Abstract
Objective: Essential tremor (ET) is a common movement disorder that has a high heritability. A number of genetic studies have associated different genes and loci with ET, but few have investigated the biology of any of these genes. STK32B was significantly associated with ET in a large genome-wide association study (GWAS) and was found to be overexpressed in ET cerebellar tissue. The objective of this study is to determine the effects of overexpressed STK32B in cerebellar DAOY cells.
Methods: Here, we overexpressed STK32B RNA in human cerebellar DAOY cells and used an RNA-Seq approach to identify differentially expressed genes (DEGs) by comparing the transcriptome profile of these cells to one of the control DAOY cells.
Results: Pathway and gene ontology enrichment identified axon guidance, olfactory signaling, and calcium-voltage channels as significant. Additionally, we show that overexpressing STK32B affects transcript levels of previously implicated ET genes such as FUS.
Conclusion: Our results investigate the effects of overexpressed STK32B and suggest that it may be involved in relevant ET pathways and genes.
Keywords: STK32B, essential tremor, transcriptome, FUS, neurology
Introduction
Essential tremor (ET) is one of the most common movement disorders and is typically characterized by a kinetic tremor in the hand or arms (Elble, 2013). The severity tends to increase with age and may involve different regions such as the head, voice, or jaw (Haubenberger and Hallett, 2018). Previous studies investigating the histology of post-mortem tissue have pinpointed the cerebellum as a region of interest for ET (Gironell, 2014). Specifically, abnormalities with Purkinje cell axons and dendrites were found in ET brains (Axelrad et al., 2008). Furthermore, the olivocerebellar circuitry has been implicated in ET pathology and many calcium voltage channels are highly expressed in this circuitry (Schmouth et al., 2014).
The genetic etiology of ET has remained largely elusive and most studies have focused on common or rare variants and on looking for genetic overlap with other disorders (Houle et al., 2017; Liao et al., 2018). Twin studies have shown that ET has a concordance of 69–93% in monozygotic twins and 27–29% in dizygotic twins, which suggests that both genetic and environmental factors drive the onset and development of this complex trait (Tanner et al., 2001). A recent genome-wide association study (GWAS) identified a significant locus in STK32B and found that ET patients overexpressed STK32B in cerebellar tissue by comparison to healthy controls, suggesting potential implications of STK32B in ET (Müller et al., 2016). STK32B is transcribed and translated into YANK2, a serine/threonine kinase, that has not been well characterized. There have been several exome-wide studies that implicated different genetic variants as causes of ET. The first ET-implicated gene found through exome sequencing was the Fused in Sarcoma gene (FUS) (Merner et al., 2012). However, it is unclear whether these “ET” genes interact or have indirect effects on the expression of each other.
To understand the effects of overexpressed STK32B mRNA and identify pathways potentially relevant to ET, we overexpressed this gene in human cerebellar DAOY cells and compared transcriptomic changes in overexpressed cells and empty-vector controls using RNA sequencing. Several interesting pathways such as axon guidance, calcium ion transmembrane transport, and olfactory transduction were significantly enriched after overexpression of STK32B. We also identified previously implicated ET genes whose expression is dysregulated through the overexpression of STK32B, suggesting that overexpressed STK32B may have relevant downstream effects.
