Abstract
Functional annotation of protein sequence with high accuracy has become one of the most important issues in modern biomedical studies, and computational approaches of significantly accelerated analysis process and enhanced accuracy are greatly desired. Although a variety of methods have been developed to elevate protein annotation accuracy, their ability in controlling false annotation rates remains either limited or not systematically evaluated. In this study, a protein encoding strategy, together with a deep learning algorithm, was proposed to control the false discovery rate in protein function annotation, and its performances were systematically compared with that of the traditional similarity-based and de novo approaches. Based on a comprehensive assessment from multiple perspectives, the proposed strategy and algorithm were found to perform better in both prediction stability and annotation accuracy compared with other de novo methods. Moreover, an in-depth assessment revealed that it possessed an improved capacity of controlling the false discovery rate compared with traditional methods. All in all, this study not only provided a comprehensive analysis on the performances of the newly proposed strategy but also provided a tool for the researcher in the fields of protein function annotation.
Keywords: protein function prediction, deep learning, prediction stability, annotation accuracy, false discovery rate
Introduction
Functional annotation of protein sequence with high accuracy (AC) has become one of the most important issues in understanding the molecular mechanism of life [1, 2] and has great biological [3–5], pathological [6–8] and pharmaceutical [9–16] implications. With the rapid accumulation of a wealth of protein sequences, the functional annotation of proteins has become increasingly challenging [17]. Particularly, only ~1% of all protein sequences in UniProt [18] have experimentally verified functions [19–22], and it is estimated that ~90% of the annotated proteins in the ontology of molecular function (MF)/biological process (BP) [23] come from only nine species [24]. Even for these nine model organisms, ~60% of their proteins have not had any experimentally determined MF/BP term [24]. Traditional methods for protein function annotation are mainly based on the experiments such as mass spectrometry, microscopy and RNA interference, which are reported as very time-consuming and resource-demanding because of the low throughput and restricted scope of methodology [25–31]. As such, the computational approaches of significantly accelerated analysis process and enhanced AC are greatly desired [32–37].
Computational methods popular in current protein function prediction [38–40] can be roughly divided into three categories: information, structure and sequence based [41]. The information-based methods [26, 42–45] are suitable for predicting the functions of interacting proteins [46–49] and proteins from coexpressed genes [41, 47, 50] but seriously restricted by the great noises in protein–protein interaction data and insufficient number of annotated proteins [17, 41, 49]. The structure-based methods [51–54] are found to be accurate [47, 55] but significantly limited by the lack of crystallized protein folds or structures [47] and the unclear relations between structural similarity and functional similarity in many cases [41, 56]. Among those three method categories, the sequence-based ones have now become the most widely applied method in protein function prediction [41] due to the relatively easy access of abundant high-quality sequence data in public database [46, 50] and its powerful ability to predict the function of remotely relevant protein and the homologous proteins of distinct functions [47, 57]. There are two types of sequence-based approaches: similarity based and de novo [41]. Particularly, similarity-based methods (like BLAST (a tool used for finding regions of similarity between biological sequences) [58] and HMMER (a tool used for searching sequence databases for sequence homologs, and for making sequence alignments) [59]) assign an unannotated protein with the function of another protein similar in sequence to that protein [41]. Since the similarity-based methods are reported to depend heavily on the sequence homology, the de novo ones are considered as an effective complement [41, 60], which is irrespective of sequence similarity and good at predicting the distantly related proteins and the homologous proteins of distinct functions [61, 62]. The de novo methods are generally based on supervised learning model, such as K-nearest neighbor (KNN) [63], probabilistic neural network (PNN) [64] and support vector machine (SVM) [62]. They are reported to be powerful in predicting the functions of proteins [65–70] and other molecules [71].
However, the high false discovery rate of the sequence-based protein function prediction remains a severe problem [72–76]. In particular, the databases adopted by similarity-based methods for searching homology often contain noise, and the relation between sequence similarity and homology is sometimes unclear [41]; the representativeness of the training data analyzed by de novo method is not always sufficient [62]. In the past few years, several pioneer efforts have been made to solve the problems [77]. On one hand, a stringent score cutoff is adopted by BLAST and HMMER to control the false discovery hits in detecting homologies [78]. On the other hand, some machine learning methods have been used to identify false homologies [79, 80], and a putative negative training data set derived from representative seed proteins of Pfam families, which has high coverage of the protein family space, is constructed to reduce the false discovery rate [62]. Recently, the deep learning algorithm is frequently applied in sequence and omics data analysis [81, 82], biomedical imaging and biomedical signal processing [83, 84], which demonstrates a remarkable performance [85]. In protein function annotation, a multitask deep neural network has been designed to predict the function of the proteins from multiclasses [83], and a deep restricted Boltzmann machine is used to annotate the proteins with Gene Ontology (GO) term in the deep position of a directed acyclic graph [23, 86]. Moreover, a multiclassification model has been constructed to predict protein functional classes [87]. Although those methods are reported to be effective in elevating protein annotation AC [88], their ability in controlling false annotation rates is either limited or not systematically evaluated [57]. Thus, the significant enhancement on controlling false discovery rate is still urgently needed, and the corresponding tool is required in the filed of protein function annotation [62].
In this study, a protein encoding strategy, together with a deep learning algorithm, was proposed to control the false discovery rate in protein function annotation, and its performances were systematically compared to that of the traditional similarity-based and de novo methods. First, the training and testing data sets with the highest and lowest similarities were separately constructed for distinguishing the performances among different methods. Second, a protein encoding strategy was proposed and integrated to deep learning-based algorithm, and its performances were compared with other traditional methods from multiple perspectives. Third, the capacity of the proposed method in controlling the false discovery rate was assessed by the comprehensive genome scanning and enrichment factor (EF). In summary, this study provided a comprehensive analysis on the performances of a newly proposed protein function annotation strategy.
Materials and methods
The functional families studied in and protein sequences collected for this analysis
In total, 20 protein families of different GO terms were collected from diverse subclasses in the MF of the GO database by maximizing their representativeness among all MF categories [23], and the GO families with different numbers of proteins were selected to enable the discussion of the effect of sample size on prediction result. The total numbers of proteins in these GO families were from ~800 to ~33 500 (after removing repeated protein sequences). As provided in Table 1, each studied GO family was indicated by a GO ID [23], and the total number of proteins (with sequence length of ≤1000) in the 20 GO families were listed (ranging from 802 to 33 178). The sequences of the proteins in these 20 families were collected from the UniProt database [89], and the repeated sequences were removed to avoid possible bias.
Table 1.
