipenem and inhibitor concentrations were varied between 0.01 and 20 μM. Enzyme and inhibitors were preincubated for 3 h before testing.
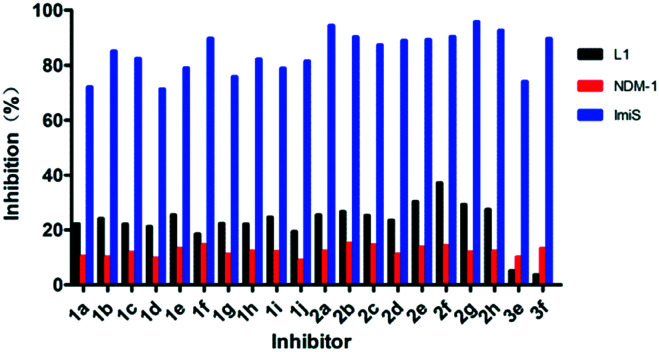
Given the best potency of sulfonamide 2g, its time- and concentration-dependent inhibitions on ImiS were assayed (Fig. 3). It can be observed that in the presence of sulfonamide at a concentration of 0.5, 1, and 5 μM, respectively, the residual activity of ImiS decreased with the increase in the inhibitor concentration (Fig. 3A). Also, the residual activity of the enzyme decreased with the increase in the inhibitor incubation time with the enzyme, and the molecule showed maximum inhibition at a concentration of around 5.0 μM and after incubation with ImiS for about 3 h. The concentration-dependent inhibition of 2g on ImiS was assayed, and the resulting inhibition curve (Fig. 3B) showed that more than 92% activity (<8% residual activity) of the enzyme was inhibited by the molecule at a concentration of around 10 μM.
Fig. 3. Time- and concentration-dependent inhibition of ImiS by benzenesulfonamide 2g (A), and the inhibition curve of ImiS by 2g at a concentration between 0.01 and 10 μM (B).
Inhibition assay of ImiS by ITC
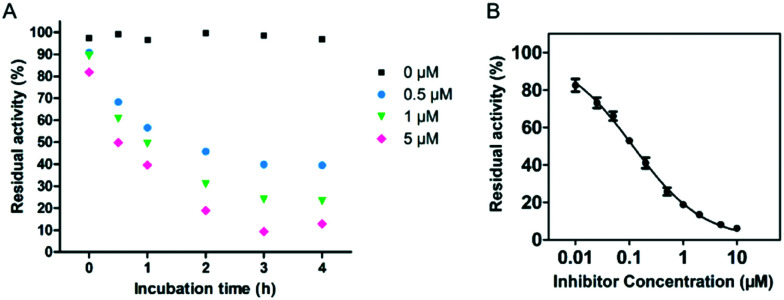
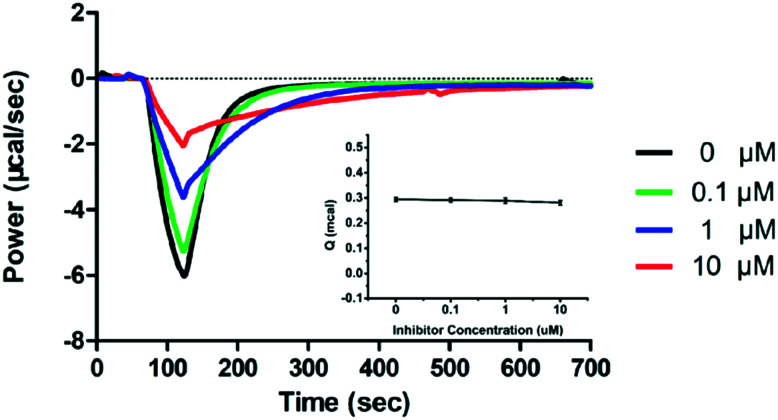
Isothermal titration calorimetry (ITC), an essential approach to measure the heat (Q) released or absorbed during ligand–protein binding by titrating one solution into another, has been applied in enzyme kinetic studies.30 A MicroCal-ITC200 was employed to investigate the inhibition of imipenem hydrolysis with ImiS by sulfonamide in single injection mode at 25 °C, as previously reported.31 The concentrations of the enzyme and imipenem were 100 nM and 0.5 mM, respectively, and the concentration of the inhibitor was varied between 0 and 10 μM. ITC was used to monitor the inhibition of imipenem hydrolysis with ImiS by 2g, as shown in Fig. 4. The thermopower (dq/dt) gradually decreased with the increase in the inhibitor concentration from 0 to 10 μM, confirming that 2g inhibited ImiS in a dose-dependent form thermodynamically. It should be noted that the addition of 2g did not result in changes in the total heat (Q) released (Fig. 4, inset), revealing that the sulfonamide reversibly inhibited ImiS, similar to the inhibition mode of d-captopril and azolylthioacetamide (ATAA) on NDM-1, as previously reported.31 This is likely due to the formation of the enzyme–inhibitor complex (EI) that slowed down substrate hydrolysis.
