Abstract
Lactobacilli commonly used as a probiotic and they can be isolated from various sources such as fermented foods and gastrointestinal tracts of humans and animals. The aims of this study were isolation and identification of lactobacilli from honey and investigation of some probiotic properties and antimicrobial effects against foodborne bacterial pathogens. A total of 88 honey samples were collected from different areas in Iran. About 1.00 g of each honey was cultured in de Man, Rogosa, and Sharpe (MRS) broth and then sub-cultured on MRS agar. The isolates were assessed for probiotic potentials such as tolerance to acid and bile. Then, antimicrobial activity of isolates against seven foodborne pathogens including Listeria monocytogenes, Shigella flexneri, Staphylococcus aureus, Salmonella enteritidis, Enteropathogenic Escherichia coli, Escherichia coli O157 H7 and Bacillus cereus was investigated. From 88 honey samples, 39 isolates were identified by 16S rDNA gene sequencing method. Fructophilic lactic acid bacteria (FLAB) with 29 (74.00%) isolates were dominant identified bacteria (27 L. kunkeei and two Fructobacillus fructosus). Also, four L. plantarum, two L. paracasei, one L. brevis, one L. rhamnosus, one L. casei and one L. fermentum were identified. Two L. kunkeei isolates and one F. fructosus isolate were resistant to acid and bile salt. Two L. rhamnosus isolates and one L. paracasei isolate inhibited all pathogens (100%). This is the first study in Iran that isolated lactobacilli from honey. The FLAB especially L. kunkeei were isolated as dominated species from honey. Some lactobacilli isolates have probiotic potential and may be useful for the prevention and treatment of infections, but more investigations are needed.
Key Words: Antimicrobial, Foodborne pathogens, Honey, Lactobacillus, Probiotic
Introduction
Lactic acid bacteria (LAB) are found in various fermented foods and gastrointestinal tracts of humans and animals.1,2 Fructophilic lactic acid bacteria (FLAB) are a narrow but special group in LAB preferring to grow in fructose-rich niches, e.g., honey, flowers, fruits and insects like honey-bees and ants such as Camponotus japonicas.3,4 The L. kunkeei and Fructobacillus spp. are two representatives for the FLAB group.5 The L. kunkeei is the only fructophilic species among lactobacilli.6 Honeybees diets are fructose-rich, so the gastrointestinal tracts of these insects are proper for FLAB growth.1 The identification of Lactobacillus species using biochemical methods is very difficult largely because of the need of many biochemical tests. In contrast to the phenotypic methods, genetic identification methods such as 16S rDNA sequencing are more consistent, rapid, reliable, and reproducible and can discriminate even between closely related species.7
Probiotics are live micro-organisms, which when consumed in an adequate amount confer health benefits to the host by altering indigenous microflora.8 Probiotic bacteria must be resistant to gastric acidity and bile salts and adhere to the intestinal epithelial cells.2 Some isolated LAB from honey can be probiotic and be able to inhibit pathogens. Other mechanisms for antimicrobial activity of honey are due to different factors for example osmolarity, acidity, hydrogen peroxide and non-peroxide compounds like flavonoids and benzoic and cinnamic acids.9 Lacto-bacillus species as common probiotics isolated from various foods can inhibit pathogens.10-12 For example, in a study, L. acidophilus isolates isolated from honey marketed in Malaysia showed antibacterial activity against multiple antibiotic-resistant Gram-positive bacteria.13 Although these bacteria can inhibit human pathogens, they also may have antimicrobial activity against bee pathogens. In another research, Endo and Salminen., isolated FLAB including L. kunkeei and F. fructosus from flowers that one of the L. kunkeei isolates showed antibacterial activity against Melissococcus plutonius, a causative pathogen of European foulbrood.1
To our knowledge, isolation, and investigation of the probiotic potential of LAB, especially FLAB from honey have not been studied yet in Iran. So, the aim of this study was isolation and identification of Lactobacillus in Iranian kinds of honey and examination of probiotic properties and antibacterial activity of Lactobacillus isolates against foodborne pathogens.