Twenty GO families collected from diverse subclasses in MF of GO database by maximizing their representativeness among all MF categories. The numbers of proteins (with the sequence length of ≤1000) in these GO families were from 802 to 33 178, and the number of Pfam families covered by each GO family was from 57 to 1092. These GO families were sorted by their total numbers of proteins included
| Functional GO families studied in this work | GO ID | No. of proteins | No. of Pfam families covered | No. of proteins in training data set | No. of proteins in testing data set |
|---|---|---|---|---|---|
| Cyclase activity | GO:0009975 | 802 | 57 | 624 | 178 |
| Cyclin-dependent protein kinase activity | GO:0097472 | 2951 | 64 | 2295 | 656 |
| Phosphoprotein phosphatase activity | GO:0004721 | 4324 | 152 | 3363 | 961 |
| Transcription coregulator activity | GO:0003712 | 4684 | 346 | 3643 | 1041 |
| Positive regulation of transferase activity | GO:0051347 | 4732 | 355 | 3680 | 1052 |
| Negative regulation of catalytic activity | GO:0043086 | 6239 | 512 | 4853 | 1386 |
| Transferase activity, transferring acyl groups | GO:0016746 | 8845 | 238 | 6879 | 1966 |
| Ubiquitin-like protein transferase activity | GO:0019787 | 9553 | 255 | 7430 | 2123 |
| Transferase activity, transferring glycosyl groups | GO:0016757 | 9694 | 278 | 7540 | 2154 |
| Structural constituent of ribosome | GO:0003735 | 10 492 | 204 | 8160 | 2332 |
| Regulation of hydrolase activity | GO:0051336 | 10 995 | 686 | 8551 | 2444 |
| Positive regulation of catalytic activity | GO:0043085 | 11 803 | 719 | 9180 | 2623 |
| Positive regulation of MF | GO:0044093 | 13 677 | 884 | 10 637 | 3040 |
| Peptidase activity | GO:0008233 | 18 665 | 597 | 14 517 | 4148 |
| DNA-binding transcription factor activity | GO:0003700 | 19 677 | 693 | 15 304 | 4373 |
| Hydrolase activity, acting on ester bonds | GO:0016788 | 22 599 | 802 | 17 578 | 5021 |
| Protein kinase activity | GO:0004672 | 23 068 | 642 | 17 942 | 5126 |
| Hydrolase activity, acting on acid anhydrides | GO:0016817 | 28 327 | 779 | 22 032 | 6295 |
| Signaling receptor activity | GO:0038023 | 28 700 | 525 | 22 322 | 6378 |
| Catalytic complex | GO:1902494 | 33 178 | 1092 | 25 805 | 7373 |
Constructing the data sets of training and testing
Since a binary classification model was constructed for the studied GO families, the proteins in each family were considered as positive data. In order to significantly enhance the representativeness of negative data (nonmembers of a GO family), a putative data set was therefore constructed by considering the following steps: (1) the Pfam family of each protein in a particular GO family was collected from the Pfam database [90], (2) the Pfam families of all proteins in that GO family were considered as the ‘positive Pfam family’ and (3) three representative seed proteins from the rest of the Pfam families (named as ‘negative Pfam family’) were collected to construct a putative negative data set (PND). Since the resulting PND was characterized by its proteins of significantly diverse Pfam families, its representativeness on those nonmembers of a GO family was substantially enhanced and could be applied for controlling false discovery [57, 62].
Moreover, the level of representativeness of the studied data sets has great impacts on the performances of the analyzed methods [62]. Particularly, the higher similarity between the data set used to construct models and that to test models could result in better functional prediction [57]. Thus, the training and testing data sets with the highest or the lowest similarity were constructed for the analyses here. First, the sequences were converted to digital vectors using the novel strategy proposed in this study (described in the following section). Second, the positive data set (containing all proteins from a particular GO family) was classified into six groups using the K-means clustering algorithm (the distance used in this clustering was Euclidean distance) [91]. On one hand, in order to construct the training and testing data sets with the highest similarity, one-third of proteins in each group of the positive data set were randomly selected out to construct the testing data set (combining six groups), and the remaining two-thirds were used to form training data set (combining six groups). On the other hand, to construct the training and testing data sets with the lowest similarity, two groups were randomly selected out from those six groups to construct the testing data set, and the remaining four were used to form training data set. For both situations, two out of those three representative seed proteins from the negative Pfam families were randomly selected out to form the training data set, and the remaining one was used to construct the testing data set. Furthermore, to evaluate the false discovery rate of those studied methods, all protein sequences in the human genome were collected from the UniProt database [89].
The methods studied and assessed in this work
The sequence homology was the basis of protein function prediction, which could be detected by similarity analysis among sequences. Generally, the sequences with higher similarity were more likely homologous. BLAST was one of the most popular tools for sequence similarity analysis [58]. Herein, the sequences of proteins in the training data set of each GO family were collected to construct a searchable database, and then any query protein was searched against this database using BLAST and annotated with the function of the most similar protein. HMMER was another popular tool for sequence similarity analysis used in this study, which was based on the hidden Markov model [59]. Similar to the BLAST, the searchable database was also constructed based on the sequences of proteins in the training data set of each GO family. Then, any protein was searched against this database using HMMER [59] and annotated based on the most similar one.
Moreover, there were three de novo methods studied and assessed in this study. The SVM tried to find a hyperplane to separate the members from nonmembers of a particular GO family by maximizing the margin defined in protein feature space [92]. The KNN predicted the class of a protein by the majority vote of its neighbors with a given distance metric [93], and the PNN was a neural network based on Bayesian decision theory [94]. These three methods were directly applied under the Python environment. As reported in the previous studies [57, 62], a method for converting the protein sequence to the digital feature vector had been successfully applied in SVM, KNN and PNN and had been shown good performance in protein function prediction. This method converted the protein sequence into the digital vector according to its properties and composition of amino acids [62, 95]. The properties adopted here included (1) hydrophobicity, (2) Van der Waals volume, (3) polarity, (4) polarizability, (5) charge, (6) secondary structure, (7) solvent accessibility and (8) surface tension [62, 96–98]. Each property was represented by three groups of descriptors: the global composition of given amino acid, frequencies with which the property changes along entire protein and distribution pattern of property along sequence. The detail on this digitalizing method could be found in previous studies [57, 62].
The deep learning algorithm used in this study was the convolutional neural network (CNN). As shown in Figure 1 (Encoding Layer), a new sequence encoding technique was first proposed to generate a 1000 × 5 binary array for any protein sequence. Particularly, each amino acid was encoded by a 5-bit binary number (from [0,0,0,0,1] for alanine to [1,0,1,0,0] for tyrosine). Only the proteins with the sequence length of no more than 1000 amino acids were analyzed in this study, which constitute the majority (>98%) of the proteins in any GO family. For the proteins of sequence length of less than 1000 amino acids, their empty amino acid positions were complemented by the 5-bit binary number [0,0,0,0,0]. Moreover, for the amino acids that were not among those 20 common ones, they were encoded by another number [1,0,1,0,1]. Second, CNN was applied, which consisted of multiple layers: a convolutional layer, a pooling layer, two fully connected layers and a softmax layer (Figure 1). The encoding array connected directly with the convolutional layer, which scanned the encoding array through an mk × 5 convolution kernel and resulted in a feature vector:
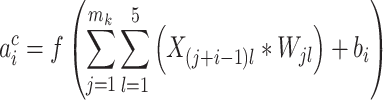 |
1 |
where 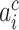 indicated the output of the ith neuron of the feature vector,
indicated the output of the ith neuron of the feature vector, 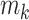 denoted the length of the kth convolution kernel,
denoted the length of the kth convolution kernel, 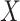 referred to input protein encoding array and
referred to input protein encoding array and 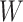 and
and 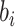 were a (mk × 5) weight array and a bias, respectively.
were a (mk × 5) weight array and a bias, respectively. 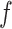 defined the ReLU activation function [99]. Third, the max pooling layer was used, and the maximum neuron output value of the feature vector was selected as the output of the pooling layer.
defined the ReLU activation function [99]. Third, the max pooling layer was used, and the maximum neuron output value of the feature vector was selected as the output of the pooling layer.
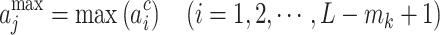 |
2 |
where 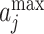 indicated the output of the jth neuron of the pooling layer. To fully extract protein features, eight different lengths of convolution kernel (for each length, there are 120 kernels) were used to scan the protein encoding array. Therefore, after the pooling layer, a vector containing 960 outputs for each protein was obtained. Fourth, using this vector, the fully connected layers generated the output for each layer:
indicated the output of the jth neuron of the pooling layer. To fully extract protein features, eight different lengths of convolution kernel (for each length, there are 120 kernels) were used to scan the protein encoding array. Therefore, after the pooling layer, a vector containing 960 outputs for each protein was obtained. Fourth, using this vector, the fully connected layers generated the output for each layer:
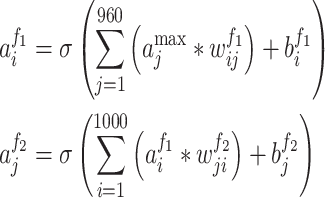 |
3 |
where 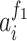 denoted the output of the ith neuron of the first fully connected layer and
denoted the output of the ith neuron of the first fully connected layer and 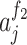 referred to the output of the jth neuron of the second fully connected layer.
referred to the output of the jth neuron of the second fully connected layer. 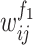 and
and 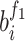 indicated the jth weight and the bias of the ith neuron of the first fully connected layer;
indicated the jth weight and the bias of the ith neuron of the first fully connected layer; 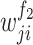 and
and 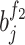 indicated the ith weight and the bias of the jth neuron of the second fully connected layer.
indicated the ith weight and the bias of the jth neuron of the second fully connected layer. 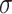 was ELU activation function [100]. Finally, the output vector of the fully connected layer was further used as the input of a softmax layer, which provided the classification probability of the query protein:
was ELU activation function [100]. Finally, the output vector of the fully connected layer was further used as the input of a softmax layer, which provided the classification probability of the query protein:
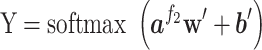 |
5 |
where 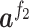 indicated the output vector of the second fully connected layer and
indicated the output vector of the second fully connected layer and 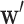 and
and 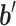 referred to weight array and bias vector, respectively. Y was the classification probability of sequence X (Figure 1).
referred to weight array and bias vector, respectively. Y was the classification probability of sequence X (Figure 1).