Fig. 4. Overlayed heat flow curves of imipenem (0.5 mM) hydrolysis by ImiS in the absence and presence of benzenesulfonamide 2g in the concentration range of 0–10 μM; the concentration of ImiS was 100 nM.
Inhibition mode of benzenesulfonamides
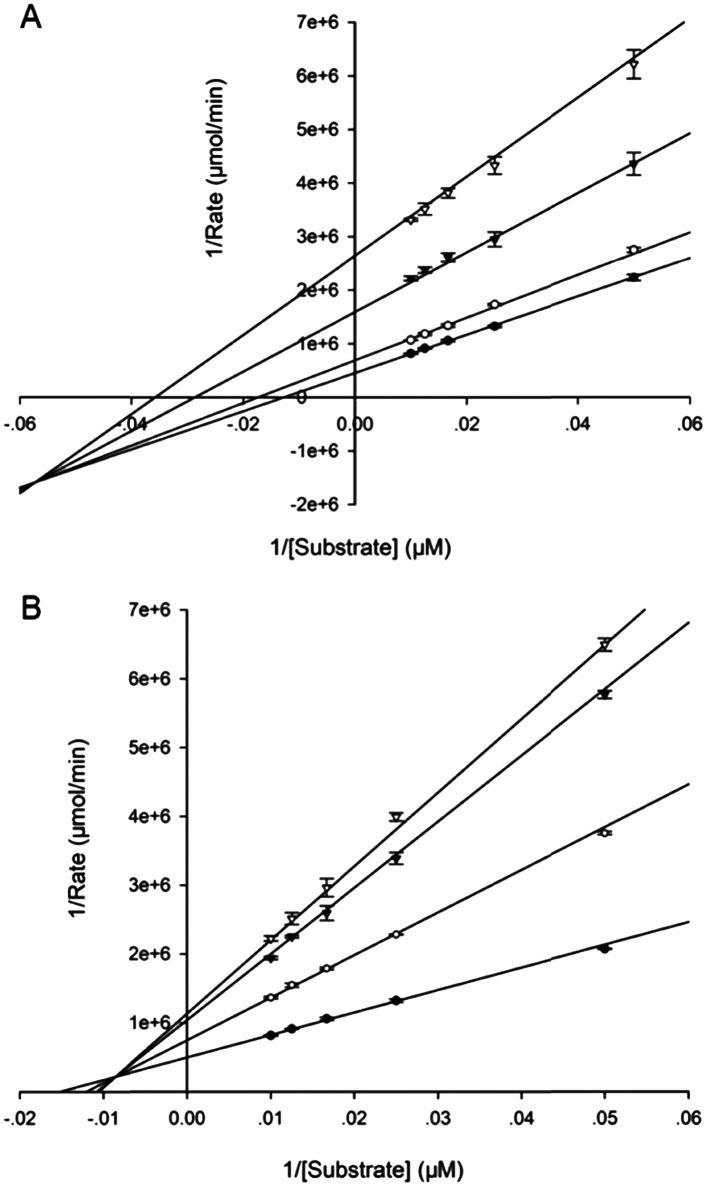
To further identify the inhibition mode of the sulfonamides against ImiS, 1g and 2g were chosen to determine the Ki values. The concentrations of the inhibitors were varied between 0 and 10 μM, and substrate (imipenem) concentrations were varied between 20 and 100 μM. All experimental hydrolytic rates were determined in triplicate. The inhibition mode was assayed by generating Lineweaver–Burk plots, and the Ki values were obtained by fitting initial velocity versus substrate concentration at each inhibitor concentration using SigmaPlot 12.0. The Lineweaver–Burk plots of imipenem hydrolysis catalyzed by ImiS in the absence and presence of two benzenesulfonamides are shown in Fig. 5. The Lineweaver–Burk plots along with slope-intercept replots (Fig. S1 and S2†) suggest that both inhibitors are partial mixed-type inhibitors, implying that the ESI† can yield the product without the release of I.32 The Ki values of 1g and 2g were determined to be 8.0 ± 0.3 and 0.55 ± 0.04 μM (average ± standard deviation of triplicates), respectively.
Fig. 5. Lineweaver–Burk plots of the ImiS-catalyzed hydrolysis of imipenem in the absence and presence of benzenesulfonamides 1g (A) and 2g (B). Inhibitor concentrations were 0 ([black circle]), 1 (○), 5 ([blacktriangledown]), and 10 μM (▽).
Antibacterial activity assays in vitro
The capability of benzenesulfonamides to restore the antibacterial activity of imipenem and meropenem against E. coli producing ImiS was investigated by measuring the minimum inhibitory concentration (MIC) changes in the antibiotics in the absence and presence of sulfonamides using a previously reported method.33 The E. coli BL21 (DE3) with and without ImiS were used for the evaluation of the inhibitors. The concentration of inhibitors used was 16 μg mL–1. The collected MIC data (Table 2) indicated that all sulfonamides tested increased the antimicrobial effect of both imipenem and meropenem on E. coli expressing ImiS, resulting in a 2–4-fold reduction in MICs, which is consistent with their inhibitory effect on ImiS. However, the inhibitors alone did not inhibit the bacterial growth at a concentration of up to 64 μg mL–1. Also, no synergistic antibacterial effect of the sulfonamides on E. coli without ImiS was observed using the same dose of inhibitor, suggesting that the capability of sulfonamides to restore antibiotic activity is due to their inhibition toward ImiS in living bacterial cells.
