Abstract
Gallbladder cancer is the most common biliary tract malignant tumor, with unfavorable patient outcomes. The present study aimed to identify potential diagnostic or prognostic biomarkers for gallbladder cancer. To do so, differentially expressed genes in the gallbladder walls and tumor tissues of patients with gallbladder cancer were analyzed via microarray. Furthermore, a protein-protein interaction network was constructed and genes with a degree score >10 were selected as hub genes. As ubiquitin conjugating enzyme E2T (UBE2T) was considered to be a hub gene, its expression was assessed via reverse transcription-quantitative (RT-q)PCR and immunohistochemistry (IHC). In addition, the association between UBE2T expression and the clinicopathological characteristics of patients with gallbladder cancer was analyzed using the χ2 test. Furthermore, all patients were divided into high- and low groups based on UBE2T expression level and overall survival analysis was performed. Univariate and multivariate Cox regression analyses were performed to determine whether UBE2T may serve as an independent risk factor for gallbladder cancer. The results demonstrated that UBE2T expression was upregulated in the gallbladder walls and tumor tissues of patients with gallbladder cancer. Furthermore, UBE2T expression level was confirmed to be upregulated following RT-qPCR, and results from IHC demonstrated that UBE2T was predominantly expressed in the cytoplasm of gallbladder cancer cells. In addition, high UBE2T expression level was associated with clinical stage, T classification, N classification and M classification. The results from Univariate and multivariate analyses indicated that UBE2T expression level may be considered as an independent risk factor for gallbladder cancer. Taken together, the findings from this study suggested that high UBE2T expression level may contribute to the poor prognosis of patients with gallbladder cancer, and that UBE2T may act as an independent prognostic biomarker for these patients.
Keywords: ubiquitin conjugating enzyme E2T, gallbladder cancer, prognosis, hub genes, microarray analysis
Introduction
Gallbladder cancer is the most common malignancy of the biliary tract, with 219,420 new cases and 165,087 mortalities cases reported in 2018 worldwide (1,2). At present, gallbladder resection is the principle treatment option for patients with gallbladder cancer (3). However, recurrence is common following complete resection. Furthermore, the recurrence risk is increased when excision surgery happens in the advanced stage (4,5). It is therefore essential for patients with gallbladder cancer to be diagnosed in the earlier stages of the disease (6). However, since patients with gallbladder cancer have no apparent symptoms in the early stage, they often miss the optimal treatment opportunity (7). The determination of potential biomarkers would therefore provide screening opportunities for patients at high risk of gallbladder cancer.
In order to develop an optimal treatment strategy that would improve the overall outcome of patients with gallbladder cancer, it is crucial to better understand the prognostic risk factors. This strategy may help identifying patients receiving preventive cholecystectomy who may be at risk of developing gallbladder cancer, and assisting oncologists to develop individualized treatment (8).
With the rapid development of microarray and RNA sequencing technology, a high amount of differentially expressed genes (DEGs) have been identified in cancer tissues compared with non-tumor tissues (9,10). A previous study identified 758 long non-coding RNAs (lncRNAs) and 1,254 mRNAs in gallbladder cancer tissues compared with adjacent normal tissues (11). However, clinical trials still lack precise biomarkers for gallbladder cancer (12). The present study identified DEGs in both gallbladder walls and tumor tissues of gallbladder cancer via microarray analysis. Microarray analysis has been extensively used to screen biomarkers of gallbladder cancer. For example, a previous study demonstrated through microarray analysis that lncRNA GCASPC can negatively regulate pyruvate carboxylase-dependent cell proliferation in gallbladder cancer (13). Furthermore, microarray analysis confirmed that CD44 overexpression is associated with the progression of gallbladder cancer, confirming its contribution to poor patient prognosis (14). However, there is little research on gene expression changes in gallbladder walls of patients with gallbladder cancer (15). Kyoto Encyclopedia of Genes and Genomics (KEGG) has been extensively used to better understand the functions of numerous biological systems, including cells (such as liver, gastric and gallbladder), organisms (such as heart, kidney and pancreas) and ecosystems (16,17). Among all hub genes identified, ubiquitin conjugating enzyme E2T (UBE2T) was firstly highlighted in a patient with Fanconi anemia (18). It has been reported that hypoxia may disrupt the Fanconi anemia pathway and sensitize tumor cells to chemotherapy by regulating UBE2T (19,20). Furthermore, UBE2T overexpression is associated with poor prognosis of patients with various types of cancer, including breast cancer and multiple myeloma (21,22). In addition, UBE2T is upregulated in hepatocellular carcinoma and exerts oncogenic activities through p53 ubiquitination (23). A previous study demonstrated that UBE2T is present in the nuclei and cytoplasm of bladder cancer cells, furthermore, silencing UBE2T may induce cell cycle arrest at the G2/M phase and may promote cell apoptosis (24). Furthermore, high UBE2T expression has been reported to promote osteosarcoma cell proliferation, migration and invasion via activation of the PI3K/Akt signaling pathway (25). However, the role of UBE2T in the development of gallbladder cancer remains unknown. The present study aimed therefore to investigate the expression and clinical characteristics of UBE2T in patients with gallbladder cancer.
Materials and methods
Patients and specimens for microarray analysis
The present study was approved by the Ethics Committee of the First Affiliated Hospital of China Medical University and written informed consent was provided by all patients prior to the study. A total of three patients with gallbladder cancer (2 men and 1 woman; mean age, 61.00) and three patients with gallbladder adenoma (2 men and 1 woman; mean age, 60.67) as the control groups were recruited from the Department of Pancreatic and Biliary Surgery of the First Affiliated Hospital of China Medical University (Shenyang, China) between September 2018 and December 2019. All patients were blindly reviewed by two independent pathologists at the First Affiliated Hospital of China Medical University. None of the patients had received radiotherapy or chemotherapy prior to gallbladder resection. Gallbladder cancer specimens and wall tissues were obtained from the same patients via biopsy. The control specimens were collected from adenoma tissues and gallbladder wall tissues from patients with gallbladder adenoma by biopsy. Following collection, fresh specimens were instantly frozen in liquid nitrogen within 15 min and subsequently stored in RNA Fixer Reagent (Thermo Fisher Scientific, Inc.) at −80°C for microarray analysis. The clinical characteristics of patients with gallbladder cancer are presented in Table I.
Table I.
Clinical characteristics of patients with gallbladder cancer (n=3).
| Characteristic | Patient, n |
|---|---|
| Age, years | |
| ≤60 | 1 |
| >60 | 2 |
| Sex | |
| Male | 2 |
| Female | 1 |
| Cholecystolithiasis | |
| Absent | 3 |
| Present | 0 |
| Diabetes | |
| Absent | 0 |
| Present | 3 |
| Jaundice | |
| Absent | 3 |
| Present | 0 |
| Pathological types | |
| Adenocarcinoma | 3 |
| Adenosquamous carcinoma | 0 |
| Papillocarcinoma | 0 |
| Degree of differentiation | |
| Poor | 3 |
| Moderate-well | 0 |
| Resection margin status | |
| Positive | 2 |
| Negative | 1 |
| T stage | |
| Tis-T1a | 0 |
| T1b-T2b | 0 |
| T3 | 2 |
| T4 | 1 |
| N stage | |
| N0 | 0 |
| N1 | 1 |
| N2 | 2 |
| Distant metastasis | |
| Absent | 2 |
| Present | 1 |
RNA isolation, quantification and quality control
Total RNA was extracted from gallbladder cancer and adjacent normal tissues using standard methods (RNA Easy; Thermo Fisher Scientific, Inc.). RNA purity and concentration was detected using NanoDrop ND-2000 Spectrophotometer (Thermo Fisher Scientific, Inc.). RNA integrity was determined using Agilent Bioanalyzer 2100 system (Agilent Technologies GmbH).
