Abstract
SARS-CoV-2 interaction with the ACE-2 receptor cannot alone explain the demography and remarkable variation in clinical progression of Covid-19 infection.
Unlike SARS-CoV, the cause of SARS, several SARS-CoV-2 spike glycans contain sialic acid residues. In contrast to the SARS secreted glycoprotein (SGP), SARS-CoV-2 SGP are thus potential ligands for Sialic acid-binding Siglecs on host immune cells, known to regulate immune function. Such SARS-CoV-2 glycoproteins would contribute to immune deviation.
CD33-related Siglecs are important immune regulators. Siglec-5 and −14 are paired receptors with opposed actions on the NLRP3 inflammasome, which is critical in early viral clearance. SGP binding in persons of Siglec-14 null genotype (30–70% in Black, Asian and Minority Ethnic (BAME) persons, 10% in North Europeans) would induce unopposed inhibitory signalling, causing viral persistence through inflammasome inhibition.
Siglec-3 (CD33) and Siglec-5 are expressed on CD33 myeloid derived suppressor cells (CD33 MDSC). Immunosuppressive CD33 MDSC populations are increased in all groups at risk of severe Covid-19 infection. CD33 expression is increased in persons with the CD33 rs3865444 CC allele, associated with Alzheimer’s disease, who would thus show enhanced susceptibility. Viral SGP ligation of CD33, potentially in conjunction with Siglec-5, would promote expansion of CD33 MDSC cells, as occurs in cancers but at much greater scale.
CD33 is expressed on CNS microglia, potentially activated by SGP penetration through the porous cribriform plate to cause anosmia.
Genotyping of severe or fatal Covid-19 cases can confirm or refute this pathophysiological mechanism. Early data have confirmed extremely high-level increase of CD33 MDSC numbers in severe Covid-19 infection, consistent with the proposed mechanism.
Susceptibility to severe Covid-19 infection
The variability of Covid-19 infection is remarkable, with responses ranging from asymptomatic clearance to death. The course of disease in symptomatic persons may be unusually prolonged.
The primary tropism of the SARS-CoV-2 virus is for angiotensin converting enzyme-2 (ACE-2), which is expressed on lung and intestinal epithelium and in heart, kidney, pancreas and brain. Widespread viral infection may thus induce multisystem inflammation, augmented by disruption of the renin-angiotensin system [1].
Several factors have emerged as associated with severe or persistent disease, including old age, diabetes, hypertension, COPD, cigarette smoking, previous or current cancer, dementia and ethnicity [2]. A theory of pathogenesis based solely on variation in ACE-2 receptor expression and targeted inflammation cannot explain many of these demographic features of Covid-19 susceptibility, the phenomenon of prolonged viral persistence or the features of ongoing disease such as late-onset cytokine storm and frequent vascular thromboses.
Potential binding of SARS-CoV-2 secreted glycans to CD33-related Siglecs
The demography of severe disease is compatible with a known viral mechanism, binding of sialylated secreted glycans to host sialic acid-binding immunoglobulin-type lectins (Siglecs) [3]. This interaction can allow immune evasion and immunosuppression [4]. Siglec binding glycans are common amongst RNA viruses, including Siglec-1 binding of macrophages by Betaarterivirus suid in Porcine Reproductive and Respiratory Syndrome [4]. Both HIV and Ebola viruses bind Siglec-1 [4]. So far, there have not been reports of viral interactions with the family of CD33-related (CD33-r) Siglecs.
CD33-r Siglecs are a subgroup of Siglecs of quite homologous structure which show evidence of unusually rapid evolution in all primate species [5], [6]. They recognise Self Associated Molecular Patterns (SAMPS), which themselves have to evolve to evade targeting by sialic acid expressing pathogens while maintaining self-recognition. There are marked inter-species differences between primates in CD33-r Siglec expression and this group of Siglec receptors have been postulated to be at the forefront of an evolutionary arms race between pathogens and hosts [6].
Pathogens expressing sialylated residues can interact with CD33-r Siglecs to modulated host immune responses. This is so far best recognised in bacteria, notably group B streptococcus (GBS), which binds to Siglecs-5 and −14 via its surface β-protein, through which it modulates neutrophil responses [4]. In humans, Siglecs 3 (CD33) and 5 are co-expressed on monocytes, macrophages and neutrophils while dendritic cells are Siglec 3+5− [4]. Siglec-14 is expressed on monocytes, macrophages and neutrophils.
CD33 is additionally expressed on CNS microglia [7], while Siglec-5 is expressed on amniotic epithelium and plays a role in GBS-induced preterm delivery [8].
Known ligands of CD33-related Siglecs
Many sialolactosamine (SLL) carbohydrate motifs have been shown to bind to CD33-r Siglecs, including 6′-SLL (Neu5Acα2-6Galβ1-4Glc), 3′-SLL (Neu5Acα2-3Galβ1-4Glc) and 3′-SLacNAc (Neu5Acα2-3Galβ1-4GlcNAc) ligands [9]. Siglec-5 binds equally to 6′-SLL, 3′-SLL and 3′-SLacNAc ligands while Siglec-3 (CD33) binds preferentially to 6′-SLL with lesser avidity for 3′-SLL and 3′-SLacNAc [9]. Thus, a single sialylated ligand can potentially signal through both CD33 and Siglec-5. Siglec-14 binding will be identical to Siglec-5 due to the common extracellular domain.