Table 2. MICs of carbapenems, imipenem (A) and meropenem (B), against E. coli BL21 (DE3) cells expressing ImiS in the absence and presence of benzenesulfonamides at a concentration of 16 μg mL–1 a .
| A | |||||||||||
| Compd. | 1a | 1b | 1c | 1d | 1e | 1f | 1g | 1h | 1i | 1j | |
| MIC | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | |
| Compd. | 2a | 2b | 2c | 2d | 2e | 2f | 2g | 2h | 3e | 3f | RS |
| MIC | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 8 | 8 | 8 |
| B | |||||||||||
| Compd. | 1a | 1b | 1c | 1d | 1e | 1f | 1g | 1h | 1i | 1j | |
| MIC | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | |
| Compd. | 2a | 2b | 2c | 2d | 2e | 2f | 2g | 2h | 3e | 3f | RS |
| MIC | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 32 | 32 | 32 |
aMICs of imipenem and meropenem alone on E. coli BL21 (DE3) with ImiS were 16 and 64 μg mL–1, respectively. The MICs of imipenem and meropenem on E. coli without ImiS in the absence and presence of benzenesulfonamides were 0.25 and 0.0375 μg mL–1, respectively.
Real-time tracking of ImiS in living bacterial cells
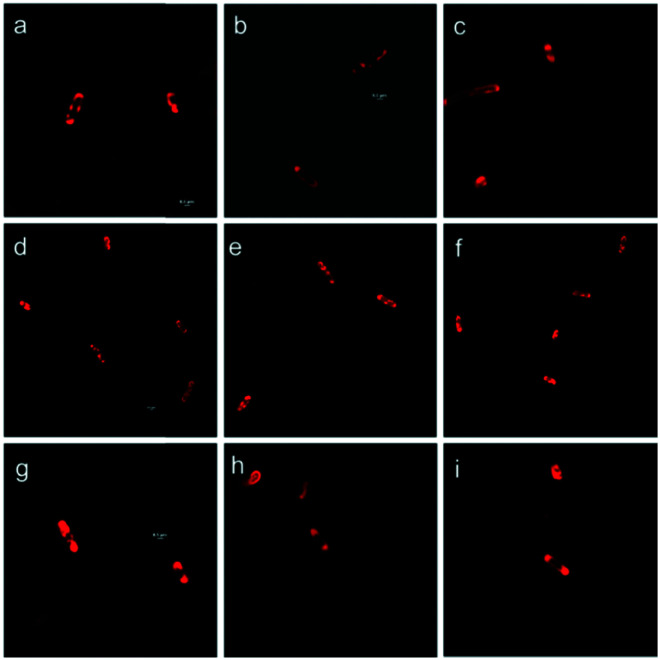
To track the ImiS inside living bacterial cells, we constructed a fluorescent sulfonamide RS (Fig. 1), and used it to perform the super-resolution fluorescence imaging of the E. coli BL21 cells harboring ImiS on a 3D-structured illumination microscopy (3D-SIM).34Fig. 6 shows the 3D-SIM images of ImiS E. coli cells incubated with RS in MHB at a dose of around 50 μM (final concentration) for different time periods. The interactions of RS with bacteria were imaged using a microscopy technique, where a clear visualization of the interactions of RS and intracellular enzyme (ImiS) was clearly demonstrated. RS was found to associate at certain sites on and in the bacteria, depending on the incubation time. On incubation for 2 h, the inhibitors initially associated with the surface of the bacteria due to electrostatic interactions (Fig. 6a–c). On incubation for 8 h, a high density of bacterial cells with either membrane-associated or uniform localization of the inhibitors distributed in the cytosol of the bacterial cells was observed (Fig. 6d–f). On incubation for 16 h, the target ImiS accumulated at the cell poles was observed (Fig. 6g–i), suggesting the formation of the inclusion bodies of the enzyme.35
Fig. 6. Confocal microscopic real-time imaging of E. coli BL21 cells harboring ImiS after treatment with RS (50 μM) for 2 (a–c), 8 (d–f) and 16 h (g–i). Scale bars, 0.5 μm.