Results
Confirming Overexpression of STK32B
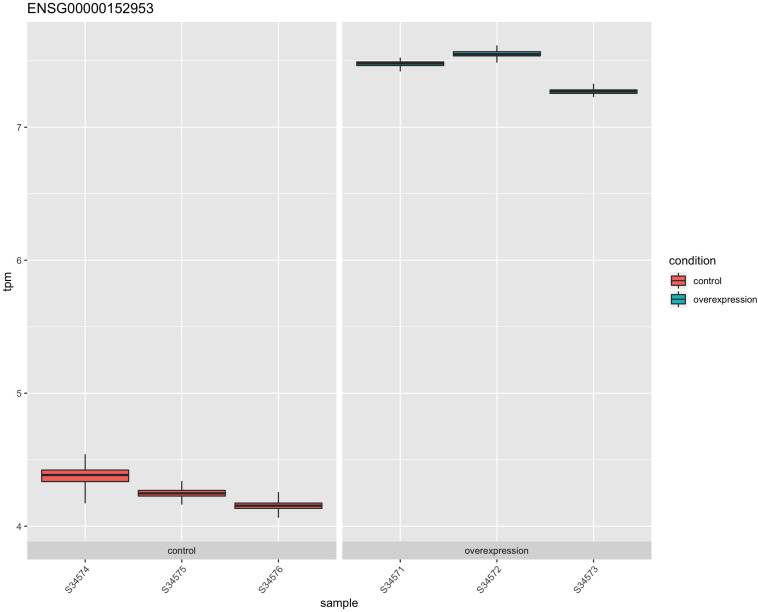
The STK32B stable cell lines had higher RNA expression of STK32B, which was detected by reverse-transcriptase qPCR (Supplementary Figure S1). RNA sequencing (RNA-Seq) also confirmed that the stable cell lines have higher STK32B RNA levels compared to controls (Figures 1, 2). The STK32B gene was the top significant differentially expressed gene (P = 7.07E-245, β = 3.1789).
FIGURE 1.
The RNA expression of STK32B overexpressed cells compared to empty-vector controls based on RNA sequencing data. Error bars show the technical variance determined with 200 bootstraps. TPM, transcript per million.
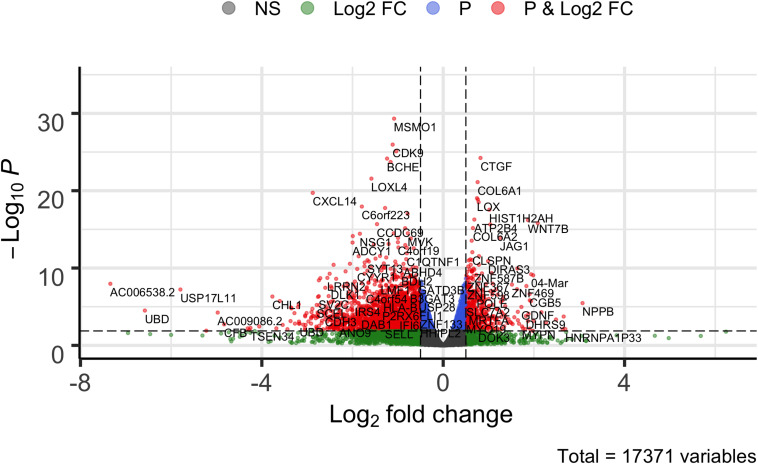
FIGURE 2.
Volcano plot of comparing overexpressed STK32B to empty-vector controls. Differentially expressed genes (Wald test, PFDR–corrected < 0.05 and β > | 0.05|) are shown in red. NS, not significant; FC, fold change.
Quality Control of RNA-Seq Analyses
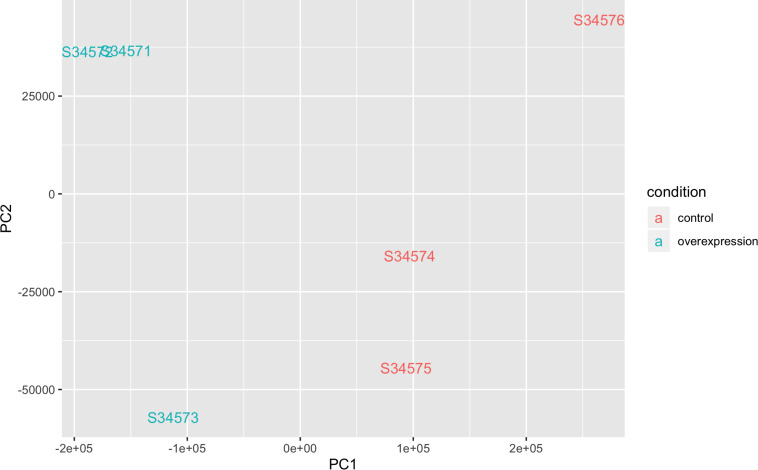
The QQ-plot for differentially expressed genes (DEGs) did not show stratification or potential biases (Supplementary Figure S2). The M-A plot showed slight enrichment of downregulated genes compared to upregulated genes (Supplementary Figure S3). A principal component (PC) plot showed separation of controls and STK32B cell lines (Figure 3). Additionally, the dendrogram with heatmap shows that controls and STK32B cell lines are distinct (Supplementary Figure S4). The mean-variance plot shows the shrinkage under the sleuth model (Supplementary Figure S5).