An additional sialylated carbohydrate motif, associated with shorter glycan chain and less microheterogeneity, is found in sialyl-Tn glycans. Both CD33 and Siglec-5 are significantly bound by sialyl-Tn [6], [9]. Sialyl Tn glycan binding is a functionally important modulator of CD33 signalling, as identified for HIV envelope glycoprotein Gp120 [3].
SARS-CoV-2 has 16 N-linked glycans between the S1 subunit, which binds the angiotensin-2 receptor, and the S2 subunit, which mediates membrane fusion [10]. Glycosylation mapping has identified N-glycans terminating in 3′-SLacNAc structures at N234 and N282 on spike-1 and N1098 on spike 2, while the O-glycan at T323 shows short sialic acid capped chains, including sialyl T antigen and di-sialyl core 1, trimmable to sialyl Tn [11], [12]. This represents a fundamental difference to SARS-CoV, where glycans show negligible sialylation.
The major SARS-CoV glycoprotein, mediating membrane fusion for viral entry in SARS, is also released into the circulation in soluble form [13]. Similar release through cleavage would allow SARS-CoV-2 sialylated secreted glycoproteins (SGP) to modulate immune response through CD33-r Siglecs. In particular, such interaction with Siglecs-3, −5 and −14 would explain specific features of the demography and pathogenesis of Covid-19 infection.
Potential consequence of Siglec 14 null polymorphism in Covid-19 infection
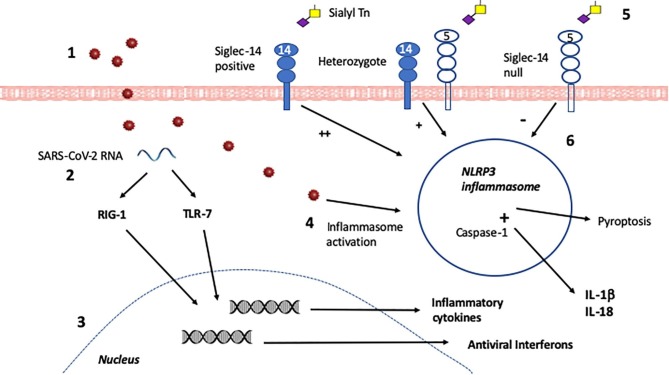
Siglec-5 and Siglec-14 are paired regulators of the Nucleotide-binding oligomerization domain, Leucine rich Repeat and Pyrin domain containing-3 (NLRP-3) inflammasome, which responds to danger signals in infection by production of inflammatory mediators (Fig. 1 ) [14]. While unregulated NLRP-3 inflammasome activation may contribute to damaging inflammatory responses in chronic infection, it is critical in the early response to infection necessary for viral eradication: viruses including Epstein-Barr virus and Zika virus have developed immune evasion strategies by inhibiting the NLRP3 inflammasome [14], [15].
Fig. 1.
Innate immunity in the initial phase of infection. The initial immune response to SARS-CoV-2 by innate immune cells, after its entry via the ACE-2 receptor on lung or gut epithelium, determines whether or not infection is swiftly controlled and minimally symptomatic. 1. SARS-CoV-2 virions are taken up by a tissue-based macrophage or circulating monocyte. 2. Within the cell, SARS-CoV-2 single-stranded RNA is recognised by the RNA-responsive intracellular sensors RIG-1 and TLR-7 and signalling pathways are activated. 3. Due to RIG-1 and TLR-7 activation, nuclear transcription of antiviral interferons and inflammatory mediators is stimulated. Antiviral interferons predominate in females and inflammatory cytokines in males after TLR7 signalling, contributing to more efficient viral clearance in females.4. Intracellular disturbance caused by whole SARS-CoV-2 virions cause activation of the NLRP3 inflammasome. This induces pyroptosis, an inflammatory programmed cell death pathway for infected cells which limits viral replication and release. In addition, pro-IL-1β and pro-IL-18 cytokines are produced, and cleaved to active IL-1β and IL-18 by Caspase-1. Such inflammasome activation is critical in an effective antiviral response. 5. NLRP3 activation is regulated by signals from cell surface Siglecs, if these are stimulated by Siglec ligands. An appropriately configured sialyl Tn or 3′SLacNac secreted glycoprotein would bind equally to Siglec-5 or Siglec-14 as their extracellular domain is almost identical. 6. In a homozygous Siglec-14 positive person such glycoprotein binding induces strong activation of the NLRP3 inflammasome, while in a Siglec-14 null person this induces strong inhibition. In a person who is heterozygous, a more modest activation response occurs. Thus, an infected person who is homozygous Siglec-14 positive is more likely to mount a successful early antiviral response than one who is Siglec-14 null, with heterozygotes making an intermediate response.