RNA extraction and microarray hybridization
Total RNA from tissue samples was analyzed on the Agilent Bioanalyzer 2100 system (Agilent Technologies GmbH) and reverse transcribed into cDNA using the PrimeScript™ RT Reagent kit (cat. no. RR037A; Takara Biotechnology Co., Ltd), according to the manufacturers' instructions. Briefly, cDNA was synthesized via first-strand synthesis, and a double-stranded DNA template was subsequently obtained via second-strand synthesis. cDNA was purified with purification beads (Beckman Coulter, Inc.) and fragmented, prior to hybridization with the chip probe. Following hybridization, the chip was automatically washed and stained (GeneChip Hybridization Wash and Stain Kit; Affymetrix) using the GeneChip Fluidics Station 450 instrument, prior to scanning to obtain the image and the Affymetrix original microarray data.
Differential expression analysis
Microarray data were extracted using Feature Extraction software [version 10.5.1.1] (26), whereas GeneSpring software (version 12.0; Agilent Technologies, Inc.) was used to normalize the quantiles of the raw data. DEGs with fold-change (|FC|)>1.5 and P<0.05 were selected between gallbladder cancer wall tissues and gallbladder adenoma wall tissues, and DEGs with FC>2.0 and P<0.05 were selected between gallbladder cancer tissues and gallbladder adenoma tissues. The DEGs from the two microarray datasets were overlapped using Bioinformatics & Evolutionary Genomics (version 1.0; http://bioinformatics.psb.ugent.be/webtools/Venn). Different FC values were implemented as the two microarray datasets contained gallbladder tumor walls vs. gallbladder adenoma walls and gallbladder tumor tissues vs. gallbladder adenoma tissues, respectively, thus, the gene expression patterns of the two datasets varied.
Protein-protein interaction (PPI) network construction and module analysis
A PPI network was constructed using the Search Tool for Retrieval of Interacting Genes (STRING) database (http://string-db.org), which provides integrated information of the known and predicted associations for protein networks (27). An interaction with a combined score >0.4 was considered to be statistically significant. Cytoscape software version 3.7.2 (28) was used to visualize the PPI network. The most significant module in the PPI network was selected by using the Molecular Complex Detection (MCODE) plug-in (29), within Cytoscape. The criteria for selection were as follows: MCODE scores >5, degree cut-off=2, node score cut-off=0.2, Max depth=100 and k-score=2.
Identification of hub genes
The genes in the key module were considered to be hub genes. Hierarchical clustering analysis of hub genes in the key module was performed by using GraphPad Prism software (version 7.0; GraphPad Software, Inc.). A co-expression network of hub genes was constructed using the cBioPortal online platform (http://www.cbioportal.org). Biological analysis of the hub genes was performed and visualized using the Biological Network Gene Ontology tool (BiNGO) plug-in (30), within Cytoscape.
Functional enrichment analysis
KEGG and Gene Oncology (GO) enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (31), in order to determine the potential biological processes and signaling pathways the DEGs were involved in. Adjusted P<0.05 was considered to indicate significantly enriched processes or signaling pathways.
Reverse transcription-quantitative (RT-q) PCR
Total RNA was extracted from 30 paired gallbladder cancer tissues and corresponding adjacent non-cancerous tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Total RNA was reverse transcribed into cDNA using the PrimeScript RT Reagent kit (RR037A, Takara), according to the manufacturers' protocol. qPCR was subsequently performed using the SYBR Green PCR kit (cat. no. A25742; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for qPCR: One cycle for 2 min at 50°C (initial denaturation); one cycle for 10 min at 95°C (denaturation); 40 cycles for 15 sec at 95°C (annealing and elongation) and 40 cycles for 1 min at 60°C (final extension). The sequences of the primers used were as follows: UBE2T, forward 5′-CAAATATTAGGTGGAGCCAACAC-3′, reverse 5′-TAGATCACCTTGGCAAAGAACC-3′; β-actin, forward 5′-AGAAAATCTGGCACCACACC-3′ and reverse 5′-TAGCACAGCCTGGATAGCAA-3′. Relative expression level was normalized to the endogenous control β-actin and was expressed as 2−ΔΔCq (32).
Western blotting
Total protein was extracted from 12 paired gallbladder cancer tissues and corresponding adjacent normal tissues using RIPA lysis buffer on ice (Nanjing KeyGen Biotech Co., Ltd.). Total protein was quantified using the bicinchoninic acid assay kit (cat. no. P0009; Beyotime Institute of Biotechnology) and 12 µg protein/lane was separated via SDS-PAGE on a 10% gel, and subsequently transferred onto a polyvinylidene difluoride membrane (EMD Millipore). Membranes were blocked with 5% skim milk at room temperature for 1 h. Subsequently, membranes were incubated with primary antibodies against UBE2T (1:500; cat. no. 12992; Cell Signaling Technology, Inc.) and β-actin (1:500; cat. no. 4970; Cell Signaling Technology, Inc.) overnight at 4°C, followed by incubation with the HRP-labeled secondary antibody (1:5,000; cat. no. SA00001-2; ProteinTech Group, Inc.) for 45 min at room temperature. Enhanced chemiluminescence reagent (Thermo Fisher Scientific, Inc.) was used to detect the signal on the membrane.
Immunohistochemistry
A total of 127 cases of formalin-fixed and paraffin-embedded gallbladder cancer tissues from patients with gallbladder cancer [40 men (31.5%) and 87 women (68.5%)] were obtained from the Department of Pancreatic and Biliary Surgery of the First Affiliated Hospital of China Medical University (Shenyang, China). All patients provided written informed consent. Clinicopathological classification and AJCC TNM classification of the samples were determined according to the criteria from the American Joint Committee on Cancer (33). Tissues were fixed with 4% paraformaldehyde overnight at room temperature. Then, tissue sections were deparaffinized and blocked with 1% bovine serum albumin (Hyclone, Inc.) in PBS at room temperature for 1 h, prior to quenching to inhibit endogenous peroxidase activity. The sections were at 20 µm thickness. Tissue sections were subsequently incubated with the rabbit anti-human-UBE2T antibody (1:500; cat. no. 12992; Cell Signaling Technology, Inc.) at 4°C overnight, followed by incubation with secondary antibody (1:5,000; cat. no. PV-9001; OriGene Technologies, Inc.) for 2 h at room temperature. Subsequently, sections were counterstained with ChemMate Hematoxylin (Dako; Agilent Technologies, Inc.). Sections were subsequently mounted using neutral gum and observed under an optical microscope (Olympus Corporation; magnification, ×200). Sections were blindly assessed by two independent pathologists at the First Affiliated Hospital of China Medical University (Shenyang, China) and were semi-quantitatively scored according to the percentage of positive cells as follows: 1, 2, 3 or 4 for 0–25, 26–50, 51–75 or 76–100% of positively stained cells, respectively. Furthermore, the sections were stained with Harris hematoxylin for 5 min and Eosin for 2 min at room temperature and scored according to the cell staining intensity as follows: 0, 1, 2 or 3 for absence for color, pale yellow color, yellow/brown color and brown color, respectively. The two scores were multiplied to provide the following composite scores: low expression (score <6) and high expression (score ≥6).