FIGURE 3.
Principle component analysis plot of RNA sequencing data of STK32B overexpressed cells and empty-vector controls. Overexpressed cells shown in blue and controls shown in red.
STK32B Overexpression Affects the Expression of Previously Identified ET-Associated Genes
A total of 3,794 genes were found to be differentially expressed with a q-value < 0.05. There were 425 genes with a β > | 0.5| and q-value < 0.05. Amongst the 425 genes, there were several potentially relevant ET genes were dysregulated such as FUS (q-value = 0.007, β = 0.812) and two calcium voltage channel genes that are enriched in the olivocerebellar circuitry, CACNA1C (q-value = 0.011, β = 0.503) and CACNA1A (q-value = 0.030, β = 0.702) (Table 1). Several pathways and gene ontology terms were enriched for DEGs due to the overexpression of STK32B, including olfactory transduction (P = 2.68E-39), axon guidance (P = 9.50E-36), and calcium ion transmembrane transport (P = 3.02E-31) (Table 2).
TABLE 1.
Effects of STK32B perturbation on ET-implicated genes.
| Genes | P-value | Q-value | Beta |
| MTHFR | 3.22E-15 | 2.15E-12 | –1.0905696 |
| SHF | 6.74E-07 | 2.72E-05 | –0.9995849 |
| MAPT | 2.15E-06 | 6.81E-05 | –1.3021309 |
| PPARGC1A | 6.87E-06 | 0.00016336 | –1.691415 |
| SNCA | 0.00071236 | 0.0054543 | –0.4253643 |
| FUS | 0.00090877 | 0.00653715 | 0.81221158 |
| CACNA1C | 0.001691143 | 0.010366605 | 0.503658389 |
| PPP2R2B | 0.00168856 | 0.01035793 | –0.7621076 |
| GSTP1 | 0.00249219 | 0.01388975 | –0.2723091 |
| GBA | 0.00326972 | 0.01712692 | –0.4716877 |
| CACNA1A | 0.00306972 | 0.03713307 | 0.70202999 |
| LINGO1 | 0.02206061 | 0.07248666 | –1.2292913 |
| HNMT | 0.05546041 | 0.14360684 | –0.3656343 |
| LRRK1 | 0.07843181 | 0.18520013 | 0.30296973 |
| VDR | 0.07963403 | 0.18738651 | 0.26752548 |
| MAPT | 0.08202636 | 0.19175911 | –1.786066 |
| HMOX1 | 0.08258822 | 0.19278723 | 0.28335731 |
| ALAD | 0.09508151 | 0.21263593 | –0.2562971 |
| LRRK2 | 0.20628994 | 0.36752893 | –0.3644185 |
| MAPT | 0.2395589 | 0.40824182 | –1.0324492 |
| NOS1 | 0.26006033 | 0.43219823 | –1.558382 |
| HMOX2 | 0.30661818 | 0.48353042 | –0.2005729 |
| IL1B | 0.65088456 | 0.78183136 | 0.19430951 |
| SLC1A2 | 0.66644945 | 0.79215448 | –0.2943051 |
TABLE 2.