Siglec-5 and −14 are paired receptors, having essentially identical extracellular binding domains but differing intracellular signalling pathways [8]. Siglec-5 associates with tyrosine phosphatase SHP-1 to send an inhibitory signal upon ligation, while Siglec-14 associates with the adapter protein DAP-12 and the tyrosine kinase Syk to send an activating signal. Thus Siglec-5 binding downregulates NLRP3 inflammasome activity, while Siglec-14 binding upregulates it [8], [16]. Importantly, there is a common polymorphism, in which Siglec-14 is replaced by a Siglec-5 like fusion protein (Siglec 14/5) signalling via the inhibitory SHP-1 pathway identically to Siglec-5 [17]. Individuals can express both Siglec-5 and −14 (Siglec-14+/+), can have one allele where Siglec-14 is replaced by 14/5 (Siglec-14+/- heterozygous) or have both alleles where Siglec-14 is replaced by 14/5 (Siglec-14-/-, known as Siglec-14 Null allele) [18].
Thus, persons with Siglec-14 Null allele receive an inhibitory input to the NLRP3 inflammasome upon Siglec-5 binding while those without have a stimulatory input, greater in Siglec-14+/+ homozygotes than heterozygotes (Fig. 1) [16], [18]. There are recognised functional consequences. Adults with the Siglec-14 null allele are at reduced risk for infective exacerbation of COPD [18] and may be less susceptible to severe tuberculosis [19]. Conversely Siglec-14 null women are at increased risk of premature delivery If infected with GBS [17].
There is substantial ethnic variation in Siglec-14 null allele, with a frequency of 50–70% in Chinese, South East Asians and Middle Eastern persons, 30–40% in Indo-Pakistanis and Sub-Saharan Africans and only 10% in Northern Europeans [9]. The pathogen drive currently or previously underlying such major variability remains so far unknown.
The NLRP3 inflammasome is a critical element in effective antiviral responses and its inhibition by pathogens an important cause of immune evasion, as seen in Zika virus infection [14], [15]. Binding of SARS-CoV-2 SGP to Siglec-5 would allow such immune evasion in persons of Siglec-14 null genotype through inflammasome inhibition. Newly recruited monocytes would be susceptible to similar inflammasome inhibition. Siglec-14 null women would also be more prone to preterm labour with Covid-19, as occurs if GBS colonised [8].
This mechanism would put persons of origin from China or the Far East, the Middle East, the Indian subcontinent and Sub-Saharan Africa at increased risk of inability to clear SARS-CoV-2 virus upon initial exposure and thus of more severe and chronic disease.
Increase of Siglec-3 (CD33) expressing MDSC in Covid-19 vulnerable groups
Both Siglec-3 (CD33) and Siglec-5 are expressed on myeloid-derived suppressor cells (MDSC), important mediators of immune evasion and immunosuppression [20], [21]. MDSC are recently recognised cells, best studied in cancer, where they have been described as the Queen Bee in the tumour microenvironment [20]. They are increased in persons with cancer, and remain increased in remission [22]. In cancer, they are induced by direct contact notably dependant on sialyl-Tn ligands, playing a central role in tumour induced suppression of the immune system. Thus sialyl-Tn monoclonal antibodies are receiving increasing attention as potential therapy for cancers [20].
CD33+ MDSC populations release Arginase-1 to cause arginine depletion, inducing decreased T cell receptor (TCR)-ζ chain expression and impaired adaptive immune responses [20], [21]. They also release the immunosuppressive cytokines TGF-β and IL-10, as well as effector molecules such as nitric oxide and reactive oxygen metabolites. In addition B cell proliferation and antibody production are diminished by MDSC activation [23].
CD33 MDSC populations are not restricted to cancer, and are increased in a number of other conditions. Indeed, in all other conditions known to be associated with increased Covid-19 mortality, circulating CD33+ MDSC are increased. Significant increase is seen in persons aged 61–76, compared to those aged 19–59, with further increase in frail elderly [22].
Increased CD33 + MDSC numbers in cancer are associated with decreased TCR-ζ expression, contributing to compromised anti-tumour responses [24]. CD33+HLA-DRlow/- MDSC cells are significantly increased in obesity, associated with decreased TCR-ζ expression [25]. Type-2 diabetes mellitus and arterial hypertension cause increased circulating CD33+ MDSC cells producing immunosuppressive TGF-β and IL-10 [26]. Circulating CD33+HLA-DR- MDSC are upregulated in smokers with or without chronic obstructive pulmonary disease (COPD) [27]. In COPD this persists after stopping smoking, associated with significant downregulation of TCR-ζexpression [27]. Stopping smoking reduces CD33 + MDSC numbers.
Persons with dementia, likely to have high age-related CD33 MDSC, have increased frequency of the CD33 rs3865444 CC allele which upregulates CD33 expression on microglia and immune cells [7]. Monocyte expression of CD33 is upregulated 7-fold in young persons carrying this allele, which explains over 70% of variance in CD33 cell expression [28]. While its putative role in dementia is that this CD33 variant impairs protective uptake of β-amyloid, the relevance of this allele in CD33 rs3865444 CC carriers of whatever age in Covid-19 infection would likely be of increased response by MDSC and monocytes to a CD33-binding viral SGP. This may therefore be a second genetic predisposition to severe Covid-19 infection.
Potential consequences of CD33 MDSC expansion by SARS-CoV-2 SGP
The excess of CD33 MDSC in persons with older age or frailty, diabetes, hypertension, COPD, cancer or previous malignancy or in those who smoke cigarettes would put them at risk of expansion of the MDSC populations by a SARS-CoV-2 Siglec-binding SGP. This would cause an inappropriate immunosuppressive response to SARS-CoV-2 infection. Persons with dementia in care homes would be at particular risk.
