Abstract
Glycosylation is an important post-translational modification of proteins. Many diseases, such as cancer have proved to be related to aberrant glycosylation. High throughput quantitative methods have gained attention recently in the study of glycomics. With the development of high-resolution mass spectrometry, the sensitivity of detection in glycomics has largely improved; however, most of the commonly used MS-based techniques are focused on relative quantitative analysis, which can hardly provide direct comparative glycomic quantitation results. In this study, we developed a novel multiplex glycomic analysis method on an LC-ESI-MS platform. Reduced glycans were stable isotopic labeled during the permethylation procedure, with the use of iodomethane reagents CH2DI, CHD2I, CD3I, 13CH3I, 13CH2DI, 13CHD2I, 13CD3I, and CH3I. Up to 8-plex glycomic profiling was possible in a single analysis by LC-MS, and a 100k mass resolution was sufficient to allow a baseline resolution of the mass differences among the 8-plex labeled glycans. The major advantages of this method are that it overcomes quantitative fluctuations caused by nanoESI, it facilitates a level of comparative quantitative glycomic analysis that accurately reflects the quantitative information in samples, and it dramatically shortens analysis time. Quantitation validation was tested on glycans released from bovine fetuin and model glycoprotein mixtures (RNase B, bovine fetuin, and IgG) with good linearity (R2=0.9884) and a dynamic range from 0.1 to 10. The 8-plex strategy was successfully applied to a comparative glycomic study of cancer cell lines. The results demonstrate that different distributions of sialylated glycans are related to the metastatic properties of cell lines and provide important clues for a better understanding of breast cancer brain metastasis.
Keywords: N-glycans, LC-MS/MS, Multiplex, Stable isotopic labeling, Permethylation
Graphical Abstract
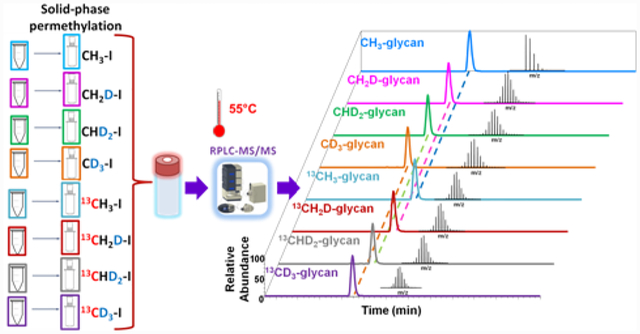
Introduction
Glycosylation is an important post-translational modification of proteins. Aberrant glycosylation has been proved to be related to many diseases such as cancer,1–4 neurodegenerative diseases,5 and immune disorder diseases.6,7 Glycans that are covalently attached to proteins participate in cell function and regulation, including cell-cell and cell-protein interactions.8,9 Glycomic profiling varies among individuals, both healthy and diseased, and it shows variance at different stages of disease development.10–13 The comparative quantitative method is a powerful tool to reveal connections between glycosylation and disease, and large numbers of samples are usually required for this purpose. Thus, the fast and efficient comparative quantitation of glycans is in great demand for clinical glycomic and disease pathogenic mechanism studies.
Mass Spectrometry (MS) based quantitation has recently become widely used in the glycomic analysis because of its high sensitivity and ability to elucidate complex glycan structures. However, MS quantitation is not as reliable as optical analysis methods. This is partly due to the limitations of MS ionization methods. Electrospray ionization (ESI) efficiency is affected by flow rate, solvent composition, sheath gas flow, and the properties of analytes.14,15 Also, the responsiveness of the instrument may fluctuate over time and among different injections.13 For nanoLC-ESI, in particular, the ionization efficiency may be largely influenced by the quality of spray. The variance among different sample injections can be as large as 30%.16 As for MALDI; ionization efficiency may vary with the spot layer thickness on the target plate, salt concentration in the samples, and the contact positions of the laser beam.16,17 In order to improve MS quantitation reliability, improvement of ionization efficiency, and reduction of the instrument’s variations are necessary. Multiplex quantitative analysis is a promising strategy that enables the elimination of instrument variance, allows reliable comparative glycomic studies; and shortening analysis time on the instrument, benefits analysis tasks with large numbers of samples.
Isobaric labeling, or tandem mass tag (TMT), is one of the most popular multiplex strategies used in the recent glycomic analysis. It was originally developed for proteomic quantitation.18 So far, only 6-plex AminoxyTMT has reported its use in the glycomic analysis.19 An advantage of TMT is that there is no retention time shifts among the six tags, and at full MS stage, labeled glycans with the same masses are isobaric; the full MS intensity is high since it is the sum of the six labeled glycans, and so sensitivity is improved. The comparative quantitation was achieved by reporter ions at the tandem MS stage. Advantages of TMT include its wide dynamic range (1:40) for MS1 along with its high throughput that allows for six glycomic samples to be analyzed simultaneously. One of the disadvantages of this method is the ratio distortion in MS2 quantitation. This problem can be solved to some extent by enhancing the yields of the reporter ions via adding sodium chloride during sample analysis,19 or by using advanced instruments to acquire high-resolution MS3 data for quantitation.20 Another issue associated with TMT is the inaccurate quantitation for glycans that contain sialylated or fucosylated structures. The quantitation results cannot truly reflect the amount of these kinds of glycans due to sialic acid loss or fucose migration during ionization and ion transmission. This problem is expected because AminoxyTMT labeling occurs only at the reducing end, and therefore cannot stabilize fragile glycan structures.21
Stable isotopic labeling is a category of multiplex methods for comparative glycomics. Stable isotopic labeling of glycans can be achieved by reductive amination derivatization.13 This type of isotopic labeling has been used in classical reductive reagents like 2-aminobenzoic acid (2-AA),22,23 aniline,24,25 and pyridine (PA)26 by introducing deuterium or 13C to the aromatic ring. However, the size of the labeling reagents limits the multiplexing capacity to 4-plex. Recently, a new strategy termed glycan reductive isotope-coded amino acid labeling (GRIAL) was developed by introducing isotopic arginine—Arg(12C6), Arg(13C6) and Arg(13C6, 15N4)—at the side chain NH2 group through reductive amination.27 Another new approach employed metallic element chelated tag labeling (MeCTL) at the reducing end. The label (p-NH2-Bn-DOTA) chelated with rare earth metal Lu/Ho achieved 3-plex glycomic analysis.28 Stable isotopes can also be introduced into glycan molecules during the enzymatic digestion process with 18O incorporation,29 or metabolically by introducing 15N to monosaccharides during cell culturing.16,30 However, the process is limited to only two samples at a time.
