Abstract
CircRNA (circular RNA) is a kind of closed circular structure of noncoding RNA molecules without 5′ hat structure and 3′ polyA, mainly located in the cytoplasm or stored in exosomes. It is not affected by RNA exonuclease, so it's stable and hard to be degraded. Proved to be widespread in a variety of eukaryotes, most circRNAs are cyclized by exons, some are lasso structures formed by intron cyclization. Recently, circRNAs have been demonstrated to play crucial roles in cardiomyocyte hypertrophy, fibrosis, autophagy and apoptosis, participating in the development of heart failure. There is increasing evidence that circRNAs may be a novel target for the treatment of heart failure.
Keywords: Circular RNA, Heart failure, Cardiomyocyte hypertrophy, Cardiomyocyte fibrosis, Apoptosis, Autophagy
With the advance of high-throughput sequencing technology and bioinformatics, more and more circRNAs have been discovered and become a new field of various diseases, such as cardiovascular disease (Wang et al., 2016; Du et al., 2017), cancers (Chen et al., 2017; Shang et al., 2016; Wan et al., 2016), neurological dysfunction (Lukiw, 2013), autoimmune diseases (Luo et al., 2018) (Li et al., 2018). Heart failure, originated in systolic and/or diastolic dysfunction of heart, leads to congestion in the venous system and low perfusion in the arterial system, and arises cardiac circulatory disorder syndrome (Warren and Grossman, 1991), it is the terminal stage of heart disease and the leading cause of death from various cardiovascular diseases (Rossignol et al., 2019). Although great progress has been made in studying the development of heart failure from molecular to cellular level, it is still a major cause of death in humans (Mudd and Kass, 2008). In recent years, increasing studies have demonstrated that circRNAs can interact with miRNA through “sponge effect” and play an important regulatory role in the development of heart failure (Wang et al., 2016; Qu et al., 2017; Fan et al., 2017), the purpose of this paper is to review the role of circRNAs in heart failure.
1. Circular RNA overview
1.1. Discovery and formation of circRNA
CircRNAs were firstly thought to be a plant-like virus, Thierry Candresse et al. found that some viroids could invade solanaceae plants and affect their growth (Diener, 1971). Such viroids were not covered by protein shells, and the genome was a small RNA molecule with single-stranded closure (Hsu and Coca-Prados, 1979). In 1979, Hsu et al. firstly observed a single-stranded covalent closed RNA molecule under an electron microscope (Sanger et al., 1976). In the following decades, with advance of high-throughput sequencing technology and bioinformatics, circRNAs were found in different organisms, including plants (Sablok et al., 2016), fish (Shen et al., 2017), and bacteria (Kjems and Garrett, 1988), etc. NIGRO et al. firstly confirmed the presence of endogenous circRNA in human body (Nigro et al., 1991). Since Hansen et al. found that cirs-7/CDR1 could inhibit the expression of mir-7 through “sponge” action in 2013 (Hansen et al., 2013), the published articles of circRNA had shown an exponential growth.
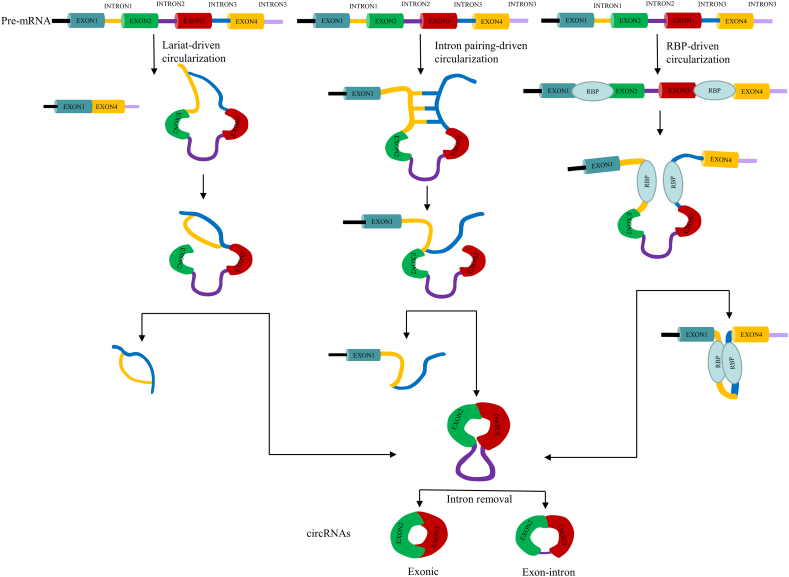
Circular RNA (circRNA) is a special kind of non-coding RNA. Different from the traditional linear RNA, circRNA are generated by backsplice, and there are two main ways of formation: exon cyclization and intron cyclization. Exon cyclization includes intron pair drive cyclization, lasso drive cyclization and RNA-binding.