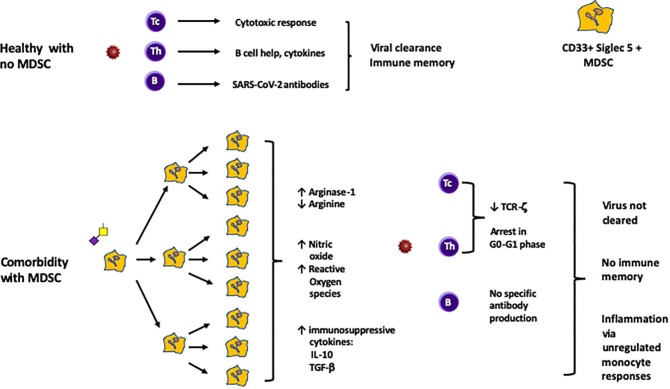
CD33 MDSC activation by SARS-CoV-2 SGP would lead to inhibition of both T cell and B cell responses through release of Arginase-1, depleting arginine, and the immunosuppressive cytokines TGF-β and IL-10. Release of reactive oxygen radicals and nitric oxide would contribute to tissue damage (Fig. 2 ). The overall impact of this phase of infection would be paralysis of adaptive immune responses.
Fig. 2.
Adaptive immunity in the second phase of infection. The response by T cells and B cells of the adaptive immune system determines whether effective immunity is induced against SARS-CoV-2. In a healthy person without comorbidities, CD4 T helper cells (Th) will expand clonally and interact with B cells to promote IgM and later IgG antibodies against SARS-CoV-2. CD8 cytotoxic T cells (Tc) will expand clonally and underpin antiviral cell mediated responses to subsequent infections. In a person with comorbidities characterised by CD33 MDSC populations, this process of immune memory would be subverted by sialyl Tn or 3′SLacNac glycoprotein released by SARS-CoV-2 virus. CD33 MDSC express both Siglec-3 (CD33) and Siglec-5, and can be of monocytic type (CD14+ ) or polymorphonuclear type (CD15+). Some comorbidities are characterised by monocytic and some by polymorphonuclear MDSC. Both types function largely identically, and both would be expanded similarly. Thus, some persons would have predominant CD14 + and others CD15 + MDSC expansion and activation, with the same functional outcome. Binding of CD33 and/or Siglec-5 by circulating secreted glycoprotein would induce massive expansion of MDSC cells (as has been confirmed in vivo in severe Covid-19 infection). Release of Arginase-1 by these cells, causing depletion of arginine, would synergise with release of the immunosuppressive cytokines IL-10 and TGF-β to impair T cell proliferation and memory cell generation, as well as prevent the effective T-B cell interaction required for antiviral antibody production. B cell proliferation and antibody production would also be impaired. Release of nitric oxide and reactive oxygen species by these cells would contribute to ongoing inflammatory responses.
There is evidence that persistent elevation of monocytic MDSC cells, identified as CD14+ HLA-DRlow, is strongly associated with worse outcome in septic shock [29]. CD14+ HLA-DRlow cells have been identified at high frequency in cases of severe Covid-19 infection with immune dysregulation [30]. Further characterisation would be required to determine how many of these cells are monocytic MDSC and how many are monocytes in which HLA-DR has been downregulated as suggested by the authors.
In addition, there is evidence that MDSC of proinflammatory potential can be released from the bone marrow in inflammation [31]. There is so far no clear consensus on their eventual phenotype but these new marrow derived cells are initially characterised as Early stage MDSC (eMDSC), which do not at first express lineage markers but do express CD33 and would thus be susceptible to SARS-CoV-2 SGP manipulation until the virus was cleared (Fig. 3 ). Proinflammatory MDSC are recognised in murine and human arthritis and in IBD, where they suppress T cell proliferation, as do mature MDSC, but also strongly promote Th17 responses [31], [32], [33].
Fig. 3.
Late-phase multisystem inflammation and vascular damage. The response by bone marrow as infection progresses may determine the clinical phenotype. Persons suffering bone marrow suppression with severe disease will not release new MDSC into the circulation. These persons will show reduced numbers of circulating monocytes and neutrophils and will tend to progress towards macrophage activation syndrome as MDSC suppression wanes. Those without bone marrow suppression maintain higher numbers of circulating cells. These include early phase MDSC, which can be stimulated through CD33 and Siglec-5. These cells are immunosuppressive but also proinflammatory, particularly promoting Th17 responses. In both groups, a separate prothrombotic pathway will be driven by SGP stimulation of Siglec-5+ cells within atherosclerotic plaques.
Analysis of immune responses in severe Covid-19 infection identified two distinct groups [30]. Around a quarter developed a macrophage activation syndrome with hypercytokinaemia and hyperferritinaemia in association with reduced total neutrophils and monocytes compared to persons with milder disease. Three quarters showed preservation of circulating cell numbers, with a state of chronic immune deviation but without macrophage activation syndrome. This association of reduced circulating cells with a hyperinflammatory response is consistent with bone marrow suppression, where release of new MDSC is prevented. By contrast, the prolonged state of immune suppression with immunopathology is explicable by viral SGP stimulation of the newly released proinflammatory eMDSC (Fig. 3).