Another stable isotopic labeling strategy has been achieved through permethylation. Permethylation increases the sensitivity of glycan detection by significantly increasing the ionization efficiency of glycans, but also stabilize acidic glycan structures and eliminate the ionization difference between neutral and acidic glycan species.13,31,32 Orlando and colleagues reported the use of 13CH3I and CH3I permethylated glycans to facilitate the comparative quantitation analysis by MS.33 They also successfully used 13CH3I and 12CH2DI during permethylation in quantifying glycomic changes during early embryogenesis.15 At the same time, Mechref, Novotny, and coworkers introduced the use of CH3I and CD3I during permethylation (C-GlycoMAP).34 This method reduced measurement discrepancies on MALDI-MS and was proven to be reproducible and reliable in the N-glycomics of human blood serum for breast cancer comparative studies. Recently, Mechreef and co-workers employed CH2DI and CD3I permethylation in the glycomic study of patients with different esophageal diseases.35 However, the studies mentioned above were limited to only 2-plex in a single analysis by MS.
To enhance quantitative multiplexing while maintaining the advantages of the multiplex permethylation method, we investigated all possible combinations of isotope replacement in iodomethane molecules. We found that the use of iodomethane reagents CH3I, CH2DI, CHD2I, CD3I, 13CH3I, 13CH2DI, 13CHD2I, and 13CD3I enabled up to eight permethylation samples to be analyzed in a single run. In the current research, we used eight iodomethane reagents for model glycan permethylation, mixed with different ratios to test the reliability of the multiplex permethylation method. This method was then applied to the glycomic analysis of both breast cancer and brain cancer cell lines.
Material and Methods
Reagents
Ribonuclease B (RNase B) and Fetuin were purchased from Sigma-Aldrich (St. Louis, MO, USA). Intact mAb was acquired from Waters Corporation. PNGase F and G7 buffer (50mM sodium phosphate buffer, pH 7.5) were from New England Biolabs (Ipswich, MA, USA). Ammonium-borane complex, CH3I, CH2DI, CHD2I, CD3I, 13CH3I, 13CH2DI, 13CHD2I, 13CD3I iodomethane reagents, Dimethyl sulfoxide (DMSO), and NaOH beads were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ethanol was obtained from PHARMCO-AAPER. HPLC-grade methanol, Acetonitrile (ACN), Formic Acid, and HPLC water were obtained from Fisher Scientific (Fair Lawn, New Jersey, USA).
Sample Preparation
Cell lines were lysed with a homogenizer, and our routine protocol was employed to extract glycoproteins.36 N-glycans from model glycoproteins and cell lines were released by PNGase F digestion, as previously reported.35 Glycans were reduced with ammonia-borane complex. Next, 8-plex permethylation was performed using CH3I, CH2DI, CHD2I, CD3I, 13CH3I, 13CH2DI, 13CHD2I, and 13CD3I iodomethane reagents and using the solid-phase permethylation protocol, as previously reported.32,36–38 Detailed sample preparation protocols are described in the Supporting Information.
Instrument Method
The 8-plex permethylated N-glycans were dissolved in a 20% ACN solution containing 0.1% formic acid before LC-MS analysis. The separation was achieved with a 3000 nano-LC system (Dionex, Sunnyvale, CA, USA), under 55°C with optimized LC conditions,35,39,40 on a reversed phase Acclaim PepMap capillary column (150 mm× 75μm i.d.) packed with 100 Å C18 bounded phase (Dionex). The mobile phase A was 98% HPLC water, 2% ACN, 0.1% formic acid; the mobile phase B was 100% ACN, and 0.1% formic acid. The flow rate was 0.35 μl/min with a gradient elution of 20% mobile phase B over 10 minutes, then increased to 42% B (10–11min), 42% to 55% B (11–48min), 55% to 90% B (48–49min), 90% B (49–54min), 90% to 20% B (54–55min), 20% B (55–60min). Full MS spectra were obtained with LTQ Orbitrap Velos (Thermo Scientific, San Jose, CA, USA), in the mass range of 700–2000 m/z using positive ionization mode. The resolution of the instrument was set to 100,000 while mass accuracy was 5 ppm.
Data Processing and Quantitation Corrections by MultiGlycan-ESI Quantification
The raw files of the artificial mixtures of model glycoproteins and the mixture of cancer cell lines acquired on LC-MS/MS were imported to MultiGlycan software for quantitation. Glycan compositions were identified by matching the measured m/z value of the monoisotopic peaks, isotopic distributions, and retention times of all MS scans with the theoretical values in the default list built into the software. The isotope envelope tolerance was set to 6 ppm, and the mass tolerance of the monoisotopic peak was set to 5 ppm.41,42 Quantitation of the isotopically permethylated glycans was further corrected by activating the “multiplex permethylation” function in the software. A binomial distribution algorithm was used to calculate the theoretical distribution of ions with a specific composition. A series of calculated isotopic distributions with different numbers of unlabeled permethylation sites was compared to the measured MS scans to estimate the purity of the iodomethane reagents. The best purity estimations were applied as the correction factor to the quantitation obtained from the previous steps. Exported quantitation results of the glycan structures were provided by merging all charge states and adducts of the ions. The workflow of the software is described in Figure S1.
Results and Discussion
Principles of 8-plex Permethylation Quantitative Glycomics
8-plex permethylation is a stable isotopic labeling method that introduces mass differences to released glycans during the permethylation procedure using eight iodomethane reagents (CH2DI, CHD2I, CD3I, 13CH3I, 13CH2DI, 13CHD2I, 13CD3I, and CH3I). Then, the isotope-coded eight samples were mixed into one sample vial for LC-MS/MS analysis. The glycan structures with the same composition in all samples can be easily assigned in the full MS. The quantitation results were given by peak area or intensity under the peak in extracted ion chromatography (EIC), adding up all ion adducts and charge states of the glycan of interest. Since peak intensities or peak areas are compared directly among eight different samples without normalization, 8-plex permethylation strategy can reflect quantitation results closer to the true glycan abundance ratio in different samples that otherwise only absolute quantitation methods could provide. Also, it dramatically saves sample analysis time; eight samples typically require at least eight hours for analysis, not including the washing process after running each sample. However, the 8-plex permethylation method can analyze the same number of samples in only one hour.