Antibacterial activity assays in vivo
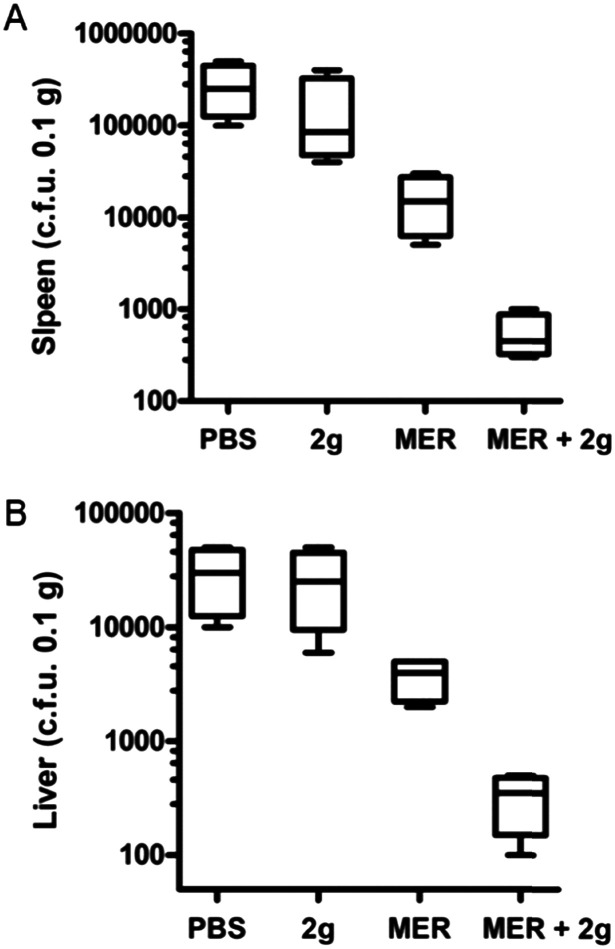
To test whether the sulfonamides were capable of restoring the activity of meropenem in vivo, 2g was chosen to perform synergistic antibacterial assays with meropenem in a mice model with a previously reported method.36 Briefly, the Kunming mice model of systemic infection was established by the intraperitoneal injection of E. coli BL21 (DE3) cells harboring ImiS. Two hours after infection with the bacteria, the infected mice were intraperitoneally injected with a single dose of the drug. The effects of meropenem or sulfonamide 2g monotherapy or antibiotic-inhibitor combination therapy on bacterial load in tissues were evaluated. The assay results are shown in Fig. 7. The bacterial load in the tissues treated with sulfonamide alone was almost unaffected. However, compared with meropenem monotherapy, the synergistic therapy of sulfonamide with meropenem significantly reduced the bacterial load in the spleen (Fig. 7A) and liver (Fig. 7B) after a single intraperitoneal dose, indicating that sulfonamide had synergistic antibacterial efficacy in vivo.
Fig. 7. Kunming mice were injected intraperitoneally with E. coli BL21 cells harboring ImiS. Groups of mice were treated with a single dose of meropenem (10 mg kg–1), inhibitor 2g (10 mg kg–1), a combination of meropenem (10 mg kg–1) and 2g (10 mg kg–1), and PBS buffer (as control) by intraperitoneal injection. The bacterial loads in the spleen (A) and liver (B) were determined by selective plating.
Docking study
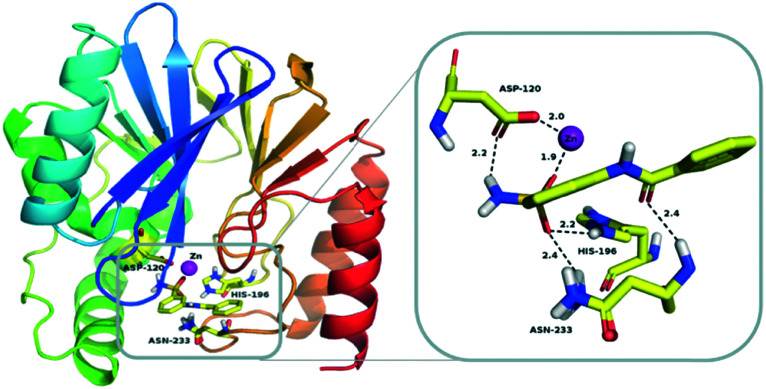
To further explore the binding of sulfonamides to the target enzyme, 2a was selected for molecular docking. Since there is no available crystal structure of ImiS, CphA (PDB code ; 2QDS) that shares 96% sequence identity with ImiS was used instead. The lowest energy-docking conformations of these clusters are shown in Fig. 8, and the binding energy of the CphA/2a complex is –6.87 kcal mol–1. The sulfonamide group acted as a Zn(ii)-binding group (ZBG) and one of the oxygen atoms was coordinated to the Zn(ii) ion at the catalytic site (1.9 Å for CphA/2a), another oxygen atom formed two hydrogen bonds with two amino acid residues, namely, His196 (2.2 Å) and ASN233 (2.4 Å), and a hydrogen atom on the amino group formed a hydrogen bond with ASP120 (2.2 Å). The amide group of the sulfonamide also interacted with the amino acid residue ASN233 (2.4 Å). As a result, the sulfonamide functional group coordinated with the Zn(ii) ion and residual amino acid within the CphA active center, tightly anchoring the inhibitor in the active site. This metal-binding mode of sulfonamide revealed by docking studies is similar to that of the azolylthioacetamide for CphA.37
Fig. 8. Lowest-energy docking conformation of sulfonamide 2a docked into the active site of CphA (PDB code ; 2QDS). The enzyme backbone is shown as a cartoon in green, and selected residues are shown as sticks colored by elements (H, white; C, yellow; N, blue; O, red; S, orange). The Zn(ii) ion is shown as a magenta sphere; 2a is shown as sticks with the same color code as amino acid residues. Characteristic short distances between the inhibitor and the protein are indicated by dashed lines. These figures were generated with PyMOL.