Figure 1.
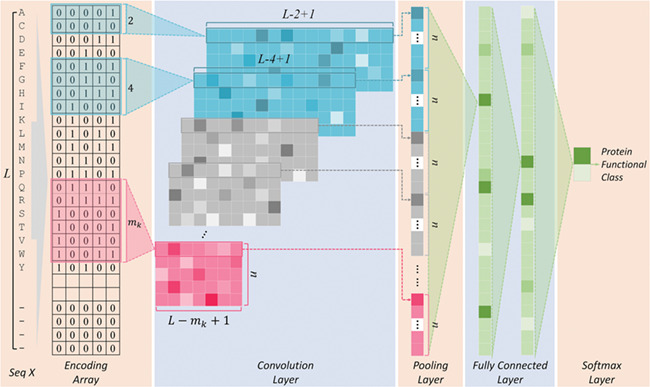
The workflow of the deep learning algorithm (CNN) applied together with the sequence encoding technique proposed in this study.
The CNN model was implemented with the Python programming language and the TensorFlow library. The binary cross-entropy loss function was adopted in all models training, and the Adam [101] optimizer (learning rate = 0.001, β1 = 0.9, β2 = 0.999 and ε = 10−8) was used for the optimization during back-propagation. The weight parameters were initialized with the He initialization method [102], and biases were initialized to zero. The batch normalization was applied in the fully connected layers before ELU activation function for accelerating the speed of convergence, and the strategy of dropout [103] was also used in the fully connected layer with a drop rate of 0.6 to randomly remove a certain number of neurons at each training step in order to prevent the overfitting of the neural network.
Assessing the AC and false discovery rate of protein function annotation
Five popular measurements, such as sensitivity (SE), specificity (SP), precision (PR), AC and Matthews correlation coefficient (MCC) [57, 104], were adopted in this study to evaluate the performance of each protein functional annotation method. Since the SP was reported to be effective in evaluating the methods’ false discovery rate [57], it was adopted in this study to assess the false discovery rate of the de novo methods and the constructed deep learning model based on the testing data set. Moreover, in order to further assess the false discovery rate of methods, a real-world application of the human genome scanning was performed with each method. For BLAST and HMMER, the training data set was used to construct the searching databases. For de novo and deep learning methods, the training data set was applied to train the models. Because it is not necessary to use the negative data set in predictions with BLAST and HMMER, the SP cannot be calculated in the genome scanning for both methods, the false discovery rate was evaluated by the EF for discovering the proteins in each GO family:
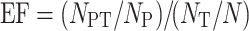 |
6 |
where 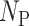 referred to the total number of proteins in the testing data set predicted by the method as the members of certain GO family,
referred to the total number of proteins in the testing data set predicted by the method as the members of certain GO family, 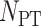 indicated the total number of predicted proteins in the testing data set truly belonging to this particular GO family,
indicated the total number of predicted proteins in the testing data set truly belonging to this particular GO family, 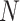 denoted the total number of proteins in the studied genome and
denoted the total number of proteins in the studied genome and 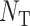 referred to the total number of proteins in the testing data set truly belonging to the studied GO family. The value of EF is no less than zero; however, only when the EF value is larger than 1, there is an enrichment. The larger the EF, the lower the false discovery rate.
referred to the total number of proteins in the testing data set truly belonging to the studied GO family. The value of EF is no less than zero; however, only when the EF value is larger than 1, there is an enrichment. The larger the EF, the lower the false discovery rate.
Results and discussion
Methods’ performances based on the training and testing data sets with the highest similarity
The performances of studied methods were first calculated and assessed based on the constructed training and testing data sets with the highest similarity (the way to construct such data sets was provided in the second section of Materials and Methods). As shown in Table 2, eight representative GO families were selected randomly from all 20 studied GO families, and the performances of three de novo (SVM, KNN and PNN) and the proposed deep learning (CNN) methods were provided based on five measurements (SE, SP, PR, AC and MCC). Taking AC as an example, it spanned from 89.0% to 98.5%, from 89.1% to 97.3%, from 84.5% to 95.1%, and from 86.7% to 96.2% for CNN, SVM, KNN and PNN, respectively. Moreover, for MCC, it ranged from 0.72 to 0.91, from 0.72 to 0.87, from 0.65 to 0.81, and from 0.62 to 0.81, respectively. As shown, the exact AC and MCC values of CNN and SVM were slightly higher than that of the remaining methods (KNN and PNN), but no significant difference was observed for AC values (the P-values for AC values between any two methods were larger than 0.05). For MCC values, CNN is better than KNN and PNN with the P-values of 0.004 and 0.011, respectively, SVM is better than KNN and PNN with the P-values of 0.012 and 0.038, respectively, and there is no significant difference between CNN and SVM.
Table 2.
The performances of three de novo (SVM, KNN and PNN) and the deep learning (CNN) methods proposed in this study on the training and testing data sets (for eight representative GO families) with the highest similarity based on popular measurements (SE, SP, PR, AC and MCC). Each GO family was denoted by its GO ID, and its corresponding GO term was shown in Table 1
| GO ID | CNN | SVM | KNN | PNN | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SE% | SP% | PR% | AC% | MCC | SE% | SP% | PR% | AC% | MCC | SE% | SP% | PR% | AC% | MCC | SE% | SP% | PR% | AC% | MCC | |
| GO:0097472 | 85.2 | 99.9 | 99.1 | 98.5 | 0.91 | 81.0 | 99.1 | 90.2 | 97.3 | 0.84 | 70.9 | 97.7 | 76.4 | 95.1 | 0.71 | 72.7 | 98.6 | 84.7 | 96.2 | 0.76 |
| GO:0004721 | 79.5 | 99.2 | 93.9 | 96.6 | 0.85 | 65.2 | 99.3 | 93.2 | 94.7 | 0.75 | 68.0 | 95.6 | 70.8 | 91.9 | 0.65 | 55.5 | 98.7 | 87.0 | 92.9 | 0.66 |
| GO:0051347 | 81.3 | 99.6 | 97.5 | 96.9 | 0.87 | 78.4 | 97.3 | 83.4 | 94.5 | 0.78 | 75.6 | 93.5 | 66.9 | 90.9 | 0.66 | 63.8 | 99.1 | 92.8 | 93.9 | 0.74 |
| GO:0003735 | 80.7 | 98.7 | 95.8 | 93.8 | 0.84 | 87.2 | 97.6 | 93.2 | 94.8 | 0.87 | 84.7 | 95.6 | 87.8 | 92.6 | 0.81 | 84.9 | 95.5 | 87.7 | 92.6 | 0.81 |
| GO:0016746 | 71.7 | 96.2 | 85.8 | 90.2 | 0.72 | 70.5 | 96.6 | 86.8 | 90.2 | 0.72 | 70.7 | 92.5 | 75.2 | 87.2 | 0.65 | 64.8 | 93.7 | 76.8 | 86.7 | 0.62 |
| GO:0043085 | 69.3 | 98.1 | 94.3 | 89.2 | 0.74 | 75.4 | 95.2 | 87.5 | 89.1 | 0.74 | 78.8 | 87.1 | 73.2 | 84.5 | 0.65 | 66.7 | 95.6 | 87.2 | 86.7 | 0.68 |
| GO:0003700 | 81.1 | 95.0 | 92.3 | 89.0 | 0.78 | 82.8 | 94.8 | 92.2 | 89.7 | 0.79 | 81.9 | 90.9 | 87.0 | 87.1 | 0.74 | 84.3 | 89.0 | 85.0 | 87.0 | 0.73 |
| GO:0044093 | 80.7 | 93.5 | 86.6 | 89.1 | 0.76 | 78.7 | 94.1 | 87.5 | 88.8 | 0.75 | 80.6 | 85.5 | 74.5 | 83.8 | 0.65 | 72.6 | 92.5 | 83.7 | 85.6 | 0.68 |
The results above indicated that, for the data sets with the highest similarity, the studied methods showed a similar performance as each other. In other words, the models trained using the high representative data sets by difference methods performed consistently well, which denoted that the training and testing data sets with the highest similarity might have a low resolution on distinguishing the performances of different methods. Therefore, in order to provide an in-depth assessment on studied methods, the training and testing data sets with the lowest similarity could be considered as more effective in providing the performance assessment of higher resolution. Moreover, in the real world of protein function annotation, it was almost impossible to have all query proteins (with function unknown) fully represented by the proteins with annotated functions. Therefore, the assessment based on the data sets with the highest similarity could only draw the upper ceiling of the methods’ performances, and the data sets in most real-world occasions were much less representative. Especially for the novel or newly discovered proteins, which attract great and broad attentions, the training data set or database could not provide any representativeness to the novel ones. Therefore, compared with the upper ceiling, a bottom line (the training and testing data sets with the lowest similarity) was expected to be capable of revealing the lower limits of the performances of all studied methods.