The 8-plex strategy combined “large” (ΔM 1–4 Da/permethylation site) and “small” (ΔM 0.00293 Da/permethylation site) mass differences. Although the isotopic iodomethane reagents alone can provide a limited mass difference, the differences would be multiplied by the number of permethylation sites on the glycan molecule. The “large” mass differences pairs that have different nominal masses would be safely apart from each other in the MS spectra.The only possible overlapping may occurr between the “small” mass differences pairs (CH2DI and 13CH3I, CHD2I and 13CH2DI, or CD3I and 13CHD2I) that have the same nominal mass. However, with the mass differences enlarged by multiple sites, the MS resolution needed to separate those peaks with closer masses is attainable on an Orbitrap mass spectrometer. Table 1 shows the mass difference, and MS resolution needed to achieve a baseline separation of glycans that are permethylated with isobaric reagents. The mass difference depends on the number of permethylation sites a glycan molecule may have. For example, the smallest mass difference among the 8-plex analysis of Man 5 is 0.0703 Da because it has 24 permethylation sites, and the minimum resolution needed to differentiate its protonated ion [M+2H]2+ is approximately 23,404. The smallest mass difference among 8-plex analysis of HexNAc5Hex6NeuAc3 is 0.1489 Da, which has 51 permethylation sites and a required resolution of approximately 25,183. The resolving power of 100,000 on the LTQ Velos Orbitrap is needed to achieve the baseline separation. While setting resolving power at 60,000, the only partial separation was obtained, as shown in Figure S2. The MS spectra with a resolving power of 100,000 of 13CH3I and CH2DI permethylated HexNAc5Hex6NeuAc3 and MS of 13CHD2I and CD3I permethylated HexNAc5Hex6NeuAc3 are displayed in Figure S3a and Figure S3b.
Table 1.
Mass differences among isotopically permethylated glycans and MS resolutions needed to obtain a baseline mass separation.
 |
 |
 |
 |
|
|---|---|---|---|---|
| Number of permethylation sites | 24 | 26 | 29 | 51 |
| M[CH2D-glycan] | 1596.9754 | 1855.1302 | 2062.2488 | 3647.1446 |
| M[13CH3-glycan] | 1596.9051 | 1855.0540 | 2062.1639 | 3646.9957 |
| ΔM | 0.0703 | 0.0761 | 0.0849 | 0.1489 |
| M[CHD2-glycan] | 1621.1260 | 1881.2933 | 2091.4308 | 3698.4647 |
| M[13CH2D-glycan] | 1621.0557 | 1881.2172 | 2091.3459 | 3698.3157 |
| ΔM | 0.0703 | 0.0761 | 0.0849 | 0.1489 |
| M[CD3-glycan] | 1645.2766 | 1907.4565 | 2120.6127 | 3749.7847 |
| M[13CHD2-glycan] | 1645.2063 | 1907.3803 | 2120.5279 | 3749.6358 |
| ΔM | 0.0703 | 0.0761 | 0.0849 | 0.1489 |
| MS resolution | 23,404 | 25,065 | 24,978 | 25,183 |
Symbols:  , N-acetylglucosamine (GlcNAc);
, N-acetylglucosamine (GlcNAc);  , Galactose (Gal);
, Galactose (Gal);  , Fucose (Fuc);
, Fucose (Fuc);  , Mannose (Man);
, Mannose (Man);  , N-acetylneuraminic acid (NeuAc/Sialic Acid).
, N-acetylneuraminic acid (NeuAc/Sialic Acid).
8-plex Permethylation Analysis of N-glycans Derived from Model Glycoproteins
A standard glycoprotein mixture composed of 15 μg RNase B, 15 μg fetuin, and 10 μg IgG was digested in one vial to ensure uniform digestion efficiency. After reduction reaction, the reduced N-glycan mixtures of the three standard glycoproteins were distributed to eight equal aliquots and 8-plex permethylated with CH3I, CH2DI, CHD2I, CD3I, 13CH3I, 13CH2DI, 13CHD2I, and 13CD3I. Finally, the eight tagged portions were mixed in one vial with three different mixing ratios (1:1:1:1:1:1:1:1, 1:2:3:4:4:3:2:1, and 4:3:2:1:1:2:3:4) (Table S1) and analyzed on LC-ESI-MS. It has been well-documented that PNGase F digestion and glycan purification steps do not introduce significant differences to isotopically permethylated glycans.33–35 Thus, herein, we only investigated the possible effect of isotopic permethylation on qualification and quantitation.
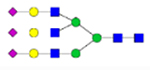
The EICs in Figure 1 indicated that different isotopic permethylation reagents could affect the retention of permethylated glycans to different extents. The deuterium replacement introduced by iodomethane reagents can reduce the hydrophobicity of the permethylated glycans and make them elute earlier. With the number of deuterium in the isotopic iodomethane increase, the hydrophobicity of the permethylated glycans would be further reduced. For example, the retention time of CH3I tagged Man 5 is 24.9 min, while CH2DI, CHD2I, and CD3I permethylated Man 5 are eluted at 24.2 min, 23.6 min, and 23.0 min respectively as shown in Figure 1a. The traces of fucosylated glycan (Figure 1b) and sialylated glycan (Figure 1c) attest to this result. Although the deuterium replacement in the iodomethane made retention times shift amongst the different isotopic permethylations, the biggest retention time shift in large glycan structure like HexNAc5Hex6NeuAc3 was 4 min. However, considering in traditional one-by-one sequential analysis, the 1st sample run and the 8th sample run can be 480 min apart, 8-plex is a big improvement in reducing the fluctuation over time. Also, it’s worth mentioning that thanks to the retention time shift by deuterium, even for the “small” mass differences pairs, the overlapping in MS (examples shown in Figure S2 and S3) was not substantial.
Figure 1.
EICs of 8-plex permethylated (a) Man 5; (b) fucosylated glycan; and (c) trisialylated triantennary glycan. The glycans were released from model glycoprotein mixture (RNaseB, fetuin, and IgG), 8-plex permethylated and mixing at a ratio of 1:1:1:1:1:1:1:1 before analysis on LC-MS. Symbols as in Table 1.
On the other hand, introducing 13C in permethylated glycans would not affect the retention. Compared to the retention time of CH3I permethylated Man 5, the retention time of the 13CH3I permethylated Man 5 remained the same at 24.9 min. Man 5 permethylated with CH2DI and 13CH2DI both eluted at 24.2 min. The traces of CHD2I and 13CHD2I permethylated Man 5, both with two deuterium atoms, also showed the same retention time of 23.6 min. The traces of CD3I and 13CD3I permethylated Man 5 with three deuterium atoms at one permethylation site again eluted at the same retention time of 23.0 min. This result was observed in other glycan species such as the fucosylated glycans (Figure 1b) and the sialylated glycans (Figure 1c). All the above demonstrated that 13C would not affect the retention time or hydrophobic property of permethylated glycans in a C18 column. This property is advantageous because glycans of the same compositions would elute at the same time, largely reducing the fluctuation of ionization due to nano-electrospray stability.