Materials and Methods
Sample collection and culture. A total of 88 honey samples were collected randomly from different areas in Iran, especially mountains, plains, and forests in Mazandaran (north of Iran). Honey samples were taken from beekeepers during spring and summer 2017. About 10.00 g of each sample was collected in a sterile container, labeled and immediately transferred to the Microbiology Laboratory of Babol University of Medical Sciences, Babol, Iran. Approximately 1.00 g of honey samples were suspended in 9.00 mL de Man, Rogosa, and Sharpe (MRS) broth (Merck, Darmstadt, Germany) and incubated at 30.00 ˚C for 3-7 days in a candle jar. Then, subcultured on MRS agar (Merck) and incubated at 30.00 ˚C for 2-5 days in a candle jar. About three to five different colonies in the size or shape of each positive culture were selected for further investigation. Gram-positive and catalase-negative rods were stored in tubes containing MRS broth with 20.00% glycerol at – 20.00 ˚C for further investigations.
Molecular identification. Lactobacillus isolates were identified to the species level by 16S rDNA gene sequencing method. Genomic DNA extraction of isolates was performed by boiling methods. A single colony from each isolate was suspended in 50.00 mL of TES buffer containing 50.00 mM Tris hydrochloride (pH = 8.00), 5.00 mM EDTA and 50.00 mM NaCl) and the suspension was heated in a boiling water bath at 95.00 ˚C for 10 min. Then, the suspension was centrifuged at 15,000 g for 3 min and the supernatant was used as a DNA template.14 The polymerase chain reaction (PCR) primer sequences were as follows: Forward primer, 5′-CTCGTTGCGGGA CTTAA-3′ and reverse primer, 5′-GCAGCAGTAGGGAATC TTC-3′ (Bioneer, Daejeon, Korea). The reaction mixture consisted of 0.25 pmol primers, 1.50 mmol MgCl2, 0.20 mmol dNTPs, 10.00 ng of genomic DNA, 1X PCR buffer, and 3.75 U of Taq DNA polymerase (Takapouzist, Tehran, Iran) in a final volume of 50.00 mL. The PCR program started with an initial denaturation at 94.00 ˚C for 5 min, followed by 35 cycles of 94.00 ˚C for 1 min, 55.00 ˚C for 1 min and 72.00 ˚C for 1 min and terminated by one cycle of 72.00 ˚C for 10 min as a final extension. The PCR products were separated by agarose gel electrophoresis (1.50%; w/v) containing safe stain (Yekta Tajhiz, Tehran, Iran). The PCR products were sequenced (Bioneer) and finally 16S rDNA sequences were compared with known sequences in GeneBank using BLAST.The L. acidophilus ATCC 4356 and L. rhamnosus GG were used as control isolates in PCR reactions. 15
Acid and bile resistance. Probiotic tests such as tolerance to acid and bile were performed. In the acid tolerance test, 1.00 mL of the fresh culture of Lactobacillus isolates in MRS broth with the concentration of 109 CFU mL-1 was transferred into 9.00 mL phosphate-buffered saline (pH 3.30) and incubated at 30.00 ˚C for 3 hr. The number of viable bacteria was determined by plating onto MRS agar at time zero and 3 hr after incubation. The Lactobacillus isolates survived with colony counting more than 106 CFU mL-1, considered as acid-tolerant. In the bile tolerance test for each isolate, two tubes with 9.00 mL MRS broth were considered, one with 0.30% (w/v) oxgall bile (Sigma, Neustadt, Germany) and another without it. Ninety microliters of the fresh culture of Lactobacillus isolates in MRS broth were inoculated in two MRS broth tubes and tubes were incubated at 30.00 ˚C for 8 hr. The growth rate of Lactobacillus isolates was evaluated by measuring the absorbance at 600 nm at time 0 and 8 hr after of incubation.2 Coefficient of inhibition (Cinh) was calculated using the following method described by Gopal et al.:16
Cinh = ( Δ T 8 -T 0 Control - Δ T 8 -T 0 Treatment)/( Δ T 8 -T 0 Control)
where, Δ represented the differences in absorbance between T0 (zero hr reading) and T8 (reading at 8th hr). The test was performed twice for each isolate. Based on calculated Cinh, isolates were classified into non-sensitive (resistant) to 0.30% bile salt (Cinh≈ 0), with retarded growth (0.20 < Cinh < 0.40) and poorly tolerant (Cinh > 0.4). The L. acidophilus ATCC 4356 was used as a control.