Methods’ performances based on the training and testing data sets with the lowest similarity
The performances of the studied methods were calculated and assessed based on the constructed data sets with the lowest similarity (the way to construct such data sets was provided in the second section of Materials and Methods). As illustrated in Table 3, the performances of three de novo and one deep learning methods on all 20 GO families were provided based on five measurements. Moreover, the statistical differences of these measurements between any two methods were shown in Figure 2. As one of the most comprehensive parameters in any category of predictors [57], the MCC reflected the stability of protein function predictor, which described the correlation between a predictive value and the actual value [62, 105, 106]. As illustrated on the left panel of Figure 2A, the variations of MCC values among methods were provided (the actual MCC value of each method was subtracted by the minimum MCC value among four different methods). As shown, the CNN method showed the consistently higher MCC values compared with the other three methods, and the MCC values of KNN method were the lowest in most of the GO families. As all GO families were ordered by their total numbers of proteins, there was no clear trend on the MCC values with the increase of protein amount. Moreover, to assess the statistical differences of MCC values between any two methods, the violin box plots based on the MCC values in Table 3 were drawn on the right panel of Figure 2A. As illustrated, there were significant differences (P < 0.01) between the MCCs of CNN and that of the rest methods, and there was no significant difference in MCCs between any two of those three de novo methods. These results demonstrated an enhanced stability of the CNN-based protein function annotation model compared with three popular de novo methods.
Table 3.
The performances of three de novo (SVM, KNN and PNN) and the deep learning (CNN) methods proposed in this study on the training and testing data sets (for 20 studied GO families) with the lowest similarity based on popular measurements (SE, SP, PR, AC and MCC). Each GO family was indicated by its GO ID, and its corresponding GO term was provided in Table 1
| GO ID | CNN | SVM | KNN | PNN | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SE% | SP% | PR% | AC% | MCC | SE% | SP% | PR% | AC% | MCC | SE% | SP% | PR% | AC% | MCC | SE% | SP% | PR% | AC% | MCC | |
| GO:0009975 | 26.4 | 99.9 | 94.0 | 97.9 | 0.49 | 11.8 | 99.8 | 67.7 | 97.4 | 0.28 | 10.1 | 98.3 | 14.4 | 95.8 | 0.10 | 16.3 | 96.3 | 11.2 | 94.1 | 0.10 |
| GO:0097472 | 67.7 | 99.9 | 99.6 | 96.9 | 0.81 | 39.0 | 99.3 | 85.1 | 93.5 | 0.55 | 43.3 | 97.8 | 67.6 | 92.6 | 0.50 | 40.2 | 98.6 | 74.8 | 93.0 | 0.52 |
| GO:0004721 | 55.6 | 99.5 | 94.5 | 93.6 | 0.70 | 28.4 | 99.2 | 84.0 | 89.7 | 0.45 | 36.4 | 95.2 | 54.2 | 87.3 | 0.38 | 32.4 | 97.3 | 64.8 | 88.5 | 0.40 |
| GO:0003712 | 32.7 | 99.7 | 94.2 | 89.9 | 0.52 | 20.4 | 98.5 | 70.2 | 87.1 | 0.33 | 35.9 | 94.5 | 52.5 | 85.9 | 0.36 | 38.1 | 92.9 | 47.8 | 84.9 | 0.34 |
| GO:0051347 | 42.5 | 99.9 | 98.7 | 91.4 | 0.62 | 26.9 | 98.8 | 79.3 | 88.2 | 0.42 | 37.4 | 92.7 | 47.0 | 84.5 | 0.33 | 37.5 | 96.0 | 62.0 | 87.4 | 0.42 |
| GO:0043086 | 35.5 | 99.1 | 90.4 | 87.1 | 0.52 | 17.4 | 98.0 | 66.4 | 82.8 | 0.28 | 28.7 | 91.8 | 44.8 | 79.9 | 0.25 | 10.1 | 99.9 | 96.6 | 83.0 | 0.28 |
| GO:0016746 | 40.0 | 97.7 | 84.9 | 83.7 | 0.51 | 27.1 | 93.7 | 58.0 | 77.5 | 0.28 | 30.0 | 86.2 | 41.1 | 72.5 | 0.18 | 9.9 | 99.4 | 84.8 | 77.7 | 0.24 |
| GO:0019787 | 79.9 | 96.2 | 87.8 | 92.0 | 0.79 | 37.7 | 96.1 | 76.9 | 81.1 | 0.44 | 34.8 | 90.3 | 55.4 | 76.0 | 0.30 | 40.1 | 89.9 | 57.9 | 77.1 | 0.34 |
| GO:0016757 | 48.9 | 97.3 | 86.5 | 84.7 | 0.57 | 22.0 | 96.2 | 68.0 | 77.0 | 0.29 | 38.5 | 84.9 | 47.2 | 72.8 | 0.25 | 32.7 | 92.7 | 61.2 | 77.1 | 0.32 |
| GO:0003735 | 87.6 | 97.3 | 92.4 | 94.6 | 0.86 | 87.9 | 96.6 | 90.8 | 94.2 | 0.85 | 86.0 | 95.0 | 86.7 | 92.6 | 0.81 | 84.4 | 93.1 | 82.2 | 90.7 | 0.77 |
| GO:0051336 | 28.3 | 98.9 | 91.2 | 78.2 | 0.43 | 23.0 | 94.4 | 63.1 | 73.5 | 0.26 | 38.6 | 84.3 | 50.5 | 70.9 | 0.25 | 46.5 | 83.2 | 53.4 | 72.4 | 0.31 |
| GO:0043085 | 44.0 | 98.0 | 90.7 | 81.3 | 0.54 | 32.9 | 94.9 | 74.3 | 75.7 | 0.37 | 54.8 | 79.8 | 54.9 | 72.1 | 0.35 | 57.8 | 80.5 | 57.0 | 73.5 | 0.38 |
| GO:0044093 | 41.6 | 96.3 | 85.4 | 77.5 | 0.48 | 29.0 | 94.6 | 73.9 | 72.0 | 0.33 | 46.7 | 80.2 | 55.4 | 68.7 | 0.28 | 23.1 | 97.3 | 81.7 | 71.7 | 0.33 |
| GO:0008233 | 62.0 | 92.9 | 86.0 | 80.2 | 0.59 | 49.2 | 92.8 | 82.7 | 74.9 | 0.48 | 67.9 | 72.6 | 63.4 | 70.7 | 0.40 | 60.6 | 80.7 | 68.7 | 72.4 | 0.42 |
| GO:0003700 | 77.7 | 94.1 | 90.7 | 87.1 | 0.74 | 62.2 | 89.1 | 81.0 | 77.6 | 0.54 | 67.0 | 85.2 | 77.1 | 77.4 | 0.53 | 68.2 | 84.5 | 76.6 | 77.5 | 0.54 |
| GO:0016788 | 53.9 | 89.1 | 81.0 | 72.8 | 0.46 | 45.9 | 86.5 | 74.5 | 67.6 | 0.36 | 64.8 | 65.0 | 61.5 | 64.9 | 0.30 | 55.9 | 78.2 | 68.9 | 67.8 | 0.35 |
| GO:0004672 | 76.8 | 95.5 | 93.7 | 86.8 | 0.74 | 49.1 | 88.0 | 78.1 | 69.9 | 0.41 | 60.3 | 71.7 | 65.0 | 66.4 | 0.32 | 50.9 | 86.6 | 76.8 | 70.0 | 0.40 |
| GO:0016817 | 60.7 | 95.0 | 92.9 | 77.1 | 0.59 | 45.3 | 84.9 | 76.5 | 64.3 | 0.33 | 68.2 | 60.8 | 65.4 | 64.7 | 0.29 | 67.1 | 65.4 | 67.8 | 66.3 | 0.32 |
| GO:0038023 | 71.3 | 91.6 | 90.0 | 81.1 | 0.64 | 59.4 | 90.6 | 87.1 | 74.5 | 0.52 | 57.2 | 83.7 | 78.9 | 70.0 | 0.42 | 57.0 | 84.7 | 79.9 | 70.4 | 0.43 |
| GO:1902494 | 49.6 | 87.2 | 83.4 | 66.0 | 0.39 | 42.8 | 79.9 | 73.4 | 59.0 | 0.24 | 58.6 | 62.5 | 66.9 | 60.3 | 0.21 | 58.9 | 66.6 | 69.6 | 62.3 | 0.25 |
Figure 2.
