Abstract
Plants can produce and emit nitrous oxide (N2O), a potent greenhouse gas, into the atmosphere, and several field-based studies have concluded that this gas is emitted at substantial amounts. However, the exact mechanisms of N2O production in plant cells are unknown. Several studies have hypothesised that plants might act as a medium to transport N2O produced by soil-inhabiting microorganisms. Contrarily, aseptically grown plants and axenic algal cells supplied with nitrate (NO3) are reported to emit N2O, indicating that it is produced inside plant cells by some unknown physiological phenomena. In this study, the possible sites, mechanisms, and enzymes involved in N2O production in plant cells are discussed. Based on the experimental evidence from various studies, we determined that N2O can be produced from nitric oxide (NO) in the mitochondria of plants. NO, a signaling molecule, is produced through oxidative and reductive pathways in eukaryotic cells. During hypoxia and anoxia, NO3 in the cytosol is metabolised to produce nitrite (NO2), which is reduced to form NO via the reductive pathway in the mitochondria. Under low oxygen condition, NO formed in the mitochondria is further reduced to N2O by the reduced form of cytochrome c oxidase (CcO). This pathway is active only when cells experience hypoxia or anoxia, and it may be involved in N2O formation in plants and soil-dwelling animals, as reported previously by several studies. NO can be toxic at a high concentration. Therefore, the reduction of NO to N2O in the mitochondria might protect the integrity of the mitochondria, and thus, protect the cell from the toxicity of NO accumulation under hypoxia and anoxia. As NO3 is a major source of nitrogen for plants and all plants may experience hypoxic and anoxic conditions owing to soil environmental factors, a significant global biogenic source of N2O may be its formation in plants via the proposed pathway.
Keywords: anoxia, hypoxia, nitrate, nitrite, nitric oxide, nitrous oxide, mitochondrion
Introduction
Nitrous oxide (N2O) is a potent greenhouse gas, and its potential to increase global warming is approximately 300-fold higher than CO2 (Tian et al., 2018). Globally, the primary sources that release N2O into the atmosphere are soil, ocean, manure application, industries, and biomass burning (Thomson et al., 2012). The nitrification and denitrification processes, mainly mediated by certain groups of soil micro-organisms (Hu et al., 2015), account for more than two-thirds of its emission into the atmosphere (Thomson et al., 2012). These processes are considerably increased by human activities, leading to an increase in the production of N2O in the soil, and thus, its concentration in the atmosphere. Increased level of N2O in the atmosphere has significantly contributed to global warming (Tian et al., 2018); therefore, understanding the pathways of N2O formation in various sources is essential for mitigating these effects.
Key pathways involved in N2O production in microbes include nitrification, nitrifier denitrification, nitrification-coupled denitrification, and denitrification (Baggs, 2011; Hu et al., 2015; Tian et al., 2018). However, there seems to be a gap between source estimation and the global N2O budget, leading to a high level of uncertainty in the budget estimations (Davidson and Kanter, 2014). This gap may be because not all sources of N2O to the atmosphere are accounted for (Syakila and Kroeze, 2011). Therefore, it is necessary to understand all sources of N2O and underlying mechanisms to elucidate its global budget. The production of N2O in axenic microalgae (Weathers, 1984; Weathers and Niedzielski, 1986; Guieysse et al., 2013; Plouviez et al., 2017) and ascetically grown plants (Goshima et al., 1999; Hakata et al., 2003) indicates that it could be produced by higher organisms and that the processes might be different from those in micro-organisms. Algae and plants are not included as sources of N2O (Syakila and Kroeze, 2011; Lenhart et al., 2019; Plouviez et al., 2019), but they might be the missing sources of N2O, causing high uncertainties in the global budget.
The roles of plants in N2O emission to the atmosphere are diverse. Plants can not only modify soil characteristics and subsequently influence N2O production in the soil (Gao et al., 2019) but also produce it in significant amounts and release it to the atmosphere (Lenhart et al., 2019). Thus, understanding the pathway of N2O formation and the contribution of plants to total emission is essential to accurately estimate the global N2O budget. Several field-based studies have hypothesised that N2O emitted by plants is produced in soil by microorganisms (Chang et al., 1998; Rusch and Rennenberg, 1998; Machacova et al., 2013; Bowatte et al., 2014; Wen et al., 2017). In this theory, plants are considered just a medium to transport N2O produced by soil microorganisms; however, laboratory-based studies have provided clear evidence that plants produce and emit N2O although the underlying mechanisms are unknown (Goshima et al., 1999; Hakata et al., 2003; Lenhart et al., 2019). Therefore, N2O emitted by plants might originate from two sources, namely, the soil microorganisms and plants.
Studies, which have hypothesised that plant-emitted N2O is produced by soil microorganisms, have only measured the fluxes from plants and concluded that plant-emitted N2O might be produced by soil microorganisms. N2O produced in plant cells might also use the same pathway, that is, transpiration, to release it to the atmosphere. This raises the question whether measuring the fluxes alone provides substantial evidence to prove the hypothesis, because flux measurement methods can just estimate the emission of N2O and cannot distinguish the sources. More robust methods such as isotope studies would provide more insights to distinguish the sources of N2O. For examples, injecting 15N-N2O into the root zone and measuring the subsequent fluxes would elucidate whether plants are a medium for N2O transport or not. However, no study has injected 15N-labeled N2O into the soil zone and measured the subsequent N2O emission from plants. Moreover, more powerful tools such as site preference (SP) measurement would provide insights to distinguish the sources of N2O emitted by plants under field conditions.
In the natural environment, if plant emitted N2O constitute significant amount of both sources (soil micro-organisms and plant cells produced N2O), it will be highly challenging to distinguish the portion of the sources. A recent field experiment reported considerably lower N2O concentrations in soil water than in tree stems (Ward et al., 2019). Similarly, plants exposed to NH4 did not emit N2O despite the high rate of N2O production in the rhizosphere (Smart and Bloom, 2001), indicating that N2O emitted by plants might not be produced by soil microorganisms and that N2O emitted through transpiration might be a less significant process than N2O production in plants. Furthermore, the hypothesis that plants are just a conduit for soil microorganisms-produced N2O is not supported by a recent study of Lenhart et al. (2019). They provided new evidence that dual isotopocule fingerprints of N2O emitted by plants differed from that produced by all known microbial or chemical processes, indicating that plant-emitted N2O is produced in plant cells.
Although plants are known to produce N2O and emit it to the atmosphere, the exact mechanisms of N2O production in plant cells are unknown (Goshima et al., 1999; Hakata et al., 2003; Lenhart et al., 2019). This might be the reason that most studies on N2O fluxes in plants (Chang et al., 1998; Rusch and Rennenberg, 1998; Machacova et al., 2013; Bowatte et al., 2014; Wen et al., 2017) have hypothesised that plant parts act as a conduit for soil-produced N2O. Studies, which have claimed that plants could produce N2O, have not elucidated a possible production pathway. Therefore, the main objective of this study was to review the possible pathway of N2O formation in plant cells.
Pathway of N2O Formation in Plant Cells
Nitrate (NO3) as a Precursor for N2O Formation in Plant Cells
Nitrogen (N) is an essential macronutrient influencing cell metabolism (O’Brien et al., 2016). Plants can use several forms of N from the soil; however, NO3 and ammonium (NH4) are the major forms of inorganic N that are readily available for plant uptake (O’Brien et al., 2016; Hachiya and Sakakibara, 2017). NO3 is a major source of N for plants in agricultural and natural soils (von Wirén et al., 2000), due to its high soil concentration and diffusion coefficients, making it readily available to plant roots (Miller and Cramer, 2005). After absorption, NO3 is directly reduced in the root, transported to the leaf for reduction (Maathuis, 2009; Hachiya and Sakakibara, 2017), or stored in the vacuoles and remobilised when the external supply is limited (van der Leij, et al., 1998; Fan et al., 2007), making it an essential macronutrient in plant metabolism.