Gene ontology and pathway analyses of differentially expressed genes.
| Gene set ID | Gene set name | P | Database |
| KEGG_OLFACTORY_ TRANSDUCTION | Olfactory transduction | 2.68E-39 | KEGG |
| KEGG_PATHWAYS_ IN_CANCER | Pathways in cancer | 2.96E-28 | |
| KEGG_FOCAL_ ADHESION | Focal adhesion | 1.19E-26 | |
| KEGG_BLADDER_ CANCER | Bladder cancer | 6.53E-25 | |
| KEGG_ECM_ RECEPTOR_ INTERACTION | ECM-receptor interaction | 8.38E-25 | |
| GO:0001650 | Fibrillar center | 1.13E-29 | GO Cellular |
| GO:0031012 | Extracellular matrix | 8.57E-27 | |
| GO:0001725 | Stress fiber | 1.24E-26 | |
| GO:0005925 | Focal adhesion | 2.82E-23 | |
| GO:0015629 | Actin cytoskeleton | 4.02E-22 | |
| REACTOME:R-HSA-381753 | Olfactory signaling pathway | 3.22E-46 | Reactome |
| REACTOME:R-HSA-418555 | G alpha (s) signaling events | 1.19E-29 | |
| REACTOME:R-HSA-3000170 | Syndecan interactions | 1.53E-27 | |
| REACTOME:R-HSA-2022090 | Assembly of collagen fibrils and other multimeric structures | 4.31E-25 | |
| REACTOME:R-HSA-1474290 | Collagen formation | 2.35E-24 | |
| GO:0050911 | Detection of chemical stimulus involved in sensory perception of smell | 1.00E-44 | GO Processes |
| GO:0007411 | Axon guidance | 9.50E-36 | |
| GO:0048146 | Positive regulation of fibroblast proliferation | 1.31E-33 | |
| GO:0070588 | Calcium ion transmembrane transport | 3.02E-31 | |
| GO:0050907 | Detection of chemical stimulus involved in sensory perception | 3.14E-31 |
Gene Network Analyses
To identify which cluster of DEGs drove the significant pathways, gene network analysis based on co-expression was done using and identified 14 different clusters within the DEGs (Supplementary Table S1). Analysis of the top 3 largest clusters for pathway enrichments found several similar pathways in different databases including axon guidance (P < 1.09E-12) in cluster 1 and calcium signaling pathway in cluster 3 (2.06E-05).
Discussion
The genetic etiology of ET is complex and likely explained by a combination of copy number variants, rare variants, gene-gene interactions and common variant drivers. One of the most significantly enriched pathways we identified was olfactory signaling and transduction. Previous reports have shown conflicting evidence about olfactory loss in ET (Applegate and Louis, 2005; Shah et al., 2008). It may be that ET patients with dysregulated olfactory signaling have overexpressed STK32B, which may contribute to the subset of ET patients with olfactory loss.
Interestingly, FUS was amongst the genes dysregulated when STK32B was overexpressed. In the exome familial ET study that identified FUS, we also found reduced mRNA levels for FUS (Merner et al., 2012). Similarly, the expression of FUS is lower when STK32B is overexpressed, suggesting an indirect relationship between the two genes. Additionally, two calcium voltage-gated channel genes, CACNA1C and CACNA1A were overexpressed. Enrichment analyses found the GO term calcium ion transmembrane transport to be highly significant in GO Biological Processes, suggesting that STK32B may play a role upstream of these genes. In the olivocerebellar circuitry, a system implicated in ET, is enriched for both of these genes (Mori et al., 1991; Schmouth et al., 2014). CACNA1A has been shown to be predominantly expressed in Purkinje cells, a cell type relevant to ET (Mori et al., 1991). Another enriched pathway was axon guidance. A previous study identified the TENM4 missense variant segregating within ET families (Hor et al., 2015). TENM4 is a regulator of axon guidance and myelination and Tenm4 knockout mice showed an ET phenotype, suggesting that axon guidance is important in ET (Hor et al., 2015). Similarly, overexpressed STK32B could be dysregulating important pathways relevant to axon guidance.