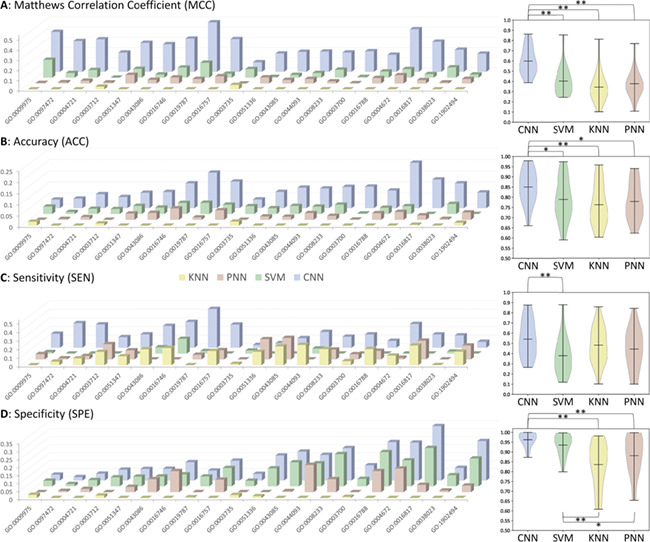
Variations (the actual value of each method subtracted by the minimum value among four different methods) of four measurements among different protein function annotation methods: (A) MCC, (B) AC, (C) SE, (D) SP. On the right side, the statistical differences between any two methods were provided as the violin box plots. * indicated great difference of P < 0.05, and ** denoted significant difference of P < 0.01. Detailed P-values are provided in Supplementary Table S1.
As another important parameter for protein function annotation assessment, the AC referred to the total number of true members (positive plus negative) divided by the number of studied proteins [57], which was essential to be compared among different methods. Herein, the variations of AC values among methods (the actual AC of each method was subtracted by the minimum ACs among four different methods) were therefore calculated and provided on the left panel of Figure 2B. Similar to MCCs, the CNN method showed the consistently higher AC values compared with other three methods, and the AC values of KNN method were also the lowest in most of those GO families (there was no clear trend on the AC values with the increase of protein amount either). Furthermore, to assess the statistical differences of the ACs between any two methods, the violin box plot based on the ACs in Table 3 was drawn on the right panel of Figure 2B. As illustrated, there were significant differences (P < 0.05) between the AC values of CNN and that of the rest methods, and there was no significant difference in ACs between any two of those three de novo methods. These results showed the elevated AC of the CNN-based protein function annotation model compared with three popular de novo methods.
Besides the stability (MCC) and AC, SE and SP were also frequently used to assess the methods’ prediction performances on positive and negative data sets, respectively. Thus, in this study, similar analyses were also conducted and shown in Figure 2C, D. Since the SP was known as an effective metric reflecting the false discovery rate, both CNN and SVM showed enhanced control on the false discovery when comparing with KNN and PNN. All in all, based on the comprehensive assessment from four different perspectives (MCC, AC, SE and SP), the CNN method proposed here was found to perform better in both prediction stability and annotation AC compared with three popular de novo methods. Although both CNN and SVM were found with enhanced control of false discovery rate, the SEs of CNN were found to be significantly enhanced (P = 0.007) compared with that of SVM.
In-depth assessment on the false discovery rates based on genome scanning
Besides the SP, the EF was one of the most popular and effective metrics for assessing the false discovery rate of any functional annotation method [57]. As known, the SP values assess the false discovery rate via only considering the prediction performance on the PND, while the EF evaluates the false discovery by fully considering the real-world true members of a particular GO family. Therefore, the EFs were applied in this study to complement the SP and further make in-depth assessment on the false discovery rate of each studied methods. In other words, in order to evaluate the false discovery rate of each method in the real world, multiple methods (CNN, BLAST, HMMER, SVM, KNN and PNN) were used to scan the human genome to identify human proteins belonging to each GO family. As shown in Table 4, the total numbers of proteins identified by different methods together with their corresponding EFs were provided. Moreover, the EFs of each method based on different GO families were illustrated in Figure 3. As shown, there were clear variations among the EFs of different methods. Particularly, as shown on the right panel of Figure 3, there were significant differences (P < 0.01) between the EFs of CNN and that of all de novo methods and BLAST. Based on the statistical analysis conducted in this study, there was no significant variation between the EFs of CNN and that of PHMM (a probabilistic model called Poisson Hidden Markov Model, which used in the HMMER), but the calculated P-value (0.054) was very close to 0.05. Furthermore, as provided in Table 4, the majority (17 out of 20, 85.0%) of the EFs of CNN were higher than that of PHMM. In conclusion, based on the information provided in Figure 3 and Table 4, the deep learning strategy CNN proposed in this study showed an improved ability to control the false discovery rate.
Table 4.