Statistical analysis
Statistical analysis was performed using SPSS software (version 22.0; IBM Corp.) and GraphPad Prism software. χ2 test was used to determine the correlation between UBE2T expression and the clinicopathological characteristics of patients with gallbladder cancer. Survival analysis was performed using Kaplan-Meier method (34) and two groups were compared using the log-rank test. Univariate and multivariate Cox regression analyses were performed. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of hub genes in tumor tissues and gallbladder walls of patients with gallbladder cancer
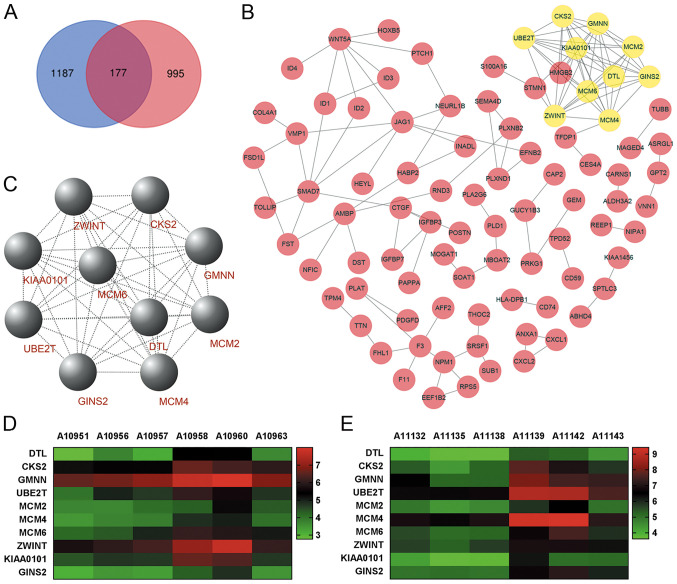
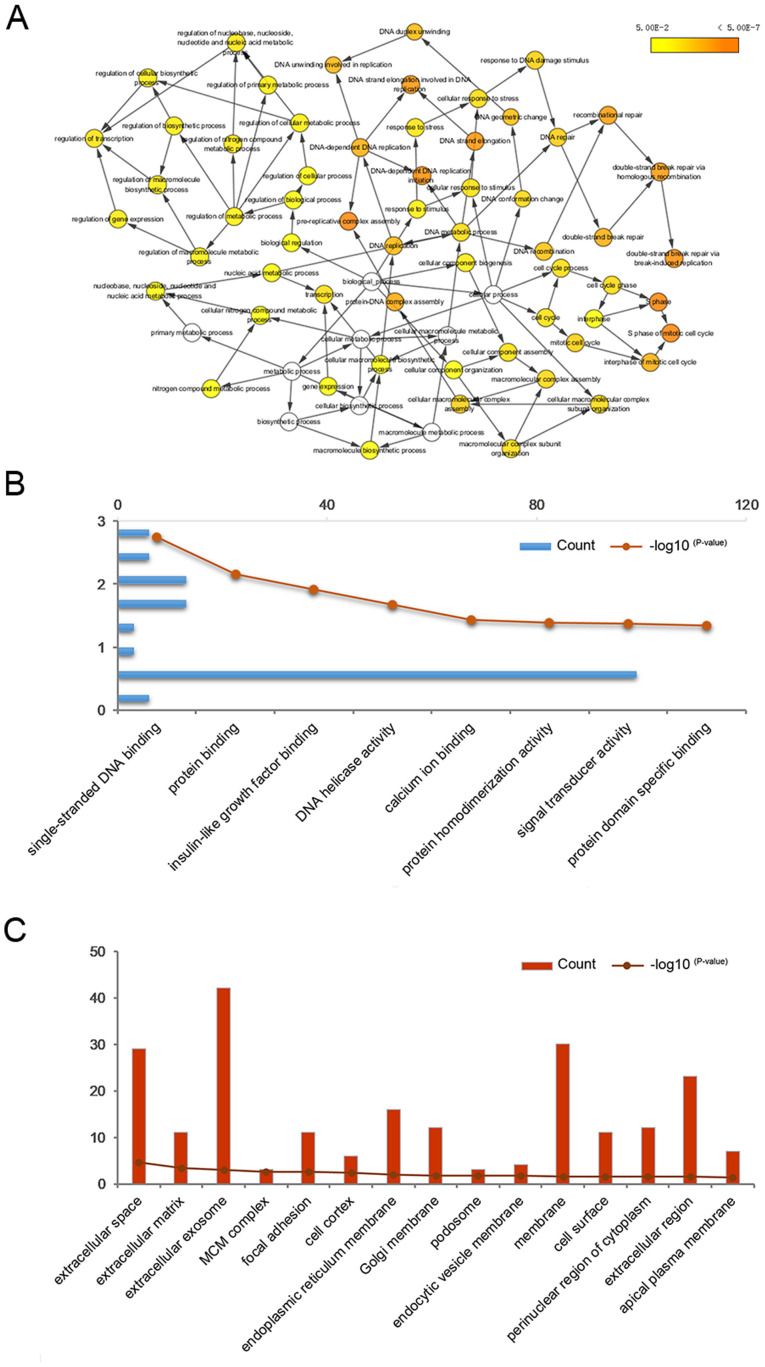
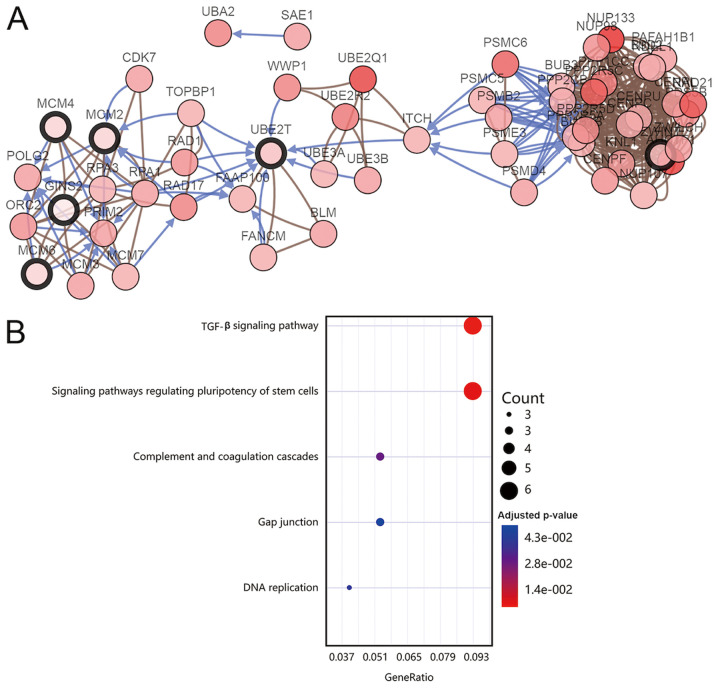
The work flowchart is presented in Fig. 1. Differential expression analysis was performed for two respective microarray datasets. First, DEGs with |FC|>1.5 and P<0.05 were selected by comparing three gallbladder cancer walls with three gallbladder adenoma walls. Furthermore, DEGs with |FC|>2 and P<0.05 were selected by comparing three gallbladder cancer tissues with three gallbladder adenoma tissues. The two datasets were overlapped to identify DEGs in both tumor tissues and gallbladder walls of patients with gallbladder cancer, resulting in the identification of 177 DEGs (Fig. 2A). Functional enrichment analyses were performed to determine the biological processes and signaling pathways enriched by the DEGs. A network of biological processes associated with the DEGs was constructed by using BiNGO (Fig. 3A). Furthermore, results from GO analysis included molecular function (Fig. 3B), cell component (Fig. 3C) and biological processes (Table II). The results from KEGG pathway analysis demonstrated that the DEGs were mainly enriched in the ‘TGF-β signaling pathway‘, ‘signaling pathways regulating pluripotency of stem cells’ and ‘complement and coagulation cascades’ (Fig. 4B). Subsequently, a PPI network of the 177 DEGs was constructed (Fig. 2B) and the key modules were selected using the MCODE plug-in within Cytoscape (Fig. 2C). A total of 10 nodes were identified as hub genes (Table III). Hierarchical clustering analysis was performed to determine the expression pattern of the 10 hub genes in the two microarray datasets (Fig. 2D and E). The co-expression network of the 10 hub genes was constructed using cBioPortal (Fig. 4A).