By sub-stratifying the overexpressed genes into different clusters that are co-expressed, several interesting pathways involving the cardiovascular system were identified. GO terms and pathways such as right ventricular cardiomyopathy in KEGG and angiogenesis in GO Biological Processes were found among the significant pathways in Table 3. In certain ET patients, beta-blockers can reduce tremor magnitude and frequency. Beta-blockers lower blood pressure and are used to treat irregular heart rhythm and other cardiovascular phenotypes. A common treatment for ET is propranolol, a beta-blocker. This was a drug developed for cardiovascular health that affects adrenergic activity, but still reduces tremor in ET individuals. This could suggest that overexpressed STK32B affects cardiovascular health, which may in turn affect or lead to the ET phenotype or that STK32B may be pleiotropic and affect the nervous and cardiovascular system differently.
TABLE 3.
Gene ontology and pathway analyses of differentially expressed gene for the three largest clusters based on co-expression.
| Gene set name | P-value | Database | Cluster |
| Regulation of actin cytoskeleton | 1.03E-13 | KEGG | 1 |
| Pathways in cancer | 1.23E-13 | ||
| Axon guidance | 1.09E-12 | ||
| Focal adhesion | 4.96E-12 | ||
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 4.63E-09 | ||
| Fibrillar center | 1.13E-29 | GO Cellular | |
| Extracellular matrix | 8.57E-27 | ||
| Stress fiber | 1.24E-26 | ||
| Focal adhesion | 2.82E-23 | ||
| Actin cytoskeleton | 4.02E-22 | ||
| Actin binding | 3.66E-15 | GO Function | |
| Laminin binding | 2.37E-13 | ||
| Insulin-like growth factor binding | 9.72E-13 | ||
| Collagen binding | 3.73E-12 | ||
| Protein heterodimerization activity | 5.54E-12 | ||
| Cell migration involved in sprouting angiogenesis | 7.07E-18 | GO Processes | |
| Cell migration | 2.94E-16 | ||
| inactivation of MAPK activity | 3.31E-15 | ||
| Cytoskeleton organization | 6.34E-15 | ||
| Negative regulation of cell migration | 9.12E-15 | ||
| Cell-extracellular matrix interactions | 7.16E-15 | Reactome | |
| Signal transduction | 1.98E-13 | ||
| Non-integrin membrane-ECM interactions | 2.58E-12 | ||
| Signaling by Rho GTPases | 1.84E-11 | ||
| EPHA-mediated growth cone collapse | 2.30E-11 | ||
| RNA polymerase | 4.57E-13 | KEGG | 2 |
| Pyrimidine metabolism | 1.37E-12 | ||
| Alzheimer’s disease | 3.55E-11 | ||
| Purine metabolism | 4.11E-11 | ||
| GnRH signaling pathway | 1.84E-09 | ||
| Nucleolus | 5.96E-19 | GO Cellular | |
| Preribosome, large subunit precursor | 2.21E-18 | ||
| Small-subunit processome | 5.13E-18 | ||
| DNA-directed RNA polymerase III complex | 5.10E-15 | ||
| DNA-directed RNA polymerase I complex | 1.85E-14 | ||
| Pseudouridine synthase activity | 7.75E-16 | GO Function | |
| RNA binding | 1.25E-15 | ||
| snoRNA binding | 4.90E-14 | ||
| rRNA binding | 3.55E-13 | ||
| ATP-dependent RNA helicase activity | 5.23E-13 | ||
| RNA secondary structure unwinding | 1.57E-16 | GO Processes | |
| tRNA modification | 7.73E-16 | ||
| Positive regulation of gene expression, epigenetic | 3.09E-15 | ||
| Ribosomal large subunit biogenesis | 3.17E-15 | ||
| Maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 3.32E-15 | ||
| rRNA modification in the nucleus and cytosol | 3.38E-18 | Reactome | |
| tRNA processing | 1.65E-17 | ||
| tRNA modification in the nucleus and cytosol | 1.66E-16 | ||