The Effect of Isotopic Iodomethane Purity on MS Quantitation and Solutions
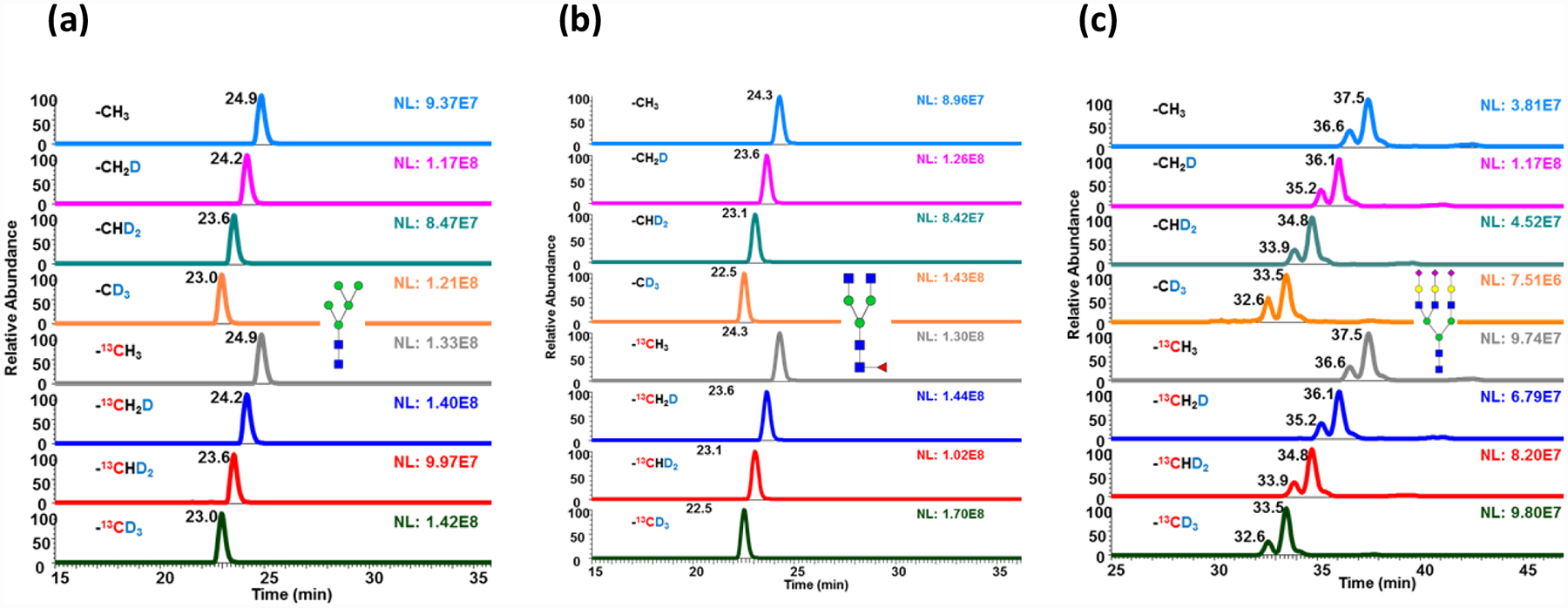
The monoisotopic peak shifts were observed in all iodomethane isotopically permethylated glycans, as examples shown in Figure 2a, 2b, 2c, and 2d. The impurity peak clusters in front of the theoretical monoisotopic peak were due to the small amount of 12C or 1H residues in the heavy iodomethane reagents. The relative intensities of the impurity peaks observed are associated with the purities of the isotopic iodomethane reagents. Compared to the 40% impurity peak abundance of the CH2DI permethylated glycan ions in Figure 2a, the 13CD3I tagged fucosylated glycan shows a relative impurity peak height around 20% (Figure 2b). Correspondingly, the merchandise provided purity of CH2DI is 98%, and 13CD3I is 99%. The MS spectrum of trisialylated glycan permethylated with CH2DI (Figure 2c) and 13CD3I (Figure 2d) also show the same thing. Also, the intensities of the impurity peaks may vary in the spectrum of different glycan compositions because the number of permethylation sites varies. For the CH2DI tagged fucosylated glycan, with 26 permethylation sites, the relative abundance of the highest impurity peak was around 40% (Figure 2a). While trisialylated triantennary glycan, with 51 permethylation sites (Figure 2c), the relative abundance of the highest impurity peak was 50%.
Figure 2.
MS spectra of (a) CH2DI permethylated fucosylated glycan, (b) 13CH3I permethylated fucosylated glycan, (c) CH2DI permethylated trisialylated triantennary glycan, and (d) 13CH3I permethylated trisialylated triantennary glycan. The glycans were released from model glycoprotein mixture (RNaseB, fetuin, and IgG), 8-plex permethylated and mixing at a ratio of 1:1:1:1:1:1:1:1 before analysis on LC-MS. Peaks with astarlikes indicate impurity peaks influenced by isotopic labeling iodomethane reagents purity. Symbols as in Table 1.
The impurity peaks must be taken into consideration because these ions were also ionized glycans. If not, the quantitation accuracy can be affected because the monoisotopic peak intensity or peak area in MS is used to quantitatively compare the abundances of glycans in the samples. Thus, the intensity of the completely heavy labeled permethylated glycans and the intensity of the partially heavy labeled permethylated glycans should be determined first, and then add them all together.
The correction principle is briefly described in Figure S4. Figure S4a is an example MS spectrum of a CH2DI permethylated fucosylated glycan. The arrow pointed peak represents the theoretical monoisotopic peak, which was calculated by assuming that all permethylated active hydrogen was derived into CH2D. The impurity peak clusters in front of the theoretical monoisotopic peak were the result of one or more permethylation sites unoccupied by CH2D. The distributions of all peaks in Figure S4a were depicted as blue columns in Figure S4b. Notice that each of the impurity peaks also has its isotopic distributions (inset in Figure S4b) and they are overlapped together. By subtracting all of the overlapped impurity peak clusters, the third subtraction, shown as yellow columns in Figure S4b, represent the 100% isotopically labeled ions. Quantitation corrections of the isotopic impurities were achieved by adding all of the subtracted partially isotopically permethylated ions together plus the 100% isotopically labeled ions. Thus far, MultiGlycan, as an efficient glycomics quantitation software, has developed an isotopic quantitation correction function for the permethylated glycans. The workflow of the software is shown in Figure S1.