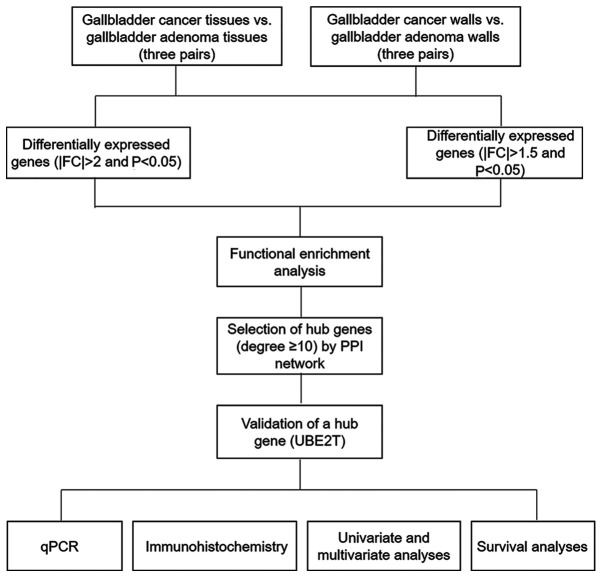
Figure 1.
Work flowchart of the present study. FC, fold-change; PPI, protein-protein interaction; UBE2T, ubiquitin conjugating enzyme E2T; qPCR, quantitative polymerase chain reaction.
Figure 2.
PPI network and the key modules of DEGs. (A) Intersection between three pairs of DEGs with FC>1.5 and P<0.05 (gallbladder tumor walls vs. gallbladder adenoma walls), and three pairs of DEGs with |FC|>2 and P<0.05 (gallbladder tumor tissues vs. gallbladder adenoma tissues). For microarray analysis, gallbladder adenoma was used as the control. Blue represents DEGs in gallbladder tumor walls and pink represents DEGs in gallbladder adenoma walls. (B) PPI network of 177 DEGs was constructed using Cytoscape software. In the PPI network, each node represents a protein and each edge represents a PPI. (C) DEGs with degrees ≥10 were selected from the PPI network as the hub genes for the key modules. Hierarchical clustering analysis of the hub genes in the datasets, (D) three pairs of gallbladder tumor walls vs. gallbladder adenoma walls and (E) three pairs of gallbladder tumor tissues vs. gallbladder adenoma tissues. A10951, A10956 and A10957; gallbladder adenoma wall samples; A10958, A10960 and A10963, gallbladder tumor wall samples; A11132, A11135 and A11138; gallbladder adenoma tissue samples; A11139, A11142 and A11143, gallbladder tumor tissue samples. PPI, protein-protein interaction; DEG, differentially expressed gene; FC, fold-change.
Figure 3.
Biological processes and GO analysis of the hub genes. (A) A network of biological processes associated with the hub genes was constructed using the Biological Network Gene Ontology tool. The node color is associated with the adjusted p-value. P<0.01 was considered to indicate significantly enriched processes. GO analysis, including (B) molecular function and (C) cell component was performed using the Database for Annotation, Visualization and Integrated Discovery. GO, Gene Ontology.
Table II.
Biological processes associated with the 177 differentially expressed genes.
| Term | Count | P-value |
|---|---|---|
| Negative regulation of osteoblast differentiation | 5 | 4.580×10−4 |
| Cellular response to transforming growth factor beta stimulus | 5 | 1.097×10−3 |
| Negative regulation of sequence-specific DNA binding transcription factor activity | 5 | 2.333×10−3 |
| DNA replication | 7 | 3.76×10−3 |
| DNA unwinding involved in DNA replication | 3 | 3.675×10−3 |
| Positive regulation of cell proliferation | 12 | 4.225×10−3 |
| Regulation of cell growth | 5 | 6.561×10−3 |
| Negative regulation of fat cell differentiation | 4 | 6.926×10−3 |
| Negative regulation of transcription, DNA-templated | 12 | 6.973×10−3 |
| Brain development | 7 | 8.699×10−3 |
| Organ morphogenesis | 5 | 1.064×10−2 |
| Cell adhesion | 11 | 1.072×10−2 |
| Positive regulation of fibroblast proliferation | 4 | 1.380×10−2 |
| Negative regulation of neuron differentiation | 4 | 1.522×10−2 |
| Negative regulation of transcription from RNA polymerase II promoter | 14 | 1.685×10−2 |
| Positive regulation of neutrophil chemotaxis | 3 | 1.754×10−2 |
| Positive regulation of T cell proliferation | 4 | 1.829×10−2 |
| Response to wounding | 4 | 2.081×10−2 |
| Cell proliferation | 9 | 2.116×10−2 |
| Positive regulation of endothelial cell proliferation | 4 | 2.638×10−2 |
| Negative regulation of apoptotic process | 10 | 2.641×10−2 |
| Epithelial cell differentiation | 4 | 2.737×10−2 |
| Positive regulation of cell migration | 6 | 2.873×10−2 |
| Ventricular septum morphogenesis | 3 | 2.955×10−2 |
| Actin cytoskeleton organization | 5 | 3.310×10−2 |
| Regulation of angiogenesis | 3 | 3.344×10−2 |
| DNA replication initiation | 3 | 3.546×10−2 |
| Diacylglycerol biosynthetic process | 2 | 3.665×10−2 |
| Semaphorin-plexin signaling pathway | 3 | 3.752×10−2 |
| Positive regulation of catalytic activity | 4 | 3.966×10−2 |
| Signal transduction | 18 | 4.163×10−2 |
| Proteolysis | 10 | 4.397×10−2 |
| Lipid catabolic process | 4 | 4.471×10−2 |
Figure 4.