NO3 is a major source for N2O formation in both soil (Thomson et al., 2012) and plants (Goshima et al., 1999; Smart and Bloom, 2001; Hakata et al., 2003; Lenhart et al., 2015; Lenhart et al., 2019). Isotope labeling methods have demonstrated that plants as well as other eukaryotic organisms emit N2O, only when supplied with NO3. For example, when 15N-labeled NO3 was supplied as a source of N to various species of plants (Goshima et al., 1999; Smart and Bloom, 2001; Lenhart et al., 2019), lichens (Lenhart et al., 2015), and animals (Stief et al., 2009), 15N-labeled N2O was emitted, but when the N source was 15N-labeled NH4, there was no N2O emission. This evidence clearly shows that NO3 is the precursor of N2O in lichens, higher plants, and animals. Therefore, if plants were just a medium of transportation of soil-produced N2O as hypothesised by many studies (Chang et al., 1998; Rusch and Rennenberg, 1998; Machacova et al., 2013; Bowatte et al., 2014; Wen et al., 2017), aseptically grown plants would not have emitted N2O when supplied with NO3 (Goshima et al., 1999; Hakata et al., 2003). Similarly, if N2O emitted by plants is produced by microorganisms (nitrifying and denitrifying bacteria), NH4 supplementation should contribute to N2O emission from plants and aseptically grown plants should not emit N2O. As N2O was not emitted when plants were supplied with NH4 and aseptically grown plants emitted N2O (Goshima et al., 1999; Hakata et al., 2003), we predicted that NO3 metabolism in a cell might play a role in N2O formation in plants. Moreover, the processes in the soil microbial communities and higher organisms may be different, or the denitrification process may be common between microorganism and plants, as NO3 is the substrate for denitrification.
Nitrite (NO2) Derived From NO3 Reduction Is the Precursor of N2O in Plant Cells
After the uptake of NO3 by plant roots, it is reduced to NO2 in plant cells by a cytosolic enzyme called nitrate reductase (NR) (Chamizo-Ampudia et al., 2017). However, in animals, NO3 from food is reduced by bacteria in the digestive tracks (Lundberg et al., 2008). Moreover, germ-free mice are reported to possess NR activity, and the activity is catalysed by xanthine oxidoreductase, which is significantly high in the gastrointestinal tissues, compared with other tissues (Jansson et al., 2008). Under normal conditions, NO3 absorbed by the roots is reduced to NO2 by the NR, and then nitrite reductase (NiR) catalyses the reduction of NO2 to NH4, which is incorporated into amino acids (Oliveira and Sodek, 2013; Plouviez et al., 2017). However, under hypoxic and anoxic conditions, root NO3 uptake increases with the activation level of NR (Botrel and Kaiser, 1997; Rockel et al., 2002; Morard et al., 2004; Horchani et al., 2010). Furthermore, NO2 accumulates in the cytoplasm of cells (Allègre et al., 2004; Morard et al., 2004), as both hypoxia and anoxia suppress the reduction of NO2 to NH4 (Botrel and Kaiser, 1997). A 15N isotope labeling study has showed that 15N-NO2 assimilation into amino acids is sharply reduced under hypoxic conditions (Oliveira and Sodek, 2013). The accumulated NO2 in the cytoplasm enters the mitochondria with the help of proteins in the chloroplast (Sugiura et al., 2007; Gupta and Igamberdiev, 2011). Moreover, mitochondrial inner membrane anion channels may import NO2 to the mitochondria (Gupta and Igamberdiev, 2011).
Not only NO3, but also NO2 is widely reported to be a precursor of N2O in eukaryotic organisms. Using 15N isotope labeling method, it has been demonstrated that NO2 is another precursor of N2O formation in plants and algal cells. For example, when 15N-labeled NO2 was supplied to aseptically grown tobacco plants (Goshima et al., 1999; Hakata et al., 2003) and algal systems (Weathers, 1984), they emitted 15N-N2O. Furthermore, axenic algae supplied with NO2 produced N2O (Guieysse et al., 2013; Plouviez et al., 2017). The enzyme NR has been proved to play a role in N2O production in plants. For example, when tobacco plants were supplied with NO3 and tungstate (NR inhibitor), N2O production was inhibited in the plants (Goshima et al., 1999). As NO2 also contributes to N2O production in plants (Goshima et al., 1999; Hakata et al., 2003), NR might indirectly be involved in N2O formation by catalysing the reduction of NO3 to NO2. A similar role of NR has been observed in algae (Plouviez et al., 2017). However, NiR-deficient transgenic plants and algae have been reported to produce N2O when supplied with NO2 (Hakata et al., 2003; Plouviez et al., 2017), and this suggests that the pathway of NO2 reduction to NH4 is not involved in N2O production in plants and algal cells. Additionally, NO3, NR, and NO2 are involved in N2O production in plants and algal cells, but NiR and NH4 are not involved in the N2O production pathway. This indicates that NO2 has to be transported to other cell organelles rather than plastid. Overall, the available evidence indicates that exogenous NO2 along with endogenous NO2 derived through NO3 reduction in the cytosol by NR plays a role in N2O formation in plant cells.
Mitochondrial Reduction of Nitrite to NO
We previously discussed NO3 reduction to NO2 in the cytosol. NO has several essential roles in plant and animal cells (Wendehenne et al., 2001), and the conversion mechanisms of NO2 to NO are well established in eukaryotic cells. In plants, NO can be produced in the chloroplast, peroxisomes, and mitochondria by either oxidative or reductive pathways (Rőszer, 2012). The oxidative pathway is dependent on L-arginine, polyamine, or hydroxylamine, whereas the reductive pathway is dependent on NO3 and NO2 (Benamar et al., 2008; Lundberg et al., 2008; Gupta et al., 2011; Gupta and Igamberdiev, 2011; Astier et al., 2018). The oxidative pathway of NO formation is dominant when the oxygen supply to cells is sufficient, whereas the reductive path is dominant under hypoxic conditions. By shifting from the oxidative to reductive pathway, the cells maintain the level of NO along with the physiological and pathological oxygen and proton gradients (Lundberg et al., 2008). It may be essential to shift processes, as plants may experience hypoxia due to soil environmental conditions.
In plant cells, NO2 assimilation to NH4 by the NiR enzyme is a well-known pathway of NO2 metabolism. As NO2 addition can lead to N2O formation in plants (Goshima et al., 1999; Hakata et al., 2003) and NiR-deficient plants can produce N2O (Hakata et al., 2003), we suggest that NO2 is metabolised by another pathway in plants to produce N2O. Although the mechanisms of NO2 transport to the mitochondria are not precise, it is evident that the mitochondria are a site of reduction of NO2 to NO. For example, the mitochondria have been reported to reduce NO2 to NO under hypoxic and anoxic conditions in fungi (Kobayashi et al., 1996; Castello et al., 2006), algae (Tischner et al., 2004; Calatrava et al., 2017), plants (Gupta et al., 2005; Planchet et al., 2005; Benamar et al., 2008; Gupta and Kaiser, 2010), and animals (Ghafourifar and Richter, 1997; Giulivi et al., 1998; Kozlov et al., 1999; Castello et al., 2006; Ascenzi et al., 2014). However, the enzymes involved in the mitochondrial reduction of NO2 to NO are not clear. For example, mitochondria that lack NiR can reduce NO2 to NO in animals and plants (Gupta and Igamberdiev, 2011). Nitric oxide synthases (NOS) have been reported to be present in plant (Guo and Crawford, 2005) and animal mitochondria (Giulivi et al., 1998). However, the NOS activity in the mitochondria of plants is questioned (Moreau et al., 2008; Gupta and Kaiser, 2010). Tischner et al. (2004) identified an alternative oxidase (AOX) in the mitochondria as a catalyser of the reduction of NO2 to NO under anoxic conditions. The mitochondrial respiratory chain is responsible for NO production using NO2 as the substrate under low pH, hypoxic, or anoxic conditions (Castello et al., 2006). Mitochondrial and bacterial electron transport chains (ETCs) are involved in NO production from NO2 under hypoxic conditions than under normoxic conditions (Horchani et al., 2011). Under hypoxic conditions, NO2 is reduced to NO at complex III in the mitochondria of pea plants (Benamar et al., 2008). Ascenzi et al. (2014) reported cytochrome c in horse heart cells and bovine heart reduced NO2 to NO, and the activity was high under anoxic and acidic conditions (Basu et al., 2008). The mitochondrial molybdopterin enzymes in the reduced form catalyse the reduction of NO2 to NO, and the rate was increased when the pH was decreased from 7.5 to 6.5 (Jakobs et al., 2014; Sparacino-Watkins et al., 2014; Maia and Moura, 2015; Bender and Schwarz, 2018). Furthermore, cytochrome reductase in tobacco plants can reduce NO2 to NO (Alber et al., 2017). Although at the molecular level, the reductive pathway for NO formation is well documented, at the field scale, the emission of NO is less documented. For instance, when plants were supplied with NO3, NO was emitted under anoxic conditions (Klepper, 1987; Rockel et al., 2002). The leaf NO2 level and NO emission under anoxic conditions were significantly higher than those under normoxic conditions (Rockel et al., 2002). These findings suggest that NO2 can be reduced to NO in the mitochondria; however, the involvement of various enzymes within the mitochondria raises the question whether these enzymes catalyse the reduction process simultaneously or they function differently under varied cell environment.