Potential contribution to neuropathy and CNS disease
There is increasing evidence that SARS-CoV-2 may invade the central nervous system [34]. This would be most likely in severe disease with blood brain barrier disruption, and SARS-CoV-2 SGP would show tropism for CD33-expressing microglia.
A common presentation of Covid-19 disease is anosmia (loss of smell) with ageusia (loss of taste). It has been postulated that the virus might gain access through the sieve-like barrier at the cribriform plate, in proximity to the olfactory bulb [34]. Penetration of a CD33 binding SGP would potentially induce activation of CD33-expressing microglia within the olfactory bulb to cause these symptoms. This would not require ingress of whole viral structures and could occur even if blood brain barrier function was intact elsewhere.
Potential contribution to ischaemia and vascular thrombosis
In severe Covid-19 infection there is a significant increase of venous and arterial thrombosis [2]. Several mechanisms are likely to contribute to this. A CD33-r Siglec binding viral SGP would provide a further and independent risk factor for vascular thrombosis.
Analysis of 274 circulating cytokines in diabetic patients with critical limb ischaemia identified soluble Siglec-5 as the outstandingly increased marker, and confirmed this in a large second cohort, which excluded confounding risk factors [35]. Soluble Siglec-5 concentrations were considerably more than an order of magnitude greater than reported previously in healthy controls and patients with SLE [36]. The source was identified as irregularly-shaped Siglec-5+ cells that were present within atherosclerotic plaques but not normal vascular endothelium [35]. These cells were not further characterised, and thus may have been either Siglec-5+ macrophages or Siglec-5+ MDSC. The mechanism by which Siglec-5 contributes to the progression of vascular ischaemia has yet to be determined, but there is clear potential for modulation of the Siglec-5+ cells within existing vascular atherosclerotic plaques by such a Siglec-5 binding viral SGP.
Potential contribution to multisystem inflammatory syndrome in children (MIS-C).
There has been recent recognition of a Covid-19 associated syndrome of multisystem inflammation in previously healthy paediatric patients, usually without the initial presentation with respiratory disease seen in adults. By contrast, gastrointestinal symptoms were noted in around 90% of cases in both the UK and USA multicentre cohorts [37], [38]. What is notable about paediatric cases of Covid-19 infection is that SARS-CoV-2 is more readily detected in faeces than the respiratory tract [39]. There is now evidence that the virus can productively infect enterocytes, which strongly express the ACE-2 receptor. [40]. I suggest that the lack of respiratory symptoms at presentation may point to preferential infection via the gastrointestinal tract in children. The subsequent mechanism of disease would then be as for complicated adult disease (Fig. 1, Fig. 2, Fig. 3), but without the dominance of respiratory symptoms seen in adult disease.
In support of this, there was evidence of ethnic predisposition in both cohorts. The UK patients showed significant overrepresentation of Afro-Caribbean and Asian children (47% and 28% of the cohort respectively, compared to 8% and 7% of the overall UK population) [37]. The large USA cohort had ethnicity data on 147 children, of whom only 35 were White, non-Hispanic [38]. While most children had no recognised comorbidities, 37% of the US children for whom data was available had diagnosed or BMI-based obesity while the UK group showed a significant increase in the observed to expected weight ratio. Thus, a significant comorbidity in children may be an increase in MDSC cells due to overweight. It is also notable that MDSC populations are increased in children with atopic rhinitis [41] and asthma [42].
Features not explained by the model
The relative susceptibility of males is not explained by this, and the mechanism is more likely to relate to a distinct antiviral immune response (Fig. 1). TLR7 responds to intracellular viral RNA in innate immune cells, and plasmacytoid dendritic cells of females produce more antiviral type-1 interferons than do males [43], related to the number of X chromosomes and serum testosterone production [44]. This may underpin sex-related symptomatic differences in RNA viral infections more generally, but may have a fundamentally more important impact with a virus as virulent as SARS-CoV-2.
Testing predictions of the model
This model of SARS-CoV-2 pathogenesis can rapidly be supported or excluded. Such interaction could be confirmed or excluded by genetic analysis of persons with severe or fatal illness compared to the less affected.
Genetic testing would be predicted to show:
-
1.
Persons with homozygous Siglec-14 null allele would have higher incidence of severe disease than those Siglec-5+ Siglec 14+, with heterozygotes intermediate. Preterm delivery would be more common in Siglec-14 null women, as in GBS infection.
-
2.
Persons of CD33 rs3865444 CC allele, when stratified by disease susceptibility, would have higher incidence of severe disease than those with CD33 rs3865444 TT allele.
Analysis of blood taken from affected persons would be predicted to show:
-
3.
Increased numbers of CD33 MDSC at presentation in those who go on to develop severe disease.
-
4.
Expanded numbers of CD33 MDSC cells in severe disease, characterised as CD33+CD11b+HLA-DRlow/-, particularly in those with immune dysregulation rather than hyperinflammatory late responses. These would be either monocytic (CD14+ ) or polymorphonuclear (CD15+) depending on the initial population undergoing expansion.
-
5.
Increased serum Arginase-1 and decreased arginine in severe disease, with increased percentage of TCR-ζ low T cells.