Co-expression network and KEGG pathway analysis of the hub genes. (A) A co-expression network was constructed using cBioPortal. Nodes with bold black outline represent the hub genes, while nodes with thin black outline represent the co-expressed genes. (B) KEGG pathway analysis of the hub genes was performed using the Database for Annotation, Visualization and Integrated Discovery. KEGG, Kyoto Encyclopedia of Gene and Genomes.
Table III.
Ten hub genes in tumor tissues and gallbladder walls of gallbladder cancer.
| Gene symbol | Full name |
|---|---|
| DTL | Denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| CKS2 | CDC28 protein kinase regulatory subunit 2 |
| GMNN | Geminin, DNA replication inhibitor |
| UBE2T | Ubiquitin conjugating enzyme E2T |
| MCM2 | Minichromosome maintenance complex component 2 |
| MCM4 | Minichromosome maintenance complex component 4 |
| MCM6 | Minichromosome maintenance complex component 6 |
| ZWINT | ZW10 interacting kinetochore protein |
| KIAA0101 | KIAA0101 |
| GINS2 | GINS complex subunit 2 (Psf2 homolog) |
UBE2T is upregulated in gallbladder cancer tissues
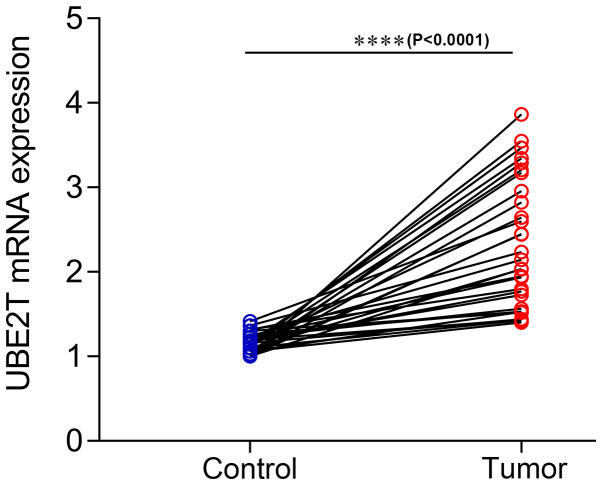
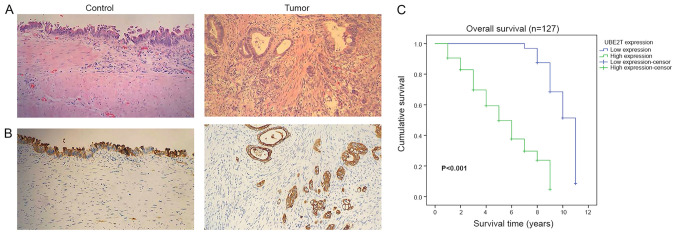
Functional enrichment analysis indicated that UBE2T is involved in several biological processes and pathways that are closely associated with gallbladder cancer, such as the TGF-β signaling pathway (35). Subsequently, UBE2T was selected for further analysis in the present study, and UBE2T expression in gallbladder cancer tissues was assessed. The results from RT-qPCR on 30 paired gallbladder cancer tissues and corresponding adjacent noncancerous tissues demonstrated that UBE2T was upregulated in gallbladder cancer tissues compared with adjacent noncancerous tissues (P<0.0001; Fig. 5). Furthermore, the results from western blotting demonstrated that UBE2T was highly expressed in 12 paired gallbladder cancer tissues compared with corresponding adjacent noncancerous tissues at the protein level (Fig. 6). The results from IHC indicated that UBE2T was mostly expressed in the cytoplasm (Fig. 7A and B). As presented in Table IV, among the 127 gallbladder cancer cases, UBE2T was downregulated in 32 cases (25.2%) and upregulated in 95 cases (74.8%). Taken together, these results demonstrated that UBE2T expression was upregulated in gallbladder cancer.
Figure 5.
Reverse transcription-quantitative PCR analysis of UBE2T expression in 30 paired gallbladder cancer tissues and adjacent noncancerous tissues. β-actin was used as the internal control. ****P<0.0001. UBE2T, ubiquitin conjugating enzyme E2T.
Figure 6.
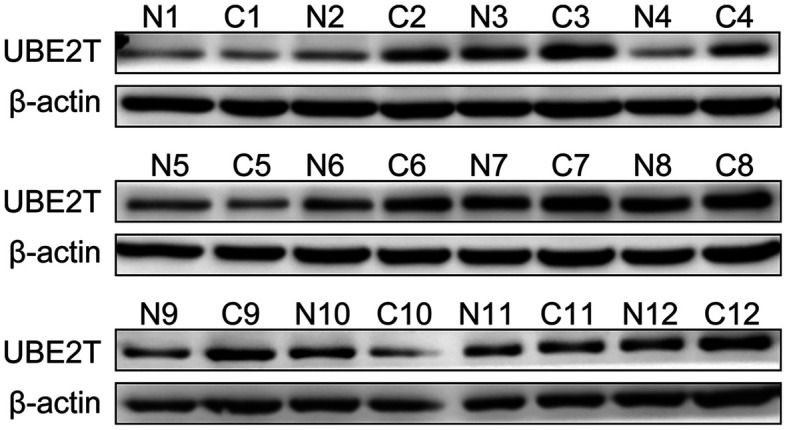
Western blot analysis of UBE2T expression in 12 pairs of gallbladder cancer tissues and adjacent noncancerous tissues. β-actin was used as the internal control. UBE2T, ubiquitin conjugating enzyme E2T; N, normal tissues; C, cancer tissues.
Figure 7.
UBE2T was upregulated and associated with poor prognosis in patients with gallbladder cancer. (A) Representative hematoxylin and eosin staining images and (B) immunohistochemistry analysis of UBE2T expression in gallbladder cancer tissues and adjacent noncancerous tissues (magnification, ×200). (C) Kaplan-Meier overall survival curves for 127 patients with gallbladder cancer, stratified by high and low UBE2T expression levels. UBE2T, ubiquitin conjugating enzyme E2T.
Table IV.
Association between UBE2T expression and the clinicopathological characteristics of patients with gallbladder cancer (n=127).
| UBE2T expression | ||||
|---|---|---|---|---|
| Characteristic | Cases, n (%) | Low (n=32) | High (n=95) | P-value |
| Age, years | ||||
| <50 | 4 (3.1) | 0 | 4 | 0.238 |
| ≥50 | 123 (96.9) | 32 | 91 | |
| Sex | ||||
| Male | 40 (31.5) | 9 | 31 | 0.635 |
| Female | 87 (68.5) | 23 | 64 | |
| Clinical stage | ||||
| I | 2 (1.6) | 2 | 0 | <0.001b |
| II | 87 (68.5) | 30 | 57 | |
| III | 23 (18.1) | 0 | 23 | |
| V | 15 (11.8) | 0 | 15 | |
| T classification | ||||
| T1 | 2 (1.6) | 2 | 0 | <0.001b |
| T2 | 88 (69.3) | 30 | 58 | |
| T3 | 32 (25.2) | 0 | 32 | |
| T4 | 5 (3.9) | 0 | 5 | |
| N classification | ||||
| N0 | 91 (71.7) | 32 | 59 | <0.001b |
| N1 | 28 (22.0) | 0 | 28 | |
| N2 | 8 (6.3) | 0 | 8 | |
| M classification | ||||
| M0 | 113 (89.0) | 32 | 81 | 0.021a |
| M1 | 14 (11.0) | 0 | 14 | |
| Histologic grade | ||||
| I | 24 (18.9) | 9 | 15 | 0.3 |
| II | 60 (47.2) | 13 | 47 | |
| III | 43 (33.9) | 10 | 33 | |
P<0.05
P<0.001. UBE2T, ubiquitin conjugating enzyme E2T.