NO Conversion to N2O in the Mitochondria
NO is a signaling molecule in cells, and several studies have focused on its formation in the mitochondria. However, studies on the reduction of NO to N2O in the mitochondria are limited, although there is a strong indication that this process exists (Gupta and Igamberdiev, 2011). The inner membrane of the mitochondria has an enzyme called cytochrome c oxidase (CcO). The primary function of CcO is to reduce O2 to H2O (Collman et al., 2007; Blomberg and Ädelroth, 2018). Moreover, CcO has several other functions, such as the oxidisation of NO formed in the mitochondria to NO2 (Brudvig et al., 1980; Zhao et al., 1995; Pearce et al., 2002; Taylor and Moncada, 2010). Furthermore, the reduced form of CcO can catalyse the reduction of NO to N2O (Brudvig et al., 1980; Zhao et al., 1995). Thus, either oxidation or reduction of NO by CcO results in the metabolism of NO with safe end products. The similar properties of O2 and NO facilitate the binding of NO to CcO, and this activity is pronounced under oxygen-limited conditions (Ghafourifar and Cadenas, 2005). The mitochondrial electron transport chain (ETC) in axenic algae (Chlamydomonas reinhardtii and Chlorella vulgaris) catalyses the reduction of NO to N2O (Guieysse et al., 2013; Plouviez et al., 2017; Plouviez et al., 2019). CcO has some rudimentary nitric oxide reductase activity, and therefore, when NO is the substrate instead of O2, two molecules of NO yield N2O and H2O (Brudvig et al., 1980; Zhao et al., 1995; Koivisto et al., 1997; Igamberdiev et al., 2010; Blomberg and Ädelroth, 2018; Poderoso et al., 2019). It has also been proven isotopically that NO is reduced to N2O by CcO in higher organisms (Brudvig et al., 1980). As mitochondrial CcO has evolved from denitrifying enzymes, under hypoxic conditions in cells, the mitochondrial CcO can still reduce NO to N2O (Saraste, 1994; Saraste and Castresana, 1994). Furthermore, another enzyme in the mitochondria, that is, quinone of the ETC catalyses NO reduction to N2O (Alegria et al., 2004; Igamberdiev and Hill, 2009; Sanchez-Cruz and Alegría, 2009). Therefore, mitochondria can be a potent site of N2O formation under oxygen-limited conditions, and it should be a focus of future research.
Similar to the observations in plants, macrofauna and earthworms are also found to emit N2O when supplied with NO3 and under O2-limited conditions (Horn et al., 2003; Stief et al., 2009). Earthworms do not produce N2O when supplied NH4 (Horn et al., 2003). Moreover, in other studies, listed in Table 1, when 15N-labeled NH4 was used as a substrate, there was no N2O emission. This shows that NO3 metabolism at the cellular level produces N2O in both plants and animals. As described above, the ETC in (Figure 1) mitochondria can reduce NO to N2O under less oxic conditions, which suggests that N2O emitted by earthworms and macrofauna (Horn et al., 2003; Stief et al., 2009) might also be produced from hypoxic mitochondria. The gut of insects has a hypoxic environment (Johnson and Barbehenn, 2000), which may explain the higher level of N2O production in the gut (Stief et al., 2009). Moreover, axenic algae supplied with NO2 produced significantly higher levels of N2O under dark conditions than under light conditions (Guieysse et al., 2013; Plouviez et al., 2017). The low emission of N2O under light conditions may be due to the supply of photosynthetic O2 to the cells.
Table 1.
Compilation of the substrates, mediums and products that used labeled N sources and their subsequent measurements of N2O emissions.
| Substrate | Medium | Product | Reference | |
|---|---|---|---|---|
| 1. |
15N labeled NO3 15N labeled NH4 |
Aseptically grown tobacco plants |
15N labeled N2O No N2O emission |
Goshima et al. (1999) |
| 2. |
15N labeled NO3 15N labeled NH4 |
Lichen |
15N labeled N2O No N2O emission |
Lenhart et al. (2015) |
| 3. |
15N labeled NO3 15N labeled NH4 |
Wheat plant |
15N labeled N2O No N2O emission |
Smart and Bloom (2001) |
| 4. | 15N labeled NO3 | Soybean plant | 15N labeled NO and N2O | Dean and Harper (1986) |
| 5. | 15N labeled NO3 | Macro fauna | 15N labeled N2O | Stief et al. (2009) |
| 6. | 15N labeled NO2 | Tobacco plant | 15N labeled N2O | Goshima et al. (1999) |
| 7. | 15N labeled NO2 | Aseptically grown tobacco plant | 15N labeled N2O | Hakata et al. (2003) |
| 8. | 15N labeled NO2 | Axenic algae | 15N labeled N2O | Weathers (1984) |
| 9. |
15N labeled NO 14N labeled NO |
Reduced form of beef heart cytochrome c oxidase (CcO) |
15N labeled N2O 14N labeled N2O |
Brudvig et al. (1980) |
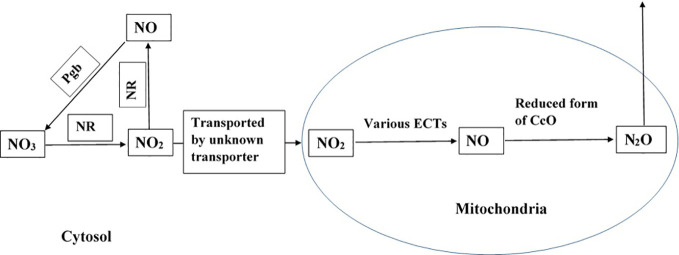
Figure 1.
Potential pathway of N2O formation in plant cells. NR represents nitrate reductase and Pgb represents phytoglobin (Brudvig et al., 1980; Zhao et al., 1995; Goshima et al., 1999; Gupta and Kaiser, 2010; Guieysse et al., 2013; Plouviez et al., 2017). The pathway is active in presence of NO3 and NO2, and when plants experience hypoxia and anoxia.
Based on experimental evidence gathered from various studies, we propose that the reductive pathway of NO formation in the mitochondria and further reduction of NO by the mitochondrial ETCs contributes to the formation of N2O (in eukaryotic cells, as presented in Figure 1). The process is catalysed by various enzymes, and it might be pronounced under hypoxic and anoxic conditions but not under normoxic conditions. The proposed pathway is further supported by the existence of a denitrifying pathway, and the associated enzymes and genes in Globobulimina species and the localisation of enzymes in the mitochondria (Woehle et al., 2018). As higher animals possess well developed respiratory and circulatory systems that transport O2, they may not experience hypoxia. However, plants lack such sophisticated systems to transport O2 (Voesenek et al., 2016), and therefore, may experience hypoxia and anoxia that favour N2O formation. Field studies have reported high N2O emission from plants under flooded conditions (Rusch and Rennenberg, 1998; Machacova et al., 2013), suggesting the role of hypoxia and anoxia in N2O formation in plants.