The total numbers of proteins and the EFs of three de novo (SVM, KNN and PNN) identified by two similarity-based (BLAST and HMMER) and the deep learning (CNN) methods proposed in this study based on the training and testing data sets (for all 20 studied GO families) of the lowest similarity. Each GO family was indicated by its GO ID, and its corresponding GO term was provided in Table 1
| GO ID | No. of proteins identified by each method | EF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CNN | SVM | KNN | PNN | BLAST | PHMM | CNN | SVM | KNN | PNN | BLAST | PHMM | |
| GO:0009975 | 56 | 235 | 657 | 1416 | 1594 | 1347 | 53.61 | 0 | 2.28 | 3.18 | 8.48 | 11.14 |
| GO:0097472 | 202 | 434 | 619 | 561 | 1598 | 923 | 44.59 | 14.65 | 8.56 | 13.22 | 8.29 | 14.92 |
| GO:0004721 | 352 | 712 | 1824 | 1402 | 2403 | 1844 | 32.73 | 12.36 | 5.17 | 7.17 | 6.28 | 8.41 |
| GO:0003712 | 637 | 1403 | 2648 | 3114 | 4408 | 3541 | 10.79 | 3.38 | 2.19 | 2.40 | 2.80 | 3.65 |
| GO:0051347 | 725 | 1689 | 3873 | 1421 | 5858 | 4521 | 9.15 | 3.37 | 1.22 | 4.17 | 2.43 | 3.22 |
| GO:0043086 | 1087 | 2291 | 3822 | 528 | 5911 | 4888 | 7.75 | 2.63 | 1.92 | 9.37 | 2.14 | 2.67 |
| GO:0016746 | 924 | 2173 | 3878 | 401 | 2076 | 1604 | 12.61 | 3.51 | 2.51 | 17.44 | 7.76 | 10.44 |
| GO:0019787 | 1954 | 2339 | 3822 | 3921 | 3328 | 2499 | 7.72 | 4.79 | 2.67 | 3.06 | 5.02 | 6.62 |
| GO:0016757 | 881 | 1265 | 4146 | 2352 | 1770 | 1451 | 11.20 | 6.05 | 2.56 | 3.88 | 8.36 | 10.20 |
| GO:0003735 | 520 | 750 | 965 | 1564 | 1207 | 1029 | 33.03 | 21.22 | 17.8 | 10.98 | 14.58 | 17.10 |
| GO:0051336 | 1436 | 4334 | 6885 | 6693 | 7187 | 5887 | 3.76 | 1.10 | 0.81 | 1.26 | 1.81 | 2.28 |
| GO:0043085 | 2152 | 4710 | 8846 | 8082 | 7984 | 6470 | 4.27 | 1.78 | 1.23 | 1.53 | 1.83 | 2.29 |
| GO:0044093 | 3458 | 5172 | 9417 | 2664 | 8654 | 7048 | 2.65 | 1.61 | 1.10 | 2.79 | 1.69 | 2.07 |
| GO:0008233 | 2850 | 3842 | 8106 | 5906 | 5409 | 4327 | 3.52 | 1.85 | 1.18 | 1.50 | 2.24 | 2.77 |
| GO:0003700 | 3877 | 6586 | 6861 | 6604 | 6537 | 5167 | 4.07 | 1.87 | 1.66 | 1.84 | 2.58 | 3.27 |
| GO:0016788 | 3975 | 4867 | 9351 | 7019 | 5338 | 4414 | 2.45 | 1.75 | 1.18 | 1.52 | 2.72 | 3.33 |
| GO:0004672 | 2904 | 5918 | 8287 | 4895 | 6621 | 4633 | 3.87 | 1.40 | 1.35 | 2.05 | 2.21 | 2.96 |
| GO:0016817 | 2207 | 5443 | 10 649 | 8903 | 6531 | 5053 | 5.15 | 1.89 | 1.24 | 1.48 | 2.43 | 3.13 |
| GO:0038023 | 4230 | 5919 | 7314 | 6684 | 5616 | 3573 | 3.06 | 1.93 | 1.79 | 2.03 | 2.60 | 3.48 |
| GO:1902494 | 4133 | 6900 | 10 017 | 8399 | 6466 | 4991 | 2.45 | 1.31 | 1.04 | 1.35 | 2.16 | 2.84 |
Figure 3.
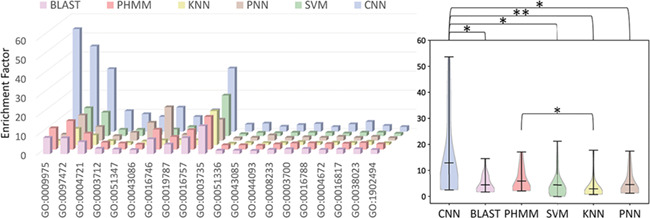
The EFs of different protein function annotation methods of the training and testing data sets (for all studied GO families) with the lowest similarity. On the right side, the statistical differences of the EFs between any two methods were provided as the violin box plots. * indicated great difference of P < 0.05, and ** denoted significant difference of P < 0.01. Detailed P-values are provided in the Supplementary Table S2.
Conclusions
Based on the comprehensive assessment using different measurements (MCC, AC, SE and SP), the CNN method together with the protein encoding strategy proposed in this study was found to perform better in both prediction stability and annotation AC compared with the popular de novo methods. Moreover, the in-depth assessment revealed that it possessed an improved capacity of controlling the false discovery rate in current protein functional annotation compared with other traditional methods. All in all, this study not only provided a comprehensive analysis on the performances of the newly proposed strategy but also provided a valuable tool for the researcher in the fields of protein function annotation.
Key Points
Functional annotation of protein sequence with high accuracy has become one of the most important issues in modern biomedical studies.
A protein encoding strategy, together with a deep learning algorithm, was proposed to control false discovery rate in protein function annotation.
The proposed strategy and algorithm were found to perform better in prediction stability, annotation accuracy and false discovery rate compared with the traditional methods.
Supplementary Material
Acknowledgments
This study was funded by the National Key Research and Development Program of China (2018YFC0910500), National Natural Science Foundation of China (81872798), Fundamental Research Funds for Central Universities (2018QNA7023, 10611CDJXZ238826, 2018CDQYSG0007 and CDJZR14468801), Innovation Project on Industrial Generic Key Technologies of Chongqing (cstc2015zdcy-ztzx120003), Key Project of Zhejiang Province Ministry of Science and Technology (2015C03055) and Key Project of National Natural Science Foundation of China (81730108).
Jiajun Hong, Yang Zhang and Junbiao Ying are doctoral, master’s and undergraduate students at the College of Pharmaceutical Sciences in Zhejiang University, China, and jointly cultivated by the School of Pharmaceutical Sciences in Chongqing University, China. They are interested in artificial intelligence.
Weiwei Xue is a professor at the School of Pharmaceutical Sciences in Chongqing University, China. He is interested in the area of computer-based drug design and molecular dynamics simulation.
Tian Xie and Lin Tao are professors at the School of Medicine in Hangzhou Normal University, China. They are interested in the area of traditional Chinese medicine, bioinformatics and machine learning.
Feng Zhu is a professor at the College of Pharmaceutical Sciences in Zhejiang University, China. He got his PhD degree from the National University of Singapore, Singapore. His research group (https://idrblab.org/) has been working in the fields of bioinformatics, omics-based drug discovery, system biology and medicinal chemistry. Welcome to visit his personal website at: https://idrblab.org/Peoples.php.