Quantitation Validation of 8-plex Permethylation as a Reliable Glycomic Method
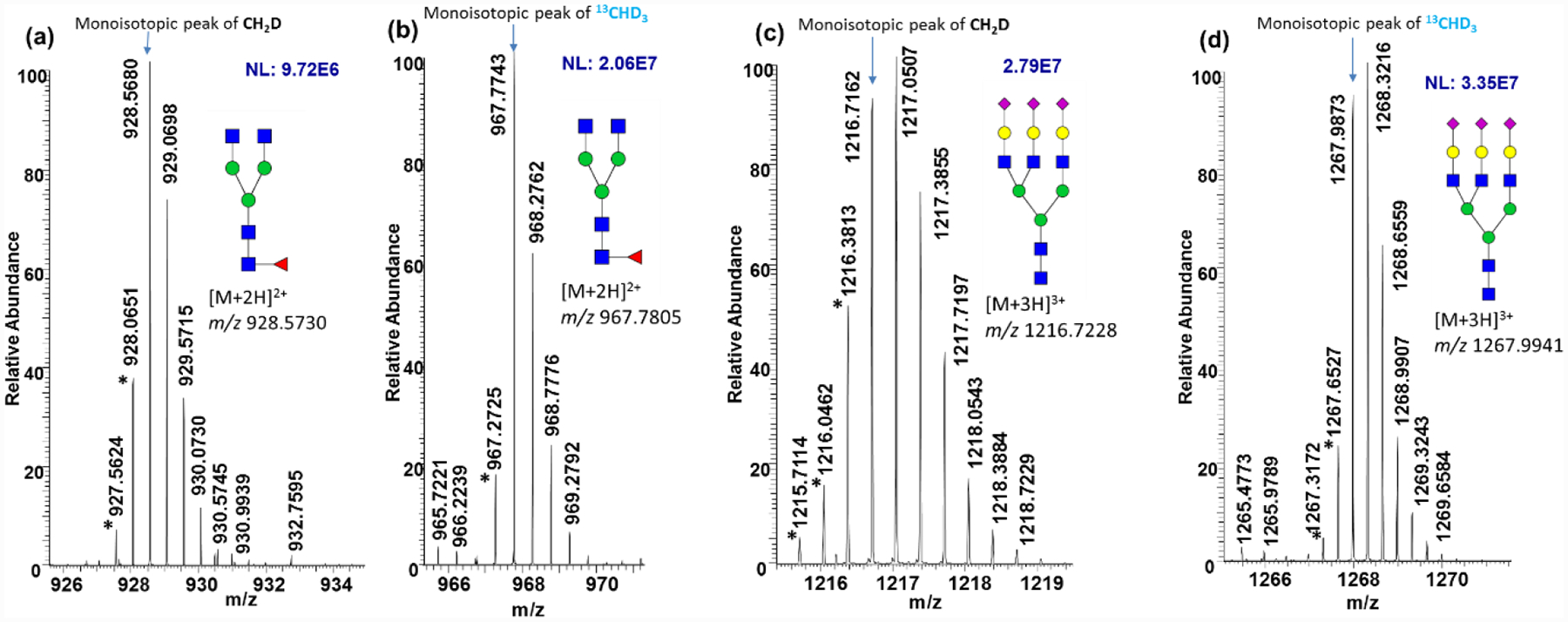
A mixture of N-glycans derived from RNase B, Fetuin, and IgG was utilized to validate the quantitation corrections. The N-glycans were released, reduced, 8-plex permethylated, and mixed at different ratios (1:1:1:1:1:1:1:1, 1:2:3:4:4:3:2:1, and 4:3:2:1:1:2:3:4) before injected on LC-MS/MS. Three preparations were analyzed for each mixing ratio. Figure 3a shows the EICs of Man 5 with a mixing ratio of eight reagents at 1:1:1:1:1:1:1:1. The peak heights reflect intensities. The bar graphs in Figure 3b show the uncorrected quantitation of Man 5. The heights of the column represent peak area added all adducts and the charge state of the ions under the corresponding peaks in Figure 3a. The peak heights in Figure 3a and the column heights in Figure 3b showed similar quantitation discrepancy among the eight reagents before corrections were conducted. After quantitation correction, the accuracy was improved, as shown in Figures 3c and 3d, presenting quantitation of Man 5 with theoretical mixing ratios 1:2:3:4:4:3:2:1 and 4:3:2:1:1:2:3:4. The experimental ratio of isotopic reagents vs. CH3 before and after corrections are shown as the “Experimental ratio” and the “Corrected ratio” respectively in Table S3. The relative errors to theoretical ratio before and after corrections are also summarized in Table S3. The linearity of the quantitation ratio was also improved with R2 > 0.9683. The specific linearities of different isotopic permethylations for various glycan structures are shown in Figure S5a.
Figure 3.
(a) Stacked EICs of 8-plex permethylated Man 5 with mixing ratio of 1:1:1:1:1:1:1; (b) Uncorrected quantitation bar graph of 8-plex permethylated Man 5 with mixing ratio 1:1:1:1:1:1:1; (c) Corrected quantitation bar graph of 8-plex Man 5 mixed with ratio of 1:2:3:4:4:3:2:1; (d) Corrected quantitation bar graph of 8-plex Man 5 mixed with ratio of 4:3:2:1:1:2:3:4. Man 5 was released from a glycoprotein mixture of RNaseB, fetuin, and IgG. (e) Variance plot of relative intensity before correction with an 8-plex mixing ratio of 1:1:1:1:1:1:1. (f) Variance plot of relative intensity after correction with an 8-plex mixing ratio of 1:1:1:1:1:1:1. Ix and I0 represent the intensity of isotopic and CH3I permethylated glycans. Dashed lines are the upper and lower 95% confidential limits. Glycan compositions of the x-axis are named according to the number of HexNAc, Hexose, Fucose, and Sialic acid in a structure.48 (Three sample preparation replicates were performed for each mixing ratio; Error bars stand for standard deviation.) Symbols as in Table 1.
Figure 3e presents the intensity variance of heavy permethylated glycans relative to CH3I permethylated glycans (Ix/I0) for all structures extracted from the standard protein mixture with mixing ratio 1:1:1:1:1:1:1:1. All the points theoretically should fall on the line of y=0; however, from the results without applying a correction (Figure 3e), we observed that all points wave around the y=0 line instead of deviating to one direction, which indicated that some variances were caused by instrument fluctuation over time. However, by focusing on the variance tendency of one particular isotopic permethylated glycan, we found that each isotopic reagent provided its fluctuation tendency, an effect that is even more obvious for glycans with more permethylation sites. This result demonstrated that part of the intensity variance came from isotopic reagent impurity. After correction, all points aligned to the y=0 line as expected (Figure 3f), which indicated that most variances were due to isotopic reagent impurity and validated our quantitation correction method.