Antimicrobial activity. Antimicrobial activity was carried out by agar well diffusion assay.17 Foodborne pathogenic bacteria including L. monocytogenes PTCC 1295, S. flexneri ATCC 12022, S. aureus ATCC 25923, Salmonella enteritidis F17, Enteropathogenic E. coli (EPEC) E2348/69, E. coli O157 H7 EDL 933 and B. cereus D14 were cultured on nutrient agar (Scharlau, Barcelona, Spain) at 37.00 ˚C for 24 hr. Then, a microbial density of about 107 CFU mL-1of each pathogen was prepared in normal saline. Lactobacillus isolates were grown in MRS broth at 30.00 ˚C for 24 hr in a candle jar. Cell-free culture supernatants (CFCSs) were obtained by centrifuging the MRS broth (10,000 g for 10 min). Finally, pathogenic bacteria were sub-cultured on nutrient agar and 100 µL of the CFCSs were placed into the wells of the nutrient agar and incubated at 37.00 ˚C for 15 hr. The diameter of the inhibition zones around the wells was measured. Isolates with clear inhibition less than 11.00 mm, 11.00-16.00 mm, 17.00-22.00 mm and more than 23.00 mm, were classified as negative (-), mild (+), strong (++) and very strong (+++) inhibitor, respectively. The L. acidophilus ATCC 4356 and L. rhamnosus GG were used as positive controls and sterile MRS broth was used as a negative control.
Antibiotic susceptibility. Antibiogram was studied by the Kirby Bauer disc diffusion method.18 Lactobacillus isolates were cultured in MRS broth and then their concentration adjusted to 0.50 McFarland turbidity standard and inoculated onto agar plates containing a mixed formulation of Mueller-Hinton agar (Scharlau) added with 10.00% (w/v) MRS broth. Antibiotic disks (MAST Diagnostics, Merseyside, UK) were placed onto the agar, and plates were incubated at 30.00 ˚C for 48 hr in a candle jar. Inhibition zone diameters were measured and results were reported as resistant (≥ 15.00 mm), moderately susceptible (16.00-20.00 mm), or susceptible (21.00 mm). Eleven antibiotic disks were used as follows: Cefotaxime (30.00 μg); nalidixic acid (30.00 μg); erythro-mycin (15.00 μg); genta-micin (10.00 μg); cotrimoxazole (25.00 μg); ampicillin (10.00 μg); streptomycin (10.00 μg); tetracycline (30.00 μg); vancomycin (30.00 μg); cipro-floxacin (5.00 μg) and amikacin (30.00 μg).
Results
Sample collection and culture. Eighty-eight collected honey samples were classified into four groups as follows: 51 (58.00%) mountain honey, 18 (20.00%) forest honey, 11 (13.00%) plain honey and 8 (9.00%) garden honey. Dominant plants in every group were a) Thymus and Astragalus, b) forest plants and types of grass, c) Medicagosativa, flowers, and vegetables and d) Citrus, respectively. From 88 samples, 16 (18.18%) had positive culture on MRS agar. Two-three different colonies from each positive culture were studied and finally, 39 Lactobacillus isolates were obtained which 21 isolates were from mountain honeys, 12 isolates were from plain honey and six isolates were from forest honey (Fig. 1).
Fig. 1.
Colonies of Lactobacillus isolates on MRS agar
Molecular identification. Biochemical methods are not sensitive enough for identification of Lactobacillus species; therefore 16S rDNA gene sequencing method was used for species identification. From the total of 39 isolates, 37 Lactobacillus isolates including 27 (69.00%) L. kunkeei, four L. plantarum, two L. paracasei, one L. brevis, one L. rhamnosus, one L. casei and one L. fermentum were identified. Furthermore, two isolates were identified as F. fructosus. The FLAB with 29 (74.00%) isolates were dominant among identified bacteria (Table 1).
Table 1.