Significance of N2O Formation via the NO3-NO2-NO Pathway in Plants
NO3 is not only a major nutrient in plant cells but also a signaling molecule (Zhao et al., 2018). Several studies have reported that NO3 plays a role in hypoxia tolerance. For example, NO3 maintains the growth of plants under oxygen-limited conditions, and its absence disturbs plant growth (Horchani et al., 2010). Anoxia tolerance of tomato plant is enhanced by nitrate reduction (Allègre et al., 2004). Moreover, anoxia strongly induces NR activity and the induced NR activity prevents pH from dropping to life-threatening levels (Allègre et al., 2004). NO3 nutrition in plants decreases the total respiration rate and reactive oxygen species levels (Wany et al., 2019), but increases ATP production under hypoxic conditions (Stoimenova et al., 2007; Wany et al., 2019). Under oxygen-limited conditions, NO3 protects the ultrastructure of mitochondria (Vartapetian et al., 2003). The addition of NO3 to the root zone of plants released significantly less amount of ethanol compared with roots supplied with NH4 under hypoxic conditions (Oliveira et al., 2013). This suggests NO3 plays an important role to decrease alcoholic fermentative metabolism in plants during hypoxia (Oliveira et al., 2013). Overall, these findings suggest that NO3 and NR play an important role to maintain the integrity of plant cells under oxygen-limited conditions.
NO2 is also reported to play important roles under oxygen-limited conditions in plants. Benamar et al. (2008) found that NO2-dependent NO production in the mitochondria can regulate surrounding O2 level. Moreover, plant mitochondria can synthesise ATP under anaerobic conditions when supplied with NO2 (Stoimenova et al., 2007; Gupta et al., 2016). The supply of NO2 decreased lipid peroxidation and reactive oxygen species formation (Gupta et al., 2016). The absence of NO2 as a terminal acceptor for ETC during hypoxia leads to mitochondrial depolarisation (Gupta et al., 2016). NO2 supplemented roots released significantly less amount of fermentative ethanol during hypoxia than NH4-supplemented roots (Oliveira et al., 2012; Oliveira et al., 2013), suggesting the vital role of NO2 in plants to survive under oxygen-limited conditions.
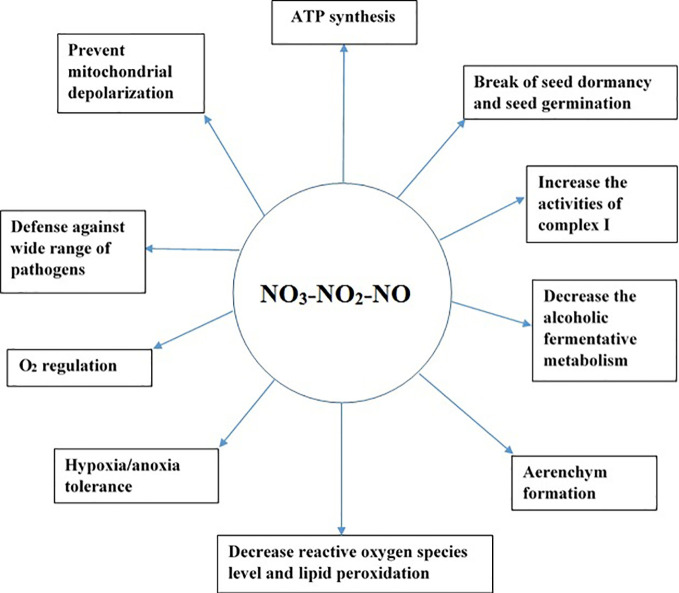
NO helps plants to cope under several environmental stresses. For example, NO is essential for the homeostasis of O2 level in plants under oxygen-limited conditions (Gupta and Igamberdiev, 2011). NO production in the mitochondria has several implications in plants as illustrated in Figure 2. For example, NO can break seed dormancy and stimulate seed germination in plants (Beligni and Lamattina, 2000; Bethke et al., 2004). Similarly, under hypoxic stress, NO is vital for the formation of aerenchyma in the roots (Wany et al., 2017). NO production in the mitochondria under low-oxygen conditions can help in ATP synthesis, preventing excessive depletion of energy (Stoimenova et al., 2007). NO3-NO2-dependent NO production in plant roots decreases fermentative ethanol production during hypoxia (Oliveira et al., 2012; Oliveira et al., 2013).
Figure 2.
Significance of the NO3-NO2-NO pathway in plants under hypoxic and anoxic conditions.
Although NO has been well established as a signaling molecule, its high concentration in cells leads to cell death (Boscá and Hortelano, 1999; Brown and Borutaite, 2002). Therefore, it is critical to regulate its concentration in cells, as a higher amount of NO is formed under hypoxic and anoxic conditions. Two mechanisms are reported to occur in the mitochondria to detoxify the high amount of NO formed, namely, oxidation of NO to NO2 during normoxia (Cooper, 2002; Taylor and Moncada, 2010) and reduction of NO to N2O during hypoxia (Cooper, 2002). As both products of NO metabolism in the mitochondria, that is, NO2 and N2O, are non-toxic, their formation might play a protective role in the mitochondria. If hypoxia-induced NO production in cells is high, it can cause DNA fragmentation, leading to cell death; however, if NO is scavenged, it can reduce DNA fragmentation (Wany et al., 2017). Therefore, scavenging of NO is essential to protect cells from high NO toxicity. Phytoglobins are reported to scavenge NO in the cytosol (Igamberdiev et al., 2010). Additionally, purified mitochondria have been reported to scavenge exogenous NO (Gupta et al., 2005; de Oliveira et al., 2008; Wulff et al., 2009; Gupta and Kaiser, 2010). Furthermore, the addition of NADH as an electron donor increased NO scavenging by the mitochondria (Gupta et al., 2005; de Oliveira et al., 2008; Wulff et al., 2009; Gupta et al., 2016), indicating that the mitochondria have a protective mechanism to detoxify the excess NO formed. NADH might act as an electron donor to reduce cytochrome c oxidase, leading to an increase in NO scavenging in purified mitochondria. As discussed in our proposed pathway of N2O formation in the mitochondria, the conversion of NO to N2O by the reduced form of CcO might be the potential pathway regulating excessive NO formed under oxygen-limited conditions in the mitochondria. Mitochondria are not only a source of NO, but also an important sink and target of NO (Igamberdiev et al., 2014), and long-term exposure of mitochondria to NO can lead to the dysfunction of mitochondria (Brown and Borutaite, 2002). Although the NO3-NO2-NO pathway has several roles (Figure 2) in plants during hypoxia and anoxia, NO accumulation at higher level is toxic to cells (Brown and Borutaite, 2002). Therefore, N2O formation in the mitochondria via the NO3-NO2-NO pathway might be a strategy to protect cells and mitochondrial components from excessive NO formed under oxygen-limited conditions. Therefore, at molecular level, further research should focus on measuring NO and N2O from isolated mitochondria to obtain more insights on mitochondria’s role in scavenging excess NO during hypoxia and anoxia.
Do Plant Cells Reduce N2O to N2?
The last two enzymes of the denitrification process, namely, nitric oxide reductase (NOR) and nitrous oxide reductase (N2OR), merged to form CcO (Saraste and Castresana, 1994; Stanton et al., 2018). Moreover, the copper site in bacterial N2OR is similar to the CuA site in CcO (Kroneck, 2018). Many catalytic properties of CcO from denitrifying bacteria (Paracoccus denitrificans) and eukaryotic organisms are similar (Ludwig, 1987; Kadenbach et al., 1991). As eukaryotic mitochondrion is considered to be evolved from P. denitrificans, a denitrifying bacterium (John and Whatley, 1975), it may still possess rudimentary denitrification properties. Although during the evolution most of genes of the bacterium transferred to the nucleus, few remained in the mitochondrial DNA including genes of CcO (Kadenbach et al., 1991). Therefore, it may be possible that CcO of higher organisms might also possess similar properties like that of its ancestor, P. denitrificans. The significant negative relationship between N2O consumption and CO2 respiration rates in plants and lichens (Machacova et al., 2017) suggests that mitochondria are the possible site of N2O consumption. This N2O consumption observed in these eukaryotes might be at the site of CcO, as this enzyme is formed from the last two enzymes of denitrification. There are also reports of emission of 15N-labeled N2 from wheat crops supplied with 15N-labeled NO2 (Vanecko and Varner, 1955), suggesting that under certain cell conditions, mitochondria may also metabolise N2O to N2. However, to date, N2 emission from plants is less reported. It may be due to the advanced systems of O2 regulation in plants, and this might inhibit the complete process of denitrification. A recent study, which measured N2O and N2 emission from soil–plant systems, showed that N2O and N2 emitted by NO3-rich soil–plant systems was three times higher than that by NH4-supplemented soil–plant systems and bare soil (Senbayram et al., 2020), and this suggests that the possible role of N2O and N2 production in plants. Further experiments at the molecular level (mitochondria) are needed to explore the reason for the significant negative relation between N2O emission and respiration rate in plants and lichen, as reported by Machacova et al. (2017).