protein (RBP)-driven circularization (Fig. 1), intron sequences on both sides of the former cyclization region are complementary in reverse, and the link formation between upstream shear receptor and downstream shear donor was promoted by efficient pairing. In the lasso drive cyclization model, exon jump mediates the connection between the upstream splice receptor and the downstream splice donor to form a lasso structure, and the introns are excised to form a circRNA composed of two exons (Jeck et al., 2013). In RBP-driven cyclization, the back-splicing event is guided by RBPs that recognize and dock on specific motifs located in the introns flanking the circularized exons (Aufiero et al., 2019). The formation of intron cyclization depends on the cyclization of the reverse complementary sequence at both ends of the intron, and its characteristic sequence includes the nucleotide GU enrichment element at the 5′end and the nucleotide C enrichment element at the adjacent branch point (Zhang et al., 2013).
Fig. 1.
Three mechanisms for circular RNA biogenesis. In the lasso drive cyclization model, exon jump mediates the connection between the upstream splice receptor and the downstream splice donor to form a lasso structure, and the introns are excised to form a circRNA composed of two exons. In intron pairing-driven cyclization model, intron sequences on both sides of the cyclization region are complementary in reverse, and the link formation between upstream shear receptor and downstream shear donor was promoted by efficient pairing. In RBP-driven cyclization, the back-splicing event is guided by RBPs that recognize and dock on specific motifs located in the introns flanking the circularized exons.
1.2. Characteristics of circRNAs
In recent decades, with the development of biotechnology and the in-depth study of circRNAs, their nature has been gradually displayed.
1.2.1. CircRNA is highly expressed in the different organs of organisms
Williamr. Jeck et al. conducted the high-throughput sequencing on ribosomal missing RNA libraries treated with or without RNA exonuclease, and identified at least 25,000 different RNAs from human fibroblasts containing non-collinear exons, then degraded and enriched them repeatedly by linear RNA exonuclease, these RNAs were finally confirmed as circRNAs (Jeck et al., 2013). Deep RNA sequencing was carried out on ribosom-deficient RNAs of human and mouse hearts, using specific bioinformatics tools, 15,318 and 3017 cardiac circRNAs were found in human and mouse, respectively. Their abundance is usually related to homologous linear RNA, but in special cases, the abundance of some circRNAs is disproportionately high (Tan et al., 2017). Li, Y.S. et al. also demonstrated that at least 400 host genes could produce more than one circRNA (Li et al., 2017).
1.2.2. CircRNA is stable in the body
Suzuki H et al. found that RNase R could completely degrade rich linear RNA and y-structure RNA, but retained the circular part of lasso RNA, when they studied the persistence of the removed introns (Suzuki et al., 2006). William R et al. treated Hs68 cells with the transcriptional inhibitor actinomycin D to extract RNA at a specific time point, and the results suggested that circRNAs such as 18S and p16INK4a showed a longer transcriptional half-life than their linear RNA (Jeck et al., 2013).
1.2.3. CircRNA is highly conserved in evolution
William R et al. observed in Hs68 cells that HIPK2 and HIPK3, two homogenous kinases, produced rich circRNAs, which were fully differentiated to form a unique map, but saved similar gene structure (Jeck et al., 2013). Tracing the existence of circRNAs in different species to their common ancestor more than a billion years ago suggests that this aspect of gene expression is either very conserved or the result of repeated converged evolution (Wang et al., 2014). Ven et al. found that 15–20% of splicing sites in circRNAs generated in the mice's brain were homogenous with those in the pigs' brains (Veno et al., 2015).