Cytotoxicity assays
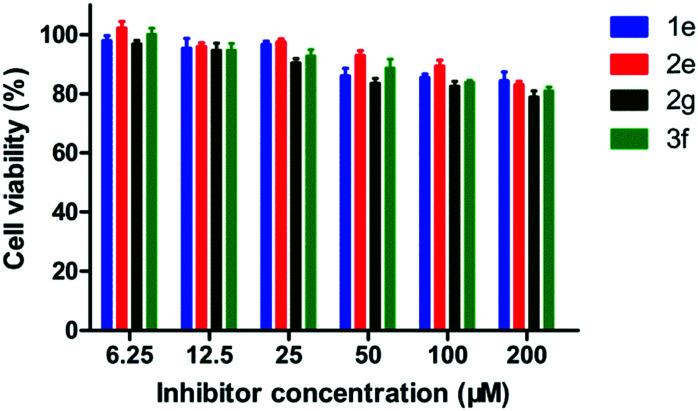
The potential toxicity of enzyme inhibitors is a principal problem for clinical medical applications. Four sulfonamides 1e, 2e, 2g and 3f with different working concentrations (6.25, 12.5, 25, 50, 100, 200 μM) were subjected to a cytotoxicity assay with mouse fibroblast cells (L929), based on a previously reported method.38 As shown in Fig. 9, more than 80% of the cells tested maintained viability in the presence of the inhibitors at a concentration of up to 200 μM, suggesting that these sulfonamides have low cytotoxicity.
Fig. 9. Percent cell viability (relative to the absence of sulfonamide) of L-929 mouse fibroblastic cells in the presence of 1e, 2e, 2g and 3f at concentrations of 6.25, 12.5, 25, 50, 100, and 200 μM, respectively.
Conclusions
Twenty-one benzenesulfonamides (1a–j, 2a–h, 3e–f and RS) were synthesized and characterized via NMR and MS. Biological assays revealed that all these compounds specifically inhibited ImiS, exhibiting IC50 values in the range of 0.11–9.3 μM, and 2g was found to be the best inhibitor (IC50 = 0.11 μM). The structure–activity relationship analysis revealed that the meta-substitutes of the phenyl ring improved the activity of sulfonamides on ImiS. ITC assays indicated that the sulfonamide reversibly inhibited ImiS. The identification of Ki showed that 1g and 2g were partially mixed enzymatic inhibitors. MIC tests demonstrated that the sulfonamides restored a 2–4-fold antimicrobial activity of imipenem and meropenem on E. coli expressing ImiS. Mouse experiments showed that 2g had synergistic efficacy with meropenem, and significantly reduced the bacterial load in the spleen and liver after a single intraperitoneal dose. Tracing the ImiS in living E. coli cells through labeling it with RS at a super-resolution level using 3D-SIM showed that the target was initially associated on the surface of cells, then there was a high density of uniform localization distributed in the cytosol of cells, and it finally accumulated in the formation of inclusion bodies at the cell poles. Docking studies suggested that the sulfonamide group acted as the zinc-binding group (ZBG) to coordinate with the Zn(ii) and the residual amino acid within the CphA active center, tightly anchoring the inhibitor at the active site. Cytotoxicity assays indicated that the sulfonamides had low cytotoxicity.
Experimental
Chemicals
Sulfanilamide, thionyl chloride and substituted benzoic acid were purchased from Sigma-Aldrich (Shanghai) Trading Co. Ltd. All other starting materials were purchased from commercial sources and were purified using standard methods. Analytical thin layer chromatography (TLC) was performed on silica gel GF254 plates with visualization by ultraviolet radiation. 1H and 13C NMR spectra were recorded on a Bruker Avance III 400 MHz NMR spectrometer. Chemical shifts are given in parts per million (ppm) on the delta scale. The peak patterns are reported as singlet (s), doublet (d), triplet (t), quartet (q), doublet (dd), and multiplet (m). The spectra were recorded with TMS as the internal standard. Coupling constants (J) were reported in Hertz (Hz). Mass spectra were recorded on a micro TOF-Q (BRUKER) mass spectrometer. The activity evaluation of inhibitors was performed on an Agilent 8453 UV-vis spectrometer.
General procedure for the preparation of benzenesulfonamides (1a–j, 2a–h, 3e–f and RS)
The acyl chlorides were synthesized as previously reported.28 To a solution of the corresponding benzoic acid (4 mmol) in SOCl2 (20 mL) DMF was added dropwise for the catalytic reaction. The reaction mixture was stirred and refluxed at 85 °C for 3 h. After the reaction was completed, the solvent was removed by rotary evaporation under a reduced pressure, and the resulting residue was dissolved in acetone (10 mL) for the next step reaction without further purification. The benzenesulfonamides were synthesized using a previously reported method.29 The as-prepared acyl chloride solution was added dropwise over 30 min to a stirred solution of sulfanilamide (2.9 mmol, 500 mg) and pyridine (5.8 mmol, 467 mL) in dry acetone (10 mL) at 0 °C. The reaction mixture was stirred at room temperature for about 5 h, and the resulting products were filtered or evaporated under reduced pressure. The crude product was purified via flash column chromatography on silica gel (ethyl acetate/petroleum ether, 2 : 1) to give the sulfonamides 1a–j, 2a–h and 3e–f.