| rRNA processing | 9.91E-15 | ||
| tRNA processing in the nucleus | 1.69E-14 | ||
| RIG-I-like receptor signaling pathway | 1.24E-10 | KEGG | 3 |
| Cytosolic DNA-sensing pathway | 1.92E-07 | ||
| O-glycan biosynthesis | 1.163E-06 | ||
| Focal adhesion | 1.36893E-06 | ||
| Calcium signaling pathway | 2.06986E-05 | ||
| Caveola | 2.00E-08 | GO Cellular | |
| Proteinaceous extracellular matrix | 7.41E-08 | ||
| Postsynaptic membrane | 2.45E-06 | ||
| Microfibril | 4.99E-06 | ||
| Extracellular matrix | 1.12E-05 | ||
| Type I interferon receptor binding | 3.64E-07 | GO Function | |
| Neuropeptide binding | 5.89E-07 | ||
| Scavenger receptor activity | 6.23E-07 | ||
| Calcium-dependent phospholipid binding | 9.44E-07 | ||
| RNA binding | 2.46E-06 | ||
| Response to mechanical stimulus | 8.42E-09 | GO Processes | |
| Lung development | 1.55E-08 | ||
| Negative regulation of neuron apoptotic process | 5.29E-08 | ||
| Cellular response to hypoxia | 7.29E-08 | ||
| Regulation of type I interferon-mediated signaling pathway | 1.11E-07 | ||
| O-glycosylation of TSR domain-containing proteins | 3.73E-10 | Reactome | |
| Defective B3GALTL causes Peters plus syndrome (PpS) | 5.15E-10 | ||
| DDX58/IFIH1-mediated induction of interferon-alpha/beta | 1.29E-09 | ||
| O-linked glycosylation | 4.91E-08 | ||
| Diseases associated with O-glycosylation of proteins | 1.04E-07 |
Due to the heterogeneity of ET, it is likely that not all ET-affected individuals have overexpression of STK32B. One limitation of this study is that DAOY cells are a cancerous cell line and may not be the ideal model for understanding the transcriptome of Purkinje cells. Future studies on the effects of STK32B overexpression could use induced pluripotent stem cells differentiated into cells with Purkinje cells properties; such cells would likely have less biological noise. However, our findings support the idea that STK32B is a gene of interest for a subset of ET cases, and further investigations into how STK32B may interact with other genes are warranted.
Materials and Methods
Cell Culture and Plasmid Construction
The DAOY cell line was cultured in Eagle’s Minimum Essential Medium (EMEM) with 10% fetal bovine serum (FBS) at 37°C and 5% CO2 with the Glico Pen-Strep-Glutamine cocktail at 1×. Cells were passaged every 2 days at 80–90% confluence and at the same time. The cDNA of STK32B (NM_001306082.1) was inserted into a pcDNA3.1(+) vector containing a neomycin resistance gene, used as a selectable marker. This transcript was picked because it had the highest expression in the cerebellum based on GTEx 53 v7. The plasmid was transformed and expanded in XL10-Gold Escherichia coli strain (Stratagene). The cDNA of the plasmid was sent for Sanger sequencing to validate the gene sequence.
Transfection and Deriving Stable Cell Lines
The cells were transfected for 48 h with 8 ug of the vector of using the jetPRIME transfection reagent (Polyplus). A green fluorescent protein (GFP) control vector was used to assess and estimate transfection efficiency and optimize transfection parameters. Stable cell lines were established by subjecting transfected cells to G418 antibiotic at 1 μg/mL for 10 days and maintained at a concentration of 400 μg/mL for an additional 2 weeks. A kill curve was done to determine the ideal antibiotic selection. In parallel, empty pcDNA3.1(+) vector control lines were also grown and underwent antibiotic selection.