1.2.4. The expression of circRNA is tissue-specific
To ascertain the spatial expression pattern of circRNAs in the central nervous system, Agnieszka RW et al. performed the cluster statistics on the circRNAs expression data in different brain regions of mice, and the results showed that there were significant differences in the expression of circRNAs in various brain regions, among which the cerebellum was most abundant, and the expression of circRNAs was more abundant than its related linear RNA (Rybak-Wolf et al., 2015). Wang, Y.H. et al. demonstrated that circRNAs were differentially-expressed in CD28 (+) CD8 (+) T cells vs CD28 (−) CD8 (+) T cells in the elderly, and CD28 (−) CD8 (+) T cells in the elderly vs in the adult when the function of circular RNA100783 was testified in chronic CD28-associated CD8 (+) T cell ageing (Wang et al., 2015).
1.3. CircRNAs function
1.3.1. CircRNAs regulate miRNA expression through its “sponge effect”
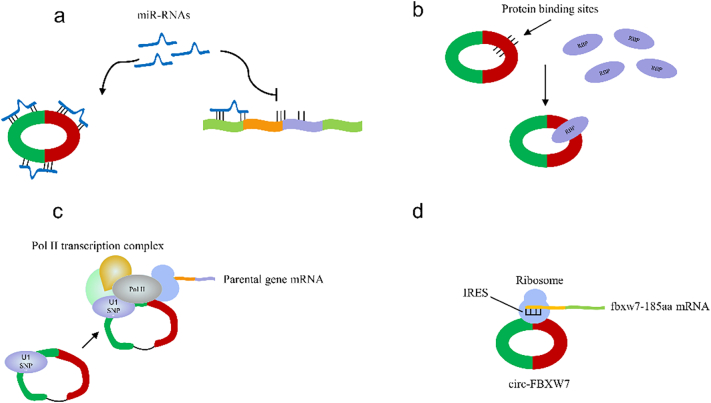
CircRNAs can competitively inhibited miRNAs binding to mRNA through sponge effect (Fig. 2a). Hansen, T.B. et al. observed the overlapping co-expression of cirs-7 and mir-7 in the brains of mice, especially in the neocortex and hippocampal neurons. Further studies found that cirs-7 in the nervous system contained more than 70 selectively conserved miRNA targets and had the negative regulatory effects on miRNAs. Furthermore, it was proved that the testicular specific circRNA and SryRNA could also negatively regulated mir-138 through sponge effect (Hansen et al., 2013). Li, Y et al. found that circPVT1, which was expressed in gastric cancer tissues and upregulated with disease development, could bind to miRNA-125 family, promoted the expression of its target gene E2F2, thus aggravated the proliferation of cancer cells (Li and Huang, 2017).
Fig. 2.
CircRNAs' function mechanisms. a. CircRNAs competitively inhibited miRNAs binding to mRNA through sponge effect. b. circRNAs can bind to RNA-binding proteins (RBPs) thus serving as a scaffold for RNA-proteins. c. EIciRNAs can regulate the expression of parent genes by interacting with U1 snRNPs. d. circ-FBXW7 can be subjected to translation in an internal ribosome entry site (IRES)-dependent and cap-independent manner, thus encoding fbxw7-185aa.
1.3.2. CircRNAs bind to proteins to form RNA-protein complexes
Circ-MBL/MBNL1 expresses in human and drosophilis and its endogenous expression is regulated by exogenous MBL due to highly conserved binding sites of muscleblind and flank introns (Ashwal-Fluss et al., 2014). Jeck, W.R. et al. proposed that some features of exon circRNAs indicated that they might bind to multiple proteins, and promote their stable interactions by increasing the potential stability of circRNAs transcripts, thus serving as a scaffold for RNA-proteins (Jeck and Sharpless, 2014) (Fig. 2b). To detect whether CDR1as is bound to miRNA, Memczak, S et al. analyzed the biochemical and transcriptome binding site data of miRNA effector protein-AGO by immunoprecipitation, the result showed that CDR1as could participate in transcriptional regulation as sponge molecules of miRNA by interacting with AGO and RNA polymerase II (Memczak et al., 2013).