High UBE2T expression is associated with poor prognosis of patients with gallbladder cancer
The results from Kaplan-Meier survival curves demonstrated that the overall survival time of patients with high UBE2T expression was significantly shorter than those with low UBE2T expression (P<0.001; Fig. 7C). The results indicated that upregulated UBE2T may be associated with poor prognosis of patients with gallbladder cancer.
UBE2T may serve as an independent risk factor of gallbladder cancer
The association between UBE2T expression and the clinicopathological characteristics of patients with gallbladder cancer was assessed (Table IV). The results demonstrated that upregulated UBE2T was significantly associated with clinical stage (P<0.001), T classification (P<0.001), N classification (P<0.001) and M classification (P=0.021). However, no significant association was observed between UBE2T expression and age (P=0.238), sex (P=0.635) and histologic grade (P=0.3). These results were consistent with Spearman's correlation analysis (Table V). Furthermore, whether UBE2T expression may be considered a a risk factor for gallbladder cancer was determined via univariate and multivariate Cox regression analyses. The results demonstrated that high UBE2T expression was significantly associated with an increased risk of gallbladder cancer (P<0.001; hazard ratio, 6.453; 95% confidence interval, 2.985–13.952; Table VI). Taken together, these results suggested that UBE2T expression may be considered as an independent risk factor for patients with gallbladder cancer.
Table V.
Spearman's correlation between UBE2T expression and the clinicopathological characteristics of patients with gallbladder cancer (n=127).
| UBE2T expression | ||
|---|---|---|
| Characteristic | Spearman's correlation | P-value |
| Age | −0.105 | 0.242 |
| Sex | −0.042 | 0.638 |
| Clinical stage | 0.402 | <0.001b |
| T classification | 0.398 | <0.001b |
| N classification | 0.362 | <0.001b |
| M classification | 0.204 | 0.021a |
| Grade | 0.09 | 0.316 |
P<0.05
P<0.001. UBE2T, ubiquitin conjugating enzyme E2T.
Table VI.
Univariate and multivariate analyses of prognostic parameters in patients with gallbladder cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Parameter | P-value | HR | 95% CI | P-value | HR | 95% CI |
| Clinical stage | <0.001a | 3.784 | 2.508–5.711 | 0.162 | 3.958 | 0.576–27.176 |
| T classification | <0.001a | 3.454 | 2.261–5.278 | 0.397 | 0.495 | 0.097–2.522 |
| N classification | <0.001a | 4.266 | 2.735–6.654 | 0.208 | 1.917 | 0.696–5.279 |
| M classification | <0.001a | 5.663 | 2.322–13.811 | 0.342 | 0.368 | 0.047–2.892 |
| UBE2T expression | <0.001a | 7.831 | 3.724–16.466 | <0.001a | 6.453 | 2.985–13.952 |
P<0.001. HR, hazard ratio; CI, confidence interval; UBE2T, ubiquitin conjugating enzyme E2T.
Discussion
Late diagnosis and poor prognosis are the main issues in the effective treatment of gallbladder cancer (36). Surgical resection remains the principle curable treatment option (37). However, >1/3 patients experience locoregional and distant recurrence following gallbladder cancer resection (5). It is therefore crucial to determine potential diagnostic or prognostic biomarkers.
Microarray technology is commonly used to assess gene expression changes in gallbladder cancer, which was proven useful in identifying novel biomarkers (38). In the present study, 177 genes were differentially expressed in gallbladder wall tissues and tumor tissues of patients with gallbladder cancer. To the best of our knowledge, not much is known about DEGs in gallbladder cancer walls. Unlike other studies, the present study used microarray analysis to identify DEGs for gallbladder cancer and gallbladder adenoma, whereas a precancerous lesion of gallbladder cancer was used as the control (35,39–41).
To the best of our knowledge, there is currently no research on the PPIs of gallbladder cancer. Subsequently, a PPI network of 177 DEGs was constructed in the present study by using the STRING database. A total of 10 genes with degree ≥10 were selected as the hub genes. The results from KEGG analysis demonstrated that the 10 hub genes were enriched in the following pathways: ‘TGF-β signaling pathway’, ‘signaling pathways regulating pluripotency of stem cells’ and ‘complement and coagulation cascades’. These pathways have been associated with the development and progression of gallbladder cancer (42–44). For example, the crosstalk between the TGF-β signaling pathway and lncRNAs plays a key role in cancer. It has been reported that several members of the TGF-β signaling pathway are targeted by certain lncRNAs (such as HOXD-AS1 and UCA1), and that the production of numerous lncRNAs is induced by TGF-β treatment in different types of cancer, such as gastric cancer (45). Furthermore, focal adhesion (focal adhesion proteins such as, vinculin, talin, zyxin, FAK, and paxillin) activation mediates cell migration and metastasis in different types of cancer, including gallbladder cancer (46). In the present study, hub genes were significantly enriched in stem cell-related pathways. As a critical characteristic of cancer stem cells, pluripotency contributes to self-renewal and chemoresistance (47). Therefore, these 10 hub genes may be involved in the development of gallbladder cancer.
The present study demonstrated that UBE2T was upregulated in gallbladder tumor and walls of patients with gallbladder cancer. The results from RT-qPCR and western blotting confirmed that UBE2T was upregulated in 30 pairs of gallbladder cancer tissues compared with adjacent normal tissues. Furthermore, results from IHC demonstrated that UBE2T was mainly expressed in the cytoplasm of gallbladder cancer cells.
Previous studies demonstrated that UBE2T serves a key role in protein ubiquitination, which is an essential post-translational modification that regulates several biological processes, including inflammation, immune response, cell differentiation and cell proliferation (48–51). The drug vulnerability of cancer cells is dependent on protein participation in ubiquitination and degradation, which allowed the development of therapeutic agents based on druggable genomic modifications (52,53). Therefore, UBE2T may be considered as a potential drug target for patients with gallbladder cancer. Numerous studies demonstrated that UBE2T is overexpressed in various types of cancer, including multiple myeloma (20), hepatocellular carcinoma (54), bladder cancer (24) and osteosarcoma (25), suggesting that it may be an attractive drug target. The results from the present study indicated that UBE2T may be considered as a prognostic factor for patients with gallbladder cancer. Furthermore, the Kaplan-Meier survival curves analysis demonstrated that high UBE2T expression was associated with worse prognosis compared with low UBE2T expression group. T stage independently affects prognosis of gallbladder cancer (55,56). The present study demonstrated that UBE2T overexpression was associated with certain clinicopathological characteristics of patients with gallbladder cancer, including clinical stage, T classification, N classification and M classification. In addition, results from univariate and multivariate Cox regression analyses indicated that high UBE2T expression may be considered as an independent risk factor for patients with gallbladder cancer. Taken together, the findings from the present study suggested that upregulated UBE2T expression may have a prognostic value for patients with gallbladder cancer.