RNA Extraction and Sequencing
Total RNA was extracted using the RNeasy Mini Kit (Qiagen). The total RNA concentration was measured using the Synergy H4 microplate reader. RNA for the top three overexpressed STK32B cell lines and three empty-vector controls was sent to Macrogen Inc., for sequencing. RT-qPCR was used to confirm overexpression in the cell lines compared to controls. Preparation of the cDNA library was done with the TruSeq Stranded Total RNA Kit (Ilumina) with Ribo-Zero depletion. Sequencing was done on the NovaSeq 6000 at 150 bp paired-end reads with a total of 200M reads. Samples were randomized for cell lysis, RNA extraction, Ribo-Zero depletion, library preparation and sequencing to account for potential batch effects.
Data Processing and Quality Control
The FASTQ files were pseudo-aligned with Salmon using the Ensembl v94 annotation of the human genome (Patro et al., 2017). The data is available on the public repository of NCBI, Gene Expression Omnibus (GEO), with the GEO accession as GSE150393. It can be accessed with the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150393. The parameters of Salmon included the following: 200 bootstraps with mapping validation, GC bias correction, sequencing bias correction, 4 range factorization bins, a minimum score fraction of 0.95 and VB optimization with a VB prior of 1e-5. The estimated counts were analyzed using sleuth to identify DEGs (Pimentel et al., 2017). A Wald test was used to get β values [log2(x + 0.5)]. Additionally, transcript-level aggregation was done against the Ensembl v94 annotation to determine gene-level differential expression. P-values were corrected using the Benjamini–Hochberg procedure to account for false discovery rate (FDR). The FDR threshold was set at 0.05. A PC plot and heatmap was done to identify clusters.
Pathway and Gene Set Enrichment Analysis
Pathway and gene set enrichment for relevant databases was done using Gene Network 2.0 (Deelen et al., 2019). To investigate genes with larger fold changes, only DEGs with a β > |0.5| were analyzed for pathway enrichments. Network clustering was done by grouping genes based on co-expression using Gene Network 2.0. Pathway analyses were also conducted with Gene Network 2.0 for the clusters using a Wilcoxon test. We included the top five significant pathways across different databases.
Data Availability Statement
The data is available on the public repository of NCBI, Gene Expression Omnibus (GEO), with the GEO accession as GSE150393. It can be accessed with the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150393.
Ethics Statement
The review board at the McGill University Health Center Research Ethics Board (MUHC REB) approved the study protocols (reference number IRB00010120).
Author Contributions
CL conducted the experiments and analyses and drafted the manuscript. FS, VV, DR, FA, SD, GH, QH, and HC helped with experiments. AL and DS helped with bioinformatic analyses. PD and GR oversaw the study and drafting of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The manuscript has been released as a pre-print at https://www.biorxiv.org/content/10.1101/552901v2.