1.3.3. Regulation of the parental gene expression
Most circRNAs do not appear to be involved in gene expression, but other studies have proved that ciRNA (intron circRNAs) plays other roles different from the exon cirRNAs “sponge effect” based on specific production patterns. CircRNAs from ANKRD52, MCM5, and SIRT7 introns are widely distributed in the nucleus, while their parental mRNAs are mainly located in the cytoplasm, In order to explore the possible role of ciRNA in the nucleus, Zhang, y. et al. mainly studied ci-ankrd52, and observed that the down-regulation of ci-ankrd52 led to a significant decrease in the expression of ankrd52mRNA, suggesting that ci-ankrd52 may play a role in the expression of local genes (Zhang et al., 2013). EIciRNAs are a special type of circRNAs related to RNAPII, consisting of exons and introns and its parent genes were down-regulated after EIciRNAs knockout (Li et al., 2015). Chuan H. further found that U1 snRNPs was related to the promoter of parent genes of EIciRNAs. AMO specific block of U1 snRNA could eliminate the influence of EIciRNAs on the regulation of parent genes at multiple levels, demonstrating that EIciRNAs regulated the expression of parent genes by interacting with U1 snRNPs (Huang and Shan, 2015) (Fig. 2c).
1.3.4. As a translation template
Most circRNAs do not translate into proteins, but some researchers suggested that some ecRNAs (exonic circRNAs) had a starting point for translation. Yang, Y.B. et al. found that circ-fbxw7, widely distributed in human brain, encoded a 21-kda protein (fbxw7-185aa) through an internal ribosome entry site (IRES) and further proved that the up-regulation of fbxw7-185aa inhibited the proliferation of cancer cells and the acceleration of cell cycle, which would also provide a new target for the treatment of glioma (Yang et al., 2018) (Fig. 2d).
2. The role of circRNAs in the regulation of heart failure
During chronic heart failure, when the myocardial contractility decreases and/or the hemodynamic load increases, the heart produces a series of adaptive changes to keep cardiac output, one of the most important adaptation is Frank - Staring mechanism (Fukuda and Granzier, 2004), that is, cardiac output is improved while cardiac preload is increased, and cardiomyocyte hypertrophy and fibrosis are the typical characteristics of cardiac remodeling in the process of resisting stress (Tham et al., 2015; Ma et al., 2014). The gradual loss of cardiomyocytes is the main reason for the decline of cardiac function (Takemura et al., 2018). Besides cell necrosis and apoptosis, autophagy also plays an important role in the loss of cardiomyocytes (Kroemer et al., 2009).
In 2016, the expression profile of circRNAs in mouse hearts was confirmed for the first time (Jakobi et al., 2016), to analyze the expression of circRNA in the heart and its changes under different conditions. Werfel S et al. performed RNA-seq analysis of libraries with ribosomal deletion in newborn and adult rats, mice (sham or TAC) and humans (fail and non-fail), 9000 circRNAs candidates were identified in each species, and they expressed significantly different in each groups of heart models (Werfel et al., 2016). Differential expression analysis also revealed 43 out of 826 commonly identified circRNAs to be differentially expressed in patients with non-ischaemic end-stage heart failure compared with control samples (Khan et al., 2016). In summary, there is increasing evidence that circRNAs play an important regulatory role in heart failure (Table 1).
Table 1.
Researches of heart failure regulated by circRNAs.
| Circular RNA | Gene ID | Function | Mechanism | Targets | Reference |
|---|---|---|---|---|---|
| HRCR | mm9-circ-012559 | Inhibits cardiomyocytes hypertrophy | miRNA sponge | miR-223 | (Wang et al., 2016) |
| circRNA_010567 | Not found | Mediates fibrosis-associated protein resection. | miRNA sponge | miR-141 | (Zhou and Yu, 2017) |
| circNFIB | mmu_circ_0011794 | Attenuates cardiac fibrosis | miRNA sponge | miR-433 | (Zhu et al., 2019) |
| CircRNA_000203 | Not found | Eliminates the antifibrotic effect of miR-26b in mouse CFs | miRNA sponge | miR-26b-5p | (Tang et al., 2017) |
| ACR | lme_circ_0001107 | Attenuates autophagy and cell death in cardiomyocytes | Protein binding | Dnmt3B | (Zhou et al., 2019) |
| MFACR | Not found | Regulates cardiomyocyte mitochondrial fission and apoptosis | miRNA sponge | miR-652-3p | (Wang et al., 2017) |
mmu, mouse;miRNA, microRNA; lme, not found.