Identification of Lactobacillus species in different Iranian kinds of honey
| Isolates | Accession number | Kind of honey |
|---|---|---|
| L. kunkeei H5 | KY494242.1 | Forest |
| L. kunkeei H9 | KY494418.1 | Plain |
| L. kunkeei H11 | KY494430.1 | Plain |
| L. kunkeei H12 | KY494855.1 | Mountain |
| L. kunkeei H18-2 | KY490703.1 | Forest |
| L. kunkeei H19 | KY486268.1 | Mountain |
| L. kunkeei H21 | KY486510.1 | Mountain |
| L. kunkeei H28 | KY486772.1 | Plain |
| L. kunkeei H29 | KY486298.1 | Plain |
| L. kunkeei H30 | KY486238.1 | Plain |
| L. kunkeei H31 | KY486297.1 | Plain |
| L. kunkeei H32 | KY485187.1 | Plain |
| L. kunkeei H34 | KY486266.1 | Plain |
| L. kunkeei H35 | KY486233.1 | Plain |
| L. kunkeei H36 | KY486235.1 | Plain |
| L. kunkeei H37 | KY486197.1 | Mountain |
| L. kunkeei H38 | KY486237.1 | Mountain |
| L. kunkeei H39 | KY486256.1 | Mountain |
| L. kunkeei H40 | KY486236.1 | Mountain |
| L. kunkeei H41-1 | KY485154.1 | Mountain |
| L. kunkeei H41-3 | KY485155.1 | Mountain |
| L. kunkeei H43 | KY486265.1 | Mountain |
| L. kunkeei H45 | KY486776.1 | Mountain |
| L. kunkeei H48 | KY486263.1 | Mountain |
| L. kunkeei H49 | KY486196.1 | Mountain |
| L .kunkeei H50 | KY486264.1 | Mountain |
| L. kunkeei H51 | KY485156.1 | Mountain |
| L. plantarum H59 | KY486194.1 | Mountain |
| L. plantarum H46 | KY486189.1 | Plain |
| L. plantarum H47 | KY486193.1 | Plain |
| L. plantarum H15 | KY494858.1 | Forest |
| L. paracasei H13 | KY485186.1 | Mountain |
| L. paracasei H14 | KY486195.1 | Mountain |
| L. brevis H8 | KY490536.1 | Mountain |
| L. rhamnosus H3 | KY486198.1 | Forest |
| L. casei H7 | KY514165.1 | Forest |
| L. fermentum H22 | KY486331.1 | Mountain |
| F. fructosus H25-2 | KY486190.1 | Mountain |
| F. fructosus H4 | KY497788.1 | Forest |
Acid and bile resistance. Probiotic bacteria must be resistant to some conditions such as the acidity of the stomach and bile salts.2 In bile resistance test only three isolates including L. kunkeei H41-1, L. kunkeei H41-3 and F. fructosus H25-2 had Cinh lower than 0.20 and considered as bile-resistant. Other isolates were sensitive to bile with Cinh more than 0.40, but in acid resistance test all 39 isolates were survived in an acidic condition (pH = 3.30), so 3 isolates (H41-1, H41-3, and H25-2) were resistant in both bile and acid tests. Antibiotic resistance in probiotic bacteria is not always a safety issue. When there is a risk of resistance transfer, it becomes a safety issue.
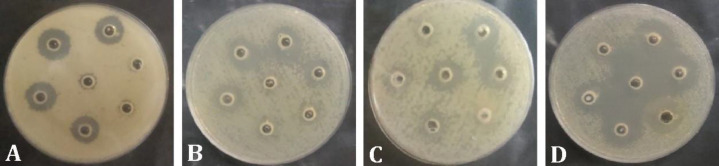
Antimicrobial activity. Fifteen isolates inhibited the growth of at least one foodborne pathogenic bacterium, but the other 24 isolates did not have an inhibitory effect (Table 2). Three isolates including L. rhamnosus H3, L. paracasei H13 and L. paracasei H14 exhibited inhibitory activity against all seven studied pathogens and three isolates of L. plantarum (H46, H47, and H59) inhibited the growth of six pathogens. In FLAB group, only seven isolates (six L. kunkeei and one F. fructosus) had inhibitory effect against one to three pathogens. The highest inhibitory effect was seen against S. flexneri and E. coli O157 H7 by 10 isolates and the lowest inhibitory effect was seen against L. monocytogenes by four isolates (Fig. 2).
Table 2.