In summary, the present study confirmed that UBE2T expression was upregulated in gallbladder tumor tissues and gallbladder walls of patients with gallbladder cancer compared with patients with gallbladder adenoma. Furthermore, high UBE2T expression was demonstrated to be associated with certain clinicopathological characteristics of patients with gallbladder cancer, including the AJCC TNM classification and clinical stage. Furthermore, high UBE2T expression was associated with poor patient prognosis. Univariate and multivariate Cox regression analyses indicated that UBE2T may serve as an independent prognostic biomarker for patients with gallbladder cancer. The present study is not without limitations. First, the patient population was heterogeneous. Thus, UBE2T should be validated in a larger cohort of patents with gallbladder cancer. Secondly, the present study only assessed the expression of UBE2T in gallbladder cancer tissues, thus lacking functional in vivo and in vitro studies on UBE2T. Prospective studies will therefore aim to further investigate the underlying mechanism of UBE2T in gallbladder cancer.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- PPI
protein-protein interaction
- RT-qPCR
reverse transcription-quantitative PCR
- KEGG
Kyoto Encyclopedia of Genes and Genomics
- GO
Gene Oncology
- FC
fold-change
- STRING
Search Tool for Retrieval of Interacting Genes
- MCODE
Molecular Complex Detection
- BiNGO
Biological Network Gene Ontology tool
- UBE2T
ubiquitin conjugating enzyme E2T
Funding
This work was funded by Natural Science Foundation of Liaoning Province (2020-BS-283).
Availability of data and materials
The datasets analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
CLG conceived and designed the present study. XZ, TL performed most of the experiments, analyzed the data and drafted the initial manuscript. XN, LJC acquired the data and helped draft the initial manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The First Affiliated Hospital of China Medical University and written informed consent was provided by all patients prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Ethun CG, Postlewait LM, Le N, Pawlik TM, Buettner S, Poultsides G, Tran T, Idrees K, Isom CA, Fields RC, et al. Association of optimal time interval to Re-resection for incidental gallbladder cancer with overall survival: A multi-institution analysis from the US extrahepatic biliary malignancy consortium. JAMA Surg. 2017;152:143–149. doi: 10.1001/jamasurg.2016.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez-Aguiar AG, Ethun CG, McInnis MR, Pawlik TM, Poultsides G, Tran T, Idrees K, Isom CA, Fields RC, Krasnick BA, et al. Association of perioperative transfusion with survival and recurrence after resection of gallbladder cancer: A 10-institution study from the US extrahepatic biliary malignancy consortium. J Surg Oncol. 2018;117:1638–1647. doi: 10.1002/jso.25086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Margonis GA, Gani F, Buettner S, Amini N, Sasaki K, Andreatos N, Ethun CG, Poultsides G, Tran T, Idrees K, et al. Rates and patterns of recurrence after curative intent resection for gallbladder cancer: A multi-institution analysis from the US extra-hepatic biliary malignancy consortium. HPB (Oxford) 2016;18:872–878. doi: 10.1016/j.hpb.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hundal R, Shaffer EA. Gallbladder cancer: Epidemiology and outcome. Clin Epidemiol. 2014;6:99–109. doi: 10.2147/CLEP.S37357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baiu I, Visser B. Gallbladder Cancer. JAMA. 2018;320:1294. doi: 10.1001/jama.2018.11815. [DOI] [PubMed] [Google Scholar]
- 8.Barreto SG, Dutt A, Chaudhary A. A genetic model for gallbladder carcinogenesis and its dissemination. Ann Oncol. 2014;25:1086–1097. doi: 10.1093/annonc/mdu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valle JW, Lamarca A, Goyal L, Barriuso J, Zhu AX. New horizons for precision medicine in biliary tract cancers. Cancer Discov. 2017;7:943–962. doi: 10.1158/2159-8290.CD-17-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renfro LA, An MW, Mandrekar SJ. Precision oncology: A new era of cancer clinical trials. Cancer Lett. 2017;387:121–126. doi: 10.1016/j.canlet.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Liu H, Shen X, Wang Y, Zhang D, Shen S, Suo T, Pan H, Ming Y, Ding K, Liu H. Long non-coding RNA expression profiles in gallbladder carcinoma identified using microarray analysis. Oncol Lett. 2017;13:3508–3516. doi: 10.3892/ol.2017.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sicklick JK, Fanta PT, Shimabukuro K, Kurzrock R. Genomics of gallbladder cancer: The case for biomarker-driven clinical trial design. Cancer Metastasis Rev. 2016;35:263–75. doi: 10.1007/s10555-016-9602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma MZ, Zhang Y, Weng MZ, Wang SH, Hu Y, Hou ZY, Qin YY, Gong W, Zhang YJ, Kong X, et al. Long Noncoding RNA GCASPC, a Target of miR-17-3p, negatively regulates pyruvate carboxylase-dependent cell proliferation in gallbladder cancer. Cancer Res. 2016;76:5361–5371. doi: 10.1158/0008-5472.CAN-15-3047. [DOI] [PubMed] [Google Scholar]
- 14.He Y, Xue C, Yu Y, Chen J, Chen X, Ren F, Ren Z, Cui G, Sun R. CD44 is overexpressed and correlated with tumor progression in gallbladder cancer. Cancer Manag Res. 2018;10:3857–3865. doi: 10.2147/CMAR.S175681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao X, Xu M, Cai Z, Yuan W, Cui W, Li MD. Identification of LIFR, PIK3R1, and MMP12 as novel prognostic signatures in gallbladder cancer using network-based module analysis. Front Oncol. 2019;9:325. doi: 10.3389/fonc.2019.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–96. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 19.Ramaekers CH, van den Beucken T, Meng A, Kassam S, Thoms J, Bristow RG, Wouters BG. Hypoxia disrupts the Fanconi anemia pathway and sensitizes cells to chemotherapy through regulation of UBE2T. Radiother Oncol. 2011;101:190–197. doi: 10.1016/j.radonc.2011.05.059. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Zhang Y, Yang Z, Liu X, Yang P, Wang J, Hu K, He X, Zhang X, Jing H. High expression of UBE2T predicts poor prognosis and survival in multiple myeloma. Cancer Gene Ther. 2019;26:347–355. doi: 10.1038/s41417-018-0070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mamrak NE, Shimamura A, Howlett NG. Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome Fanconi anemia. Blood Rev. 2017;31:93–99. doi: 10.1016/j.blre.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Peña J, Corrales-Sánchez V, Amir E, Pandiella A, Ocana A. Ubiquitin-conjugating enzyme E2T (UBE2T) and denticleless protein homolog (DTL) are linked to poor outcome in breast and lung cancers. Sci Rep. 2017;7:17530. doi: 10.1038/s41598-017-17836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu LP, Yang M, Peng QZ, Li MY, Zhang YS, Guo YH, Chen Y, Bao SY. UBE2T promotes hepatocellular carcinoma cell growth via ubiquitination of p53. Biochem Biophys Res Commun. 2017;493:20–27. doi: 10.1016/j.bbrc.2017.09.091. [DOI] [PubMed] [Google Scholar]