-
6.
Increased numbers of early stage CD33 + Siglec-5 + MDSC (CD14+/-CD15+/-) producing proinflammatory cytokines in persons with late-phase responses with immune deviation, in association with a Th17-skewed response. Conversely, these cells would be at low level in persons with macrophage activation syndrome and hyperferritinaemia.
-
7.
Increased circulating Siglec-5 in persons with Covid-19 induced vascular disease.
Glycobiological testing of Siglec binding
Proceeding from the direction of assessing in vitro SGP binding to cell lines or tissues suffers from the disadvantage that glycan expression is known to be modulated by enzymes within the Golgi apparatus of host cells during viral replication [12]. This may lead to impaired glycan maturation resulting in shorter glycan chains [12]. Thus, predicted glycans may not be representative of those functioning in vivo to mediate pathogenesis. However, detection of the footprints of Siglec-defined immunopathology would provide imperative for such extensive studies to be undertaken in order to characterise binding sites.
Supporting data
Prediction 4 appears to have been fulfilled, as MDSC populations (CD33+ but including both CD14+ monocytic and CD15+ granulocytic MDSC) were increased from 0.3% (IQR 0.13–2.13) in healthy donors to 47.5% (IQR 28.4–65.6%) in Covid-19 infection [45]. Further characterisation by the same group has demonstrated massive expansion of MDSC in 18 patients, making up to 90% of total peripheral blood mononuclear cells (PBMC) in severe disease and 25% in mild disease [46]. The frequency of these MDSC, largely CD15+ in this cohort, declined during recovery, with reduction of TGF-β. These cells demonstrated potent in vitro suppressive function on T cell activation and cytokine production.
This extraordinary percentage of MDSC in Covid-19 infection contrasts with the increase of MDSC to 5–10% of PBMC in dengue fever, where they have been implicated in disease outcome [47]. This is a strong pointer towards a specific mechanism of induction, unique to Covid-19 infection.
Prediction 6 is supported by evidence that proinflammatory MDSC of immature phenotype occur in human and murine arthritis, and strongly promote Th17 type responses [31], [32]. The cytokine storm in severe Covid-19 infection has indeed been characterised to be of Th17 type [48], with over 30% of total CD4 cells being of Th17 phenotype (CD4+CCR6+) in one well-studied case with fatal outcome [49]. The patient died on day 14 of illness, suggesting that such proinflammatory MDSC are released within the first two weeks of infection.
Therapeutic implications
If predictions based on this model are confirmed, inhibition of SARS-CoV-2 secreted glycoprotein interaction with host CD33-related Siglecs should be of clinical benefit. If the testing predictions are confirmed, the next stage would be to characterise the binding, which may be mediated by a single SGP or by two.
If the CD33-related Siglec-binding SGP is of sialyl Tn type, there may well be an existing sialyl Tn monoclonal that serendipitously blocks SGP binding to Siglecs. This would potentially be therapeutic.
If not, characterisation of SGP-Siglec binding would allow design of a small molecule that blocks interaction (possibly two), which would then be suitable for creating blocking immunoglobulin- (Ig)-fusion protein(s) for clinical use. In due course, this might allow persons who fail to respond to immunisation to be pre-treated to block Siglec binding sites, allowing them to be exposed to small infecting doses of SARS-Cov-2 in clinically controlled circumstances and thus generate a broad immune response, including to the pathogenic glycan determinants.
Conclusion
The clinical course of Covid-19 infection is highly variable. The currently enigmatic determinants of disease severity can be fully explained on the basis of viral manipulation of host CD33-related Siglecs. There is evidence that both CD33 and Siglec-5 (thus also Siglec-14) bind similar sialylated ligands, and that sialylated Tn ligands induce expansion and activation of CD33 MDSC. Both virally induced NLRP3 inflammasome inhibition and expansion of CD33 MDSC have been identified in other viruses. However, the magnitude of the latter response is dramatically greater in Covid-19 infection and this appears to be the dominant innate immune response. Confirmation of this mechanism through simple and accessible testing would provide a clear target for therapeutic manipulation. It is possible that specific therapeutic agents already exist, because of the increasing recognition of the importance of MDSC immunosuppression in the pathogenesis of cancers.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
I confirm that I have no conflicts of interest relating to the article entitled Common determinants of severe Covid-19 infection are explicable by SARS-CoV-2 secreted glycoprotein interaction with the CD33-related Siglecs, Siglec-3 and Siglec-5/14. Simon Murch PhD FRCP FRCPCH
References