Then, we utilized N-glycans derived from bovine fetuin to evaluate the quantitation dynamic range of the 8-plex permethylation method. CH3I was used as a reference, and the theoretical mixing ratios of 0.1, 0.5, 1, 5, and 10 (isotopic labeling reagents vs. CH3I) for each isotopic labeling reagent were investigated (Table S2). The experimental ratios acquired before and after quantitation correction are shown in Table S4. In the mixing ratio ranging from 0.1 to 10, glycans with relative abundances higher than 18% in the fetuin samples can be accurately quantified after correction conducted. The correlation and linearity of the experimental ratios after correction vs. the theoretical ratio for each isotopic reagent are plotted in Figure S5b. All isotopic reagents demonstrated good linearity in the mixing ratios, ranging from 0.1 to 10 with every R2 being better than 0.9990. In addition, the N-glycan profiles of 8-plex permethylation of each channel in glycoprotein mixtures were consistent with the results acquired from a pure glycoprotein (fetuin alone) analysis, as displayed in Figure S6.
Application of 8-plex Permethylation to Study Breast Cancer Brain Metastasis
The 8-plex permethylation was utilized on the glycomic analysis of five breast cancer cell lines (MDA-MB-231BR, MDA-MB-231, MDA-MB-361, HTB131, and HTB22) and one brain cancer cell line (CRL-1620) to investigate the mechanism of breast cancer brain metastasis. Each cell line has different specifications and metastatic properties (information related to these cell lines is available at the American Type Culture Collection website). Among them, the brain seeking cell line MDA-MB-231BR (231BR) is the sub-line of MDA-MB-231 (231).43 Cell line 231 was initially injected into the left heart ventricle of nude mice and the subsequent brain metastatic tumor cells were isolated and injected to other nude mice. After six repeated cycles, 231BR was generated, which had a 100% tendency of brain metastasis and could not migrate to any other organs.44,45 In contrast, 231 and other breast cancer cell lines can migrate to other organs such as bone, lungs, and ovaries, but the brain is not the first target. The brain metastatic capacity of 231BR makes it essential in the interpretation of brain metastasis. The modifications in 231BR may contribute to the penetration through the blood-brain barrier that facilitating brain metastasis. Also, aberrant glycosylation has been recently reported to be associated with brain metastasis.46 Thus, herein we considered 231BR as the baseline to compare with the other cell lines. The unique differential glycan expression of 231BR may provide clues to a deeper insight into breast cancer brain metastasis.
In the glycomic analysis of five breast cancer cell lines (361, 231BR, 231, HTB131, and HTB22) and one brain cancer cell line (CRL), released N-glycans were isotopically permethylated with CH3I, CH2DI, CHD2I, 13CH3I, 13CD3I, and CD3I, respectively (Only six cell lines were accessible in this study, thus 6-plex experiment was performed. The five isotopic iodomethane reagents with higher purities and the normal iodomethane were selected). Then, based on a protein assay of cell lysates, equal protein amounts of each 6-plex glycan sample were mixed in one vial for LC-MS analysis.
In total, 59 N-glycan structures were identified and quantified in 231BR. The relative abundances of the glycan structures from 231BR were used as a reference group to compare with the other cell lines. Overall 52, 45, 44, 34, and 33 structures were identified from 361, 231, CRL, HTB22, and HTB131, respectively. Among them, 19, 18, 20, 24, and 17 glycan expressions exhibited significant alteration compared to 231BR. The quantitation results of all glycans are listed in Table S5. Significant alterations can be observed among the cell lines. The EICs of HexNAc4Hex5Fuc1 in different cell lines is presented in Figure S7 as an example.
The N-glycan profiling of six cell lines with significant differences (p-value <0.05) are shown in Figure 4. Comparing the individual structures in different cell lines, highly sialylated glycans, such as HexNAc4Hex5DeoxyHex1NeuAc2 and HexNAc5Hex6DeoxyHex1NeuAc3, were observed to be overexpressed in 231BR (Figure 4). Because of the ultimate brain metastatic capacity of 231BR, the expression of these highly sialylated glycans may play important roles in the penetration of breast cancer cells through the blood-brain barrier, thus facilitating brain metastasis. Also, a clearer view was shown when the glycan structures were compared by categories. The distributions of different types of glycan structures among the six cell lines are shown in Figure S8. 231 and 231BR exhibit the highest percentage of sialylated structures while HTB22 show the least. Generally, 231 and 231BR are considered to be the most invasive brain cancer cell lines, while HTB22 is considered the least invasive.45,47 Moreover, the sialylation level in 231BR was even higher than in 231, which suggested that the sialylation level might contribute to breast cancer cell invasion and is even more essential to breast cancer brain metastasis.
Figure 4.
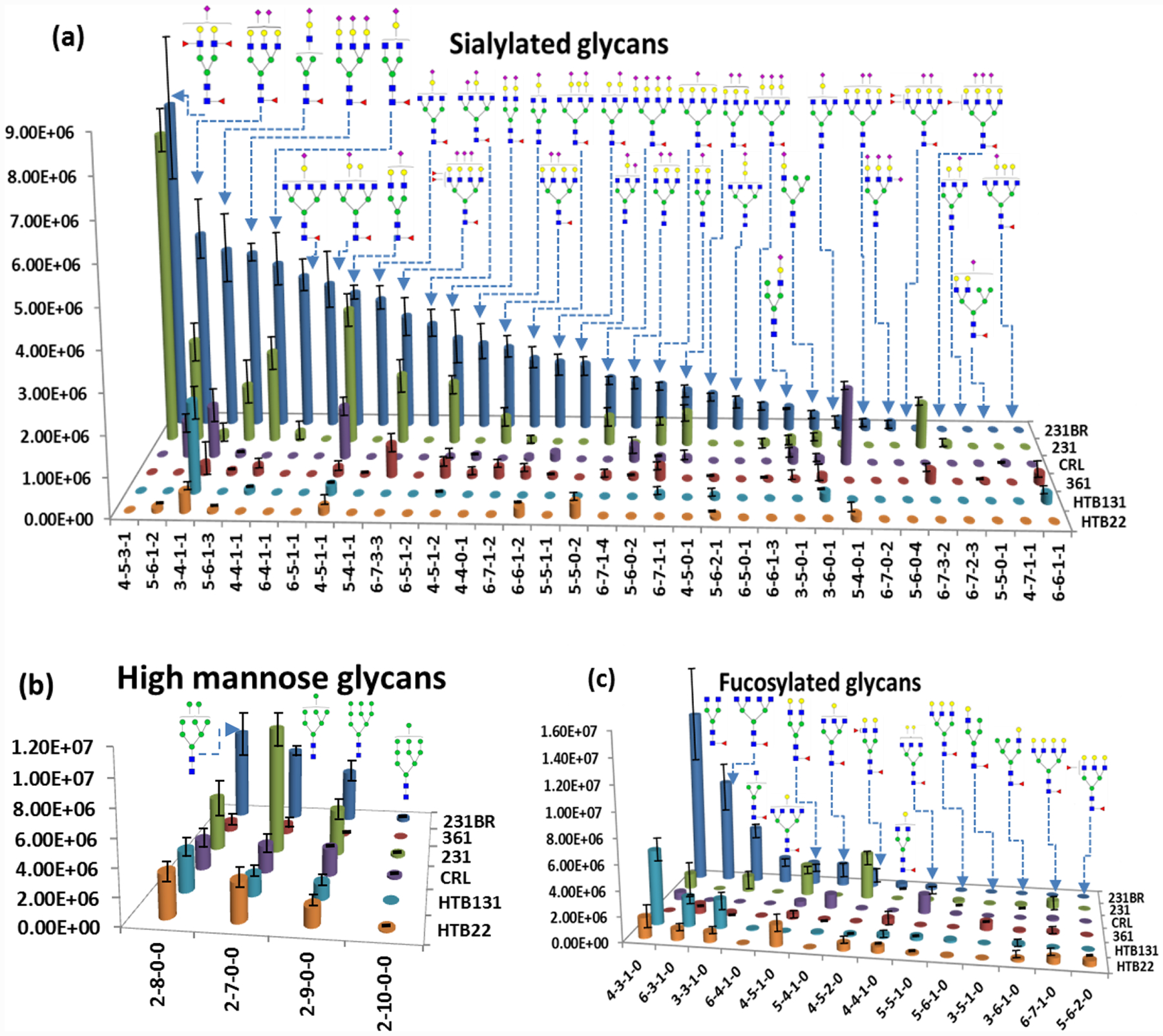
Quantitation bar graphs of glycan structures that are significantly different (P <0.05) in cancer cell lines (231BR, 231, 361, HTB131, and HTB22 and CRL). (Two sample preparation replicates were performed; Error bars stand for standard deviation.) Symbols as in Table 1. Naming of glycan compositions as in Figure 3.