2.1. CircRNAs and cardiomyocyte hypertrophy
When individual cardiomyocytes were stimulated by ranges of neurohumoral factors or increased ventricular wall tension, they would undergo hypertrophy as an adaptive response, therefore, persistent cardiac hypertrophy was an important index to predict the occurrence of heart failure (Oka et al., 2014). More and more intracellular signaling pathways are considered to be important transducers of hypertrophy response, including G protein-coupled receptor, epinephrine, angiotensin, and endovascular peptide receptor, etc (Tham et al., 2015) (Molkentin and Dorn, 2001). However, the detailed pathologic mechanisms of heart failure are poorly understood.
In recent years, a new regulatory pathway consisted of HRCR, mir-223 and ARC has been revealed. The study found that mir-223 transgenic mice had an increased risk of cardiac hypertrophy related to heart failure. While the myocytes were induced by isoproterenol with overexpression of mir-223, the expression of the marker proteins associated with myocardial hypertrophy was significantly increased and the cell volume significantly increased, but the same myocyte hypertrophy index was significantly inhibited in the myocytes with mir-223 knockout, suggesting that mir-223 was a positive regulator of cardiac hypertrophy. WB analysis found that ARC was the downstream target of mir-223 and Target Protector Technology proved that the regulation of mir-223 on ARC was specific. Further observation revealed that in the absence of ARC, the inhibition of mir-223 knockout on cardiac hypertrophy response was reduced, suggesting that ARC would be a target for downstream regulation of cardiac hypertrophy by mir-223. At the same time, the expression of mm9-circ-012559 (cardiac-related circRNA, HRCR) was significantly decreased in cardiomyocytes treated with ISO or TAC. RNA Hybrid showed that HRCR contained 6 binding sites of mir-223, which could be used as an endogenous mir-223 sponge inhibitor, further indicated that HRCR inhibited cardiomyocyte hypertrophy by regulating the expression of mir-223 and ARC, thereby blocked heart failure (Wang et al., 2016).
2.2. CircRNAs and cardiomyocyte fibrosis
Lack of oxygen causes cardiac cell death in myocardial infarction, dead cells release inflammatory mediators, leading to an inflammatory response in the injured area, the inflammatory response sets up a cascade of wound healing that eventually deposits fibrous scar tissue in the infarcted area, resulting in excessive deposition of the extracellular matrix and myocardial fibrosis, which further leads to heart sclerosis, ventricular dysfunction, and heart failure (Biswas and Longmore, 2016).
Zhou, B et al. used circRNA chip to detect the circRNA expression profile of myocardial fibrosis in the diabetic db/db mouse model. Heat maps and volcanic maps suggested that 43 circRNAs were expressed differently, among them circRNA_010567 was significantly up-regulated, further studies revealed that the expression of fibroblast-related proteins such as Co II, Col III and col-SMA were significantly inhibited in the myocardial fibroblasts with intervention of AngII and circRNA_010567 knocked out, then the important role of circRNA_010567/mir-141/TGF-β axis in the regulation of myocardial fibrosis in diabetic mice was confirmed (Zhou and Yu, 2017). CircRNAs regulate myocyte fibrosis by acting as sponges with miRNA. Zhu, Y.J. et al. observed that circNFIB was significantly down-regulated in mice hearts with overexpression of TGF–β or myocardial infarction, and luciferase reporter gene test confirmed that mir-433 was a direct target of circNFIB, Up-regulation of circNFIB inhibited the proliferation of cardiac fibroblasts, while down-regulation of circNFIB reversed the anti-proliferation effect of mir-433 inhibitor. It was further confirmed that circNFIB could regulate the process of myocardial fibrosis by endogenous antagonism against mir-433, and the circNFIB/mir-433/AZIN1 axis also provided a novel treatment of myocardial fibrosis (Zhu et al., 2019).