Conclusions
To cope with the problems of global warming and ozone layer depletion, a good understanding of N2O formation processes in various source is critical. Therefore, the N2O formation process in plants is a matter of concern. The reductive pathway of NO formation in the mitochondria along with further reduction of NO by ETC is a possible pathway of N2O formation in plants. Considering available evidence, we conclude that there is strong possibility that plant cells produce N2O in the mitochondria under hypoxic and anoxic conditions. The theory that plants are only a conduit for N2O produced by soil-inhabiting microorganisms might be an ambiguous explanation. The root zone may sense hypoxia and anoxia due to the soil environmental conditions, which may favour N2O formation in the root mitochondria. As some studies have shown that N2O emission from tree stems is higher than that from soils in natural habitats (Welch et al., 2019), the proposed pathway of N2O formation in plants might play a significant role in understanding N cycling in eukaryotic organisms and the global N2O budget. Furthermore, we have highlighted the reduction of NO to N2O in the mitochondria, and therefore, it would be valuable to reassess the role of mitochondrial ETC under both hypoxic and anoxic conditions. Although, N2O is a potent greenhouse gas ,its formation in the mitochondria might help to protect the integrity of the mitochondria and protect cells from the toxicity of NO accumulation during hypoxia.
Author Contributions
AT wrote the manuscript. CH supervised the whole work. CZ, BP, FB, and WD commented on the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (no. 41530859) and the National Key Research and Development Program of China (2016YFD0800102-4 and 2018YFC0213300-01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
AT is grateful to CAS-TWAS President’s Fellowship Program for granting the Ph.D. fellowship. We are thankful to Prof. Chengcai Chu from the Institute of Genetics and Developmental Biology, Beijing, China, for thoughtful comments. Moreover, we are grateful to Prof. Oene Oenema and Prof. Jiafa Luo for their valuable comments.
References
- Alber N. A., Sivanesan H., Vanlerberghe G. C. (2017). The occurrence and control of nitric oxide generation by the plant mitochondrial electron transport chain. Plant Cell Environ. 40, 1074–1085. 10.1111/pce.12884 [DOI] [PubMed] [Google Scholar]
- Alegria A. E., Sanchez S., Quintana I. (2004). Quinone-enhanced ascorbate reduction of nitric oxide: role of quinone redox potential. Free Radical Res. 38, 1107–1112. 10.1080/10715760400009852 [DOI] [PubMed] [Google Scholar]
- Allègre A., Silvestre J., Morard P., Kallerhoff J., Pinelli E. (2004). Nitrate reductase regulation in tomato roots by exogenous nitrate: a possible role in tolerance to long-term root anoxia. J. Exp. Bot. 55, 2625–2634. 10.1093/jxb/erh258 [DOI] [PubMed] [Google Scholar]
- Ascenzi P., Marino M., Polticelli F., Santucci R., Coletta M. (2014). Cardiolipin modulates allosterically the nitrite reductase activity of horse heart cytochrome c. J. Biol. Inorg. Chem. 19, 1195–1201. 10.1007/s00775-014-1175-9 [DOI] [PubMed] [Google Scholar]
- Astier J., Gross I., Durner J. (2018). Nitric oxide production in plants: an update. J. Exp. Bot. 69, 3401–3411. 10.1093/jxb/erx420 [DOI] [PubMed] [Google Scholar]
- Baggs E. M. (2011). Soil microbial sources of nitrous oxide: recent advances in knowledge, emerging challenges and future direction. Curr. Opin. Env. Sust. 3, 321–327. 10.1016/j.cosust.2011.08.011 [DOI] [Google Scholar]
- Basu S., Azarova N. A., Font M. D., King S. B., Hogg N., Gladwin M. T., et al. (2008). Nitrite reductase activity of cytochrome c. J. Biol. Inorg. Chem. 283, 32590–32597. 10.1074/jbc.M806934200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni M. V., Lamattina L. (2000). Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 210 (2), 215–221. 10.1007/PL00008128 [DOI] [PubMed] [Google Scholar]
- Benamar A., Rolletschek H., Borisjuk L., Avelange-Macherel M. H., Curien G., Mostefai H. A., et al. (2008). Nitrite–nitric oxide control of mitochondrial respiration at the frontier of anoxia. BBA-Bioenergetics 1777, 1268–1275. 10.1016/j.bbabio.2008.06.002 [DOI] [PubMed] [Google Scholar]
- Bender D., Schwarz G. (2018). Nitrite-dependent nitric oxide synthesis by molybdenum enzymes. FEBS Lett. 592, 2126–2139. 10.1002/1873-3468.13089 [DOI] [PubMed] [Google Scholar]
- Bethke P. C., Gubler F., Jacobsen J. V., Jones R. L. (2004). Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta 219, 847–855. 10.1007/s00425-004-1282-x [DOI] [PubMed] [Google Scholar]
- Blomberg M. R., Ädelroth P. (2018). Mechanisms for enzymatic reduction of nitric oxide to nitrous oxide-A comparison between nitric oxide reductase and cytochrome c oxidase. BBA-Bioenergetics 1859, 1223–1234. 10.1016/j.bbabio.2018.09.368 [DOI] [PubMed] [Google Scholar]
- Boscá L., Hortelano S. (1999). Mechanisms of nitric oxide-dependent apoptosis: involvement of mitochondrial mediators. Cell Signal. 11, 239–244. 10.1016/S0898-6568(98)00064-3 [DOI] [PubMed] [Google Scholar]
- Botrel A., Kaiser W. M. (1997). Nitrate reductase activation state in barley roots in relation to the energy and carbohydrate status. Planta 201, 496–501. 10.1007/s004250050094 [DOI] [PubMed] [Google Scholar]
- Bowatte S., Newton P. C., Theobald P., Brock S., Hunt C., Lieffering M., et al. (2014). Emissions of nitrous oxide from the leaves of grasses. Plant Soil 374, 275–283. 10.1007/s11104-013-1879-6 [DOI] [Google Scholar]
- Brown G. C., Borutaite V. (2002). Nitric oxide inhibition of mitochondrial respiration and its role in cell death. Free Radical Bio. Med. 33, 1440–1450. 10.1016/S0891-5849(02)01112-7 [DOI] [PubMed] [Google Scholar]
- Brudvig G. W., Stevens T. H., Chan S., II (1980). Reactions of nitric oxide with cytochrome c oxidase. Biochemistry 19, 5275–5285. 10.1021/bi00564a020 [DOI] [PubMed] [Google Scholar]
- Calatrava V., Chamizo-Ampudia A., Sanz-Luque E., Ocaña-Calahorro F., Llamas A., Fernandez E., et al. (2017). How Chlamydomonas handles nitrate and the nitric oxide cycle. J. Exp. Bot. 68, 2593–2602. 10.1093/jxb/erw507 [DOI] [PubMed] [Google Scholar]
- Castello P. R., David P. S., McClure T., Crook Z., Poyton R. O. (2006). Mitochondrial cytochrome oxidase produces nitric oxide under hypoxic conditions: implications for oxygen sensing and hypoxic signaling in eukaryotes. Cell Metab. 3, 277–287. 10.1016/j.cmet.2006.02.011 [DOI] [PubMed] [Google Scholar]
- Chamizo-Ampudia A., Sanz-Luque E., Llamas A., Galvan A., Fernandez E. (2017). Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 22, 163–174. 10.1016/j.tplants.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Chang C., Janzen H. H., Nakonechny E. M., Cho C. M. (1998). Nitrous oxide emission through plants. Soil Sci. Soc Am. J. 62, 35–38. 10.2136/sssaj1998.03615995006200010005x [DOI] [Google Scholar]