- 24.Gong YQ, Peng D, Ning XH, Yang XY, Li XS, Zhou LQ, Guo YL. UBE2T silencing suppresses proliferation and induces cell cycle arrest and apoptosis in bladder cancer cells. Oncol Lett. 2016;12:4485–4492. doi: 10.3892/ol.2016.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Leng H, Chen H, Wang L, Jiang N, Huo X, Yu B. Knockdown of UBE2T inhibits osteosarcoma cell proliferation, migration, and invasion by suppressing the PI3K/Akt signaling pathway. Oncol Res. 2016;24:361–369. doi: 10.3727/096504016X14685034103310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zahurak M, Parmigiani G, Yu W, Scharpf RB, Berman D, Schaeffer E, Shabbeer S, Cope L. Pre-processing Agilent microarray data. BMC Bioinformatics. 2007;8:142. doi: 10.1186/1471-2105-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41((Database issue)):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bandettini WP, Kellman P, Mancini C, Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY, Arai AE. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: A clinical validation study. J Cardiovasc Magn Reson. 2012;14:83. doi: 10.1186/1532-429X-14-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 31.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 34.Miller RG., Jr What price Kaplan-Meier? Biometrics. 1983;39:1077–1081. doi: 10.2307/2531341. [DOI] [PubMed] [Google Scholar]
- 35.Qiu Y, Luo X, Kan T, Zhang Y, Yu W, Wei Y, Shen N, Yi B, Jiang X. TGF-β upregulates miR-182 expression to promote gallbladder cancer metastasis by targeting CADM1. Mol Biosyst. 2014;10:679–685. doi: 10.1039/c3mb70479c. [DOI] [PubMed] [Google Scholar]
- 36.Sharma A, Sharma KL, Gupta A, Yadav A, Kumar A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: Recent update. World J Gastroenterol. 2017;23:3978–3998. doi: 10.3748/wjg.v23.i22.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ethun CG, Postlewait LM, Le N, Pawlik TM, Buettner S, Poultsides G, Tran T, Idrees K, Isom CA, Fields RC, et al. A novel pathology-based preoperative risk score to predict locoregional residual and distant disease and survival for incidental gallbladder cancer: A 10-institution study from the U.S. Extrahepatic biliary malignancy consortium. Ann Surg Oncol. 2017;24:1343–1350. doi: 10.1245/s10434-016-5637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen H, He M, Lin R, Zhan M, Xu S, Huang X, Xu C, Chen W, Yao Y, Mohan M, Wang J. PLEK2 promotes gallbladder cancer invasion and metastasis through EGFR/CCL2 pathway. J Exp Clin Cancer Res. 2019;38:247. doi: 10.1186/s13046-019-1250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin Y, Zhou Y, Ge A, Chang L, Shi H, Fu Y, Luo Q. Overexpression of SNORA21 suppresses tumorgenesis of gallbladder cancer in vitro and in vivo. Biomed Pharmacother. 2019;118:109266. doi: 10.1016/j.biopha.2019.109266. [DOI] [PubMed] [Google Scholar]
- 40.Yang L, Gao Q, Wu X, Feng F, Xu K. Long noncoding RNA HEGBC promotes tumorigenesis and metastasis of gallbladder cancer via forming a positive feedback loop with IL-11/STAT3 signaling pathway. J Exp Clin Cancer Res. 2018;37:186. doi: 10.1186/s13046-018-0847-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niu J, Li Z, Li F. Overexpressed microRNA-136 works as a cancer suppressor in gallbladder cancer through suppression of JNK signaling pathway via inhibition of MAP2K4. Am J Physiol Gastrointest Liver Physiol. 2019;317:G670–G681. doi: 10.1152/ajpgi.00055.2019. [DOI] [PubMed] [Google Scholar]
- 42.Manohar R, Li Y, Fohrer H, Guzik L, Stolz DB, Chandran UR, LaFramboise WA, Lagasse E. Identification of a candidate stem cell in human gallbladder. Stem Cell Res. 2015;14:258–269. doi: 10.1016/j.scr.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong L, Wu Q, Zhao L, Ye J, Li N, Yang H. Identification of messenger and long noncoding RNAs associated with gallbladder cancer via gene expression profile analysis. J Cell Biochem. 2019;120:19377–19387. doi: 10.1002/jcb.28953. [DOI] [PubMed] [Google Scholar]
- 44.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Shao N, Ding X, Tan B, Song Q, Wang N, Jia Y, Ling H, Cheng Y. Crosstalk between transforming growth factor-β signaling pathway and long non-coding RNAs in cancer. Cancer Lett. 2016;370:296–301. doi: 10.1016/j.canlet.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Banning A, Babuke T, Kurrle N, Meister M, Ruonala MO, Tikkanen R. Flotillins Regulate Focal Adhesions by Interacting with α-Actinin and by Influencing the Activation of Focal Adhesion Kinase. Cells. 2018;7:28. doi: 10.3390/cells7040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharif T, Martell E, Dai C, Kennedy BE, Murphy P, Clements DR, Kim Y, Lee PW, Gujar SA. Autophagic homeostasis is required for the pluripotency of cancer stem cells. Autophagy. 2017;13:264–284. doi: 10.1080/15548627.2016.1260808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang L, Guo W, Zhang S, Wang G. Ubiquitination-proteasome system: A new player in the pathogenesis of psoriasis and clinical implications. J Dermatol Sci. 2018;89:219–225. doi: 10.1016/j.jdermsci.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Qiu J, Sheedlo MJ, Yu K, Tan Y, Nakayasu ES, Das C, Liu X, Luo Z-Q. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature. 2016;533:120–124. doi: 10.1038/nature17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kao SH, Wu HT, Wu KJ. Ubiquitination by HUWE1 in tumorigenesis and beyond. J Biomed Sci. 2018;25:67. doi: 10.1186/s12929-018-0470-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alpi AF, Chaugule V, Walden H. Mechanism and disease association of E2-conjugating enzymes: Lessons from UBE2T and UBE2L3. Biochem J. 2016;473:3401–3419. doi: 10.1042/BCJ20160028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pagliarini R, Shao W, Sellers WR. Oncogene addiction: Pathways of therapeutic response, resistance, and road maps toward a cure. EMBO Rep. 2015;16:280–296. doi: 10.15252/embr.201439949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garraway LA, Verweij J, Ballman KV. Precision oncology: An overview. J Clin Oncol. 2013;31:1803–1805. doi: 10.1200/JCO.2013.49.4799. [DOI] [PubMed] [Google Scholar]
- 54.Tao Y, Li R, Shen C, Li J, Zhang Q, Ma Z, Wang F, Wang Z. SENP1 is a crucial promotor for hepatocellular carcinoma through deSUMOylation of UBE2T. Aging (Albany NY) 2020;12:1563–1576. doi: 10.18632/aging.102700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aloia TA, Jarufe N, Javle M, Maithel SK, Roa JC, Adsay V, Coimbra FJ, Jarnagin WR. Gallbladder cancer: Expert consensus statement. HPB (Oxford) 2015;17:681–690. doi: 10.1111/hpb.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goetze TO, Paolucci V. Adequate extent in radical re-resection of incidental gallbladder carcinoma: Analysis of the German Registry. Surg Endosc. 2010;24:2156–2164. doi: 10.1007/s00464-010-0914-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the present study are available from the corresponding author upon reasonable request.