2.3. CircRNAs and autophagy
Autophagy has been identified as an evolutively conserved important process in eukaryotes for the turnover of intracellular materials. In this process, some damaged proteins or organelles are wrapped with a double membrane structure of autophagic vesicles,and then sent to lysosomes or vacuoles for degradation and recycling (Mizushima, 2007; Galluzzi et al., 2017). More and more evidences have shown that autophagy is related to heart failure (Maejima et al., 2013).
CircRNA microarray analysis using mmu_circRNA_006636 as the internal reference gene in mice heart induced by ischemia-reperfusion injury showed that autophagy-related circRNA (ACR) was significantly down-regulated. To further explore the relationship between ACR and cardiomyocyte autophagy, a mouse model of ACR overexpression was constructed, data analysis suggested that ACR overexpression in mice reduced the level of lc3-II after myocardial ischemia/reperfusion injury, and significantly improved the vacuolization in ventricular tissue. In addition, exogenous ACR can also improve myocardial dysfunction induced by ischemia/reperfusion injury. Further research revealed that ACR can activate Pink1 expression by its binding to Dnmt3B and blocking Dnmt3B mediated DNA methylation of Pink1 promoter, which activates the expression of its downstream target FAM65B and inhibits autophagy of cardiomyocytes in the process of heart failure (Zhou et al., 2019).
2.4. CircRNAs and apoptosis
Apoptosis refers to the spontaneous and orderly death of cells controlled by genes in order to maintain a stable internal environment (Savitskaya and Onishchenko, 2015). Wencker, D et al. found in the mouse model that inhibition of myocardial cell apoptosis largely prevented the development of cardiac dilatation and systolic dysfunction, suggesting that myocardial cell apoptosis may be a causal mechanism for heart failure (Wencker et al., 2003).
In recent years, circRNAs have been shown to regulate heart failure by participating in the process of myocardial cell apoptosis. The heart has a large number of mitochondria that provide enough energy to keep cardiomyocytes function, and the dysfunction of mitochondrial fission can lead to a variety of heart diseases, such as myocardial infarction and heart failure (Wang et al., 2017). MTP18 is a nuclear-encoded mitochondrial membrane protein involved in the mitochondrial fission of mammalian cells (Tondera et al., 2005). Wang, K et al. demonstrated that MTP18 mediated mitochondria fission and apoptosis of myocardial cells via constructing models of primary cardiac myocytes of mice and ischemia-reperfusion injury in mice liver, at the same time, they found that Mir-652-3p regulated the expression of MTP18 at the translation level but not change of the expression of MTP18 mRNA, further studies showed that a circRNA, mm9-circ-016597 (MFACR), which was significantly highly expressed in ischemia-reperfusion mice heart, could function as a mir-652-3p sponge and regulate the expression and activity of MTP18, thus, a regulatory pathway of mitochondrial fission and apoptosis in cardiomyocytes composed of MFACR/mir-652-3p/MTP18 was obtained (Wang et al., 2017). Persistent myocardial inflammation promotes damage to cardiomyocytes, which eventually leads to heart failure or death. Shengwei, S et al. found that the expression of circANKRD36 in LPS-treated H9c2 cells was significantly up-regulated, while silencing it could reduce LPS-induced apoptosis and inflammatory damage, luciferase reporter assay suggested that mir-138 was a direct target for circANKRD36, revealing the regulation of the circANKRD36/mir-138/p38MAPK/NF-B signaling pathway on LPS-induced apoptosis and inflammatory injury in H9c2 cells (Shi et al., 2019).
3. Expectation
Heart failure is the terminal stage of cardiovascular disease, although great progress has been made in the development of it in recent decades, the advance of the clinical therapy in heart failure is still limited. CircRNA have become a hotspot in the treatment of cardiovascular diseases, the specific mechanisms are stayed in their infancy, and most researches are settled at the cellular and animal levels. If the regulation effect of circRNA pathways in heart failure is confirmed in human, the new drugs can be designed for circRNAs as a target, and a novel chapter will be coming in the circRNA field.