The fluorescent sulfonamide RS was synthesized via a previously reported method.19 To a stirred solution of 4-hydroxybenzenesulfonamide (2.0 eq.) in DCM was added DCC (1.3 eq.), DMAP (0.13 eq.) and rhodamine B (1.0 eq.) at 0 °C. The reaction mixture was stirred at 0 °C for 10 min, and then at room temperature for 4 h; the solvent was removed under vacuum and extracted with DCM (3 × 10 mL). The combined organic phase was dried over anhydrous Na2SO4, distilled under reduced pressure, and the crude product was purified via flash column chromatography on silica gel (CHCl3/CH3OH, 10 : 1) to afford RS.
Determination of IC50
MβLs from subclasses B1 (NDM-1), B2 (ImiS) and B3 (L1) were over-expressed and purified using a previously described method,39–41 and the details are provided in the ESI.† The inhibitor concentration causing 50% decrease in the enzyme activity (IC50) was determined at 25 °C using imipenem as the substrate for ImiS. The sulfonamides 1a–j, 2a–h, 3e–f and RS were dissolved in a small volume of DMSO and diluted with 30 mM Tris (pH 7.0), and ImiS sample and imipenem were mixed in 30 mM Tris (pH 7.0). The final concentrations of DMSO in inhibition experiments were below 0.5%, and control experiments verified that 0.5% DMSO had no inhibition against ImiS. All inhibitors were assayed at six different concentrations between 0 and 20 μM, and substrate concentration was 40 μM. The hydrolysis of imipenem was monitored at 300 nm on an Agilent UV8453 spectrometer, and the hydrolytic rates were determined in triplicate. The IC50 values were calculated by plotting the average percentage inhibition against inhibitor concentration and the linear fitting of the data.
Determination of Ki
The inhibition mode of sulfonamides was determined by generating Lineweaver–Burk plots, and the Ki values were determined by fitting initial velocity versus substrate concentration at each inhibitor concentration using SigmaPlot 12.0. The concentrations of imipenem used were 20, 40, 60, 80 and 100 μM, respectively. The initial hydrolysis rate of the substrate was determined by the method for measuring IC50, as described above. All hydrolytic rates were determined in triplicate.
ITC assays
Isothermal titration calorimetry (ITC) experiments were performed on a Malvern MicroCal iTC 200 instrument at 25 °C using a single injection mode. During all experiments, the reference cell was filled with degassed water. The MβLs, substrates, and inhibitor 2g were prepared in 30 mM Tris with 0.5% addition of DMSO. The concentrations of ImiS and substrate were 100 nM and 0.5 mM, respectively, and the concentrations of the inhibitor were varied between 0 and 10 μM. Before starting measurements, the enzyme sample was pre-incubated with different concentrations of 2g for 3 h and then delivered into the sample cell (210 μL). The reaction in the absence of inhibitor was also monitored as the control. The stirring rate of the syringe was set to 750 rpm. The system was equilibrated at the desired 25 °C. Each reaction was automatically initiated by adding imipenem (0.5 mM) in the form of a single 38 μL injection into the enzyme-containing sample cell. Heat flow was recorded as a function of time. Data were collected every 1 s until the signal reached the baseline and continued to be recorded for the appropriate time to generate the final baseline. The progress curves of imipenem hydrolysis catalyzed by ImiS in the absence and presence of 2g at different concentrations are shown in Fig. 5.
Determination of MIC
A single colony of E. coli BL21 (DE3) containing plasmids pET26b-ImiS on LB agar plates was transferred to 5 mL of a Mueller-Hinton (MH) liquid medium. Strains grown in the MH medium to OD600 of 0.5 were used as inoculum after an 84-fold dilution to 1 × 105 CFU mL–1 in the MH medium. MIC values were determined using the Clinical and Laboratory Standards Institute (CLSI) macrodilution (tube) broth method.31 The antibiotic was dissolved in the MH medium to prepare 128, 64, 32, 16, 8, 4, 2, and 1 μg mL–1 stock solutions, respectively. The sulfonamides 1a–j, 2a–h, 3e–f and RS were dissolved in the DMSO and MH media to prepare 64 μg mL–1 (initial concentration) stock solutions. The as-prepared solutions with different antibiotic concentrations (50 μL) and inhibitor solution (50 μL) were added to a 96-hole plate, and then 100 μL inoculum was added sequentially to the as-prepared solutions. MIC was interpreted as the lowest concentration of the drug that completely inhibited the visible growth of bacteria after incubating plates at 37 °C for at least 16 h. Each measurement was performed in triplicate in at least two independent experiments and the highest MIC value was reported.