References
- 1. Chang YC, Hu Z, Rachlin J, et al. COMBREX-DB: an experiment centered database of protein function: knowledge, predictions and knowledge gaps. Nucleic Acids Res 2016;44:D330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sahraeian SM, Luo KR, Brenner SE. SIFTER search: a web server for accurate phylogeny-based protein function prediction. Nucleic Acids Res 2015;43:W141–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goldstrohm AC, Hall TMT, McKenney KM. Post-transcriptional regulatory functions of mammalian Pumilio proteins. Trends Genet 2018;34:972–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qiao W, Akhter N, Fang X, et al. From mutations to mechanisms and dysfunction via computation and mining of protein energy landscapes. BMC Genomics 2018;19:671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Woods RJ. Predicting the structures of glycans, glycoproteins, and their complexes. Chem Rev 2018;118:8005–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shiihashi G, Ito D, Yagi T, et al. Mislocated FUS is sufficient for gain-of-toxic-function amyotrophic lateral sclerosis phenotypes in mice. Brain 2016;139:2380–94. [DOI] [PubMed] [Google Scholar]
- 7. Skrlj B, Konc J, Kunej T. Identification of sequence variants within experimentally validated protein interaction sites provides new insights into molecular mechanisms of disease development. Mol Inform 2017;36:00017. [DOI] [PubMed] [Google Scholar]
- 8. Seneviratne U, Nott A, Bhat VB, et al. S-nitrosation of proteins relevant to Alzheimer's disease during early stages of neurodegeneration. Proc Natl Acad Sci U S A 2016;113:4152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li B, Tang J, Yang Q, et al. NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res 2017;45:W162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li B, Tang J, Yang Q, et al. Performance evaluation and online realization of data-driven normalization methods used in LC/MS based untargeted metabolomics analysis. Sci Rep 2016;6:38881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lai AC, Crews CM. Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov 2017;16:101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang J, Fu J, Wang Y, et al. Simultaneous improvement in the precision, accuracy and robustness of label-free proteome quantification by optimizing data manipulation chains. Mol Cell Proteomics 2019; doi:10.1074/mcp.RA118.001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li YH, Li XX, Hong JJ, et al. Clinical trials, progression-speed differentiating features and swiftness rule of the innovative targets of first-in-class drugs. Brief Bioinform 2019; doi:10.1093/bib/bby130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Y, Ying JB, Hong JJ, et al. How does chirality determine the selective inhibition of histone deacetylase 6? A lesson from trichostatin a enantiomers based on molecular dynamics. ACS Chem Nerosci 2019;10:2467–80. [DOI] [PubMed] [Google Scholar]
- 15. Li X, Li X, Li Y, et al. What makes species productive of anti-cancer drugs? Clues from drugs' species origin, druglikeness, target and pathway. Anticancer Agents Med Chem 2018;19:194–203. [DOI] [PubMed] [Google Scholar]
- 16. Han Z, Xue W, Tao L, et al. Identification of key long non-coding RNAs in the pathology of Alzheimer's disease and their functions based on genome-wide associations study, microarray, and RNA-seq data. J Alzheimers Dis 2019;68:339–55. [DOI] [PubMed] [Google Scholar]
- 17. Zhao B, Hu S, Li X, et al. An efficient method for protein function annotation based on multilayer protein networks. Hum Genomics 2016;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res 2017;45:D158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Das S, Orengo CA. Protein function annotation using protein domain family resources. Methods 2016;93:24–34. [DOI] [PubMed] [Google Scholar]
- 20. You R, Zhang Z, Xiong Y, et al. GOLabeler: improving sequence-based large-scale protein function prediction by learning to rank. Bioinformatics 2018;34:2465–73. [DOI] [PubMed] [Google Scholar]
- 21. Tang J, Fu J, Wang Y, et al. ANPELA: analysis and performance assessment of the label-free quantification workflow for metaproteomic studies. Brief Bioinform 2019; doi:10.1093/bib/bby127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li S, Li J, Ning L, et al. In silico identification of protein S-palmitoylation sites and their involvement in human inherited disease. J Chem Inf Model 2015;55:2015–25. [DOI] [PubMed] [Google Scholar]
- 23. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clark WT, Radivojac P. Analysis of protein function and its prediction from amino acid sequence. Proteins 2011;79:2086–96. [DOI] [PubMed] [Google Scholar]
- 25. Frasca M, Cesa-Bianchi N. Multitask protein function prediction through task dissimilarity. IEEE/ACM Trans Comput Biol Bioinform 2017; doi:10.1109/TCBB.2017.2684127. [DOI] [PubMed] [Google Scholar]
- 26. Cao R, Cheng J. Integrated protein function prediction by mining function associations, sequences, and protein–protein and gene–gene interaction networks. Methods 2016;93:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schnoes AM, Ream DC, Thorman AW, et al. Biases in the experimental annotations of protein function and their effect on our understanding of protein function space. PLoS Comput Biol 2013;9:e1003063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li YH, Yu CY, Li XX, et al. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res 2018;46:D1121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang H, Qin C, Li YH, et al. Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res 2016;44:D1069–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu F, Shi Z, Qin C, et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res 2012;40:D1128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhu F, Han B, Kumar P, et al. Update of TTD: therapeutic target database. Nucleic Acids Res 2010;38:D787–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cao R, Freitas C, Chan L, et al. ProLanGO: protein function prediction using neural machine translation based on a recurrent neural network. Molecules 2017;22:1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu F, Han L, Zheng C, et al. What are next generation innovative therapeutic targets? Clues from genetic, structural, physicochemical, and systems profiles of successful targets. J Pharmacol Exp Ther 2009;330:304–15. [DOI] [PubMed] [Google Scholar]
- 34. Xu J, Wang P, Yang H, et al. Comparison of FDA approved kinase targets to clinical trial ones: insights from their system profiles and drug-target interaction networks. Biomed Res Int 2016;2016:2509385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fu J, Tang J, Wang Y, et al. Discovery of the consistently well-performed analysis chain for SWATH-MS based pharmacoproteomic quantification. Front Pharmacol 2018;9:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu F, Li XX, Yang SY, et al. Clinical success of drug targets prospectively predicted by in silico study. Trends Pharmacol Sci 2018;39:229–31. [DOI] [PubMed] [Google Scholar]
- 37. Xue W, Yang F, Wang P, et al. What contributes to serotonin-norepinephrine reuptake inhibitors' dual-targeting mechanism? The key role of transmembrane domain 6 in human serotonin and norepinephrine transporters revealed by molecular dynamics simulation. ACS Chem Nerosci 2018;9:1128–40. [DOI] [PubMed] [Google Scholar]
- 38. Jain A, Kihara D. Phylo-PFP: improved automated protein function prediction using phylogenetic distance of distantly related sequences. Bioinformatics 2019;35:753–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang C, Freddolino PL, Zhang Y. COFACTOR: improved protein function prediction by combining structure, sequence and protein–protein interaction information. Nucleic Acids Res 2017;45:W291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wan S, Duan Y, Zou Q. HPSLPred: an ensemble multi-label classifier for human protein subcellular location prediction with imbalanced source. Proteomics 2017;17:1700262. [DOI] [PubMed] [Google Scholar]
- 41. Cruz LM, Trefflich S, Weiss VA, et al. Protein function prediction. Methods Mol Biol 1654;2017:55–75. [DOI] [PubMed] [Google Scholar]
- 42. Piovesan D, Giollo M, Ferrari C, et al. Protein function prediction using guilty by association from interaction networks. Amino Acids 2015;47:2583–92. [DOI] [PubMed] [Google Scholar]
- 43. Lv Q, Ma W, Liu H, et al. Genome-wide protein–protein interactions and protein function exploration in cyanobacteria. Sci Rep 2015;5:15519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mateos A, Dopazo J, Jansen R, et al. Systematic learning of gene functional classes from DNA array expression data by using multilayer perceptions. Genome Res 2002;12:1703–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huttenhower C, Hibbs M, Myers C, et al. A scalable method for integration and functional analysis of multiple microarray datasets. Bioinformatics 2006;22:2890–7. [DOI] [PubMed] [Google Scholar]
- 46. Hawkins T, Chitale M, Kihara D. New paradigm in protein function prediction for large scale omics analysis. Mol Biosyst 2008;4:223–31. [DOI] [PubMed] [Google Scholar]
- 47. Tiwari AK, Srivastava R. A survey of computational intelligence techniques in protein function prediction. Int J Proteomics 2014;2014:845479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vazquez A, Flammini A, Maritan A, et al. Global protein function prediction from protein–protein interaction networks. Nat Biotechnol 2003;21:697–700. [DOI] [PubMed] [Google Scholar]
- 49. Peng W, Wang J, Cai J, et al. Improving protein function prediction using domain and protein complexes in PPI networks. BMC Syst Biol 2014;8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nariai N, Kolaczyk ED, Kasif S. Probabilistic protein function prediction from heterogeneous genome-wide data. PLoS One 2007;2:e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hwang H, Dey F, Petrey D, et al. Structure-based prediction of ligand–protein interactions on a genome-wide scale. Proc Natl Acad Sci U S A 2017;114:13685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sillitoe I, Lewis TE, Cuff A, et al. CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res 2015;43:D376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lam SD, Dawson NL, Das S, et al. Gene3D: expanding the utility of domain assignments. Nucleic Acids Res 2016;44:D404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res 2010;38:W545–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maghawry HA, Mostafa MG, Gharib TF. A new protein structure representation for efficient protein function prediction. J Comput Biol 2014;21:936–46. [DOI] [PubMed] [Google Scholar]