Abbreviation
- CircRNAs
circular RNAs
- mmu
mouse
- miRNA
microRNA
Author contributions
Li Jiang: Writing - Original draft preparation, Methodology, Software. Xiaoyan Wang: Data curation. Xiaopeng Zhan: Writing - Original draft preparation. Sheng Kang: Supervision. Haibo Liu: Software, Validation. Yu Luo: Writing - Reviewing and Editing. Li Lin: Writing - Reviewing and Editing.
Declaration of funding source
National Natural Science Foundation of China (81570237, 81870197, 81770350, 81870247), Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (PWZxq2017-05), Top-level Clinical Discipline Project of Shanghai Pudong District (PWYgf2018-02).
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled.
Contributor Information
Yu Luo, Email: wangyily1839@126.com.
Li Lin, Email: linli777@126.com.
References
- Ashwal-Fluss R. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Aufiero S. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019;16(8):503–514. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- Biswas H., Longmore G.D. Action of SNAIL1 in cardiac myofibroblasts is important for cardiac fibrosis following hypoxic injury. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0162636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- Diener T.O. Potato spindle tuber “virus”. IV. A replicating, low molecular weight RNA. Virology. 1971;45(2):411–428. doi: 10.1016/0042-6822(71)90342-4. [DOI] [PubMed] [Google Scholar]
- Du W.W. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38(18):1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- Fan X. Circular RNAs in cardiovascular disease: an overview. Biomed. Res. Int. 2017;2017:5135781. doi: 10.1155/2017/5135781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda N., Granzier H. Role of the giant elastic protein titin in the Frank-Starling mechanism of the heart. Curr. Vasc. Pharmacol. 2004;2(2):135–139. doi: 10.2174/1570161043476357. [DOI] [PubMed] [Google Scholar]
- Galluzzi L. Activating autophagy to potentiate immunogenic chemotherapy and radiation therapy. Nat. Rev. Clin. Oncol. 2017;14(4):247–258. doi: 10.1038/nrclinonc.2016.183. [DOI] [PubMed] [Google Scholar]
- Hansen T.B. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hsu M.T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- Huang C., Shan G. What happens at or after transcription: insights into circRNA biogenesis and function. Transcription. 2015;6(4):61–64. doi: 10.1080/21541264.2015.1071301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobi T. Profiling and validation of the circular RNA repertoire in adult murine hearts. Genomics Proteomics Bioinformatics. 2016;14(4):216–223. doi: 10.1016/j.gpb.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W.R. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.A. RBM20 regulates circular RNA production from the titin gene. Circ. Res. 2016;119(9):996–1003. doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- Kjems J., Garrett R.A. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell. 1988;54(5):693–703. doi: 10.1016/s0092-8674(88)80014-x. [DOI] [PubMed] [Google Scholar]
- Kroemer G. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16(1):3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Huang S. Response to comment on response to “Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer,” Cancer Lett. 2017 Mar 1; 388(2017): 208-219. Cancer Lett. 2017;411:64. doi: 10.1016/j.canlet.2017.09.006. [DOI] [PubMed] [Google Scholar]
- Li Z. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- Li Y. Dynamic organization of lncRNA and circular RNA regulators collectively controlled cardiac differentiation in humans. EBioMedicine. 2017;24:137–146. doi: 10.1016/j.ebiom.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin. Chim. Acta. 2018;480:17–25. doi: 10.1016/j.cca.2018.01.026. [DOI] [PubMed] [Google Scholar]
- Lukiw W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD) Front. Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q. Identification of circular RNAs hsa_circ_0044235 in peripheral blood as novel biomarkers for rheumatoid arthritis. Clin. Exp. Immunol. 2018;194(1):118–124. doi: 10.1111/cei.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. Myofibroblasts and the extracellular matrix network in post-myocardial infarction cardiac remodeling. Pflugers Arch. 2014;466(6):1113–1127. doi: 10.1007/s00424-014-1463-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima Y. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat. Med. 2013;19(11):1478–1488. doi: 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Molkentin, J.D. and G.W. Dorn, 2nd, Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu. Rev. Physiol., 2001. 63: p. 391–426. [DOI] [PubMed]