Antimicrobial activity of cell-free culture supernatants of lactic acid bacteria isolates against foodborne pathogenic bacteria
| Bacteria |
B. cereus
D14 |
S. enteritidis
F17 |
EPEC
E2348/69 |
E. coli
O157 H7 EDL 933 |
S. flexneri
ATCC 12022 |
S. aureus
ATCC 25923 |
L. monocytogenes
PTCC 1295 |
|---|---|---|---|---|---|---|---|
| L. rhamnosus H3 | 15 (+) * | 13 (+) | 15 (+) | 12 (+) | 23 (+++) | 18 (++) | 18 (++) |
| L. paracasei H13 | 14 (+) | 14 (+) | 11 (+) | 11 (+) | 19 (++) | 18 (++) | 13 (+) |
| L. paracasei H14 | 15 (+) | 15 (+) | 16 (+) | 15 (+) | 16 (+) | 16 (+) | 16 (+) |
| L. plantarum H46 | 13 (+) | 13 (+) | 10 (-) | 14 (+) | 19 (++) | 14 (+) | 15 (+) |
| L. plantarum H47 | 12 (+) | 13 (+) | 11 (+) | 16 (+) | 20 (++) | 16 (+) | 10 (-) |
| L. plantarum H59 | 13 (+) | 20 (++) | 14 (+) | 21 (+) | 26 (+++) | 20 (++) | 10 (-) |
| F. fructosus H4 | 0 (-) | 10 (-) | 0 (-) | 0 (-) | 11 (+) | 0 (-) | 0 (-) |
| F. fructosus H25-2 | 0 (-) | 10 (-) | 0 (-) | 9 (-) | 11 (+) | 8 (-) | 0 (-) |
| L. kunkeei H21 | 0 (-) | 10 (-) | 0 (-) | 9 (-) | 11 (+) | 0 (-) | 0 (-) |
| L. kunkeei H32 | 0 (-) | 7 (-) | 0 (-) | 8 (-) | 11 (+) | 7 (-) | 0 (-) |
| L. kunkeei H34 | 0 (-) | 11 (+) | 0 (-) | 0 (-) | 0 (-) | 6 (-) | 0 (-) |
| L. kunkeei H41-1 | 0 (-) | 11 (+) | 7 (-) | 11 (+) | 8 (-) | 0 (-) | 0 (-) |
| L. kunkeei H41-3 | 0 (-) | 11 (+) | 7 (-) | 11 (+) | 8 (-) | 0 (-) | 0 (-) |
| L. kunkeei H48 | 0 (-) | 0 (-) | 11 (+) | 12 (+) | 0 (-) | 18 (++) | 0 (-) |
| L. fermentum H22 | 8 (-) | 0 (-) | 0 (-) | 13 (+) | 6 (-) | 0 (-) | 0 (-) |
| L. rhamnosus GG | 16 (+) | 14 (+) | 15 (+) | 15 (+) | 20 (++) | 18 (++) | 11 (+) |
| L. acidophilus ATCC 4356 | 13 (+) | 13 (+) | 12 (+) | 13 (+) | 19 (++) | 12 (+) | 11 (+) |
*Interpretation of zone inhibition diameter. -, less than 11 mm; +, 11-16 mm; ++, 17-22 mm and +++, more than 23 mm.
Fig. 2.
Antimicrobial activity of CFCS of Lactobacillus isolates against some foodborne pathogens. A) Bacillus cereus D14; B) Salmonella enteritidis F17; C) Enteropathogenic E. coli E2348/69; D) Shigella flexneri ATCC 12022
Antibiotic susceptibility. The LAB used as probiotic bacteria should not harbor transmissible antibiotic resistance genes.2 Antibiotic susceptibility of isolates was investigated by 11 different antibiotics. Isolates showed the highest resistance to vancomycin (100%), nalidixic acid (100%), cotrimoxazole (97.40%), streptomycin (97.40%), ciprofloxacin (92.30%), gentamicin (82.00%) and tetracycline (53.80%). Also, the lowest resistance was seen to ampicillin (0.00%), erythromycin (0.00%), cefo-taxime (7.60%) and amikacin (38.40%).
Discussion
The FLAB are a new group in LAB have been more investigated in the last few years. To our knowledge, this is the first study in Iran isolating lactobacilli from honey. In the present study, FLAB especially L. kunkeei were isolated as dominated species from honey about 74.00% of the total identified species which this result is following previous studies. For example, Endo and Salminen have isolated 66 isolates from honey, bees, flowers, and larvae in Fenland in 2013.1 All of these isolates were FLAB consisting of 63 L. kunkeei and three F. fructosus. Also, Endo et al. in another study in 2012 have investigated nine Lactobacillus isolates previously isolated from honey, flowers, and wine in different countries and identified all of them as obligatory FLAB.19 Asama et al. have also isolated 78 isolates from whole guts and honey stomachs in bees and nine isolates from bee bread in Japan in 2015. Their results showed that all isolates were L. kunkeei.20 Aween et al. have isolated six L. acidophilus isolates from 13 marketed honey in Malaysia in 2012, but they did not report any isolate of L. kunkeei.13 Our study like others showed that L. kunkeei is the most frequent species in fructose-rich niches such as honey and bees.