Footnotes
Funding. This work was supported by a Canadian Institutes of Health Research Foundation Scheme grant (#332971). GR holds a Canada Research Chair in Genetics of the Nervous System and the Wilder Penfield Chair in Neurosciences. CL is a recipient of the Frederick Banting and Charles Best Canada Graduate Scholarship from the Canadian Institutes of Health Research (CIHR). FA is funded by the Fonds de Recherche du Québec-Santé (FRQS). GH is funded by the Canadian Institutes of Health Research (CIHR; FRN 159279).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00813/full#supplementary-material
References
- Applegate L. M., Louis E. D. (2005). Essential tremor: mild olfactory dysfunction in a cerebellar disorder. Parkinsonism Relat. Disord. 11 399–402. 10.1016/j.parkreldis.2005.03.003 [DOI] [PubMed] [Google Scholar]
- Axelrad J. E., Louis E. D., Honig L. S., Flores I., Ross G. W., Pahwa R., et al. (2008). Reduced Purkinje cell number in essential tremor: a postmortem study. Arch. Neurol. 65 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen P., van Dam S., Herkert J. C., Karjalainen J. M., Brugge H., Abbott K. M., et al. (2019). Improving the diagnostic yield of exome- sequencing by predicting gene–phenotype associations using large-scale gene expression analysis. Nat. Commun. 10:2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R. J. (2013). What is essential tremor? Curr. Neurol. Neurosci. Rep. 13:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gironell A. (2014). The GABA hypothesis in essential tremor: lights and shadows. Tremor. Hyperkinet. Mov. 4:254 10.5334/tohm.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubenberger D., Hallett M. (2018). Essential TREMOR. N. Engl. J. Med. 378 1802–1810. [DOI] [PubMed] [Google Scholar]
- Hor H., Francescatto L., Bartesaghi L., Ortega-Cubero S., Kousi M., Lorenzo-Betancor O., et al. (2015). Missense mutations in TENM4, a regulator of axon guidance and central myelination, cause essential tremor. Hum. Mol. Genet. 24 5677–5686. 10.1093/hmg/ddv281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle G., Schmouth J.-F., Leblond C. S., Ambalavanan A., Spiegelman D., Laurent S. B., et al. (2017). Teneurin transmembrane protein 4 is not a cause for essential tremor in a Canadian population. Mov. Disord. 32 292–295. 10.1002/mds.26753 [DOI] [PubMed] [Google Scholar]
- Liao C., Houle G., He Q., Laporte A. D., Girard S. L., Dion P. A., et al. (2018). Investigating the association and causal relationship between restless legs syndrome and essential tremor. Parkinsonism Relat. Disord. 61 238–240. 10.1016/j.parkreldis.2018.10.022 [DOI] [PubMed] [Google Scholar]
- Merner N. D., Girard S. L., Catoire H., Bourassa C. V., Belzil V. V., Rivière J. B., et al. (2012). Exome sequencing identifies FUS mutations as a cause of essential tremor. Am. J. Hum. Genet. 91 313–319. 10.1016/j.ajhg.2012.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y., Friedrich T., Kim M.-S., Mikami A., Nakai J., Ruth P., et al. (1991). Primary structure and functional expression from complementary DNA of a brain calcium channel. Nature 350 398–402. 10.1038/350398a0 [DOI] [PubMed] [Google Scholar]
- Müller S. H., Girard S. L., Hopfner F., Merner N. D., Bourassa C. V., Lorenz D., et al. (2016). Genome-wide association study in essential tremor identifies three new loci. Brain 139 3163–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H., Bray N. L., Puente S., Melsted P., Pachter L. (2017). Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14 687–690. 10.1038/nmeth.4324 [DOI] [PubMed] [Google Scholar]
- Schmouth J.-F., Dion P. A., Rouleau G. A. (2014). Genetics of essential tremor: From phenotype to genes, insights from both human and mouse studies. Prog. Neurobiol. 11 1–19. 10.1016/j.pneurobio.2014.05.001 [DOI] [PubMed] [Google Scholar]
- Shah M., Muhammed N., Findley L. J., Hawkes C. H. (2008). Olfactory tests in the diagnosis of essential tremor. Parkinsonism Relat. Disord. 14 563–568. 10.1016/j.parkreldis.2007.12.006 [DOI] [PubMed] [Google Scholar]
- Tanner C. M., Goldman S. M., Lyons K. E., Aston D. A., Tetrud J. W., Welsh M. D., et al. (2001). Essential tremor in twins: an assessment of genetic vs environmental determinants of etiology. Neurology 57 1389–1391. 10.1212/wnl.57.8.1389 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is available on the public repository of NCBI, Gene Expression Omnibus (GEO), with the GEO accession as GSE150393. It can be accessed with the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150393.