Sample preparation for cell imaging
The cell imaging was performed on a Nikon N-SIM instrument at 25 °C. E. coli BL21 cells harboring ImiS (OD600 = 0.5) were incubated with the fluorescent sulfonamide RS at a dose of around 50 μM (final concentration) in tubes at 37 °C for different times. After incubation, the cells were washed five times with PBS buffer (10 000 rpm, 5 min) to remove any adsorbed RS probe on the cell surface. The cells were re-suspended in PBS to give the final samples for confocal microscopic imaging.
Mouse experiments
Healthy male and female KM mice, 20 ± 2 g, were supplied by the Experimental Animal Center, Medical College of Xi'an Jiaotong University, China. Mouse experiments were performed in the Xi'an Jiaotong University Health Science Center. The animals were provided with a standard commercial mouse pellet diet and ultrapure water ad libitum. Mice were randomly divided into four groups with six mice in each group, with half being male and the other half female. All mice were treated with 200 μL bacteria suspension (OD600 = 1.0) via intraperitoneal injection. After infection for 2 h, the infected mice in groups 1, 2, 3 and 4 were subjected to intraperitoneal treatment with PBS, 10 mg kg–1 meropenem, and 10 mg kg–1 sulfonamide 2g, 10 mg kg–1 meropenem plus 10 mg kg–12g, respectively. All experimental mice were euthanized 24 h post-infection, and 0.1 g each of the spleen and liver were collected and homogenized into 1 mL of sterile PBS. Organ homogenates were then serially diluted in PBS, and plated on LB-agar for c.f.u. enumeration.
Ethical statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Xi'an Jiaotong University and experiments were approved by the Animal Ethics Committee of Xi'an Jiaotong University, (approval ID: NO. XJTULAC2019-1234).
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by Grant (21572179) from the National Natural Science Foundation of China and Grant (2019KW-068) from the Shaanxi Province International Cooperation Project.
Footnotes
†Electronic supplementary information (ESI) available: 1H, 13C NMR and MS characterization, Over-expression and purification of MβLs, Docking studies, cytotoxicity assay. See DOI: 10.1039/c9md00455f
References
- Drawz S. M., Bonomo R. A. Clin. Microbiol. Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demain A. L., Elander R. P. Antonie Van Leeuwenhoek. 1999;75:5–19. doi: 10.1023/a:1001738823146. [DOI] [PubMed] [Google Scholar]
- Medeiros A. A. Clin. Infect. Dis. 1997;24(Suppl 1):S19. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- King D. T., Sobhanifar S., Strynadka N. C. J. Protein Sci. 2016;25:787–803. doi: 10.1002/pro.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambler R. P. Philos. Trans. R. Soc., B. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Galleni M., Lamotte-Brasseur J., Raquet X., Dubus A., Monnaie D., Knox J. R. Biochem. Pharmacol. 1995;49:1171–1178. doi: 10.1016/0006-2952(94)00502-d. [DOI] [PubMed] [Google Scholar]
- Mojica M. F., Bonomo R. A., Fast W. Curr. Drug Targets. 2016;17:1029–1050. doi: 10.2174/1389450116666151001105622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garau G., Garcia-Saez I., Bebrone C., Anne C., Mercuri P., Galleni M. Antimicrob. Agents Chemother. 2004;48:2347–2349. doi: 10.1128/AAC.48.7.2347-2349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp-Wallace K. M., Endimiani A., Taracila M. A., Bonomo R. A. Antimicrob. Agents Chemother. 2011;55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmann D. E., Jahic H., Ross P. L., Gu R. F., Hu J., Durand-Reville T. F. J. Biol. Chem. 2013;288:27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornaglia G., Giamarellou H., Rossolini G. M. Lancet Infect. Dis. 2011;11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- Docquier J. D., Mangani S. Drug Resist. Updates. 2018;36:13–29. doi: 10.1016/j.drup.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Chang Y. N., Ge Y., Kang J. S., Zhang Y. L., Liu X. L. Bioorg. Med. Chem. Lett. 2017;27:5225–5229. doi: 10.1016/j.bmcl.2017.10.038. [DOI] [PubMed] [Google Scholar]
- Zhai L., Zhang Y. L., Kang J. S., Oelschlaeger P., Xiao L., Nie S. S. ACS Med. Chem. Lett. 2016;7:413–417. doi: 10.1021/acsmedchemlett.5b00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González M. M., Kosmopoulou M., Mojica M. F., Castillo V., Hinchliffe P., Pettinati I. ACS Infect. Dis. 2015;1:544–554. doi: 10.1021/acsinfecdis.5b00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y., Eto M., Mano Y., Tateda K., Yamaguchi K. Zhongguo Ganran Yu Hualiao Zazhi. 2011;54:3625–3629. [Google Scholar]