- Collman J. P., Devaraj N. K., Decréau R. A., Yang Y., Yan Y. L., Ebina W., et al. (2007). A cytochrome c oxidase model catalyzes oxygen to water reduction under rate-limiting electron flux. Science 315, 1565–1568. 10.1126/science.1135844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C. E. (2002). Nitric oxide and cytochrome oxidase: substrate, inhibitor or effector? Trends Biochem. Sci. 27, 33–39. 10.1016/S0968-0004(01)02035-7 [DOI] [PubMed] [Google Scholar]
- Davidson E. A., Kanter D. (2014). Inventories and scenarios of nitrous oxide emissions. Environ. Res. Lett. 9:105012. 10.1088/1748-9326/9/10/105012 [DOI] [Google Scholar]
- de Oliveira H. C., Wulff A., Saviani E. E., Salgado I. (2008). Nitric oxide degradation by potato tuber mitochondria: evidence for the involvement of external NAD(P)H dehydrogenases. BBA-Bioenergetics 1777, 470–476. 10.1016/j.bbabio.2008.02.006 [DOI] [PubMed] [Google Scholar]
- Dean J. V., Harper J. E. (1986). Nitric oxide and nitrous oxide production by soybean and winged bean during the in vivo nitrate reductase assay. Plant Physiol. 82, 718–723. 10.1104/pp.82.3.718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Jia L., Li Y., Smith S. J., Miller A. J., Shen Q. (2007). Comparing nitrate storage and remobilization in two rice cultivars that differ in their nitrogen use efficiency. J. Exp. Bot. 58 (7), 1729–1740. 10.1093/jxb/erm033 [DOI] [PubMed] [Google Scholar]
- Gao G. F., Li P. F., Zhong J. X., Shen Z. J., Chen J., Li Y. T., et al. (2019). Spartina alterniflora invasion alters soil bacterial communities and enhances soil N2O emissions by stimulating soil denitrification in mangrove wetland. Sci. Total Environ. 653, 231–240. 10.1016/j.scitotenv.2018.10.277 [DOI] [PubMed] [Google Scholar]
- Ghafourifar P., Cadenas E. (2005). Mitochondrial nitric oxide synthase. Trends Pharmacol. Sci. 26, 190–195. 10.1016/j.tips.2005.02.005 [DOI] [PubMed] [Google Scholar]
- Ghafourifar P., Richter C. (1997). Nitric oxide synthase activity in mitochondria. FEBS Lett. 418, 291–296. 10.1016/S0014-5793(97)01397-5 [DOI] [PubMed] [Google Scholar]
- Giulivi C., Poderoso J. J., Boveris A. (1998). Production of nitric oxide by mitochondria. J. Biol. Chem. 273, 11038–11043. 10.1074/jbc.273.18.11038 [DOI] [PubMed] [Google Scholar]
- Goshima N., Mukai T., Suemori M., Takahashi M., Caboche M., Morikawa H. (1999). Emission of nitrous oxide (N2O) from transgenic tobacco expressing antisense NiR mRNA. Plant J. 19, 75–80. 10.1046/j.1365-313X.1999.00494.x [DOI] [PubMed] [Google Scholar]
- Guieysse B., Plouviez M., Coilhac M., Cazali L. (2013). Nitrous Oxide (N2O) production in axenic Chlorella vulgaris microalgae cultures: evidence, putative pathways, and potential environmental impacts. Biogeosciences 10, 6737–6746. 10.5194/bg-10-6737-2013 [DOI] [Google Scholar]
- Guo F. Q., Crawford N. M. (2005). Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell 17, 3436–3450. 10.1105/tpc.105.037770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K. J., Igamberdiev A. U. (2011). The anoxic plant mitochondrion as a nitrite: NO reductase. Mitochondrion 11, 537–543. 10.1016/j.mito.2011.03.005 [DOI] [PubMed] [Google Scholar]
- Gupta K. J., Kaiser W. M. (2010). Production and scavenging of nitric oxide by barley root mitochondria. Plant Cell Physiol. 51, 576–584. 10.1093/pcp/pcq022 [DOI] [PubMed] [Google Scholar]
- Gupta K. J., Stoimenova M., Kaiser W. M. (2005). In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO, in vitro and in situ. J. Exp. Bot. 56, 2601–2609. 10.1093/jxb/eri252 [DOI] [PubMed] [Google Scholar]
- Gupta K. J., Fernie A. R., Kaiser W. M., van Dongen J. T. (2011). On the origins of nitric oxide. Trends Plant Sci. 16, 160–168. 10.1016/j.tplants.2010.11.007 [DOI] [PubMed] [Google Scholar]
- Gupta A. K., Kumari A., Mishra S., Wany A., Gupta K. J. (2016). “The functional role of nitric oxide in plant mitochondrial metabolism,” in Advances in Botanical Research, vol. 77 (Cambridge, MA: Academic Press; ), 145–163. 10.1016/bs.abr.2015.10.007 [DOI] [Google Scholar]
- Hachiya T., Sakakibara H. (2017). Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants. J. Exp. Bot. 68, 2501–2512. 10.1093/jxb/erw449 [DOI] [PubMed] [Google Scholar]
- Hakata M., Takahashi M., Zumft W., Sakamoto A., Morikawa H. (2003). Conversion of the nitrate nitrogen and nitrogen dioxide to nitrous oxides in plants. Acta Biotechnol. 23, 249–257. 10.1002/abio.200390032 [DOI] [Google Scholar]
- Horchani F., Aschi-Smiti S., Brouquisse R. (2010). Involvement of nitrate reduction in the tolerance of tomato (Solanum lycopersicum L.) plants to prolonged root hypoxia. Acta Physiol. Plant 32, 1113–1123. 10.1007/s11738-010-0503-0 [DOI] [Google Scholar]
- Horchani F., Prévot M., Boscari A., Evangelisti E., Meilhoc E., Bruand C., et al. (2011). Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol. 155, 1023–1036. 10.1104/pp.110.166140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M. A., Schramm A., Drake H. L. (2003). The earthworm gut: an ideal habitat for ingested N2O-producing microorganisms. Appl. Environ. Microb. 69, 1662–1669. 10.1128/AEM.69.3.1662-1669.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H. W., Chen D., He J. Z. (2015). Microbial regulation of terrestrial nitrous oxide formation: understanding the biological pathways for prediction of emission rates. FEMS Microbiol. Rev. 39, 729–749. 10.1093/femsre/fuv021 [DOI] [PubMed] [Google Scholar]
- Igamberdiev A. U., Hill R. D. (2009). Plant mitochondrial function during anaerobiosis. Ann. Bot. 103, 259–268. 10.1093/aob/mcn100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev A. U., Bykova N. V., Shah J. K., Hill R. D. (2010). Anoxic nitric oxide cycling in plants: participating reactions and possible mechanisms. Physiol. Plantarum 138, 393–404. 10.1111/j.1399-3054.2009.01314.x [DOI] [PubMed] [Google Scholar]
- Igamberdiev A. U., Ratcliffe R. G., Gupta K. J. (2014). Plant mitochondria: source and target for nitric oxide. Mitochondrion 19, 329–333. 10.1016/j.mito.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Jakobs H. H., Froriep D., Havemeyer A., Mendel R. R., Bittner F., Clement B. (2014). The mitochondrial amidoxime reducing component (mARC): involvement in metabolic reduction of N-oxides, oximes and N-hydroxyamidinohydrazones. ChemMedChem 9, 2381–2387. 10.1002/cmdc.201402127 [DOI] [PubMed] [Google Scholar]
- Jansson E.Å., Huang L., Malkey R., Govoni M., Nihlén C., Olsson A., et al. (2008). A mammalian functional nitrate reductase that regulates nitrite and nitric oxide homeostasis. Nat. Chem. Biol. 4, 411. 10.1038/nchembio.92 [DOI] [PubMed] [Google Scholar]
- John P., Whatley F. R. (1975). Paracoccus denitrificans and the evolutionary origin of the mitochondrion. Nature 254, 495. 10.1038/254495a0 [DOI] [PubMed] [Google Scholar]
- Johnson K. S., Barbehenn R. V. (2000). Oxygen levels in the gut lumens of herbivorous insects. J. Insect Physiol. 46, 897–903. 10.1016/S0022-1910(99)00196-1 [DOI] [PubMed] [Google Scholar]