- 56. Pearson WR. Protein function prediction: problems and pitfalls. Curr Protoc Bioinformatics 2015;51:4.12.1–8. [DOI] [PubMed] [Google Scholar]
- 57. Yu CY, Li XX, Yang H, et al. Assessing the performances of protein function prediction algorithms from the perspectives of identification accuracy and false discovery rate. Int J Mol Sci 2018;19:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinformatics 2009;10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Potter SC, Luciani A, Eddy SR, et al. HMMER web server: 2018 update. Nucleic Acids Res 2018;46:W200–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhao B, Wang J, Wu FX. Computational methods to predict protein functions from protein–protein interaction networks. Curr Protein Pept Sci 2017;18:1120–31. [DOI] [PubMed] [Google Scholar]
- 61. Peled S, Leiderman O, Charar R, et al. De-novo protein function prediction using DNA binding and RNA binding proteins as a test case. Nat Commun 2016;7:13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li YH, Xu JY, Tao L, et al. SVM-Prot 2016: a web-server for machine learning prediction of protein functional families from sequence irrespective of similarity. PLoS One 2016;11:e0155290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lan L, Djuric N, Guo Y, et al. MS-kNN: protein function prediction by integrating multiple data sources. BMC Bioinformatics 2013;14:S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gonzalez-Camacho JM, Crossa J, Perez-Rodriguez P, et al. Genome-enabled prediction using probabilistic neural network classifiers. BMC Genomics 2016;17:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Khan ZU, Hayat M, Khan MA. Discrimination of acidic and alkaline enzyme using Chou's pseudo amino acid composition in conjunction with probabilistic neural network model. J Theor Biol 2015;365:197–203. [DOI] [PubMed] [Google Scholar]
- 66. Hayat M, Khan A. Predicting membrane protein types by fusing composite protein sequence features into pseudo amino acid composition. J Theor Biol 2011;271:10–7. [DOI] [PubMed] [Google Scholar]
- 67. Naveed M, Khan A. GPCR-MPredictor: multi-level prediction of G protein-coupled receptors using genetic ensemble. Amino Acids 2012;42:1809–23. [DOI] [PubMed] [Google Scholar]
- 68. Nath N, Mitchell JB. Is EC class predictable from reaction mechanism? BMC Bioinformatics 2012;13:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shen HB, Yang J, Chou KC. Fuzzy KNN for predicting membrane protein types from pseudo-amino acid composition. J Theor Biol 2006;240:9–13. [DOI] [PubMed] [Google Scholar]
- 70. Xue W, Wang P, Tu G, et al. Computational identification of the binding mechanism of a triple reuptake inhibitor amitifadine for the treatment of major depressive disorder. Phys Chem Chem Phys 2018;20:6606–16. [DOI] [PubMed] [Google Scholar]
- 71. Li H, Yap CW, Ung CY, et al. Machine learning approaches for predicting compounds that interact with therapeutic and ADMET related proteins. J Pharm Sci 2007;96:2838–60. [DOI] [PubMed] [Google Scholar]
- 72. Hernandez C, Mella C, Navarro G, et al. Protein complex prediction via dense subgraphs and false positive analysis. PLoS One 2017;12:e0183460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Brylinski M. Unleashing the power of meta-threading for evolution/structure-based function inference of proteins. Front Genet 2013;4:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Brandes N, Ofer D, Linial M. ASAP: a machine learning framework for local protein properties. Database 2016;2016:baw133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zheng G, Yang F, Fu T, et al. Computational characterization of the selective inhibition of human norepinephrine and serotonin transporters by an escitalopram scaffold. Phys Chem Chem Phys 2018;20:29513–27. [DOI] [PubMed] [Google Scholar]
- 76. Wang P, Zhang X, Fu T, et al. Differentiating physicochemical properties between addictive and nonaddictive ADHD drugs revealed by molecular dynamics simulation studies. ACS Chem Nerosci 2017;8:1416–28. [DOI] [PubMed] [Google Scholar]
- 77. Pearson WR, Li W, Lopez R. Query-seeded iterative sequence similarity searching improves selectivity 5-20-fold. Nucleic Acids Res 2017;45:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fokkens L, Botelho SM, Boekhorst J, et al. Enrichment of homologs in insignificant BLAST hits by co-complex network alignment. BMC Bioinformatics 2010;11:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fujimoto MS, Suvorov A, Jensen NO, et al. Detecting false positive sequence homology: a machine learning approach. BMC Bioinformatics 2016;17:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wei L, Zou Q. Recent progress in machine learning-based methods for protein fold recognition. Int J Mol Sci 2016;17:2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang ZQ, Zhao Y, Liao XK, et al. Deep learning in omics: a survey and guideline. Brief Funct Genomics 2019;18:41–57. [DOI] [PubMed] [Google Scholar]
- 82. Zou Q, Xing PW, Wei LY, et al. Gene2vec: gene subsequence embedding for prediction of mammalian N-6-methyladenosine sites from mRNA. RNA 2019;25:205–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fa R, Cozzetto D, Wan C, et al. Predicting human protein function with multi-task deep neural networks. PLoS One 2018;13:e0198216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zeng NY, Zhang H, Song BY, et al. Facial expression recognition via learning deep sparse autoencoders. Neurocomputing 2018;273:643–9. [Google Scholar]
- 85. Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinform 2017;18:851–69. [DOI] [PubMed] [Google Scholar]
- 86. Zou X, Wang G, Yu G. Protein function prediction using deep restricted Boltzmann machines. Biomed Res Int 2017;2017:1729301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Seo S, Oh M, Park Y, et al. DeepFam: deep learning based alignment-free method for protein family modeling and prediction. Bioinformatics 2018;34:i254–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zou Q, Wan S, Ju Y, et al. Pretata: predicting TATA binding proteins with novel features and dimensionality reduction strategy. BMC Syst Biol 2016;10:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res 2018;46:2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. El-Gebali S, Mistry J, Bateman A, et al. The Pfam protein families database in 2019. Nucleic Acids Res 2019;47:D427–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Brusco MJ, Shireman E, Steinley D. A comparison of latent class, K-means, and K-median methods for clustering dichotomous data. Psychol Methods 2017;22:563–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Noble WS. What is a support vector machine? Nat Biotechnol 2006;24:1565–7. [DOI] [PubMed] [Google Scholar]
- 93. Jiang Y, Kang J, Wang X. RRAM-based parallel computing architecture using k-nearest neighbor classification for pattern recognition. Sci Rep 2017;7:45233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Basant N, Gupta S, Singh KP. Predicting the acute neurotoxicity of diverse organic solvents using probabilistic neural networks based QSTR modeling approaches. Neurotoxicology 2016;53:45–52. [DOI] [PubMed] [Google Scholar]
- 95. Han LY, Cai CZ, Ji ZL, et al. Predicting functional family of novel enzymes irrespective of sequence similarity: a statistical learning approach. Nucleic Acids Res 2004;32:6437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Karchin R, Karplus K, Haussler D. Classifying G-protein coupled receptors with support vector machines. Bioinformatics 2002;18:147–59. [DOI] [PubMed] [Google Scholar]
- 97. Dobson PD, Doig AJ. Distinguishing enzyme structures from non-enzymes without alignments. J Mol Biol 2003;330:771–83. [DOI] [PubMed] [Google Scholar]
- 98. Bock JR, Gough DA. Predicting protein–protein interactions from primary structure. Bioinformatics 2001;17:455–60. [DOI] [PubMed] [Google Scholar]
- 99. Eckle K, Schmidt-Hieber J. A comparison of deep networks with ReLU activation function and linear spline-type methods. Neural Netw 2019;110:232–42. [DOI] [PubMed] [Google Scholar]
- 100. Chen Y, Mai Y, Xiao J, et al. Improving the antinoise ability of DNNs via a bio-inspired noise adaptive activation function rand softplus. Neural Comput 2019;31:1215–33. [DOI] [PubMed] [Google Scholar]
- 101. Hamm CA, Wang CJ, Savic LJ, et al. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol 2019;29:3338–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kim J, Calhoun VD, Shim E, et al. Deep neural network with weight sparsity control and pre-training extracts hierarchical features and enhances classification performance: evidence from whole-brain resting-state functional connectivity patterns of schizophrenia. Neuroimage 2016;124:127–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sato M, Horie K, Hara A, et al. Application of deep learning to the classification of images from colposcopy. Oncol Lett 2018;15:3518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wang J, Yang B, An Y, et al. Systematic analysis and prediction of type IV secreted effector proteins by machine learning approaches. Brief Bioinform 2017; doi:10.1093/bib/bbx164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Cui X, Yang Q, Li B, et al. Assessing the effectiveness of direct data merging strategy in long-term and large-scale pharmacometabonomics. Front Pharmacol 2019;10:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Li XX, Yin J, Tang J, et al. Determining the balance between drug efficacy and safety by the network and biological system profile of its therapeutic target. Front Pharmacol 2018;9:1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