Conclusion
In this study, we proved that the 8-plex permethylation is a valid comparative glycomic analysis approach. We observed that the inclusion of deuterium in the permethylated glycans could decrease the hydrophobicity of the molecules, but 13C has no such effect. We also found that the purities of isotopic iodomethane reagents would influence quantitation accuracy. However, mathematical corrections were achieved to guarantee quantitation reliability and can be included in the software to realize automatic data processing. We applied the 8-plex permethylation method in the study of cancer cell lines, which showed the capacity of utilizing this method to complex sample comparative glycomic analysis and proved the association of sialylated N-glycans with breast cancer brain metastasis.
Supplementary Material
Acknowledgments
This work was supported by an NIH grant (1R01GM112490-04)
Footnotes
Supporting Information
The supporting Information is available free of charge on the ACS Publications website at DOI:
Supplementary methods, Glycan reduction and solid-phase permethylation protocol; Table S1, protein amount of the model glycoprotein cocktail (RNase B: fetuin: IgG=1.5:1.5:1) in one injection. Numbers in brackets stand for specific protein species amount (RNase B, fetuin, and IgG); Table S2, protein amount of fetuin for different 8-plex mixing ratios in one injection; Table S3, experimental ratios of isotopic reagents vs. CH3I before and after quantitation correction for glycan mixture; Table S4, experimental ratios of isotopic reagents vs. CH3I before and after quantitation correction conducted for glycans from fetuin; Table S5, quantitation of 8-plex permethylated glycans derived from breast cancer cell lines (MDA-MB-231BR, MDA-MB-231, MDA-MB-361, HTB131, and HTB22) and brain cancer cell line (CRL-1620); Figure S1, processing workflow of MultiGlycan; Figure S2, MS spectra of CH2DI and 13CH3I permethylated Man 5, spectra acquired at a resolution of (a) 60k and (b) 100k; Figure S3, MS spectrum of HexNAc5Hex6NeuAc3 derivatized with (a) 13CH3I and CH2DI, and (b) 13CHD2I and CD3I; Figure S4, (a) MS isotopic peak distributions of CH2DI permethylated fucosylated glycan. (b) Principle of correcting quantitative error caused by the purity of isotopic iodomethane reagents. Inset is the theoretical isotopic distribution of CH2DI permethylated fucosylated glycan; Figure S5, (a) Plots of experimental ratio after correction against theoretical mixing ratio. The glycans were released from model glycoprotein mixture (RNaseB, fetuin and IgG), 8-plex permethylated and mixing at ratio of 1:1:1:1:1:1:1:1, 1:2:3:4:4:3:2:1, and 4:3:2:1:1:2:3:4 before analysis on LC-MS. (b) Plots of experimental ratio after correction against theoretical mixing ratio. Glycans were released from fetuin, 8-plex permethylated, and mixed at ratios (Reagent vs. CH3I) of 0.1, 0.5, 1, 5, and 10 in an injection; Figure S6, bar graph of relative abundances of N-glycans in fetuin (3 μg) vs. in glycoprotein mixture (RNase B: fetuin: IgG=1.5:1.5:1); Figure S7, (a) EICs of multiplex permethylated HexNAc4Hex5Fuc1. (b) EICs of multiplex permethylated Man 8 derived from breast cancer cell lines (MDA-MB-231BR, MDA-MB-231, MDA-MB-361, HTB131, and HTB22) and brain cancer cell line (CRL-1620); Figure S8, quantitative distribution of different types of N-glycans in cell lines as shown in pie charts (a) cell line 361, (b) 231, (c) CRL, (d) HTB 131, (e) HTB 22, and (f) 231 BR. (PDF)
The authors declare no conflict of interest.
Research data used in this article are available from the corresponding author on request.