- Kadenbach B., Stroh A., Hüther F. J., Reimann A., Steverding D. (1991). Evolutionary aspects of cytochromec oxidase. J. Bioenerg. Biomembr. 23 (2), 321–334. 10.1007/BF00762225 [DOI] [PubMed] [Google Scholar]
- Klepper L. A. (1987). Nitric oxide emissions from soybean leaves during in vivo nitrate reductase assays. Plant Physiol. 85, 96–99. 10.1104/pp.85.1.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Matsuo Y., Takimoto A., Suzuki S., Maruo F., Shoun H. (1996). Denitrification, a novel type of respiratory metabolism in fungal mitochondrion. J. Biol. Chem. 271, 16263–16267. 10.1074/jbc.271.27.1626 [DOI] [PubMed] [Google Scholar]
- Koivisto A., Matthias A., Bronnikov G., Nedergaard J. (1997). Kinetics of the inhibition of mitochondrial respiration by NO. FEBS Lett. 417, 75–80. 10.1016/S0014-5793(97)01258-1 [DOI] [PubMed] [Google Scholar]
- Kozlov A. V., Staniek K., Nohl H. (1999). Nitrite reductase activity is a novel function of mammalian mitochondria. FEBS Lett. 454, 127–130. 10.1016/S0014-5793(99)00788-7 [DOI] [PubMed] [Google Scholar]
- Kroneck P. M. (2018). Walking the seven lines: binuclear copper A in cytochrome c oxidase and nitrous oxide reductase. J. Biol. Inorg. Chem. 23, 27–39. 10.1007/s00775-017-1510-z [DOI] [PubMed] [Google Scholar]
- Lenhart K., Weber B., Elbert W., Steinkamp J., Clough T., Crutzen P., et al. (2015). Nitrous oxide and methane emissions from cryptogamic covers. Global Change Biol. 21, 3889–3900. 10.1111/gcb.12995 [DOI] [PubMed] [Google Scholar]
- Lenhart K., Behrendt T., Greiner S., Steinkamp J., Well R., Giesemann A., et al. (2019). Nitrous oxide effluxes from plants as a potentially important source to the atmosphere. New Phytol. 221, 1398–1408. 10.1111/nph.15455 [DOI] [PubMed] [Google Scholar]
- Ludwig B. (1987). Cytochrome c oxidase in prokaryotes. FEMS Microbiol. Rev. 3 (1), 41–56. 10.1111/j.1574-6968.1987.tb02451.x [DOI] [Google Scholar]
- Lundberg J. O., Weitzberg E., Gladwin M. T. (2008). The nitrate–nitrite–nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discovery 7, 156. 10.1038/nrd2466 [DOI] [PubMed] [Google Scholar]
- Maathuis F. J. (2009). Physiological functions of mineral macronutrients. Curr. Opin. Plant Biol. 12, 250–258. 10.1016/j.pbi.2009.04.003 [DOI] [PubMed] [Google Scholar]
- Machacova K., Papen H., Kreuzwieser J., Rennenberg H. (2013). Inundation strongly stimulates nitrous oxide emissions from stems of the upland tree Fagus sylvatica and the riparian tree Alnus glutinosa. Plant Soil 364, 287–301. 10.1007/s11104-012-1359-4 [DOI] [Google Scholar]
- Machacova K., Maier M., Svobodova K., Lang F., Urban O. (2017). Cryptogamic stem covers may contribute to nitrous oxide consumption by mature beech trees. Sci. Rep. 7, 13243. 10.1038/s41598-017-13781-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia L. B., Moura J. J. (2015). Nitrite reduction by molybdoenzymes: a new class of nitric oxide-forming nitrite reductases. J. Biol. Inorg. Chem. 20, 403–433. 10.1007/s00775-014-1234-2 [DOI] [PubMed] [Google Scholar]
- Miller A. J., Cramer M. D. (2005). “Root nitrogen acquisition and assimilation,” in Root Physiology: from Gene to Function. (Dordrecht: Springer; ), 1–36. [Google Scholar]
- Morard P., Silvestre J., Lacoste L., Caumes E., Lamaze T. (2004). Nitrate uptake and nitrite release by tomato roots in response to anoxia. J. Plant Physiol. 161, 855–865. 10.1016/j.jplph.2003.11.003 [DOI] [PubMed] [Google Scholar]
- Moreau M., Lee G., II, Wang Y., Crane B. R., Klessig D. F. (2008). AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 283, 32957–32967. 10.1074/jbc.M804838200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira H. C., Sodek L. (2013). Effect of oxygen deficiency on nitrogen assimilation and amino acid metabolism of soybean root segments. Amino Acids 44, 743–755. 10.1007/s00726-012-1399-3 [DOI] [PubMed] [Google Scholar]
- Oliveira H. C., Salgado I., Sodek L. (2012). Involvement of nitrite in the nitrate-mediated modulation of fermentative metabolism and nitric oxide production of soybean roots during hypoxia. Planta 237, 255–264. 10.1007/s00425-012-1773-0 [DOI] [PubMed] [Google Scholar]
- Oliveira H. C., Salgado I., Sodek L. (2013). Nitrite decreases ethanol production by intact soybean roots submitted to oxygen deficiency: a role for mitochondrial nitric oxide synthesis? Plant Signal. Behav. 8, e23578. 10.4161/psb.23578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J. A., Vega A., Bouguyon E., Krouk G., Gojon A., Coruzzi G., et al. (2016). Nitrate transport, sensing, and responses in plants. Mol. Plant 9, 837–856. 10.1016/j.molp.2016.05.004 [DOI] [PubMed] [Google Scholar]
- Pearce L. L., Kanai A. J., Birder L. A., Pitt B. R., Peterson J. (2002). The catabolic fate of nitric oxide the nitric oxide oxidase and peroxynitrite reductase activities of cytochrome oxidase. J. Biol. Chem. 277, 13556–13562. 10.1074/jbc.M109838200 [DOI] [PubMed] [Google Scholar]
- Planchet E., Gupta K. J., Sonoda M., Kaiser W. M. (2005). Nitric oxide emission from tobacco leaves and cell suspensions: rate limiting factors and evidence for the involvement of mitochondrial electron transport. Plant J. 41, 732–743. 10.1111/j.1365-313X.2005.02335.x [DOI] [PubMed] [Google Scholar]
- Plouviez M., Wheeler D., Shilton A., Packer M. A., McLenachan P. A., Sanz-Luque E., et al. (2017). The biosynthesis of nitrous oxide in the green alga Chlamydomonas reinhardtii. Plant J. 91, 45–56. 10.1111/tpj.13544 [DOI] [PubMed] [Google Scholar]
- Plouviez M., Shilton A., Packer M. A., Guieysse B. (2019). Nitrous oxide emissions from microalgae: potential pathways and significance. J. Appl. Phycol. 31, 1–8. 10.1007/s10811-018-1531-1 [DOI] [Google Scholar]
- Poderoso J. J., Helfenberger K., Poderoso C. (2019). The effect of nitric oxide on mitochondrial respiration. Nitric. Oxide 88, 61–72. 10.1016/j.niox.2019.04.005 [DOI] [PubMed] [Google Scholar]
- Rockel P., Strube F., Rockel A., Wildt J., Kaiser W. M. (2002). Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 53, 103–110. 10.1093/jexbot/53.366.103 [DOI] [PubMed] [Google Scholar]
- Rőszer T. (2012). The biology of subcellular nitric oxide. Dordrecht: Springer. 10.1007/978-94-007-2819-6 [DOI] [Google Scholar]
- Rusch H., Rennenberg H. (1998). Black alder (Alnus glutinosa (L.) Gaertn.) trees mediate methane and nitrous oxide emission from the soil to the atmosphere. Plant Soil 201, 1–7. 10.1023/A:1004331521059 [DOI] [Google Scholar]
- Sanchez-Cruz P., Alegría A. E. (2009). Quinone-enhanced reduction of nitric oxide by xanthine/xanthine oxidase. Chem. Res. Toxicol. 22, 818–823. 10.1021/tx800392j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M., Castresana J. (1994). Cytochrome oxidase evolved by tinkering with denitrification enzymes. FEBS Lett. 341, 1–4. 10.1016/0014-5793(94)80228-9 [DOI] [PubMed] [Google Scholar]