- 1.Ni W., Yang X., Yang D., Bao J., Li R., Xiao Y. Role of angiotensin- converting enzyme 2 (ACE2) in COVID-19. Crit Care. 2020;24:422. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng Z., Peng F., Xu B., Zhao J., Liu H., Peng J. Risk factors of critical & mortal COVID-19 cases: A systematic literature review and meta-analysis. J Infect. 2020 Apr 23;S0163–4453(20):30234–30236. doi: 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang Y.C., Nizet V. Siglecs at the Host-Pathogen Interface. Adv Exp Med Bio. 2020;1204:197–214. doi: 10.1007/978-981-15-1580-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varki A., Angata T. Siglecs–the major subfamily of I-type lectins. Glycobiology. 2006;16:1R–27R. doi: 10.1093/glycob/cwj008. [DOI] [PubMed] [Google Scholar]
- 5.Bornhöfft K.F., Goldammer T., Rebl A., Galuska S.P. Siglecs: A journey through the evolution of sialic acid-binding immunoglobulin-type lectins. Dev Comp Immunol. 2018;86:219–231. doi: 10.1016/j.dci.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Padler-Karavani V., Hurtado-Ziola N., Chang Y.C., Sonnenburg J.L., Ronaghy A., Yu H. Rapid evolution of binding specificities and expression patterns of inhibitory CD33-related Siglecs in primates. FASEB J. 2014;28:1280–1293. doi: 10.1096/fj.13-241497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malik M., Simpson J.F., Parikh I., Wilfred B.R., Fardo D.W., Nelson P.T. CD33 Alzheimer's risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci. 2013;33:13320–13325. doi: 10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ali S.R., Fong J.J., Carlin A.F., Busch T.D., Linden R., Angata T. Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J Exp Med. 2014;211:1231–1242. doi: 10.1084/jem.20131853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinkman-Van der Linden E.C., Varki A. New aspects of siglec binding specificities, including the significance of fucosylation and of the sialyl-Tn epitope.J. Biol Chem. 2000;275 doi: 10.1074/jbc.275.12.8625. [DOI] [PubMed] [Google Scholar]
- 10.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shajahan A, Supekar NT, Gleinich AS, Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 2020 May 4. pii: cwaa042. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed]
- 12.Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020 May 4. pii: eabb9983. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed]
- 13.Xiao X., Dimitrov D.S. The SARS-CoV S glycoprotein. Cell Mol Life Sci. 2004;61:2428–2430. doi: 10.1007/s00018-004-4257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao C., Zhao W. NLRP3 Inflammasome-A Key Player in Antiviral Responses. Front Immunol. 2020;18(11):211. doi: 10.3389/fimmu.2020.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taxman D.J., Huang M.T., Ting J.P. Inflammasome inhibition as a pathogenic stealth mechanism. Cell Host Microbe. 2010;8:7–11. doi: 10.1016/j.chom.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai C.M., Riestra A.M., Ali S.R., Fong J.J., Liu J.Z., Hughes G. Siglec-14 Enhances NLRP3-Inflammasome Activation in Macrophages. J Innate Immun. 2020;12:333–343. doi: 10.1159/000504323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamanaka M., Kato Y., Angata T., Narimatsu H. Deletion polymorphism of SIGLEC14 and its functional implications. Glycobiology. 2009;19:841–846. doi: 10.1093/glycob/cwp052. [DOI] [PubMed] [Google Scholar]
- 18.Angata T., Ishii T., Motegi T., Oka R., Taylor R.E., Soto P.C. Loss of Siglec-14 reduces the risk of chronic obstructive pulmonary disease exacerbation. Cell Mol Life Sci. 2013;70:3199–3210. doi: 10.1007/s00018-013-1311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graustein A.D., Horne D.J., Fong J.J., Schwarz F., Mefford H.C., Peterson G.J. The SIGLEC14 null allele is associated with Mycobacterium tuberculosis- and BCG-induced clinical and immunologic outcomes. Tuberculosis. 2017;104:38–45. doi: 10.1016/j.tube.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tesi R.J. MDSC; the Most Important Cell You Have Never Heard Of. Trends Pharmacol Sci. 2019;40:4–7. doi: 10.1016/j.tips.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Poschke I., Kiessling R. On the armament and appearances of human myeloid-derived suppressor cells. Clin Immunol. 2012;144:250–268. doi: 10.1016/j.clim.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Verschoor C.P., Johnstone J., Millar J., Dorrington M.G., Habibagahi M., Lelic A. Blood CD33(+)HLA-DR(-) myeloid-derived suppressor cells are increased with age and a history of cancer. J Leukoc Biol. 2013;93:633–637. doi: 10.1189/jlb.0912461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lelis F.J.N., Jaufmann J., Singh A., Fromm K., Teschner A.C., Pöschel S. Myeloid-derived suppressor cells modulate B-cell responses. Immunol Lett. 2017;188:108–115. doi: 10.1016/j.imlet.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Dar A.A., Patil R.S., Pradhan T.N., Chaukar D.A., D'Cruz A.K., Chiplunkar S.V. Myeloid-derived suppressor cells impede T cell functionality and promote Th17 differentiation in oral squamous cell carcinoma. Cancer Immunol Immunother. 2020;69:1071–1086. doi: 10.1007/s00262-020-02523-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bao Y., Mo J., Ruan L., Li G. Increased monocytic CD14+HLADRlow/- myeloid-derived suppressor cells in obesity. Mol Med Rep. 2015;11:2322–2328. doi: 10.3892/mmr.2014.2927. [DOI] [PubMed] [Google Scholar]