References
- (1).Abd Hamid UM; Royle L; Saldova R; Radcliffe CM; Harvey DJ; Storr SJ; Pardo M; Antrobus R; Chapman CJ; Zitzmann N; Robertson JF; Dwek RA; Rudd PM Glycobiology 2008, 18, 1105–1118. [DOI] [PubMed] [Google Scholar]
- (2).Meany DL; Chan DW Clin. Proteom 2011, 8, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wooding KM; Peng W; Mechref Y Curr. Pharm. Biotechnol 2016, 17, 788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Veillon L; Fakih C; Abou-El-Hassan H; Kobeissy F; Mechref Y ACS Chem Neurosci 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Abou-Abbass H; Abou-El-Hassan H; Bahmad H; Zibara K; Zebian A; Youssef R; Ismail J; Zhu R; Zhou S; Dong X; Nasser M; Bahmad M; Darwish H; Mechref Y; Kobeissy F Electrophoresis 2016, 37, 1549–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Veillon L; Huang Y; Peng W; Dong X; Cho BG; Mechref Y Electrophoresis 2017, 38, 2100–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Goulabchand R; Vincent T; Batteux F; Eliaou JF; Guilpain P Autoimmun. Rev 2014, 13, 742–750. [DOI] [PubMed] [Google Scholar]
- (8).Zhu R; Zacharias L; Wooding KM; Peng W; Mechref Y Methods Enzymol. 2017, 585, 397–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Varki A; Kannagi R; Toole BP In Essentials of Glycobiology, A V, Ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor (NY), 2009. [Google Scholar]
- (10).Adamczyk B; Tharmalingam T; Rudd PM Biochim. Biophys. Acta 2012, 1820, 1347–1353. [DOI] [PubMed] [Google Scholar]
- (11).Mechref Y; Hu Y; Garcia A; Hussein A Electrophoresis 2012, 33, 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Tsai TH; Wang M; Di Poto C; Hu Y; Zhou S; Zhao Y; Varghese RS; Luo Y; Tadesse MG; Ziada DH; Desai CS; Shetty K; Mechref Y; Ressom HW J. Proteome Res 2014, 13, 4859–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Mechref Y; Hu Y; Desantos-Garcia JL; Hussein A; Tang H Mol Cell Proteomics 2013, 12, 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Page JS; Kelly RT; Tang K; Smith RD J Am Soc Mass Spectrom 2007, 18, 1582–1590. [DOI] [PubMed] [Google Scholar]
- (15).Atwood JA 3rd; Cheng L; Alvarez-Manilla G; Warren NL; York WS; Orlando R J Proteome Res 2008, 7, 367–374. [DOI] [PubMed] [Google Scholar]
- (16).Zhou S; Tello N; Harvey A; Boyes B; Orlando R; Mechref Y Electrophoresis 2016, 37, 1489–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chen H; He M Journal of the American Society for Mass Spectrometry 2005, 16, 100–106. [DOI] [PubMed] [Google Scholar]
- (18).Huang F-K; Zhang G; Lawlor K; Nazarian A; Philip J; Tempst P; Dephoure N; Neubert TA J. Proteome Res 2017, 16, 1121–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhou S; Hu Y; Veillon L; Snovida SI; Rogers JC; Saba J; Mechref Y Anal. Chem 2016, 88, 7515–7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Ting L; Rad R; Gygi SP; Haas W Nat. Methods 2011, 8, 937–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Zhou S; Veillon L; Dong X; Huang Y; Mechref Y Analyst 2017, 142, 4446–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Hitchcock AM; Costello CE; Zaia J Biochemistry 2006, 45, 2350–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Hitchcock AM; Yates KE; Shortkroff S; Costello CE; Zaia J Glycobiology 2007, 17, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lawrence R; Olson SK; Steele RE; Wang L; Warrior R; Cummings RD; Esko JD J. Biol. Chem 2008, 283, 33674–33684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Xia B; Feasley CL; Sachdev GP; Smith DF; Cummings RD Anal. Biochem 2009, 387, 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Yuan J; Hashii N; Kawasaki N; Itoh S; Kawanishi T; Hayakawa TJ Chromatogr. A 2005, 1067, 145–152. [DOI] [PubMed] [Google Scholar]
- (27).Cai Y; Jiao J; Bin Z; Zhang Y; Yang P; Lu H Chem. Commun 2015, 51, 772–775. [DOI] [PubMed] [Google Scholar]
- (28).Yang L; Peng Y; Jiao J; Tao T; Yao J; Zhang Y; Lu H Anal. Chem 2017, 89, 7470–7476. [DOI] [PubMed] [Google Scholar]
- (29).Zhang W; Wang H; Tang H; Yang P Anal. Chem 2011, 83, 4975–4981. [DOI] [PubMed] [Google Scholar]
- (30).Orlando R; Lim JM; Atwood JA 3rd; Angel PM; Fang M; Aoki K; Alvarez-Manilla G; Moremen KW; York WS; Tiemeyer M; Pierce M; Dalton S; Wells LJ Proteome Res. 2009, 8, 3816–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Dong X; Zhou S; Mechref Y Electrophoresis 2016, 37, 1532–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Abou-Abbass H; Bahmad H; Abou-El-Hassan H; Zhu R; Zhou S; Dong X; Hamade E; Mallah K; Zebian A; Ramadan N; Mondello S; Fares J; Comair Y; Atweh S; Darwish H; Zibara K; Mechref Y; Kobeissy F Electrophoresis 2016, 37, 1562–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Alvarez-Manilla G; Warren NL; Abney T; Atwood J 3rd; Azadi P; York WS; Pierce M; Orlando R Glycobiology 2007, 17, 677–687. [DOI] [PubMed] [Google Scholar]
- (34).Kang P; Mechref Y; Kyselova Z; Goetz JA; Novotny MV Anal. Chem 2007, 79, 6064–6073. [DOI] [PubMed] [Google Scholar]
- (35).Hu Y; Desantos-Garcia JL; Mechref Y Rapid Commun. Mass Spectrom 2013, 27, 865–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Zhou S; Huang Y; Dong X; Peng W; Veillon L; Kitagawa DAS; Aquino AJA; Mechref Y Anal. Chem 2017, 89, 6590–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kang P; Mechref Y; Klouckova I; Novotny MV Rapid Commun. Mass Spectrom 2005, 19, 3421–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Mechref Y; Kang P; Novotny MV Methods Mol. Biol 2009, 534, 53–64. [DOI] [PubMed] [Google Scholar]
- (39).Hu Y; Mechref Y Electrophoresis 2012, 33, 1768–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zhou S; Hu Y; Mechref Y Electrophoresis 2016, 37, 1506–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Hu Y; Zhou S; Yu CY; Tang H; Mechref Y Rapid Commun. Mass Spectrom 2015, 29, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Yu CY; Mayampurath A; Hu Y; Zhou S; Mechref Y; Tang H Bioinformatics 2013, 29, 1706–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Gupta P; Adkins C; Lockman P; Srivastava SK PLoS One 2013, 8, e67278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Yoneda T; Williams PJ; Hiraga T; Niewolna M; Nishimura RJ Bone Miner Res. 2001, 16(8), 1486–1495. [DOI] [PubMed] [Google Scholar]
- (45).Peng W; Zhang Y; Zhu R; Mechref Y Electrophoresis 2017, 38, 2124–2134. [DOI] [PubMed] [Google Scholar]
- (46).Bos PD; Zhang XH; Nadal C; Shu W; Gomis RR; Nguyen DX; Minn AJ; van de Vijver MJ; Gerald WL; Foekens JA; Massague J Nature 2009, 459, 1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Chen W; Smeekens JM; Wu R Chem. Sci 2015, 6, 4681–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Huang Y; Zhou S; Zhu J; Lubman DM; Mechref Y Electrophoresis 2017, 38, 2160–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