- Saraste M. (1994). Structure and evolution of cytochrome oxidase. Anton. Leeuw. Int. J. 65, 285–287. 10.1007/BF00872214 [DOI] [PubMed] [Google Scholar]
- Senbayram M., Well R., Shan J., Bol R., Burkart S., Jones D. L., et al. (2020). Rhizosphere processes in nitrate-rich barley soil tripled both N2O and N2 losses due to enhanced bacterial and fungal denitrification. Plant Soil 448, 509–522. 10.1007/s11104-020-04457-9 [DOI] [Google Scholar]
- Smart D. R., Bloom A. J. (2001). Wheat leaves emit nitrous oxide during nitrate assimilation. P. Natl. Acad. Sci. 98, 7875–7878. 10.1073/pnas.131572798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparacino-Watkins C. E., Tejero J., Sun B., Gauthier M. C., Thomas J., Ragireddy V., et al. (2014). Nitrite reductase and nitric-oxide synthase activity of the mitochondrial molybdopterin enzymes mARC1 and mARC2. J. Biol. Chem. 289, 10345–10358. 10.1074/jbc.M114.555177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton C. L., Reinhard C. T., Kasting J. F., Ostrom N. E., Haslun J. A., Lyons T. W., et al. (2018). Nitrous oxide from chemodenitrification: A possible missing link in the Proterozoic greenhouse and the evolution of aerobic respiration. Geobiology 16, 597–609. 10.1111/gbi.12311 [DOI] [PubMed] [Google Scholar]
- Stief P., Poulsen M., Nielsen L. P., Brix H., Schramm A. (2009). Nitrous oxide emission by aquatic macrofauna. P. Natl. Acad. Sci. 106, 4296–4300. 10.1073/pnas.0808228106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoimenova M., Igamberdiev A. U., Gupta K. J., Hill R. D. (2007). Nitrite-driven anaerobic ATP synthesis in barley and rice root mitochondria. Planta 226, 465–474. 10.1007/s00425-007-0496-0 [DOI] [PubMed] [Google Scholar]
- Sugiura M., Georgescu M. N., Takahashi M. (2007). A nitrite transporter associated with nitrite uptake by higher plant chloroplasts. Plant Cell Physiol. 48, 1022–1035. 10.1093/pcp/pcm073 [DOI] [PubMed] [Google Scholar]
- Syakila A., Kroeze C. (2011). The global nitrous oxide budget revisited. GGMM 1, 17–26. 10.3763/ghgmm.2010.0007 [DOI] [Google Scholar]
- Taylor C. T., Moncada S. (2010). Nitric oxide, cytochrome C oxidase, and the cellular response to hypoxia. Arterioscl. Throm. Vas. 30, 643–647. 10.1161/ATVBAHA.108.181628 [DOI] [PubMed] [Google Scholar]
- Thomson A. J., Giannopoulos G., Pretty J., Baggs E. M., Richardson D. J. (2012). Biological sources and sinks of nitrous oxide and strategies to mitigate emissions. Philos. T. R. Soc B. 367, 1157–1168. 10.1098/rstb.2011.0415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Yang J., Lu C., Xu R., Canadell J. G., Jackson R. B., et al. (2018). The global N2O model intercomparison project. B. Am. Meteorol. Soc. 99, 1231–1251. 10.1175/BAMS-D-17-0212.1 [DOI] [Google Scholar]
- Tischner R., Planchet E., Kaiser W. M. (2004). Mitochondrial electron transport as a source for nitric oxide in the unicellular green alga Chlorella sorokiniana. FEBS Lett. 576, 151–155. 10.1016/j.febslet.2004.09.004 [DOI] [PubMed] [Google Scholar]
- van der Leij M., Smith S., Miller A. (1998). Remobilisation of vacuolar stored nitrate in barley root cells. Planta 205, 64–72. 10.1007/s004250050297 [DOI] [Google Scholar]
- Vanecko S., Varner J. E. (1955). Studies on nitrite metabolism in higher plants. Plant Physiol. 30:388. 10.1104/pp.30.4.388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartapetian B. B., Andreeva I. N., Generozova I. P., Polyakova L., II, Maslova I. P., Dolgikh Y., II, et al. (2003). Functional electron microscopy in studies of plant response and adaptation to anaerobic stress. Ann. Bot. 91, 155–172. 10.1093/aob/mcf244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek L. A., Sasidharan R., Visser E. J., Bailey-Serres J. (2016). Flooding stress signaling through perturbations in oxygen, ethylene, nitric oxide and light. New Phytol. 209, 39–43. 10.1111/nph.13775 [DOI] [PubMed] [Google Scholar]
- von Wirén N., Gazzarrini S., Gojon A., Frommer W. B. (2000). The molecular physiology of ammonium uptake and retrieval. Curr. Opin. Plant Biol. 3, 254–261. 10.1016/S1369-5266(00)80074-6 [DOI] [PubMed] [Google Scholar]
- Wany A., Kumari A., Gupta K. J. (2017). Nitric oxide is essential for the development of aerenchyma in wheat roots under hypoxic stress. Plant Cell Environ. 40, 3002–3017. 10.1111/pce.13061 [DOI] [PubMed] [Google Scholar]
- Wany A., Gupta A. K., Kumari A., Mishra S., Singh N., Pandey S., et al. (2019). Nitrate nutrition influences multiple factors in order to increase energy efficiency under hypoxia in Arabidopsis. Ann. Bot. 123, 691–705. 10.1093/aob/mcy202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward N. D., Indivero J., Gunn C., Wang W., Bailey V., McDowell N. G. (2019). Longitudinal gradients in tree stem greenhouse gas concentrations across six Pacific Northwest coastal forests. J. Geophys. Res.- Biogeo. 124, 1401–1412. 10.1029/2019JG005064 [DOI] [Google Scholar]
- Weathers P. J., Niedzielski J. J. (1986). Nitrous oxide production by cyanobacteria. Arch. Microbiol. 146, 204–206. 10.1007/BF00402352 [DOI] [Google Scholar]
- Weathers P. J. (1984). N2O evolution by green algae. Appl. Environ. Microb. 48, 1251–1253. 10.1128/AEM.48.6.1251-1253.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch B., Gauci V., Sayer E. J. (2019). Tree stem bases are sources of CH4 and N2O in a tropical forest on upland soil during the dry to wet season transition. Global Change boil. 25, 361–372. 10.1111/gcb.14498 [DOI] [PubMed] [Google Scholar]
- Wen Y., Corre M. D., Rachow C., Chen L., Veldkamp E. (2017). Nitrous oxide emissions from stems of alder, beech and spruce in a temperate forest. Plant Soil 420, 423–434. 10.1007/s11104-017-3416-5 [DOI] [Google Scholar]
- Wendehenne D., Pugin A., Klessig D. F., Durner J. (2001). Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends Plant Sci. 6, 177–183. 10.1016/S1360-1385(01)01893-3 [DOI] [PubMed] [Google Scholar]
- Woehle C., Roy A. S., Glock N., Wein T., Weissenbach J., Rosenstiel P., et al. (2018). A novel eukaryotic denitrification pathway in foraminifera. Curr. Biol. 28, 2536–2543. 10.1016/j.cub.2018.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff A., Oliveira H. C., Saviani E. E., Salgado I. (2009). Nitrite reduction and superoxide-dependent nitric oxide degradation by Arabidopsis mitochondria: influence of external NAD (P) H dehydrogenases and alternative oxidase in the control of nitric oxide levels. Nitric. Oxide 21, 132–139. 10.1016/j.niox.2009.06.003 [DOI] [PubMed] [Google Scholar]
- Zhao X. J., Sampath V., Caughey W. S. (1995). Cytochrome c oxidase catalysis of the reduction of nitric oxide to nitrous oxide. Biochem. Bioph. Res. Co. 212, 1054–1060. 10.1006/bbrc.1995.2076 [DOI] [PubMed] [Google Scholar]
- Zhao L., Liu F., Crawford N. M., Wang Y. (2018). Molecular regulation of nitrate responses in plants. Int. J. Mol. Sci. 19, 2039. 10.3390/ijms19072039 [DOI] [PMC free article] [PubMed] [Google Scholar]