- Mudd J.O., Kass D.A. Tackling heart failure in the twenty-first century. Nature. 2008;451(7181):919–928. doi: 10.1038/nature06798. [DOI] [PubMed] [Google Scholar]
- Nigro J.M. Scrambled exons. Cell. 1991;64(3):607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- Oka T. Angiogenesis and cardiac hypertrophy: maintenance of cardiac function and causative roles in heart failure. Circ. Res. 2014;114(3):565–571. doi: 10.1161/CIRCRESAHA.114.300507. [DOI] [PubMed] [Google Scholar]
- Qu S. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol P. Heart failure drug treatment. Lancet. 2019;393(10175):1034–1044. doi: 10.1016/S0140-6736(18)31808-7. [DOI] [PubMed] [Google Scholar]
- Rybak-Wolf A. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Sablok G., Zhao H., Sun X. Plant circular RNAs (circRNAs): transcriptional regulation beyond miRNAs in plants. Mol. Plant. 2016;9(2):192–194. doi: 10.1016/j.molp.2015.12.021. [DOI] [PubMed] [Google Scholar]
- Sanger H.L. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitskaya M.A., Onishchenko G.E. Mechanisms of apoptosis. Biochemistry (Mosc) 2015;80(11):1393–1405. doi: 10.1134/S0006297915110012. [DOI] [PubMed] [Google Scholar]
- Shang X. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular carcinoma development. Medicine (Baltimore) 2016;95(22) doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Guo X., Wang W. Identification and characterization of circular RNAs in zebrafish. FEBS Lett. 2017;591(1):213–220. doi: 10.1002/1873-3468.12500. [DOI] [PubMed] [Google Scholar]
- Shi S. Silencing circANKRD36 protects H9c2 cells against lipopolysaccharide-induced injury via up-regulating miR-138. Exp. Mol. Pathol. 2019;111:104300. doi: 10.1016/j.yexmp.2019.104300. [DOI] [PubMed] [Google Scholar]
- Suzuki H. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34(8):e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura G. Anti-apoptosis in nonmyocytes and pro-autophagy in cardiomyocytes: two strategies against postinfarction heart failure through regulation of cell death/degeneration. Heart Fail. Rev. 2018;23(5):759–772. doi: 10.1007/s10741-018-9708-x. [DOI] [PubMed] [Google Scholar]
- Tan W.L. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017;113(3):298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- Tang C.M. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci. Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham Y.K. Pathophysiology of cardiac hypertrophy and heart failure: signaling pathways and novel therapeutic targets. Arch. Toxicol. 2015;89(9):1401–1438. doi: 10.1007/s00204-015-1477-x. [DOI] [PubMed] [Google Scholar]
- Tondera, D., et al., The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J. Cell Sci., 2005. 118(Pt 14): p. 3049–59. [DOI] [PubMed]
- Veno M.T. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/beta-catenin pathway. Biomed. Res. Int. 2016;2016:1579490. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.L. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9(6):e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.H. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun. Ageing. 2015;12:17. doi: 10.1186/s12979-015-0042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37(33):2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- Wang K. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24(6):1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, S.E. and W. Grossman, Prognosis in heart failure: is systolic or diastolic dysfunction more important? Herz, 1991. 16 Spec No 1: p. 324–9. [PubMed]
- Wencker D. A mechanistic role for cardiac myocyte apoptosis in heart failure. J. Clin. Invest. 2003;111(10):1497–1504. doi: 10.1172/JCI17664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werfel S. Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell. Cardiol. 2016;98:103–107. doi: 10.1016/j.yjmcc.2016.07.007. [DOI] [PubMed] [Google Scholar]
- Yang Y. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110(3) doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- Zhou B., Yu J.W. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem. Biophys. Res. Commun. 2017;487(4):769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- Zhou L.Y. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/FAM65B pathway. Cell Death Differ. 2019;26(7):1299–1315. doi: 10.1038/s41418-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. Upregulation of circular RNA CircNFIB attenuates cardiac fibrosis by sponging miR-433. Front. Genet. 2019;10:564. doi: 10.3389/fgene.2019.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]