- 26.Fernández-Ruiz J.C., Galindo-De Ávila J.C., Martínez-Fierro M.L., Garza-Veloz I., Cervantes-Villagrana A.R., Valtierra-Alvarado M.A. Myeloid-Derived Suppressor Cells Show Different Frequencies in Diabetics and Subjects with Arterial Hypertension. J Diabetes Res. 2019;2019:1568457. doi: 10.1155/2019/1568457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scrimini S., Pons J., Agustí A., Soriano J.B., Cosio B.G., Torrecilla J.A. Differential effects of smoking and COPD upon circulating myeloid derived suppressor cells. Respir Med. 2013;107:1895–1903. doi: 10.1016/j.rmed.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Bradshaw E.M., Chibnik L.B., Keenan B.T., Ottoboni L., Raj T., Tang A. CD33 Alzheimer's disease locus: altered monocyte function and amyloid biology. Nat Neurosci. 2013;16:848–850. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waeckel L., Venet F., Gossez M., Monard C., Rimmelé T., Monneret G. Delayed persistence of elevated monocytic MDSC associates with deleterious outcomes in septic shock: a retrospective cohort study. Crit Care. 2020;24:132. doi: 10.1186/s13054-020-02857-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020;27(992–1000) doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H., Wang S., Huang Y., Wang H., Zhao J., Gaskin F. Myeloid-derived suppressor cells are proinflammatory and regulate collagen-induced arthritis through manipulating Th17 cell differentiation. Clin Immunol. 2015;157:175–186. doi: 10.1016/j.clim.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo C., Hu F., Yi H., Feng Z., Li C., Shi L. Myeloid-derived suppressor cells have a proinflammatory role in the pathogenesis of autoimmune arthritis. Ann Rheum Dis. 2016;75:278–285. doi: 10.1136/annrheumdis-2014-205508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y.J., Chang S.Y., Ko H.J. Myeloid-derived suppressor cells in inflammatory bowel disease. Intest Res. 2015;13:105–111. doi: 10.5217/ir.2015.13.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das G., Mukherjee N., Ghosh S. Neurological Insights of COVID-19 Pandemic. ACS Chem Neurosci. 2020;11:1206–1209. doi: 10.1021/acschemneuro.0c00201. [DOI] [PubMed] [Google Scholar]
- 35.Li J.Y., Yang X.Y., Wang X.F., Jia X., Wang Z.J., Deng A.P. Siglec-5 is a novel marker of critical limb ischemia in patients with diabetes. Sci Rep. 2017;7:11272. doi: 10.1038/s41598-017-11820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J., Lee J., Baek S., Koh J.H., Kim J.W. Soluble siglec-5 is a novel salivary biomarker for primary Sjogren's syndrome. J Autoimmun. 2019;100:114–119. doi: 10.1016/j.jaut.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Davies P, Evans C, Kanthimathinathan HK, Lillie J, Brierley J, Waters G, et al. Intensive care admissions of children with paediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2 (PIMS-TS) in the UK: a multicentre observational study.Lancet Child Adolesc Health 2020 Jul 9: S2352-4642(20)30215-7. [DOI] [PMC free article] [PubMed]
- 38.Feldstein L.R., Rose E.B., Horwitz S.M., Collins J.P., Newhams M.M., Son M.B.F. Multisystem Inflammatory Syndrome in U.S. Children and Adolescents. N Engl J Med. 2020;383:334–346. doi: 10.1056/NEJMoa2021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zahran A.M., Saad K., Elsayh K.I., Abdelmoghny A., Aboul-Khair M.D., Sobhy A. Myeloid-Derived Suppressor Cells and Costimulatory Molecules in Children With Allergic Rhinitis. Ann Otol Rhinol Laryngol. 2019;128:128–134. doi: 10.1177/0003489418812902. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y.L., Luan B., Wang X.F., Qiao J.Y., Song L., Lei R.R. Peripheral blood MDSCs, IL-10 and IL-12 in children with asthma and their importance in asthma development. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0063775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berghöfer B., Frommer T., Haley G., Fink L., Bein G., Hackstein H. TLR7 ligands induce higher IFN-alpha production in females. J Immunol. 2006;177:2088–2096. doi: 10.4049/jimmunol.177.4.2088. [DOI] [PubMed] [Google Scholar]
- 44.Webb K., Peckham H., Radziszewska A., Menon M., Oliveri P., Simpson F. Sex and Pubertal Differences in the Type 1 Interferon Pathway Associate With Both X Chromosome Number and Serum Sex Hormone Concentration. Front Immunol. 2019;9:3167. doi: 10.3389/fimmu.2018.03167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bordoni V, Sacchi A, Cimini E, Notari S, Grassi G, Tartaglia E, et al. An inflammatory profile correlates with decreased frequency of cytotoxic cells in COVID-19. Clin Infect Dis 2020 May 15. pii: ciaa577. doi: 10.1093/cid/ciaa577. [DOI] [PMC free article] [PubMed]
- 46.Agrati C., Sacchi A., Bordoni V., Cimini E., Notari S., Grassi G. Expansion of myeloid-derived suppressor cells in patients with severe coronavirus disease (COVID-19) Cell Death Differ. 2020 Jun;8:1–12. doi: 10.1038/s41418-020-0572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo P.-L., Li L.-H., Li W.-L., Zhao J.-C., Hu F.-Y., Zhang F.-C. The clinical significance of myeloid-derived suppressor cells in dengue fever patients. BMC Infect Dis. 2019;19:926. doi: 10.1186/s12879-019-4574-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. J Microbiol Immunol Infect. 2020;53:368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]