- King A. M., Reid-Yu S. A., Wang W., King D. T., De Pascale G., Strynadka N. C. Nature. 2014;510:503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proschak A., Kramer J., Proschak E., Wichelhaus T. A. J. Antimicrob. Chemother. 2017;73:425–430. doi: 10.1093/jac/dkx403. [DOI] [PubMed] [Google Scholar]
- Chen C., Xiang Y., Yang K. W., Zhang Y., Wang W. M., Su J. P. Chem. Commun. 2018;54:4802–4805. doi: 10.1039/c8cc01067f. [DOI] [PubMed] [Google Scholar]
- Everett M., Sprynski N., Coelho A. Antimicrob. Agents Chemother. 2018;62:00074. doi: 10.1128/AAC.00074-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. L., Yang K. W., Zhou Y. J. ChemMedChem. 2015;9:2445–2448. doi: 10.1002/cmdc.201402249. [DOI] [PubMed] [Google Scholar]
- Costello A. L., Sharma N. P., Yang K. W., Crowder M. W., Tierney D. L. Biochemistry. 2006;45:13650–13658. doi: 10.1021/bi061547e. [DOI] [PubMed] [Google Scholar]
- Yang S. K., Kang J. S., Oelschlaeger P., Yang K. W. ACS Med.ACS Med. Chem. Lett.Chem. Lett. 2015;6:455–460. doi: 10.1021/ml500534c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei F., Yang K. W., Zhou L. S., Xiao J. M., Yang X., Zhai L. Bioorg. Med. Chem. Lett. 2012;22:5185–5189. doi: 10.1016/j.bmcl.2012.06.074. [DOI] [PubMed] [Google Scholar]
- Groot T. D., Sinke A. P., Kortenoeven M. L. A., Alsady M., Baumgarten R., Devuyst O. J. Am. Soc. Nephrol. 2015;27:2082–2091. doi: 10.1681/ASN.2015070796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaškevičienė I., Paketurytė V., Pajanok N., Sapijanskaitė B., Kantminienė K., Mickevičius V. Bioorg. Med. Chem. 2018;27:322–337. doi: 10.1016/j.bmc.2018.12.011. [DOI] [PubMed] [Google Scholar]
- Abo-Ashour M. F., Eldehna W. M., Alessio N., Ibrahim H. S., Silvia B., Abou-Seri S. M. Eur. J. Med. Chem. 2018;157:28–36. doi: 10.1016/j.ejmech.2018.07.054. [DOI] [PubMed] [Google Scholar]
- Kumar M., Ramasamy K., Mani V., Mishra R. K., Majeed A. B. A., Clercq E. D. Arabian J. Chem. 2014;7:396–408. doi: 10.1016/j.arabjc.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammazzalorso A., Carrieri A., Verginelli F., Bruno I., Carbonara G., D'Angelo A. Eur. J. Med. Chem. 2016;114:191–200. doi: 10.1016/j.ejmech.2016.02.064. [DOI] [PubMed] [Google Scholar]
- Draczkowski P., Tomaszuk A., Halczuk P., Strzemski M., Matosiuk D., Jozwiak K. Biochim. Biophys. Acta. 2016;1860:967–974. doi: 10.1016/j.bbagen.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Zhang Y. J., Wang W. M., Oelschlaeger P., Chen C., Lei J. E., Lv M. ACS Infect. Dis. 2018;4:1671–1678. doi: 10.1021/acsinfecdis.8b00147. [DOI] [PubMed] [Google Scholar]
- Segal I. H., Enzyme Kinetics, Wiley-Interscience, New York, 1993, ISBN: 0-471-30309-7. [Google Scholar]
- Cockerill F. R., Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: Approved standard, Clinical and Laboratory Standards Institute, 2000. [Google Scholar]
- Shu J. L., O'Briensimpson N. M., Pantarat N., Sulistio A., Wong E. H. H., Chen Y. Y. Nat. Microbiol. 2016;1:16162. doi: 10.1038/nmicrobiol.2016.162. [DOI] [PubMed] [Google Scholar]
- Rokney A., Shagan M., Kessel M., Smith Y., Rosenshine I., Oppenheim A. B. J. Mol. Biol. 2009;392:589–601. doi: 10.1016/j.jmb.2009.07.009. [DOI] [PubMed] [Google Scholar]
- King A. M., Reid-Yu S. A., Wenliang W., King D. T., Gianfranco D. P., Strynadka N. C. Nature. 2014;510:503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Li A., Guan L., Zhang X., Wang L. Y. Dyes Pigm. 2016;129:163–173. [Google Scholar]
- Yang S. K., Kang J. S., Oelschlaeger P., Yang K. W. ACS Med. Chem. Lett. 2015;6:455–460. doi: 10.1021/ml500534c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Aitha M., Hetrick A. M., Richmond T. K., Tierney D. L., Crowder M. W. Biochemistry. 2012;51:3839–3847. doi: 10.1021/bi300056y. [DOI] [PubMed] [Google Scholar]
- Crawford P. A., Sharma N., Chandrasekar S., Sigdel T., Walsh T. R., Spencer J. Protein Expression Purif. 2004;36:272–279. doi: 10.1016/j.pep.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Crowder M. W., Walsh T. R., Banovic L., Pettit M., Spencer J. Antimicrob. Agents Chemother. 1998;42:921–926. doi: 10.1128/aac.42.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.