Our results showed that among all isolates, L. rhamnosus, L. paracasei, and L. plantarum had a very good inhibitory effect on the most of studied foodborne pathogens, but FLAB species had an inhibitory effect against few pathogens. It means other LAB species can inhibit the growth of pathogens stronger than FLAB species. The antimicrobial activity of FLAB has not been studied yet. It seems that the present study is the first investigation about antimicrobial activity of FLAB especially L. kunkeei isolated from honey. Aween et al. have reported that L. acidophilus isolates from honey have good antimicrobial activity against S. aureus.13
In the present study, isolates showed the highest resistance to vancomycin, nalidixic acid, cotrimoxazole, streptomycin, ciprofloxacin, gentamicin, and tetracycline. Antibiotic resistance in LAB is not always a safety issue. When there is a risk of resistance transfer, it becomes a safety issue. The origin of antibiotic resistance in probiotics can be intrinsic, acquired as a result of mutations in the chromosome, or acquired by horizontal gene transfer. In intrinsic resistance or acquired resistance due to chromosomal mutations, the transfer risk is considered to be very low, but in horizontally transferred antibiotic resistance, genes such as transposons and plasmids can spread mainly by conjugation and the transfer risk is high.21 For example, high levels of resistance to vancomycin or aminoglycosides such as streptomycin have been reported in several studies.22-24 It has been suggested that the resistance to vancomycin and aminoglycosides is mostly intrinsic.25,26 The presence of antibiotic resistance determinants in a probiotic's genome must be systematically screened before usage.
In conclusion, different species of Lactobacillus present in Iranian's kinds of honey which FLAB mainly L. kunkeei are dominant Lactobacillus among them. Some isolates showed good probiotic properties such as resistance to acid and bile or antimicrobial activity against foodborne pathogens. These results suggest that Lactobacillus isolates from honey may be useful for the prevention or treatment of foodborne infections, but more studies are still required.
Acknowledgments
We are grateful to Dr. Mahdi Rajabnia for his support in Babol University of Medical Sciences, Babol, Iran. This work was supported by the Vice-Chancellor of Tehran University of Medical Sciences, Tehran, Iran under Grant number 32129.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Endo A, Salminen S. Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst Appl Microbiol. 2013;36(6):444–448. doi: 10.1016/j.syapm.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Davoodabadi A, Soltan Dallal MM, Foroushani AR, et al. Antibacterial activity of Lactobacillus spp isolated from the feces of healthy infants against enteropathogenic bacteria. Anaerobe. 2015;34:53–58. doi: 10.1016/j.anaerobe.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Endo A, Futagawa-Endo Y, Dicks LM. Isolation and characterization of fructophilic lactic acid bacteria from fructose-rich niches. Syst Appl Microbiol. 2009;32(8):593–600. doi: 10.1016/j.syapm.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 4.He H, Chen Y, Zhang Y, et al. Bacteria associated with gut lumen of Camponotusjaponicus Mayr. Environ Entomol. 2011;40(6):1405–1409. doi: 10.1603/EN11157. [DOI] [PubMed] [Google Scholar]
- 5.Endo A, Futagawa-Endo Y, Sakamoto M, et al. Lactobacillusflorum sp nov a fructophilic species isolated from flowers. Int J Syst Evol Microbiol. 2010;60(Pt 10):2478–2482. doi: 10.1099/ijs.0.019067-0. [DOI] [PubMed] [Google Scholar]
- 6.Endo A. Fructophilic lactic acid bacteria inhabit fructose-rich niches in nature. Microb Ecol Health Dis. 2012;23(1):18563. doi: 10.3402/mehd.v23i0.18563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh S, Goswami P, Singh R, et al. Application of molecular identification tools for Lactobacillus, with a focus on discrimination between closely related species: A review. LWT-Food Sci Technol. 2009;42(2):448–457. [Google Scholar]
- 8.Ouwehand AC, Kirjavainen PV, Shortt C, et al. Probiotics: Mechanisms and established effects. Int Dairy J. 1999;9(1):43–52. [Google Scholar]
- 9.Weston RJ. The contribution of catalase and other natural products to the antibacterial activity of honey: A review. Food Chem. 2000;71(2):235–239. [Google Scholar]
- 10.Halder D, Shyamapada M. Antibacterial potentiality of commercially available probiotic lactobacilli and curd lactobacilli strains, alone and in combination, against human pathogenic bacteria. Transl Biomed. 2016;7(2):1–7. [Google Scholar]
- 11.Soltan Dallal MM, Davoodabadi A, Abdi M, et al. Inhibitory effect of Lactobacillusplantarum and Lb fermentum isolated from the feces of healthy infants against nonfermentative bacteria causing nosocomial infections. New Microbes New Infect. 2017;15:9–13. doi: 10.1016/j.nmni.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zare Mirzaei E, Lashani E, Davoodabadi A. Anti-microbial properties of lactic acid bacteria isolated from traditional yogurt and milk against Shigella strains. GMS Hyg Infect Control. 2018;13:1–5. doi: 10.3205/dgkh000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aween MM, Hassan Z, Muhialdin BJ, et al. Antibacterial activity of Lactobacillus acidophilus strains isolated from honey marketed in Malaysia against selected multiple antibiotic resistant (MAR) Gram-positive bacteria. J Food Sci. 2012;77(7):364–371. doi: 10.1111/j.1750-3841.2012.02776.x. [DOI] [PubMed] [Google Scholar]
- 14.Antonsson M, Molin G, Ardö Y. Lactobacillus strains isolated from Danbo cheese as adjunct cultures in a cheese model system. Int J Food Microbiol. 2003;85(1-2):159–169. doi: 10.1016/s0168-1605(02)00536-6. [DOI] [PubMed] [Google Scholar]
- 15.Yun JH, Lee KB, Sung YK, et al. Isolation and characterization of potential probiotic lactobacilli from pig feces. J Basic Microbiol. 2009;49(2):220–226. doi: 10.1002/jobm.200800119. [DOI] [PubMed] [Google Scholar]
- 16.Gopal PK, Prasad J, Smart J, et al. In vitro adherence properties of Lactobacillusrhamnosus DR20 and Bifidobacteriumlactis DR10 strains and their anta-gonistic activity against an enterotoxigenic Escherichiacoli. Int J Food Microbiol. 2001;67(3):207–216. doi: 10.1016/s0168-1605(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 17.Rammelsberg M, Radler F. Antibacterial polypeptides of Lactobacillus species. J Appl Microbiol. 1990;69(2):177–184. [Google Scholar]
- 18.Hashemi SMB, Shahidi F, Mortazavi SA, et al. Potentially probiotic Lactobacillus strains from traditional Kurdish cheese. Probiotics Antimicrob Proteins. 2014;6(1):22–31. doi: 10.1007/s12602-014-9155-5. [DOI] [PubMed] [Google Scholar]
- 19.Endo A, Irisawa T, Futagawa-Endo Y, et al. Characterization and emended description of Lactobacillus kunkeei as a fructophilic lactic acid bacterium. Int J Syst Evol Microbiol. 2012;62(Pt 3):500–504. doi: 10.1099/ijs.0.031054-0. [DOI] [PubMed] [Google Scholar]
- 20.Asama T, Arima TH, Gomi T, et al. Lactobacillus kunkeei YB38 from honeybee products enhances IgA production in healthy adults. J Appl Microbiol. 2015;119(3):818–826. doi: 10.1111/jam.12889. [DOI] [PubMed] [Google Scholar]
- 21.Gueimonde M, Sánchez B, Clara G, et al. Antibiotic resistance in probiotic bacteria. Front Microbiol. 2013;4:1–6. doi: 10.3389/fmicb.2013.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirtzalidou E, Pramateftaki P, Kotsou M, et al. Screening for lactobacilli with probiotic properties in the infant gut microbiota. Anaerobe. 2011;17(6):440–443. doi: 10.1016/j.anaerobe.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J, Pillidge C, Gopal P, et al. Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifido-bacterium strains. Int J Food Microbiol. 2005;98(2):211–217. doi: 10.1016/j.ijfoodmicro.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 24.Arici M, Bilgin B, Sagdic O, et al. Some characteristics of Lactobacillus isolates from infant faeces. Food Microbiol. 2004;21(1):19–24. [Google Scholar]
- 25.Saarela M, Mogensen G, Fonden R, et al. Probiotic bacteria: Safety, functional and technological properties. J Biotech. 2000;84(3):197–215. doi: 10.1016/s0168-1656(00)00375-8. [DOI] [PubMed] [Google Scholar]
- 26.Ammor MS, Flórez AB, Mayo B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 2007;24(6):559–570. doi: 10.1016/j.fm.2006.11.001. [DOI] [PubMed] [Google Scholar]